Abstract
Purpose
Colorectal cancer (CRC) can develop via a hypermutagenic pathway characterized by frequent somatic DNA base-pair mutations. Alternatively, the immunogenicity of tumor cells themselves may influence the anticancer activity of the immune effector cells. Impaired DNA repair mechanisms drive mutagenicity, which then increase the neoantigen load and immunogenicity. However, no studies have analyzed immune checkpoint protein expression, particularly programmed death-1 (PD-1)/programmed death-ligand 1 (PD-L1), in adenoma–carcinoma progression and its relationship with the emergence of other DNA repair gene mutation.
Materials and Methods
We investigated mutations of 10 genes involved in DNA repair function: XRCC1, TP53, MLH1, MSH, KRAS, GSTP, UMP, MTHF, DPYD, and ABCC2. We performed sequencing to determine mutations and immunohistochemistry of immune checkpoints in clinical samples and determined changes in XRCC1 expression during progression through the adenoma–carcinoma pathway. We further investigated the prognostic associations of gene XRCC1 according to the expression, mutational profile, and immune profile using The Cancer Genome Atlas-colon adenocarcinoma (TCGA-COAD) dataset.
Results
From clinical samples, XRCC1 mutation demonstrated the strongest association with adenomas with a mutation frequency of 56.2% in adenomas and 34% in CRCs (p =0.016). XRCC1 was abnormally expressed and altered by mutations contributing to adenoma carcinogenesis. High expression of XRCC1, CD4, FOXP3, and PD-1/PD-L1 showed an overall upward trend with increased lesion severity (all p < 0.01). PD-1/PD-L1 expression and CD4+ intraepithelial lymphocytes (IELs) correlated with cytological dysplasia progression, specifically in patients with wild-type XRCC1 (all p < 0.01), whereas FOXP3 expression was independently associated with adenoma–carcinoma progression. From TCGA-COAD analysis, XRCC1 expression was associated with patients survival, tumor-infiltrating lymphocytes and immune marker expression.
Conclusion
Increased IEL density and PD-1/PD-L1 expression correlate with cytological dysplasia progression and specifically with the XRCC1 mutation status in CRC. Our findings support a stepwise dysplasia-carcinoma sequence of adenoma carcinogenesis and an XRCC1 hypermutated phenotypic mechanism of lesions.
Keywords: adenoma–carcinoma, immune environment, PD-1/PD-L1, tumor-infiltrating lymphocytes, XRCC1
Plain Language Summary
Colorectal cancer progresses through a well‑defined series of transformations from normal colonic epithelial cells to precursor adenoma lesions that eventually evolve into increasingly more invasive and malignant stages. An improved understanding of the genetic and molecular drivers of colorectal cancer, especially the progression of adenoma to carcinoma, is necessary for developing effective prognostic biomarkers and personalized treatment strategies. Using publicly available data of The Cancer Genome Atlas-colon adenocarcinoma database and original data derived from 153 clinical samples from our institution, we identified a new molecular mechanism underlying the progression of colorectal adenoma to carcinoma. This mechanism involves a specific mutation of the DNA repair system gene XRCC1. Specifically, a “mutator phenotype” of XRCC1 might change the protein levels and immunogenicity, thereby worsening the tumor biology and patient outcomes. This mechanism may additionally stimulate a host immune response characterized by rapid intraepithelial lymphocyte infiltration and immune checkpoint (PD-1/PD-L1) expression, which are known to be associated with cancer progression and the development of immunotherapy resistance. Our results support XRCC1 as a potential biomarker and target for the personalized treatment of colorectal cancer.
Introduction
Colon cancer progression occurs through a well‑defined series of transformations from normal colonic epithelial cells to precursor adenoma lesions that eventually evolve into increasingly more invasive and malignant stages.1 The following three molecular carcinogenesis pathways have been recognized: (1) chromosomal instability,2 (2) microsatellite instability (MSI),3,4 and (3) CpG island methylator phenotype.5 Morphologically, this progression initially manifests as low-grade dysplasia (LGD) with the development of cytologic adenomatous dysplasia, and subsequent stepwise progression into high-grade dysplasia (HGD), ultimately leading to the development of colorectal cancer (CRC).6 The extensive succession of multiple somatic mutations confers a growth advantage to a mutant cell, which can aid in the establishment of a putative model of cancer that implicates chance as well as causality.
The tumor microenvironment (TME) is an important barrier for tumor immune escape, and the core of the TME is involved in the underlying mechanism of immune tolerance.7 The programmed death-1 (PD-1)/programmed death-ligand 1 (PD-L1) pathway plays a critical role in regulating T-cell tolerance.8 Regulatory T cells (Tregs) are a subset of immunosuppressive CD4+ T lymphocytes, which are actively engaged in the maintenance of immunological self-tolerance by suppressing self-reacting T cells and preventing autoimmunity.8 Only few studies have examined the immune environment in the precancerous lesions of CRC to date.9,10 Rau et al9 were the first to study the immune environment in serrated precancerous lesions, with an emphasis on tumor-infiltrating lymphocytes (TILs). However, no study has yet focused on immune checkpoint protein expression, particularly PD-1/PD-L1, in the colorectal adenoma–carcinoma progression or its relationship with the emergence of abnormal DNA repair gene status.
Impaired DNA repair and the associated genomic instability not only increases mutagenicity/carcinogenicity but can also increase the neoantigen load on the tumor cell surface, resulting in increased immunogenicity.11 Mutations in X-ray repair cross-complementary 1 (XRCC1), a limiting factor in the base excision repair pathway, in a constitutively active state have been shown to promote tumor growth and angiogenesis.12 Clinical evidence demonstrates that the interplay between DNA repair and the PD-1/PD-L1 pathway promotes aggressive tumor phenotypes and enables XRCC1-directed personalization of immune checkpoint inhibitor therapy in breast cancer.11 However, the role of XRCC1 in precancerous CRC lesions has not been elucidated, and whether it exerts a significant influence in the early stages of colorectal carcinogenesis, particularly on the immune response in CRC, has not been determined thus far.
To resolve these questions, in this study, we first conducted sequencing analyses of mutations in 10 selected genes involving in DNA repair function in colorectal adenomas. Then we selected XRCC1 as the focus of this study, and performed immunohistochemical assays of immune checkpoints and XRCC1 in clinical samples of hyperplastic polyps (HPs), LGDs, HGDs, and CRCs from patients at our institute. In addition, we investigated the prognostic associations of gene XRCC1, and its expression, mutational profile, and immune profiles based on data available in The Cancer Genome Atlas-colon adenocarcinoma (TCGA-COAD) database. Overall, the objective of this study was to examine the density of intraepithelial lymphocytes (IELs), expression of PD-1/PD-L1 in infiltrating lymphocytes, expression of XRCC1 in the lesion epithelium, and the XRCC1 mutation status at various stages of progression through the adenoma–carcinoma pathway.
