Summary
The functional heterogeneity of resident stem cells that support adult organs is incompletely understood. Here, we directly visualize the corneal limbus in the eyes of live mice and identify discrete stem cell niche compartments. By recording the lifecycle of individual stem cells and their progeny, we directly analyze their fates and show that their location within the tissue can predict their differentiation status. Stem cells in the inner limbus undergo mostly symmetric divisions and are required to sustain the population of transient progenitors that support corneal homeostasis. Using in situ photo-labeling, we captured their progeny exiting the niche before moving centripetally in unison. The long-implicated slow-cycling stem cells are functionally distinct and display local clonal dynamics during homeostasis, but can contribute to corneal regeneration after injury. This study demonstrates how the compartmentalized organization of functionally diverse stem cell populations support the maintenance and regeneration of an adult organ.
Graphical Abstract
eTOC blurb
Farrelly et al. capture dynamic cell behaviors across the ocular surface epithelium of live mice during homeostasis and regeneration. This study shows that the limbus harbors functionally discrete stem cell populations that display distinct clonal activities.
Introduction
Stratified squamous epithelia undergo continuous regeneration. During this process, terminally differentiated cells are shed from their surface and are replenished by keratinocytes from the basal layer that display stem cell properties (Marques-Pereira & Leblond 1965), (Green 1980). Due to constant cell loss, stratified squamous epithelia must exist in a state of dynamic equilibrium to maintain their tissue structure and function (Belokhvostova et al. 2018). Diseases that affect surface epithelia are characterized by abnormal cell proliferation or differentiation, yet it is not fully understood how the fate of individual stem cells is coordinated organ-wide to achieve tissue homeostasis. Studies in the epidermis and the esophagus have supported a model whereby basal layer keratinocytes have equal potential, and likely regulate their fates based on local interactions with neighboring stem cells (Clayton et al. 2007), (Mascré et al. 2012), (Doupé et al. 2012), (Rompolas et al. 2016), (Piedrafita et al. 2020), (Mesa et al. 2018). The cornea is a unique example of a squamous stratified epithelium that has typical histological characteristics and a barrier function, but display a distinct stem cell organization (Schermer et al. 1986), (Cotsarelis et al. 1989), (Pellegrini et al. 1999), (Notara et al. 2010).
The cornea lines the anterior ocular surface and is critical for vision, acting as a protective barrier for the eye and refracting light towards the retina. In contrast to the epidermis or the esophagus, stem cells in the corneal epithelium are proposed to be hierarchically organized and reside in a specific niche in the periphery of the tissue called the limbus (Lavker et al. 2004), (West et al. 2015). Evidence suggests that the cornea is self-contained and is anatomically and functionally separated from the more posteriorly located conjunctiva, with which they form a continuous surface. Long-lived stem cells, located at the border between the two epithelia, generate the more differentiated progeny that replenish the entire corneal tissue (Collinson et al. 2002), (Mort et al. 2009), (Di Girolamo et al. 2015), (Amitai-Lange, Altshuler, et al. 2015), (Dorà et al. 2015), (Kasetti et al. 2016). This hierarchical organization implies two distinct “fluxes” of cells: one within the basal layer, from the limbus to the center of the cornea and a second, from the proliferative cells throughout the basal layer to the terminally differentiated, desquamated cells at the surface of the tissue (West et al. 2018), (Lobo et al. 2016). To expand upon this knowledge, we set out to directly test these models by resolving the dynamics of corneal stem cells and their progeny in the live mouse cornea.
A common feature of stem cells in regenerating organs is their localization within discrete niches, which are critical for establishing their identity and regulating their distinct behaviors (Schofield 1978), (Ohlstein et al. 2004), (Moore & Lemischka 2006), (Scadden 2014). Experimental and clinical evidence suggest that the limbus harbors bona fide stem cells that regenerate the cornea, and is also the location of slow-cycling, label-retaining cells, long presumed to be part of the same corneal stem cell hierarchy (Collinson et al. 2002), (A. J. Huang & Tseng 1991), (Lavker et al. 1991), (Rama et al. 2010). Slow-cycling ability, a hallmark of stem cells in various tissues, is also associated with distinct reserve populations (Cotsarelis et al. 1990), (Tumbar et al. 2004), (Wilson et al. 2008), (Takeda et al. 2011). The behavior of individual stem cells in the limbus has not been directly visualized in vivo, and therefore the functional heterogeneity and requirement of these cells for corneal homeostasis and regeneration remain unresolved.
In recent years, 2-photon microscopy has been successfully used to visualize stem cell behavior in regenerating organs, including the skin, intestine and hematopoietic system (Celso et al. 2009), (Rompolas et al. 2012), (Ritsma et al. 2014), (S. Huang & Rompolas 2017). Such studies have offered critical insight into the cellular mechanisms that govern tissue regeneration by capturing stem cell activity in their native tissue environment. The cornea is an ideal tissue for intravital imaging due its accessibility, optical transparency, defined topology, and relatively simple organization. Widefield fluorescent imaging combined with in vivo lineage tracing has provided critical information for the contribution of limbal stem cells to corneal homeostasis and regeneration (Di Girolamo et al. 2015), (Amitai-Lange, Altshuler, et al. 2015), (Amitai-Lange, Berkowitz, et al. 2015), (Richardson et al. 2017), (Park, Richardson, Pandzic, Lobo, Whan, et al. 2019), (Nasser et al. 2018). Building on this paradigm, we developed genetic tools and a 2-photon microscopy-based intravital imaging system to capture the activity of corneal stem cells and their progeny, in real-time as well as long-term, in the intact eyes of live mice.
Results
Intravital imaging of the mouse corneal limbus by 2-photon microscopy
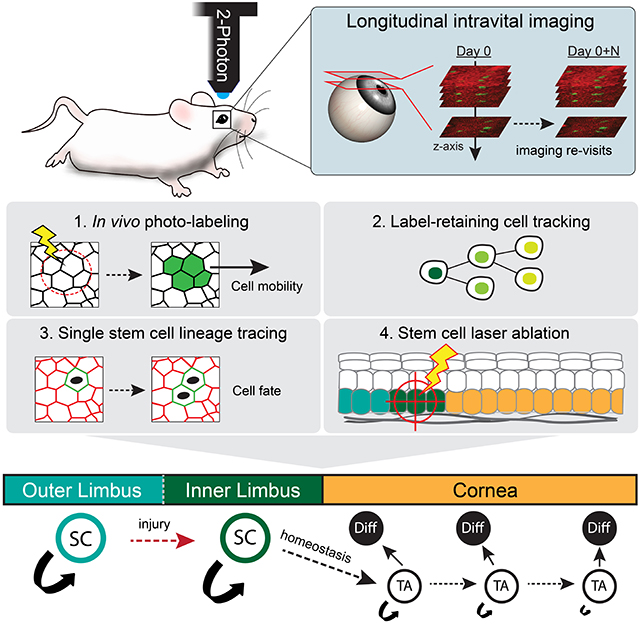
To interrogate the clonal dynamics that enable the homeostatic maintenance of the cornea and determine the precise location of stem cells in the limbal niche, we devised a 2-photon microscopy-based system to visualize the live mouse limbus, at single-cell resolution, without compromising the structural integrity or physiology of the eye (Fig. 1A; Fig. S1A; Movie S1; Movie S2). We combined this with a Cre recombinase-based in vivo lineage tracing strategy to resolve the activity and contribution of individual stem cells by longitudinal live imaging (Fig. 1B). Several molecular markers have been proposed to distinguish bona fide stem cells in the mouse and human limbus, but there remains a lack of consensus regarding whether any one marker is expressed exclusively in this population (Guo et al. 2018). To address this challenge, we hypothesized that we could identify all the potential stem cell populations within the limbus retrospectively by labeling single cells without bias and directly tracing their respective lineage over time by intravital imaging.
Figure 1. Live imaging of the limbal niche reveals distinct clonal growth patterns.
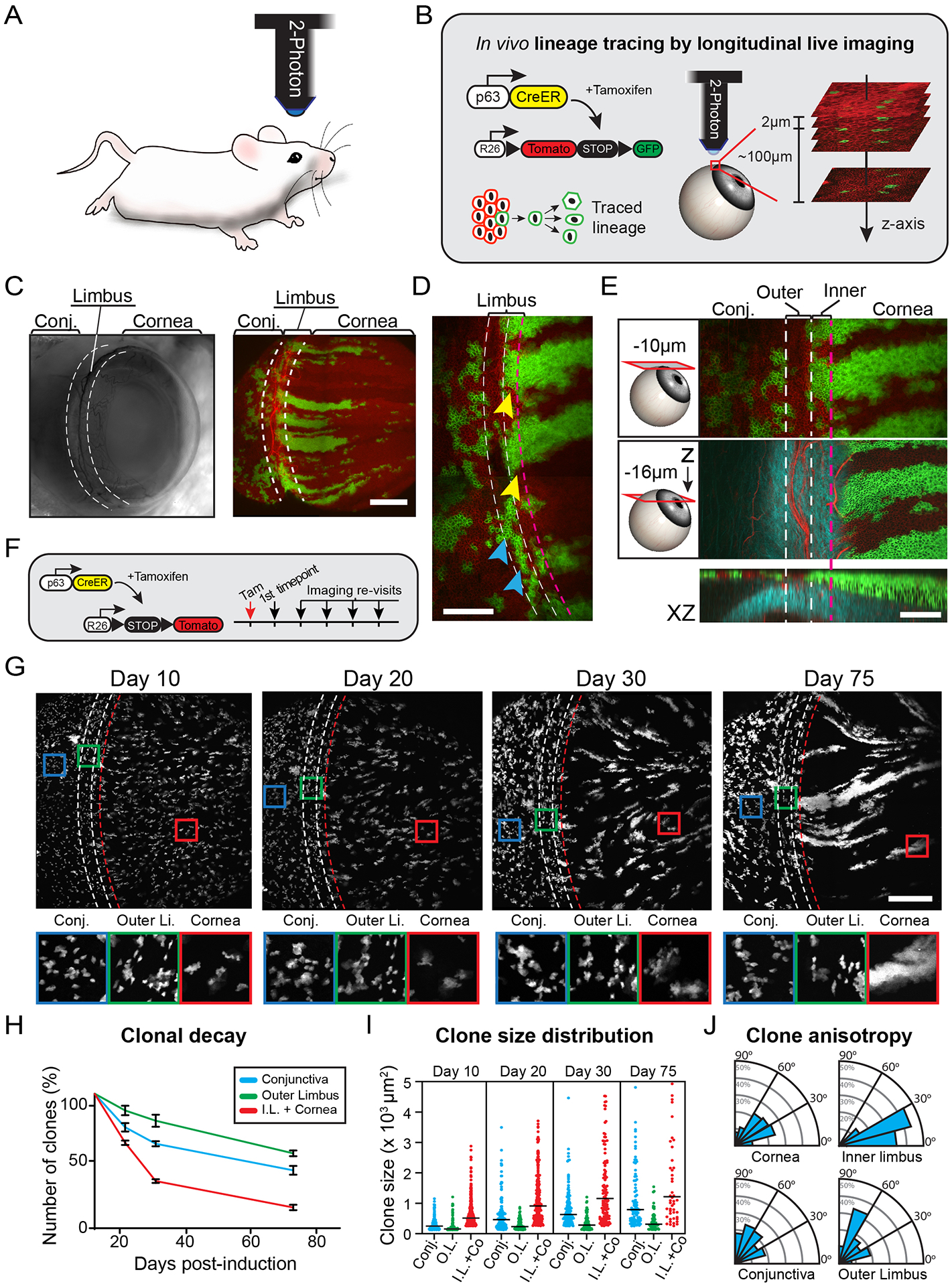
(A) Intravital imaging of the mouse eye is performed at single-cell resolution with 2-photon microscopy. (B) Experimental strategy to resolve stem cell dynamics in the ocular surface epithelium by in vivo longitudinal lineage tracing. Also see Movies S1, S2 and S3. (C) Examples of live mouse eyes imaged by brightfield (left) and 2-photon (right) microscopy. (D) Expanded view of the limbus after three months of lineage tracing illustrating the distinct organization and relative position of clones in the outer limbus (cyan arrows) and inner limbus (yellow arrows). (E) Two representative optical planes of the limbus, at the indicated depth from the surface, and a reconstructed side view (XZ). (F) Experimental strategy and genetic alleles for in vivo lineage tracing by longitudinal live imaging. (G) Lineage tracing time series from re-imaging the same eye at the indicated time points after induction. Representative magnified views of indicated areas in the conjunctiva, outer limbus and cornea (lower panels). (H) Quantification of clonal decay measured as a fraction of clones that persist within the indicated compartments at each re-imaging time point. For these quantifications, the inner limbus is considered part of the cornea due to the uniform origin of centripetally expanding corneal clones from this compartment (n = 725 traced clones in 6 re-sampled areas from 2 mice, p = 0.0024; 2-way ANOVA). (I) Quantification of size distribution of clones in each epithelial compartment across the different time points (n = 1775 clones in 12 randomly sampled areas from 2 mice p < 0.0001; Nested 1-way ANOVA). (J) Radial graphs show quantification of clonal growth anisotropy five days after induction, measured as the angle between each clone’s Feret’s diameter and the radial line that connects the limbus with the center of the cornea (n = 306 clones in 4 randomly sampled areas per compartment, from 2 mice; ****p < 0.0001). Panels C, D, E and G show tiled images of the cornea and limbus constructed from multiple fields-of-view. Dotted lines indicate the margins of the outer (white) and inner (red) limbus. Scale bars: 500 μm (C, G), 200 μm (D, E).
