Abstract
Ultra‐high‐field magnetic resonance imaging (MRI) enables sub‐millimetre resolution imaging of the human brain, allowing the study of functional circuits of cortical layers at the meso‐scale. An essential step in many functional and structural neuroimaging studies is segmentation, the operation of partitioning the MR images in anatomical structures. Despite recent efforts in brain imaging analysis, the literature lacks in accurate and fast methods for segmenting 7‐tesla (7T) brain MRI. We here present CEREBRUM‐7T, an optimised end‐to‐end convolutional neural network, which allows fully automatic segmentation of a whole 7T T1w MRI brain volume at once, without partitioning the volume, pre‐processing, nor aligning it to an atlas. The trained model is able to produce accurate multi‐structure segmentation masks on six different classes plus background in only a few seconds. The experimental part, a combination of objective numerical evaluations and subjective analysis, confirms that the proposed solution outperforms the training labels it was trained on and is suitable for neuroimaging studies, such as layer functional MRI studies. Taking advantage of a fine‐tuning operation on a reduced set of volumes, we also show how it is possible to effectively apply CEREBRUM‐7T to different sites data. Furthermore, we release the code, 7T data, and other materials, including the training labels and the Turing test.
Keywords: 3D image analysis, brain MRI segmentation, convolutional neural networks, weakly supervised learning
We here present CEREBRUM‐7T, an optimised end‐to‐end convolutional neural network, that allows fully automatic segmentation of a whole 7T T1w magnetic resonance imaging (MRI) brain volume at once, neither partitioning the volume, pre‐processing, nor aligning it to an atlas. The trained model is able to produce accurate multi‐structure segmentation masks on six different classes plus background in only a few seconds. The experimental part, a combination of objective numerical evaluations and subjective analysis, confirms that the proposed solution outperforms the training labels it was trained on and is suitable for neuroimaging studies, such as layer functional MRI studies.

1. INTRODUCTION
Image segmentation of brain magnetic resonance imaging (MRI) scans is an essential quantitative analysis step for assessing both healthy brain anatomy and pathophysiological conditions. The segmentation of brain structures is necessary for monitoring anatomical variations during the development of neuro‐degenerative processes, psychiatric disorders and neurological diseases. In addition, segmentation is an essential step in functional MRI (fMRI) studies to isolate specific brain regions and to investigate brain activity patterns. For example, the accurate segmentation of inner and outer grey matter (GM) boundaries is critical for cortex‐based analysis in laminar fMRI studies.
The advent of 7‐tesla (7T) scanners, together with improvements in acquisition methods, increased the imaging resolution to a sub‐millimetre level (Duyn, 2012), thus enabling functional imaging of different cortical depths and columns with high spatial specificity and the visualisation of structures with an unprecedented definition (e.g., hippocampal substructures). However, these innovative systems come with new technical challenges. One of the most relevant issues is the need for intensity‐based pipelines specific for 7T data, due to the lack of standardisation across 7T sites (Clarke et al., 2020).
Segmentation tools available for 3T data—such as FreeSurfer (Fischl, 2012)—which usually apply atlas‐based or multi‐atlas‐based segmentation strategies, are not maximally effective on 7T volumes, due to increased data complexity, voxel intensity inhomogeneity (intra‐/inter‐sites) and site‐specific artefacts. Even the latest version of FreeSurfer (v7), which has been improved for ultra‐high‐field data, can fail in performing a fully automatic segmentation due to the high inhomogeneity of 7T volumes, requiring expert knowledge and multiple iterative steps by the user. Consequently, in‐house and site‐specific pipelines are commonplace, which account for a sequence of several manual or semi‐manual operations, such as in the example shown in Figure 1a.
FIGURE 1.
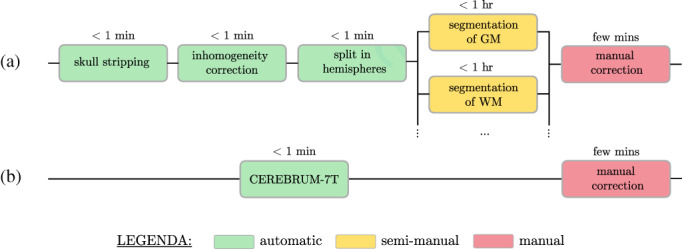
CEREBRUM‐7T aims to replace (a) complex and site‐specific segmentation pipelines which combine automatic and semi‐manual methods with (b) a unique automatic segmentation step which mimics expert knowledge
Automating and optimising segmentation and other processes by artificial intelligence (AI) methods could improve clinical care with high speed and low additional cost in healthcare settings (Esteva et al., 2019), especially since intra‐ and inter‐expert variability remains a severe limitation in medical (as well as in brain) imaging. Furthermore, an increasing number of brain imaging projects rely on pre‐collected data, with thousands of structural scans (Lu et al., 2021) available on open platforms such as OpenNeuro, 1 EBRAINS, 2 or the Human Connectome Project (Van Essen et al., 2013). This unprecedented data availability demands a new class of fast, robust, reliable and reproducible tools that provide high‐accuracy output and are fully automatic and scanner independent. Accurate and shared analysis workflows would advance the opportunities offered by the huge quantity of available shared data and could help to minimise otherwise negative effects on the scientific conclusions of the entire field (Botvinik‐Nezer et al., 2020).
