Abstract
Klebsiella pneumoniae spp ozaenae is a versatile bacterial species able to acquire antimicrobial resistance; the species presents a higher antimicrobial resistance profile compared to Klebsiella pneumoniae spp pneumoniae. Carbapenemase and extended spectrum β-lactamase (ESBL)-producing bacteria commonly arise in clinical settings where antimicrobial stewardship is limited. Our study aims to report the phenotypical and genetic characteristics of nosocomial Klebsiella pneumoniae spp ozaenae isolates associated with mortality collected from a tertiary-level hospital in Panama City. In October 2020, 11 consecutive multidrug-resistant Gram-negative isolates were recovered from secretions and blood cultures from hospitalized patients. Nearly 90% (10/11) of these patients died, and bacteria was obtained from six patients for investigation. Biochemical evaluation of the six isolates revealed the presence of multidrug-resistant Klebsiella pneumoniae spp ozaenae. Phenotypic evaluation indicated resistance to carbapenemase and EBSL. In contrast, genetic evaluation by PCR showed that only 30% (2/6) were resistant to CTX-M-1 (CTX-M group 1), whereas 60.7% (4/6) presented carbapenemase resistance genes, and 33.3% (2/6) presented New Delhi metallo-β-lactamase (NDM) resistance genes. Klebsiella pneumoniae ST258 was identified in 83.3% (5/6) of the isolates. Phylogenetic analysis using 16S revealed low homology among the six isolates. These results suggest that antibiotic resistance genes may have been incorporated into these Klebsiella pneumoniae spp ozaenae isolates within the hospital environment. We recommend strengthening the antimicrobial stewardship program and antibiotic control policy, as well as heightened infection control and prevention measures, such as ward sanitation and increased hand washing frequency.
Keywords: Gram-negative, Klebsiella pneumoniae spp ozaenae, multidrug resistant, Panama, ST258
Introduction
The Klebsiella genera remains the most rapid producer of extended spectrum β-lactamase (ESBL) worldwide.1–3 The production of ESBLs results in resistance to most common antibiotics, including ampicillin, ticarcillin, piperacillin, and cephalosporins. 4 This resistance greatly impacts clinical practice, especially since carbapenems are included among the broad-spectrum antibiotics used to manage patients during extended hospitalizations. 5 Unfortunately, point mutations in Klebsiella have resulted in the production of carbapenemase enzymes, which directly counteracts this treatment strategy. Between 2010 and 2013, Klebsiella pneumoniae carbapenemase (KPC) emerged as major drug-resistant strain in hospitals worldwide. Thus, the characterization of carbapenemase-producing Klebsiella is essential for reinforcing antimicrobial stewardship efforts and assuring adequate patient management. 6
Risk stratification using rapid diagnostic laboratory workflows is crucial for selecting appropriate therapies in accordance with patient characteristics and severity of infection. 7 The updated VITEK platform both identifies Klebsiella pneumoniae spp ozaenae and performs antimicrobial susceptibility testing. 8 Multilocus Sequence Typing (MLST) is a DNA sequence-based method that characterizes genetic relationships among bacterial isolates. The MLST approach provides unambiguous data that allow the construction of international multiuser databases. 9 These molecular epidemiology tools have been used to characterize Klebsiella pneumoniae ST512, ST101, ST307 and ST258. These ST are described as public health concerns with high potential for causing hospital outbreaks and a high mortality burden. The genetic characteristics of these clones may contribute to their adaptation to hospital environments and human hosts and favor the spread of antibiotic resistance.10,11
Antimicrobial stewardship remains the main strategy to manage increasing bacterial resistance. Several countries have adopted antimicrobial stewardship among hospital and clinical settings. Moreover, strong regulations have been set in place to decrease the general population’s access to antibiotics. Finally, intensive communication campaigns have been launched to complement these strategies. The overall goal is to prevent the dissemination of resistant gene cassettes, plasmids and other genetic fragments between bacteria and contain the emergence of resistant super bugs. However, Klebsiella pneumoniae spp ozaenae, which has a higher antimicrobial resistance profile than Klebsiella pneumoniae spp pneumoniae, has recently escaped such containment efforts. In fact, these bacteria harbor both carbapenemase- and ESBL-resistance genes and have become one of the most dangerous pathogens in healthcare and hospital settings. 12 Here, we report the phenotypic and genetic characterization of six Klebsiella pneumoniae spp ozaenae isolates from an urban hospital setting. We found that most of the isolates belong to the ST258 clonal lineage and are not closely related genetically, indicating that in-hospital evolutionary divergence has occurred.
