Abstract
Glioma is the most common primary intracranial tumor. Abnormal expression of CBX2 (ChromoBox2) is associated with tumorigenesis and tumor development. TCGA data in UALCAN showed that CBX2 was overexpressed in glioma tissue. To confirm the role of CBX2 in glioma, we regulated the level of CBX2 and conducted colony formation, Transwell, and CCK-8 assays to verify the effect of CBX2. The results showed that CBX2 knockdown reduced glioma cell proliferation and invasion and that the cells were less tumorigenic. CBX2 overexpression induced glioma cell proliferation and invasion and glioma stem cell self-renewal. The animal experiments showed that CBX2 knockdown inhibited glioma growth and improved survival time. CBX2 knockdown inhibited activation of the Akt/PI3K pathway. epidermal growth factor rescued the effects of CBX2. CBX2 could induce the growth and invasion of glioma cells via the Akt/PI3K pathway.
Keywords: glioma, CBX2, Akt, PI3K
Introduction
Glioma is the most common primary intracranial tumor and is characterized by aggressive growth, the difficulty of complete resection, and easy relapse. 1 Most gliomas are characterized by invasive growth. Surgical resection combined with regular radiotherapy and chemotherapy is still the main treatment, but the treatment efficacy is poor, and relapse easily occurs. 2 It has been reported that the occurrence of gliomas is closely related to the abnormal expression of various genes and is usually accompanied by abnormally high expression of oncogenes and the deletion or low expression of tumor suppressor genes. 3 Therefore, it has become a hot topic in the field of medical research to further explore the root causes of glioma occurrence and development at the genetic level.
ChromoBox (CBX) proteins include 8 members and are crucial proteins associated with gene expression and development.4,5 Five members make up the PRC1 complex: CBX2, CBX4, CBX6, CBX7, and CBX8. 6 Studies have shown that CBX2 can bind H3K9me3 or H3K27me3, and CBX4 is associated with H3K9me3. 6 CBX proteins play important roles in the progression of tumor development. CBX4 overexpression is associated with overall survival in colorectal carcinoma, CBX6 downregulation occurs in glioblastoma, and CBX7 is overexpressed in gastric and prostate cancers.7–10
In this study, we found that CBX2 was overexpressed in glioma compared to normal brain tissue. However, the mechanism was not clear. To confirm the role of CBX2 in the proliferation and invasion of glioma cells, we conducted an in vitro and in vivo study.
Methods
The Cancer Genome Atlas (TCGA) Dataset Analysis
Glioma and normal brain tissue data were obtained from the TCGA database in UALCAN (http://ualcan.path.uab.edu/index.html).
Clinical Samples
Glioma tissues were obtained from patients with glioma from Tianjin Medical University. And normal brain tissues were obtained from patients with cerebral hemorrhage. The brain tissue was collected from June 1, 2019, to December 31, 2019. All samples were collected and used with the consent of patients and their families, and the written informed consent was signed. And the research was approved by the Ethical Committee of Tianjin Medical University General Hospital in Scientific Research Building on May 3, 2019, ethical number: IRB2019-WZ-083. And the manuscript was approved by the Ethical Committee of Tianjin Medical University General Hospital in Scientific Research Building on July 19, 2020, ethical number: IRB2020-WZ-091.
Cell Culture
The glioma cell lines U87, U251, HEB, and LN229 were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C with 5% CO2. The epidermal growth factor (EGF) was used in the study and the concentration was 100 ng/mL. 11
Transfection
U87 cells were transfected with CBX2-shRNA to knock down the expression of CBX2, and the negative control group was transfected with Scramble-shRNA. The sequences are shown in Supplemental Table S1. LN229 cells were transfected with the overexpression plasmid. The overexpression plasmid, pcDNA3.1-C-(k)DKY encoding CBX2 protein.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA isolation was followed by reverse transcription, and SYBR Green master mix (Promega) was used for qRT-PCR. The CBX2 primer sequences were as follows: forward: 5′-GCCCAGCACTGGACAGAAC-3′ and reverse: 5′-CACTGTGACGGTGATGAGGTT-3′. GAPDH was used as the reference gene, and the primer sequences for GAPDH were as follows: forward: 5′-TGTGGGCATCAATGGATTTGG-3′ and reverse: 5′-ACACCATGTATTCCGGGTCAAT-3′. The 2−△△Cq method was used to calculate the relative expression.