To test this hypothesis, we used an inducible p63CreERT2 genetic driver because of its ubiquitous activity in the basal layer of stratified epithelia (Pellegrini et al. 2001), (Truong et al. 2006), (Senoo et al. 2007), (Lee et al. 2014). Using the p63CreERT2 driver, in conjunction with a dual-fluorescent ROSA26LoxP-tdTomato-STOP-LoxP-EGFP Cre-reporter (R26-mTmG), we marked basal stem/progenitor cells across the entire surface epithelium, comprised of the cornea, limbus and conjunctiva (Fig. 1C). To directly track the emergence of long-lived lineages generated from labeled stem cells, we acquired high-resolution, full-thickness serial optical sections of the whole anterior eye, including the limbus, and then re-imaged the same eyes over time using identical acquisition parameters (Fig. S1B). Several weeks after induction, only clones that were sustained by stem cells were present in the tissue (Fig. 1C, D; Fig. S1B).
Close examination of the limbus revealed heterogeneity in the appearance, localization and behavior of the long-lived clones emerging from the niche (Fig. 1D, E; Fig. S1B). In line with previous studies, we observed cells that gave rise to lineages that exited the niche and expanded centripetally towards the central cornea. Almost all of the centripetal limbal clones had a common line of origin where the tissue transitions from the multi-layered cornea to the 3-layered limbal epithelium (Fig. 1E). Surprisingly, immediately adjacent to the line of origin of the radial lineages, we observed discrete limbal clones that only expanded within their original location and never exited the niche (Fig. 1D; Fig. S1B). R26-mTmG ubiquitously labels the membrane of every cell in the live tissue. This reporter, combined with second harmonic generation (SHG) microscopy, which was used to visualize the extracellular matrix, enabled us to resolve the histological organization of the live tissue, and determine the relative location of labeled cells within the niche (Chen et al. 2012) (Fig. 1E; Movie S3). We observed that the stationary limbal clones co-localized and oriented parallel to the circumferentially aligned collagen fibers and blood vessels of the underlying limbal stroma (Fig. S1C, D). Using additional in vivo reporters, we found that larger bundles of corneal nerves and immune cells of the myeloid lineage also localized to this area of the limbus (Fig. S1C, E). Based on these findings, we hypothesized that the limbus is organized into at least two distinct stem cell compartments. We used topological criteria to define these compartments as the “outer” and “inner” limbus, according to their proximity to the cornea.
Spatially distinct clonal dynamics reveal a compartmentalized limbal niche
To further resolve the functional heterogeneity of the stem cells in the limbus, we performed lineage tracing by longitudinal live imaging and analyzed the growth characteristics of the emerging long-lived clones. For these experiments, we used the ROSA26LoxP-STOP-LoxP-tdTomato (R26-tdTom) Cre-reporter because it allowed us to precisely control the extent of recombination necessary to label a representative fraction of spatially separated basal cells, including all the potential limbal stem cell populations (Fig. 1F; Fig. S2A). Based on the lineage tracing analysis, clones were classified into three major categories according on their location, dimension, and orientation (Fig. 1G–J; Fig. S2B). In the limbus, we found evidence of at least two distinct stem cell activities. We observed large, radially oriented clones that emanated from the inner limbus shortly after induction and expanded centripetally over time (Fig. 1G; S2B). These progressively replaced the smaller, randomly distributed short-lived clones in the central cornea, which conforms to the definition of a bona fide corneal stem cell population. However, we also consistently identified smaller clones that formed exclusively in the outer limbus (Fig. 1G). These smaller clones remained spatially separated from their larger counterpart throughout the entire imaging time course.
Based on quantitative clonal analysis, the number of clones originally labeled within the cornea showed the most rapid decrease over time (Fig. 1H). This decrease was balanced by the robust growth of a few expanding clones emanating from the inner limbus, which was reflected by the bifurcation in the size of the centripetally expanding corneal clones over the imaging time course (Fig. 1I). Interestingly, clones in the outer limbus showed the slowest decline in their overall numbers and the slowest change in their average size compared to the other populations (Fig. 1H, I). Progenitors marked in the conjunctiva displayed no apparent polarization in their overall growth patterns. Analysis of the anisotropy of the growth dynamics in the emerging clones in the outer limbus showed that they orient perpendicularly to the radial line between the limbus and the center of the cornea (Fig. 1J).
Corneal progenitors that exit the limbal niche display uniform centripetal mobility
We next tested the hypothesis that basal progenitors in the cornea have a limited lifespan and are replenished exclusively by stem cells that reside in the inner limbus. For this, we used a globally expressed ROSA26LoxP-STOP-LoxP-PAGFP (R26-PAGFP) photo-activatable mouse reporter combined with live imaging. Our goal was to selectively mark basal cells in the cornea in situ, and directly capture their behavior over time (Fig. 2A; Movie S4). Photo-labeling of quiescent cells in the limbal stroma and corneal endothelium provided stable reference points to evaluate the mobility of progenitors in the epithelium (Fig. 2A). We found that throughout the cornea, all basal cells move unidirectionally and in concert towards the center, at defined rates (Fig. 2B, C; Fig. S3A). Based on the ubiquitous centripetal mobility of basal cells, we conclude that, at least in vivo and in the absence of injury, corneal progenitors have limited potential and transit in unison towards the center of the cornea until they are exhausted and terminally differentiate.
Figure 2. Photo-labeling and tracking of stem cell dynamics in the live eye.
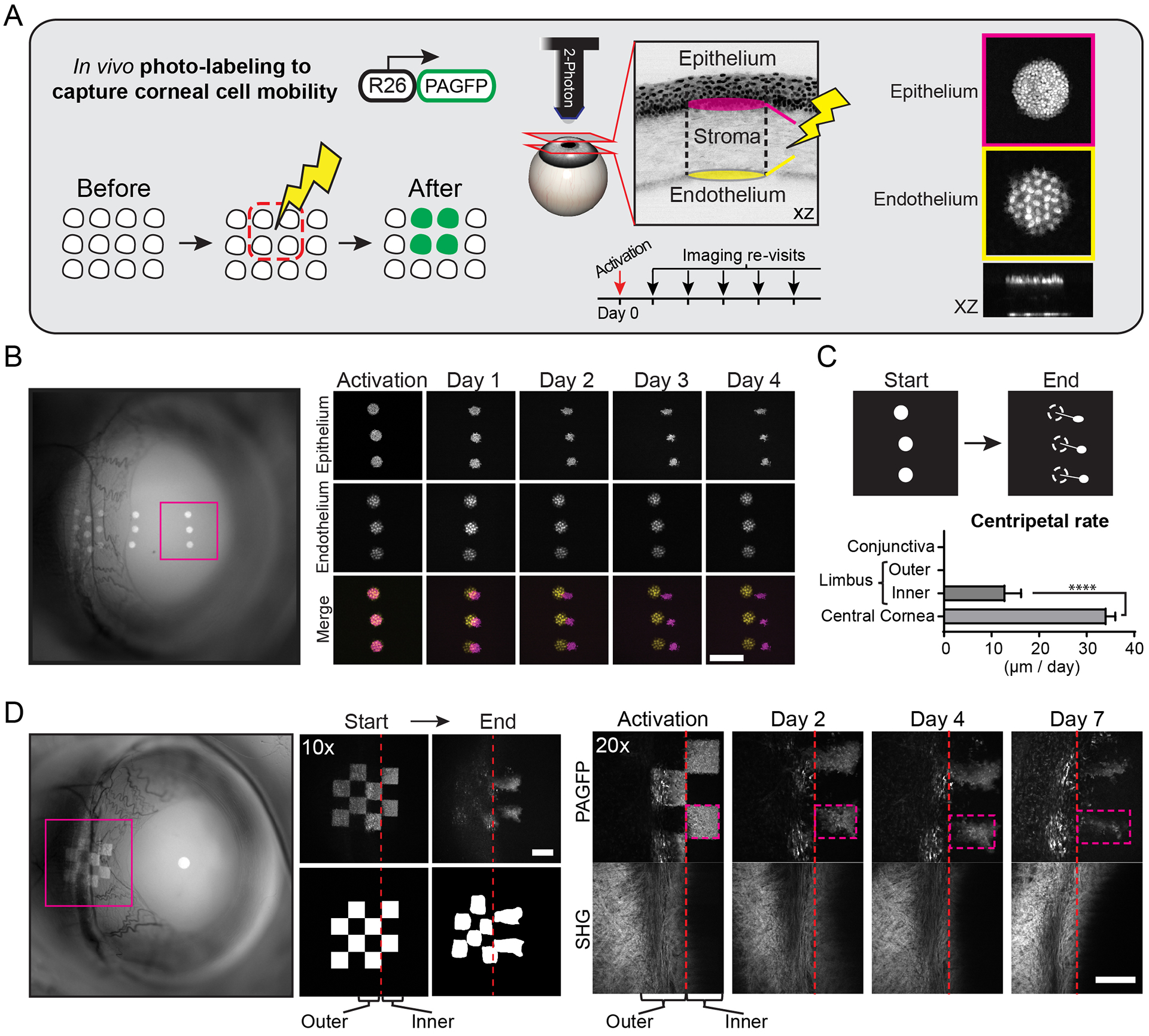
(A) Experimental strategy to directly capture the mobility and fate of stem/progenitor cells in the ocular surface epithelium. All basal cells within the selected area are marked in situ by laser scanning a globally-expressed, photo-activatable reporter (PAGFP). Post-mitotic corneal endothelial cells are used as a reference. The movement and fate of marked cells are tracked over time by re-imaging the same areas of the eye. (B) Low magnification, widefield fluorescent image of the eye showing the location of photo-labeled cells (left panel). Full-thickness projections of serial optical sections acquired by 2-photon microscopy (right panels). Basal corneal progenitors (magenta) co-localize with endothelial cells (yellow) immediately after photo-activation, but show uniform centripetal translocation over time. Also see Movie S4. (C) Quantification of cellular translocation rates across the ocular surface epithelium (n = 12 tracked groups of labeled cells from 3 mice, ****p < 0.0001). (D) Basal cells in the limbal area are marked in a checkered pattern to capture local cellular dynamics. Pattern deformation shows centripetal expansion from the inner limbus, but not the outer limbus or conjunctiva. The collagen fiber organization (SHG) in the underlying limbal stroma is shown for positional reference. Scale bars: 200 μm.
Photo-labeled groups of cells in the limbus displayed two distinct behaviors. Marked cells in the outer limbus did not display lateral mobility or expansion (Fig. 2D). However, the group of marked cells in the inner limbus retained their presence at their original location but also expanded centripetally (Fig. 2D). To quantify the cellular turnover within each compartment, we measured the rate of label dilution due to proliferation and differentiation (Fig. S3B, C). Basal cells labeled in the outer limbus showed a slower overall turnover compared to those in the inner limbus (Fig. S3C). Taken together, our data directly demonstrate distinct cellular dynamics within the limbal niche. These results further indicate that stem cells in the inner limbus are likely the sole source of basal cells that populate the rest of the cornea during homeostasis and that the corneal progenitors exiting the niche are short-lived and have finite contributions to tissue maintenance.
Slow-cycling stem cells reside in the outer limbus
Although slow-cycling stem cell populations have been reported in various regenerating organs, including the cornea, their activity in vivo remains unresolved (Amitai-Lange, Altshuler, et al. 2015), (Parfitt et al. 2015), (Sartaj et al. 2017). Therefore, we next used our 2-photon imaging system to visualize individual label-retaining cells to define their precise location within the limbus and determine their respective behaviors by directly tracking them over time. For this, we devised a genetic system that combined inducible Cre-LoxP and Doxycycline alleles (ROSA26-LoxP-STOP-LoxP-tTA and TetO-H2BGFP) (Fig. 3A) (Farrelly et al. 2019). Using the p63CreERT2 driver, we first induced the expression of the H2B-GFP fusion reporter in all basal cells in the epithelium. We then imaged the eyes to validate that basal cells in all epithelial compartments expressed the reporter at equivalent levels of intensity (Fig. 3B). After this initial imaging time point, Doxycycline was introduced to the diet to suppress the expression of the H2B-GFP. During the chase period, we continued to re-image the same eyes at regular intervals and quantified the gradual dilution of the H2B-GFP signal in the actively cycling epithelial populations (Fig. 3B, C). After a one month chase period, only cells that divided the least were still visible.