Materials and methods
A total of 1373 Klebsiella spp. strains were isolated during 2020 from the Complejo Hospitalario Metropolitano Dr. Arnulfo Arias Madrid, Caja de Seguro Social (CHMDrAAM-CSS) of Panamá; 169 (12.3%) strains were identified as Klebsiella pneumoniae spp ozaenae. In one month of the same year, 11 patients were identified with multidrug-resistant Klebsiella pneumoniae spp ozaenae, and 90% (10/11) of these patients died. We were able to analyze six consecutive isolates from positive blood cultures for their phenotypic and genetic elements of resistance. The six bacterial isolates were obtained as part of the hospital’s standard of care and stored in a laboratory freezer. None of the authors were involved in obtaining the patient samples, nor had access to the patients’ full medical information. The authors analyzed an anonymized bacterial strain collection. This information was anonymized by hospital laboratory staff, who work independently of the author team.
Standard microbiological procedures were employed to identify the six isolates using the automated VITEK 2 Compact system. Antimicrobial susceptibility testing was also performed using the VITEK® 2 AST (Cat—N250, bioMerieux, Madrid, Spain). The antimicrobials included Nalidixic acid (NA), Amikacin (AMK), Cefepime (FEP), Cefotaxin (CTX), Ceftazidime (CAZ), Cefuroxima acetil (CXM-A), Ciprofloxacin (CIP), Gentamicin (GEN), Imipenem (IMP), Meropenem (MEM), Nitrofurantoin (NIT), Piperacillin-Tazobactam (TZP), Trimethoprim-sulfamethoxazole (SXT), Cephalothin (CEF), and Cefazolin (CFZ). We also investigated the presence of carbapenem resistance using Xpert Carba-R (Cat. GXCARBAR-CE-10, Cepheid, USA). Both commercial kits were used according to the manufacturer’s instructions.
DNA extraction from the six isolates was performed using a method previously described by Van Soolingen et al. 13 The extracted DNA was stored at 4°C. Molecular typing analyses were performed using Pasteur’s (Multi Locus Sequence Type (MLST) scheme. We conducted MLST schemes using a standardized protocol specific for Klebsiella pneumoniae.14,15 The sequencing fragments of seven housekeeping genes (i.e. rpoB, gapA, mdh, pgi, phoE, infB, and tonB) were amplified from chromosomal DNA of the Klebsiella pneumoniae spp ozaenae strains. Sequencing of polymerase chain reaction (PCR) products was performed using the services of Macrogen (Macrogen Inc.; Seoul, Korea). Sequences of the genes were analyzed by Geneious prime v. 2020.5 (Biomatters, Ltd.; Auckland, New Zealand) and the allelic profile was determined using Klebsiella pneumoniae MLST databases. 16
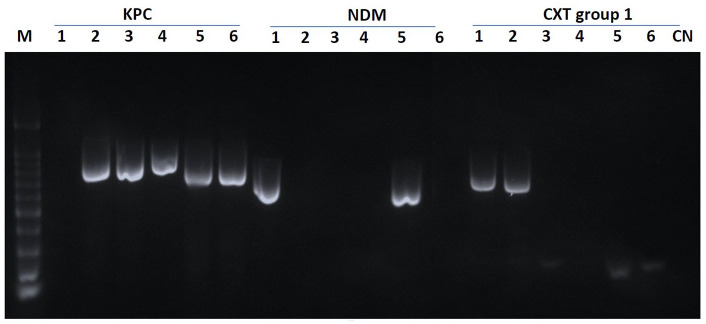
The carbapenemases and CTX-M genes were not sequenced. Instead, we detected these resistant genes among the six isolates using 11 specific PCR primers as described by Ding et al. 17 The PCR end point products were screened for carbapenemase genes (blaKPC, blaVIM, blaIMP, blaNDM, and blaOXA-48) and extended-spectrum β-lactamase (ESBL)-encoding genes (blaTEM, blaSHV, blaOXA-1-4-30, and blaCTX-M group 1, 2, 9). 3 The lengths of the PCR products were analyzed by gel electrophoresis in 2% agarose and visualized under ultraviolet (UV) light (Figure 1).
