Abstract
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is unprecedented in human history. As a major structural protein, nucleocapsid protein (NPro) is critical to the replication of SARS-CoV-2. In this work, 17 NPro-targeting phenanthridine derivatives were rationally designed and synthesized, based on the crystal structure of NPro. Most of these compounds can interact with SARS-CoV-2 NPro tightly and inhibit the replication of SARS-CoV-2 in vitro. Compounds 12 and 16 exhibited the most potent anti-viral activities with 50% effective concentration values of 3.69 and 2.18 μM, respectively. Furthermore, site-directed mutagenesis of NPro and Surface Plasmon Resonance (SPR) assays revealed that 12 and 16 target N-terminal domain (NTD) of NPro by binding to Tyr109. This work found two potent anti-SARS-CoV-2 bioactive compounds and also indicated that SARS-CoV-2 NPro-NTD can be a target for new anti-virus agents.
Keywords: Phenanthridine, Nucleocapsid protein, SARS-CoV-2, N-terminal domain
Graphical abstract
Abbreviations
- COVID-19
Corona Virus Disease 2019
- SARS-COV-2
severe acute respiratory syndrome coronavirus 2
- HCoV-OC43
human coronavirus strains OC43
- MERS-CoV
Middle East respiratory syndrome coronavirus
- SARS-CoV
severe acute respiratory syndrome coronavirus
- MHV
mouse hepatitis virus
- IBV
avain infectious bronchitis virus
- NPro
nucleocapsid protein
- RNP
ribonucleoprotein
- NTD
N-terminal domain
- CTD
C-terminal domain
- N-NTD
N-terminal domain of nucleocapsid protein
- CADD
computer-aided drug design
- SPR
surface plasmon resonance
- KD
equilibrium dissociation constant
- CCK8
Cell Counting Kit-8
- CC50
half cytotoxic concentration
- EC50
half effective concentration
- RT-qPCR
quantitative reverse transcription PCR
- MOI
multiplicity of infection
- DMSO
dimethyl sulfoxide
- PBS
phosphate buffer saline
- RU
response units
- EDC
N-Ethyl-N’-(3-dimethylaminopropyl) carbodiimide
- NHS
N-hydroxysuccinimide
- DMF
N,N-dimethylformamide
- THF
tetrahydrofuran
- DPPA
diphenyl azidophosphate
1. Introduction
The current COVID-19 pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) of the betacoronavirus family [1,2]. Thanks to the unremitting efforts of the vast number of scientists, several vaccines were invented and gradually controlled the spread of the epidemic [3,4]. However, mutations in spike protein of SARS-CoV-2 render it a global healthcare challenge [5]. As a result, agents against SARS-CoV-2 infection are urgently needed for the clinical treatment of COVID-19.
Previous works have identified that in some coronavirues, such as HCoV-OC43, MERS-CoV and SARS-CoV, nucleocapsid protein (NPro) is a major structural protein that serves multiple purposes [6]. Its primary functions are to recognize and wrap viral RNA into ribonucleoprotein (RNP), which insulates the viral genome from external environment and is essential to maintain the RNA in an ordered conformation for replication and transcription. The common domain architectures of coronavirus NPro consists of two highly conserved parts, a N-terminal domain (NTD) and a C-terminal dimerization domain (CTD), connected with a disordered central Ser/Arg-rich linker [[7], [8], [9]]. In some coronavirues, such as MHV, IBV and SARS-CoV, the NTD of NPro (NPro-NTD) has been found to associate with the 3’ end of the viral RNA genome, possibly through electrostatic interactions, and some identified critical residues in the NTD of coronavirus NPro for RNA binding could be a potential cross-strain target for drug development [[10], [11], [12]].
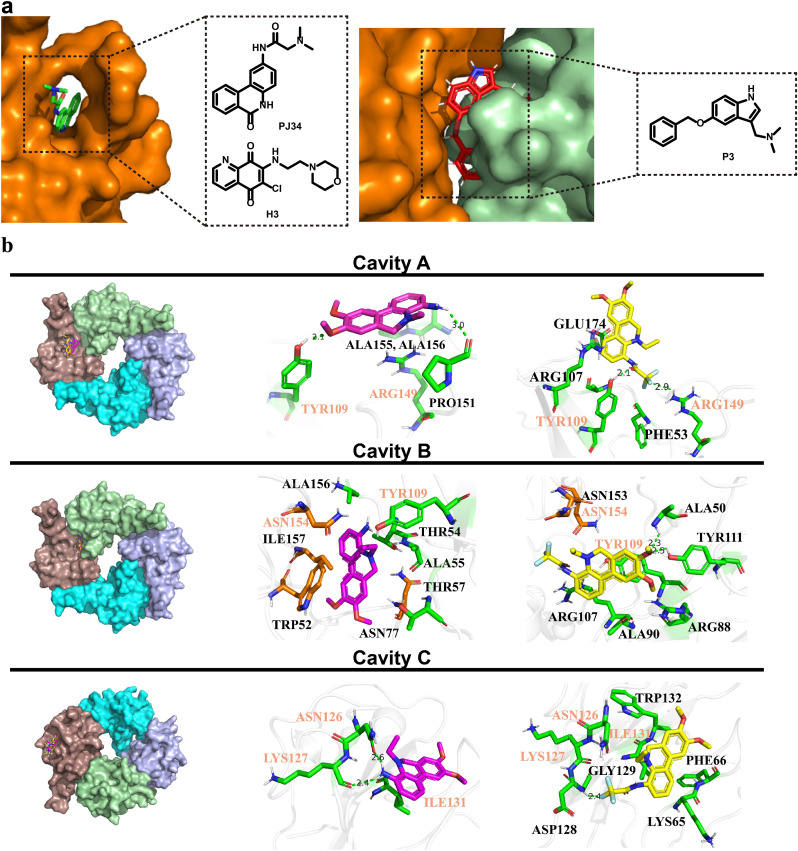
Due to the important role that NPro-NTD plays in the process of virus replication, it could be expected that small active molecules binding with NPro-NTD would interfere with the combination of SARS-CoV-2 NPro and viral RNA genome to reduce virus replication. Previous research on HCoV-OC43 NPro-NTD reported a phenanthridine competitive NPro inhibitor (Fig. 1 a) that efficiently suppressed the HCoV-OC43. This aroused our interest since we have conducted a lot of research on phenanthridine compounds previously and collected many structure-activity relationship data of phenanthridine derivatives [[13], [14], [15]]. With the purpose to explore new NPro-NTD targeted bioactive compounds for inhibiting SARS-CoV-2, a series of phenanthridine derivatives was rationally designed with computer-aided drug design (CADD) and synthesized. The in vitro antiviral activities of these compounds were evaluated and the possible mechanism of inhibition was verified by Surface Plasmon Resonance (SPR) and site-directed mutagenesis on SARS-CoV-2 NPro.
Fig. 1.
Molecular Docking analysis of phenanthridine and SARS-CoV-2 NPro. a. X-ray crystallographic structure of HCoV-OC43 and MERS-CoV with representative inhibitors (PJ34, H3 and P3) targetting the CoVs N-NTD [13,20,23]; b. Molecular docking of compound 12 (purple sticks) and 16 (yellow sticks) bound to the crystal structure of the NTD of SARS-Cov-2 NPro (PDB 6VYO) indicated are three predicted pockets.
2. Results and discussion
2.1. Design of potential SARS-CoV-2 NPro inhibitors
The design of SARS-CoV-2 NPro inhibitors was based on the reported crystal structure of SARS-CoV-2 NPro (PDB:6VYO) [16]. The molecular docking of phenanthridine core X0 to SARS-CoV-2 NPro-NTD identified that phenanthridine core can form a variety of interactions with the residues which imply that phenanthridine X0 can bind to the SARS-CoV-2 NPro-NTD if groups that can be hydrogen bonded with receptor are added to X0 (Fig. S1). This analysis suggested that hydrogen bond acceptors can be introduced at C-3, C-4, N-5 and C-6 positions in X0. These positions could be substituted with amido groups as hydrogen bond acceptors or further reduced in size amino groups. In addition, hydrogen bonds acceptors at C-8 and C-9 would be also beneficial to the binding energy. These two positions could be substituted with oxygen-containing functional groups, whose oxygen atom might be a hydrogen bond acceptor (Fig. S1).