Figure 3. Capturing the activity of slow-cycling cells in the live limbal niche.
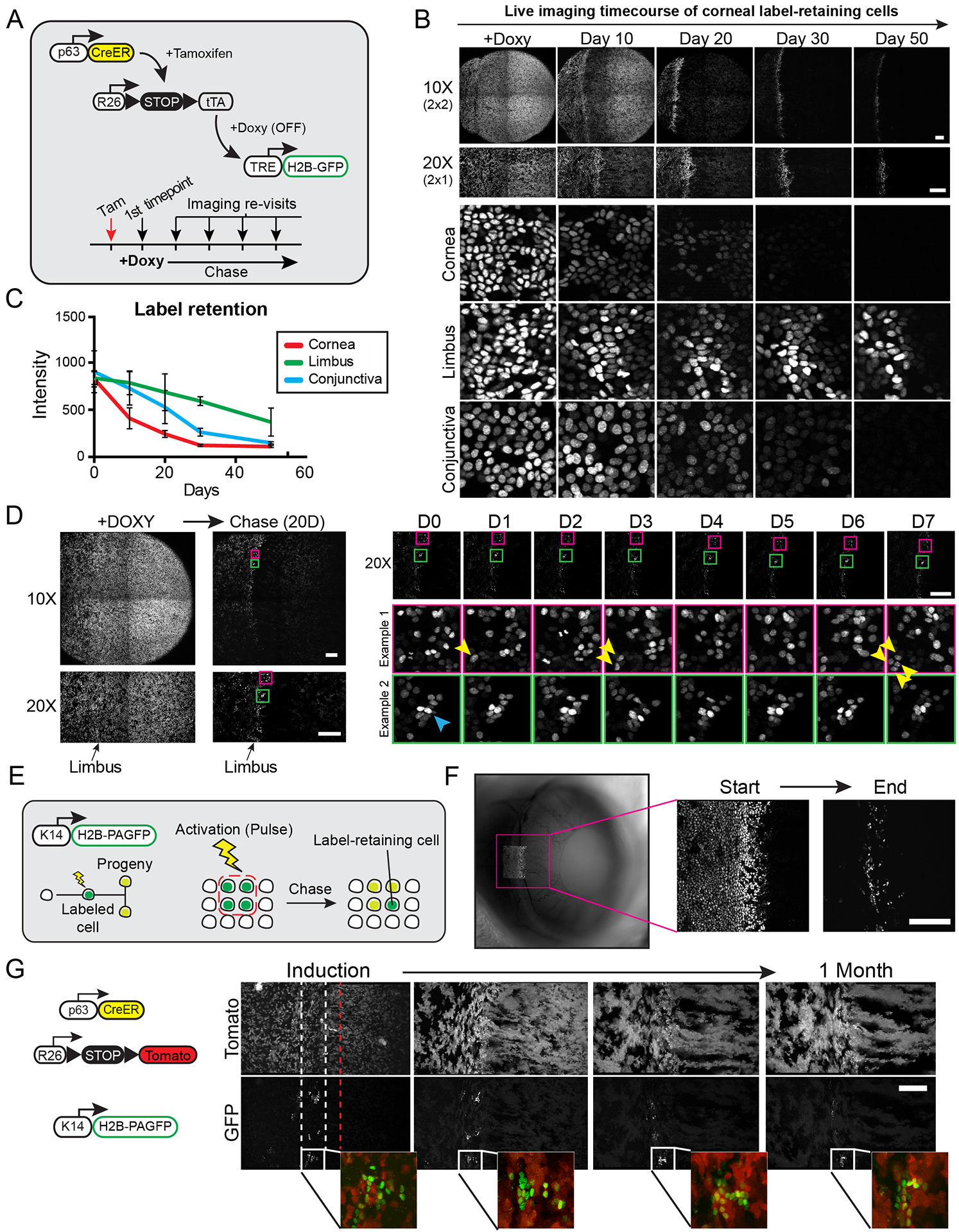
(A) Experimental strategy and genetic alleles used to visualize slow-cycling, label-retaining cells in the anterior ocular epithelium. Tamoxifen induces the uniform expression of the H2B-GFP fusion reporter in all basal cells. Addition of Doxycycline to the diet suppresses the expression of the reporter that is subsequently diluted among daughter cells after each cell division. (B) Global and high-magnification views of the eye imaged at the indicated time points following the addition of Doxycycline. (C) Quantification of label retention in cells within the indicated epithelial compartments (n = 15 sampled images per time point from 2 mice, p < 0.001; 2-way ANOVA). (D) Examples capturing the activity of slow-cycling limbal cells in real time by live imaging. Label-retaining cells in the limbus are imaged at single-cell resolution after a 20-day chase period, and the same cells are re-imaged at daily intervals. Yellow arrows show a label-retaining cell undergoing two symmetric cell divisions. The cyan arrow indicates a cell that remains quiescent during the entire time course. Also see Movie S5. (E) Experimental strategy and genetic alleles to visualize label-retaining cells in the limbus using an in vivo photo-activatable reporter. H2B-PAGFP expressing cells in the limbal area are marked by photo-activation and the same cells are re-imaged after a chase period. (F) Low magnification, widefield fluorescent image of the eye immediately after photo-labeling cells in the limbal area (left panel). High magnification views of the same photo-labeled cells at the beginning and end of the chase period acquired by 2-photon live imaging (right panels). (G) Representative images of the limbal area taken at weekly intervals after Tamoxifen induction. Prior to induction, cells in the limbus were photo-labeled and chased to reveal label-retaining cells. Dotted lines indicate the margins of the outer (white) and inner (red) limbus. Panels B, D, F and G show tiled images of the cornea and limbus constructed from multiple fields-of-view. Scale bars: 200 μm.
The ability to directly visualize the slow-cycling, label-retaining cells allowed us to capture their behavior in vivo and analyze their fates in real time. For this, we repeated the pulse-chase regimen to visualize the label-retaining cells and then re-imaged the same areas of the limbus in 24-hour intervals for up to one week (Fig. 3D; Movie S5). Analysis of the imaging data revealed heterogeneity within the behaviors of the label-retaining population. For instance, we observed label-retaining cells undergo cell divisions where invariably, the daughter cells remained in the same position and did not undergo terminal differentiation (Fig. 3D; Movie S5). This indicated that these cell divisions were symmetric in terms of fate. Other label-retaining cells remained quiescent and stationary throughout the imaging period (Fig. 3D; Movie S5). Furthermore, we captured a few label-retaining cells spontaneously undergo terminal differentiation and shedding from the tissue. Our analysis did not reveal any measurable mobility of the label-retaining cells towards the cornea.
A still untested hypothesis is whether the slow-cycling, label-retaining cells in the limbus are the source of the progenitors that maintain corneal homeostasis. To directly test this, we designed an experiment to co-localize label-retaining cells with the stem cells at the limbal origin of the long-lived radial clones in the cornea. For this, we engineered transgenic mice that express a photo-activatable GFP fused to Histone-H2B under the control of a Keratin 14 promoter (K14-H2B-PAGFP), which shows particularly high activity within the limbus (Park et al. 2019), (Pajoohesh-Ganji et al. 2016). Photo-activating the reporter produces a robust nuclear signal that is stable and is diluted equally among daughter cells after each cell division (Fig. 3E). After a 50-day chase period, we confirmed that GFP+, label-retaining cells were present only in the limbus (Fig. 3F). We then combined the H2B-PAGFP photo-activatable reporter with the p63Tom lineage tracing alleles. Consistent with our previous results, we observed centripetally expanding clones that began to emerge from the inner limbus soon after tamoxifen induction (Fig. 3G). Photo-activated, label-retaining cells co-localized with the small, stationary clones in the outer limbus and were not part of the lineages that emerged from the inner limbus (Fig. 3G). Therefore, our data thus far support a previously unrecognized bi-compartmentalized organization of the limbal niche and show that during homeostasis, label-retaining cells are likely functionally distinct from stem cells that support corneal maintenance.
Compartmentalization of corneal cell fates revealed at the single-stem-cell level
Evidence from the skin and other organs support the hypothesis that the compartmentalization of stem cell fates is critical for homeostatic maintenance and tissue regeneration (Fleming et al. 1993), (Ito et al. 2005), (Levy et al. 2005), (Clevers 2009). To test whether the cornea conforms to this model, we analyzed individual cell fate choices in the limbus and cornea by directly capturing the lifecycle of stem cells and their progeny at the single-cell level. This required the use of a genetic driver that preferentially labels stem cells in the limbus, even at very low induction levels. To accomplish this, we tested proposed limbal stem cell markers by in vivo lineage tracing and longitudinal live imaging (Fig. S4A–D). The Keratin 15 (K15CrePR) driver marked cells in the limbus and conjunctiva, but most of the labeled clones quickly committed to terminal differentiation and disappeared from the tissue within one month after induction (Fig. S4A). Keratin 19 (K19CreER) also marked cells in the limbus and conjunctiva, with a small number of clones persisting in the outer limbus after one month, indicating stem cell potential (Fig. S4B). However, both drivers failed to mark the stem cells in the inner limbus that generate the long-lived, radial corneal lineages. Lrig1 marks stem cells in the epidermis and evidence suggests that it is also expressed in limbal stem and progenitor cells (Jensen et al. 2009), (Nakamura et al. 2014), (Kaplan et al. 2019). Lineage tracing using the Lrig1CreER driver confirmed higher specificity, even at low induction levels, for marking cells in the outer and inner limbus that produced long-lived clones (Fig. S4C).
Validation of the Lrig1 marker allowed us to label single basal cells in the limbus and cornea that were well separated and easily identified in consecutive live imaging sessions. We first evaluated the stem cell potential of the labeled cells by directly following them over several weeks and visualizing their ability to self-renew and generate differentiating progeny. Consistent with our previous data, with this strategy, we confirmed that among the marked cells were cells in the outer limbus that were slow-cycling and generated small clones that remained within their compartment (Fig. S5). Single Lrig1+ cells were also marked in the inner limbus. These continued to self-renew, while retaining their original point of origin in the niche and generated progeny that gradually expanded into the cornea (Fig. S5). Most importantly, our live imaging approach enabled us to capture the sequential steps in the lifecycle of these cell populations and use these data to retrospectively analyze the fate decisions of the respective founder stem or progenitor cell and the fates of their immediate progeny (Fig. 4A).
Figure 4. Direct quantitative fate analysis of single limbal stem cells.
