Abstract
N6-methyladenosine (m6A) methylation has been reported to play a role in type 2 diabetes (T2D). However, the key component of m6A methylation has not been well explored in T2D. This study investigates the biological role and the underlying mechanism of m6A methylation genes in T2D. The Gene Expression Omnibus (GEO) database combined with the m6A methylation and transcriptome data of T2D patients were used to identify m6A methylation differentially expressed genes (mMDEGs). Ingenuity pathway analysis (IPA) was used to predict T2D-related differentially expressed genes (DEGs). Gene ontology (GO) term enrichment and the Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to determine the biological functions of mMDEGs. Gene set enrichment analysis (GSEA) was performed to further confirm the functional enrichment of mMDEGs and determine candidate hub genes. The least absolute shrinkage and selection operator (LASSO) regression analysis was carried out to screen for the best predictors of T2D, and RT-PCR and Western blot were used to verify the expression of the predictors. A total of 194 overlapping mMDEGs were detected. GO, KEGG, and GSEA analysis showed that mMDEGs were enriched in T2D and insulin signaling pathways, where the insulin gene (INS), the type 2 membranal glycoprotein gene (MAFA), and hexokinase 2 (HK2) gene were found. The LASSO regression analysis of candidate hub genes showed that the INS gene could be invoked as a predictive hub gene for T2D. INS, MAFA,and HK2 genes participate in the T2D disease process, but INS can better predict the occurrence of T2D.
Keywords: N6-methyladenosine methylation, type 2 diabetes, INS, MAFA, HK2, bioinformatics
Introduction
N6-methyladenosine (m6A) is one of the most prevalent and abundant methylations in mRNA and viral RNA. The dysregulation of RNA modification has been linked to obesity, diabetes, metabolic syndrome, cancer, and other human diseases (1, 2, 3).
Diabetes is a chronic disease in which the blood glucose level elevates due to either suboptimal production of insulin by the pancreas or insulin resistance in peripheral tissues. It is a major global health threat and was the 15th most important cause of lives lost in 2015. Despite the efforts made by the World Health Organization to reduce the burden of diabetes, its prevalence is increasing and is likely to cause further mortality (4). In China, the incidence of type 2 diabetes (T2D) has rapidly increased over the recent decades, and T2D has become a leading public health challenge (5). Therefore, it is urgent to explore new therapeutic methods for T2D. Moreover, recent research reported that m6A content was decreased in T2D patients, which suggested that mRNA m6A methylation may play a role in the pathogenesis of T2D (6, 7).
In the dynamic regulation of m6A modification, m6A methyltransferases and demethylases play crucial roles. Recent evidence indicates that m6A can be demethylated by fat mass- and obesity-associated protein and alpha-ketoglutarate-dependent dioxygenase alkB homolog 5 protein (6). In T2D, m6A methylation may have an important role in regulating the biological function of human β-cells (8). Nevertheless, the exact role of m6A methylation in T2D has not been explored.
In this study, bioinformatics is used to investigate the biological role and underlying mechanism of m6A methylation genes in T2D, and the INS gene was screened out as the hub gene for T2D, providing a new research direction for T2D molecular research.
Materials and methods
Data acquisition and processing
The raw data of GSE120024, including 30 samples obtained from 7 T2D patients and 8 non-diabetic controls, were downloaded from the GEO database, which is an openly accessible database.
For the quality control of the RNA-seq data, the quality control tool FastQC was used to perform on the raw data. At the same time, the data output of each sample was recorded. After testing, a total of 755,188M clean reads were obtained, and the base quality value Q of each sample was greater than 30, which met the needs of subsequent analysis.
For sequence alignment, after quality control, the clean data were aligned to the reference human genome (Homo sapiens, GRCh38), using the alignment tool hierarchical indexing for spliced alignment of transcripts 2 (HISAT2), and then analyzed. HISAT2 is an efficient comparison system for aligning both DNA and RNA sequences using a graph Ferragina Manzini index (9). The comparison efficiency of reads and the reference genome of each sample were above 95.34%, so biometric analysis could be performed.
The analysis of m6A methylation
To observe the m6A methylation of RNA in T2D patients and healthy human pancreatic islet tissues, the samples were divided into the healthy group and the T2D group. At the same time, each group was divided into the input group (transcriptome sequencing) and the IP group (m6A immunoprecipitation sequencing), and a total of 30 sequencing samples were analyzed for the m6A methylation process.
For obtaining m6A methylated call peaks, model-based analysis of ChIP-Seq (MACS) (https://github.com/taoliu/MACS) is the most popular peak calling software. It was originally designed for the ChIP data of transcription factors. In its latest version, aptamers for histone modifications have been added (10). In this study, MACS2 was used to detect the m6A methylation peak region on RNA to obtain the peak count matrix of 15 patients.
For further analysis of m6A methylation peaks, the R package DESeq2 was used to analyze the difference between groups, and the screening criteria were |log2FC| ≥ 1 and P-value ≤ 0.05 (11).
The analysis of DEGs in the input group
featureCounts is a tool in the subread software package. It is mainly used to calculate the results of the Subread comparison and count reads (12). It can not only support gene quantification but also supports exon, gene bodies, genomic bins, and quantification of chromosomal locations (12). For DEG analysis, the R package DESeq2 was used for differential expression analysis and screened for DEGs by setting the screening criteria (|log2FC| ≥ 1 and adj. P-value ≤ 0.05).
Pathway and network analysis using IPA
The list of DEGs in the GSE120024 dataset, containing gene identifiers and corresponding expression values, was uploaded into the ingenuity pathway analysis (IPA) software. The core analysis function included in the software was used to interpret the differentially expressed data, which comprised biological processes, canonical pathways, upstream transcriptional regulators, and gene networks (13). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathway Knowledge Base.
GO and KEGG pathway functional enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and GO enrichment analyzes were performed using clusterProfiler and default settings. The bubble diagrams were constructed using ggplot2. Protein–protein functional networks were constructed using the online resource Search Tool for the Retrieval of Interacting Genes (STRING).