2.2. Biomolecular simulations
According to aforementioned analyses, six model molecules (M1-6) were designed to evaluate the effects on binding between the substituent groups of phenanthridine and the target protein (Fig. S1). By analyzing the binding mode of the six model phenanthridines (M1-6) and SARS-CoV-2 NPro-NTD, it can be indicated that six compounds showed similar binding modes. The amino acid composition of this binding site includes Thr54, Ala55, Thr57, Arg107, Tyr109, Arg149, Pro151, Ala155, Ala156 and Glu174. The phenolic hydroxyl from the Tyr109 side chain is 2.0 Å from the methoxybenzene group on the M1, indicating that a hydrogen bond may form. The methoxy group moiety of M3 at C-9 also forms a hydrogen bond with Arg149 with a distance of 2.2 Å. Substituted with amide groups contribute to hydrogen bond formation, for instance, M2 is amidated at N-5 to form a hydrogen bond with Arg149. And M4 with an amide group substituted at C-4 also has a hydrogen bond with Tyr109. The phenanthridine aromatic ring on the M1-M6 participates in pi-Cation or pi-Anion interaction with the Arg107, Arg149 or Glu174. Methyl or ethyl substituted in M1-M6 formed lipophilic interactions with Ala55 and Pro151. Residues Thr54, Ala55, Pro151, Ala155 and Ala156 involved in the forming of hydrophobic interaction networks both in M1-M6. It can be concluded that a) the phenanthridine core can form a variety of interactions with the target protein; b) the introduction of methoxy groups (at C-8 and C-9) and C-4 side chain could be of advantage to bind; c) the alkyl substituted at different positions is favorable for the formation of lipophilic interactions with target protein.
2.3. Chemistry
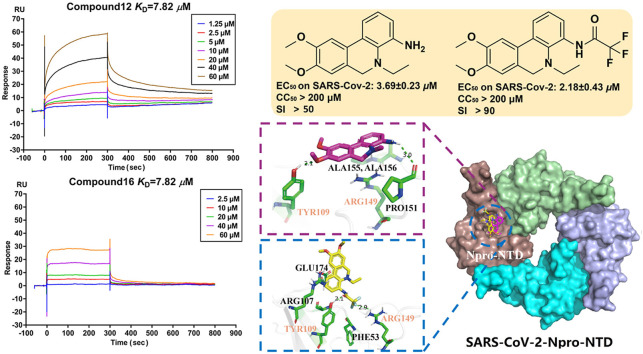
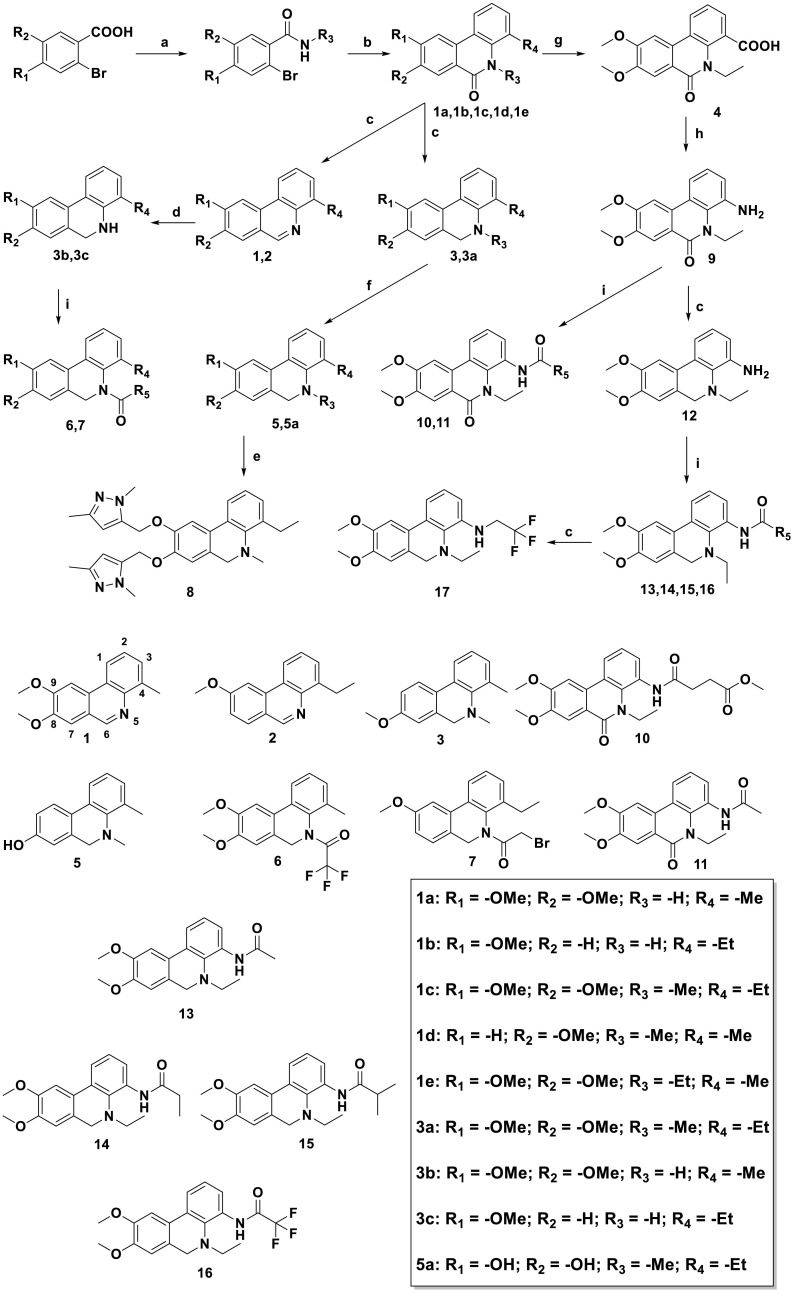
Therefore, 17 phenanthridine derivatives were designed and synthesized (Fig. 1b, Scheme 1 ). 2-bromo-benzoic acids analogues firstly condensed with various amines and then coupled with 2-methyliodobenzene or 2-ethyliodobenzene by palladium catalyzing to yield 1a-1e, followed by the reduction of the C-6 carbonyl to yield 1, 2, 3 or 3a. Compound 1 and 2 were further reacted with sodium borohydride to yield 3b and 3c. By reacting with commercially available anhydride or acyl halide reagents, 3b and 3c were converted to 6 and 7. Compound 5 or 5a were respectively obtained by the deprotection of 3 or 3a in the presence of boron tribromide. 5a was nucleophilic substituted with pyrazole derivative to obtain compound 8. In order to prepare C-4-modified molecules, compound 1e was oxidized to afford compound 4, then converted to compound 9 by curtius rearrangement. 9 condensed with various acylation reagents to get compounds 10 and 11. In addition, dihydrophenanthridine 12 was converted from 9 in the presence of borane. Following, analogues of compound 12 bearing amide functionalities at C-4 position (compounds 13-16) were synthesized. Finally, compound 17 was obtained via reduction of 16. All of these compounds were modified at C-4 to form more hydrogen bond interactions or N-5 acylated or alkylated to tertiary amines instead of being hydrogen bond donors. Moreover, introducing methoxy groups at C-8 and C-9 positions allows more opportunities to generate interactions. Fig. 1b reveals the detailed interactions with compound 12 and 16. In pocket A, the phenolic hydroxyl from the Tyr109 side chain is 2.1 Å from the methoxybenzene group on the compound 12, indicating that a hydrogen bond formed between Tyr109 and compound 12. The hydrogen bonds also formed between the backbone carbonyl group of Pro151 with amino of compound 12. Compound 16 with an amide group substituted at C-4 formed a hydrogen bond with Tyr109. In addition, The Fluorine atom in the 16 also formed a single hydrogen bond with the Arg149 side chain of the target protein with a distance of 2.9 Å. The phenanthridine core of the 12 and 16 participates in pi-Cation or pi-Anion interaction with the Arg107, Arg149 or Glu174. And Residues Thr54, Ala55, Ala155 and Ala156 formed hydrophobic pockets both in 12 and 16. Pocket B is located between two monomers, most interactions were hydrophobic contacts. Compound 12 formed lipophilic interactions with residues Thr54, Tyr109 or Ala156 in the first monomer, and formed similar interactions with Trp52, Thr54 and Ile157 in the second monomer. Besides, the Asn 154 close to the amino group of 12 may form electrostatic interactions. Methoxy group of 16 has a 2.3 Å or 2.5 Å distance from Ala50 or Tyr111 in the first monomer, indicating that hydrogen bonds may be formed between them. Compound 16 also has lipophilic interactions with Ala90, Arg107 and Tyr109 in the first monomer. Furthermore, the Fluorine atom in the 16 might form Halogen interactions with Asn153 and Asn154 of the second monomer. The docking poses of 12 and 16 in Pocket C were almost superimposed. Both of them have hydrogen bonds or lipophilic interactions with Asn126, Lys127 and Ile131.
Scheme 1.
Synthesis of Phenanthridine Derivatives. Reagents and conditions:(a) SOCl2, CH2Cl2, 50 °C, 5 h, then R3-NH2, H2O, 0 °C, 2 h yield 80–90%; (b) Iodobenzene-R4, Pd(OAc)2, Norbornene, TFP, K2CO3, DMF, 105 °C, 8 h yield 75–85%; (c) BH3-THF, THF, 60 °C, 6 h yield 65–80%; (d) NaBH4, MeOH, r.t., 2 h yield 55–60%; (e) K2CO3, 5-(bromomethyl)-1,3-dimethyl-1H-pyrazole, CH3CN, 80 °C, 6 h. yield 80%; (f) BBr3, CH2Cl2, -78 °C, 4 h yield 65–80%; (g) KMnO4, py, 80 °C, 6 h yield 80%; (h) TEA, DPPA, THF, 60 °C, 6 h yield 70%; (i) NaHCO3, R5COX, CH2Cl2, r.t., 2 h yield 70–85%.
