Abstract
Purpose
Growing evidence has demonstrated an indispensable role for N 6‐methyladenosine (m6A) in human diseases, but the copy number variations (CNVs) of m6A regulatory genes in bladder cancer (BLCA) remains largely unknown.
Methods
We investigated the CNVs on all known m6A regulatory genes using the Cancer Genome Atlas (TCGA) database. The association between CNV events and clinicopathological as well as molecular characteristics of BLCA patients were explored. Gene set enrichment analysis (GSEA) was implemented to reveal relative cellular processes. Association between m6A regulatory genes and immune infiltrates was analyzed by The Tumor Immune Estimation Resource (TIMER) database.
Results
CNV events of m6A regulatory genes were frequently observed in BLCA. CNVs of METTL3, METTL14, and METTL16 correlated with molecular characteristics of BLCA patients including TP53 mutation. CNVs of METTL3 associated with the overall survival (OS) of BLCA patients. METTL3 was also associated with several cancer‐related cellular processes, including mitotic spindle assembly, G2/M checkpoint, and E2F targets signaling pathway. Besides, the CNVs of m6A regulatory genes were correlated with specific kinds of immune infiltrates.
Conclusions
There are significant correlations between m6A regulatory genes with CNVs and clinicopathological characteristics. METTL3 with CNVs were associated with the immune infiltrates and performed as a prognostic marker in BLCA.
Keywords: bladder cancer, copy number variation, immune cells, N 6‐methyladenosine, prognosis
We investigated the CNV events on all known m6A regulatory genes which were obtained from the Cancer Genome Atlas (TCGA) database through bioinformatics analysis of prognosis as well as immune infiltrates of BLCA patients. We found that CNVs of METTL3 associated with the overall survival (OS). Furthermore, RNA methyltransferases may affect the outcome of tumor immunotherapy via regulating immune infiltrates.
1. INTRODUCTION
Bladder cancer (BLCA) is one of the most common tumors with high morbidity and mortality worldwide. 1 BLCA is the eighth most frequently diagnosed cancer in men in 2018, accounting for approximately 4% of all cancer‐related deaths in the United States. 2 There are two major classifications of BLCA. According to histological features, BLCA is classified as urothelial carcinomas, squamous cell carcinoma, small‐cell carcinoma, and adenocarcinoma. 3 Urothelial carcinomas account for approximately 90% of all diagnosed patients. 4 There is another classification according to whether the tumors invade the detrusor muscle. They are classified as non‐muscle‐invasive bladder cancer (NMIBC, approximately 75%) and muscle‐invasive bladder cancer (MIBC, approximately 25%). 5 The latter classification is widely used in clinical practice relatively 1 since the prognosis of two subtypes varies greatly. Usually, NMIBC is treated with transurethral resection, and followed by intravesical Bacillus Calmette‐Guerin (BCG) or intravesical chemotherapy. While MIBC is typically treated with radical cystectomy and neoadjuvant chemotherapy because of higher rates of progression and recurrence. 6 Despite the progress in surgical techniques and adjuvant therapy, the 5‐year survival rate of bladder cancer with metastasis is about 8%. 7 Last decade, immunotherapy had been proved to be a favorable regimen for both early and late stages in BLCA. 8 , 9 For example, Nivolumab, an immunotherapy drug targeting programmed cell death protein 1 (PD‐1), was confirmed to induce neoplastic cell death in urothelial bladder cancers with metastases. 10 Therefore, exploring immunotherapeutic target is promising for the diagnosis as well as treatment of BLCA.
The genetic and epigenetic alterations of DNA, such as gene mutations and copy number variations, are frequently reported in BLCA. 11 It has been reported that mutations of specific genes like TP53 may lead to tumor progression by dysregulation of cell cycle and DNA damage response. 12 Copy number variations (CNVs) of several genes like CCNE1 were also reported to have good diagnostic and prognostic value. 13 Apart from these alterations, emerging evidence has revealed that RNA modifications are important for post‐transcriptional regulation of gene expression. 14 N 6‐methyladenosine (m6A) modification is one of the most common RNA modifications in mammalian systems. 15 , 16 It is regulated dynamically by methyltransferases (METTL3, METTL14, METTL16, WTAP, RBM15, ZC3H13, and KIAA1429), binding proteins (YTHDC1, YTHDC2, YTHDF1, YTHDF2, and HNRNPC) and demethylases (ALKBH5 and FTO). Recent studies indicated that among these regulators, RNA methyltransferases (METTL3, METTL14, and METTL16) play indispensable roles in progression of BLCA. For instance, METTL3 promotes the expression of ITGA6, resulting in increased growth and progression of BLCA, 17 while METTL14 inhibits bladder tumorigenesis by reducing mRNA stability of NOTCH1. 18 It was indicated that m6A RNA methylation regulators contributed to the malignant progression of BLCA. 19 Jin et al reported that m6A modification of ITGA6 promoted the development and progression of BLCA. 17 Furthermore, CNVs were enriched in specific molecular subtypes of BLCA. 11 While some novel molecular subtypes of BLCA were found based on immune‐cell‐associated CpG sites. 20 Elizabeth et al found that copy number variations of FGFR‐3 were negatively correlated with Fibroblast growth factor 2 (FGF‐2) while positively associated with NMIBC. 21 Recent studies also revealed the significantly association between tumorigenesis and copy number variations of m6A‐related genes in head and neck squamous cell carcinoma (HNSCC) 22 and clear cell renal cell carcinoma (ccRCC). 23 However, the copy number variations, expression and prognostic value of m6A regulators in bladder cancer remain largely unknown.