Western Blotting
Total protein was extracted and mixed with 5 × SDS loading buffer, separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to PVDF membranes (Millipore). The membranes were incubated with primary antibodies overnight and then incubated with HRP-conjugated secondary antibodies. Primary antibody for CBX2 (cat. no. ab235305, dilution ratio1:1000 for Western Blot) and for p-GSK-3β (cat. no. ab75745, dilution ratio1:1000 for Western Blot) were purchased from Abcam. Primary antibodies for N-cadherin (cat. no. 13116, dilution ratio 1:1000 for Western Blot), E-cadherin (cat. no. 14472, dilution ratio 1:1000 for Western Blot), Slug (cat. no.9585, dilution ratio 1:1000 for Western Blot), Snail (cat. no. 3879, dilution ratio 1:1000 for Western Blot), GSK-3β (cat. no. D5C5Z, dilution ratio 1:1000 for Western Blot), Akt (cat. no.4691, dilution ratio 1:1000 for Western Blot), and p-Akt (cat. no.4060, dilution ratio 1:1000 for Western Blot) were purchased from CST. Protein expression was visualized using an enhanced chemiluminescence kit (Millipore). The bands were analyzed with ImageJ software.
Transwell Invasion Assay
Matrigel and DMEM were diluted at a ratio of 1:2 and served as the extracellular matrix, and the Matrigel solidified after being incubated for 30 min at 37°C. After transfection for 48 h, the cells were cultured in the wells with Matrigel under general conditions for an additional 48 h. The invaded cells were detected by staining the cells with crystal violet, and the cells were counted and imaged with a microscope at 200 × magnification.
Cell Counting Kit-8 Assay
The transfected cells were cultured in 96-well plates at 2000 cells per well. Twenty microliters of CCK-8 solution (Solarbio) were added to each well and incubated at 37°C for 4 h after 24, 48, 72, and 96 h of culture. The results were measured via spectrophotometry at 450 nm (BioTek Instruments, Inc.).
Colony Formation Assay
After transection for 48 h, the cells were seeded in 6-well plates at a density of 1000 cells/well and cultured for 14 days. The colonies were fixed with paraformaldehyde (4% w/v) and stained with crystal violet (0.5% w/v). The number of colonies was counted with a microscope (Olympus Corporation).
Extreme Limiting Dilution Analysis
Glioma cells were seeded into 96-well plates and cultured with 100 μL of serum-free DMEM/F12 medium containing B27, insulin, FGF, and EGF. After 14 days, GSCs were detected by extreme limiting dilution analysis. 12
Animal Experiments
To verify the effect of CBX2 knockdown, the U87 cells transfected with CBX2-shRNA1 were injected into the brains of BALB/c-A nude mice by intracranial inoculation. And scramble-shRNA1 were injected into the brains of mice in the scramble group. All mice were purchased from the Animal Center of the Cancer Institute, Chinese Academy of Medical Science. There were 10 mice in each group and there were 20 mice used in the study. The age of the mice was 5 to 6 weeks and the sex of the mice was male, the weight was about 20 g. Transduced U87-luciferase and CBX2-shRNA (2 × 105) cells were injected in the right frontal lobe of the mouse brains by stereotaxic intracranial injection. Body weights were measured every 2 days. The tumor volumes of mice were imaged for Fluc activity using bioluminescence imaging per 7 days. We recorded the weights of the mice and their survival until the mice die. The survival curve analysis uses the Kaplan-Meier method. Log-rank method was used. The mice were sacrificed by cervical dislocation. And the research was approved by the Ethical Committee of Tianjin Medical University General Hospital in Scientific Research Building on May 3, 2019, ethical number: IRB2021-DW-24. We had made efforts to minimize the number of animals utilized and to decrease their suffering. And this study conforms to ARRIVE 2.0 guidelines. The animals were given adequate care in the study. 13
Immunohistochemical Staining
The paraffin-embedded sections were used for immunohistochemical staining detected with the avidin-biotin complex method. The sections were incubated with antibodies specific for CBX2, Ki67, N-cadherin, E-cadherin, slug, and snail overnight at 4°C, followed by secondary antibody incubation at 37°C for 1 h. Primary antibody for CBX2 (cat. no. ab235305, 1:100 for immunohistochemistry [IHC]) was purchased from Abcam. Primary antibodies for N-cadherin (cat. no. 13116, dilution ratio 1:100 for IHC), E-cadherin (cat. no. 14472, dilution ratio 1:100 for IHC), slug (cat. no.9585, dilution ratio 1:100 for IHC), snail (cat. no. 3879, dilution ratio 1:100 for IHC), and Ki-67 (cat. no. 9449, dilution ratio 1:100 for IHC) were purchased from CST (USA). The expression of CBX2, Ki67, N-cadherin, E-cadherin, slug, was determined by coloration with DAB.
