Abstract
Background:
The noninvasive study of the structure and functions of the brain using neuroimaging techniques is increasingly being used for its clinical and research perspective. The morphological and volumetric changes in several regions and structures of brains are associated with the prognosis of neurological disorders such as Alzheimer’s disease, epilepsy, schizophrenia, etc. and the early identification of such changes can have huge clinical significance. The accurate segmentation of three-dimensional brain magnetic resonance images into tissue types (i.e., grey matter, white matter, cerebrospinal fluid) and brain structures, thus, has huge importance as they can act as early biomarkers. The manual segmentation though considered the “gold standard” is time-consuming, subjective, and not suitable for bigger neuroimaging studies. Several automatic segmentation tools and algorithms have been developed over the years; the machine learning models particularly those using deep convolutional neural network (CNN) architecture are increasingly being applied to improve the accuracy of automatic methods.
Purpose:
The purpose of the study is to understand the current and emerging state of automatic segmentation tools, their comparison, machine learning models, their reliability, and shortcomings with an intent to focus on the development of improved methods and algorithms.
Methods:
The study focuses on the review of publicly available neuroimaging tools, their comparison, and emerging machine learning models particularly those based on CNN architecture developed and published during the last five years.
Conclusion:
Several software tools developed by various research groups and made publicly available for automatic segmentation of the brain show variability in their results in several comparison studies and have not attained the level of reliability required for clinical studies. The machine learning models particularly three dimensional fully convolutional network models can provide a robust and efficient alternative with relation to publicly available tools but perform poorly on unseen datasets. The challenges related to training, computation cost, reproducibility, and validation across distinct scanning modalities for machine learning models need to be addressed.
Keywords: Neuroimaging, automatic brain segmentation, FreeSurfer, FSL, SPM, CNN, machine learning
Introduction
The advancements in the field of neuroimaging particularly structural magnetic resonance imaging (MRI) had resulted in the availability of high-resolution three-dimensional (3D) imaging data of the human brain. Several large-population studies using MRI datasets have found different morphometric and volumetric changes in normal vs. various neurological conditions such as Alzheimer’s disease, mild cognitive impairment, etc.1, 2
The study of different areas of the brain, cortical and subcortical regions and structures, changes in grey matter (GM), white matter (WM), hippocampus, thalamus, amygdala, etc., has found greater interest in neuroscience studies for research and clinical intervention. Medical image segmentation is used for addressing a wide range of such biomedical research problems. 3 The accurate segmentation of MRI images thus becomes a necessary prerequisite for the quantitative analysis of the study of neurological diseases including their diagnosis, progression, and treatment monitoring in a wide variety of neurological disorders such as Alzheimer’s, dementia, focal epilepsy, multiple sclerosis, etc. 4
Apart from whole-brain segmentation into GM, WM, cerebrospinal fluid (CSF), etc. and parcellation of brain structures, several regions, and structures have received added focus on the account of their association with important cognitive functions. The hippocampus which is a part of the limbic system that is involved in learning and memory is one such important structure. The MRI can be used to monitor morphological changes that occur in the hippocampus in diseases such as Alzheimer’s, schizophrenia, epilepsy, depression, etc. and can act as biomarkers for several brain disorders such as Alzheimer’s disease, schizophrenia, epilepsy, etc.5–8 The amygdala is another brain structure of the limbic system that is involved in emotion, learning, memory, attention, etc. It is also associated with negative emotions and fear. 9 The caudate nucleus and putamen also have importance in several neurological disorders and diseases such as Parkinson’s disease, Huntington’s disease, Alzheimer’s disease, depression, schizophrenia, etc.10–13
The manual segmentation is considered the “gold standard” for anatomical segmentation. It involves the demarcation of structure, grey and white matter along the anatomical boundaries for each layer of the region of interest (ROI) by experienced anatomic tracers 14 utilizing specific software tools. A single MRI scan can have up to hundreds of layers depending upon the scan resolution, and thus manual segmentation is a time-consuming, subjective, laborious process and is not suitable for large-scale neuroimaging studies.15, 16
The automatic segmentation techniques attempt to address the above-discussed limitations associated with manual segmentation. The advancement in the algorithms and computation resources over the years has further helped the development of various segmentation techniques. Some of the hugely used publicly available software tools for neuroimaging studies that are used for the automatic segmentation of brain regions and structure are (a) FreeSurfer, (b) FMRIB Software Library (FSL), and (c) Statistical Parametric Mapping (SPM). The machine learning approaches based on CNN architecture are also being increasingly used for automated segmentation.
The remaining portion of the review will discuss the models for automatic segmentation techniques, publicly available neuroimaging tools, and emerging machine learning models for automatic segmentation and their comparison.
Methods
The study outlines the literature review of different publicly available segmentation methodologies that are extensively used for automatic human brain segmentation along with comparison studies for segmentation accuracy between different methods during the last five years. The recent machine learning models, particularly those based upon deep learning/CNN architectures that have been proposed by various research groups for efficient and accurate segmentation, have also been covered. The discussion covers the advantages and disadvantages of traditional software tools and deep learning machine models and emphasizes future challenges in the area.
The preprocessing steps which are commonly applied before any segmentation algorithm are detailed as follows.