GESA
Gene set enrichment analysis (GSEA) was used to assess the related pathways and molecular mechanisms in mMDEGs from the T2D patients. The collapse data set to gene symbols was set to true, the permutation type was set to phenotype, the enrichment statistic was set to weight, and the Signal2Noise metric was used for ranking genes. GSEA calculated a gene set running enrichment score, which analyzed genes that were enriched in the biological signal, and the mean normalization method was utilized to obtain the normalized enrichment score (NES) in the gene sets. Enriched gene sets with a nominal P-value < 0.05 were accepted as statistically significant.
LASSO regression analysis
The least absolute shrinkage and selection operator (LASSO) regression analysis is a compression estimation method. It obtains a more refined model by constructing a penalty function, which makes it compress some coefficients and set some coefficients to zero (14). Therefore, the advantage of subset shrinkage is retained, and it is a biased estimate for processing multicollinearity data (15). The degree of LASSO regression complexity adjustment is controlled by the parameter λ. Larger the λ, greater the penalty for the linear model with more variables, so that a model with fewer variables is finally obtained (16).
In this study, a LASSO regression model based on gene expression combined with the R package glmnet was constructed, taking the occurrence of disease as the dependent variable (control = 0; T2D = 1) and different genes as dependent variables. The type was Gaussian, the number of predictive models was lambda = 1000, and the standard parameter of the LASSO regression was alpha = 1.
Sample collection
A total of 30 samples was obtained from 11 T2D patients and 20 non-diabetic controls. The present study was approved by the ethics committee of The Second Hospital Affiliated to Kunming Medical University. The study was consistent with the Declaration of Helsinki of 1964 (including later amendments) and related ethical standards. The samples were collected after written informed consent was obtained.
RNA isolation and real-time PCR
High-quality total RNA (>200 nucleotides) was isolated with the standard Trizol reagent (Invitrogen), according to the manufacturer’s instructions. The RNA quality and quantity were analyzed using a NanoDrop 1000 Spectrophotometer (Thermo Fisher) and then used for RT using the high-capacity cDNA synthesis kit (Applied Biosciences). cDNA syntheses were performed with PrimeScript RT Master Mix (Takara Bio) and amplified with SYBR pre-mix EX Taq (Takara Bio). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The primer sequence was as follows: GAPDH, forward ACAACTTTGGTATCGTGGAAGG, reverse GCCATCACGCCACAGTTTC; INS, forward CAGCCTTTGTGAACCAACACC, reverse TGTTCCACAATGCCACGCT. PCR was performed based on an unpaired approach with 20 healthy samples and 11 T2D samples. It should be noted that the data from the paired group of control 8 and patient 4 were found to be abnormal, so we excluded the data from this group. Therefore, the PCR data presented now had a total of 19 samples in the control group and 11 samples in the T2D group.
Protein isolation and Western blotting
The total protein was collected from peripheral blood mononuclear cell lysates of 20 healthy samples and 11 T2D samples using M-PER protein extraction reagent (Thermo Fisher) supplemented with proteinase and phosphatase inhibitors (Sigma), in line with the manufacturer’s protocols Protein quantitative processing was performed with a bicinchoninic acid assay kit (Sigma). Protein samples were separated on 10% SDS_PAGE and blotted to PVDF membranes, followed by standard western immunoblotting of proteins using different primary antibodies (anti-INS (ab181547, Abcam) and anti-β-actin (4970, Cell Signaling)) at 4°C overnight, and the secondary antibody at room temperature for 2 h. The blots were developed using the Odyssey infrared imaging system (LI-COR Bioscience, Lincoln, NE) and quantified with Image J software. In this study, an unpaired approach was used to combine the healthy and diseased samples.
Statistical analysis
All of the statistical tests were done with R 3.3.1 and GraphPad Prism (version: 8.3.0). All the analyses performed in the Wilcoxon rank-sum test, which is a non-parametric statistical hypothesis test mainly used for comparisons between two groups, were two-sided. A Student's t-test was performed to analyze the RT-PCR and Western blot data, and statistical significance was established at a P-value < 0.05.
Results
A difference analysis of m6A methylation peaks
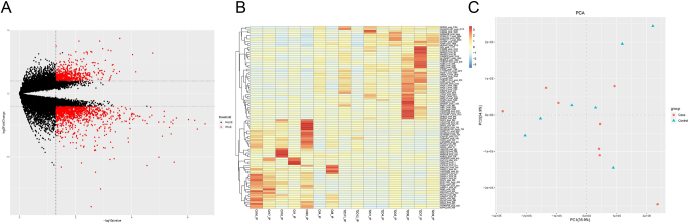
To observe the m6A methylation of RNA on the pancreatic islet tissues of T2D patients and healthy subjects, MACS2 was used to detect the m6A methylation peak region in RNA and the peak count matrices of 15 patients were obtained using the test. A total of 48,706 m6A methylation peaks were obtained, involving a total of 20,157 genes. Then, the R package DESeq2 tool was employed to analyze the differences between groups using the following cut-off values: |log2FC| ≥ 1and P-value ≤ 0.05. As a result, a total of 1912 differential peaks (gene 1719) were identified, including 1290 demethylated peaks (gene 1202) and 622 hypermethylated peaks (gene 517). A volcano map (Fig. 1A) was used to demonstrate the overall distribution of the differential peaks of T2D and control subjects. The top 50 up- and downregulated differential peak genes are clustered together in Fig. 1B, where the red-colored genes are upregulated and the blue-colored genes are downregulated. Moreover, the m6A methylome segregates disease status in a relatively homogeneous manner in the T2D samples (Fig. 1C).
Figure 1.
(A) Volcano map of the overall distribution of 1912 differential peaks between T2D and control. (B) The top 50 up- and downregulated differential peaks genes from A. The red-colored genes were upregulated and the blue-colored genes were downregulated. (C) Principle component analysis (PCA) for the m6A methylome between T2D and control. Roundness pointed T2D samples and triangle pointed control samples.