Statistical Analysis
The significance of Kaplan-Meier analysis was determined by the log-rank test. Multivariate analysis was performed with a multivariate Cox regression model. The data in the study are presented as the mean ± SD. A value of P < .05 was determined to be statistically significant. Significance was set at *P < .05, **P < .01, ***P < .001. SPSS 17.0 (SPSS, Chicago, IL) was used in the present study.
Results
The Level of CBX2 was Higher in Glioma Tissues than in Normal Brain Tissue
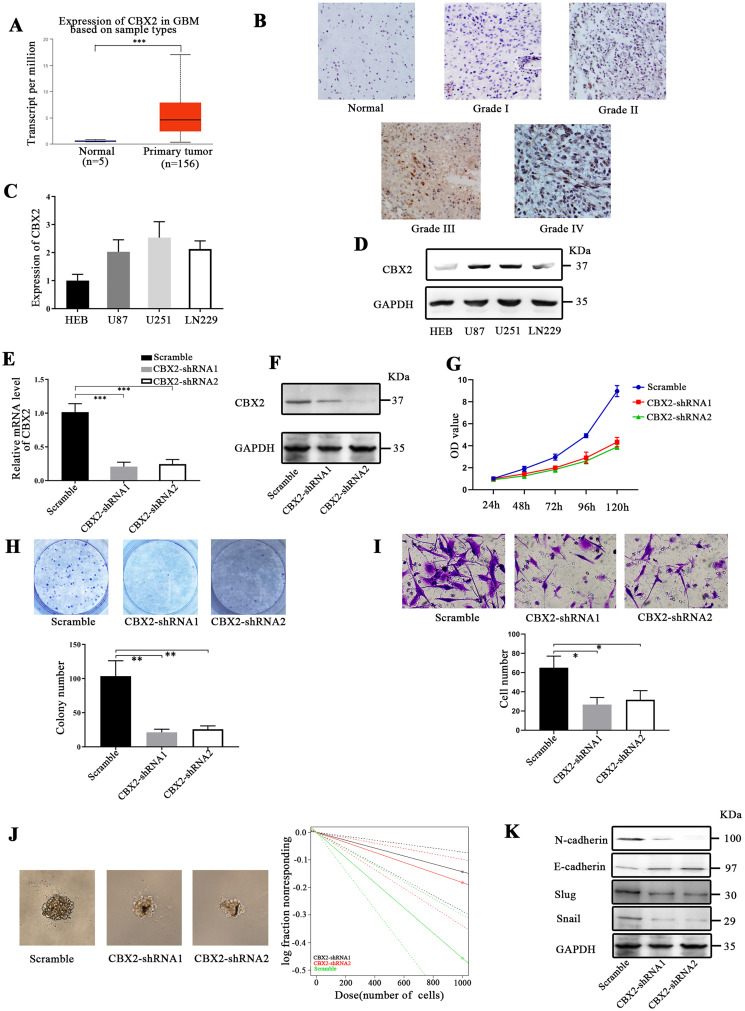
The data obtained from UALCAN demonstrated that CBX2 was overexpressed in glioma compared to normal brain tissues (Figure 1A, P < .001). There was no association between the level of CBX2 and the stage of glioma. Then we detected the expression of CBX2 in human normal brain tissue and glioma tissue. The results showed that the level of CBX2 was higher in glioma tissues. And with the increase of glioma grade, the expression of CBX2 increased, the expression of CBX2 in Grade IV glioma tissues was the highest. It means that the elevated CBX2 expression in glioma tissues was relative to the degree of malignancy (Figure 1B).
Figure 1.
Downregulation of CBX2 inhibited glioma cell proliferation, invasion, and self-renewal. (A) The data (UALCAN, http://ualcan.path.uab.edu/index.html) showed that the expression of CBX2 was higher than that in normal brain tissue (P < .001). (B) The level of CBX2 was higher in glioma tissues. And with the increase of glioma grade, the expression of CBX2 increased, the expression of CBX2 in Grade IV glioma tissues was the highest. (C and D) The expression of CBX2 was analyzed in glioma cell lines by RT-PCR and western blotting. (E and F) CBX2-shRNA transfection efficiencies were verified by RT-PCR and western blotting (P < .001). (G and H) CCK-8 and colony formation assays demonstrated that CBX2-shRNA-mediated knockdown of CBX2 decreased the proliferation of glioma cells (P < .01). (I) Transwell assays showed that CBX2 downregulation reduced invasion compared to that in the scramble group. (J) Sphere formation analysis showed that the downregulation of CBX2 decreased the glioma stem cell frequencies (P < .05). (K) Knockdown of CBX2 reduced the levels of N-cadherin, slug, and snail and increased the level of E-cadherin.