Figure 1.
Agarose gel electrophoresis of resistance genes (bp). Lines 1–6: ordered sample results (S-475386, S-476139, S-475143, S-472907, S-475338, S-477315); CN, control water; M, DNA ladder (100-bp DNA ladder).
DNA sequencing was performed for amplification of the 16S rRNA gene. The six isolates were analyzed using the universal primers 27F and 1492R, and DNA sequencing was determined. The results obtained were further analyzed against the BLAST database. The identity and maximum coverage were compared with a partial sequence of Klebsiella pneumoniae spp ozaenae strain ATCC 11296 16S ribosomal RNA (NCBI Ref.: NR_041750.1) as well as other three Klebsiella sp were used as reference. Alignment of the obtained and downloaded sequences was established using MEGA X software version 10.2.5. A phylogenetic tree was constructed using MEGA software employing the Neighborhood-Joining method based on the Maximum Composite Likelihood model. The Bootstrap test was set to 1000 replicates in order to increase the reliability of the tree. The optimal phylogenetic Neighbor-Joining tree for the five isolates is shown in Figure 2. Strain 465386 was not sequenced. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.18,19 The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are expressed as the number of base substitutions per site. 20 This analysis involved nine nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). A total of 1530 positions were included in the final dataset. Evolutionary analyses were conducted using MEGA X. 21
Results
Ten out of 11 patients with a confirmed Gram-negative bacteria infection died. Strains from only six patients were recovered for further identification and drug susceptibility testing. We were unable to re-grow the remaining strains after storage in the laboratory. Four of these patients succumbed to sepsis; one died with sepsis and coronavirus disease 19 (COVID-19); and one patient survived. The six strains isolated from the clinical specimens were identified as Klebsiella pneumoniae spp ozaenae by the VITEK-2 Compact System (BioMérieux, Madrid, Spain). In addition, these six Klebsiella pneumonia spp ozaenae isolates were found to be resistant to most of the clinical antimicrobials tested, including Nalidixic acid, Cefepime, Cefotaxin, Ceftazidime, Cefuroxime acetil, Ciprofloxacin, Imipenem, Meropenem, Nitrofurantoin, Piperacillin-tazobactam, Trimethoprim-sulfamethoxazole, Cephalothin and Cefazolin. Of these six isolates, four were sensitive to Gentamicin (GEN) and five were sensitive to Amikacin (AMK).
The antibiotic resistance genes were detected by PCR amplification using specific primers. 17 Four isolates had KPC genes present (S-475338, S-472907, S-475142, and S-477315), while two presented NDM genes (S-475338 and S-465386); however, no resistance genes were detected (blaVIM, blaIMP, Oxa-48) (Table 1). With regards to extended-spectrum β-lactamase genes, just two isolates showed resistance genes for the CTX-M group 1 (S-475386, S-476139), while no PCR products were observed for the remaining genes (blaTEM, blaSHV, blaOXA-1-4-30, and blaCTX-M group 1, 2, 9) (Figure 1).1,22 Table 1 describes the results of molecular typing using Pasteur’s MLST technique. Klebsiella pneumoniae spp ozaenae ST258 was the most frequently identified sequence type (ST) and was present in five of the six isolates (83.3%).
Table 1.