Data Samples
The segmentation is performed on MRI scans of individual subjects which are acquired on an MRI scanner, and a single structural acquisition can take up to 10 to 30 min based upon the precision, sequence, nature of studies, scanner characteristics, etc. Most studies for the evaluation of the automatic segmentation methods, however, use datasets from existing available institutional neuroimaging datasets or publicly available neuroimaging repositories. Several huge neuroimaging datasets created as a part of the Human Connectome Project, Alzheimer’s Disease Neuroimaging Initiative (ADNI), Open Access Series of Imaging Studies (OASIS), etc. have been extensively used for standardizing segmentation techniques and finding the correlation between morphometric/anatomical and volumetric changes in different regions of the brain. The T1-weighted (T1w) MRI sequence is used for anatomical characterization; however, other sequences like T2w, FLAIR, etc. are also used by certain algorithms. The methods which are discussed and evaluated in this review have greatly benefitted from the availability of such datasets.
Preprocessing
The acquired MRI images coming from a magnetic resonance (MR) scanner require a series of steps to improve the quality of brain scans for the application of segmentation algorithms. These preprocessing steps are also implemented in several publicly available neuroimaging tools which are often utilized for carrying out the preprocessing. Some of the preprocessing steps are discussed as follows.
Bias-Field Correction
The bias-field or illumination artifact arises on account of a lack of radio-frequency (RF) homogeneity. 17 Although this is not significantly noticeable on visual examination, but it can seriously degrade the volumetric quantification of MR volume upon applying the automatic segmentation algorithms that use intensity levels. 17 Several methods for bias-field correction during and after acquisition are in use. The phantom-based calibration, multi-coil imaging, and special sequences tend to improve the acquisition process and can be referred to as prospective methods, whereas retrospective methods and algorithms such as filtering, masking, intensity, gradient, etc. are used postacquisition for bias-field correction. 18
Brain Extraction
The brain extraction or “removal of nonbrain tissue” such as skull, neck, muscles, bones, fat, etc. having overlapping intensities is employed before the segmentation processing. The extraction process classifies the image voxels into the brain or nonbrain; the brain regions can then be extracted or a binary mask can be created for the brain region. 19 The commonly used methods make use of probabilistic atlas creating a deformable template registered with the image for the removal of nonbrain tissue using the brain mask 20 . The brain extraction tool which is a part of the FMRIB Software Library and which uses the center of gravity of the brain, inflating a sphere until the brain boundary is found, provides a fast and efficient alternative21-23. Pincram uses an atlas-based label propagation method for brain extraction.24
Several software tools, however, do not require separate preprocessing, bias correction, brain extraction, etc. as they are impliedly included in their segmentation processing.
Figure 1. A Schematic Representation of Different Stages of Brain Segmentation as Obtained from FreeSurfer Tutorial Data and Visualized in FreeSurfer-Freeview.
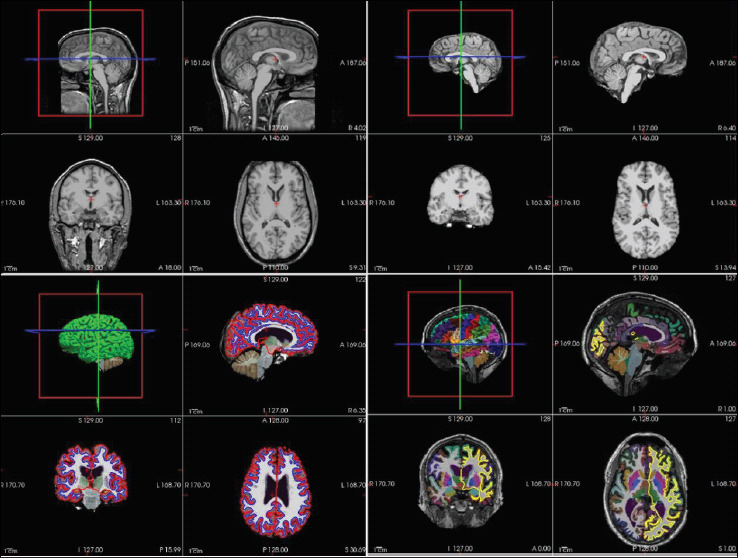
Automatic Segmentation Approaches
The MRI scan of the brain provides a 3D image of the brain scanned in x, y, z space at an appropriate slice of thickness usually ranging from 1 to 2 mm (e.g., a slice thickness of 1 mm × 1 mm × 1 mm is considered quite good). The slice thickness need not be isometric and will depend upon the MR scanner, gradient coil, channels, scan time, scanning sequence, and protocol. Every point in this 3D image “I” represents a voxel “I (x, y, z).” A higher spatial resolution will bring greater precision but require a longer scan time and may not be convenient for the patient/subject apart from the possibilities of the introduction of further noise from head movements during the longer scan session. The 3D brain structure can also be represented as a sequence of 2D images in (x, y), (y, z), or (x, z) planes. Every voxel has an intensity value typically represented in grayscale from 0 to 255. The images may also require to be resampled to a standard space for applying segmentation algorithms.
Segmentation Algorithms
The segmentation problem requires classifying every voxel into a specific tissue class such as GM, WM, and CSF, and also to identify and describe it into a specific anatomical structure by assigning a label corresponding to each voxel. Several computational algorithms for automatic segmentation have been proposed and used in isolation or conjunction with each other. The basic methods such as thresholding, fuzzy c-means, etc. which are used to segment the brain among different tissue classes (GM, WM, CSF) are not suitable for fine-grain segmentation. The deformation models, multi-atlas segmentation, model-based segmentation, and machine learning models, particularly those based upon CNN, are increasingly being used for fine-grain segmentation of the whole brain or ROIs such as the hippocampus, cerebellum, amygdala, etc.