Abbreviations: CBX, ChromoBox; CCK-8, cell counting kit-8; qRT-PCR, quantitative real-time PCR.
CBX2 Induced Glioma Cells Proliferation and Invasion
We measured the level of CBX2 in glioma cell lines by RT-PCR and western blotting (Figure 1C and D). The expression of CBX2 in U87, U251, and LN229 cells was higher than that in the HEB cell line. The U87 cell line was used for CBX2 knockdown, and the LN229 cell line was used for CBX2 overexpression. The efficiencies were verified by RT-PCR and western blotting (Figure 1E and F, P < .001). To analyze the effect of CBX2 on the proliferation, invasion, and stem cell self-renewal of glioma cells, CCK-8, colony formation, Transwell, and extreme limiting dilution assays were conducted. The outcomes showed that downregulation reduced the growth of glioma cells (Figure 1G and H, P < .01). The number of colonies in the scramble group was 103.7 ± 18.4, and the average number of colonies in the CBX2-shRNA1 and CBX2-shRNA2 groups was 21.3 ± 3.7 and 25.6 ± 4.1, respectively. In the Transwell assays, the number of cells that crossed the membrane in the 3 groups were 65.0 ± 9.9, 26.7 ± 6.0, and 31.6 ± 7.7, and the number of cells was decreased following transfection with CBX2-shRNA1 and CBX-shRNA2 (Figure 1I, P < .05). To verify the tumorigenic properties of glioma cells after CBX2 knockdown, the formation of tumor spheres was examined after 14 days. We found that the cells in the scramble group were more tumorigenic than the cells that were transfected with CBX2-shRNA (P < .05 and P < .05, respectively, Figure 1J).
Furthermore, we conducted western blotting to measure the changes in EMT (epithelial to mesenchymal transition)-related proteins after CBX2 knockdown. The expression of N-cadherin, slug, and snail was decreased, and that of E-cadherin was increased. These results indicate that CBX2 knockdown repressed EMT (Figure 1K).
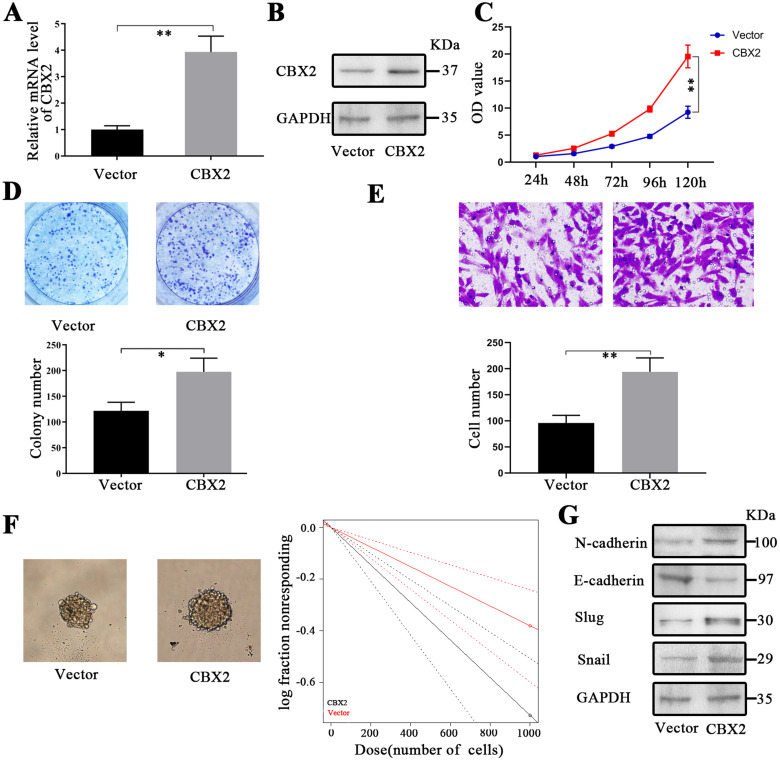
To further confirm the role of CBX2, the level of CBX2 in glioma cells was upregulated by transfection with the CBX2 plasmid. To confirm the overexpression of CBX2 in LN229 cells, RT-PCR and western blotting were conducted after transfection with the CBX2 plasmid (Figure 2A and B). The results of the CCK-8 and colony formation assays showed that the proliferation of glioma cells was improved after transfection with the CBX2 plasmid (Figure 2C and D). The colony numbers in the vector and CBX2 groups were 122.0 ± 11.6 and 197.7.0 ± 18.9, respectively. Then, we measured cell invasion. The number of cells that crossed the membrane in the vector group was 95.7 ± 10.5 and 194.0 ± 18.9 in the CBX2 group, and the number of cells was increased following transfection with the CBX2 plasmid (Figure 2E, P < .01). The tumor spheres were cultured after transfection with the CBX2 plasmid. The cells that were transfected with CBX2 were more tumorigenic than those in the vector group (Figure 2F, P < .01). We conducted western blotting to measure changes in EMT-related proteins after the upregulation of CBX2 (Figure 2G). The levels of N-cadherin, slug, and snail were increased, and the expression of E-cadherin was decreased. These results indicate that the upregulation of CBX2 induced EMT.