2.4. Experimental binding of the 17 synthesized phenanthridine compounds to NPro
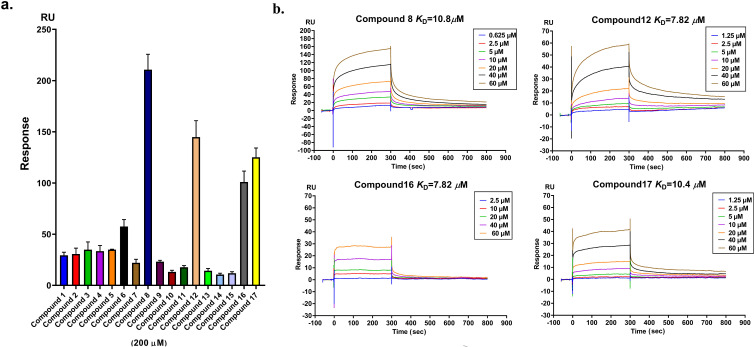
The binding activities of the compounds to SARS-CoV-2 NPro were determined by a SPR assay. The compounds with the top 4 signal responses were selected for affinity determination (Fig. 2 a, Fig. S2). They were subsequently assayed in a dose-response experiment to determine the binding affinity. The compounds with different concentrations (0.625–60 μM) were flowed over the surface of the chip immobilized with NPro. Four compounds (8, 12, 16 and 17) bind to NPro with low KD values of 10.8 μM, 7.82 μM, 7.82 μM, and 10.4 μM, respectively (Fig. 2b), suggesting that they are potent NPro ligands.
Fig. 2.
SPR assay of synthesized compounds. a. Binding level screen of single concentration SPR assay of all synthesized compounds at 200 μM; b. The sensorgrams for compounds 8, 12, 16, and 17 bound to SARS-Cov-2 Npro-His. Compounds with increasing concentrations (0.625–60 μM) were injected over the surface. Contact time and dissociation time were 120 and 180 s, respectively. Data was analyzed with Biacore S200 evaluation software (version 1.0) and automatically fitted to the 1:1 binding model.
2.5. Anti-SARS-CoV-2 activity in vitro
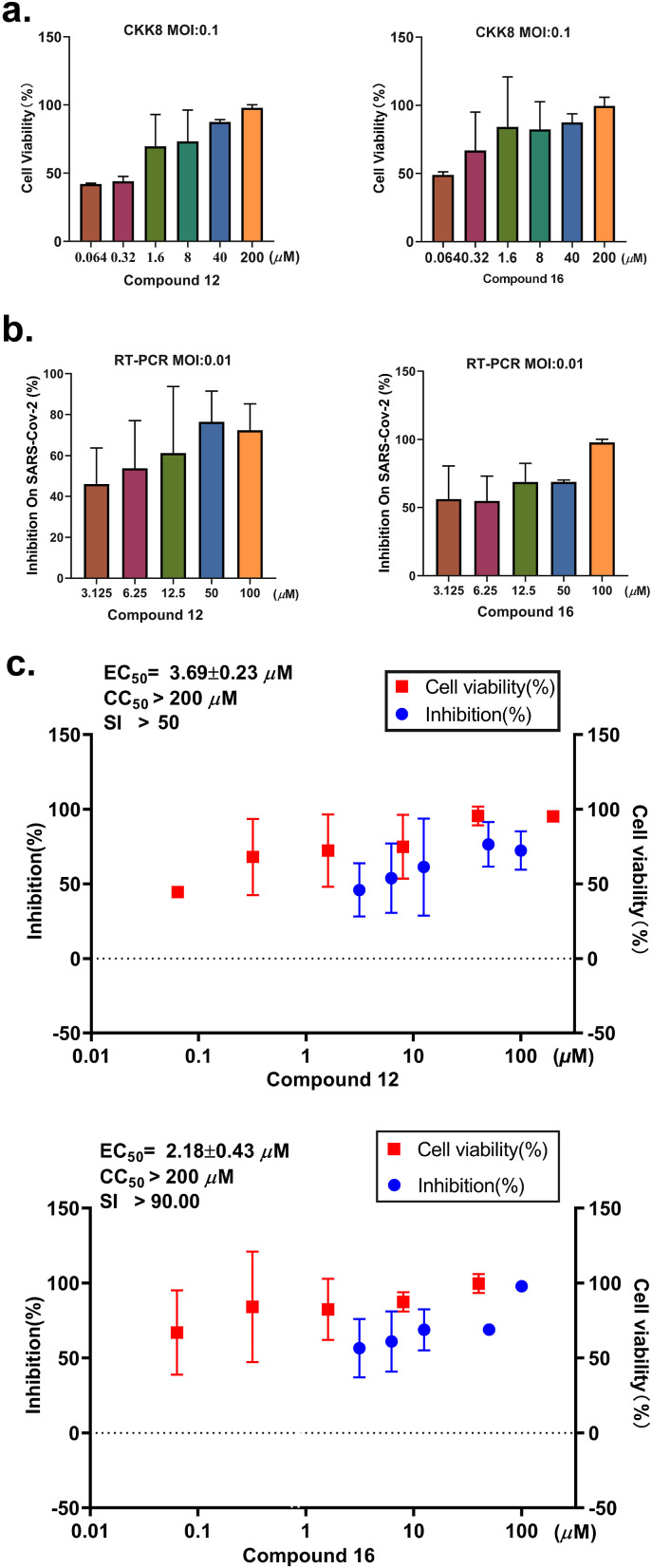
Compounds 12 and 16 with K D < 10 μM in the SPR assay were chosen to examine their cytotoxicity and cellular antiviral activity. First, the cytotoxicity of these compounds was evaluated using the Cell Counting Kit-8 (CCK8) assay and all compounds showed no cytotoxicity (CC50 > 200 μM) in the Vero E6 cell line tested. Next, the cellular antiviral activity was examined by a cell protection assay. In this assay, the viability of SARS-CoV-2 infected Vero E6 cells with or without compounds treatment was assessed using CCK8. Compounds 12 and 16 showed nearly 100% protection on cells from death mainly caused by viral infection at the high concentration of 200 μM (Fig. 3 a, Table S2) and exhibited dose-dependently protective effects on cells with EC50 values of 42.04 μM and 43.41 μM, respectively. Of note, cell viability assay is subject to some interference such as cytotoxicity of compounds. To further corroborate the antiviral potency of compounds 12 and 16, quantitative reverse transcription polymerase chain reaction (RT-qPCR) was conducted in Vero E6 cell in different concentrations (Table S3) and the results revealed that compounds 12 and 16 could clearly inhibit SARS-CoV-2 virus replication in Vero E6 cells with EC50 values of 3.69 and 2.18 μM, respectively (Fig. 3b and c, Table S4) while the positive control remdesivir showed EC50 values of 1.21 μM. These results indicated that compound 12 and 16 inhibit the replication of SARS-Cov-2 in vitro.
Fig. 3.
Antiviral activity of compound 12 and 16 against SARS-CoV-2 in cell-based assays. a. Vero E6 cells were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.1 and treated with different concentrations of test compounds (0.064 μM, 0.32 μM, 1.6 μM, 8 μM, 40 μM and 200 μM). At 3 dpi the cytopathic effect caused by SARS-CoV-2 infection was quantitatively analyzed using CCK8, according to the manufacturer's protocol. Data are means ± SD; n = 3 biological replicates. b. Vero E6 cells were infected with SARS-CoV-2 at an MOI of 0.01 and treated with different concentrations of test compounds (3.125 μM, 6.25 μM, 12.5 μM, 25 μM, 50 μM, and 100 μM). At 2 dpi, viral RNA copies (per ml) were quantified from cell culture supernatants by RT-qPCR. Data are means ± SD; n = 3 biological replicates. c. The EC50, CC50 and SI of compounds 12 and 16.