Clinical deaths caused by Klebsiella pneumoniae ssp ozaenae bloodstream infections.
| ID | Age (years) /sex | Sample (culture) | Antimicrobial susceptibility testing by Vitek-2 method | Sequence typing | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NA | AMK | FEP | CTX | CAZ | CXM-A | CXM-S | CIP | GEN | IPM | MEM | NIT | TZP | STX | CEF | CFZ | Carbapenemase β-Lactamases | ST | ||||
| S-475338 | 62/F | Peripheral blood | R | S | R | R | R | R | R | R | S | R | R | R | R | R | – | – | NDM/KPC | 258 | ND |
| S-472907 | 65/F | Tracheal secretion | R | S | R | R | R | R | R | R | S | R | R | R | R | R | – | – | KPC | 258 | ND |
| S-475142 a | 64/M | Blood | R | S | R | R | R | R | R | R | S | R | R | R | R | R | – | – | KPC | 258 | ND |
| S-477315 | 63/F | Urine | R | INT | R | R | R | R | R | R | S | R | R | R | R | R | R | – | KPC | 258 | ND |
| S-476139 b | 65/F | Central venous catheter blood | R | S | R | R | R | R | R | R | R | R | R | R | R | R | – | – | CTX-M-group-1 | 258 | ND |
| S-465386 c | 73/M | Tracheal secretion | – | S | R | R | R | R | R | R | R | R | R | – | R | R | – | R | NDM CTX-M-group-1 |
N/D | ND |
AMK, Amikacin; CAZ, ceftazidime; CEF, cephalothin; CFZ, cefazolin; CIP, ciprofloxacin; CTX, Cefotaxin; CXM-S, Cefuroxime; IPM, Imipenem; CXM-A, Cefuroxima acetil; FEP, Cefepime; GEN, gentamicin; INT, Intermedium; KPC, Klebsiella pneumoniae carbapenemase; MEM, meropenem; NA, Nalidixic acid; ND, No detected; N/D, not determined; NDM, New Delhi metallo-β-lactamase; NIT, nitrofurantoin; R, resistance; s, sensitive; TZP, piperacillin-tazobactam; –, no data.
Isolated both Klebsiella pneumoniae spp ozaenae and P. aeruginosa.
Isolated both Klebsiella pneumoniae spp ozaenae and K. ssp pneumoniae.
Klebsiella pneumoniae ssp pneumone.
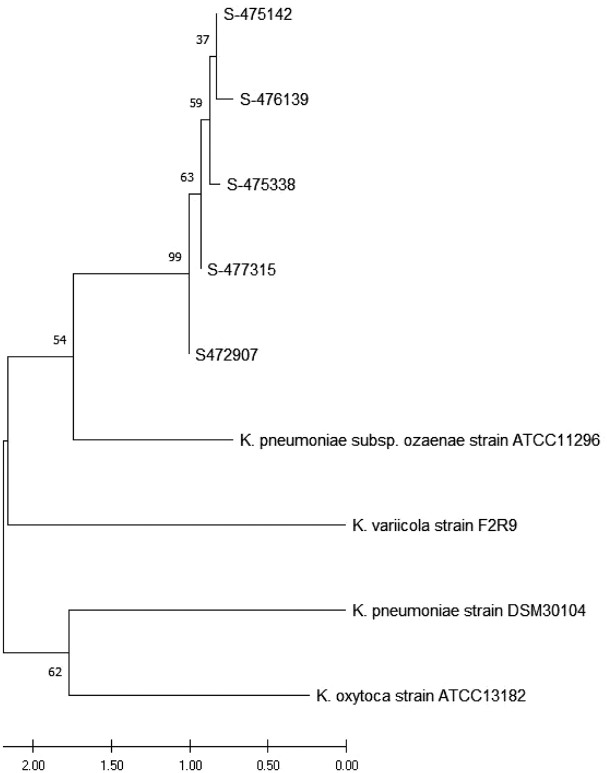
Based on their biological and biochemical characteristics, and the results of 16S rRNA gene sequencing, the six isolates were identified as Klebsiella pneumoniae spp ozaenae. The 16 S rRNA analysis of five isolates was amplified and sequenced using cut-off points of 80% or higher. These strains shared 99% sequence similarity with the reference strain, Klebsiella pneumoniae spp ozaenae ATCC 11296 (NCBI Ref: NR_041750.1) Figure 2.
Figure 2.