Identification of T2D-related DEGs
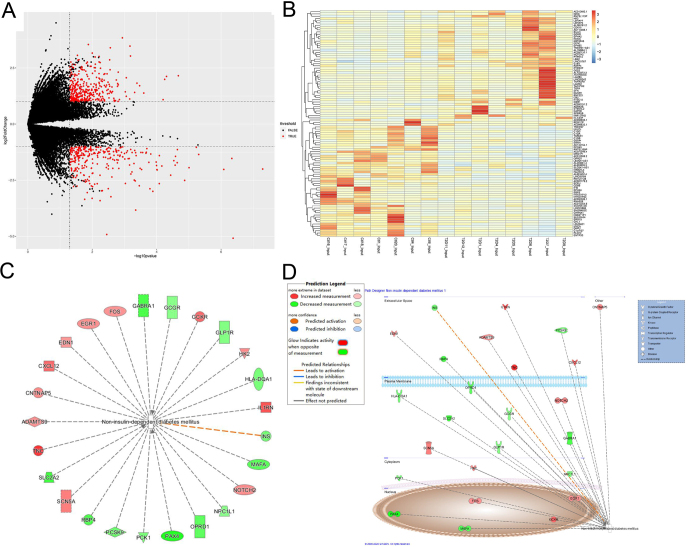
To gain further insight into the difference in gene expression between T2D patients and healthy people, conventional transcriptome sequencing data (input group) were selected in the sequencing samples for differential expression analysis. Using |log2FC| ≥ 1 and adj. P-value ≤ 0.05 as cut-off criteria, a total of 673 DEGs were detected, of which 386 were markedly upregulated and 287 were downregulated. The volcano plot shows up- and downregulated genes in the GSE120024 dataset (Fig. 2A). The expression level of the principal 100 DEGs (50 up- and 50 down-regulated) with −log (P-value) is displayed in Fig. 2B. For an in-depth understanding of the impact of gene expression on diseases and an initial identification of disease-related genes, the DEGs of the T2D group and the control group were combined to perform IPA core analysis. The predicted T2D-related genes and their networks are displayed in Fig. 2C and D.
Figure 2.
(A) Volcano plot of up- and down-regulated genes using |log2FC| ≥ 1 and adj. P-value ≤ 0.05 as cut-off criteria. (B) The expression level of the principle 100 DEGs (50 upregulated and 50 downregulated) with −log (P-value). (C and D) IPA core analysis for the combined DEGs between the T2D group and the control group to predict T2D-related genes and their networks.
GO terms and KEGG pathway enrichment analyses of mMDEGs
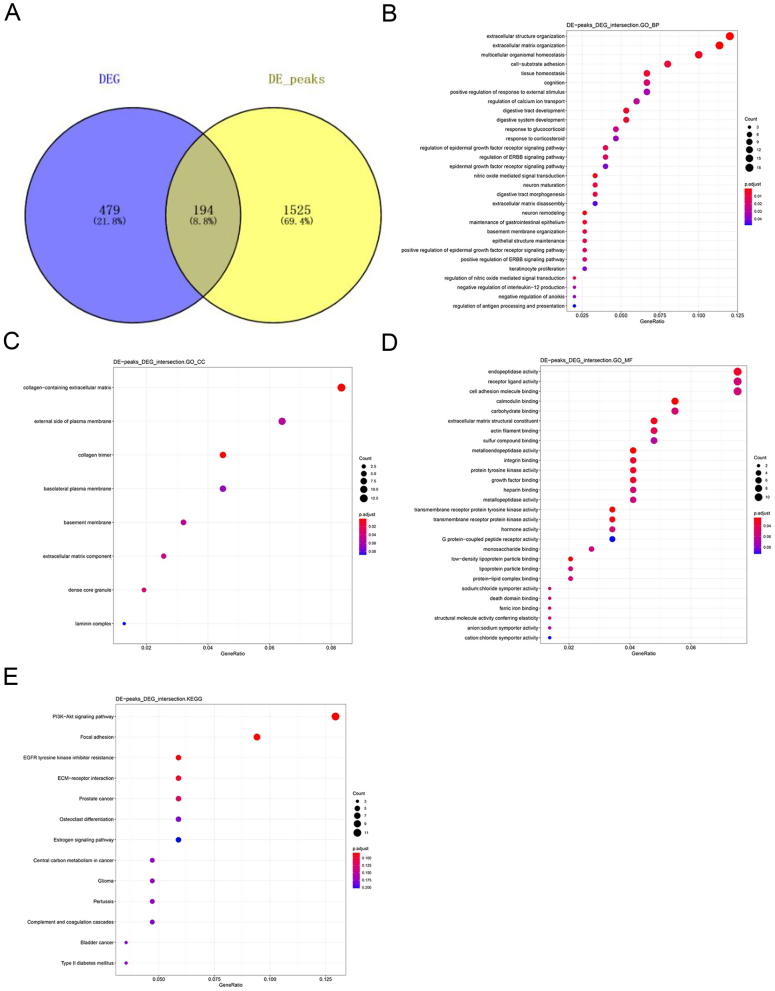
To further understand and explore the role of m6A methylation in T2D, the results of m6A methylation analysis and transcriptome difference analysis were intersected to find the mMDEGs. A total of 194 overlapping mMDEGs were detected after performing an integrated analysis of the above two results (Fig. 3A).
Figure 3.
(A) Venn diagram of intersecting the results of m6A methylation analysis and transcriptome difference ananlysis. (B, C and D) GO terms enrichment analysis of mMDEGs. (B) The top 30 significantly enriched BPs of mMDEGs. (C) All CCs of mMDEGs. (D) The top 10 MFs of mMDEGs. (E) KEGG pathway analysis of mMDEGs.