A growing amount of evidence reveals that immune infiltrate plays an essential role in many kinds of tumors and correlates with the effect of tumor immunotherapy. 24 , 25 Immune infiltrate is a part of the complex tumor microenvironment. 26 A recent study has reported that tumor‐infiltrating immune cells are prognostic factors of lung cancer, and the infiltration status is also correlated with expressions of specific m6A regulatory genes including METTL3. 27 Tumor‐infiltrating immune cells (TIICs) are composed of T cells, macrophages, dendritic cells, neutrophils, and mast cells. BLCA is an immune sensitive tumor infiltrated by TIICs. 28 Two most widely applied immunotherapies in BLCA are Bacillus Calmette–Guerin (BCG) intravesical instillation and anti‐PD‐1/PD‐L1 immune checkpoint blockade. 29
It should be noted that m6A methylation majorly regulated immune response in tumor immune microenvironment (TIME). For example, the low level of m6A increased infiltration of immune cells in the TIME, thus enhancing antitumor immunity and sensitivity to anti PDL‐1 immunotherapy in gastric cancer. 30 However, the associations between the CNVs of m6A regulators and immune infiltrates in BLCA were yet to be clarified.
In this work, we systematically analyzed the alterations of m6A RNA methylation regulators in 409 bladder cancer from the Cancer Genome Atlas (TCGA) database. The association between genetic alterations of m6A regulators and clinicopathological characteristics of the bladder cancer cohort was investigated. We also identified the associated pathways in bladder cancer. Finally, the correlations between copy number variations and tumor infiltration levels were computed. Generally, we provided evidence that m6A regulators might play crucial roles in bladder cancer and serve as potential prognostic biomarkers.
2. MATERIALS AND METHODS
2.1. Data processing
The mRNA expression and clinical information were downloaded from the TCGA database (http://cancergenome.nih.gov/), which was based on Illumina HiSeq RNA‐Seq platform. The histological types of BLCA containing urothelial carcinomas, squamous cell carcinoma, small‐cell carcinoma, and adenocarcinoma were applied in further analysis. Only 363 patients had CNV data and all of them were included for further analysis. The mutation data of m6A regulators were obtained from the TCGA program by cBioportal platform, 31 and a total of 87 patients were found to have mutation data.
2.2. Selection of m6A methylation regulators
First, we made a list of m6A regulators from published studies. Totally sixteen regulators were found. 16 , 32 , 33 Due to lack of CNV data, only thirteen representative genes were finally selected with CNV. Second, we compared the CNV events of these regulators in BLCA with the clinicopathological features. The data of frequently mutated gene in BLCA was provided by Department of Urology, the Seventh Affiliated Hospital of Sun Yat‐sen University.
2.3. Bioinformatics analysis
To clarify the functions of m6A RNA methylation regulators in BLCA, we performed a gene set enrichment analysis (GSEA) under the condition of high expression METTL3 (defined as the mRNA level of METTL3 higher than the median level) and low expression METTL3 (defined as the mRNA level of METTL3 lower than the median level). 34 , 35 GSEA was provided by the JAVA program with MSigDB v6.1, which was downloaded from the Broad Institute. 36 Hallmark gene set “h.all.v6.0.symbols.gmt” was used in this study. 37 Gene sets with normalized p‐value <0.05, and the false discovery rate (FDR) <0.25 were considered to be significantly enriched. 23 , 38
2.4. Immune infiltrate signature analysis
The correlations between copy number variations and tumor infiltration levels were generated by the Tumor Immune Estimation Resource (TIMER). 39 We used box plots to show the distributions of each immune subset at each copy number status in specific cancer types. The infiltration level for each category is compared with the normal, and two‐sided Wilcoxon rank sum test was used for analysis. The correlations between expression levels and the infiltration levels of six cell types (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, dendritic cells) were computed, 40 and the results were generated by TIMER.
2.5. Statistical analysis
One‐way ANOVA was used to compare the expression level of m6A regulators with different CNV patterns in BLCA. Chi‐square test was applied to describe the association between CNV of m6A regulators and clinical characteristics and the association between CNV patterns and typical genes mutation in BLCA. Kaplan–Meier method with a two‐sided log‐rank test was used to compare the OS of BLCA patients in the deletion‐ and gain‐risk groups. Cox proportional hazards regression analysis was used to identify the relationship between m6A regulatory genes and BLCA patients’ OS. Results with a p‐value <0.05 were considered significant. All statistical analyses were performed using R v3.6.1 (https://www.r‐project.org/) and Prism 8.0.1 (GraphPad Software Inc.).
3. RESULT
3.1. Mutations and CNVs of m6A regulators in BLCA patients
A flowchart of the study design is shown in Figure 1. Considering m6A regulators played important role in bladder cancer, we performed a comprehensive bioinformatics analysis to explore the relationships between genomics abnormalities of m6A regulators and clinical characteristics of BLCA based on the TCGA database. Among the 87 patients who have mutation data obtained from cBioportal platform, mutations of the thirteen m6A regulatory genes were seldom detected (Table 1). However, CNV events of m6A regulators were frequently observed in 363 samples of BLCA with CNV data in TCGA database (Figure 2 and Table 2). Among three RNA methyltransferases, CNVs of METTL3 (192/363, 52.89%) and METTL16 (243/363, 66.94%) were found in more than half of the samples and those of METTL14 (181/363, 49.86%) in nearly half of the samples. Among these 363 samples, most of the CNV events of m6A regulatory genes led to a loss of copy number (1586/2635).
FIGURE 1.