Neighbor-joining phylogenetic tree inferred from a 16 S rRNA dataset. A 50% majority-rule consensus tree was deduced from the 16 S rRNA gene sequences; posterior probabilities are shown at each node. Klebsiella pneumoniae spp ozaenae strain ATCC 11296 (NCB: NR_041750.1), Klebsiella oxytoca strain ATCC 13182, Klebsiella variicola strain F2R9 16 S, Klebsiella pneumoniae strain DSM 30104, Klebsiella pneumoniae strain ATCC 13883 were used as the reference. The percentage of replicate trees in which the associated taxa clustered together using the bootstrap test (1000 replicates) is shown next to each branch. The scale bar represents 0.050 substitutions per nucleotide position.
Discussion and conclusion
Klebsiella pneumoniae remains the most frequently isolated bacteria in this hospital center and acquires antimicrobial resistance faster than other Gram-negative pathogens. We focused our efforts on Klebsiella pneumoniae spp ozaenae because this spp has emerged with a higher resistance profile, producing severe infections that are difficult to control. In fact, animal studies have shown that mice infected with Klebsiella pneumoniae spp pneumoniae demonstrate transient signs of infection but then fully recover. In contrast, those infected with Klebsiella pneumoniae spp ozaenae developed pneumonia within 24 h and died between 48 and 72 h after infection. 23 Thus, the identification and characterization of Klebsiella pneumoniae spp ozaneae is especially significant for antimicrobial stewardship efforts. Our results confirm that the six Klebsiella pneumoniae spp ozaenae isolates were genetically diverse. 24 Also, we found that 5/6 (83.3%) had either ESBL resistance genes or at least one of the carbapenem resistance genes evaluated, defining these as multidrug-resistant Klebsiella pneumoniae spp ozaenae isolates.
The global dissemination of resistant Klebsiella pneumoniae carbapenemases has been predominantly associated with high-risk clones, with ST258 being the most commonly identified in North and South America. 25 Horizontal gene transfer between Enterobacteriaceae colonizing the human gut is frequent; for example, from Klebsiella pneumoniae to Escherichia coli. Recently, in Panama, the Escherichia coli ST131 clone has been identified in both inpatients and outpatients. 26 Both Escherichia coli ST131 and Klebsiella pneumoniae ST258 clones are recognized for their high risk of transmission, virulence and nosocomial morbi-mortality, which in the current context of the COVID-19 pandemic poses an extraordinary threat. 27 Reservoirs in the form of healthcare workers, patients, hospital settings and the community further contribute to lateral gene exchange between these high-risk clones, increasing the risk to patients.
The Klebsiella pneumoniae ST258 clone shows high plasticity, leading to high genetic variability. 28 The above is supported by the 16 S rRNA gene phylogenetic analysis of this study, which shows high genetic diversity among the strains. Such diversity suggests that a process of in-hospital evolutionary divergence has occurred, as has been demonstrated in other studies. 29 Moreover, we conclude that the Klebsiella pneumoniae spp ozaenae isolates from this study may have incorporated antibiotic resistance genes in the hospital setting.
Our study analyzes a small sample set including six clinical isolates. Although this limits the span of our conclusions, our findings highlight the need to reinforce antimicrobial stewardship in clinical practice. Investigating additional cases with fatal outcomes can alert clinicians to the existence of other uncommon resistance mechanisms within these multidrug-resistant strains. Appropriately monitoring and surveilling the appearance of such superbugs would allow clinicians to provide appropriate therapies to decrease mortality. For now, we support the notion that clinical antimicrobial stewardship should determine the antibiotic decision algorithm in any given hospital setting.
Multidrug-resistant bacteria remain difficult to treat. 30 Further characterization using a pan-genomic approach would contribute to a more complete depiction of the genes conferring the phenotypic resistance pattern. 31 We also recommend strengthening microbiological surveillance to assure a complete description of these bacterial communities within hospitals. Well-structured local and national plans for infection prevention, antimicrobial stewardship and comprehensive antibiotic control policies are warranted to limit the spread of these superbugs. The implementation of hand washing procedures as well as decontamination and sanitization measures will further decrease bacterial transmission.