The theoretical concepts behind various segmentation methodologies have already been detailed in the literature.19, 25, 26 The present work focuses on developed models, machine learning methods, and tools which have been applied to address the automatic segmentation problem.
Major Software Tools and Their Methodology
Several software tools have been developed over the years by different scientific groups applying various algorithms for automatic segmentation. A list of several publicly available neuroimaging software and tools is available in Table 1 with their brief descriptions. The list is not exhaustive. The important ones are discussed as follows.
Table 1. Important Publicly Available Software and Tools for Neuroimaging Studies.
| Software Tools | Features |
| FreeSurfer |
|
| Statistical parameter metric (SPM) |
|
| FMRIB Software Library (FSL) |
|
| volBrain |
|
| Multi-atlas propagation with enhanced registration (MAPER) |
|
| Multi-atlas-based multi-image segmentation (MABMIS) |
|
| Automatic segmentation of hippocampus subfield (ASHS) |
|
FreeSurfer
FreeSurfer 27 is an open-source software suite developed by the Laboratory of Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. The software package is freely available from its website https://surfer.nmr.mgh.harvard.edu and is used for a complete range of analysis of structural and functional imaging data and its visualization. The latest version of the software is version 7.1.0 (released in May 2020). The segmentation pipeline of FreeSurfer can be run in a fully automatic manner using the “recon-all all” script. It uses image intensity and probabilistic atlas with local spatial relationships between subcortical structures for carrying out the segmentation. 28 The default pipeline can segment and assign 40 different labels to corresponding voxels in an automated manner. The segmentation has been extended to further segment 9 amygdala nuclei 29 and 13 hippocampus subfields 30 that have been incorporated in the new statistical atlas based on Bayesian inference build using a postmortem specimen at high resolution and has been added in FreeSurfer version 6.0 onward. This allows the automatic simultaneous segmentation of amygdala nuclei and hippocampus subfields using standard resolution structural MR images. The software is freely available for Linux and Mac platforms, and it provides graphic as well as command line options.
FMRIB Software Library (FSL)
FSL23,31 is a comprehensive library of neuroimaging tools for structural, functional, and diffusion tensor imaging (DTI) studies. The software has been created and maintained by FMRIB Analysis Group at the University of Oxford and is available at https://fsl.fmrib.ox.ac.uk/fsl; the current version of the software is version 6.0. The tissue segmentation is done using FMRIB Automated Segmentation Tool (FAST), 32 which uses Markov random field model along with the expectation-maximization algorithm. FAST can be invoked through the command line or GUI in the brain-extracted image volumes. The segmentation pipeline of FSL can also correct RF inhomogeneity and classify the brain among GM, WM, and CSF tissue types. The probabilistic and partial volume tissue segmentation used for tissue volume calculation can also be calculated. The subcortical segmentation is performed using FMRIB Integrated Registration and Segmentation Tool (FIRST). 33 FIRST provides model-based segmentation using deformation models. The construction of the model was based upon a manually labeled dataset; the labels were parameterized as surface meshes modeled as a point distribution model. It first registers the images to MNI152 space by performing affine registration. The “run_first_all” script does the automatic segmentation of subcortical structures into 15 different labels.
Statistical Parametric Mapping (SPM)
SPM 34 is a package developed for the analysis of neuroimaging data coming from several imaging modalities like functional-MRI, positron emission tomography, magnetoencephalography, electroencephalography, etc. This is also a freely available software tool from its website https://www.fil.ion.ucl.ac.uk/spm/ at Wellcome Centre for Human Neuroimaging but requires MATLAB platform, even though a compiled version of SPM also exists. The tissue classification methodology in SPM considers the brightness information of voxels along with tissue probability maps and the position of voxel during classification. The latest version is SPM12 and is last updated in January 2020. The segmentation process of SPM can be further extended using SPM toolboxes; the VBM is a toolbox which has been used in several studies and has now been replaced with the CAT12 toolbox. In a recent study, it was noticed that CAT12 can contribute better in volumetric analysis than VBM8; this is also on account of normalization and segmentation improvements in SPM12. 35 The automated anatomical labeling atlas 3 (AAL3) is another toolbox of SPM which is a refinement of its previous versions and can parcellate the brain among 166 labels. 36
volBrain (Online Web Platform)
volBrain 37 is a web-based pipeline for MRI brain volumetry. The pipeline can be accessed from the https://volbrain.upv.es/. The webserver platform allows researchers to submit their MRI scans in NIFTI format and the online pipeline generates the volumetric measurements which are sent on the registered email id after the completion of the job; the same can also be downloaded from the volBrain system. The user is required to register with the volBrain online system and there is a limit on the number of simultaneous jobs that can be submitted. The entire processing remains a black box from the user’s point of view; the theoretical aspects have been described in published papers. The volBrain system is primarily based on a multi-atlas, patch-based segmentation method and utilizes training libraries as implemented in the volBrain online platform. The volBrain web platform has also added several brain structure segmentation methods such as CERES (CEREbellum Segmentation), 38 HIPS (HIPpocampus Segmentation), 39 and pBrain (Parkinson related deep nucleus segmentation), 40 which are all available from the volBrain platform.
Multi-Atlas Propagation With Enhanced Registration (MAPER)
The multi-atlas propagation with enhanced registration (MAPER) is an automatic segmentation tool for structural MRI images into corresponding anatomical sub regions by using a database of multiple atlases as knowledge base. 41 The standard MAPER pipeline requires brain extraction and tissue class segmentation using pincram and FSL-FAST, respectively.