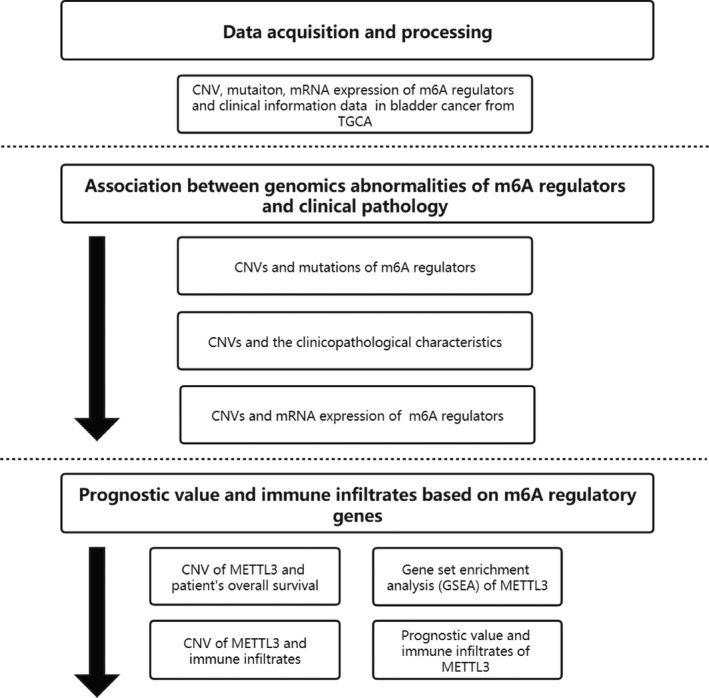
The flowchart of the study design
TABLE 1.
Mutations count of m6A regulatory genes in 87 BLCA patients
| M6A regulatory genes | Mutations count |
|---|---|
| METTL3 | 18 |
| METTL14 | 5 |
| METTL16 | 1 |
| WTAP | 7 |
| RBM15 | 14 |
| ZC3H13 | 16 |
| FTO | 4 |
| ALKBH5 | 5 |
| YTHDF1 | 4 |
| YTHDF2 | 9 |
| YTHDF3 | 3 |
| YTHDC1 | 8 |
| YTHDC2 | 13 |
Mutation count represented the number of patients with corresponding gene mutations.
FIGURE 2.
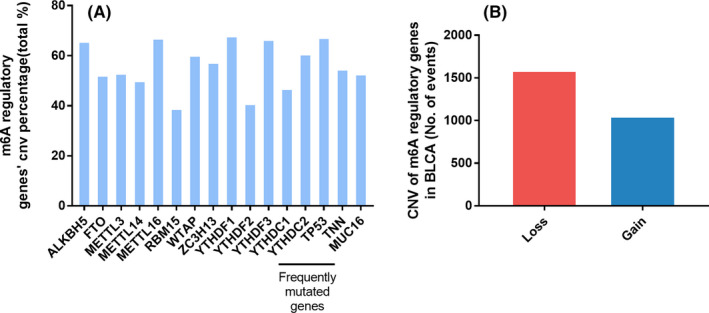
CNVs of m6A regulatory genes and three genes that were frequently mutated in BLCA. (A) Percentage of BLCA samples with CNV events of the m6A regulatory genes. (B) CNV events of copy number gain and loss in BLCA samples
TABLE 2.
Different CNV patterns of m6A regulatory genes and three frequently mutated genes occurred in BLCA samples (n = 363)
| Gene | Diploid | Deep deletion | Shallow deletion | Copy number gain | Amplification | CNV sum | Percentage | |
|---|---|---|---|---|---|---|---|---|
| Eraser | ALKBH5 | 125 | 9 | 177 | 49 | 3 | 238 | 65.56% |
| FTO | 174 | 3 | 115 | 69 | 2 | 189 | 52.07% | |
| Writer | METTL3 | 171 | 4 | 110 | 73 | 5 | 192 | 52.89% |
| METTL14 | 182 | 1 | 147 | 30 | 3 | 181 | 49.86% | |
| METTL16 | 120 | 7 | 190 | 45 | 1 | 243 | 66.94% | |
| RBM15 | 222 | 1 | 59 | 76 | 5 | 141 | 38.84% | |
| WTAP | 145 | 4 | 181 | 33 | 0 | 218 | 60.06% | |
| ZC3H13 | 155 | 20 | 118 | 68 | 2 | 208 | 57.30% | |
| Reader | YTHDF1 | 117 | 0 | 23 | 220 | 3 | 246 | 67.77% |
| YTHDF2 | 215 | 0 | 79 | 66 | 3 | 148 | 40.77% | |
| YTHDF3 | 122 | 0 | 28 | 200 | 13 | 241 | 66.39% | |
| YTHDC1 | 193 | 1 | 116 | 46 | 7 | 170 | 46.83% | |
| YTHDC2 | 143 | 6 | 187 | 25 | 2 | 220 | 60.61% | |
| Mutated genes | TP53 | 119 | 10 | 192 | 41 | 1 | 244 | 67.22% |
| TNN | 165 | 1 | 26 | 149 | 22 | 198 | 54.55% | |
| MUC16 | 172 | 1 | 121 | 65 | 4 | 191 | 52.62% |
3.2. Association between CNV events of m6A regulators and clinicopathological as well as molecular characteristics
We previously investigated the most frequently mutated genes in bladder cancer in our center, and the top 3 of these genes (TP53, TNN, MUC16) were used for analysis (Table S1). We found that the presence of CNV events of m6A regulatory genes, especially CNV events of METTL3, METTL14, and METTL16 were significantly correlated with mutations of TP53 (Figure 3 and Table 3). The samples were also divided into two groups: one consisted of samples with mutations and/or CNVs of m6A regulatory genes while the other consisted of samples without mutations or CNVs. The relationship between these two groups and the clinicopathological characteristics of BLCA patients was assessed. Advanced age (p = 0.019), pathological TNM stage (p = 0.026), pathological N stage (p = 0.02) and histological grade. (p < 0.001) of BLCA were found significantly related to the occurrence of mutations and/or CNV events (Table 4).
FIGURE 3.