2.6. Confirmation of the target protein of compounds 12 and 16
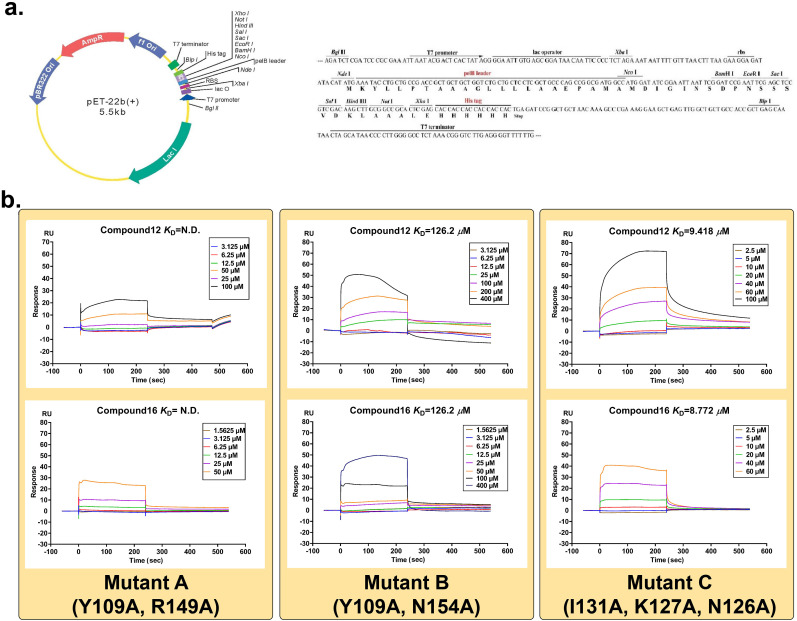
Following, directed mutagenesis was employed to confirm the anti-SARS-CoV-2 target protein of phenanthridine derivatives. The forementioned SPR assay has proved that compound 12 and 16 interact with NPro. However, the cavity that active molecules bind to NPro is still uncertain. After analyzing the crystal structure of NPro and employing the AutoDock vina program [18], three cavities A, B and C on NPro-NTD were regarded as potential binding sites. For each pocket, about 20 conformers of compound 12 or 16 outputted from vina have been analyzed and the binding pose with lower estimated free energy of binding was selected. Three pairs of residues (Tyr109 and Arg149; Tyr109 and Asn154; Ile131, Lys127 and Asn126) separately in cavities A, B and C, could make contacts with compounds 12 and 16 through electrostatic or hydrophobic interactions. To verify the importance of these residues, the site-directed mutagenesis has been conducted to generate mutant A (Y109A, R149A), mutant B (Y109A, N154A) and mutant C (I131A, K127A and N126A) (Fig. 4 a, Fig. S3). Then the affinity between compounds and mutants was evaluated by SPR. As the model predicted, two mutant proteins (mutant A and mutant B) showed significantly reduced interactions with compounds 12 and 16. As for mutant A, the Response Units (RU) value was too low to calculate K D value of compounds. For mutant B, the K D value of compounds 12 and 16 was 126.2 μM and 275.4 μM, respectively. On the other hand, 12 and 16 could still tightly bind mutant C protein with K D values 9.42 μM and 8.77 μM, respectively, which were similar to those of wild-type protein (7.82 μM and 7.82 μM) (Fig. 4b, Fig. S4). By analyzing the residues in cavities, it can be observed that Tyr109 exists in both cavity A and B, implying that Tyr109 is the critical residue for the interactions of compounds and NPro-NTD. These results suggested that compounds target the NTD of SARS-Cov-2 NPro by mainly binding with Tyr109, and thus inhibit the replication of SARS-CoV-2.
Fig. 4.
Compound 12 and 16 target SARS-Cov-2 NPro by binding to NTD. a. Site-directed mutagenesis were used to construct mutant A (Y109A, R149A), B (Y109A, N154A) and C (I131A, K127A, N126A). b. SPR analysis showed that mutant A and mutant B within the NTD showed significant decrease in combinations with compounds 12 and 16 at the concentrations from 1.5625 μM −400μM by comparing to their effects in wild-type NPro.
3. Conclusion
In this work, novel phenanthridine anti-SARS-CoV-2 agents were developed and shown to be potent anti-SARS-CoV-2 agents by combining with NPro. Mechanism studies revealed that NTD of NPro is involved in replication of SARS-CoV-2 and phenanthridine compounds could bind this cavity to inhibiting the replication of SARS-CoV-2 replication. In vitro assays indicated that the phenanthridine derivatives may offer a new strategy for anti-SARS-CoV-2 chemotherapeutic agents as supplements to vaccine therapy. It is also worth mentioning that this work proved that NPro could be a potential anti-viral target and further imply that NTD could be a critical active domain of NPro.
4. Experimental section
4.1. General methods of chemistry
Unless otherwise indicated, all commercially available solvents and reagents were purchased directly from commercial suppliers and used without further purification. The 1H NMR and 13C NMR spectra were recorded on the Bruker 500 and 600 MHz spectrometers with TMS as the internal standard. HRESIMS were acquired by Agilent 6500 Q-TOF mass spectrometer. Column chromatography was performed on silica gel (60–80 mesh, 200–300 mesh, 300–400 mesh, Qingdao Haiyang Chemical Co. Ltd., Qingdao, China). Pre-coated silica gel 60 GF254 (Merck, Darmstadt, Germany) was used for TLC analyses. The purities of all compounds used in biological assays exceeded 95%, as determined by HPLC. HPLC Conditions: Agilent 1200 Series; Column: YMC-Pack ODS-A (S-5 μm, 12 nm; 250 × 4.6 mml. D; AA12S05-2546WT; Ser. No.110YA60116); Eluent: MeCN:H2O, Flow rate: 1.0 mL/min; Detection: UV 254 nm; The synthesized procedures and characterization for intermediates were available in Supporting Information.
4.2. Design of target compounds
Molecular Operating Environment 2019 (MOE) software was employed to structure-based drug design. Initially, “Ligand R-Vectors” was employed to explore where ligand substituents can be introduced. The green arrows from the X0 heavy atoms toward their hydrogens showed the positions where the hydrogen atom could be replaced by a substituent no bigger than a methyl group in order not to introduce energy consuming van der Waals repulsions. Exit vectors substituted at C-8 and C-9 of X0 would enhance the interaction. Then “Electrostatic Maps” were used to calculate the favorable locations of the neutral, positive, and negative features of the binding site, thus predicting suitable substitutions in X0.
4.3. Synthesis of target compounds
Syntheses of compounds 1–3. To a stirred solution of 1a, 1b or 1d (1 mmol) in THF (2 mL) was added 1 M BH3-THF (2 mL, 2 mmol) dropwise. After a 6 h stirring in a 60 °C oil bath and then quenched using H2O (5 mL), phases were separated and aqueous phase was extracted with EtOAc (2 × 20 mL). The combined organic phases were washed with brine, dried over MgSO4, filtered, and concentrated in vacuum. Crude product was purified by flash chromatography on silica gel to give 1–3 as colorless oil. (yield 65%–80%).
8,9-Dimethoxy-4-methylphenanthridine (1).1H NMR (500 MHz, CDCl3) δ H 9.19 (s, 1H), 8.45–8.14 (m, 1H), 7.90 (s, 1H), 7.66–7.45 (m, 2H), 7.36 (s, 1H), 4.14 (s, 3H), 4.08 (s, 3H), 2.87 (s, 3H). 13C NMR (125 MHz, CDCl3) δ C 152.9 (C), 150.4 (CH), 149.9 (C), 142.7 (C), 137.7 (C), 128.6 (CH), 128.6 (CH), 126.2 (C), 123.7 (C), 121.6 (C), 119.7 (CH), 107.7 (CH), 102.0 (CH), 56.2 (CH3), 56.1 (CH3), 18.8 (CH3). HRMS/EI: calcd for C16H16NO2 + [M+H]+ 254.1176, found 254.1169.
4-Ethyl-9-methoxyphenanthridine (2).1H NMR (500 MHz, CDCl3) δ H 9.20 (s, 1H), 8.38 (dd, J = 7.8, 1.8 Hz, 1H), 7.97 (d, J = 8.7 Hz, 1H), 7.92 (d, J = 2.2 Hz, 1H), 7.76–7.48 (m, 2H), 7.30 (dd, J = 8.7, 2.4 Hz, 1H), 4.05 (s, 3H), 3.36 (q, J = 7.5 Hz, 2H), 1.42 (dd, J = 9.1, 6.0 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 161.6 (C), 151.4 (CH), 143.6 (C), 135.0 (C), 130.5 (CH), 128.0 (CH), 126.3 (CH), 123.8 (C), 121.4 (C), 120.1 (CH), 117.8 (CH), 102.7 (CH), 55.6 (CH3), 25.2 (CH2), 15.4 (CH3). HRMS/EI: calcd for C16H16NO+ [M+H]+ 238.1226, found 238.1229.