Comparison of Segmentation Accuracy in Automatic Methods
Several studies have been made to judge the relative comparison of various publicly available software tools and algorithms. The FreeSurfer, SPM, and FSL are among the most used tools for neuroimaging studies and are not limited to segmentation alone. The other software tools are built for specific applications. The comparison studies are generally performed on specific datasets with default options and provide mixed results which cannot be generalized for all situations. Table 2 summarizes the recent comparison of several publicly available tools for their accuracy in automatic segmentation.
Table 2. Comparison of Some Publicly Available Methods for Automatic Segmentation.
| Research Citation | Automatic Segmentation Tools | Dataset Utilized | Conclusion/Findings |
| Yaakub et al. |
|
|
|
| Palumbo et al. |
|
|
|
| Bartel et al. |
|
|
|
| Velasco-Annis et al. |
|
OASIS datasetn = 20 (scanned twice), 1.5T |
|
| Zandifar et al. |
|
ADNI database |
|
| Perlaki et al. |
|
30 healthy young Caucasian subjectsn = 30, 3T |
|
| Naess-Schmidt et al. |
|
|
|
| Grimm et al. |
|
92 participants in the age range of (18–34, mean:21.64)n = 92, 1.5T |
|
| Fellhauer et al. |
|
n = 115, 1.5T, MPRAGE(60 MCI, 34 AD, 32 healthy) |
|
Abbreviations: MCI, mild cognitive impairment;
Evolving Machine Learning Models for Automatic Segmentation
Several machine learning models have been developed over the years for the automatic segmentation of brain tissue and anatomical structures of specific structures like the cerebellum and hippocampus. Fine-grain segmentation of the whole brain has also been implemented in several methods. A summary of some recent machine learning models applied for automatic brain segmentation is given in Table 3.
Table 3. Summary of Recent Brain Segmentation Methods Utilizing Machine Learning Models.
| Method | Model/Segmentation Area | Principle | Remarks |
| A. Whole-Brain Segmentation Models for Classifying Among Different Anatomical Labels | |||
| AssemblyNet | CNN/Brain automatic segmentation | Utilizes two assemblies of 125 3D U-Nets processing different overlapping brain areas of the whole brain. | Competitive performance in comparison with U-Net, joint label fusion, and SLANT. |
| SLANT | CNN/Fine-grain segmentation >100 structures | 3D–FCN, addresses memory issues using multiple spatially distributed overlapping network tiles of U-Nets. | Training and testing can be optimized by providing 27 GPUs for SLANT-27 and 8 for SLANT-8. |
| QuickNAT | CNN/Brain segmentation(segments 27 structures) | Fully convolutional and densely connected,pretraining using existing segmentation software (FreeSurfer),fine-tuning to rectify errors using manual labels. | Posttraining, the model achieves superior computational performance in comparison with patch-based CNN and atlas-based approaches. Also compared well with FSL and FreeSurfer. |
| Bayesian QuickNAT | CNN/Brain segmentation(segments 33 structures) | F-CNN approach (of QuickNAT) with Bayesian inference for segmentation quality. | The model has been compared with QuickNAT and FreeSurfer with manual annotations. |
| 3DQ | CNN/Brain segmentation(segment 28 structures) | 3D F-CNN with model compression up to 16 times without affecting performance. Useful for storage critical applications. | Integrates training scalable factors and normalization parameter.Increases learning while maintaining compression. |
| DeepNAT | CNN/Brain segmentation of 25 structures | 3D-CNN patch-based model, the first network removes the background, second classifies brain structure. | Uses three CNN layers for pooling, normalization, and nonlinearities.Comparable with other state-of-the-art models. |
| BrainSegNet | CNN/Whole-brain segmentation | 2D/3D CNN patches | Does not require registration, saving on computational cost. |
| B. Whole-Brain Segmentation Models for Classification Among GM, WM, and CSF Only | |||
| HyperDense-Net | CNN /Brain segmentation | Fully connected 3D-CNN using multiple modalities. | Successfully participated in iSEG-2017 and MRbrainS-2013 challenge. |
| VoxResNet | CNN/Brain segmentation | Voxel-wise residual network with 25 layers utilizing CNN. | Successfully competed in MRbrainS-2015 challenge. |
| C. Brain Segmentation Models for Specific Brain Structures (Hippocampus, Cerebellum) | |||
| HippMapp3r | CNN/Hippocampus segmentation | CNN architecture based upon U-Net. Initial training on the whole brain, the output was trained again with reduced FOV on the same network architecture. | Validation is done against FreeSurfer, FSL-First, volBrain, SBHV, and HippoDeep. Algorithm and trained model are made publicly available. |
| CAST | CNN/Hippocampus subfield segmentation | Multi-scale 3D CNN with T1w and T2w imaging modalities as input. | Hippocampus subfield segmentation. |
| ACA-PULCO | CNN/Cerebellum segmentation | Alternative CNN design using U-Net with locally constrained optimization. | Applied on MPRAGE images. |
| HippoDeep | CNN/Hippocampus automatic segmentation | Deep learned appearance model based on CNN. | The training utilizes multiple cohorts and label derived from FreeSurfer output along with synthetic data. |
The regular machine learning models do not generalize well and are not suitable for complex imaging modalities; the deep learning models having multiple layers are increasingly being used to address neuroimaging challenges which have benefitted from the advancements in graphical processing unit (GPU) processing power. The deep learning models like CNN are preferred owing to their application in medical imaging problems. A typical CNN architecture contains several layers and components such as (a) input layers receiving raw image data, (b) convolutional layers applying filter (kernel) and producing feature maps, (c) activation function layer applying the activation function to the output coming from the convolution layer like rectified linear unit, (d) pooling layers for downsampling the output of the preceding layer, and (e) fully connected layer for applying weights to feature analysis to predict the label. The typical CNN models are extended in various CNN architectures such as U-Nets. 53 3D U-Nets 54 is an extension of traditional U-Net architecture for application in 3D biomedical imaging.