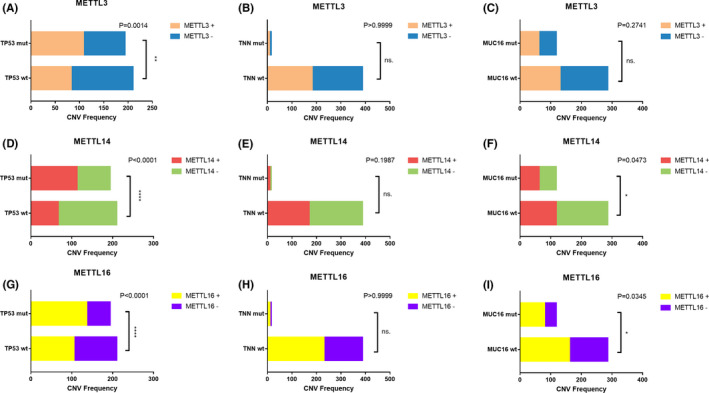
Relationship between molecular characteristics and CNVs in (A‐C) METTL3, (D‐F) METTL4, and (G‐I) METTL16, respectively. Pearson Chi‐square test was applied for analysis, and p < 0.05 was considered significant. METTL3+ METTL3 with CNVs, METTL3‐ METTL3 without CNVs, METTL14+ METTL14 with CNVs, METTL14‐ METTL14 without CNVs, METTL16+ METTL16 with CNVs, METTL16‐ METTL16 without CNVs. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns. not significant
TABLE 3.
Relationship between molecular characteristics and CNV events of m6A regulatory genes in BLCA patients (n = 409)
| Without mutation nor CNV* | With mutation and/or CNV* | χ² | p | |||
|---|---|---|---|---|---|---|
| TP53 | n = 409 | wt | 35 | 177 | 20.552 | <0.001 |
| alteration | 6 | 191 | ||||
| TNN | n = 409 | wt | 32 | 181 | 12.315 | <0.001 |
| alteration | 9 | 187 | ||||
| MUC16 | n = 409 | wt | 37 | 254 | 8.094 | 0.004 |
| alteration | 4 | 114 |
Chi‐square test was applied for analysis and p < 0.05 was considered significant.
With mutation and/or CNV: Cases have mutant or CNV or mutant and CNV, confirmed through TCGA database. Without mutant nor CNV: Cases with neither mutant nor CNV, confirmed through TCGA database. Ambiguous variables (Nx, Mx, N/A and Gx) were excluded from chi‐square test or non‐parametric test.
TABLE 4.
Clinical pathological parameters of BLCA patients with or without mutation or CNV of m6A regulatory genes
| With mutation and/or CNV* | Without mutation nor CNV* | p | ||
|---|---|---|---|---|
| Age | <=60 | 90 | 17 | 0.019 |
| >60 | 278 | 24 | ||
| Gender | Female | 97 | 9 | 0.541 |
| Male | 271 | 32 | ||
|
Pathological TNM Stages |
Ⅰ | 1 | 1 | 0.026 |
| Ⅱ | 112 | 18 | ||
| Ⅲ | 124 | 15 | ||
| Ⅳ | 129 | 7 | ||
| N/A | 2 | 0 | ||
| T stage | T1 | 2 | 1 | 0.125 |
| T2 | 103 | 15 | ||
| T3 | 174 | 18 | ||
| T4 | 56 | 2 | ||
| Tx | 29 | 3 | ||
| M stage | M0 | 166 | 27 | 0.993 |
| M1 | 10 | 1 | ||
| Mx | 187 | 11 | ||
| N stage | N0 | 204 | 31 | 0.02 |
| N1 | 44 | 1 | ||
| N2 | 73 | 3 | ||
| N3 | 7 | 0 | ||
| Nx | 33 | 3 | ||
| Histological Grade | low | 13 | 8 | <0.001 |
| high | 352 | 33 | ||
| N/A | 3 | 0 |
A chi‐square test was applied for analysis and p < 0.05 was considered significant.
With mutation and/or CNV: Cases have mutant or CNV or mutant and CNV, confirmed through TCGA database. Without mutant nor CNV: Cases with neither mutant nor CNV, confirmed through TCGA database. Ambiguous variables (Nx, Mx, N/A and Gx) were excluded from chi‐square test or non‐parametric test.
The most mutation counts were merely 18 (Table 1), which indicated the few mutations of BLCA. 23 Furthermore, the occurrence of mutations and/or CNVs of m6A regulatory genes were significantly associated with the prognosis of BLCA (Table 4). Thus, we paid more attention to whether CNVs contributed to the change of m6A level in BLCA. We next evaluated the effect of CNV events on mRNA expression of m6A regulatory genes. Among all the regulatory genes, CNV events of most genes were significantly correlated with their mRNA expression, respectively (Figure 4 and Figure S1). Copy number gain and amplification (AMP) were associated with higher expression levels, while deep deletion and shallow deletion led to lower expression levels.
FIGURE 4.
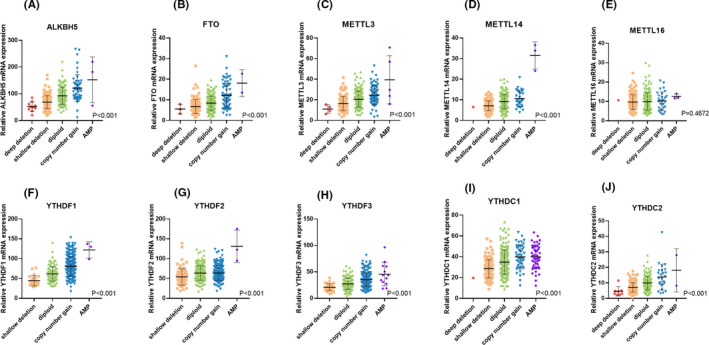
Correlation between different CNV events (deep deletion, shallow deletion, diploid, copy number gain, AMP) of m6A regulatory genes mRNA expression levels (A–J). One‐way ANOVA was applied to determine the significance, and p < 0.05 was considered significant
3.3. CNVs of METTL3 associated with overall survival of BLCA patients
Survival analysis was then performed to explore the prognostic value of CNVs in m6A regulators among BLCA patients. A separate analysis of these ten genes revealed that only CNVs in METTL3 were significantly correlated with the OS of the BLCA cohort. Patients with copy number gain and AMP of METTL3 had an inferior OS (Figure 5 and Figure S2). However, no significant correlation was found between OS and CNVs of either METTL14 or METTL16. Further univariate Cox regression analyses revealed that CNVs of METTL3 was related to OS, similar to other clinical covariates such as age and tumor grade. However, none of these variates was independent factors of survival status of BLCA patients according to the result of multivariate Cox regression (Table 5).