Figure 2.
Upregulation of CBX2 induced glioma cell proliferation, invasion, and self-renewal. (A and B) CBX2 plasmid transfection efficiencies were verified by RT-PCR and western blotting (P < .01). (C and D) CCK-8 and colony formation assays demonstrated that CBX2 plasmid-mediated overexpression of CBX2 induced the proliferation of glioma cells (P < .01, P < .05). (E) Transwell assays showed that CBX2 upregulation induced invasion compared to that of the scramble group (P < .01). (F) The glioma stem cell frequency was increased following transfection with the CBX2 plasmid, and the effects were analyzed by sphere formation assays. (G) The upregulation of CBX2 reduced the level of E-cadherin and increased the expression of N-cadherin, slug, and snail.
Abbreviations: CBX, ChromoBox; CCK-8, cell counting kit-8; qRT-PCR, quantitative real-time PCR.
CBX2 Increased Tumor Growth in the Animal Model
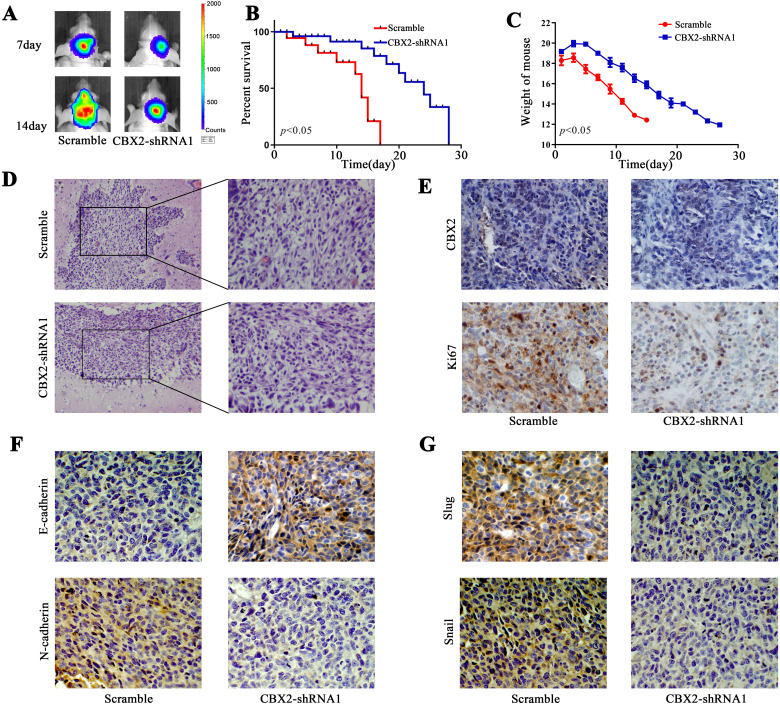
The orthotopic mouse model was established with U87 cells. The tumors were analyzed by Fluc bioluminescence imaging, and the tumors were suppressed in the CBX-shRNA1 group (Figure 3A). CBX2 knockdown improved the survival time compared to that of the control group (Figure 3B, P < .05). Mouse body weight was measured, and we found that weight loss in the control group was obvious (Figure 3C, P < .05). The expression of CBX2 and Ki67 in the tumor tissue was determined by immunohistochemical staining. The level of CBX2 was reduced in the CBX2-shRNA1 group. The level of Ki67 indicates the proliferation of cells, and Ki67 was decreased following CBX2 knockdown (Figure 3E). The level of EMT markers, N-cadherin, slug, and snail was decreased after the transfection of CBX2-shRNA1. The level of E-cadherin was increased after the transfection of CBX2-shRNA1 (Figure 3F and G).