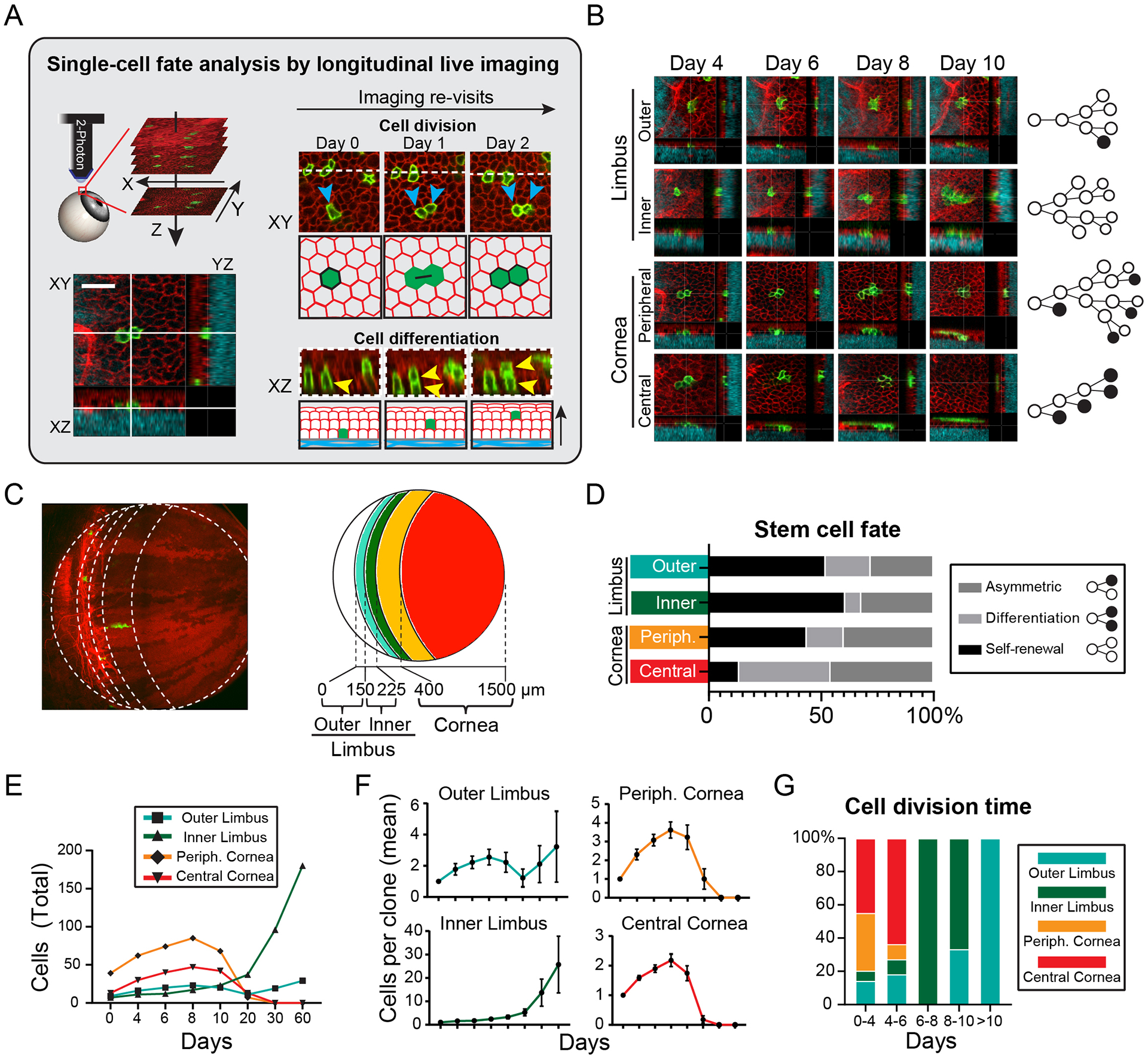
(A) Experimental strategy to capture and quantify fates at the single-cell level by live imaging. The area where individually marked stem cells are located is imaged with a series of optical sections capturing all the layers of the epithelium. The exact same areas are then re-imaged at regular time intervals and the fate decisions of each stem cell is directly captured and analyzed. (B) Representative examples of tracking the fates of single cells in the limbus and cornea. Lineage trees are constructed from analyzing the cell division and differentiation events between each time point. (C) Diagram showing the compartmentalization of the limbus and cornea based on the stratification of the clonal analysis data. (D) Quantification of the relative distribution of captured cell fates within the indicated compartments of the limbus and cornea (n = 167 fate events analyzed in 68 lineage trees from 2 mice, p < 0.002; 2-way ANOVA). (E) Quantification of aggregate cell number in followed lineages over the entire tracking period (n = 544 cells in 68 clones, p < 0.001; 2-way ANOVA). (F) Quantification of average cell number per tracked clone. (n = 9,7, 13, 39 clones analyzed per cell compartment). (G) Quantification of the relative distribution of cell division time within the indicated compartments of the limbus and cornea (n = 66 cell divisions, p < 0.001, 2-way ANOVA). Panel C shows a tiled image of the cornea and limbus constructed from multiple fields-of-view. Scale bar: 20 μm.
We analyzed the lifecycle of around 70 individual stem and basal progenitor cells in the limbus and cornea and constructed detailed lineage trees by quantifying the cell division and terminal differentiation events within their progeny (Fig. 4B). Stratification of the fate analysis data, based on the distance of the founder cell from the limbal border with the conjunctiva, showed four distinct behaviors (Fig. 4C, D). Stem cells in the outer limbus self-renew and persist long-term, but display limited growth (Fig. 4D–F). Stem cells in the inner limbus undergo mostly symmetric cell divisions and exhibit the greatest potential for long-term growth (Fig. 4D–F). We also found differences between the transient corneal progenitors. Those closer to the periphery showed higher propensity to divide symmetrically and expand, while progenitors in the central cornea were more likely to terminally differentiate and exhaust (Fig. 4D–F). The differences between these populations were also reflected in the average time between each cell division. Stem cells in the outer limbus were the slowest cycling and progenitors in the peripheral cornea were the most active (Fig. 4G). Overall, this analysis revealed that the fates of individual cells can be accurately predicted depending on their original location within the tissue, confirming our hypothesis that the cornea and its limbal stem cell niche is functionally compartmentalized.
The corneal epithelium displays a unique mode of terminal differentiation
Our data thus far have demonstrated differences in the fate choices of cells across the cornea, where more symmetric divisions, and therefore more basal cells, are generated in the periphery versus increased differentiation towards the center. This then raised the question: how does the tissue coordinate the terminal differentiation process to supply the necessary suprabasal layers that perform the critical barrier function for the tissue? To resolve the cellular process of terminal differentiation for committed corneal progenitors, we performed 3-dimensional analysis of the transit trajectories of differentiating cells from the point of their delamination and departure from the basal layer until their sloughing from the most superficial layer. We found that the transit time, which averaged around 4–5 days, did not vary significantly between differentiating cells in all corneal compartments (Fig. 5A). However, the fewer number of suprabasal layers and the lower overall cell density in the limbus indicates that the effective rate of stratification is slower in the limbus compared to the rest of the cornea (Fig. 5A, B). Analysis of the vertical paths of the terminally differentiated cells transiting through the suprabasal layers revealed a previously undescribed behavior. While terminally differentiated cells in the limbus transited in a direct upwards trajectory, those in the cornea followed an arced path through the suprabasal layers in an unexpected centrifugal direction (Fig. 5C; Movie S6).
Figure 5. Differentiation dynamics of limbal stem cells and corneal progenitors.
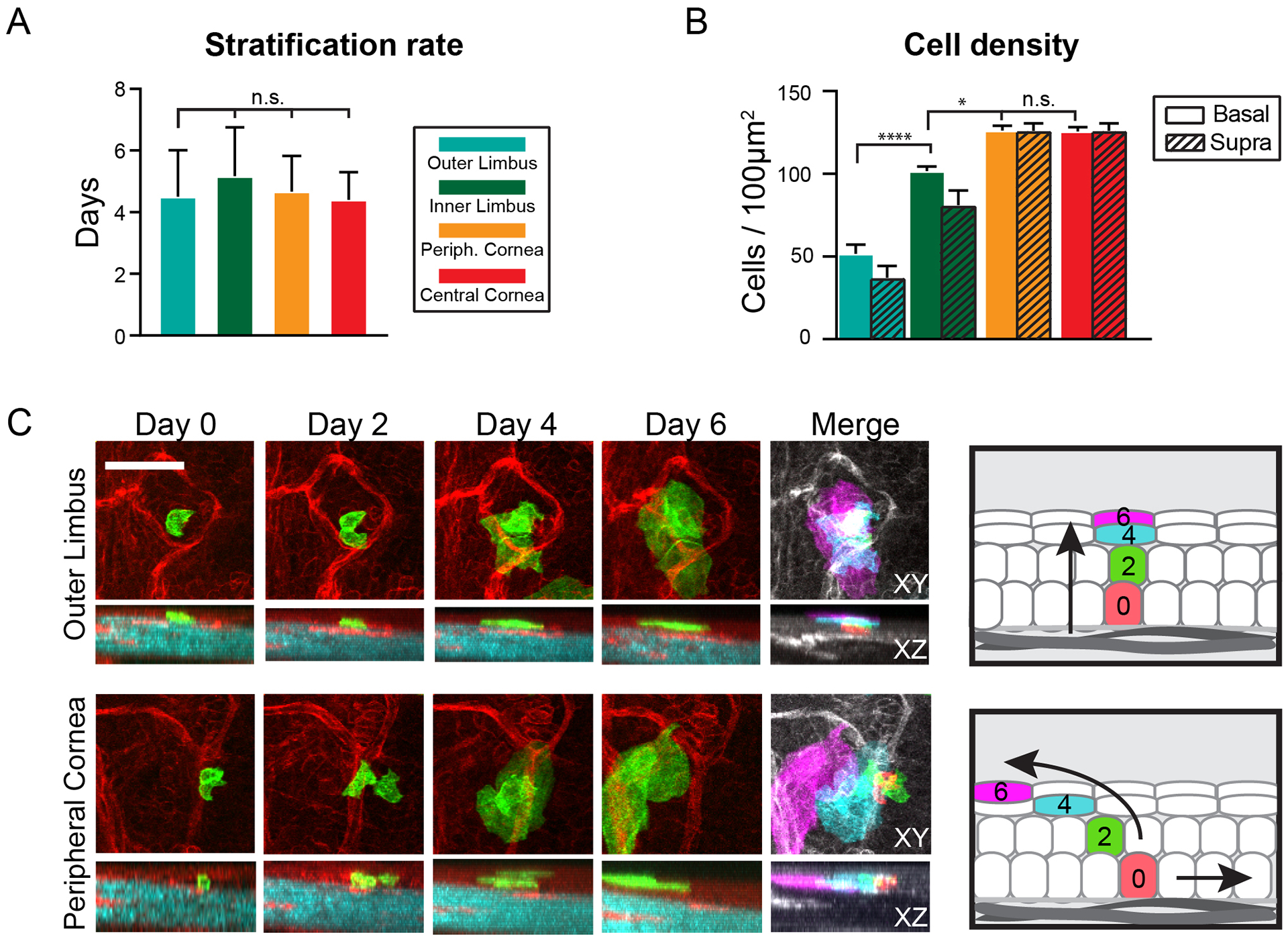
(A) Quantification of stratification rates of terminally differentiated cells in the ocular surface epithelium. The time between the departure from the basal layer and desquamation of a terminally differentiating cell is measured and averaged for each epithelial compartment (n = 32 tracked cells from 2 mice, p = 0.75; 1-way ANOVA). (B) Quantification of the total number of basal and suprabasal cells per area, measured for each epithelial compartment (n = 24 images analyzed from 3 mice, p < 0.001; 2-way ANOVA). (C) Representative examples of directly tracking terminally differentiating cells as they transit across the suprabasal layers before desquamation. The pseudo-colored images are projections of all time points and are used to emphasize the relative lateral and vertical positions of the tracked cells during all stages of terminal differentiation. Also see Movie S6. Scale bars: 50 μm.
Diverse contributions of limbal stem cells to corneal maintenance and wound healing
To this point, our experiments provided evidence to support the existence of at least two functionally distinct stem cell populations in the limbus. Furthermore, these results indicated that stem cells in the inner limbus are the likely the sole source of the transient progeny during corneal homeostasis. To evaluate the requirement of stem cells in the inner limbus to support the turnover of corneal progenitors, we utilized a femtosecond pulsed laser to specifically ablate the stem cells in situ, while preserving the integrity of the surrounding niche and preventing a tissue-wide injury response (Fig. 6A; Fig. S6). For this experiment, we first performed lineage tracing for several weeks to generate robust radial clones that extended all the way to the center of the cornea. After imaging the eyes at high resolution to capture the position of all the long-lived lineages, we proceeded to specifically ablate cells of the labeled radial clones located within the inner limbus (Fig. 6A, B, S6A). After ablation, we re-imaged the same eyes regularly over the course of several weeks and observed that the corneal clones failed to sustain growth and regressed centripetally following the removal of their stem cells (Fig. 6B, C, S6A, B; Movie S7). In complementary experiments, we also ablated basal progenitors away from the inner limbus, which did not impede the growth of their respective corneal lineage (Fig. S6C, D). Surprisingly, the centripetal motion of the progeny downstream of the ablation site continued until those cells were eventually exhausted through terminal differentiation (Fig. S6C, D).
Figure 6. Testing the requirement of limbal stem cells for corneal homeostasis and wound healing.