8-Methoxy-4,5-dimethyl-5,6-dihydrophenanthridine (3).1H NMR (500 MHz, CDCl3) δ H 7.70 (d, J = 8.6 Hz, 1H), 7.61 (dd, J = 7.4, 1.7 Hz, 1H), 7.17–7.13 (m, 1H), 7.13–7.07 (m, 1H), 6.93 (dd, J = 8.5, 2.7 Hz, 1H), 6.81 (d, J = 2.7 Hz, 1H), 4.09 (s, 2H), 3.87 (s, 3H), 2.52 (s, 3H), 2.43 (s, 3H). 13C NMR (125 MHz, CDCl3) δ C 159.3 (C), 145.9 (C), 134.2 (C), 133.3 (C), 129.5 (CH), 129.0 (C), 125.2 (C), 124.3 (CH), 124.2 (CH), 120.9 (CH), 113.0 (CH), 112.0 (CH), 55.4 (CH2), 55.2 (CH3), 40.4 (CH3), 17.5 (CH3). HRMS/EI: calcd for C16H18NO+ [M+H]+ 240.1383, found 240.1384.
5-Ethyl-8,9-dimethoxy-6-oxo-5,6-dihydrophenanthridine-4-carboxylic acid (4). Compound 1e (298 mg, 1 mmol) was dissolved in Pyridine (10 mL). The mixture was heated to reflux and KMnO4 (1.2 g, 5 mmol) was added. The reaction mixture was heated for 6 h at 100 °C and the solution was filtered and washed several times with boiling water. The aqueous solution was made acidic and the product was extracted into EtOAc (20 mL). The organic phase was dried over Na2SO4 and the solvent removed in vacuo to provide the desired product as colorless solid. (250 mg, yield 80%). 1H NMR (500 MHz, MeOD) δH 8.45 (dd, J = 8.2, 1.4 Hz, 1H), 7.80 (d, J = 8.4 Hz, 2H), 7.65 (dd, J = 7.4, 1.5 Hz, 1H), 7.39–7.34 (m, 1H), 4.44 (q, J = 7.0 Hz, 2H), 4.05 (s, 3H), 3.97 (s, 3H), 1.26 (t, J = 7.0 Hz, 3H). 13C NMR (125 MHz, MeOD) δC 173.4 (C), 163.9 (C), 155.5 (C), 151.8 (C), 135.3 (C), 130.7 (CH), 129.9 (C), 126.7 (C), 126.4 (CH), 123.4 (CH), 122.9 (C), 120.3 (C), 109.4 (CH), 104.6 (CH), 56.8 (CH3), 56.5 (CH3), 43.7 (CH2), 13.9 (CH3). HRMS/EI: calcd for C18H16NO5 − [M-H]- 326.1034, found 326.1034.
4,5-Dimethyl-5,6-dihydrophenanthridin-8-ol (5). Compound 3 (239 mg, 1 mmol) was dissolved in CH2Cl2 (5 mL). The reaction solution was then cooled to −78 °C and BBr3 (0.4 mL, 4 mmol) was added dropwise. After a 6 h stirring at room temperature and then quenched using saturated aqueous NaHCO3 (10 mL), phases were separated and aqueous phase was extracted with CH2Cl2 (2 × 20 mL). The combined organic phases were washed with water, dried over MgSO4, filtered, and concentrated in vacuum. Crude product was purified by flash chromatography on silica gel to give 5 as yellow solid. (190 mg, yield 80%). 1H NMR (500 MHz, CDCl3) δ H 7.63 (d, J = 8.4 Hz, 1H), 7.58 (dt, J = 9.7, 4.8 Hz, 1H), 7.17–7.08 (m, 2H), 6.83 (dd, J = 8.4, 2.6 Hz, 1H), 6.72 (d, J = 2.5 Hz, 1H), 4.04 (s, 2H), 2.50 (s, 3H), 2.42 (s, 3H). 13C NMR (125 MHz, CDCl3) δ C 155.6 (C), 145.5 (C), 134.2 (C), 133.2 (C), 129.7 (CH), 129.1 (C), 125.1 (C), 124.6 (CH), 124.5 (CH), 121.0 (CH), 114.7 (CH), 113.7 (CH), 55.2 (CH2), 40.3 (CH3), 17.6 (CH3). HRMS/EI: calcd for C15H16NO+ [M+H]+ 226.1226, found 226.1229.
1-(8,9-Dimethoxy-4-methylphenanthridin-5(6H)-yl)-2,2,2-trifluoroethan-1-one (6). To a stirred solution of 3b (255 mg, 1 mmol) and NaHCO3 (168 mg, 2 mmol) in CH2Cl2 (5 mL) was added trifluoroacetic anhydride (1 mL) dropwise. After a 4 h stirring at room temperature and then quenched using H2O (5 mL), phases were separated and aqueous phase was extracted with CH2Cl2 (2 × 20 mL). The combined organic phases were washed with water, dried over MgSO4, filtered, and concentrated in vacuum. Crude product was purified by flash chromatography on silica gel to give 6 as colorless solid (280 mg, yield 80%). 1H NMR (500 MHz, CDCl3) δ H 7.61 (d, J = 7.7 Hz, 1H), 7.36 (t, J = 7.7 Hz, 1H), 7.32 (d, J = 3.6 Hz, 1H), 7.25 (d, J = 7.6 Hz, 1H), 6.82 (s, 1H), 4.98 (d, J = 15.6 Hz, 1H), 4.43 (d, J = 15.5 Hz, 1H), 3.99 (s, 3H), 3.96 (s, 3H), 2.28 (s, 3H). 13C NMR (125 MHz, CDCl3) δ C 155.0 (C, q, J = 36.3 Hz), 149.5 (C), 149.3 (C), 134.4 (C), 130.5 (C), 129.8 (CH), 127.8 (CH), 126.4 (C), 125.0 (C), 121.5 (CH), 117.7 (C), 115.4 (C), 108.3 (CH), 107.3 (CH), 56.2 (CH3), 56.2 (CH3), 48.7 (CH2), 18.6 (CH3). HRMS/EI: calcd for C18H17F3NO3 + [M+H]+ 352.1155, found 352.1158.
2-Bromo-1-(4-ethyl-9-methoxyphenanthridin-5(6H)-yl)ethan-1-one (7). By referencing the synthesis method of compound 6, compound 7 was conducted by amidating of compound 3c (239 mg, 1 mmol) at the presence of bromoacetyl bromide (1 mL) in a yield of 85%.1H NMR (500 MHz, CDCl3) δ H 7.62 (t, J = 7.5 Hz, 1H), 7.37 (t, J = 7.8 Hz, 1H), 7.33 (t, J = 3.6 Hz, 1H), 7.30 (d, J = 7.8 Hz, 1H), 7.23 (t, J = 7.1 Hz, 1H), 6.87–6.82 (m, 1H), 5.64 (d, J = 14.7 Hz, 1H), 3.99 (t, J = 14.7 Hz, 1H), 3.87 (s, 3H), 3.78 (d, J = 11.4 Hz, 1H), 3.68 (t, J = 11.1 Hz, 1H), 2.69–2.54 (m, 2H), 1.25 (t, J = 7.3, Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 167.1 (C), 159.5 (C), 139.0 (C), 133.4 (C), 131.9 (C), 128.9 (CH), 128.0 (CH), 127.7 (C), 127.4 (CH), 122.8 (CH), 113.2 (CH), 109.8 (CH), 55.5 (CH3), 46.8 (CH2), 27.2 (CH2), 24.5 (CH2), 15.1 (CH3). HRMS/EI: calcd for C18H19BrNO2 + [M+H]+ 360.0594, found 360.0592.
8,9-Bis((1,3-dimethyl-1H-pyrazol-5-yl)methoxy)-4-ethyl-5-methyl-5,6-dihydrophenanthridine (8). To a stirred solution of 5a (255 mg, 1 mmol) and K2CO3 (552 mg, 4 mmol) in CH3CN (5 mL) was added 5-bromomethyl-1,3-dimethyl-1H-pyrazole (472 mg, 2.5 mmol). After a 6 h stirring in a 80 °C oil bath, the solvent was removed by evaporation. The residue was added H2O (10 mL) and extracted with EtOAc (2 × 20 mL). The combined organic phases were washed with brine, dried over MgSO4, filtered, and concentrated in vacuum. Crude product was purified by flash chromatography on silica gel to give 8 as colorless solid (375 mg, 80%). 1H NMR (500 MHz, CDCl3) δ H 7.48 (t, J = 8.9 Hz, 1H), 7.36 (s, 1H), 7.26 (s, 2H), 7.21–7.12 (m, 2H), 6.84 (s, 1H), 6.05 (d, J = 2.3 Hz, 2H), 5.04 (s, 2H), 5.02 (s, 2H), 4.00 (s, 2H), 3.82 (s, 3H), 3.81 (s, 3H), 2.80 (q, J = 7.5 Hz, 2H), 2.47 (s, 3H), 2.24 (d, J = 1.8 Hz, 6H), 1.31 (t, J = 7.5 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 148.5 (C), 147.9 (C), 148.5 (C), 147.3 (C), 147.2 (C), 145.6 (C), 139.7 (C), 138.1 (C), 137.9 (C), 128.8 (C), 128.1 (CH), 127.3 (C), 126.9 (C), 124.7 (CH), 121.0 (CH), 114.2 (CH), 111.7 (CH), 106.9 (CH), 106.9 (CH), 62.6 (CH2), 62.2 (CH2), 54.8 (CH2), 41.4 (CH3), 36.4 (CH3), 36.3 (CH3), 23.2 (CH2), 14.9 (CH3), 13.4 (2CH3). HRMS/EI: calcd for C28H33N5O2 + [M+H]+ 472.2634, found 472.2635.