The recent methods of automatic machine learning models of segmentation are discussed as follows.
AssemblyNet 55
This model uses a large assembly of CNNs, each processing different overlapping regions. The framework is arranged in the form of two assemblies of U-Nets, each having 125 3D U-Nets (i.e., a total of 250 compact 3D U-Nets). The method showed competitive performance in respect of U-Net, patch-based joint label fusion, and SLANT-27 methods. The segmentation accuracy was improved with the use of nonlinearly registered “Atlas prior” for expected classification, transfer learning to initialize spatially nearest U-Net and multi-scale cascade to communicate between the two assemblies for refinement. The training time was shown to be 7 days in the case of AssemblyNet but the classification time is just 10 min.
SLANT 56
The SLANT pipeline first registers the target image to the MNI305 template using affine registration, and this is followed by bias-field correction and normalization. Since the whole volume cannot fit into the GPU memory using the FCN network, the entire MNI space is divided into k independent 3D U-Nets. The SLANT-8 divides the volume into eight nonoverlapped subspaces with each subspace of size 86 × 110 × 78 voxels, whereas SLANT-27 divides the volume into 27 overlapped network tiles with each subspace of 96 × 128 × 88 voxels. The overlapped SLANT-27 thus requires majority voting for label fusion. The entire pipeline is available as a docker image. It provides fine-grain segmentation of the whole brain in more than 100 ROIs. The training time could be as high as 109 h for SLANT-27 on 5,111 training scans using a single GPU (NVIDIA Titan 12 GB), which can be reduced to 4 h with 27 GPUs, and the testing time is roughly 15 min.
Figure 2. An Illustration of AssemblyNet Model Using U-Net 55 .
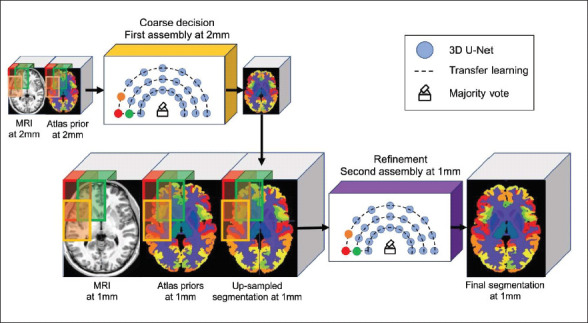
Source: Reprinted with permission from Elsevier.
Figure 3. An Illustration of SLANT-27 (27-Network Tiles) Model Using U-Net 56 (Reprinted with permission from Elsevier).
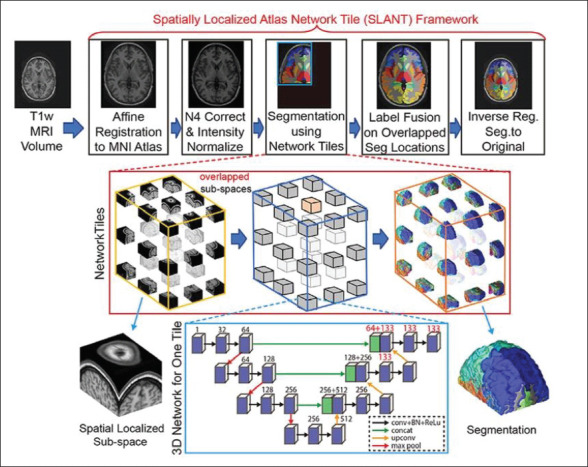
Source: Reprinted with permission from Elsevier.
3DQ 57
This is a generalizable method that can be applied to various 3D F-CNN architecture such as 3D U-Nets, MALC, and “V-Net on MALC” for providing model compression of 16 times to address the memory issues, and it incorporates quantization mechanism for integrating the trainable scaling factor and the normalization parameter, which not only maintains compression but also increases the learning capacity of the model. The model was successfully applied for 3D whole-brain segmentation and achieved comparable performance with up to 16 times model compression.
QuickNAT 58
QuickNAT tries to address the limitation of limited manually curated data for training by first utilizing the existing publicly available software tools such as FreeSurfer to segment the data which is then used to train the network; this pretrained network is then further improved by training with limited manually annotated data to achieve high segmentation accuracy. The model does not utilize 3D F-CNN but instead uses three two-dimensional F-CNN, each operating in separate planes (i.e., coronal, axial, and sagittal), followed by aggregation resulting in final segmentation and labels corresponding to 27 brain structures.
The details of other recent methods and architecture have been given in Table 3.