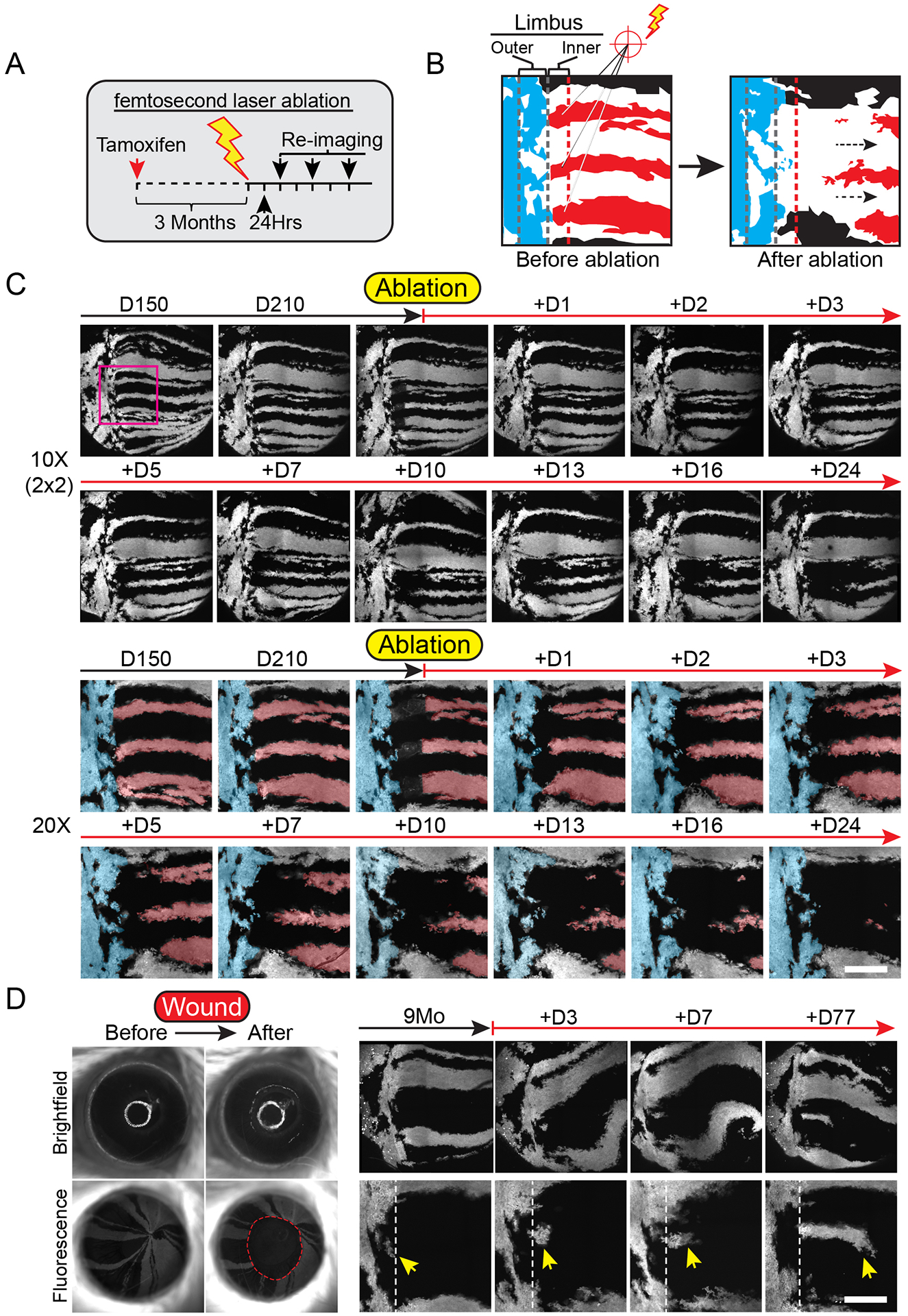
(A) Experimental strategy to test the requirement of stem cells in the inner limbus in sustaining the homeostatic maintenance of their corneal-fated progeny. (B) Diagram depicting a high magnification view of the limbus before and after ablating inner limbal clones (red). Outer limbal clones are shown in blue, intact inner limbal clones are shown in black. (C) Regression of corneal lineages after ablation of their respective stem cells in the inner limbus. Example shows global and high-magnification live views of the eye and limbus, imaged at the indicated time points, before and after the ablation of stem cells in the inner limbus. A pseudo-colored overlay is used to demarcate labeled clones in the outer limbus (blue), as well as the ablated (red) and intact (black) clones in the cornea. Also see Movie S7. (D) Imaging time course after corneal epithelial debridement wound. Yellow arrow indicates cells in the outer limbus entering the cornea. Panels C and D show tiled images of the cornea and limbus constructed from multiple fields-of-view. Scale bars: 20 μm.
Under these conditions, outer limbal stem cells did not cross over into the cornea, but cells from proximal inner limbal lineages compensated for the loss of their ablated counterparts (Fig. 6C; Fig. S6E). We then tested the hypothesis that stem cells in the outer limbus may contribute to the regeneration of the cornea after extensive epithelial injury. For this, we performed a scrape wound in the central cornea and tracked the response of cells located in the outer limbus. We found evidence that clones from the outer limbus exited their compartment and gradually expanded into the cornea during the wound healing process (Fig. 6D; Fig. S7). These radially expanding lineages that emerged from the outer limbus, persisted in the cornea for several weeks after wounding (Fig. 6D). Taken together, our data demonstrate the cellular activities that sustain corneal homeostasis and support a model for the compartmentalization of the limbus into at least two distinct stem cell populations with diverse function in corneal maintenance and wound healing (Fig. 7).
Figure 7. Model for the spatiotemporal organization of stem cell activity during corneal homeostasis.
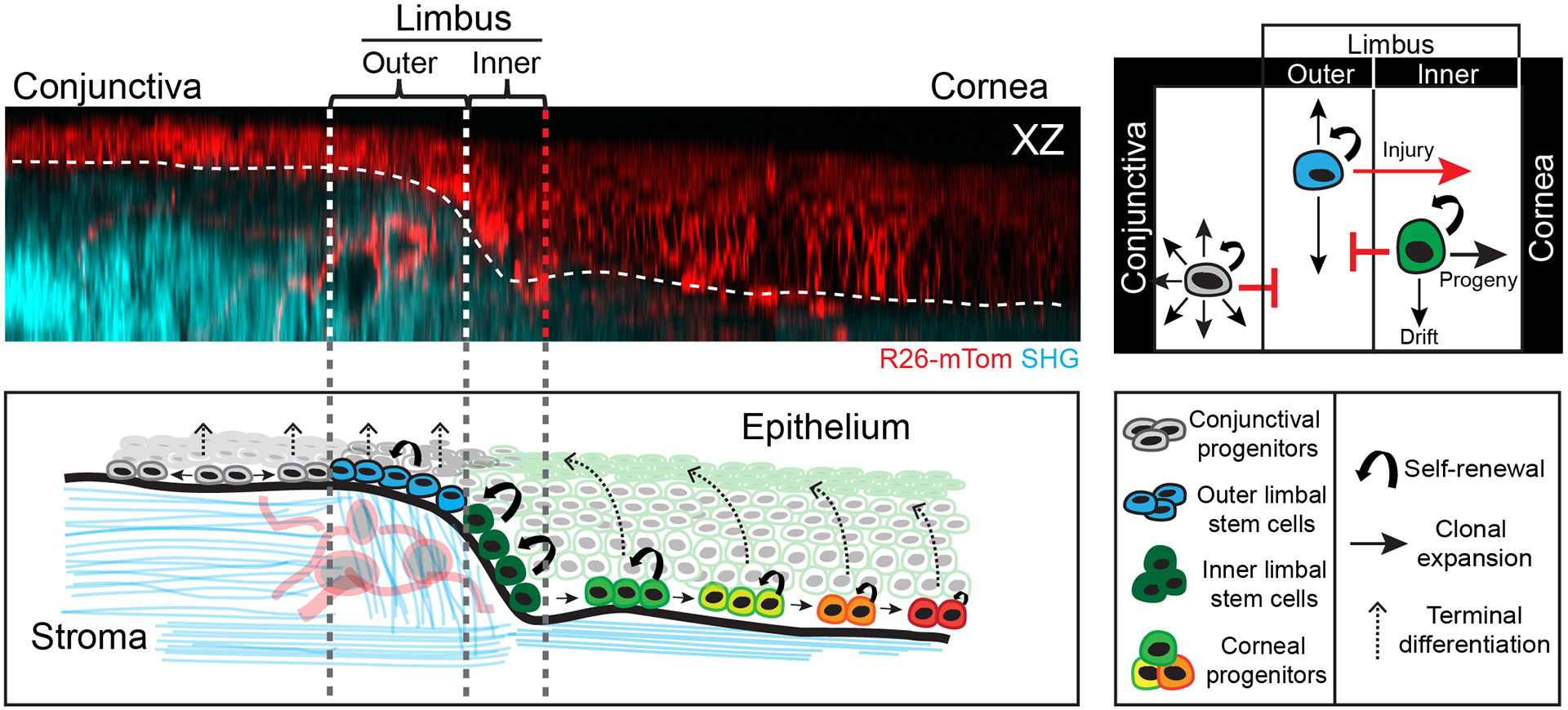
Proposed model of the bi-compartmentalized stem cell organization of the limbal niche based on direct visualization of their long-term clonal dynamics, the quantification of individual fate choices and their respective contributions to corneal homeostasis. Stem cells in the outer limbus are slow-cycling and display local clonal dynamics. Stem cells in the inner limbus undergo primarily symmetric cell divisions and are required to sustain the progenitors that support the homeostatic maintenance of the corneal epithelium. Progenitors that exit the niche transit centripetally within the basal layer, in unison, switching to mostly asymmetric cell divisions as they move closer to central cornea. Terminally differentiating cells in the cornea leave the basal layer and transit upwards in a centrifugal trajectory before they are shed from the tissue.
Discussion
Surface epithelia exhibit a continuous turnover of cells generated in the basal layer that replace those shed from the surface. This process generates vertical columns of cells with common clonal origin (Ghazizadeh & Taichman 2001). In the epidermis and the esophagus, lineage tracing experiments show that the size of these columns may fluctuate over time due to neutral cellular competition, but their overall topology remains unchanged (Clayton et al. 2007), (Mascré et al. 2012), (Doupé et al. 2012), (Rompolas et al. 2016), (Piedrafita et al. 2020). In striking contrast, corneal lineages additionally exhibit centripetal expansion, which indicates a secondary flow of cells within the basal layer from the periphery towards the center of the tissue (Collinson et al. 2002), (Mort et al. 2009), (Di Girolamo et al. 2015), (Amitai-Lange, Altshuler, et al. 2015), (Dorà et al. 2015), (Kasetti et al. 2016), (West et al. 2018). Our data show an apparent imbalance between the fates of basal cells located in the periphery of the tissue versus those at the center. Stem cells in the periphery are more likely to divide symmetrically and increase their number, creating positive pressure for lateral expansion within the basal layer. We also find that basal progenitors transiting towards the center of the cornea are more likely to undergo asymmetric cell divisions or terminally differentiate. This effectively equalizes the pressure from the periphery and maintains balance in the overall cell number across the epithelium. However, it is still unclear what prevents clones in the inner limbus from expanding in both lateral directions, and into the conjunctiva as well. It is possible that cells in the outer limbus are required to maintain a barrier between the cornea and conjunctiva, since such functionally distinct stem cell populations appear to be common in transition areas between epithelia in other organs (Runck et al. 2010). Nonetheless, further work is necessary to understand the intrinsic and extrinsic mechanisms that dictate the compartmentalization of different cell fates.
Since the discovery of slow-cycling, label-retaining cells in the limbus, there has been intense interest to identify these cells and investigate their behaviors in vivo. With the use of RNA sequencing technologies, several studies have curated lists of genes that are enriched in this slow-cycling population (Parfitt et al. 2015), (Sartaj et al. 2017). Yet there has been limited insight into how these cells behave in their native environment. Here, we find that these cells self-renew within the outer limbus and do not appear to contribute to the formation of the lineages that maintain the corneal epithelium during homeostasis. This is in contrast to what is observed in other tissues, such as the hair follicle, where cells in the bulge remain slow cycling, but active, during the growth phase of the hair cycle and directly contribute to progeny that are needed for hair follicle regeneration, or the hematopoietic system, where rare hematopoietic stem cells occasionally give rise to multipotent progenitors during unperturbed hematopoiesis (Cotsarelis et al. 1990), (Tumbar et al. 2004), (Rompolas et al. 2013), (Wilson et al. 2008), (Foudi et al. 2009), (Busch et al. 2015), (Rodriguez-Fraticelli et al. 2018). Though it is tempting to devise a similar cohesive model for the cornea, where largely quiescent cells in the outer limbus give rise to cells in the inner limbus at low rates, this does not appear to take place under normal physiologic conditions. Based on the topology of the lineages in our long-term tracing experiments, we find that clones in the outer and inner limbus remain well-segregated during homeostasis. However, there is sufficient evidence, including data from this study, to suggest that the compartmentalization of the limbus breaks down during extensive corneal injury. Slow-cycling cells in the outer limbus appear to display plasticity and may serve as a reserve stem cell population, where they enter the cell cycle and exit their niche to aid in corneal repair (A. J. Huang & Tseng 1991), (Richardson et al. 2017), (Park, Richardson, Pandzic, Lobo, Lyons, et al. 2019), (Pellegrini & De Luca 2014).