4-Amino-5-ethyl-8,9-dimethoxyphenanthridin-6(5H)-one (9). To a stirred solution of 4 (327 mg, 1 mmol) and Et3N (0.4 mL) in THF (5 mL) was added DPPA (0.5 mL, 2 mmol). After a 6 h stirring in a 60 °C oil bath, H2O (1 mL) was added and the reaction mixture was refluxed for 2 h. The solvent was removed in vacuo and the residue was treated with saturated aqueous K2CO3 (10 mL), diluted with H2O (20 mL), and extracted with EtOAc (2 × 20 mL). The combined organic phases were washed with water, dried over MgSO4, filtered, and concentrated in vacuum. Crude product was purified by flash chromatography on silica gel to give 1 as colorless solid (210 mg, 70%). 1H NMR (500 MHz, CDCl3) δ H 7.86 (s, 1H), 7.63–7.57 (m, 1H), 7.50 (d, J = 5.6 Hz, 1H), 7.13–7.08 (m, 1H), 6.84 (dd, J = 7.7, 1.3 Hz, 1H), 4.61–4.50 (m, 2H), 4.06 (s, 3H), 4.00 (s 3H), 1.47–1.42 (m, 3H). 13C NMR (125 MHz, CDCl3) δ C 164.1 (C), 153.2 (C), 149.7 (C), 136.3 (C), 129.4 (C), 128.8 (C), 123.4 (CH), 122.6 (C), 120.0 (C), 118.3 (CH), 114.0 (CH), 108.8 (CH), 103.1 (CH), 56.2 (CH3), 56.1 (CH3), 42.3 (CH2), 15.7 (CH3). HRMS/EI: calcd for C17H18N2O3Na+ [M+Na]+ 321.1210, found 321.1210.
4.3.1. Syntheses of compounds 10 and 11. By referencing the synthesis method of compound 6, compound 10 and 11 was conducted by amidating of compound 9 (1 mmol) at the presence of methyl-4-chloro-4-oxobutyrate or Ac2O (1 mL) in a yield of over 80%
Methyl-4-((5-ethyl-8,9-dimethoxy-6-oxo-5,6-dihydrophenanthridin-4-yl)amino) -4-oxobutanoate (10).1H NMR (500 MHz, CDCl3) δ H 7.93 (d, J = 7.8 Hz, 1H), 7.81 (s, 1H), 7.69 (s, 1H), 7.53 (d, J = 7.7 Hz, 1H), 7.40 (s, 1H), 4.38 (q, J = 6.9 Hz, 2H), 4.04 (s, 3H), 3.98 (s, 3H), 3.69 (s, 3H), 2.68 (dd, J = 9.8, 4.0 Hz, 2H), 2.62 (dd, J = 10.2, 4.4 Hz, 2H), 1.39 (t, J = 7.0 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ C 176.7 (C), 172.7 (C), 163.4 (C), 153.4 (C), 149.9 (C), 133.5 (C), 129.5 (CH), 128.2 (C), 125.1 (C), 122.9 (CH), 122.4 (C), 121.2 (CH), 119.3 (C), 108.6 (CH), 102.7 (CH), 56.2 (CH3), 56.1 (CH3), 52.0 (CH3), 43.0 (CH2), 31.5 (CH2), 28.7 (CH2), 15.2 (CH3). HRMS/EI: calcd for C22H25N2O6 + [M+H]+ 413.1707, found 413.1710.
N-(5-Ethyl-8,9-dimethoxy-6-oxo-5,6-dihydrophenanthridin-4-yl)acetamide (11).1H NMR (500 MHz, CDCl3) δ H 7.89 (d, J = 8.0 Hz, 1H), 7.63 (s, 1H), 7.58 (s, 1H), 7.53 (d, J = 7.5 Hz, 1H), 7.35 (s, 1H), 7.24 (t, J = 6.2 Hz, 1H), 4.31 (q, J = 6.9 Hz, 2H), 4.02 (s, 3H), 3.95 (s, 3H), 2.32 (s, 3H), 1.42 (t, J = 7.0 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 169.2 (C), 163.4 (C), 153.3 (C), 149.8 (C), 133.9 (C), 129.6 (CH), 128.1 (C), 125.2 (CH), 122.9 (C), 122.4 (CH), 121.2 (C), 119.2 (CH), 108.5 (CH), 102.6 (C), 56.1 (CH3), 56.1 (CH3), 43.4 (CH2), 24.0 (CH3), 15.3 (CH3). HRMS/EI: calcd for C19H21N2O4 + [M+H]+ 341.1496, found 341.1504.
5-Ethyl-8,9-dimethoxy-5,6-dihydrophenanthridin-4-amine (12). By referencing the synthesis method of compound 3, compound 12 was conducted by reduction of compound 9 (299 mg, 1 mmol) at the presence of 1 M BH3 –THF (2 mL, 2 mmol) in a yield of 75%. 1H NMR (500 MHz, CDCl3) δ H 7.26 (s, 1H), 7.15–7.09 (m, 1H), 7.01 (t, J = 7.8 Hz, 1H), 6.72 (s, 1H), 6.68 (dd, J = 7.8, 1.2 Hz, 1H), 4.05 (s, 2H), 3.95 (s, 3H), 3.93 (s, 3H), 2.68 (q, J = 7.0 Hz, 2H), 1.09 (t, J = 7.1 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 148.8 (C), 148.4 (C), 143.4 (C), 141.6 (C), 129.7 (C), 125.7 (C), 125.4 (C), 124.9 (CH), 114.0 (CH), 112.8 (CH), 109.9 (CH), 106.6 (CH), 56.1 (CH3), 56.0 (CH3), 49.8 (CH2), 44.3 (CH2), 13.9 (CH3). HRMS/EI: calcd for C17H21N2O2 + [M+H]+ 285.1598, found 285.1605.
Syntheses of compounds 13–16. By referencing the synthesis method of compound 6, compound 13–16 was conducted by amidating of compound 12 (1 mmol) at the presence of Ac2O, propionyl chloride, isobutyryl chloride or trifluoroacetic anhydride (1 mL) in a yield of over 80%.
N-(5-Ethyl-8,9-dimethoxy-5,6-dihydrophenanthridin-4-yl)acetamide(13).1H NMR (600 MHz, CDCl3) δ H 8.45 (s, 1H), 8.27 (d, J = 8.0 Hz, 1H), 7.40 (d, J = 7.5 Hz, 1H), 7.20 (t, J = 8.0 Hz, 1H), 6.73 (s, 1H), 4.04 (s, 2H), 3.97 (s, 3H), 3.93 (s, 3H), 2.68–2.57 (m, 2H), 2.24 (s, 3H), 1.10 (t, J = 7.2 Hz, 3H). 13C NMR (150 MHz, CDCl3) δ C 168.2 (C), 149.1 (C), 148.6 (C), 135.6 (C), 133.4 (C), 129.5 (C), 125.4 (CH), 125.2 (C), 124.6 (C), 118.0 (CH), 117.6 (CH), 110.0 (CH), 106.4 (CH), 56.1 (CH3), 56.0 (CH3), 50.0 (CH2), 46.1 (CH2), 24.9 (CH3), 13.9 (CH3). HRMS/EI: calcd for C19H21N2O3 + [M+H]+ 325.1547, found 325.1550.
N-(5-Ethyl-8,9-dimethoxy-5,6-dihydrophenanthridin-4-yl)propionamide(14). 1H NMR (500 MHz, CDCl3) δ H 8.51 (s, 1H), 8.31 (d, J = 8.1 Hz, 1H), 7.40 (dd, J = 7.9, 1.2 Hz, 1H), 7.20 (t, J = 8.0 Hz, 1H), 6.73 (s, 1H), 4.04 (s, 2H), 3.97 (s, 3H), 3.93 (s, 3H), 2.62 (q, J = 7.2 Hz, 2H), 2.47 (q, J = 7.6 Hz, 2H), 1.30 (t, J = 7.6 Hz, 3H), 1.12–1.08 (m, 3H). 13C NMR (125 MHz, CDCl3) δ C 171.9 (C), 149.1 (C), 148.7 (C), 133.5 (C), 129.5 (C), 125.5 (C), 125.2 (CH), 124.7 (C), 120.7 (C), 118.0 (CH), 117.6 (CH), 110.0 (CH), 106.5 (CH), 56.1 (CH3), 56.0 (CH3), 50.0 (CH2), 46.0 (CH2), 31.2 (CH2), 13.9 (CH3), 9.8 (CH3). HRMS/EI: calcd for C20H25N2O3 + [M+H]+ 341.1860, found 341.1863.