FIGURE 5.
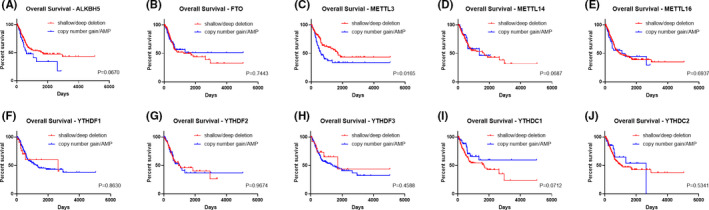
Overall survival of BLCA patients with different CNV types (deep deletion, shallow deletion, diploid, copy number gain, AMP) of m6A regulatory genes (A–J). Kaplan‐Meier method was used for analyses and p < 0.05 was considered significant
TABLE 5.
Univariate and multivariate Cox regression analysis of different factors in BLCA patients
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Age (>60 vs <=60) | 2.107 (1.374–3.231) | 0.001 | 0.824 | |
| Tumor grade (I‐II vs III‐IV) | 2.681 (1.758–4.087) | <0.001 | 0.897 | |
| M (M1 vs M0) | 3.018 (1.372–6.639) | 0.006 | 0.025 | |
| N (N1 vs N0) | 2.134 (1.350–3.372) | 0.001 | 0.829 | |
| T (T3‐T4 vs T1‐T2) | 2.438 (1.603–3.708) | <0.001 | 0.982 | |
| METTL3 (copy number gain/AMP vs shallow/deep deletion) | 1.712 (1.097–2.672) | 0.018 | 0.331 | |
p < 0.05 was considered significant.
3.4. Gene set enrichment analysis (GSEA) of METTL3
Given the significance of METTL3 in the m6A modification and the surprising results we found, we explored the effect of dysregulated m6A on the pathogenesis of BLCA. GSEA was performed to identify the gene sets enriched in these samples with different mRNA expression levels of METTL3. We found that high expression level of METTL3 associated with several cancer‐related biological processes, including mitotic spindle formation, G2‐M checkpoint signaling, and E2F targets signaling pathway (Figure 6 and Table S2). High expression of METTL14 was also correlated with several pathways, and the results were showed in Table S3.
FIGURE 6.
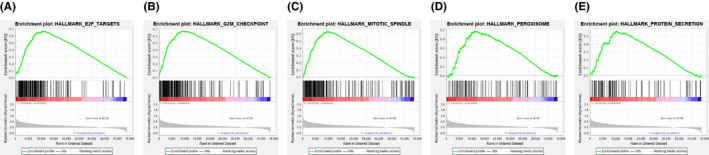
GSEA results of different expression level of METTL3. Gene set enrichment plots of (A) E2F targets, (B) G2‐M checkpoint, (C) mitotic spindle, (D) peroxisome, and (E) protein secretion were shown
3.5. METTL3 associated with the immune infiltrates in BLCA
We finally computed the infiltration levels of different immune cell types including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. The results highlighted the correlation between RNA methyltransferase and immune infiltrates (Figure 7 and Figure S3). The CNV events of both METTL3 were significantly correlated with the downregulated infiltration levels of CD4+ T cells, neutrophils, and dendritic cells. On the other hand, the expression level of METTL3 was significantly associated with most kinds of immune infiltrates. The correlation between METTL14 and immune infiltrates was also computed and showed in Figure S3.
FIGURE 7.
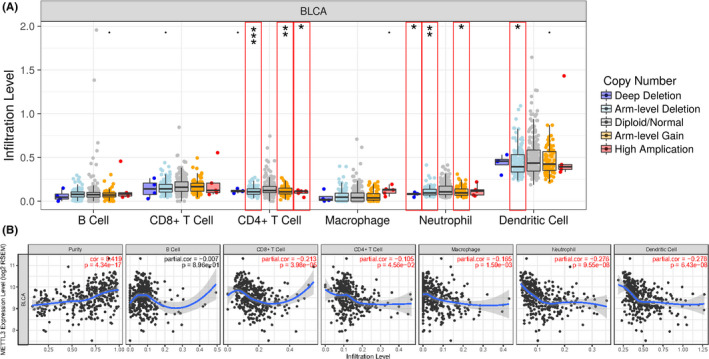
TIMER revealed that both CNV events and expression of METTL3 were associated with immune infiltrates. (A) CNVs of METTL3 were significantly correlated with immune infiltrates of CD4+ T cells, Neutrophils and Dendritic cells. *p < 0.05, **p < 0.01, ***p < 0.001. (B) METTL3 expression was significantly associated with almost all kinds of immune infiltrates. p < 0.05 was considered significant and highlighted in red
4. DISCUSSION
In this study, we discovered high frequency of CNV events of m6A regulatory genes in BLCA, which were associated with clinicopathological as well as molecular characteristics, the prognosis, oncogenic signaling pathways, and immune infiltrates.