Materials and Methods
Patients and Samples
A total of 153 clinical samples, including 103 precancerous lesions of the colon (30 HPs, 44 LGDs, 29 HGDs) and 50 CRCs, were retrospectively analyzed from archives at the Zhejiang Provincial People’s Hospital collected from January 2019 to September 2021. HPs, LGDs, and HGDs were combined into a single precancerous lesions group. Demographic and clinical characteristics such as age at initial diagnosis, sex, lesion location, and tumor stage were also collected from clinical records. All samples were resected via endoscopic mucosal resection, endoscopic submucosal dissection, or surgery. This retrospective study was performed in accordance with the Declaration of Helsinki. Approval was obtained from the Institutional Review Board of Zhejiang Provincial People’s Hospital (protocol 2021QT329). Informed consent was waived due to this study’s retrospective nature and the anonymized processing of patient data.
Single Nucleotide Polymorphism (SNP) Selection, DNA Isolation, and Genotyping
The polymorphisms in XRCC1 (rs25487), TP53 (rs1042522, rs12947788, rs17880604, and rs17884306), MLH1 (rs28930073), MSH2 (rs1800152, rs1802577, and rs12476364), KRAS (rs112445441, rs121913529, and rs121913530), GSTP1 (rs1695), UMPS (rs1801019), MTHFR (rs1801131, rs1801133), DPYD (rs1801159), and ABCC2 (rs2273697) were selected for genotype comparison. Genomic DNA was extracted from paraffin-embedded tissues with at least 15 slides per sample. A pathologist (X. Zhang) assessed the normal and tumor areas and the percentage of tumor cells based on hematoxylin and eosin (H&E)-stained slides. Only samples with at least 70% tumor tissue present were included for analysis. DNA was isolated using High Pure FFPET DNA Isolation Kit (Roche Applied Science, Penzberg, Germany) according to the manufacturers’ instructions. Genomic DNA concentrations, and OD260/280 and OD260/230 ratios were measured with the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). High-quality genomic DNA samples were then used for genotyping through the Sanger sequencing method on an ABI 3730 XL instrument (Thermo Fisher Scientific, Tempe, AZ, USA). The sequencing results were analyzed using Chromas Lite v2.1.
Immunohistochemical Staining
The tissue sections were deparaffinized, rehydrated, and heated in a microwave oven with ethylenediaminetetraacetic acid antigen repair solution (ORIGENE, Beijing, China) at pH 8 for 15 min. After the sections were cooled, they were placed in phosphate-buffered saline for decolorization and were incubated for 20 min at 18–25°C. Negative and positive (with omission of the primary antibody and IgG-matched serum, respectively) controls were included for each marker in each experiment. The primary HP, LGD, HGD, and CRC samples were sectioned (5 μm thick) and stained with H&E per routine procedures. Immunostaining was performed manually using anti-XRCC1 (sc-56254; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-CD4 (ab183685; Abcam, Cambridge, UK), anti-FOXP3 (ab215206; Abcam), anti-PD-1 (GT228107; Genetech, Shanghai, China), and anti-PD-L1 (GT228007; Genetech). IEL density, corresponding to the highest density area of CD4-positive cells, and infiltrating lymphocyte expression of PD-1 per 200 epithelial cells were separately scored for areas of HP, LGD, HGD, and CRC. The number of PD-1-positive cells in a high-power microscopic field (hpf) was counted and scored as 0 (not detectable), 1 (1–2 cells/hpf), 2 (3–5 cells/hpf), 3 (6–10 cells/hpf), or 4 (>10 cells/hpf).10 The number of cells exhibiting a nuclear reaction to FOXP3 and the percentage of cells showing a membrane reaction to CD4 were determined based on 10 randomly chosen hpfs (400×) and the average levels were calculated. The intensity of PD-L1 staining of the infiltrating lymphocytes was semi-quantitatively scored from 0 to 4 according to the percentage of positive cells (0 = 0%; 1 = ≥1%; 2 = ≥5%; 3 = ≥10%; 4 = ≥50%).13 Only cases with staining intensity scores of 3 or 4 were considered as high-expression cases. The percentage of stained cells was evaluated separately by two pathologists.
Gene Expression and Survival Analysis
Transcriptome RNA-sequencing data of samples from 514 COAD patients (41 normal samples and 473 tumor samples) and corresponding clinical data were downloaded from TCGA database (https://portal.gdc.cancer.gov/). In the presented study, all COAD samples were grouped into XRCC1 high-expression group and XRCC1 low-expression group compared with the XRCC1 median expression. The R language “survival” and “survminer” packages were used for survival analysis. Additional survival data for 446 tumor samples derived from 473 patients with COAD were available were used for survival analysis. The Kaplan–Meier method was used to plot the survival curve, and the log-rank was used for assessing the statistical significance test; p < 0.05 was considered significant.
Genetic Alteration Analysis
The genetic alteration characteristics of XRCC1 were obtained from the cBioPortal website, including the alteration frequency, mutation type, and copy number alterations in COAD, from the “Cancer Types Summary” module. The mutated site information of XRCC1 in the protein structure or the three-dimensional diagram structure was obtained from the “Mutations” module. We also used the “Comparison” module to obtain data on the survival differences for COAD patients with or without XRCC1 genetic alteration. Kaplan-Meier plots were also generated for these data along with P-values obtained from the Log rank test.
Tumor Mutational Burden (TMB), MSI, and DNA Mismatch Repair (MMR) System Gene Mutation Analysis
Abnormalities in the TMB, MSI, and MMR system can lead to tumorigenesis. Therefore, we evaluated the relationship of XRCC1 expression levels with TMB, MSI, and MMR via Pearson correlation analysis based on TCGA database. Data on the mutation levels of five MMR-related genes (MLH1, MSH2, MSH6, PMS2, and EPCAM) were obtained from TCGA database.
Tumor-Infiltrating Immune Cell Profiles and Immune Correlation Analysis
The CIBERSORT computational method was applied for estimating the abundance of tumor-infiltrating immune cells in TCGA-COAD samples. Spearman correlation analysis was used to evaluate the correlation of XRCC1 mutation with the infiltrating immune cell scores of COAD downloaded from the TIMER database. Moreover, the correlation between XRCC1 and immune checkpoint marker levels was assessed from the TISIDB database (http://cis.hku.hk/TISIDB/).
Statistical Analysis
All statistical analyses were performed using SPSS v26.0 (IBM Corp. Armonk, NY, USA) and R software (R version 4.0.4). Significant differences among categorical variables were analyzed by the chi-square test (followed by Fisher’s exact test if necessary). The correlation of gene expression was evaluated by Spearman correlation coefficient. Bivariate and multivariate binary logistic regression analyses performed with 95% confidence intervals (CIs) were used to test the factors associated with adenoma–carcinoma progression. A two-sided P-value < 0.05 was considered statistically significant.