Acknowledgments
The authors would like to acknowledge the support of the nurses, physicians, and laboratory staff involved in the diagnosis and management of the patients where these strains were obtained. The authors thank Colleen Goodridge for her critical review of this manuscript and valuable suggestions. We also thank Axel Villalobos from Instituto de Innovación Agropecuarias de Panama—(IDIAP) for his support with Molecular Biology Laboratory (LABMA).
Footnotes
Author contributions: SV, FA, AG, VNS and IL conceptualized the study, FA, JG, MM and GPP conducted experiments in the laboratory. SV, FA, AG, VNS, IL analyzed the data, SV, FA, AG, VNS, IL wrote and drafted the manuscript, all author drafted, edited and approved the mansucript
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: S.V., A.G., V.N.S. and I.L. were supported by the Sistema Nacional de Investigación (SNI) of Panama’s Secretaría Nacional de Ciencia, Tecnología e Innovación (SENACYT).
Ethicals statement: Ethical approval was not required for this case series, as it was based on anonymized information. The authors analyzed an existing, anonymized bacterial strain collection, which was already stored in the hospital laboratory freezer, as part of the local standard of care. None of the authors were involved in obtaining the patient samples, nor had access to the patient’s full medical information. This information was anonymized by hospital laboratory staff, who work independently from the author team.
ORCID iD: Amador Goodridge  https://orcid.org/0000-0003-3910-0482
https://orcid.org/0000-0003-3910-0482
Contributor Information
Silvio Vega, Laboratorio Clínico, Complejo Hospitalario Metropolitano Dr. Arnulfo Arias Madrid, Caja de Seguro Social (CHMDrAAM-CSS), Panama City, Panama.
Fermín Acosta, Tuberculosis Biomarker Research Unit at Centro de Biología Molecular y Celular de Enfermedades (CBCME) del Instituto de Investigaciones Científicas y Servicios de Alta Tecnología (INDICASAT-AIP), City of Knowledge, Panama.
Iván Landires, Instituto de Ciencias Médicas, Las Tablas, Panama.
Mitchelle Morán, Tuberculosis Biomarker Research Unit at Centro de Biología Molecular y Celular de Enfermedades (CBCME) del Instituto de Investigaciones Científicas y Servicios de Alta Tecnología (INDICASAT-AIP), City of Knowledge, Panama.
Johanna Gonzalez, Laboratorio Clínico, Complejo Hospitalario Metropolitano Dr. Arnulfo Arias Madrid, Caja de Seguro Social (CHMDrAAM-CSS), Panama City, Panama.
Gumercindo Pimentel-Peralta, Instituto de Ciencias Médicas, Las Tablas, Panama.
Virginia Núñez-Samudio, Instituto de Ciencias Médicas, P.O. Box 0710-00043, Las Tablas, Los Santos, Panama.
Amador Goodridge, Tuberculosis Biomarker Research Unit, Centro de Biología Molecular y Celular de Enfermedades (CBCME) del Instituto de Investigaciones Científicas y Servicios de Alta Tecnología (INDICASAT-AIP), P.O. Box 0843-01103, City of Knowledge, Panama.