N6 ‐methyladenosine (m6A) is one of the most abundant internal RNA modifications in mammalian systems. 41 In recent years, a number of studies chose to evaluate the altered expression of m6A regulatory genes to portrait a unique expression pattern of specific pathological process, indirectly exploring the m6A level and biological significance in human diseases. Because of the distinct m6A status of different kinds of tumors, we aimed to explore the CNV events of m6A “writers”, “erasers” and “readers”. In BLCA cohort, the frequency of CNV events of thirteen m6A regulatory genes was higher than that reported in ccRCC 23 and AML, 32 especially m6A “writers”, suggesting that m6A might play a more indispensable role in BLCA. Furthermore, CNV data showed that all three kinds of m6A regulatory genes had a high frequency of CNV events, indicating that the regulation of m6A modification in BLCA was so complicated that further investigations are needed to demonstrate the m6A regulatory mechanism in BLCA.
After a general understanding of CNV events in BLCA patients, we evaluated the association between alterations of m6A regulators and clinicopathological and molecular characteristics of BLCA cohort. Our results showed that alterations of these genes were significantly related to the age of patients, pathological TNM stage, and histological grade of BLCA. These results suggested that CNV alterations may associate with histology transformation, similar to ccRCC, 23 making it possible for CNV alterations of these genes to serve as biomarkers predicting the grade of bladder cancer. Furthermore, alterations of m6A regulators were found significantly correlated with alterations of TP53, TNN, and MUC16, which were frequently detected as mutated genes, 42 similar to the results of AML. TP53 was the most frequently altered genes among these three genes. And most of the CNVs led to a loss of copy number (shallow/deep deletion: copy number gain/AMP = 202:42). 32 It was also reported that altering the expression of METTL3 would significantly affect the expression level and alternative splicing patterns of certain genes, resulting in modulation of downstream targets of the TP53 signaling pathway such as P21, FAS, and BAX. 43 These effects would lead to inhibition of apoptosis and tumorigenesis. Together, these findings suggested that alterations of m6A regulatory genes and TP53 signaling pathway may synergistically play important roles in BLCA pathogenesis and progression.
In our study, METTL3 showed slightly increase in deletion than copy number gain and amplification. However, recent studies had demonstrated that expression level of METTL3 was significantly upregulated in BLCA. 44 , 45 It has been proven that METTL3 could promote BLCA progression via AFF4/NF‐κb/MYC signaling network. 44 METTL3 was also shown capable of promoting BLCA proliferation by accelerating pri‐miR221/222 maturation in an m6A‐dependent manner. 45 These findings suggested that although a significant association between CNV patterns and mRNA expression levels was found in most of the m6A regulatory genes including METTL3, the expression levels of these genes may not be directly predicted by CNV patterns. Still, further studies on regulation mechanisms between CNV and gene expression and the following functions are needed to clarify the relationship between CNV and human diseases.
The effect of CNV events of m6A regulatory genes on the survival of BLCA patients was also evaluated. Among all these regulatory genes, only the CNVs on METTL3 were significantly associated with the OS of BLCA patients. It is noteworthy that although most of its CNVs led to a loss of copy number (shallow/deep deletion: copy number gain/AMP = 114:78), copy number gain and AMP of METTL3 were linked to poor OS. Despite the fact that METTL3 and other clinical covariates such as age, pathological TNM stage were associated with OS according to the results of univariate analysis, none of them showed the consistent result in multivariate analysis. However, we should still attach importance to the effect of METTL3 alteration on survival as prior studies have established that alteration of METTL3 could promote tumor proliferation and migration, which are related to higher T stage and tumor grade of BLCA. 17 , 44 Further analyses of regulation mechanisms of METTL3 alterations on other clinical covariates are needed to illustrate this contradictory phenomenon.
Several cancer‐related biological processes were found to be dysregulated in BLCA. We found in this BLCA cohort that downregulated expression of METTL3 was associated with some pathways including mitotic spindle assembly, G2‐M checkpoint signaling and E2F targets signaling pathway, which are all important cellular processes in tumorigenesis. 46 , 47 These findings suggest that these processes may also play indispensable roles in BLCA pathogenesis, and it is likely that alterations of m6A regulatory genes can regulate BLCA progression via these pathways. However, the specific regulation mechanisms of METTL3 alteration on these pathways is yet to be clarified.
Given the latest finding of the association between m6A regulatory genes and immune infiltrates, 27 we finally computed that both CNV events and expression of METTL3 were negatively correlated with immune infiltration levels of CD4+ T cells, neutrophils, and dendritic cells. These findings were also partly confirmed in lung squamous cell carcinoma. 27 Previous studies have established that immune infiltrates in BLCA were composed mainly by CD4+ T cells, 48 and the infiltration level was negatively correlated with BLCA stage and risk of recurrence. 49 Under some aspects of cell pathology including tumorigenesis, several signaling pathways would be activated for immune cell recruitment, such as CCL2/CCR2 pathway 50 and CCL5‐related pathway. 51 Collectively, these findings suggested that RNA methyltransferases may regulate BLCA immune infiltrates in an m6A‐dependent manner via specific signaling pathways, and CD4+ T cells could be the main targets. However, the relationship between m6A and immune infiltrates was hardly reported, and these results highlighted the need for more in‐depth studies.