Results
Findings in the Clinical Samples
Genotypes of 10 Genes in Clinical Adenoma Samples
The genotypes of 18 SNPs in 10 genes were detected in eight HGD and eight LGD colorectal lesions. Among the 10 genes, we found that 8 genes had SNPs in LGD and HGD. The associations of 5 of these genes (TP53,14 KRAS,15 GSTP1,16 MTHFR,17 and ABCC218) with colonic adenoma have previously been reported in several studies, whereas no such association of XRCC1, UMPS, or DPYD has been reported to date (Supplementary Table 1). In addition, we selected XRCC1 as the focus of this study given previous results from a meta-analysis demonstrating a relationship between the XRCC1 rs25487 polymorphism and sensitivity to platinum-based chemotherapy drugs in CRC patients.19 Therefore, we further focused on the potential contribution of the XRCC1 rs25487 mutation in the adenoma–carcinoma pathway of CRC by evaluating the mutation status in all 153 samples.
Clinicopathological Features of the Patients
Basic characteristics of all patients are shown in Table 1. The mean age of all patients was 60 years, with a mean age of 57.6 years for patients with HPs, 57.1 years for patients with LGDs, 58.1 years for patients with HGDs, and 65.0 years for patients with CRCs; thus, the vast majority of patients were younger than 65 years (67.3%; p < 0.001, Table 1). Additionally, 97 of the 153 patients were male (p = 0.814) and 119 of the 153 lesions were present in the left side of the colon (p = 0.020) (Table 1).
Table 1.
Clinical Characteristics and Marker Expression of the Studied Lesions
| Characteristic | CRC | HGD | HP | LGD | Total | P |
|---|---|---|---|---|---|---|
| Mean Age (Range) | 65.0 (31–84) | 58.1 (30–75) | 57.6 (28–79) | 57.1 (29–76) | 60.0 (28–84) | |
| Age (Y) | ||||||
| ≤65 | 23 (46.0%) | 19 (65.5%) | 25 (83.3%) | 36 (81.8%) | 103 (67.3%) | <0.001 |
| >65 | 27 (54.0%) | 10 (34.5%) | 5 (16.7%) | 8 (18.2%) | 50 (32.7%) | |
| Sex | ||||||
| Men | 30 (60.0%) | 19 (65.5%) | 21 (70.0%) | 27 (61.4%) | 97 (63.4%) | 0.814 |
| Women | 20 (40.0%) | 10 (34.5%) | 9 (30.0%) | 17 (38.6%) | 56 (36.6%) | |
| Location | ||||||
| Right side | 11 (22.0%) | 16 (55.2%) | 18 (60.0%) | 25 (56.8%) | 70 (45.8%) | 0.020 |
| Left side | 45 (90.0%) | 24 (82.8%) | 19 (63.3%) | 31 (70.5%) | 119 (77.8%) | |
| Markers | ||||||
| PD-1 high | 24 (48.0%) | 5 (17.2%) | 0 (0.0%) | 7 (15.9%) | 36 (23.5%) | <0.001 |
| PD-1 low | 26 (52.0%) | 24 (82.8%) | 30 (100.0%) | 37 (84.1%) | 117 (76.5%) | |
| PD-L1 high | 21 (42.0%) | 11 (37.9%) | 1 (3.3%) | 3 (6.8%) | 36 (23.5%) | <0.001 |
| PD-L1 low | 29 (58.0%) | 18 (62.1%) | 29 (96.7%) | 41 (93.2%) | 117 (76.5%) | |
| XRCC1 high | 28 (56.0%) | 5 (17.2%) | 1 (3.3%) | 10 (22.7%) | 44 (28.8%) | <0.001 |
| XRCC1 low | 22 (44.0%) | 24 (82.8%) | 29 (96.7%) | 34 (77.3%) | 109 (71.2%) | |
| FOXP3 high | 35 (70.0%) | 7 (24.1%) | 1 (3.3%) | 8 (18.2%) | 51 (33.3%) | <0.001 |
| FOXP3 low | 15 (30.0%) | 22 (75.9%) | 29 (96.7%) | 36 (81.8%) | 102 (66.7%) | |
| CD4 high | 34 (68.0%) | 16 (55.2%) | 3 (10.0%) | 15 (34.1%) | 68 (44.4%) | <0.001 |
| CD4 low | 16 (32.0%) | 13 (44.8%) | 27 (90.0%) | 29 (65.9%) | 85 (55.6%) | |
| XRCC1 rs25487 status | ||||||
| Wild | 33 (66.0%) | 13 (44.8%) | 12 (40.0%) | 19 (43.2%) | 77 (50.3%) | 0.059 |
| Mutant | 17 (34.0%) | 16 (55.2%) | 18 (60.0%) | 25 (56.8%) | 76 (49.7%) |
Note: Bold values, the statistically significant (P value < 0.05).
Correlation of PD-1/PD-L1 Expression with Adenoma–Carcinoma Progression
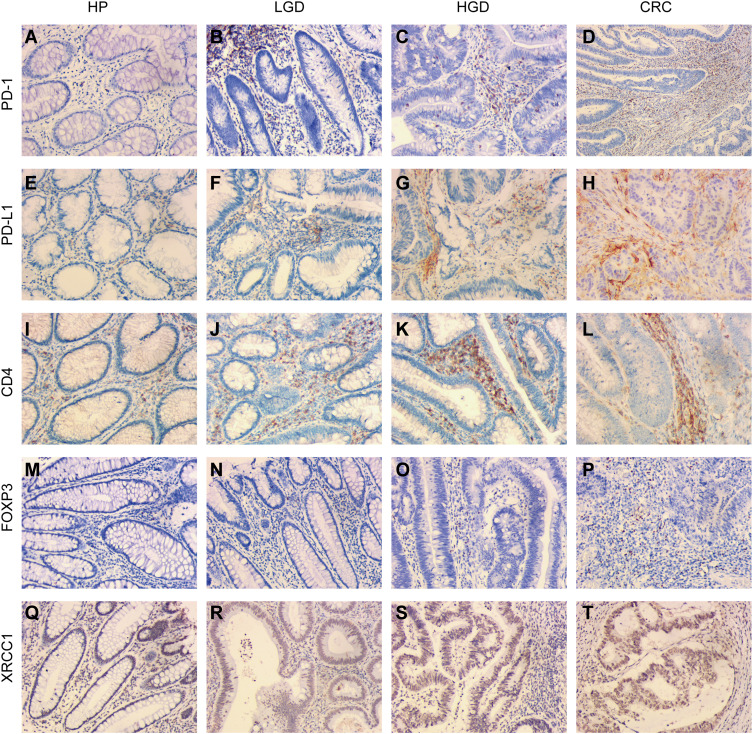
PD-1 and PD-L1 expression within the IELs, corresponding to the highest IEL-density area assessed via CD4 immunostaining, was scored separately for each histomorphological lesion subtype. PD-1 expression increased with stepwise progression through the adenoma–carcinoma pathway (Figure 1A–D, Table 1). None of the 30 cases of HPs assessed showed PD-1 expression, whereas the percentage of high-expression samples increased to 15.9%, 17.2%, and 48.0% in LGDs, HGDs, and CRCs, respectively (p < 0.001). The extent of PD-L1 expression showed a similar trend, with percentages of 3.3%, 6.8%, 37.9%, and 42.0% in HPs, LGDs, HGDs, and CRCs, respectively (p < 0.001) (Figure 1E–H, Table 1).
Figure 1.