References
- 1. Pishtiwan AH, Khadija KM. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumoniae and Escherichia coli isolated from thalassemia patients in Erbil, Iraq. Mediterr J Hematol Infect Dis 2019; 11: e2019041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moghnieh RA, Kanafani ZA, Tabaja HZ, et al. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. Lancet Infect Dis 2018; 18: e379–e394. [DOI] [PubMed] [Google Scholar]
- 3. Ferreira RL, da Silva BCM, Rezende GS, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and beta-lactamase encoding genes in a Brazilian Intensive Care Unit. Front Microbiol 2018; 9: 3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sugumar M, Kumar KM, Manoharan A, et al. Detection of OXA-1 beta-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS ONE 2014; 9: e91800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antunes NT, Fisher JF. Acquired class D beta-lactamases. Antibiotics 2014; 3: 398–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lopez-Vinau T, Penalva G, Garcia-Martinez L, et al. Impact of an antimicrobial stewardship program on the incidence of carbapenem resistant Gram-negative bacilli: an interrupted time-series analysis. Antibiotics 2021; 10: 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bassetti M, Peghin M. How to manage KPC infections. Ther Adv Infect Dis 2020; 7: 2049936120912049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanders CC, Peyret M, Moland ES, et al. Potential impact of the VITEK 2 system and the advanced expert system on the clinical laboratory of a university-based hospital. J Clin Microbiol 2001; 39: 2379–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skurnik D, Clermont O, Guillard T, et al. Emergence of antimicrobial-resistant Escherichia coli of animal origin spreading in humans. Mol Biol Evol 2016; 33: 898–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Venditti C, Butera O, Meledandri M, et al. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin Microbiol Infect 2021; 27: 1040.e1041–1040.e1046. [DOI] [PubMed] [Google Scholar]
- 11. Loconsole D, Accogli M, De Robertis AL, et al. Emerging high-risk ST101 and ST307 carbapenem-resistant Klebsiella pneumoniae clones from bloodstream infections in Southern Italy. Ann Clin Microbiol Antimicrob 2020; 19: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khan HA, Baig FK, Mehboob R. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian Pac J Trop Biomed 2017; 7: 478–482. [Google Scholar]
- 13. Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 1997; 35: 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brisse S, Fevre C, Passet V, et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS ONE 2009; 4: e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diancourt L, Passet V, Verhoef J, et al. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005; 43: 4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pasteur I. Institut Pasteur MLST and whole genome MLST databases, https://bigsdb.pasteur.fr/klebsiella/klebsiella.html (2021, accessed 10 September 2021).
- 17. Ding L, Yang Z, Lu J, et al. Characterization of phenotypic and genotypic traits of Klebsiella pneumoniae from lung cancer patients with respiratory infection. Infect Drug Resist 2020; 13: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 19. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 2004; 101: 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018; 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delgado DY, Barrigas ZP, Astutillo SG, et al. Detection and molecular characterization of beta-lactamase genes in clinical isolates of Gram-negative bacteria in Southern Ecuador. Braz J Infect Dis 2016; 20: 627–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Renois F, Jacques J, Guillard T, et al. Preliminary investigation of a mice model of Klebsiella pneumoniae subsp. ozaenae induced pneumonia. Microbes Infect 2011; 13: 1045–1051. [DOI] [PubMed] [Google Scholar]
- 24. Rodriguez I, Novais A, Lira F, et al. Antibiotic-resistant Klebsiella pneumoniae and Escherichia coli high-risk clones and an IncFII(k) mosaic plasmid hosting Tn1 (blaTEM-4) in isolates from 1990 to 2004. Antimicrob Agents Chemother 2015; 59: 2904–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee CR, Lee JH, Park KS, et al. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 2016; 7: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nunez-Samudio V, Pecchio M, Pimentel-Peralta G, et al. Molecular epidemiology of Escherichia coli clinical isolates from Central Panama. Antibiotics 2021; 10: 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Canton R, Gijon D, Ruiz-Garbajosa P. Antimicrobial resistance in ICUs: an update in the light of the COVID-19 pandemic. Curr Opin Crit Care 2020; 26: 433–441. [DOI] [PubMed] [Google Scholar]
- 28. Garcia-Fulgueiras V, Magallanes C, Reyes V, et al. In vivo high plasticity of multi-drug resistant ST258 Klebsiella pneumoniae. Microb Drug Resist 2021; 27: 1126–1130. [DOI] [PubMed] [Google Scholar]
- 29. Marsh JW, Mustapha MM, Griffith MP, et al. Evolution of outbreak-causing carbapenem-resistant Klebsiella pneumoniae ST258 at a Tertiary Care Hospital over 8 years. mBio 2019; 10: e01945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Senard O, Bouchand F, Deconinck L, et al. Efficacy of cefoxitin for the treatment of urinary tract infection due to extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates. Ther Adv Infect Dis 2019; 6: 2049936118811053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caputo A, Merhej V, Georgiades K, et al. Pan-genomic analysis to redefine species and subspecies based on quantum discontinuous variation: the Klebsiella paradigm. Biol Direct 2015; 10: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]