N-(5-Ethyl-8,9-dimethoxy-5,6-dihydrophenanthridin-4-yl)isobutyramide(15).1H NMR (500 MHz, CDCl3) δ H 8.57 (s, 1H), 8.32 (d, J = 8.1 Hz, 1H), 7.39 (dd, J = 7.9, 1.2 Hz, 1H), 7.20 (t, J = 8.0 Hz, 1H), 6.73 (s, 1H), 4.04 (s, 2H), 3.96 (s, 3H), 3.92 (s, 3H), 2.64–2.56 (m, 3H), 1.31 (s, 3H), 1.29 (s, 3H), 1.11 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 175.1 (C), 149.1 (C), 148.7 (C), 135.7 (C), 133.5 (C), 129.5 (C), 125.5 (CH), 125.1 (C), 124.7 (C), 118.0 (CH), 117.6 (CH), 110.0 (CH), 106.5 (CH), 56.1 (CH3), 56.04 (CH3), 49.8 (CH2), 45.8 (CH2), 37.1 (CH), 19.7 (CH3), 19.7 (CH3), 13.8 (CH3). HRMS/EI: calcd for C21H27N2O3 + [M+H]+ 355.2016, found 355.2023.
N-(5-Ethyl-8,9-dimethoxy-5,6-dihydrophenanthridin-4-yl)-2,2,2-trifluoroacetamide (16).1H NMR (500 MHz, CDCl3) δ H 9.45 (s, 1H), 8.22 (dd, J = 8.0, 1.7 Hz, 1H), 7.52 (dd, J = 8.0, 1.1 Hz, 1H), 7.28–7.24 (m, 2H), 6.75 (s, 1H), 4.06 (s, 2H), 3.97 (s, 3H), 3.94 (s, 2H), 2.62 (q, J = 7.2 Hz, 2H), 1.12 (t, J = 7.2 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 154.6 (C, q, J = 37.5 Hz), 149.5 (C), 148.8 (C), 136.7 (C), 131.0 (C), 130.1 (C), 125.8 (CH), 125.2 (C), 124.0 (C), 119.7 (CH), 118.1 (CH), 110.2 (CH), 106.4 (CH), 56.2 (CH3), 56.1 (CH3), 49.9 (CH2), 46.4 (CH2), 13.6 (CH3). HRMS/EI: calcd for C19H20F3N2O3 + [M+H]+ 381.1421, found 328.1427.
5-Ethyl-8,9-dimethoxy-N-(2,2,2-trifluoroethyl)-5,6-dihydrophenanthridin-4-amine (17). By referencing the synthesis method of compound 1, compound 17 was conducted by reduction of compound 16 (38 mg, 0.1 mmol) at the presence of 1 M BH3 -THF (1 mL, 1 mmol) in a yield of 65%. 1H NMR (500 MHz, CDCl3) δ H 7.25 (s, 1H), 7.16–7.07 (m, 2H), 6.73 (s, 1H), 6.63 (d, J = 7.1 Hz, 1H), 5.23 (brs, 1H), 4.11 (s, 2H), 3.96 (s, 3H), 3.93 (s, 3H), 3.81 (q, J = 8.8 Hz 2H), 2.61 (q, J = 7.1 Hz, 2H), 1.09 (t, J = 7.1 Hz, 3H). 13C NMR (125 MHz, CDCl3) δ C 149.0 (C), 148.5 (C), 141.4 (C), 133.7 (C), 129.6 (C), 126.3 (C), 125.3 (CH), 125.1 (C), 124.1 (C), 112.7 (CH), 110.0 (CH), 108.7 (CH), 106.7 (CH), 56.1 (CH3), 56.0 (CH3), 49.8 (CH2), 45.8 (CH2), 44.6 (CH2 q, J = 33.8 Hz), 13.8 (CH3). HRMS/EI: calcd for C19H22F3N2O2 + [M+H]+ 367.1628, found 367.1633.
4.4. Molecular docking
Three-dimensional structures of ligands were built and optimized using ChemBioOffice package (version 2014). The Gasteiger charges were calculated, and all nonpolar hydrogen atoms were merged with the carbon atoms by using Autodock Tools program (version 1.5.6) [19]. All the X-ray crystal structure involved in this research (PDB ID:4KXJ, 6KL6 and 6VYO) [13,16,20] were downloaded from the Protein Data Bank and used for protein preparation. 4KXJ and 6KL6, the complexes of NPro-NTD and their inhibitors in HCoV-OC43 and MERS CoV, are used to analyze the pockets where the inhibitor is located. 6VYO has the highest resolution (1.7 Å) in the crystal structures of SARS-CoV-2 NPro- NTD and hence used to docking. All the solvent and organic molecules were removed from the crystal structure by using the Pymol program (version 2.1.0). Then, polar hydrogens were added to the structure, and the Gasteiger charges were computed with Autodock Tools program (version 1.5.6).
Docking of Phenanthridine core (X0) and six model molecules (M1-6) into SARS-CoV-2 N-NTD (PDB: 6VYO) were performed by AutoDock Vina program [18] (version 1.1.2). The 25 × 25 × 25 Å grid box with 1.000 Å grid spacing was used and the grid center coordinates were placed at x = −3.657, y = −2.908 and z = 2.375. The maximum number of binding modes to be generated were set to 50. The obtained lower binding affinity poses were visualized and analyzed using PyMOL (version 2.1.0) and MOE (version 2019).
To confirm the binding site of two active compounds 12 and 16, three potential pockets were analyzed. All the grid boxes were set to 25 × 25 × 25 Å with 1.000 Å grid spacing. And the grid center was placed at three different coordinates (x, y, z) = (−3.657, −2.908, 2.375), (x, y, z) = (−11.994, 2.636, 9.097) and (x, y, z) = (6.647, 40.037, 11.357). The maximum number of binding modes to be generated were set to 50, the pose with the lowest docking score in each potential pocket was selected to observe the interaction with residues. And select the amino acids that both interact with 12 and 16, of which the importance will be further evaluated by site-directed mutation.
4.5. Screening and kinetic analysis
All experiments were performed with Biacore S200 (Washington, DC, USA) instrument, at a flow rate of 10 μL/min. The data was analyzed using the Biacore S200 Evaluation software. An PBS-P buffer (1 mM PBS, 0.05% P20, 5% DMSO) was used as the running buffer. The wild and mutant SARS-CoV-2 NPro were immobilized on a CM5 sensor chip (GE Healthcare Life Sciences, Pittsburgh, PA, USA) by amide coupling chemistry. The carboxylic acid groups were first activated by placing them in a mixture of N-Ethyl-N’-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS). Then, proteins (20 μg/mL) dissolved in sodium acetate solution (pH 5.0) were immobilized on the CM5 chip, respectively. Finally, the chip was blocked with ethanolamine. To reduce nonspecific interactions, the cell immobilized without protein was used as a reference [21].
“Amine coupling” was employed as the protein immobilization method. The NPro was captured as a ligand and synthesized phenanthridine derivatives were used as analytes. According to a recently reported study [17], all the compounds were examined through the Binding Level Screen to identify compounds that could bind to the targets of interest (Table S1). Since the target involved in this study is relatively novel, there is no positive control compounds at present. For the Binding Level Screen, compounds were screened at 200 μM on a surface of >10000 RU of proteins, with a 120 s contact time and a 180 s dissociation time. Running buffer was chosen as the negative control during screening. The solvent correction sample (eight point) that includes the 5% DMSO used in the assay was prepared according to the GE Healthcare Laboratory Guideline. For data analysis, solvent correction was applied and the compound that lay outside the range was not corrected and noted. Binding Level Screen hits were then progressed into the Affinity Screen, with a concentration series in running buffer (0.625–60 μM). The contact time was 120 s and dissociation time was 180 s. Dose–response data was collected in the traditional multicycle format. The data was automatically fitted to the 1:1 binding model for both kinetics and steady-state affinity.
4.6. Viruses and cells
The SARS-CoV-2 (strain 107) was provided by Guangdong Provincial Center for Disease Control and Prevention (Guangzhou, China). This virus was propagated and titrated on African green monkey kidney epithelial cells (Vero E6) (ATCC, no. 1586), which were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin–streptavidin (Gibco). The cell line used in this study was cultured at 37 °C in a humidified 5% CO2 atmosphere. Mycoplasma testing was performed at regular intervals and no mycoplasma contamination was detected. All the infection experiments were performed at biosafety level-3 (BSL3) conditions at the Key Laboratory of Animal Models and Human Disease Mechanisms of the Chinese Academy of Sciences, Kunming Institute of Zoology (Kunming, China) [22].
4.7. Cytotoxicity assay
Vero E6 cells were seeded in 96 well plates and grown overnight. Various concentrations of compounds were then added to each well. After incubation for 72 h, the cell viability was evaluated using CCK8 (Beyotime) according to the manufacturer's protocol. All experiments were performed in triplicate [22].