Immunohistochemical expression of XRCC1, CD4, FOXP3, PD-1, and PD-L1 in clinical samples of HPs, LGDs, HGDs, and CRCs (all images: original magnification, ×200). (A–D) PD-1 epithelial expression in infiltrating lymphocytes. (E–H) PD-L1 epithelial expression in infiltrating lymphocytes. (I–L) CD4-positive intraepithelial lymphocyte density. (M–P) FOXP3-positive intraepithelial lymphocyte density. (Q–T) XRCC1 nuclei expression in colonic epithelial cells and tumor cells.
Correlation of IEL Density with Adenoma–Carcinoma Progression
The frequency of CD4-positive cells ranged from 1% to 70% with a mean of 25.6%, and the frequency FOXP3-positive cells ranged from 1 to 250 cells/hpf with a mean of 46.1 cells/hpf. The samples were grouped into high- and low-abundance groups for each cell type using the median as the cut-off value. As expected, a high IEL density correlated with the progression of precancerous lesions to CRCs through the adenoma-to-carcinoma sequence (Figure 1I–L, Table 1). The percentage of high CD4-positive cells increased to 10% in HPs, 34.1% in LGDs, 55.2% in HGDs, and 68.0% in CRCs, representing a statistically significant trend (p < 0.001) (Table 1). A high FOXP3-positive IEL density was found in 3.3% of HPs, 18.2% of LGDs, 24.1% of HGDs, and 70% of CRCs, which also represented a statistically significant increasing trend (p < 0.001) (Figure 1M–P, Table 1). Examples of the CD4-positive and FOXP3-positive IEL distribution are shown in Figure 1I–P.
Correlation of XRCC1, IEL, and Immune Checkpoint Expression
Lesions with high expression for XRCC1 in tumor nuclei in 3.3% of HPs, 22.7% of LGDs, 17.2% of HGDs, and 56% of CRCs (p < 0.001) (Figure 1Q–T, Table 1); we observed an overall upward trend in expression with increasing lesion severity from precancerous lesions to CRCs. There was a positive correlation between the expression level of XRCC1 and the levels of PD-1, PD-L1, FOXP3, and CD4, respectively (p < 0.001) (Table 2). Taken together, these data suggested that high XRCC1 expression was associated with an aggressive phenotype and TME in the adenoma-to-carcinoma progression.
Table 2.
Relationship of XRCC1 Expression with Immune Marker Expression and IELs of the Studied Lesions
| Markers | Correlation Coefficient | P value |
|---|---|---|
| PD-1 | 0.491 | <0.001 |
| PD-L1 | 0.394 | <0.001 |
| FOXP3 | 0.543 | <0.001 |
| CD4 | 0.314 | <0.001 |
Note: Bold values, the statistically significant (P value < 0.05).
Correlation of XRCC1 Mutation with Adenoma–Carcinoma Progression
A total of 76 of the 153 (49.7%) cases showed the mutant T allele in the rs25487 position of XRCC1 (Table 1). Among the 76 total mutant cases, there were 18 HPs (23.7%), 25 LGDs (32.9%), 16 HGDs (21.1%), and 17 CRCs (22.3%) (Table 1). Furthermore, we investigated the XRCC1 mutation in adenomas (LGDs and HGDs) and CRCs; the XRCC1 mutation was associated with a tumor location on the right side of the colon (p = 0.004) (Supplementary Table 2). The emergence of the XRCC1 mutation exhibited the strongest association with adenomas, with a mutation frequency of 56.2% in adenomas and 34.0% in CRCs (p =0.016) (Supplementary Table 2).
Factors Associated with Adenoma–Carcinoma Progression
In the bivariate model, eight factors, including age; location of lesions; expression of PD-1, PD-L1, FOXP3, CD4, and XRCC1; and the mutation status of XRCC1, were all significantly correlated with CRC (all p < 0.05) (Table 3). The multivariate logistic regression analysis indicated that CRC was 0.14 times [odds ratio (OR): 0.14; 95% CI 0.05–0.44] less likely to occur in individuals aged below 65 years (Table 3). By contrast, lesions with the XRCC1 rs25487 wild-type (CC) (OR: 3.38; 95% CI 1.15–9.95), high FOXP3 expression (OR: 11.66; 95% CI 2.74, 36.35), and high XRCC1 expression (OR: 3.84; 95% CI 1.37–10.78) were positively associated with CRC (Table 3). Taken together, these data suggested that age, XRCC1 mutation status, high XRCC1 expression, and a high density of FOXP3-positive IELs may be involved in the adenoma–carcinoma progression.
Table 3.
Association Between Predictor Variables and Colorectal Cancer (CRC) Determined via Bivariate and Multivariate Logistics Regression Analysis
| Variables | CRCs | Adenomas | Bivariate OR (95% CI) | p value | Multivariate OR (95% CI) | p value | |
|---|---|---|---|---|---|---|---|
| Age (Y) | ≤65 | 23 | 55 | 0.28 (0.13,0.6) | 0.001 | 0.14 (0.05,0.44) | 0.001 |
| >65 | 27 | 18 | Ref | Ref | |||
| Location | Right side | 5 | 18 | 0.34 (0.12,0.99) | 0.047 | 0.54 (0.13,2.27) | 0.396 |
| Left side | 45 | 55 | Ref | Ref | |||
| Sex | Male | 30 | 55 | 0.88 (0.42,1.84) | 0.736 | – | – |
| Female | 20 | 18 | Ref | ||||
| PD-1 | High | 24 | 12 | 4.69 (2.04,10.77) | <0.001 | 1.46 (0.45,4.77) | 0.530 |
| Low | 26 | 61 | Ref | Ref | |||
| PD-L1 | High | 21 | 14 | 3.05 (1.36,6.85) | 0.007 | 1.61 (0.49,5.23) | 0.431 |
| Low | 29 | 59 | Ref | Ref | |||
| XRCC1 | High | 28 | 15 | 4.92 (2.22,10.91) | <0.001 | 3.84 (1.37,10.78) | 0.011 |
| Low | 22 | 58 | Ref | Ref | |||
| FOXP3 | High | 35 | 15 | 9.02 (3.94,20.68) | <0.001 | 11.66 (3.74,36.35) | <0.001 |
| Low | 15 | 58 | Ref | Ref | |||
| CD4 | High | 34 | 31 | 2.88 (1.35,6.12) | 0.006 | 1.68 (0.56,5.09) | 0.357 |
| Low | 16 | 42 | Ref | Ref | |||
| XRCC1 rs25487 | Wild | 33 | 32 | 2.49 (1.18,5.24) | 0.017 | 3.38 (1.15,9.95) | 0.027 |
| Mutant | 17 | 41 | Ref | Ref | |||
Notes: Bold values, the statistically significant (P-value < 0.05).
Abbreviation: OR, adjusted odds ratio.