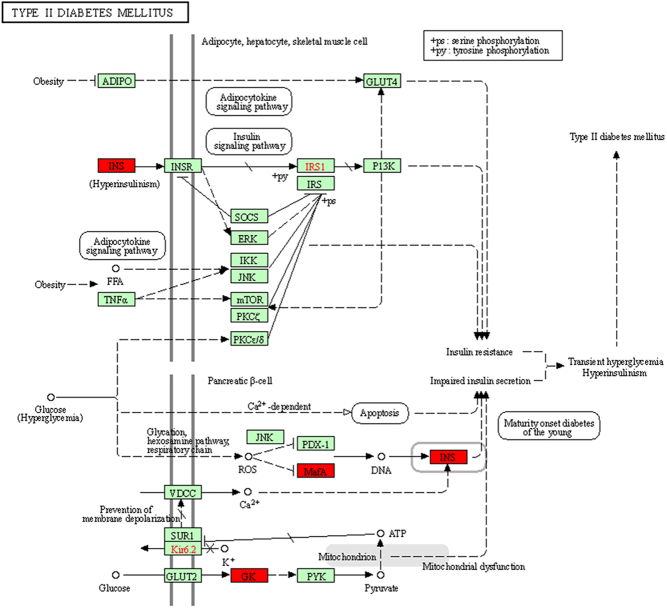
To investigate the biological significance of m6A methylation in human T2D, GO, and KEGG pathway analyses of mMDEGs were performed. GO analysis classifies genes into three functional groups: biological process (BP), cellular component (CC), and molecular function (MF). Figure 3B shows the top 30 significantly enriched BPs. All CCs and top 10 MFs of mMDEGs are shown in Fig. 3C and D. For the KEGG pathway analysis, it was found that mMDEGs were significantly associated with the ‘T2D’ signaling pathway (Fig. 3E). The mMDEGs participating in this pathway were INS, MAFA, and HK2. Thus, in combination with the previous results, it was concluded that the genes INS, MAFA, and HK2 participate in the T2D disease process. The expression of MAFA and INS was down-regulated relative to the healthy group, and the expression of HK2 was up-regulated, which was consistent with the previous analysis. The metabolites of genes HK2 and MAFA participate in the disease process intracellularly, while the INS gene participates in the form of extracellular metabolites. The down-regulation of INS promoted the pathogenesis of T2D, but the specific regulatory effects of HK2 and MAFA in the disease were unclear (Fig. 2C and D). In addition, the online tool varElect was used at the same time to perform disease association analysis on the 194 mMDEGs to understand whether the above genes were tied to the disease process. It was found that a total of 33 genes were directly related to the disease, among which the main three related genes were INS, MAFA, and HK2.
Related research has shown that INS is an insulin-encoding gene that regulates carbohydrate and lipid metabolism in the body (17). Therefore, the downregulation of INS can directly lead to the occurrence and development of diabetes (18). The HK2 gene is related to the insulin response in the body (19). The pathways related to the metabolism of this gene include glucose metabolism (20). It appears that the onset of diabetes comes from a carbohydrate metabolism disorder (21). MAFA, as a transcriptional regulator, can be bound to RIPE3b (enhancer) to regulate the expression of the INS gene (22). These results indicated that the above three genes play a critical role in T2D, so focus shifted to the T2D pathway and its related pathways and genes.
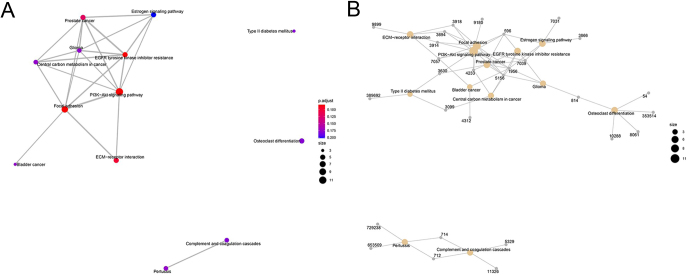
An analysis of related pathways and genes of ‘T2D’
In order to clarify the pathways and genes related to ‘T2D’, in-depth observations were conducted, and it was found that this pathway was independently distributed, indicating that it had no interaction with other enriched pathways (Fig. 4A). It is worth noting that, as shown in Fig. 4B, T2D and the PI3K-Akt signaling pathway share the gene INS (3630). Furthermore, HK2 (3099) is the shared gene of T2D and central carbon metabolism in cancer. MAFA was not noted in other pathways.
Figure 4.
(A and B) Analysis of related pathways (A) and genes (B) of ‘T2D’.
It was concluded that ‘T2D’ is the strategic pathway for mMDEGs to participate in T2D, and the genes involved: INS, MAFA, and HK2 are candidate hub genes. The specifically related mechanism of this pathway is presented in Fig. 5.
Figure 5.
The mechanism of regarding ‘T2D’ as the strategic pathway for mMDEGs.
GSEA for mMDEGs
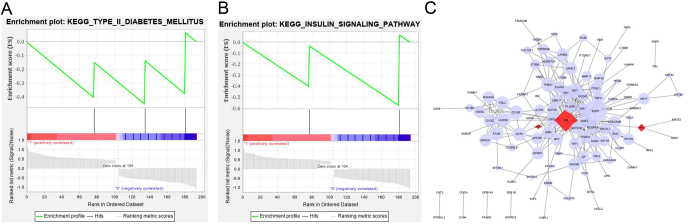
GSEA was utilized to further confirm the functional enrichment of mMDEGs. The enrichment consequences showed pathways closely related to T2D, including T2D (Fig. 6A) and insulin signaling pathway (Fig. 6B). Through observation and confirmation, the genes engaged in the above two T2D-related pathways include INS, HK2, and MAFA, of which INS and HK2 are involved in the two pathways. GSEA analysis also confirmed that the candidate hub genes INS, HK2, and MAFA participate in T2D-related pathways.
Figure 6.
(A and B) GSEA for mMDEGs, including T2D (A) and insulin signaling pathway (B). (C) The PPI network combined Cytoscape. The red diamonds represented T2D candidate hub genes, and the light purple circles represented ordinary mMDEGs.
The PPI network was constructed from the STRING database, and the results showed that the network involved 156 genes, 240 pairs of interactions, and an average link degree of 3.08. Meanwhile, as shown in Fig. 6C, Cytoscape was used to display the network, the red diamonds representing T2D candidate hub genes, and the light purple circles representing ordinary mMDEGs.