Reliability and Reusability of Automated Segmentation Methods
The automated fine-grain segmentation of the whole brain including specific ROIs has profound usage in neuroscience research. This, however, depends on the quality of MRI acquisitions, preprocessing, choice of analysis pipelines, and many other factors. A retrospective longitudinal study has shown a lack of reproducibility arising at the level of acquisition in the MRI scanner and the amount of variability being different for different scanners, 68 and this is a cause of concern having a direct bearing on subsequent analysis pipelines. The effects of automated pipelines which are often poorly documented and lack standardization have also been analyzed in a recent study in the context of functional neuroimaging analysis where the same data were provided to 70 different research groups, but none of the teams chose identical pipelines, and the results were also variable across groups, thus emphasizing the need of validation and sharing of complex workflows. 69
Discussion and Conclusion
The automatic segmentation methods have been quite successful in morphometric and volumetric measurements of brain tissue and structures in finer details. The publicly available methods such as FreeSurfer, FSL, SPM, etc. are sufficiently resilient with respect to noise and artifacts introduced at the acquisition stage and have performed consistently across different datasets and are being extensively used by the neuroimaging community; however, they are still far from being accepted at par with manual segmentation. Several studies have compared the relative performance of various publicly available methods which have provided mixed results and have been discussed in Table 2.
The machine learning methods particularly those based on CNN have been shown to perform better than the publicly available software tools, but their performance directly depends on the amount and type of training data and thus may not be reproducible across different unseen datasets whose acquisition and protocols vary significantly from the training data. The insufficient amount of manually labeled training data also affects the performance of such machine learning models. QuickNAT attempts to improve upon this limitation by first pretraining the network using auxiliary labels generated using FreeSurfer and then refining the pretrained network with the limited manually labeled data.
The 2D F-CNN methods which train the network by using images slice by slice have an inherent disadvantage of failure to fully utilize the contextual information from neighboring slices. The 3D F-CNN, on the other hand, faces the memory limitation of the available GPUs in handling millions of parameters associated with high-resolution clinical MRI imaging volumes. Several techniques have been developed to address the computational cost and memory limitations; for example, SLANT-27 breaks the original image volume into 27 overlapping network tiles which can be executed in parallel on 27 GPUs or can run sequentially in the available lesser resources; AssemblyNet goes a step further and uses two assemblies of 125 3D U-Nets processing different overlapping regions; 3DQ method attempts to provide around 16 times compression without affecting performance; other methods such as ACA-PULCO and CAST focus on specific regions such as cerebellum and hippocampus, respectively, rather than whole-brain segmentation.
Other issues with the machine learning models are with regard to insufficient documentation in the literature; only some such methods are publicly made available, even then the training modalities of some of them especially those relying on 3D-FCN have a huge computational and memory bottleneck associated with them and cannot be easily reproduced. There is a need to implement such machine learning models through computational webservers in a standardized manner as was the case with volBrain for their effective use and validation.
Acknowledgments
The authors like to thank Professor Pankaj Seth, National Brain Research Centre for his guidance, support, and help, including proofreading of the manuscript. The support of Dr D. D. Lal, Mr Sukhkir, and DBT e-Library Consortium (DeLCON) in making available the research articles is also duly acknowledged. The work has also benefitted from the infrastructure facilities under DIC project of BTISNet programme and Dementia Science programme, both funded by the Department of Biotechnology, Government of India.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Footnotes
ORCID iDs: Mahender Kumar Singh  https://orcid.org/0000-0001-9617-6112
https://orcid.org/0000-0001-9617-6112
Krishna Kumar Singh  https://orcid.org/0000-0003-3849-5945
https://orcid.org/0000-0003-3849-5945
Author Contributions
MKS conceived the study and was responsible for overall study direction and planning; KKS provided direction, suggestion, and supervision. Both the authors discussed the final manuscript.
Ethical Statement
Not Applicable.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1.Driscoll I, Davatzikos C, An Y,. et al. Longitudinal pattern of regional brain volume change differentiates normal aging from MCI. Neurology 2009; 72: 1906–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franke K and Gaser C.. Longitudinal changes in individual BrainAGE in healthy aging, mild cognitive impairment, and Alzheimer’s disease. GeroPsych 2012; 25: 235–245. [Google Scholar]
- 3.Raghuprasad MS and Manivannan M.. Volumetric and morphometric analysis of pineal and pituitary glands of an Indian inedial subject. Ann Neurosci 2018; 25: 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giorgio A and de Stefano N.. Clinical use of brain volumetry. J Magn Reson Imaging 2013; 37: 1–14. [DOI] [PubMed] [Google Scholar]
- 5.Wu W-C, Huang C-C, Chung H-W,. et al. Hippocampal alterations in children with temporal lobe epilepsy with or without a history of febrile convulsions: Evaluations with MR volumetry and proton MR spectroscopy. AJNR Am J Neuroradiol 2005; 26: 1270–1275. [PMC free article] [PubMed] [Google Scholar]