Correlation of XRCC1 Mutation with the Immune Microenvironment
The correlations of XRCC1 mutation status with immune markers across adenomas and CRCs are summarized in Table 4. High expression of PD-1, PD-L1, FOXP3, and CD4 was associated with CRC in lesions harboring wild-type XRCC1 (all p < 0.05), whereas only high FOXP3 expression was significantly associated with CRC in lesions with mutant XRCC1 (p < 0.001). These results suggested that FOXP3 might be an independent factor in the carcinogenesis of CRC, since high FOXP3 expression was a significant factor in both analyses based on the XRCC1 mutation status.
Table 4.
Immune Marker Expression and IELs in Colorectal Cancer (CRC) and Adenomas Based on the XRCC1 Gene Mutation Status
| XRCC1 rs25487 Status | Markers | CRCs | Adenomas (LGDs+HGDs) | Total | P value |
|---|---|---|---|---|---|
| Wild | PD-1 high | 17 (51.5%) | 3 (9.4%) | 20 (30.8%) | <0.001 |
| PD-1 low | 16 (48.5%) | 29 (90.6%) | 45 (69.2%) | ||
| PD-L1 high | 15 (45.5%) | 5 (15.6%) | 20 (30.8%) | 0.009 | |
| PD-L1 low | 18 (54.5%) | 27 (84.4%) | 45 (69.2%) | ||
| FOXP3 high | 22 (66.7%) | 6 (18.8%) | 28 (43.1%) | <0.001 | |
| FOXP3 low | 11 (33.3%) | 26 (81.3%) | 37 (56.9%) | ||
| CD4 high | 21 (63.6%) | 9 (28.1%) | 30 (46.2%) | 0.004 | |
| CD4 low | 12 (36.4%) | 23 (71.9%) | 35 (53.8%) | ||
| Mutant | PD-1 high | 7 (41.2%) | 9 (22.0%) | 16 (27.6%) | 0.122 |
| PD-1 low | 10 (58.8%) | 32 (78.0%) | 42 (72.4%) | ||
| PD-L1 high | 6 (35.3%) | 9 (22.0%) | 15 (25.9%) | 0.231 | |
| PD-L1 low | 11 (64.7%) | 32 (78.0%) | 43 (74.1%) | ||
| FOXP3 high | 13 (76.5%) | 9 (22.0%) | 22 (37.9%) | <0.001 | |
| FOXP3 low | 4 (23.5%) | 32 (78.0%) | 36 (62.1%) | ||
| CD4 high | 13 (76.5%) | 22 (53.7%) | 35 (60.3%) | 0.106 | |
| CD4 low | 4 (23.5%) | 19 (46.3%) | 23 (39.7%) |
Note: Bold values, the statistically significant (P value < 0.05).
Findings Based on TCGA Data
Correlation of XRCC1 Expression with Survival and Clinicopathological Characteristics in COAD
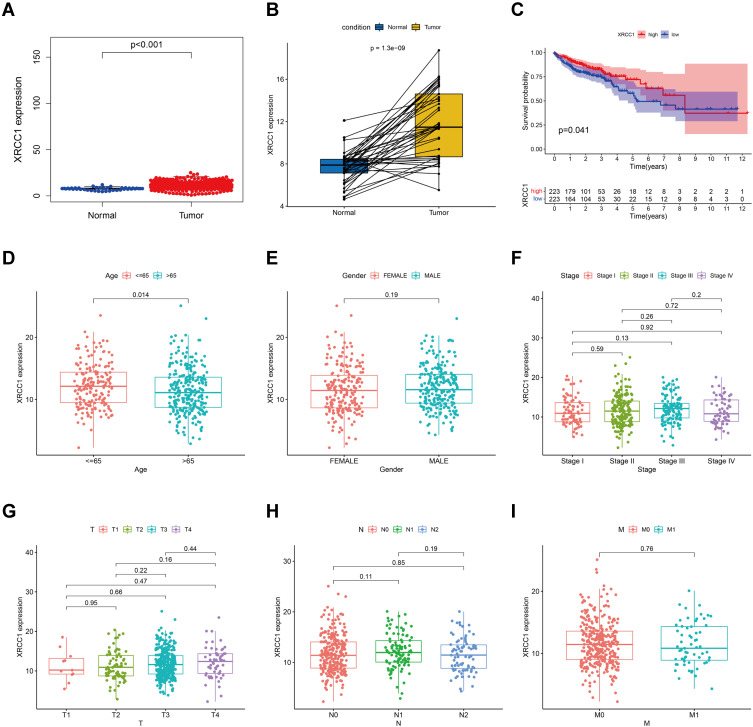
The expression of XRCC1 in the tumor samples was significantly higher than that in the normal samples from controls (Figure 2A). Similar results were observed in the comparison between paired normal and tumor tissues derived from the same patients (Figure 2B). All TCGA-COAD samples were grouped into the XRCC1 high-expression group and XRCC1 low-expression group. Survival analysis showed that COAD patients with low expression of XRCC1 had longer survival than those with high expression of XRCC1 (Figure 2C). Moreover, the expression of XRCC1 exhibited a correlation with the age of patients (Figure 2D), but no significant correlation with the gender, tumor stage and TNM of patients (Figure 2E–I). These results indicated that the expression of XRCC1 may significantly correlate with the prognosis of COAD patients.
Figure 2.
Correlation of XRCC1 expression with the survival and clinicopathological characteristics in TCGA-COAD cohort. (A) XRCC1 expression in COAD and normal tissues based on TCGA database. (B) Paired XRCC1 expression in COAD and normal tissues based on TCGA database. (C) Relationship of XRCC1 expression with survival in COAD patients. (D–I) Relationship of XRCC1 expression with clinicopathological characteristics (age, gender, tumor stage, and TNM) in COAD. The statistically significant (P value < 0.05).
Correlation of XRCC1 Genetic Alterations with Survival and Clinicopathological Characteristics of COAD Patients
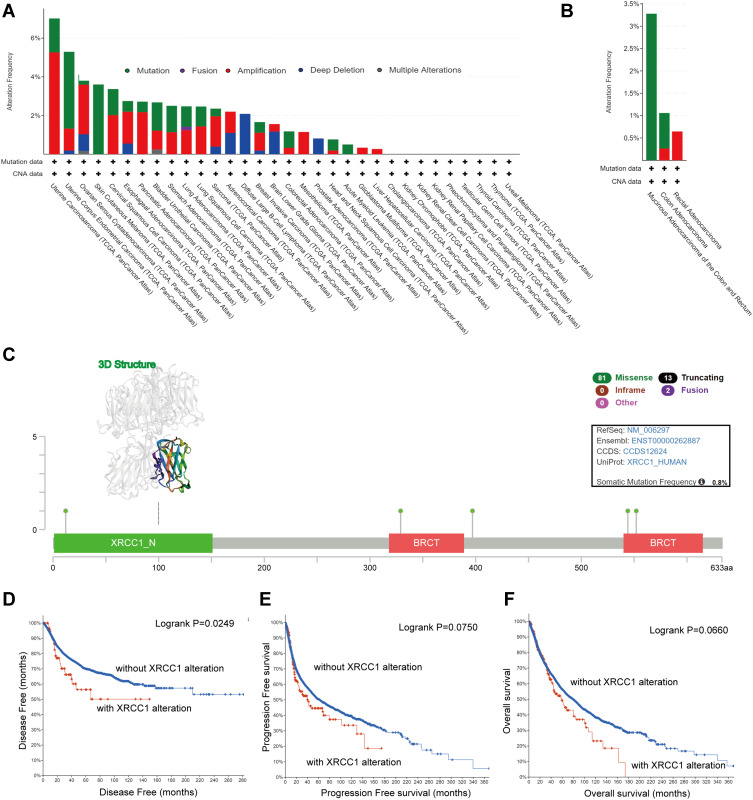
We observed the genetic alteration status of XRCC1 in different tumor samples of TCGA cohorts. As shown in Figure 3A, the overall alteration frequency of XRCC1 was 1.18% in 594 cases, with 0.84% mutations and 0.34% amplifications. The genetic alteration status of XRCC1 was further explored in different pathological types of COAD, showing a remarkably higher alteration frequency in mucinous adenocarcinoma of the colon and rectum (3.28% of 61 cases) (Figure 3B). The types, sites, and case numbers of the XRCC1 genetic alterations in COAD are further presented in Figure 3C. Missense mutation of XRCC1 was the main type of genetic alteration observed, with five mutations found in TCGA dataset (Supplementary Table 3).