Correlation analysis of candidate hub genes methylation and transcription expression
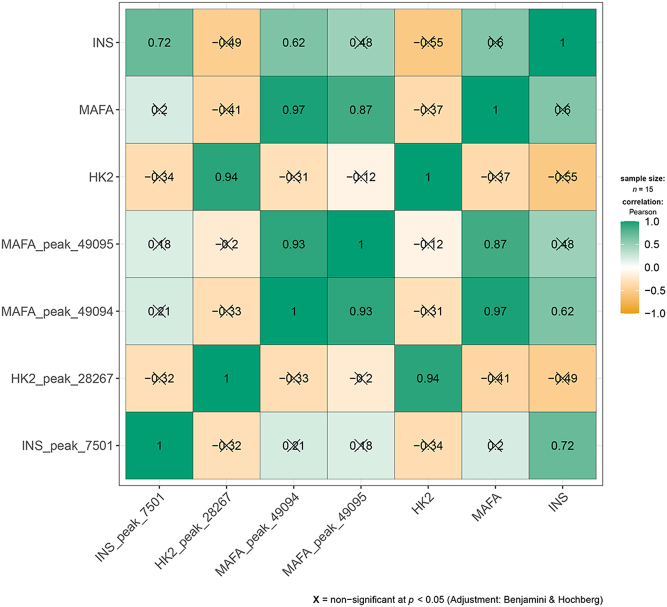
To further confirm and explore the relationship between the methylation of the candidate hub genes (INS, HK2, MAFA) and gene expression, the R package ggstatsplot was used to analyze the correlation between the m6A methylation matrix and the gene expression matrix of these three genes. Using the Pearson correlation analysis method, the significance threshold was set to P-value < 0.05, and the results are shown in Fig. 7. It was found that the m6A methylation of the three candidate hub genes was positively correlated with their gene expression, and the correlation coefficients were all greater than 0.7. For example, the INS gene and peak_7501 (r = 0.72, P = 0.01), HK2 and peak_28267 (r = 0.94, P < 0.0001), MAFA and peak_49094 (r = 0.97, P < 0.0001) and peak_49095 (r = 0.87, P = 0.0001). It was also observed that there was a significant correlation between different m6A methylation peaks in genes, such as peak_49095 and peak_49094 of the MAFA gene, and the correlation coefficient was as high as 0.93. It was also observed that the expression correlation between the three genes was not strong, nor was the m6A methylation correlation between the three genes. The m6A methylation information of the three genes is outlined in Table 1.
Figure 7.
Correlation analysis between candidate hub genes methylation and transcription expression. Using the Pearson correlation analysis method, the significance threshold was set to P < 0.05.
Table 1.
The m6A methylation information of the three genes.
| Parameter1 | Parameter2 | r | ci_low | ci_high | t | df | P | Method | Nobs |
|---|---|---|---|---|---|---|---|---|---|
| INS_peak_7501 | HK2_peak_28267 | −0.316476451 | −0.713127661 | 0.233668532 | −1.202900726 | 13 | 0.349619348 | Pearson | 15 |
| INS_peak_7501 | MAFA_peak_49094 | 0.214230527 | -0.334770853 | 0.654650495 | 0.790778555 | 13 | 0.521568637 | Pearson | 15 |
| INS_peak_7501 | MAFA_peak_49095 | 0.175242834 | −0.370258237 | 0.630872047 | 0.64177839 | 13 | 0.558779284 | Pearson | 15 |
| INS_peak_7501 | HK2 | −0.343126666 | −0.727513673 | 0.205205333 | −1.317124549 | 13 | 0.337048673 | Pearson | 15 |
| INS_peak_7501 | MAFA | 0.201734992 | −0.346316682 | 0.647123252 | 0.742634314 | 13 | 0.521568637 | Pearson | 15 |
| INS_peak_7501 | INS | 0.715356743 | 0.320565255 | 0.898400217 | 3.69119233 | 13 | 0.011404115 | Pearson | 15 |
| HK2_peak_28267 | MAFA_peak_49094 | −0.333340517 | −0.722270007 | 0.215766108 | −1.274785784 | 13 | 0.337048673 | Pearson | 15 |
| HK2_peak_28267 | MAFA_peak_49095 | −0.201296436 | −0.646857481 | 0.346718913 | −0.740951626 | 13 | 0.521568637 | Pearson | 15 |
| HK2_peak_28267 | HK2 | 0.942139515 | 0.83087715 | 0.980965689 | 10.13341078 | 13 | 1.62E-06 | Pearson | 15 |
| HK2_peak_28267 | MAFA | −0.413498624 | −0.763943063 | 0.125306678 | −1.637432976 | 13 | 0.239601756 | Pearson | 15 |
| HK2_peak_28267 | INS | −0.488492627 | −0.800452853 | 0.031703625 | −2.018507634 | 13 | 0.145868776 | Pearson | 15 |
| MAFA_peak_49094 | MAFA_peak_49095 | 0.926282716 | 0.787859265 | 0.975615698 | 8.862792024 | 13 | 5.01E-06 | Pearson | 15 |
| MAFA_peak_49094 | HK2 | −0.306591012 | −0.707705101 | 0.243992048 | −1.161359103 | 13 | 0.349619348 | Pearson | 15 |
| MAFA_peak_49094 | MAFA | 0.973242682 | 0.919304843 | 0.991291258 | 15.27148558 | 13 | 2.32E−08 | Pearson | 15 |
| MAFA_peak_49094 | INS | 0.621868607 | 0.160844378 | 0.860128698 | 2.863130273 | 13 | 0.04661101 | Pearson | 15 |
| MAFA_peak_49095 | HK2 | −0.116309418 | −0.593227196 | 0.42103937 | −0.4222252 | 13 | 0.679759997 | Pearson | 15 |
| MAFA_peak_49095 | MAFA | 0.867522089 | 0.63941472 | 0.955265557 | 6.288507873 | 13 | 0.000146744 | Pearson | 15 |
| MAFA_peak_49095 | INS | 0.481066106 | -0.041398719 | 0.796937753 | 1.978486975 | 13 | 0.145868776 | Pearson | 15 |
| HK2 | MAFA | −0.370149421 | −0.741763645 | 0.175365057 | −1.436633596 | 13 | 0.305287625 | Pearson | 15 |
| HK2 | INS | −0.552290623 | −0.829790755 | −0.055820363 | −2.388662182 | 13 | 0.086028849 | Pearson | 15 |
| MAFA | INS | 0.59697638 | 0.122031997 | 0.849465258 | 2.682960805 | 13 | 0.056383685 | Pearson | 15 |
Parameter 1 and parameter 2 represent different correlation factors; R stands for Pearson correlation coefficients; ci_Low represents the lower limit of correlation coefficient (confidence low); 'ci_high' represents the upper limit of correlation coefficient (confidence high); DF represents degrees of freedom; P represents the probability P value; T represents the t-test value of correlation coefficient; 'Method' represents the statistical method used for correlation analysis, that is, Pearson; 'Nobs' stands for number of observations, that is, the number of samples.