Recent studies indicated that METTL3 promoted the expression of ITGA6, resulting in the growth and progression of BLCA. 17 In our study, we found that CNVs of METTL3 were significantly correlated with the OS of the BLCA cohort. Besides, CNVs of METTL3 were essentially associated with the immune infiltrates in BLCA. The CNV events of METTL3 were significantly correlated with the downregulated infiltration levels of CD4+ T cells, neutrophils, and dendritic cells. Furthermore, CD4+ T cells that highly expressed the forkhead box P3 (FOXP3) suppressed the anti‐tumor immunity and suggested a poorer prognosis in colorectal cancers. 52 While the high infiltration level of neutrophils was also associated with a poorer prognosis in most tumors. 53 Moreover, it suggested that the immunoregulation of dendritic cells might be a prognostic indicator in breast cancer 54 Besides, METTL3‐mediated mRNA m6A methylation was associated with the immunocompetence of neutrophils 55 and dendritic cells. 56 Hence, we proposed that METTL3 might affect the prognosis of BLCA by regulating the immune infiltration level, which indicated that METTL3 might be a target to regulate the immune response of BLCA. In addition, we found significant correlations between CNVs of m6A regulatory genes and mutations of TP53. And TP53 mutations in tumors leading to enhanced expression of cell cycle progression genes and proteins was related a poor prognosis. 57 Hence, we speculated that m6A regulatory genes and signaling pathways involving TP53 may synergistically contribute to BLCA pathogenesis and progression. It suggested that the CNVs of m6A regulators might be an early diagnostic indicator in the future. Moreover, Shi et al.’s study 58 reported that patients with CNVs of m6A regulators genes were significantly associated with inferior OS in non‐small cell lung cancer, which was consistent with our findings in BLCA. Since the significant correlation between METTL3 and OS in BLCA patients, it indicated that CNVs of METTL3 might perform as a prognostic marker for BLCA. However, further mechanism on CNVs of METTL3 was yet to be explored.
5. CONCLUSION
There are significant correlations between m6A regulatory genes with CNVs and clinicopathological characteristics. METTL3 with CNVs were associated with the immune infiltrates and performed as a prognostic marker in BLCA.
CONFLICTS OF INTERESTS
The authors declare no conflict of interests.
AUTHORS’ CONTRIBUTIONS
Conceptualization: DCH and XZC; Data curation: WXS, YJW and HYD; Formal analysis: WXS, CJB HYD, and DZF; Investigation: WXS, CJB, HYD, and TSQ; Methodology: WXS, DZF, and HHY; Resources: CJB, TSQ, and HYY; Software: WXS and HYD; Supervision: WXS, DCH, and XZC; Validation: WXS and CJB; Visualization: DZF, TSQ and HYY; Writing—original draft, WXS and YJW; Writing—review and editing, WXS, DCH, and XZC. All authors have read and approved the manuscript.
Supporting information
Fig S1
Fig S2
Fig S3
Table S1‐S3
Xiaoshuai Wang, Jingwei Yu, and Jinbao Chen contributed equally to this work.
Changhai Ding is the co‐corresponding author.
REFERENCES
- 1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96‐108. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 3. Sanli O, Dobruch J, Knowles MA, et al. Bladder cancer. Nat Rev Dis Primers. 2017;3:17022. [DOI] [PubMed] [Google Scholar]
- 4. Roupret M, Babjuk M, Comperat E, et al. European association of urology guidelines on upper urinary tract urothelial carcinoma: 2017 update. Eur Urol. 2018;73(1):111‐122. [DOI] [PubMed] [Google Scholar]
- 5. Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388(10061):2796‐2810. [DOI] [PubMed] [Google Scholar]
- 6. DeGeorge KC, Holt HR, Hodges SC. Bladder cancer: diagnosis and treatment. Am Fam Physician. 2017;96(8):507‐514. [PubMed] [Google Scholar]
- 7. Abdollah F, Gandaglia G, Thuret R, et al. Incidence, survival and mortality rates of stage‐specific bladder cancer in United States: a trend analysis. Cancer Epidemiol. 2013;37(3):219‐225. [DOI] [PubMed] [Google Scholar]
- 8. Zheng YQ, Naguib YW, Dong Y, Shi YC, Bou S, Cui Z. Applications of bacillus Calmette–Guerin and recombinant bacillus Calmette–Guerin in vaccine development and tumor immunotherapy. Expert Rev Vaccines. 2015;14(9):1255‐1275. [PMC free article] [PubMed] [Google Scholar]
- 9. Katz H, Wassie E, Alsharedi M. Checkpoint inhibitors: the new treatment paradigm for urothelial bladder cancer. Med Oncol. 2017;34(10):170. [DOI] [PubMed] [Google Scholar]
- 10. Felsenstein KM, Theodorescu D. Precision medicine for urothelial bladder cancer: update on tumour genomics and immunotherapy. Nat Rev Urol. 2018;15(2):92‐111. [DOI] [PubMed] [Google Scholar]
- 11. Choi W, McConkey D. Reply to Joshua A. Linscott, Angela B. Smith, and Jesse D. Sammon's Letter to the Editor re: Woonyoung Choi, Andrea Ochoa, David J. McConkey, et al. Genetic Alterations in the Molecular Subtypes of Bladder Cancer: Illustration in the Cancer Genome Atlas Dataset. Eur Urol 2017;72:354–65. Eur Urol. 2018;73(4):e104‐e105. [DOI] [PubMed] [Google Scholar]
- 12. Liu Y, Kwiatkowski DJ. Combined CDKN1A/TP53 mutation in bladder cancer is a therapeutic target. Mol Cancer Ther. 2015;14(1):174‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song BN, Kim SK, Chu IS. Bioinformatic identification of prognostic signature defined by copy number alteration and expression of CCNE1 in non‐muscle invasive bladder cancer. Exp Mol Med. 2017;49(1):e282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187‐1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang S, Chai P, Jia R, Jia R. Novel insights on m(6)A RNA methylation in tumorigenesis: a double‐edged sword. Mol Cancer. 2018;17(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang S, Sun C, Li J, et al. Roles of RNA methylation by means of N(6)‐methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017;408:112‐120. [DOI] [PubMed] [Google Scholar]