Figure 3.
Mutation features of XRCC1 in TCGA-COAD cohort. (A) Alteration frequency of XRCC1 in pan-cancer data. (B) Alteration frequency of XRCC1 in different pathological types of COAD. (C) Alteration frequency of XRCC1 according to mutation type and the mutation site, with the 3D structure of XRCC1. (D–F) Relationship of XRCC1 mutation status with disease-free, progression-free, and overall survival in COAD patients. The statistically significant (P value < 0.05).
Additionally, we explored the potential association between genetic alteration of XRCC1 and the clinical survival prognosis of COAD patients. As shown in Figure 3D–F, COAD cases without altered XRCC1 were associated with a better prognosis with respect to disease-free survival (p = 0.025) (Figure 3D), but not with respect to progression-free survival, (p = 0.075) (Figure 3E) or overall survival (p = 0.066) (Figure 3F), when compared with those of XRCC1-altered cases. These results indicated that the mutation status of XRCC1 might exert an influence on the prognosis of CRC.
Correlation of XRCC1 Expression with TMB, MSI, and MMR in COAD
Based on TCGA-COAD cohort data, XRCC1 expression was correlated with both TMB and MSI in COAD (p < 0.001 and p = 0.037, respectively; Supplementary Figure 1A and B). The results further suggested that XRCC1 expression might be negatively related to the mutation levels of five MMR genes (MLH1, MSH2, MSH6, PMS2, and EPCAM) in COAD (Supplementary Figure 1C), indicating a potential role of XRCC1 in tumorigenesis.
Correlation of XRCC1 Expression with Immune Infiltration and Immune Checkpoint Markers in COAD
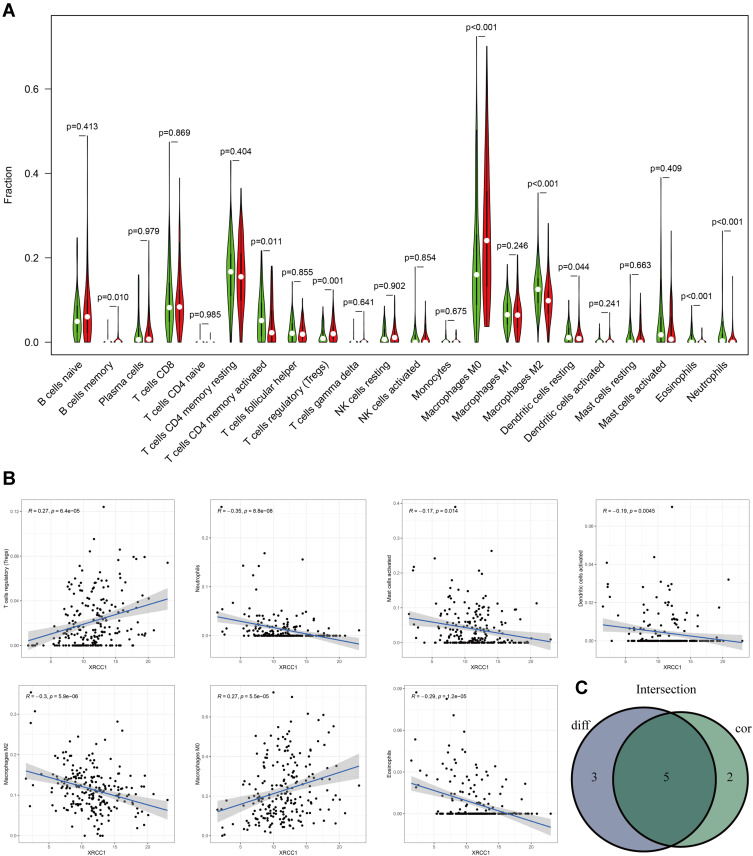
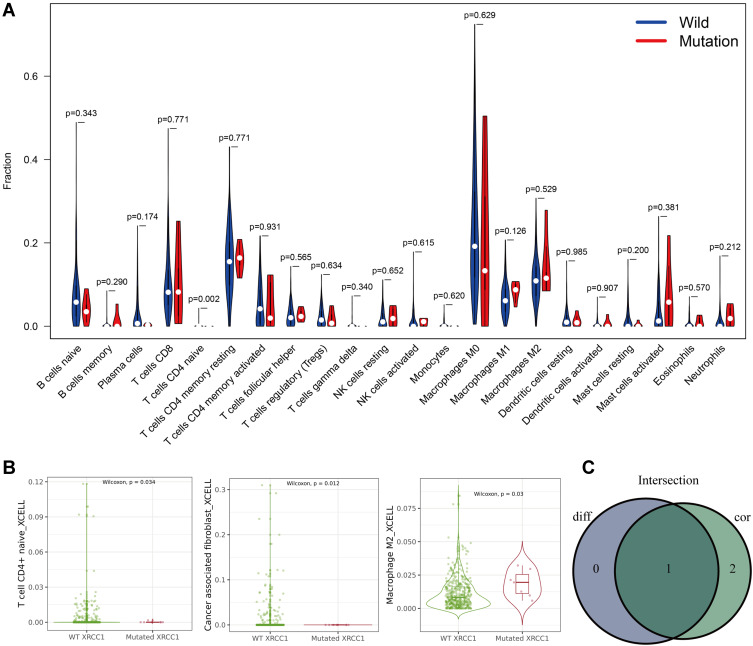
Five types of tumor-infiltrating immune cells were correlated with the expression of XRCC1 in COAD, including a positive correlation with Tregs and M0 macrophages, and a negative correlation with M2 macrophages, eosinophils, and neutrophils (Figure 4A–C). Moreover, we observed a significant negative correlation between CD4-naive T cells and XRCC1 mutant status (Figure 5A–C). We next investigated the correlation between XRCC1 expression and the expression levels of 69 common immune checkpoint genes, including 24 immunoinhibitors (Supplementary Figure 2A) and 45 immunostimulators (Supplementary Figure 2B) in COAD from the TISIDB database. Among these genes, PD-1 (PDCD1) and PD-L1 (CD274) was correlated with XRCC1 expression (Supplementary Figure 2A and C), but not with respect to XRCC1 mutation status (Supplementary Figure 2D). Collectively, these results further supported that XRCC1 expression might affect the immune activity of the TME in COAD.