In summary, it is speculated that the m6A methylation of the T2D candidate hub genes has a direct or indirect effect on its expression, and there are interactions and effects between different methylation peaks within the genes. Since the expression of the three genes or m6A methylation do not significantly affect each other, they may be a compensation mechanism and perform different functions in the disease.
LASSO regression analysis of candidate hub genes
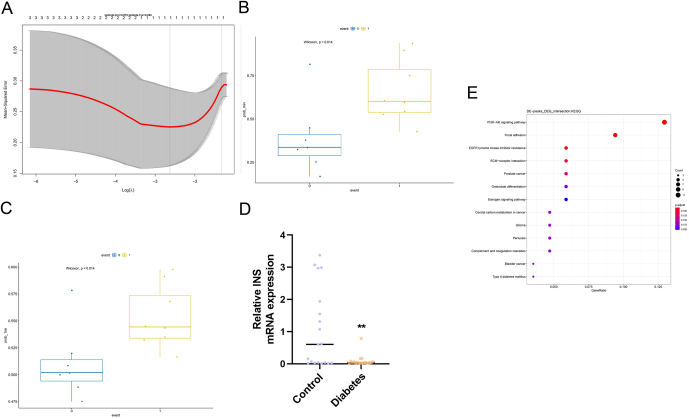
To observe and predict the impact of the three candidate hub genes on T2D, a LASSO regression model was constructed based on the gene expression combined with the R package glmnet. The ultimate goal of this analysis was to algorithmically predict the correlation between the gene expression and the disease. First, the appropriate model was selected. The two dashed lines in Fig. 8A indicated two special λ values, one was lambda.min and the other was lambda.1se. The lambda.min = 0.072 and lambda.1se = 0.263 were obtained in this analysis, so the actual event value (T2D = 1; control = 0) was combined to determine the predicted effect of the model under these two parameters (Fig. 8B and C). The parameters of the lambda.min and lambda.1se were able to predict the occurrence of T2D, and their P-values were both equal to 0.014. At the same time, it was confirmed that the independent variables (genes) under these two prediction models were INS. Therefore, it was concluded that INS could be a predictive hub gene for T2D.
Figure 8.
(A, B and C) LASSO regression analysis of candidate hub genes. (A) The appropriate model. The two dashed lines in the figure indicated two special λ values, one was lambda.min and the other was lambda.1se. (B and C) The predicted effect of the model under these two parameters combining the actual event value (T2D= 1; control= 0). (D and E) Experimental validation of INS. (D) The result of RT-PCR. (E) The result of Western blot.
Experimental validation of INS
To further test the importance of the hub genes, INS was selected for RT-PCR verification. The relative expression of INS mRNA was significantly lower (P = 0.0019) in the T2D group (n = 11) compared to the control group (n = 19) as indicated by t-test analysis (Fig. 8D), while the Western blot indicated that the INS protein levels in T2D were significantly suppressed (Fig. 8E).
Discussion
m6A methylation regulates physiology and metabolism (1) and participates in multiple cellular biological processes, such as immune modulation, fat metabolism, biological rhythm, and reproductive development. Thus, its disorders can cause many kinds of disease (23). One study (24) shows that compared to the healthy controls, the content of m6A mRNA in peripheral blood samples isolated from T2D patients is much reduced, which indicates that there is a connection between m6A and T2D.
In the GO enrichment analysis of mMDEGs, in terms of the biological process, mMDEGs were found to be enriched in extracellular structure organization, extracellular matrix organization, cell-substrate adhesion, digestive tract development, and digestive system development, whereas with regard to cellular component, mMDEGs were enriched in the collagen-containing extracellular matrix, collagen, and trimer. For molecular function, mMDEGs were enriched in endopeptidase activity, calmodulin binding, extracellular matrix structural constituent, and protein tyrosine kinase activity. One study shows that in peripheral tissues, such as muscle, liver, and adipose tissue, the extracellular matrix and integrins are the key regulators of insulin action, which provides new potential targets for the treatment of T2D insulin resistance (25).
For the KEGG pathway analysis, it was found that mMDEGs were significantly associated with the T2D signaling pathway, and the mMDEGs that were confirmed to participate in this pathway were INS, HK2, and MAFA. Since INS was the shared gene of T2D and the PI3K-Akt signaling pathway, and HK2 was the shared gene of T2D and central carbon metabolism in cancer, the PI3K-Akt signaling pathway and central carbon metabolism in cancer was focused on. Many studies have shown the correlation between the PI3K-Akt signaling pathway and T2D. In the study of Gao et al. (26), it was found that D-pinitol played a positive role in regulating insulin-mediated glucose uptake in the liver through the translocation and activation of the PI3K/Akt signaling pathway in T2D rats. They also found that D-chiro-inositol played a positive role in regulating insulin-mediated glucose uptake through the PI3K/Akt signaling pathway in T2D rats (27). There is evidence (28) that the skin blood flow oscillation extract played a positive role in alleviating T2D through the PI3K/Akt signaling pathway in HepG2 cells, and it is known that obesity and T2D are related to insulin resistance (29). Most insulin actions were carried out by activating two main signaling pathways: the PI3K/Akt pathway and the MAPK/Ras pathway. In the matter of the PI3K/Akt pathway, the activation of the Akt kinase led to phosphorylation of enzymes, transcription factors, cell-cycle regulating proteins, and so on, which means that the Akt kinase plays a key role in insulin signaling. Also, the decrease in PI3K and Akt kinases activity is one of the most common insulin resistance alterations (30). However, there are few studies on the correlation between central carbon metabolism in cancer, and it is not yet possible to confirm whether this pathway is related to T2D.