4.8. Cellular antiviral activity assay
Assays of cellular antiviral activity were performed using CCK8 and RT-qPCR methods. For the CCK8 method, Vero E6 cells were seeded in 96-well plates and grown overnight. The cells were then infected with SARS-CoV-2 at an MOI of 0.1. At the same time, the test compounds were added to the wells with different concentrations. After incubation for 2 h, the medium was replaced with fresh drug-containing medium. In 72 h, the cytopathic effect caused by SARS-CoV-2 infection was quantitatively analyzed using CCK8 (Beyotime) according to the manufacturer's protocol [22].
For the RT-qPCR method, Vero E6 cells were seeded in 48-well plates (200 μL/well) at 8 × 105 cells/well and grown overnight. Cells were infected with SARS-CoV-2 at an MOI of 0.01. Then the infected cells were treated with different concentrations of the test compounds. After 1 h of incubation at 37 °C, the virus-drug mixture was removed and replaced with fresh medium containing compounds. In 48 h, the cell supernatants were collected to extract viral RNA, which was subjected to RT-qPCR analysis. TaqMan primers for SARS-CoV-2 are 5′-GGGGAACTTCTCCTGCTAGAAT-3′ and 5′-CAGACATTTTGCTCTCAAGCTG-3′ with SARS-CoV-2 probe FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3'. The EC50 values were calculated by using a dose-response model in GraphPad Prism 7 software. All experiments were performed for two times in triplicate, and the values are presented as mean ± SD.
4.9. Cloning, expression and purification
The codon optimized genes (SARS-CoV-2 NPro, Mutant A, B and C) were synthesized and inserted into pET-22b (+) for sequencing, pET28a (+) vector for expression. Then reconstructed genes with an N-terminal 6-histidine tag were overexpressed in E. coli BL21 (DE3) POSITIVE. A single colony was incubated in LB medium with ampicillin resistance. Expression was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM at 25 °C.
Cell cultures were harvested by centrifugation at 8000 rpm for 15 min (GL-10MD, XiangYi) and resuspended in bacterial lysis buffer (20 mM Tris-HCl, 300 mM NaCl, pH 8.0). The resuspended pellet was sonicated (JY92-IIN, SCIENTZ) on ice for 2s ON/4s OFF and 30% power with 30 min between the cycles to lyse. Cell debris was removed by centrifugation (8000 rpm, 20 min, 4 °C) to obtain soluble fractions. Target protein was purified utilizing affinity chromatography on a nickel-NTA column and verified by SDS-PAGE as well as Western Blot (Fig. S3).
4.10. Statistical analysis
Results were expressed as means ± SD and analyzed with GraphPad Prism 7. Data were combined from at least three independent experiments unless otherwise stated.
Author contributions
a Y.-T. W., X.-Y. L and X. D. contributed equally to this work.Y.-T. W. and B.-J. Y. designed and performed the chemical synthesis; J.-Y. C. and X.-F. Z. purified all the compounds; X.-Y. L., R.-H. L. and Y.-T. Z. evaluated the antivirus activities. Y.-T. W. and X. D. conducted SPR assay of all the compounds under the guidance of D.-Z. C. and Y.-T. Z.; D.-Z. C. and Y.-T. W. wrote the manuscript with advice from all of the authors; D.-Z. C. and X.-J. H. guided all aspects of this work.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (81773610, 81973212); the Youth Innovation Promotion Association of CAS (2018429); the Central Asian Drug Discovery and Development Center of Chinese Academy of Sciences (CAM202103); Natural Science Foundation of Yunnan (202101AT070169).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2021.113966.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Yin W.C., Mao C.Y., Luan X.D., Shen D.D., Shen Q.Y., Su H.X., Wang X.X., Zhou F.L., Zhao W.F., Gao M.Q., Chang S.H., Xie Y.C., Tian G.H., Jiang H.W., Tao S.C., Shen J.S., Jiang Y., Jiang H.L., Xu Y.C., Zhang S.Y., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva L.R., Santos P.F.D., Brandao J.D., Anderson L., Bassi E.J., de Araujo J.X., Cardoso S.H., da Silva E.F. Druggable targets from coronaviruses for designing new antiviral drugs. Bioorg. Med. Chem. 2020;28:115745–115767. doi: 10.1016/j.bmc.2020.115745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voysey M., Clemens S.A.C., Madhi S.A. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng Y.Y., Liao G.R., Lien A., Hsu W.L. Current concepts in the development of therapeutics against human and animal coronavirus diseases by targeting NP. Comput. Struct. Biotechnol. J. 2021;19:1072–1080. doi: 10.1016/j.csbj.2021.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C.K., Lo S.C., Wang Y.S., Hou M.H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today. 2016;21:562–572. doi: 10.1016/j.drudis.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang S.S., Yang M., Hong Z.S., Zhang L.P., Huang Z.X., Chen X.X., He S.H., Zhou Z.L., Zhou Z.C., Chen Q.Y., Yan Y., Zhang C.S., Shan H., Chen S.D. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., Urquiza J., Ramirez D., Alonso C., Campillo N.E., Martinez A. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 9.Peng Y., Du N., Lei Y.Q., Dorje S., Qi J.X., Luo T.R., Gao G.F., Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39:1–12. doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saikatendu K., Joseph J., Subramanian V., Neuman B., Buchmeier M., Stevens R., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81 doi: 10.1128/JVI.02236-06. 23-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossoehme N.E., Li L., Keane S.C., Liu P., Iii C., Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S.Y., Liu C.L., Chang Y.M., Zhao J.C., Perlman S., Hou M.H. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J. Med. Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D.Z., Cai J.Y., Yin J.L., Jiang J.D., Jing C.X., Zhu Y.P., Cheng J.J., Di Y.T., Zhang Y., Cao M.M., Li S.L., Peng Z.G., Hao X.J. Lycorine-derived phenanthridine downregulators of host Hsc70 as potential hepatitis C virus inhibitors. Future Med. Chem. 2015;7:561–570. doi: 10.4155/fmc.15.14. [DOI] [PubMed] [Google Scholar]
- 15.Chen D.Z., Fan S.R., Yang B.J., Yao H.C., Wang Y.T., Cai J.Y., Jing C.X., Pan Z.H., Luo M., Yuze Y.Q., Liu G.J., Hao X.J. Phenanthridine derivative host heat shock cognate 70 down-regulators as Porcine Epidemic Diarrhea Virus inhibitors. J. Nat. Prod. 2021;84:1175–1184. doi: 10.1021/acs.jnatprod.0c01252. [DOI] [PubMed] [Google Scholar]
- 16.C. Chang, K. Michalska, R. Jedrzejczak, N. Maltseva, M. Endres, A. Godzik, Y. Kim, A. Joachimiak, Crystal structure of RNA Binding Domain of Nucleocapsid Phosphoprotein from SARS Coronavirus 2, (To be published ) 10.2210/pdb2216VYO/pdb.
- 17.Zhu Z.L., Qiu X.D., Wu S., Liu Y.T., Zhao T., Sun Z.H., Li Z.R., Shan G.Z. Blocking effect of demethylzeylasteral on the interaction between human ACE2 protein and SARS-CoV-2 RBD protein discovered Using SPR technology. Molecules. 2021;26:57–68. doi: 10.3390/molecules26010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trott O., Olson A.J. Software news and update AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin S.M., Lin S.C., Hsu J.N., Chang C.K., Chien C.M., Wang Y.S., Wu H.Y., Jeng U.S., Kehn-Hall K., Hou M.H. Structure-Based stabilization of non-native protein-protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J. Med. Chem. 2020;63:3131–3141. doi: 10.1021/acs.jmedchem.9b01913. [DOI] [PubMed] [Google Scholar]
- 21.Bergsdorf C., Wright S.K. A Guide to run affinity screens using differential Scanning Fluorimetry and Surface Plasmon Resonance assays. Methods Enzymol. 2018;610:135–165. doi: 10.1016/bs.mie.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Qiao J.X., Li Y.S., Zeng R., Liu F.L., Luo R.H., Huang C., Wang Y.F., Zhang J., Quan B.X., Shen C.J., Mao X., Liu X.L., Sun W.N., Yang W., Ni X.C., Wang K., Xu L., Duan Z.L., Zou Q.C., Zhang H.L., Qu W., Long Y.H.P., Li M.H., Yang R.C., Liu X.L., You J., Zhou Y.L., Yao R., Li W.P., Liu J.M., Chen P., Liu Y., Lin G.F., Yang X., Zou J., Li L.L., Hu Y.G., Lu G.W., Li W.M., Wei Y.Q., Zheng Y.T., Lei J., Yang S.Y. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C.K., Jeyachandran S., Hu N.J., Liu C.L., Lin S.Y., Wang Y.S., Chang Y.M., Hou M.H. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. Biosyst. 2016;12:59–66. doi: 10.1039/c5mb00582e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.