Figure 3.
Knockdown of CBX2 repressed tumor growth in vivo. (A) Fluc bioluminescence images showing the tumors in the nude mouse xenograft model. (B) Overall survival analysis demonstrated that CBX2 downregulation improved the survival time (P < .05). (C) The mouse body weights were evaluated. (D) Images of HE-stained tumor tissue. (E) The expression of CBX2 and Ki67 was analyzed by IHC staining. (F and G) The expression of EMT markers, E-cadherin, N-cadherin, slug, and slug were detected with IHC.
Abbreviations: CBX, ChromoBox; HE, hematoxylin and eosin; EMT, epithelial to mesenchymal transition; IHC, immunohistochemistry.
CBX2 Induced Glioma Cell Proliferation and Invasion by Regulating the Activity of the Akt/PI3K Signaling Pathway
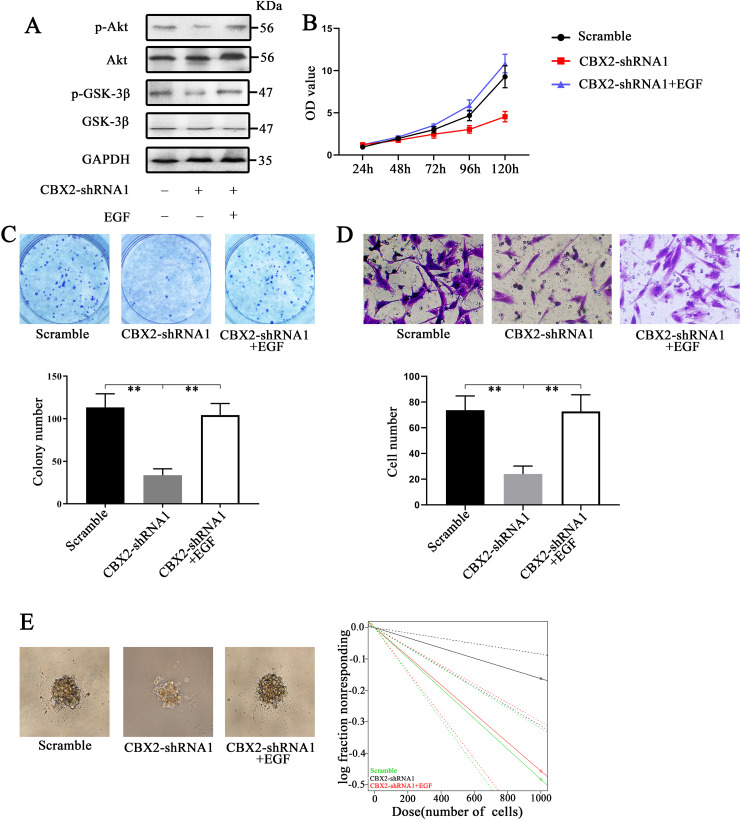
In this study, we treated CBX2-knockdown cells with EGF (an agonist of the Akt/PI3K pathway, 100 ng/mL) to rescue the effects of CBX2 inhibition. 11 We found that EGF upregulated the levels of p-Akt and p-GSK-3β (Figure 4A). CCK-8 and colony formation assays were also performed. The colony numbers in the 3 groups were 113.3 ± 11.3, 33.7 ± 5.3, and 104.3 ± 9.6 (Figure 4C, P < .01). The Transwell assay results demonstrated that the number of cells that crossed the membrane in the 3 groups were 73.7 ± 7.9, 24.0 ± 4.4, and 72.6 ± 9.2 (Figure 4D, P < .01). In addition, the cells were more tumorigenic than those in the CBX2-shRNA1 group (Figure 4E, P < .01). These results showed that EGF increased cell growth, invasion, and stem cell self-renewal in CBX2-shRNA1 cells. We concluded that CBX2 induced glioma cell proliferation and invasion via the Akt/PI3K pathway.
Figure 4.