It is not fully clear how the tissue adapts when there are varying degrees of damage to the limbus. A recent paper showed that after disruption of the limbus, including the underlying stroma, corneal-committed cells can dedifferentiate and move from the corneal epithelium back into the limbal niche (Nasser et al. 2018). In our laser-induced cell ablation experiments, we did not observe de-differentiation of p63+ corneal progenitors or the reversal of their typical centripetal movement. Instead, after local ablation of stem cells in the inner limbus, only cells from separate, but proximal lineages enter the ablation site and begin to grow towards the center of the eye. The differences we observe may be due to the techniques used to deplete stem cells in the limbus, where here, we employed a more precise and localized method of ablation that causes minimal damage to the stromal niche, and therefore an attenuated injury response. This then raises the question of what signals the stromal niche provides to maintain stem cell identity and fate. Future work will be necessary to delve deeper into what occurs when the strict compartmentalization of the anterior ocular surface collapses and what signals specific stem cell populations to compensate for these changes.
One unexpected finding from our study was the unconventional way terminally differentiating cells in the cornea transit towards the suprabasal layers. Early work in the epidermis described the three-dimensional organization of the stratified epithelium, and based on detailed histological analyses, proposed that the tissue is composed of discrete vertical epidermal proliferative units (EPU) (Ghazizadeh & Taichman 2001), (Potten 1974), (Mackenzie 1997). More recent experiments that used intravital imaging to visualize the mouse skin amended this theory, showing that the progenitors at the base of these units behave stochastically (Rompolas et al. 2013). However, upon commitment to terminal differentiation, these cells transit through the suprabasal layers using existing columnar units. The cornea appears to diverge from this organization, as we find no evidence for the presence of anatomical structures that resemble discrete columns of suprabasal cells. This is further confirmed by our observation that the upward trajectory of the terminally differentiated cells is curved towards the periphery of the cornea. Though this behavior seems counterintuitive, it illustrates the fluid nature of the differentiated corneal layers and also implies dynamic adhesions between cells, considering their critical barrier function. The functional significance of this differentiation behavior is unclear. One hypothesis is that this results from mechanical forces generated from the unique biomechanics of the cornea (Eberwein & Reinhard 2015), (Gouveia et al. 2019). Alternatively, this may be to satisfy the demand for suprabasal cells in the periphery of the cornea, where basal progenitors are more likely to divide than terminally differentiate.
Ocular trauma and diseases affecting the cornea are a major cause of blindness worldwide (Whitcher et al. 2001), (Burton 2009), (Oliva et al. 2012). Currently, corneal graft or transplant is the only curative treatment for patients with advanced corneal disease and for some, these treatment options are not viable or are met with complications (Singh et al. 2019). While the etiology of ocular surface disease is complicated and includes a multitude of infectious and inflammatory diseases, it predominately manifests through abnormal stem cell proliferation and/or differentiation. Ongoing efforts to harness the potential of limbal stem cells for cell-based therapies hold immense promise (Rama et al. 2010), (Rama et al. 2017), (Stern et al. 2018). A comprehensive understanding of how stem cells coordinate their fate and interact with their surrounding microenvironment to support tissue homeostasis is necessary to continue developing more effective therapies that address corneal blindness. This study provides a paradigm for the intergrative use of live imaging to elucidate fundamental mechanisms of stem cell biology in the mouse corneal epithelium and reconciles longstanding hypotheses for the role of limbal stem cells in corneal homeostasis and regeneration. Furthermore, our study provides a resource for future mechanistic work to elucidate the molecular regulation of limbal stem cells and uncover how changes in their activity lead to disease.
Limitations of the Study
Using 2-photon microscopy to image stem cell niches in live adult mouse organs is a powerful, but relatively new tool. Therefore, certain considerations must be taken due to the complexity and inherent limitations of this technique. One significant challenge is the ability to maintain a complete cohort of animals that were treated and imaged in tandem using identical experimental parameters over the entire longitudinal imaging time course. This results in a relatively small number of biological replicates. This issue is mitigated by conducting supplementary experiments with separate animal cohorts using similar experimental conditions. Inter-mouse variability in the behavior of cell populations likely exists, but to an extent that has not been possible to ascertain in this study. The potential leakiness of an inducible Cre mouse line in the absence of Tamoxifen induction is a general limitation of Cre-recombinase-based lineage tracing experiments. This issue was addressed by imaging the eyes before induction and matching pre-existing labeled clones across time points to exclude them from the analysis.
STAR Methods Text
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Panteleimon Rompolas (rompolas@pennmedicine.upenn.edu).
Materials Availability
Mouse lines generated in this study are available upon request to the Lead Contact.
Data and Code Availability
This study did not generate new datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All procedures involving animal subjects were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania and were consistent with the guidelines set forth by the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. p63CreER mice were created by J. Xu (Baylor College of Medicine) and obtained from A. Vaughan (University of Pennsylvania). K15CrePR mice were obtained from G. Cotsarelis (University of Pennsylvania). K19CreERT mice were created by G. Gu (Vanderbilt University) and obtained from B. Stanger (University of Pennsylvania). Lrig1CreERT2 mice were obtained from S. Millar (University of Pennsylvania). R26loxp-stop-loxp-tdTom, R26loxp-tdTom-stop-loxp-EGFP, R26loxp-nTom-stop-loxp-nGFP, TrpV1Cre, LysMCreERT2, R26loxP-stop-loxP-tTA and TetOH2BGFP mice were obtained from The Jackson Laboratory. R26PAGFP reporter mice were generated by crossing the E2aCre with R26loxp-stop-loxp-PAGFP lines, obtained from The Jackson Laboratory, to achieve germline transmission of the recombined allele and ubiquitous expression of the PAGFP reporter. K14H2B-PAGFP mice were generated by the Center for Animal Transgenesis and Germ Cell Research at the School of Veterinary Medicine of the University of Pennsylvania. All mice that were used in this study were bred for multiple generations into a Crl:CD1(ICR) mixed background. For lineage tracing experiments, Cre activation was induced with a single intraperitoneal injection of Tamoxifen in corn oil (0.1–2 mg per 20 g body weight). Mice were induced between 6–8 weeks old and subsequent experiments were conducted at the indicated times after induction. Experiments included equal representation of males and females. Mice were housed in a temperature and light-controlled environment and received food and water ad libitum. Up to 5 mice of the same sex and similar age were housed in a cage. Mice were provided Bed-o’Cobs (The Andersons Lab Bedding), a porous cob material, as bedding and Shred-n’Rich nestlets (The Andersons Lab Bedding) for nesting and enrichment.
METHOD DETAILS
Generation of K14H2B-PAGFP mice
An H2BPAGFP coding sequence was obtained from the pACAGW-H2B-PAGFP-AAV plasmid by PCR amplification and subcloned into the pG3Z-K14-H2B vector obtained from E. Fuchs (The Rockefeller University) by restriction enzyme digestion with BamHI and XbaI and ligation with T4 Ligase. The final K14-H2BPAGFP transgene was obtained from the resultant plasmid by digestion with KpnI and SphI and injected into blastocysts by the Center for Animal Transgenesis and Germ Cell Research, at the School of Veterinary Medicine of the University of Pennsylvania. Resulting mice were first screened by whole-mount imaging and photo-activation of a freshly obtained ear punch biopsy using our 2-photon microscope. Positive genotypes were confirmed using specific primers for the K14 cassette. A single founder was then selected and crossed with a Crl:CD1(ICR) mixed background breeder to establish the transgenic mouse line.
Intravital imaging of the mouse eye
Preparation of the mice for intravital imaging of the eye was performed with the following amendments to the previously described protocol (Rompolas et al. 2016). Mice were initially anesthetized with IP injection of ketamine/xylazine cocktail (0.1 ml / 20 g body weight; 87.5 mg / kg Ketamine, 12.5 mg / kg Xylazine). A deep plane of anesthesia was verified by checking pedal reflexes. The mouse head was stabilized with a custom-made stereotaxic apparatus that includes palate bar and nose clamp but no ear bars. Precision, 3-axis micro-manipulators are used to adjust the head tilt so that the eye to be imaged is facing up. A drop of eye gel (0.3 % Hypromellose) was used as an optically neutral interface between the eye and a glass coverslip, and to prevent dryness and irritation to the tissue during the anesthesia and imaging procedure. After preparation and mounting is complete, the stage is placed on the microscope platform under the objective lens. A heating pad is used to keep a stable body temperature and vaporized isoflurane is delivered through a nose cone to maintain anesthesia for the duration of the imaging process. After each imaging session, the eyes were rinsed with PBS and the mice were monitored and allowed to recover in a warm chamber before returned to the housing facility.
Imaging equipment and acquisition settings
Image acquisition was performed with an upright Olympus FV1200MPE microscope, equipped with a Chameleon Vision II Ti:Sapphire laser. The laser beam was focused through 10X, 20X or 25X objective lenses (Olympus UPLSAPO10X2, N.A. 0.40; UPLSAPO20X, N.A. 0.75; XLPLN25XWMP2, N.A. 1.05). Emitted fluorescence was collected by two multi-alkali and two gallium arsenide phosphide (GaAsP) non-descanned detectors (NDD). The following wavelengths were collected by each detector: NDD1 419–458 nm; NDD1 458–495 nm; GaAsP-NDD1 495–540 nm; GaAsP-NDD2 575–630 nm. GFP and Tomato reporters were excited at 930 nm and their signal was collected by GaAsP-NDD1 and GaAsP-NDD2, respectively. Second harmonic generation (SHG) signal was generated using 850 nm or 930 nm excitation wavelengths and detected by NDD1 or NDD2, respectively. Serial optical sections were acquired in 2–5 μm steps, starting from the surface of the eye and capturing the entire thickness of the cornea (epithelium ~40 μm, stroma/endothelium ~80 μm). Expanded views of the cornea and limbus were obtained by acquiring a grid of sequential optical fields-of-view that were automatically stitched into one high-resolution tiled image using the microscope manufacturer software. Multi-day tracing experiments were done by re-imaging the same field-of-view or the entire eye at the indicated times after the initial acquisition. For each time point, inherent landmarks within the cornea, including the organization of the vasculature and collagen fibers (SHG), were used to consistently identify the limbus and navigate back to the original regions. Macroscopic images of the mouse eye were acquired under brightfield and fluorescence with an Olympus MVX10 Fluorescent Macro Zoom microscope fitted with Hamamatsu Orca CCD camera for digital imaging.
Photo-labeling
Photo-labeling experiments with the K14H2BPAGFP and R26PAGFP reporter mice were carried out with the same equipment and imaging setup as used for acquisition. The pre-activated form of the H2B-PAGFP and PAGFP fluorescent proteins was visualized by exciting with 850 nm wavelength and emission signal was collected in GaAsP-NDD1 (495–540nm). Excitation with 930 nm verified that no signal is emitted by the reporters before activation. Photo-labeling was achieved by scanning a defined region-of-interest (ROI) at the plane of the basal layer of the epithelium, with the laser tuned to 750nm wavelength, for 5–10 sec, using 5–10% laser power. The z-plane was then moved down to the corneal endothelium, which served as a reference, and the same ROI was used for photo-labeling cells in that layer. Immediately after photo-activation, a series of optical sections, with a range that includes the entire thickness of the cornea, was acquired using the same acquisition settings as for GFP. Visualizing the signal of the activated form of PAGFP only within the ROI confirmed the successful photo-labeling of basal epithelial or endothelial cells. Following the initial image acquisition immediately after photo-labeling, the same eyes were re-imaged at the indicated times to evaluate the changes of the labeled epithelial population and their movements compared to the endothelial reference cell group.
Laser cell ablation
In vivo laser-induced cell ablation was performed using the same femtosecond Ti:Sapphire laser used for fluorescence excitation and imaging. For maximum specificity, a 25X objective lens (XLPLN25XWMP2, N.A. 1.05) was used to focus the laser beam to the basal layer of the ocular epithelium. The laser was guided to scan a small area (~100 μm2), targeting the cells within for ablation with the following parameters: 800 nm wavelength, 100% laser power, 1s exposure. Immediately after ablation, the microscope was switched to imaging mode and a series of serial optical sections was collected to visualize the effect of the laser ablation.