- 17. Jin H, Ying X, Que B, et al. N(6)‐methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. Ebiomedicine. 2019;47:195‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu C, Wang Z, Zhou N, et al. Mettl14 inhibits bladder TIC self‐renewal and bladder tumorigenesis through N(6)‐methyladenosine of Notch1. Mol Cancer. 2019;18(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen M, Nie ZY, Wen XH, Gao YH, Cao H, Zhang SF. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci Rep. 2019;39(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo Q, Vogeli TA. A methylation‐based reclassification of bladder cancer based on immune cell genes. Cancers (Basel). 2020;12(10):3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McNiel EA, Tsichlis PN. Analyses of publicly available genomics resources define FGF‐2‐expressing bladder carcinomas as EMT‐prone, proliferative tumors with low mutation rates and high expression of CTLA‐4, PD‐1 and PD‐L1. Signal Transduct Target Ther. 2017;2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou X, Han J, Zhen X, et al. Analysis of genetic alteration signatures and prognostic values of m6A regulatory genes in head and neck squamous cell carcinoma. Front Oncol. 2020;10:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Wang J, Hong B, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma ‐ a retrospective study using TCGA database. Aging. 2019;11(6):1633‐1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borcoman E, De La Rochere P, Richer W, et al. Inhibition of PI3K pathway increases immune infiltrate in muscle‐invasive bladder cancer. OncoImmunology. 2019;8(5):e1581556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kardos J, Chai S, Mose LE, et al. Claudin‐low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight. 2016;1(3):e85902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeng D, Li M, Zhou R, et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res. 2019;7(5):737‐750. [DOI] [PubMed] [Google Scholar]
- 27. Xu F, Zhang H, Chen J, Lin L, Chen Y. Immune signature of T follicular helper cells predicts clinical prognostic and therapeutic impact in lung squamous cell carcinoma. Int Immunopharmacol. 2020;81:105932. [DOI] [PubMed] [Google Scholar]
- 28. Wu Z, Zhu K, Liu Q, et al. Profiles of immune infiltration in bladder cancer and its clinical significance: an integrative genomic analysis. Int J Med Sci. 2020;17(6):762‐772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shriwas O, Mohapatra P, Mohanty S, Dash R. The impact of m6A RNA modification in therapy resistance of cancer: implication in chemotherapy, radiotherapy, and immunotherapy. Front Oncol. 2021;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator‐mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616‐624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang C, Xu B, Lu S, Zhao Y, Liu P. HN1 contributes to migration, invasion, and tumorigenesis of breast cancer by enhancing MYC activity. Mol Cancer. 2017;16(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang N, Zhang Y, Zhang P, et al. Overexpression of annexin A5 might guide the gemtuzumab ozogamicin treatment choice in patients with pediatric acute myeloid leukemia. Ther Adv Med Oncol. 2020;12:431424621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739‐1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joly JH, Lowry WE, Graham NA. Differential gene set enrichment analysis: a statistical approach to quantify the relative enrichment of two gene sets. Bioinformatics. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor‐infiltrating immune cells. Cancer Res. 2017;77(21):e108‐e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6(12):863‐865. [DOI] [PubMed] [Google Scholar]
- 42. Sjodahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377‐3386. [DOI] [PubMed] [Google Scholar]
- 43. Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature. 2012;485(7397):201‐206. [DOI] [PubMed] [Google Scholar]
- 44. Cheng M, Sheng L, Gao Q, et al. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF‐κB/MYC signaling network. Oncogene. 2019;38(19):3667‐3680. [DOI] [PubMed] [Google Scholar]
- 45. Han J, Wang JZ, Yang X, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri‐miR221/222 maturation in m6A‐dependent manner. Mol Cancer. 2019;18(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19(6):326‐338. [DOI] [PubMed] [Google Scholar]
- 47. Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12(6):385‐392. [DOI] [PubMed] [Google Scholar]
- 48. Solinas C, Chanza NM, Awada A, Scartozzi M. The immune infiltrate in prostate, bladder and testicular tumors: an old friend for new challenges. Cancer Treat Rev. 2017;53:138‐145. [DOI] [PubMed] [Google Scholar]
- 49. Liakou CI, Narayanan S, Ng TD, Logothetis CJ, Sharma P. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human bladder cancer. Cancer Immun. 2007;7:10. [PMC free article] [PubMed] [Google Scholar]
- 50. Li X, Yao W, Yuan Y, et al. Targeting of tumour‐infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66(1):157‐167. [DOI] [PubMed] [Google Scholar]
- 51. Wang X, Lang M, Zhao T, et al. Cancer‐FOXP3 directly activated CCL5 to recruit FOXP3(+)Treg cells in pancreatic ductal adenocarcinoma. Oncogene. 2017;36(21):3048‐3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saito T, Nishikawa H, Wada H, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22(6):679‐684. [DOI] [PubMed] [Google Scholar]
- 53. Ocana A, Nieto‐Jimenez C, Pandiella A, Templeton AJ. Neutrophils in cancer: prognostic role and therapeutic strategies. Mol Cancer. 2017;16(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Michea P, Noel F, Zakine E, et al. Adjustment of dendritic cells to the breast‐cancer microenvironment is subset specific. Nat Immunol. 2018;19(8):885‐897. [DOI] [PubMed] [Google Scholar]
- 55. He J, Zhou M, Yin J, et al. METTL3 restrains papillary thyroid cancer progression via m6A/c‐Rel/IL‐8‐mediated neutrophil infiltration. Mol Ther. 2021;29(5):1821‐1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang H, Hu X, Huang M, et al. Mettl3‐mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun. 2019;10(1):1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Donehower LA, Soussi T, Korkut A, et al. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 2019;28(5):1370‐1384.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi H, Zhao J, Han L, et al. Research paper retrospective study of gene signatures and prognostic value of m6A regulatory factor in non‐small cell lung cancer using TCGA database and the verification of FTO. Aging. 2020;12(17):17022‐17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1‐S3