Knockdown of CBX2 inhibited glioma cell proliferation and invasion and glioma stem cell self-renewal via the downregulation of the Akt/PI3K pathway. (A) Knockdown of CBX2 inhibited the levels of p-Akt and p-GSK-3β, reducing the activity of the Akt/PI3K pathway. The use of EGF, which is an Akt/PI3K pathway agonist, increased the levels of p-Akt and p-GSK-3β. (B and C) CCK-8 and colony formation assays showed that EGF rescued the proliferation of glioma cells. (D) Sphere formation assays showed that the cells in the CBX2-shRNA1 + EGF group were more tumorigenic than those in the CBX2-shRNA1 group (P < .05).
Abbreviations: CBX, ChromoBox; CCK-8, cell counting kit-8; EGF, epidermal growth factor.
Discussion
Recently, studies have shown that the expression of CBX2 is closely related to tumorigenesis and tumor development. Some studies have shown that the expression of CBX2 in osteosarcoma tissue is significantly increased, and the degree of the increase is closely related to metastasis, postoperative recurrence, chemotherapy reaction, and adverse prognosis. 14 In advanced serous ovarian cancer, the expression of CBX2 was significantly increased. Knockdown of CBX2 significantly inhibited nonanchored proliferation and promoted anoikis in tumor cells, increasing the sensitivity of tumor cells to pods. 15 Studies have shown that abnormally high expression of CBX2 in breast cancer, prostate cancer, and liver cancer is closely related to the degree of malignancy, lymph node metastasis, TNM stage, poor patient prognosis, and sensitivity to chemotherapy drugs.16–18 And the mechanisms were related to the Akt/PI3K pathway and miR-185 to 5p axis. There was study showed that CBX mainly targets PRC1 to chromatin, and the C-terminal polycomb receptor box of CBX2 can specifically recognize H3K27me3, thus enhancing the inhibition of gene expression. 19
The expression of CBX2 in glioma, its effect on glioma, and its mechanism have not been reported. In our study, we found that the level of CBX2 was higher in glioma tissue than in normal brain tissue. Using CCK-8, colony formation, and Transwell assays, we found that CBX2 played an important role in glioma progression. Moreover, we found that CBX2 affected the biological function of glioma cells via the Akt/PI3K pathway. The Akt/PI3K signaling pathway is an important signal transduction pathway in cells that participates in the regulation of many important biological processes, blocks cell apoptosis, and promotes the occurrence and development of various human tumors. 20 Many cytokines can activate the PI3K/Akt signaling pathway in many kinds of tumors, such as liver cancer and breast cancer.21,22 In our study, we found that CBX2 knockdown decreased the expression of p-Akt and p-GSK-3β, which are core proteins of the PI3K/Akt pathway. And the knockdown of CBX2 decreased the expression of N-cadherin, slug, and snail, which were the biomarkers of the EMT process. Cancer cells that have undergone EMT are more aggressive, displaying increased invasiveness, stem-like features, and resistance to apoptosis. So we considered that CBX2 promoting proliferation and invasion of glioma cells through the Akt/PI3K pathway acting on the EMT process. However, the detailed action mechanism of CBX2 needs more work to research as tumorigenesis is a very complex process.
In conclusion, compared with that of normal tissues, the level of CBX2 in gliomas was significantly increased. In vitro and in vivo experiments confirmed that CBX2 knockdown could significantly inhibit the formation of glioma. Further studies showed that CBX2 knockdown decreased the expression of p-Akt and p-GSK-3β, suggesting that CBX2 may play a role as an oncogene by activating the PI3K/Akt signaling pathway.
Supplemental Material
Supplemental material, sj-xlsx-1-tct-10.1177_15330338211045831 for CBX2 Induces Glioma Cell Proliferation and Invasion Through the Akt/PI3K Pathway by Le Wang, Bingcheng Ren, Hao Zhuang, Yue Zhong and Yang Nan in Technology in Cancer Research & Treatment
Acknowledgments
Le Wang, Yang Nan, and Yue Zhong designed this study and provide guidance for research. Le Wang and Bingcheng Ren conducted the word and analyzed the data. Le Wang wrote the manuscript. All authors have approved the manuscript.
Abbreviations
- CBX
ChromoBox
- CCK-8
cell counting kit-8
- DMEM
Dulbecco's modified Eagle's medium
- EMT
epithelial to mesenchymal transition
- FBS
fetal bovine serum
- qRT-PCR
quantitative real-time PCR
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TCGA
The Cancer Genome Atlas.