In this study, three candidate hub genes were obtained that were closely related to T2D, namely INS, HK2, and MAFA. The INS gene was more meaningful for T2D prediction, and it appears to be closely related to diabetes. For instance, INS gene promoter mutations have been shown to lead to both remitting/relapsing and permanent neonatal diabetes mellitus (31). Mouzaki et al. (32) thought that the INS gene was one of the most important genes in the pathogenesis of type 1 diabetes (T1D). Methylation may play a role in the relationship between INS variation and T1D development (33). Although there are currently few studies on the correlation between the INS gene and T2D, our research suggests that the INS gene can accurately predict the occurrence of T2D in the LASSO regression model. HK2 is a subtype of hexokinase and a predominant isoform in insulin-sensitive tissues such as the heart, skeletal muscle, and adipose tissues (19). Moreover, abnormal expression of the HK2 gene can affect glucose metabolism, such as glycolysis (34). This means that the HK2 gene may drive diabetes by changing the concentration of glycolysis intermediates. MAFA (v-maf musculoaponeurotic fibrosarcoma oncogene homologue A) is the only transcriptional activator present exclusively in islet β-cells (35). MAFA controls many β-cell-specific genes, such as insulin, Glut2, Pdx-1, Nkx6.1, GLP-1R, PC-1/3, and pyruvate carboxylase (36). Studies have shown that MAFA is an important regulator of glucose-stimulated insulin secretion (37), and it can drive insulin expression by binding to the enhancer/promoter region of the INS gene (38). One study pointed out that the deletion of MAFA resulted in the decreased transcription of INS1, INS2, and Pdx1 in adult mice, which further led to impaired glucose tolerance and glucose-stimulated insulin secretion (39). In light of this, MAFA can be considered to play an important role in T2D.
In conclusion, multiple biological information analyses were conducted on the 30 samples, and 48,706 m6A methylation peaks (involving a total of 20,157 genes) were obtained from the 30 samples and a total of 1912 differential peaks (involving 1719 genes) were identified. To further understand the effect of m6A methylation in T2D, mMDEGs were obtained and then GO and KEGG pathway analysis and a GSEA of mMDEGs were performed. The signaling pathways were found to be significantly related to the mMDEGs, and there were three candidate hub genes (INS, MAFA, and HK2). Finally, a LASSO regression model was constructed, and it was found that INS could be invoked as a predictive hub gene for T2D disease, which was also confirmed by the RT-PCR and Western blot. Since this has not been verified by cell or animal experiments, this line of research will be followed up in the future.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This study was funded by Natural Science Foundation of China (Nos. 81860145, 81860144). A special fund for high-level talents cultivation in health and family planning in Yunnan (No. D-2017027). Applied Basic Research Project of Science and Technology Department of Yunnan Province (202001AT070008).
References
- 1.Wu J, Frazier K, Zhang J, Gan Z, Wang T, Zhong X. Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obesity Reviews 202021 e12942. ( 10.1111/obr.12942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, Dolan K, Zhu X, Hubert N, Tao Yet al. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Reports 2018251816–1828.e4. ( 10.1016/j.celrep.2018.10.068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng X, Su R, Feng X, Wei M, Chen J. Role of N6-methyladenosine modification in cancer. Current Opinion in Genetics and Development 2018481–7. ( 10.1016/j.gde.2017.10.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bommer C, Sagalova V, Heesemann E, Manne-Goehler J, Atun R, Bärnighausen T, Davies J, Vollmer S. Global economic burden of diabetes in adults: projections from 2015 to 2030. Diabetes Care 201841963–970. ( 10.2337/dc17-1962) [DOI] [PubMed] [Google Scholar]
- 5.Hu C, Jia W. Diabetes in China: epidemiology and genetic risk factors and their clinical utility in personalized medication. Diabetes 2018673–11. ( 10.2337/dbi17-0013) [DOI] [PubMed] [Google Scholar]
- 6.Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, Jia GF, Chen J, Feng YQ, Yuan BFet al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. Journal of Clinical Endocrinology and Metabolism 2015100E148–E154. ( 10.1210/jc.2014-1893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Shen F, Huang W, Qin S, Huang JT, Sergi C, Yuan BF, Liu SM. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. Journal of Clinical Endocrinology and Metabolism 2019104665–673. ( 10.1210/jc.2018-00619) [DOI] [PubMed] [Google Scholar]
- 8.De Jesus DF, Zhang Z, Kahraman S, Brown NK, Chen M, Hu J, Gupta MK, He C, Kulkarni RN. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nature Metabolism 20191765–774. ( 10.1038/s42255-019-0089-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology 201937907–915. ( 10.1038/s41587-019-0201-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature Protocols 201271728–1740. ( 10.1038/nprot.2012.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 201415 550. ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 201430923–930. ( 10.1093/bioinformatics/btt656) [DOI] [PubMed] [Google Scholar]