Wounding assay
Corneal epithelial debridement wounds were generated as previously described (Chan & Werb 2015). Mice were initially anesthetized with IP injection of ketamine/xylazine cocktail and their eyes imaged under brightfield and fluorescence microscopy. Mice where then placed on a heating pad and observed under an Olympus SZ61 dissecting microscope. After topical application of Proparacaine, the epithelium from the central part of the cornea was removed with an Algerbrush II ophthalmic brush to generate a scrape wound of 1.5 mm in diameter. The eyes were imaged again before the mice were allowed to recover.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative image analysis
Raw digital files from 2-photon imaging were acquired and saved in the OIB format using the microscope manufacturer’s software (FLUOVIEW, Olympus USA). To capture extended fields-of-view that encompass the entire ocular surface epithelium, including the cornea, limbus and conjunctiva, a tiling method was used to reconstruct a single image from multiple full-thickness serial optical sections using the microscope acquisition software. Typically, to image the entire eye using the 10X objective lens, the microscope defines a square area consisting of 2 × 2 (XY) field-of-view with 10% overlap between them. Using the motorized platform, the microscope automatically acquires the four fields-of-view in a sequential pattern and uses information from the overlapping margins to stitch the individual field-of-view into a single image. Raw image files were imported into ImageJ/Fiji (NIH) using Bio-Formats or to Imaris (Bitplane) for further analysis. For cell counts and quantitative clonal analyses, supervised image segmentation and blob detection was performed on individual optical sections. Identified blobs were manually validated and their number, size and signal intensity as mean grey values were measured.
To quantify population clonal dynamics, following induction, high-resolution optical sections were obtained sequentially and used to construct 3-dimensional tiled views of the entire half of the ocular surface, including parts of the conjunctiva, limbus and central cornea. The same eyes were then re-imaged using identical acquisition parameters. From each time point, the equivalent areas of the conjunctiva, outer limbus and inner limbus+cornea were sampled and processed by supervised segmentation to quantify the number and dimensional parameters of the labeled clones within. Clonal growth anisotropy was analyzed by measuring Feret’s Diameter and Angles from the outlines of individual clones after image segmentation. To quantify the centripetal rate of mobility of photo-labeled basal cells in the cornea, serial optical sections were projected onto a single plane and the distance between the centroids of the epithelial group from the endothelial reference group was measured for each time point. To measure the rates of label retention, the equivalent areas of the conjunctiva, limbus and corneal were sampled for each time point and the mean grey value was measured. For single-cell lineage tracing, individual high-magnification serial optical sections were obtained for each traced clone and 3-dimensional analysis was performed once the entire imaging time course was competed to quantify the number of basal and suprabasal cells in each time point. Departure of a cell from the basal layer and subsequent upward transit was scored as differentiation, while continuous increase in the basal cell number was scored as self-renewal. Clone measurements and tracings for the cell ablation experiments were performed manually. Images shown in figures typically represent maximum projections or single optical sections selected from the z-stacks unless otherwise specified.
Statistical analysis
Sample sizes were not pre-determined, but are similar with what were reported previously (Rompolas et al. 2016). Data were collected and quantified randomly, and their distribution was assumed normal, but this was not formally tested. Lineage tracing, photo-labeling, laser cell-ablation and wounding experiments were successfully reproduced under similar conditions using different mouse cohorts. The data presented in the figures are from a single cohort of at least two mice, imaged in tandem, using identical experimental parameters. The values of “n” (sample size) refer to data points obtained from all mice within the cohort, unless otherwise indicated, and are provided in the figure legends. Statistical calculations and graphical representation of the data were performed using the Prism software package (GraphPad). Data are expressed as percentages or mean ± S.E.M and unpaired Student’s t-test was used to analyze data sets with two groups, unless otherwise stated in the figure legends. For all analyses, p-values < 0.05 were designated as significant and symbolized in figure plots as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, with precise values supplied in figure legends. No data were excluded from the analysis.
Supplementary Material
Movie S1. Intravital 2-photon imaging of the mouse eye. Example of a mouse eye imaged live by 2-photon microscopy. Serial optical sections are obtained in sequential fields of view and the entire anterior segment of the eye can be reconstructed in two or three dimensions at single-cell resolution. High resolution serial optical sections of the limbus and cornea imaged live using the R26mTmG reporter mouse line, which illuminates the membrane of all cells in the tissue, illustrate the fine histological organization of these compartments. Refers to Figure 1A.
Movie S2. Characterization of cellular number and density in the eye by live imaging. Example of serial optical sections of the conjunctiva, limbus and cornea imaged live using the R26nTnG reporter mouse line that illuminates the nucleus of all cells in the tissue. Refers to Figure 1A.
Movie S3. Characterization of extracellular matrix organization in the eye by live imaging. Example of serial optical sections of the conjunctiva, limbus and cornea imaged live using the second harmonic generation imaging (SHG), which illuminates the collagen fibers in the tissue. Refers to Figure 1E.
Movie S4. Visualizing and tracking dynamic cellular behaviors by in vivo photo-labeling. Example of a group of corneal basal progenitors marked by photo-activating the R26PAGFP reporter by 2-photon microscopy. Serial optical sections immediately after labeling show the group of epithelial cells located directly above a similarly sized group of corneal endothelial cells. Four days later, the group of epithelial cells has moved towards the center of the cornea. Refers to Figure 2B.
Movie S5. Visualizing the activity of slow-cycling limbal cells in real time by live imaging. After a chase period to reveal their presence, label-retaining cells in the limbus are imaged at single-cell resolution and the same cells are re-imaged at daily intervals. Yellow arrows show a label retaining cell undergoing two symmetric cell divisions. Blue arrow indicates a cell that remains quiescent during the entire time course. Refers to Figure 3D.
Movie S6. Terminal differentiation trajectories of limbal stem cells and corneal progenitors. Refers to Figure 5C.
Movie S7. Regression of corneal progeny following ablation of the stem cells in the inner limbus. Refers to Figure 6C.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma | Cat# T5648 |
| Ketamine | Midwest Veterinary Supply | Cat# 17033-100-10 |
| Xylazine | Midwest Veterinary Supply | Cat# 310-01150-3 |
| Proparacaine | BAUSCH & LOMB | Cat# 24208-730-06 |
| Genteal Tears Severe (Hydromellose 0.3%) | Alcon Laboratories | NDC- 0065-8064-01 |
| Experimental Models: Organisms/Strains | ||
| Mouse: CD-1 | Charles River | RRID:IMSR_CRL:022 |
| Mouse: p63-CreER | (Lee et al. 2014) | N/A |
| Mouse: Keratin15-CrePR | (Morris et al. 2004) | RRID:IMSR_JAX:005249 |
| Mouse: Keratin19-CreERT | (Means et al. 2008) | RRID:IMSR_JAX:026925 |
| Mouse: Lrig1-CreERT2 | (Powell et al. 2012) | RRID:IMSR_JAX:018418 |
| Mouse: Rosa26-stop-tdTomato | (Madisen et al. 2010) | RRID:IMSR_JAX:007908 |
| Mouse: Rosa26-tdTomato-stop-EGFP | (Muzumdar et al. 2007) | RRID:IMSR_JAX:007676 |
| Mouse: Rosa26-nTomato-stop-nGFP | The Jackson Laboratory | RRID:IMSR_JAX:023035 |
| Mouse: TrpV1-Cre | (Cavanaugh et al. 2011) | RRID:IMSR_JAX:017769 |
| Mouse: LysM-CreERT2 | (Canli et al. 2017) | RRID:IMSR_JAX:031674 |
| Mouse: Rosa26-stop-tTA | (Wang et al. 2008) | RRID:IMSR_JAX:008600 |
| Mouse: TetO-H2BGFP | (Tumbar et al. 2004) | RRID:IMSR_JAX:005104 |
| Mouse: E2a-Cre | (Lakso et al. 1996) | RRID:IMSR_JAX:003724 |
| Mouse: Rosa26-stop-PAGFP | (Peter et al. 2013) | RRID:IMSR_JAX:021071 |
| Mouse: Keratin14-H2B-PAGFP | This study | N/A |
| Recombinant DNA | ||
| Plasmid: pACAGW-H2B-PAGFP-AAV | (Lien & Scanziani 2011) | RRID:Addgene_33000 |
| Plasmid: pG3Z-K14-H2B | Fuchs lab | N/A |
| Plasmid: Keratin 14-H2B-PAGFP | This study | N/A |
| Software and Algorithms | ||
| GraphPad Prism 8 | GraphPad Software | N/A |
| ImageJ | https://imagej.nih.gov/ij/ | N/A |
| Imaris | Oxford Instruments | N/A |
| Fluoview | Olympus | N/A |
| Other | ||
| Algerbrush II fitted with a 0.5mm burr | Accutome | Cat# AM0100KL |
Highlights.
The corneal limbus is organized into two distinct stem cell compartments
Inner limbal stem cells are required to maintain transient corneal progenitors
Outer limbal stem cells display only local clonal dynamics during homeostasis
After injury, outer limbal stem cells contribute to corneal regeneration
Acknowledgments
We thank Michael Rendl, Christopher Lengner, Kenneth Zaret and Yana Kamberov for our invaluable discussions and their critical comments on the manuscript. We are especially grateful to George Cotsarelis for many helpful conversations that guided this study. We thank Elaine Fuchs for donating the K14-cassette vector. We are grateful to Steve Prouty and the Penn Skin Biology Disease Resource Center for their technical support. Funding: O.F. was supported by training grant T32HD083185 from NIH/NICHD. Y.S.H. was supported by training grant T32AR00746536 from NIH/NIAMS. G.R. was supported by training grant T32GM007229 from NIH/NIGMS. P.K. was supported by the American Association for Cancer Research-John and Elizabeth Leonard Family Foundation Basic Cancer Research Fellowship. V.L. was supported by a grant from NIH/NEI (K08 EY025742). P.R. was supported by grants from NIH/NEI (R01EY030599) and from the American Cancer Society (RSG1803101DCC). Penn SBDRC is supported by center core grant P30AR069589 from NIH/NIAMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Amitai-Lange A, Altshuler A, et al. , 2015. Lineage tracing of stem and progenitor cells of the murine corneal epithelium. Stem cells (Dayton, Ohio), 33(1), pp. 230–239. [DOI] [PubMed] [Google Scholar]
- Amitai-Lange A, Berkowitz E, et al. , 2015. A Method for Lineage Tracing of Corneal Cells Using Multi-color Fluorescent Reporter Mice. Journal of visualized experiments : JoVE, (106), p. e53370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belokhvostova D et al. , 2018. Homeostasis, regeneration and tumour formation in the mammalian epidermis. The International journal of developmental biology, 62(6-7-8), pp. 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton MJ, 2009. Prevention, treatment and rehabilitation. Community eye health, 22(71), pp. 33–35. [PMC free article] [PubMed] [Google Scholar]
- Busch K et al. , 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature, 518(7540), pp. 542–546. [DOI] [PubMed] [Google Scholar]
- Canli Ö et al. , 2017. Myeloid Cell-Derived Reactive Oxygen Species Induce Epithelial Mutagenesis. Cancer cell, 32(6), pp. 869–883.e5. [DOI] [PubMed] [Google Scholar]
- Cavanaugh DJ et al. , 2011. Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. The Journal of neuroscience : the official journal of the Society for Neuroscience, 31(13), pp. 5067–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celso Lo, C. et al. , 2009. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature, 457(7225), pp. 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MF & Werb Z, 2015. Animal Models of Corneal Injury. Bio-protocol, 5(13), p. e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X et al. , 2012. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nature Protocols, 7(4), pp. 654–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E et al. , 2007. A single type of progenitor cell maintains normal epidermis. Nature, 446(7132), pp. 185–189. [DOI] [PubMed] [Google Scholar]
- Clevers H, 2009. Searching for adult stem cells in the intestine. EMBO molecular medicine, 1(5), pp. 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson JM et al. , 2002. Clonal analysis of patterns of growth, stem cell activity, and cell movement during the development and maintenance of the murine corneal epithelium. Developmental dynamics : an official publication of the American Association of Anatomists, 224(4), pp. 432–440. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G et al. , 1989. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell, 57(2), pp. 201–209. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT & Lavker RM, 1990. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell, 61(7), pp. 1329–1337. [DOI] [PubMed] [Google Scholar]
- Di Girolamo N et al. , 2015. Tracing the fate of limbal epithelial progenitor cells in the murine cornea. Stem cells (Dayton, Ohio), 33(1), pp. 157–169. [DOI] [PubMed] [Google Scholar]
- Dorà NJ et al. , 2015. Lineage tracing in the adult mouse corneal epithelium supports the limbal epithelial stem cell hypothesis with intermittent periods of stem cell quiescence. Stem cell research, 15(3), pp. 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé DP et al. , 2012. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science (New York, N.Y.), 337(6098), pp. 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwein P & Reinhard T, 2015. Concise reviews: the role of biomechanics in the limbal stem cell niche: new insights for our understanding of this structure. Stem cells (Dayton, Ohio), 33(3), pp. 916–924. [DOI] [PubMed] [Google Scholar]
- Farrelly O, Kuri P & Rompolas P, 2019. In Vivo Genetic Alteration and Lineage Tracing of Single Stem Cells by Live Imaging. Methods in molecular biology (Clifton, N.J.), 1879(Chapter 172), pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming WH et al. , 1993. Functional heterogeneity is associated with the cell cycle status of murine hematopoietic stem cells. The Journal of cell biology, 122(4), pp. 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A et al. , 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nature biotechnology, 27(1), pp. 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh S & Taichman LB, 2001. Multiple classes of stem cells in cutaneous epithelium: a lineage analysis of adult mouse skin. The EMBO journal, 20(6), pp. 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia RM et al. , 2019. Assessment of corneal substrate biomechanics and its effect on epithelial stem cell maintenance and differentiation. Nature communications, 10(1), pp. 1496–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, 1980. The keratinocyte as differentiated cell type. Harvey lectures, 74, pp. 101–139. [PubMed] [Google Scholar]