Footnotes
Authors’ Note: Le Wang and Bingcheng Ren contributed equally to this work, and the BALB/c nude mice were purchased from the Animal Center of the Cancer Institute, Chinese Academy of Medical Science. And the study was approved by the Animal Ethical and Welfare Committee of Tianjin Medical University General Hospital. Ethical number: IRB2021-DW-24. The experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The glioma samples were selected from Tianjin Medical University. And the research was approved by the Ethical Committee of Tianjin Medical University General Hospital, ethical number: IRB2020-WZ-091. Informed consent for the study was obtained from each patient.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The study was supported by Tianjin Natural Science Foundation (18JCZDJC98600); Science and Technology Project of Tianjin Binhai New Area Health Commission (2018BWKY010), Science and Technology Fund of Tianjin Binhai New Area Health and Family Planning Commission (2018BWKZ003); Tianjin Medical University General Hospital Incubation Funding (grant no. 303071902101).
ORCID iD: Le Wang https://orcid.org/0000-0003-2475-5312
Supplemental Material: Supplemental material for this article is available online.
References
- 1.McNeill KA. Epidemiology of brain tumors. Neurol Clin. 2016;34(4):981‐998. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Reardon DA. Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12(2):69‐70. [DOI] [PubMed] [Google Scholar]
- 3.Soomro SH, Ting LR, Qing YY, Ren M. Molecular biology of glioblastoma: classification and mutational locations. J Pak Med Assoc. 2017;67(9):1410‐1414. [PubMed] [Google Scholar]
- 4.Gil J, Bernard D, Martínez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6(1):67‐72. [DOI] [PubMed] [Google Scholar]
- 5.Sun D, Cao X, Wang C. Polycomb chromobox Cbx2 enhances antiviral innate immunity by promoting Jmjd3-mediated demethylation of H3K27 at the Ifnb promoter. Protein Cell. 2019;10(4):285‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aranda S, Mas G, Di Croce L. Regulation of gene transcription by polycomb proteins. Sci Adv. 2015;1(11):e1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Li L, Wu Y, et al. CBX4 suppresses metastasis via recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma. Cancer Res. 2016;76(24):7277‐7289. [DOI] [PubMed] [Google Scholar]
- 8.Li G, Warden C, Zou Z, et al. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One. 2013;8(11):e80970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang XW, Zhang L, Qin W, et al. Oncogenic role of the chromobox protein CBX7 in gastric cancer. J Exp Clin Cancer Res. 2010;29(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard D, Martinez-Leal JF, Rizzo S, et al. CBX7 Controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2015;24(36):5543‐5551. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Li Z, Song Y, et al. AURKA Induces EMT by regulating histone modification through Wnt/β-catenin and PI3K/Akt signaling pathway in gastric cancer. Oncotarget. 2016;7(22):33152‐33164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Cao C, Liu X, et al. BRCA1-associated protein inhibits glioma cell proliferation and migration and glioma stem cell self-renewal via the TGF-β/PI3K/AKT/mTOR signalling pathway. Cell Oncol (Dordr). 2020;43(2):223‐235. [DOI] [PubMed] [Google Scholar]
- 13.Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. Br J Pharmacol. 2020;177(16):3617‐3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Q, Li C, Cao Y, et al. CBX2 Is a functional target of miRNA let-7a and acts as a tumor promoter in osteosarcoma. Cancer Med. 2019;8(8):3981‐3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler LJ, Watson ZL, Qamar L, et al. CBX2 Identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis. 2018;7(11):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen WY, Zhang XY, Liu T, Liu Y, Zhao YS, Pang D. Chromobox homolog 2 protein: a novel biomarker for predicting prognosis and taxol sensitivity in patients with breast cancer. Oncol Lett. 2017;13(3):1149‐1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clermont PL, Crea F, Chiang YT, et al. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin Epigenetics. 2016;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao J, Tian Y, Wang C, et al. CBX2 regulates proliferation and apoptosis via the phosphorylation of YAP in hepatocellular carcinoma. J Cancer. 2019;10(12):2706‐2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma RG, Zhang Y, Sun TT, Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B. 2014;15(5):412‐428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8(8):627‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang JL, Cao SW, Ou QS, et al. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer. 2018;17(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ippen FM, Grosch JK, Subramanian M, et al. Targeting the PI3K/Akt/mTOR pathway with the pan-Akt inhibitor GDC-0068 in PIK3CA-mutant breast cancer brain metastases. Neuro Oncol. 2019;21(11):1401‐1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-tct-10.1177_15330338211045831 for CBX2 Induces Glioma Cell Proliferation and Invasion Through the Akt/PI3K Pathway by Le Wang, Bingcheng Ren, Hao Zhuang, Yue Zhong and Yang Nan in Technology in Cancer Research & Treatment