- 6.Apostolova LG, Dinov ID, Dutton RA,. et al. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer’s disease. Brain 2006; 129: 2867–2873. [DOI] [PubMed] [Google Scholar]
- 7.Tanskanen P, Veijola JM, Piippo UK,. et al. Hippocampus and amygdala volumes in schizophrenia and other psychoses in the Northern Finland 1966 birth cohort. Schizophr Res 2005; 75: 283–294. [DOI] [PubMed] [Google Scholar]
- 8.Bremner JD, Narayan M, Anderson ER,. et al. Hippocampal volume reduction in major depression. Am J Psychiatry 2000; 157: 115–118. [DOI] [PubMed] [Google Scholar]
- 9.Baxter MG and Murray EA. The amygdala and reward. Nat Rev Neurosci 2002; 3: 563–573. [DOI] [PubMed] [Google Scholar]
- 10.Sacchet MD, Livermore EE, Iglesias JE,. et al. Subcortical volumes differentiate major depressive disorder, bipolar disorder, and remitted major depressive disorder. J Psychiatr Res 2015; 68: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker FO. Huntington’s disease. The Lancet 2007; 369: 218–228. [DOI] [PubMed] [Google Scholar]
- 12.Hokama H, Shenton ME, Nestor PG,. et al. Caudate, putamen, and globus pallidus volume in schizophrenia: A quantitative MRI study. Psychiatry Res Neuroimaging 1995; 61: 209–229. [DOI] [PubMed] [Google Scholar]
- 13.Sterling NW, Du G, Lewis MM,. et al. Striatal shape in Parkinson’s disease. Neurobiol Aging 2013; 34: 2510–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen L, Firpi HA, Saykin AJ,. et al. Parametric surface modeling and registration for comparison of manual and automated segmentation of the hippocampus. Hippocampus 2009; 19: 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherbuin N, Anstey KJ, Réglade-Meslin C. et al. In vivo hippocampal measurement and memory: A comparison of manual tracing and automated segmentation in a large community-based sample. PLoS ONE 2009; 4: e5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenger E, Mårtensson J, Noack H. et al. Comparing manual and automatic segmentation of hippocampal volumes: Reliability and validity issues in younger and older brains. Hum Brain Mapp 2014; 35: 4236–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gispert JD, Reig S, Pascau J,. et al. Method for bias field correction of brain T1-weighted magnetic images minimizing segmentation error. Hum Brain Mapp 2004; 22: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song S, Zheng Y, and He Y.. A review of methods for bias correction in medical images. Biomed Eng Rev 2017; 3: 1–10. [Google Scholar]
- 19.Despotović I, Goossens B, and Philips W.. MRI segmentation of the human brain: Challenges, methods, and applications. Comput Math Methods Med; 2015 ; 2015: 450341. DOI: 10.1155/2015/450341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue H, Srinivasan L, Jiang S,. et al. Automatic segmentation and reconstruction of the cortex from neonatal MRI. NeuroImage 2007; 38: 461–477. [DOI] [PubMed] [Google Scholar]
- 21.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002; 17: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battaglini M, Smith SM, Brogi S,. et al. Enhanced brain extraction improves the accuracy of brain atrophy estimation. NeuroImage 2008; 40: 583–589. [DOI] [PubMed] [Google Scholar]
- 23.Jenkinson M, Beckmann CF, Behrens TEJ,. et al. FSL. NeuroImage 2012; 62: 782–790. [DOI] [PubMed] [Google Scholar]
- 24.Heckemann RA, Ledig C, Gray KR,. et al. Brain extraction using label propagation and group agreement: Pincram. PLoS ONE 2015; 10: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazdani S, Yusof R, Karimian A,. et al. Image segmentation methods and applications in MRI brain images. IETE Tech Rev 2015; 32: 413–427. [Google Scholar]
- 26.Iglesias JE and Sabuncu MR.. Multi-atlas segmentation of biomedical images: A survey. Med Image Anal 2015; 24: 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischl B. FreeSurfer. NeuroImage 2012; 62: 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischl B, Salat DH, Busa E,. et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 2002; 33: 341–355. [DOI] [PubMed] [Google Scholar]
- 29.Saygin ZM, Kliemann D, Iglesias JE,. et al. High-resolution magnetic resonance imaging reveals nuclei of the human amygdala: Manual segmentation to automatic atlas. NeuroImage 2017; 155: 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iglesias JE, Augustinack JC, Nguyen K,. et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage 2015; 115: 117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith SM, Jenkinson M, Woolrich MW,. et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 2004; 23: S208–S219. DOI: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Brady M, and Smith S.. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 2001; 20: 45–57. [DOI] [PubMed] [Google Scholar]
- 33.Patenaude B, Smith SM, Kennedy DN,. et al. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage 2011; 56: 907–922. DOI: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner J. SPM: A history. NeuroImage 2012; 62: 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farokhian F, Beheshti I, Sone D,. et al. Comparing CAT12 and VBM8 for detecting brain morphological abnormalities in temporal lobe epilepsy. Front Neurol 2017; 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolls ET, Huang CC, Lin CP,. et al. Automated anatomical labelling atlas 3. NeuroImage 2020; 206: 116189. [DOI] [PubMed] [Google Scholar]
- 37.Manjón JV and Coupé P.. volBrain: An online MRI brain volumetry system. Front Neuroinform 2016; 10: 30. DOI: 10.3389/fninf.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero JE, Coupé P, Giraud R. et al. CERES: A new cerebellum lobule segmentation method. NeuroImage 2017; 147: 916–924. [DOI] [PubMed] [Google Scholar]
- 39.Romero JE, Coupé P, and Manjón J v. HIPS: A new hippocampus subfield segmentation method. NeuroImage 2017; 163: 286–295. [DOI] [PubMed] [Google Scholar]
- 40.Manjón J v, Bertó A, Romero JE. et al. pBrain: A novel pipeline for Parkinson related brain structure segmentation. NeuroImage Clin 2020; 25: 102184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heckemann RA, Keihaninejad S, Aljabar P,. et al. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. NeuroImage 2010; 51: 221–227. [DOI] [PubMed] [Google Scholar]