Figure 4.
Correlation of TILs with XRCC1 expression based on TCGA-COAD cohort. (A) Violin plot showing the ratio differentiation of 22 types of immune cells between COAD tumor samples with high or low XRCC1 expression. (B) Scatter plot showing seven types of TILs correlated with XRCC1 expression. (C) Venn plot displaying five types of TILs correlated with XRCC1 expression co-determined via difference and correlation tests illustrated as violin and scatter plots, respectively. The statistically significant (P value < 0.05).
Figure 5.
Correlations of TILs with XRCC1 mutation status based on TCGA-COAD cohort. (A) Violin plot showing the ratio differentiation of 22 types of immune cells between COAD tumor samples with XRCC1 wild or mutant status. (B) Scatter plot showing three types of TILs correlated with XRCC1 mutation status. (C) Venn plot displaying one type of TILs correlated with XRCC1 mutation status co-determined via difference and correlation tests illustrated as violin and scatter plots, respectively. The statistically significant (P value < 0.05).
Discussion
In this study, we found that the XRCC1 mutation occurred in the precancerous stage of CRC, which, to our knowledge, has not been previously reported to date. Based on this finding, we further evaluated the immune features of the adenoma–carcinoma pathway and their correlations with XRCC1 expression and mutation status. Additionally, this is the first study that specifically analyzed IELs and PD-1/PD-L1 in HPs, LGDs, HGDs, and CRCs, and to elucidate their associations with carcinogenesis progression and XRCC1 status.
The XRCC1 rs25487 mutation occurs at the position chr19:43551574, and the molecular consequence is missense variant, which was consistent with the mutation information we obtained from TCGA database (Figure 3). Deficiency at this position of XRCC1 results in the alteration of amino acids, with glutamine mutated to arginine, which may affect the function of the XRCC1 protein and thus its DNA repair function.20,21 Our results based on TCGA data showed that XRCC1 was abnormally expressed and altered by XRCC1 mutations that could lead to the poor survival of CRC patients. Furthermore, we found that XRCC1 expression was associated with TILs and immune marker expression in COAD. These results strongly suggested that XRCC1 might serve as a prognostic biomarker and might affect the development of tumor immunity. Thus, more in-depth experimental evidence is needed to determine whether the mutation and expression of XRCC1 play essential roles in the initiation of the CRC, or whether it is merely the result of resisting tumor changes in normal tissues.
Colorectal carcinogenesis is driven by the sequential genetic and epigenetic alterations occurring in tumor cells.22 Adenomas occur when normal mechanisms regulating DNA repair and cell proliferation are altered.23 CRC can develop via a hypermutagenic pathway characterized by frequent somatic DNA base-pair mutations, resulting in the formation of discrete adenomas as the mutant cells advance toward the colonic lumen.23 Another possibility is that the immunogenicity of tumor cells themselves may influence the immune effector cell anticancer activity. Emerging data support that tumors with multiple somatic mutations accumulate neoantigens and are highly immunogenic.24 A key determinant of the mutation load is the DNA repair capacity in cancer cells. For example, MMR-deficient CRCs are characterized by the expression of highly immunogenic neoantigen peptides and high somatic mutations, phenomena that stimulate lymphocytic infiltration as well as the upregulated expression of inflammatory cytokines.25 Several studies have shown that the presence of high XRCC1 expression levels in somatic tumors is associated with the development of aggressive breast cancers and poor survival.11,26 Our results showed that the XRCC1 expression was correlated with TMB, MSI, and MMR, and the frequency of mutant XRCC1 was significantly higher in the precancerous lesions, suggesting that impairment of other less-understood DNA repair factors might exert an influence on the progression of carcinogenesis and immune cell infiltration.
Many studies have reported that PD-L1 and PD-1 are expressed in advanced CRC cases and are associated with the density of CD8+ T cells, indicating an adaptive immune resistance mechanism in CRC.27 The abundance of TILs is also associated with specific molecular features of CRC, including MSI-high.28 Rubio et al29 analyzed the TIL density in colorectal conventional adenomas and reported that the upregulated expression of CD3-T and MHC class II molecules in the IELs of neoplastic colorectal lesions indicated that they were activated and cytotoxic. Acosta-Gonzalez et al10 reported that an increased number of IELs and PD-1/PD-L1 expression correlate with the sequential progression of sessile serrated adenomas, through the development of cytologic dysplasia up to CRC and an MSI-H status. Additionally, a recent study evaluated XRCC1 and immune checkpoint expression levels via immunohistochemistry in breast cancer, and found that the interplay between XRCC1, CD8, PD-L1, and PD-1 could promote the development of aggressive tumor phenotypes. XRCC1-directed personalization of immune checkpoint inhibitor therapy may thus be feasible in breast cancer.11
Our results showed that tumor XRCC1 expression was positively correlated with IEL density and PD-1/PD-L1 expression, which all increase as the lesions progress through the sequence of adenoma to carcinoma. We also found a strong correlation of increased FOXP3+ expression with XRCC1 mutant status. FOXP3 might be deemed an independent factor in the carcinogenesis, since high FOXP3 expression was a significant factor in CRCs in multiple analyses based on XRCC1 mutation status. Interestingly, we found that XRCC1 mutation could be observed prior to the development of the immune TME. Thus, the infiltration of lymphocytes and upregulation of PD-1/PD-L1 expression in precancerous lesions may not be solely dependent on the generation of immunogenic neoantigens, but the occurrence may be dependent on earlier molecular mechanisms that remain undefined.
Conclusion
In conclusion, our results show that increased IEL density and PD-1/PD-L1 expression may correlate with the cytological dysplasia progression from LGD to HGD to CRC, and may specifically correlate with the XRCC1 expression and mutation status. Our findings support a stepwise and sequential progression of dysplasia-carcinoma of adenoma to carcinoma, and an underlying XRCC1 hypermutated phenotype-based mechanism of lesions, which collectively stimulate a host immune response characterized by rapid lymphocyte infiltration offset by the upregulation of immune checkpoint expression in tumor cells.
Funding Statement
This study was supported by Zhejiang Medicine Key Scientific and Technology (project no: 2018258924), Zhejiang Medicine Scientific and Technology (project no. 2019RC094); Natural Science Foundation (no. 82001173), Natural Science Foundation of Zhejiang Province (no. LQ20H160061), and Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (nos. 2021443144, 2020ky034).
Abbreviations
CRC, colorectal cancer; MSI, microsatellite instability; LGD, low-grade dysplasia; HGD, high-grade dysplasia; TME, tumor microenvironment; Tregs, regulatory T cells; TILs, tumor-infiltrating lymphocytes; XRCC1, X-ray repair cross-complementary 1; TCGA, The Cancer Genome Atlas; COAD, colon adenocarcinoma; HP, hyperplastic polyps; IELs, intraepithelial lymphocytes; MMR, mismatch repair system.