- 13.Krämer A, Green J, Pollard Jr J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 201430523–530. ( 10.1093/bioinformatics/btt703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian H, Han YP, Zhang YC, Zhao Y, Yan S, Li QF, Wang BC, Wang JJ, Meng W, Yang Jet al. Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Molecular Oncology 2019132227–2245. ( 10.1002/1878-0261.12557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng D, Zhou R, Yu Y, Luo Y, Zhang J, Sun H, Bin J, Liao Y, Rao J, Zhang Yet al. Gene expression profiles for a prognostic immunoscore in gastric cancer. British Journal of Surgery 20181051338–1348. ( 10.1002/bjs.10871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Zhou Y, Qu W, Deng M, Zhang C. A Lasso regression model for the construction of microRNA-target regulatory networks. Bioinformatics 2011272406–2413. ( 10.1093/bioinformatics/btr410) [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Chan L. Monogenic diabetes: what it teaches us on the common forms of type 1 and type 2 diabetes. Endocrine Reviews 201637190–222. ( 10.1210/er.2015-1116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu J, Dai X, Plummer G, Suzuki K, Bautista A, Githaka JM, Senior L, Jensen M, Greitzer-Antes D, Manning Fox JEet al. Kv2.1 clustering contributes to insulin exocytosis and rescues human beta-cell dysfunction. Diabetes 2017661890–1900. ( 10.2337/db16-1170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan VP, Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy 201511963–964. ( 10.1080/15548627.2015.1042195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, Chandel N, Laakso M, Muller WJ, Allen ELet al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 201324213–228. ( 10.1016/j.ccr.2013.06.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay J, Luscombe-Marsh ND, Thompson CH, Noakes M, Buckley JD, Wittert GA, Yancy Jr WS, Brinkworth GD. Comparison of low- and high-carbohydrate diets for type 2 diabetes management: a randomized trial. American Journal of Clinical Nutrition 2015102780–790. ( 10.3945/ajcn.115.112581) [DOI] [PubMed] [Google Scholar]
- 22.Shao C, Cobb MH. SUMOylation regulates the transcriptional activity of MafA in pancreatic beta cells. Journal of Biological Chemistry 20092843117–3124. ( 10.1074/jbc.M806286200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. International Journal of Biological Sciences 2020161929–1940. ( 10.7150/ijbs.45231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napoli C, Benincasa G, Schiano C, Salvatore M. Differential epigenetic factors in the prediction of cardiovascular risk in diabetic patients. European Heart Journal: Cardiovascular Pharmacotherapy 20206239–247. ( 10.1093/ehjcvp/pvz062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams AS, Kang L, Wasserman DH. The extracellular matrix and insulin resistance. Trends in Endocrinology and Metabolism 201526357–366. ( 10.1016/j.tem.2015.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Zhang M, Wu T, Xu M, Cai H, Zhang Z. Effects of D-pinitol on insulin resistance through the PI3K/Akt signaling pathway in type 2 diabetes mellitus rats. Journal of Agricultural and Food Chemistry 2015636019–6026. ( 10.1021/acs.jafc.5b01238) [DOI] [PubMed] [Google Scholar]
- 27.Gao YF, Zhang MN, Wang TX, Wu TC, Ai RD, Zhang ZS. Hypoglycemic effect of D-chiro-inositol in type 2 diabetes mellitus rats through the PI3K/Akt signaling pathway. Molecular and Cellular Endocrinology 201643326–34. ( 10.1016/j.mce.2016.05.013) [DOI] [PubMed] [Google Scholar]
- 28.Gao S, Guo Q, Qin C, Shang R, Zhang Z. Sea buckthorn fruit oil extract alleviates insulin resistance through the PI3K/Akt signaling pathway in type 2 diabetes mellitus cells and rats. Journal of Agricultural and Food Chemistry 2017651328–1336. ( 10.1021/acs.jafc.6b04682) [DOI] [PubMed] [Google Scholar]
- 29.Huang X, Liu G, Guo J, Su Z. The PI3K/AKT pathway in obesity and type 2 diabetes. International Journal of Biological Sciences 2018141483–1496. ( 10.7150/ijbs.27173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutiérrez-Rodelo C, Roura-Guiberna A, Olivares-Reyes JA. Mecanismos Moleculares de la Resistencia a la Insulina: una Actualización [Molecular mechanisms of insulin resistance: an update]. Gaceta Medica de Mexico 2017153214–228. [PubMed] [Google Scholar]
- 31.Demiral M, Demirbilek H, Çelik K, Okur N, Hussain K, Ozbek MN. Neonatal diabetes due to homozygous INS gene promoter mutations: highly variable phenotype, remission and early relapse during the first 3 years of life. Pediatric Diabetes 2020211169–1175. ( 10.1111/pedi.13079) [DOI] [PubMed] [Google Scholar]
- 32.Mouzaki K, Kotanidou EP, Fragou A, Kyrgios I, Giza S, Kleisarchaki A, Tsinopoulou VR, Serbis A, Tzimagiorgis G, Galli-Tsinopoulou A. Insulin gene promoter methylation status in Greek children and adolescents with type 1 diabetes. Biomedical Reports 202013 31. ( 10.3892/br.2020.1338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carry PM, Vanderlinden LA, Johnson RK, Dong F, Steck AK, Frohnert BI, Rewers M, Yang IV, Kechris K, Norris JM. DNA methylation near the INS gene is associated with INS genetic variation (rs689) and type 1 diabetes in the diabetes autoimmunity study in the young. Pediatric Diabetes 202021597–605. ( 10.1111/pedi.12995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabbani N, Thornalley PJ. Hexokinase-2 glycolytic overload in diabetes and ischemia-reperfusion injury. Trends in Endocrinology and Metabolism 201930419–431. ( 10.1016/j.tem.2019.04.011) [DOI] [PubMed] [Google Scholar]
- 35.Matsuoka TA, Kaneto H, Stein R, Miyatsuka T, Kawamori D, Henderson E, Kojima I, Matsuhisa M, Hori M, Yamasaki Y. MafA regulates expression of genes important to islet beta-cell function. Molecular Endocrinology 2007212764–2774. ( 10.1210/me.2007-0028) [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia 200750348–358. ( 10.1007/s00125-006-0490-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa Ket al. MafA is a key regulator of glucose-stimulated insulin secretion. Molecular and Cellular Biology 2005254969–4976. ( 10.1128/MCB.25.12.4969-4976.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Liu Q, Zhou Z, Ikeda Y. PDX1, neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Research and Therapy 20178 240. ( 10.1186/s13287-017-0694-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiafukaiti G, Maimaiti S, Ogata K, Kuno A, Kudo T, Shawki HH, Oishi H, Takahashi S. MafB is important for pancreatic β-cell maintenance under a MafA-deficient condition. Molecular and Cellular Biology 201939 e00080-19. ( 10.1128/MCB.00080-19) [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a