- 42.Jia H, Yap P-T, and Shen D.. Iterative multi-atlas-based multi-image segmentation with tree-based registration. NeuroImage 2012; 59: 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yushkevich PA, Pluta JB, Wang H,. et al. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp 2015; 36: 258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaakub SN, Heckemann RA, Keller SS,. et al. On brain atlas choice and automatic segmentation methods: a comparison of MAPER & FreeSurfer using three atlas databases. Sci Rep 2020; 10: 2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palumbo L, Bosco P, Fantacci ME,. et al. Evaluation of the intra- and inter-method agreement of brain MRI segmentation software packages: A comparison between SPM12 and FreeSurfer v6.0. Physica Medica 2019; 64: 261–272. [DOI] [PubMed] [Google Scholar]
- 46.Bartel F, Vrenken H, van Herk M. et al. Fast segmentation through surface fairing (FastSURF): A novel semi-automatic hippocampus segmentation method. PLoS ONE 2019; 14: e0210641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velasco-Annis C, Akhondi-Asl A, Stamm A. et al. Reproducibility of brain MRI segmentation algorithms: Empirical comparison of local MAP PSTAPLE, FreeSurfer, and FSL-FIRST. J Neuroimaging 2018; 28: 162–172. [DOI] [PubMed] [Google Scholar]
- 48.Zandifar A, Fonov V, Coupé P. et al. A comparison of accurate automatic hippocampal segmentation methods. NeuroImage 2017; 155: 383–393. [DOI] [PubMed] [Google Scholar]
- 49.Perlaki G, Horvath R, Nagy SA,. et al. Comparison of accuracy between FSL’s FIRST and Freesurfer for caudate nucleus and putamen segmentation. Sci Rep 2017; 7: 2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Næss-Schmidt E, Tietze A, Blicher JU. et al. Automatic thalamus and hippocampus segmentation from MP2RAGE: Comparison of publicly available methods and implications for DTI quantification. Int Comput Assist Radiol Surg 2016; 11: 1979–1991. [DOI] [PubMed] [Google Scholar]
- 51.Grimm O, Pohlack S, Cacciaglia R,. et al. Amygdalar and hippocampal volume: A comparison between manual segmentation, Freesurfer and VBM. J Neurosci Methods 2015; 253: 254–261. [DOI] [PubMed] [Google Scholar]
- 52.Fellhauer I, Zöllner FG, Schröder J. et al. Comparison of automated brain segmentation using a brain phantom and patients with early Alzheimer’s dementia or mild cognitive impairment. Psychiatry Res Neuroimaging 2015; 233: 299–305. [DOI] [PubMed] [Google Scholar]
- 53.Ronneberger O, Fischer P, and Brox T.. U-Net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, et al., eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 . Springer International Publishing, 2015: pp. 234–241. [Google Scholar]
- 54.Çiçek Ö, Abdulkadir A, Lienkamp SS. et al. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu MR, et al., eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016 . Springer International Publishing, 2016: pp. 424–432. [Google Scholar]
- 55.Coupé P, Mansencal B, Clément M. et al. AssemblyNet: A large ensemble of CNNs for 3D whole brain MRI segmentation. NeuroImage 2020; 219: 117026. [DOI] [PubMed] [Google Scholar]
- 56.Huo Y, Xu Z, Xiong Y,. et al. 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage 2019; 194: 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paschali M, Gasperini S, Roy AG,. et al. 3DQ: Compact quantized neural networks for volumetric whole brain segmentation. In Shen D. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science, vol 11766. Springer, Cham, 2019: pp. 438–446. [Google Scholar]
- 58.Guha Roy A, Conjeti S, Navab N. et al. QuickNAT: A fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage 2019; 186: 713–727. [DOI] [PubMed] [Google Scholar]
- 59.Roy AG, Conjeti S, Navab N,. et al. Bayesian QuickNAT: Model uncertainty in deep whole brain segmentation for structure-wise quality control. NeuroImage 2019; 195: 11–22. [DOI] [PubMed] [Google Scholar]
- 60.Wachinger C, Reuter M, and Klein T.. DeepNAT: Deep convolutional neural network for segmenting neuroanatomy. NeuroImage 2018; 170: 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehta R, Majumdar A, and Sivaswamy J.. BrainSegNet: A convolutional neural network architecture for automated segmentation of human brain structures. J Med Imaging 2017; 4: 24003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dolz J, Gopinath K, Yuan J,. et al. HyperDense-Net: A hyper-densely connected CNN for multi-modal image segmentation. IEEE Trans Med Imaging 2019; 38: 1116–1126. [DOI] [PubMed] [Google Scholar]
- 63.Chen H, Dou Q, Yu L,. et al. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 2018; 170: 446–455. [DOI] [PubMed] [Google Scholar]
- 64.Goubran M, Ntiri EE, Akhavein H,. et al. Hippocampal segmentation for brains with extensive atrophy using three-dimensional convolutional neural networks. Hum Brain Mapp 2020; 41: 291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z, Zhuang X, Mishra V,. et al. CAST: A multi-scale convolutional neural network based automated hippocampal subfield segmentation toolbox: CAST: Segmenting hippocampal subfields. NeuroImage 2020; 218: 116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han S, Carass A, He Y,. et al. Automatic cerebellum anatomical parcellation using U-Net with locally constrained optimization. NeuroImage 2020; 218: 116819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thyreau B, Sato K, Fukuda H,. et al. Segmentation of the hippocampus by transferring algorithmic knowledge for large cohort processing. Med Image Anal 2018; 43: 214–228. [DOI] [PubMed] [Google Scholar]
- 68.Yang CY, Liu HM, Chen SK,. et al. Reproducibility of brain morphometry from short-term repeat clinical MRI examinations: A retrospective study. PLoS ONE 2016; 11: e0146913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Botvinik-Nezer R, Holzmeister F, Camerer CF. et al. Variability in the analysis of a single neuroimaging dataset by many teams. Nature 2020; 582: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]