Ethics Approval
This retrospective study was performed in accordance with the Declaration of Helsinki. Approval was obtained from the Institutional Review Board of Zhejiang Provincial People’s Hospital (protocol 2021QT329). Informed consent was waived due to this study’s retrospective nature and the anonymized processing of patient data.
Author Contributions
All authors made a significant contribution to the work reported, whether in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all mentioned areas. All authors participated in drafting, revision, or critical review of the article, provided final approval of the version to be published, have agreed upon the journal to which the article has been submitted, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Poturnajova M, Furielova T, Balintova S, Schmidtova S, Kucerova L, Matuskova M. Molecular features and gene expression signature of metastatic colorectal cancer. Oncol Rep. 2021;45(4). doi: 10.3892/or.2021.7961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0 [DOI] [PubMed] [Google Scholar]
- 3.Lynch HT, Cristofaro G, Rozen P, et al. History of the international collaborative group on hereditary non polyposis colorectal cancer. Fam Cancer. 2003;2(Suppl 1):3–5. doi: 10.1023/a:1025001714023 [DOI] [PubMed] [Google Scholar]
- 4.Gryfe R, Kim H, Hsieh ET, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342(2):69–77. doi: 10.1056/NEJM200001133420201 [DOI] [PubMed] [Google Scholar]
- 5.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosman F, Yan P. Molecular pathology of colorectal cancer. Pol J Pathol. 2014;65(4):257–266. doi: 10.5114/pjp.2014.48094 [DOI] [PubMed] [Google Scholar]
- 7.Di J, Liu M, Fan Y, et al. Phenotype molding of T cells in colorectal cancer by single-cell analysis. Int J Cancer. 2020;146(8):2281–2295. doi: 10.1002/ijc.32856 [DOI] [PubMed] [Google Scholar]
- 8.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611 [DOI] [PubMed] [Google Scholar]
- 9.Rau TT, Atreya R, Aust D, et al. Inflammatory response in serrated precursor lesions of the colon classified according to WHO entities, clinical parameters and phenotype-genotype correlation. J Pathol Clin Res. 2016;2(2):113–124. doi: 10.1002/cjp2.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acosta-Gonzalez G, Ouseph M, Lombardo K, Lu S, Glickman J, Resnick MB. Immune environment in serrated lesions of the colon: intraepithelial lymphocyte density, PD-1, and PD-L1 expression correlate with serrated neoplasia pathway progression. Hum Pathol. 2019;83:115–123. doi: 10.1016/j.humpath.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green AR, Aleskandarany MA, Ali R, et al. Clinical impact of tumor DNA repair expression and T-cell infiltration in breast cancers. Cancer Immunol Res. 2017;5(4):292–299. doi: 10.1158/2326-6066.CIR-16-0195 [DOI] [PubMed] [Google Scholar]
- 12.Menoni H, Wienholz F, Theil AF, et al. The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. 2018;46(15):7747–7756. doi: 10.1093/nar/gky579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moller K, Blessin NC, Hoflmayer D, et al. High density of cytotoxic T-lymphocytes is linked to tumoral PD-L1 expression regardless of the mismatch repair status in colorectal cancer. Acta Oncol. 2021;60(9):1210–1217. doi: 10.1080/0284186X.2021.1933585 [DOI] [PubMed] [Google Scholar]
- 14.Michel M, Kaps L, Maderer A, Galle PR, Moehler M. The role of p53 dysfunction in colorectal cancer and its implication for therapy. Cancers. 2021;13(10):2296. doi: 10.3390/cancers13102296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bui VM, Mettling C, Jou J, Sun HS. Genomic amplification of chromosome 20q13.33 is the early biomarker for the development of sporadic colorectal carcinoma. BMC Med Genomics. 2020;13(Suppl 10):149. doi: 10.1186/s12920-020-00776-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HC, Roh SA, Ga IH, Kim JS, Yu CS, Kim JC. CpG island methylation as an early event during adenoma progression in carcinogenesis of sporadic colorectal cancer. J Gastroenterol Hepatol. 2005;20(12):1920–1926. doi: 10.1111/j.1440-1746.2005.03943.x [DOI] [PubMed] [Google Scholar]
- 17.Ho V, Ashbury JE, Taylor S, Vanner S, King WD. Genetic and epigenetic variation in the DNMT3B and MTHFR genes and colorectal adenoma risk. Environ Mol Mutagen. 2016;57(4):261–268. doi: 10.1002/em.22010 [DOI] [PubMed] [Google Scholar]
- 18.Andersen V, Vogel LK, Kopp TI, et al. High ABCC2 and low ABCG2 gene expression are early events in the colorectal adenoma-carcinoma sequence. PLoS One. 2015;10(3):e0119255. doi: 10.1371/journal.pone.0119255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhang W. Effects of XRCC1 Arg399Gln gene polymorphism on platinum-based chemotherapy sensitivity and clinical prognosis in patients with colorectal cancer: a meta-analysis. Zhejiang Med. 2020;14. doi: 10.12056/j.issn.1006-2785.2020.42.14.2019-3737 [DOI] [Google Scholar]
- 20.Whitehouse CJ, Taylor RM, Thistlethwaite A, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104(1):107–117. doi: 10.1016/s0092-8674(01)00195-7 [DOI] [PubMed] [Google Scholar]
- 21.Gong L, Luo M, Sun R, Qiu L, Chen C, Luo Z. Significant association between XRCC1 expression and its rs25487 polymorphism and radiotherapy-related cancer prognosis. Front Oncol. 2021;11:654784. doi: 10.3389/fonc.2021.654784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.IJspeert JE, Vermeulen L, Meijer GA, Dekker E. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol. 2015;12(7):401–409. doi: 10.1038/nrgastro.2015.73 [DOI] [PubMed] [Google Scholar]
- 23.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis JA, Reyes-Uribe L, Chang K, Lipkin SM, Vilar E. Immune activation in mismatch repair-deficient carcinogenesis: more than just mutational rate. Clin Cancer Res. 2020;26(1):11–17. doi: 10.1158/1078-0432.CCR-18-0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sultana R, Abdel-Fatah T, Abbotts R, et al. Targeting XRCC1 deficiency in breast cancer for personalized therapy. Cancer Res. 2013;73(5):1621–1634. doi: 10.1158/0008-5472.CAN-12-2929 [DOI] [PubMed] [Google Scholar]
- 27.Mei Z, Liu Y, Liu C, et al. Tumour-infiltrating inflammation and prognosis in colorectal cancer: systematic review and meta-analysis. Br J Cancer. 2014;110(6):1595–1605. doi: 10.1038/bjc.2014.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222(4):350–366. doi: 10.1002/path.2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubio CA, Jacobsson B, Castanos-Velez E. Cytotoxic intraepithelial lymphocytes in colorectal polyps and carcinomas. Anticancer Res. 1999;19(4B):3221–3227. [PubMed] [Google Scholar]