- Guo ZH et al. , 2018. An Insight into the Difficulties in the Discovery of Specific Biomarkers of Limbal Stem Cells. International journal of molecular sciences, 19(7), p. 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AJ & Tseng SC, 1991. Corneal epithelial wound healing in the absence of limbal epithelium. Investigative Opthalmology & Visual Science, 32(1), pp. 96–105. [PubMed] [Google Scholar]
- Huang S & Rompolas P, 2017. Two-photon microscopy for intracutaneous imaging of stem cell activity in mice. Experimental dermatology, 26(5), pp. 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M et al. , 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature Medicine, 11(12), pp. 1351–1354. [DOI] [PubMed] [Google Scholar]
- Jensen KB et al. , 2009. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell stem cell, 4(5), pp. 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N et al. , 2019. Single-Cell RNA Transcriptome Helps Define the Limbal/Corneal Epithelial Stem/Early Transit Amplifying Cells and How Autophagy Affects This Population. Investigative Opthalmology & Visual Science, 60(10), pp. 3570–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasetti RB et al. , 2016. Study of corneal epithelial progenitor origin and the Yap1 requirement using keratin 12 lineage tracing transgenic mice. Scientific Reports, 6(1), p. 35202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M et al. , 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proceedings of the National Academy of Sciences, 93(12), pp. 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavker RM et al. , 1991. Relative proliferative rates of limbal and corneal epithelia. Implications of corneal epithelial migration, circadian rhythm, and suprabasally located DNA-synthesizing keratinocytes. Investigative Opthalmology & Visual Science, 32(6), pp. 1864–1875. [PubMed] [Google Scholar]
- Lavker RM, Tseng SCG & Sun T-T, 2004. Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Experimental Eye Research, 78(3), pp. 433–446. [DOI] [PubMed] [Google Scholar]
- Lee D-K et al. , 2014. The prostate basal cell (BC) heterogeneity and the p63-positive BC differentiation spectrum in mice. International journal of biological sciences, 10(9), pp. 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy V et al. , 2005. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Developmental cell, 9(6), pp. 855–861. [DOI] [PubMed] [Google Scholar]
- Lien AD & Scanziani M, 2011. In vivo Labeling of Constellations of Functionally Identified Neurons for Targeted in vitro Recordings. Frontiers in neural circuits, 5, p. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo EP et al. , 2016. Self-organized centripetal movement of corneal epithelium in the absence of external cues. Nature communications, 7(1), pp. 12388–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IC, 1997. Retroviral transduction of murine epidermal stem cells demonstrates clonal units of epidermal structure. The Journal of investigative dermatology, 109(3), pp. 377–383. [DOI] [PubMed] [Google Scholar]
- Madisen L et al. , 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience, 13(1), pp. 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Pereira JP & Leblond CP, 1965. Mitosis and differentiation in the stratified squamous epithelium of the rat esophagus. The American journal of anatomy, 117(1), pp. 73–87. [DOI] [PubMed] [Google Scholar]
- Mascré G et al. , 2012. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature, 489(7415), pp. 257–262. [DOI] [PubMed] [Google Scholar]
- Means AL et al. , 2008. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis (New York, N.Y. : 2000), 46(6), pp. 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa KR et al. , 2018. Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell stem cell, 23(5), pp. 677–686.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KA & Lemischka IR, 2006. Stem cells and their niches. Science (New York, N.Y.), 311(5769), pp. 1880–1885. [DOI] [PubMed] [Google Scholar]
- Morris RJ et al. , 2004. Capturing and profiling adult hair follicle stem cells. Nature biotechnology, 22(4), pp. 411–417. [DOI] [PubMed] [Google Scholar]
- Mort RL et al. , 2009. Mosaic analysis of stem cell function and wound healing in the mouse corneal epithelium. BMC developmental biology, 9(1), pp. 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD et al. , 2007. A global double-fluorescent Cre reporter mouse. Genesis (New York, N.Y. : 2000), 45(9), pp. 593–605. [DOI] [PubMed] [Google Scholar]
- Nakamura T et al. , 2014. LRIG1 inhibits STAT3-dependent inflammation to maintain corneal homeostasis. The Journal of clinical investigation, 124(1), pp. 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser W et al. , 2018. Corneal-Committed Cells Restore the Stem Cell Pool and Tissue Boundary following Injury. Cell Reports, 22(2), pp. 323–331. [DOI] [PubMed] [Google Scholar]
- Notara M et al. , 2010. IL6 and the human limbal stem cell niche: a mediator of epithelial-stromal interaction. Stem cell research, 5(3), pp. 188–200. [DOI] [PubMed] [Google Scholar]
- Ohlstein B et al. , 2004. The stem cell niche: theme and variations. Current opinion in cell biology, 16(6), pp. 693–699. [DOI] [PubMed] [Google Scholar]
- Oliva MS, Schottman T & Gulati M, 2012. Turning the tide of corneal blindness. Indian journal of ophthalmology, 60(5), pp. 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoohesh-Ganji A et al. , 2016. K14 + compound niches are present on the mouse cornea early after birth and expand after debridement wounds. Developmental dynamics : an official publication of the American Association of Anatomists, 245(2), pp. 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt GJ et al. , 2015. Immunofluorescence Tomography of Mouse Ocular Surface Epithelial Stem Cells and Their Niche Microenvironment. Investigative Opthalmology & Visual Science, 56(12), pp. 7338–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Richardson A, Pandzic E, Lobo EP, Lyons JG, et al. , 2019. Peripheral (not central) corneal epithelia contribute to the closure of an annular debridement injury. Proceedings of the National Academy of Sciences of the United States of America, 116(52), pp. 26633–26643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Richardson A, Pandzic E, Lobo EP, Whan R, et al. , 2019. Visualizing the Contribution of Keratin-14+ Limbal Epithelial Precursors in Corneal Wound Healing. Stem cell reports, 12(1), pp. 14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G & De Luca M, 2014. Eyes on the prize: limbal stem cells and corneal restoration. Cell stem cell, 15(2), pp. 121–122. [DOI] [PubMed] [Google Scholar]
- Pellegrini G et al. , 1999. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. The Journal of cell biology, 145(4), pp. 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G et al. , 2001. p63 identifies keratinocyte stem cells. Proceedings of the National Academy of Sciences, 98(6), pp. 3156–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M et al. , 2013. Transgenic mouse models enabling photolabeling of individual neurons in vivo. Maravall M, ed. PLoS ONE, 8(4), p. e62132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedrafita G et al. , 2020. A single-progenitor model as the unifying paradigm of epidermal and esophageal epithelial maintenance in mice. Nature communications, 11(1), pp. 1429–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, 1974. The epidermal proliferative unit: the possible role of the central basal cell. Cell and tissue kinetics, 7(1), pp. 77–88. [DOI] [PubMed] [Google Scholar]
- Powell AE et al. , 2012. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell, 149(1), pp. 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rama P et al. , 2010. Limbal stem-cell therapy and long-term corneal regeneration. The New England journal of medicine, 363(2), pp. 147–155. [DOI] [PubMed] [Google Scholar]
- Rama P, Ferrari G & Pellegrini G, 2017. Cultivated limbal epithelial transplantation. Current opinion in ophthalmology, 28(4), pp. 387–389. [DOI] [PubMed] [Google Scholar]
- Richardson A et al. , 2017. Keratin-14-Positive Precursor Cells Spawn a Population of Migratory Corneal Epithelia that Maintain Tissue Mass throughout Life. Stem cell reports, 9(4), pp. 1081–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsma L et al. , 2014. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature, 507(7492), pp. 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli AE et al. , 2018. Clonal analysis of lineage fate in native haematopoiesis. Nature, 553(7687), pp. 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P et al. , 2012. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature, pp. 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P et al. , 2016. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science (New York, N.Y.), 352(6292), pp. 1471–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR & Greco V, 2013. Spatial organization within a niche as a determinant of stem-cell fate. Nature, 502(7472), pp. 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runck LA et al. , 2010. Identification of epithelial label-retaining cells at the transition between the anal canal and the rectum in mice. Cell cycle (Georgetown, Tex.), 9(15), pp. 3039–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartaj R et al. , 2017. Characterization of slow cycling corneal limbal epithelial cells identifies putative stem cell markers. Scientific Reports, 7(1), p. 3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden DT, 2014. Nice neighborhood: emerging concepts of the stem cell niche. Cell, 157(1), pp. 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermer A, Galvin S & Sun TT, 1986. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. The Journal of cell biology, 103(1), pp. 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R, 1978. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood cells, 4(1–2), pp. 7–25. [PubMed] [Google Scholar]
- Senoo M et al. , 2007. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell, 129(3), pp. 523–536. [DOI] [PubMed] [Google Scholar]
- Singh R et al. , 2019. Corneal transplantation in the modern era. The Indian journal of medical research, 150(1), pp. 7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JH et al. , 2018. Regenerating Eye Tissues to Preserve and Restore Vision. Stem Cell, 22(6), pp. 834–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N et al. , 2011. Interconversion between intestinal stem cell populations in distinct niches. Science (New York, N.Y.), 334(6061), pp. 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB et al. , 2006. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes & development, 20(22), pp. 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T et al. , 2004. Defining the epithelial stem cell niche in skin. Science (New York, N.Y.), 303(5656), pp. 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L et al. , 2008. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiology of disease, 29(3), pp. 400–408. [DOI] [PubMed] [Google Scholar]
- West JD et al. , 2018. Computer simulation of neutral drift among limbal epithelial stem cells of mosaic mice. Stem cell research, 30, pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JD, Dorà NJ & Collinson JM, 2015. Evaluating alternative stem cell hypotheses for adult corneal epithelial maintenance. World journal of stem cells, 7(2), pp. 281–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcher JP, Srinivasan M & Upadhyay MP, 2001. Corneal blindness: a global perspective. Bulletin of the World Health Organization, 79(3), pp. 214–221. [PMC free article] [PubMed] [Google Scholar]
- Wilson A et al. , 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell, 135(6), pp. 1118–1129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Intravital 2-photon imaging of the mouse eye. Example of a mouse eye imaged live by 2-photon microscopy. Serial optical sections are obtained in sequential fields of view and the entire anterior segment of the eye can be reconstructed in two or three dimensions at single-cell resolution. High resolution serial optical sections of the limbus and cornea imaged live using the R26mTmG reporter mouse line, which illuminates the membrane of all cells in the tissue, illustrate the fine histological organization of these compartments. Refers to Figure 1A.
Movie S2. Characterization of cellular number and density in the eye by live imaging. Example of serial optical sections of the conjunctiva, limbus and cornea imaged live using the R26nTnG reporter mouse line that illuminates the nucleus of all cells in the tissue. Refers to Figure 1A.
Movie S3. Characterization of extracellular matrix organization in the eye by live imaging. Example of serial optical sections of the conjunctiva, limbus and cornea imaged live using the second harmonic generation imaging (SHG), which illuminates the collagen fibers in the tissue. Refers to Figure 1E.
Movie S4. Visualizing and tracking dynamic cellular behaviors by in vivo photo-labeling. Example of a group of corneal basal progenitors marked by photo-activating the R26PAGFP reporter by 2-photon microscopy. Serial optical sections immediately after labeling show the group of epithelial cells located directly above a similarly sized group of corneal endothelial cells. Four days later, the group of epithelial cells has moved towards the center of the cornea. Refers to Figure 2B.
Movie S5. Visualizing the activity of slow-cycling limbal cells in real time by live imaging. After a chase period to reveal their presence, label-retaining cells in the limbus are imaged at single-cell resolution and the same cells are re-imaged at daily intervals. Yellow arrows show a label retaining cell undergoing two symmetric cell divisions. Blue arrow indicates a cell that remains quiescent during the entire time course. Refers to Figure 3D.
Movie S6. Terminal differentiation trajectories of limbal stem cells and corneal progenitors. Refers to Figure 5C.
Movie S7. Regression of corneal progeny following ablation of the stem cells in the inner limbus. Refers to Figure 6C.
Data Availability Statement
This study did not generate new datasets or code.