1.1. Aims and contributions
In this work we introduce CEREBRUM‐7T: an AI tool which, mimicking expert knowledge in segmentation, encapsulates all automatic and semi‐manual segmentation modules into a unique fully automatic step, as shown in Figure 1b. CEREBRUM‐7T is, to the best of our knowledge, the first fully automatic deep learning (DL) solution for brain MRI segmentation on out‐of‐the‐scanner 3 7T data. By extending the previous work on 3T data (Bontempi et al., 2020), CEREBRUM‐7T acts in a fully 3D fashion on brain volumes and produces a segmentation using the labelling strategy proposed by the Medical Image Computing and Computer Assisted Intervention Society (MICCAI) challenge (Mendrik et al., 2015): GM, white matter (WM), cerebrospinal fluid (CSF), ventricles, cerebellum, brainstem and basal ganglia, as shown in the method overview in Figure 2. The volumetric processing of the whole brain is made possible by a light model architecture and by distributing different parts of the model on different GPUs. Processing the entire volume at once enables the tool to learn and incorporate all steps performed during manual segmentation, like inhomogeneity correction or skull stripping, and obtain a full brain segmentation in a few seconds, compared to several hours of other currently used non‐DL methods for 7T data. Furthermore, exploiting the ability of DL methods to efficiently learn internal representations of data, brain segmentation happens with neither the support of ad hoc pre‐processing, nor the alignment to reference atlases. The model is trained in a weakly‐supervised fashion by exploiting a labelling with errors, which in the following we will refer to as inaccurate ground truth (iGT) obtained using a combination of AFNI‐3dSeg (Cox, 1996) and methods as in Fracasso et al. (2016).
FIGURE 2.
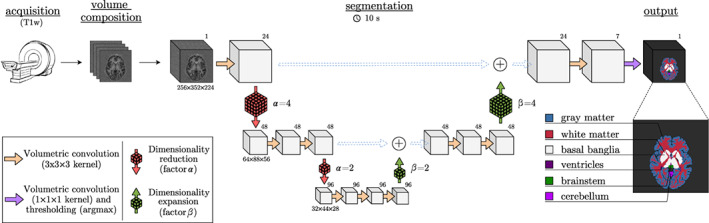
Method overview. The principal database is composed of T1w volumes (MP2RAGE at 7T, 0.63 mm3‐iso) and an inaccurate ground truth (iGT) is obtained using a combination of AFNI‐3dSeg (Cox, 1996) and methods as in Fracasso et al. (2016). Segmentation results are provided in only few seconds (~10 on a desktop CPU). Data dimensions are provided below each volume, while the numbers of filters are in the top‐right corners
CEREBRUM‐7T is tested in three different experimental scenarios. In the first one, the model trained from scratch on a large, but site‐specific data set, is compared against the reference training masks and with other state‐of‐the‐art solutions by a combination of objective numerical evaluations and subjective analysis carried out by experienced neuroscientists. In the second and third scenarios, we test the practical portability of the trained model for researchers working on scans from different MR sites, especially in conditions of limited data availability. In particular, the second scenario explores the condition when only few brain MRI 7T scans are available (e.g., less than 40): under such hypothesis, we show how a researcher can augment data and fine‐tune the pre‐trained CEREBRUM‐7T model starting from a few automatic segmentations. The last scenario simulates instead an extreme data scarcity condition (less than five scans): in such situation we demonstrate the practical portability of the pre‐trained CEREBRUM‐7T model with a fine‐tuning procedure involving very few, but accurately (i.e., manually) segmented volumes. As a last contribution, we make publicly available through the project website set of 142 7T MR scans from Glasgow (UK), 4 the segmentation masks, all the code necessary to train and fine‐tune CEREBRUM‐7T and to perform tests.
2. METHODS
2.1. State‐of‐the‐art
Due to the lack of accurate fully automatic methods, manual segmentation protocols (Wenger et al., 2014; Berron et al., 2017), although time consuming (Zhan et al., 2018; Koizumi et al., 2019), are still a common practise for 7T data. To partially reduce the laboursome process of manual segmentation, the solution proposed by Gulban et al. (2018) combines manual and semi‐automatic segmentation, by adopting a multi‐dimensional transfer function to single out non‐brain tissue voxels in 7T MRI data of nine volunteers. Other semi‐automated methods developed in the past for generic MRI data, such as ITK‐SNAP (Yushkevich et al., 2006), have been adapted by Archila‐Meléndez et al. (2018) for tackling also ultra‐high‐field brain imaging.
Often, given the lack of harmonised neuroimaging analysis protocols, multiple 7T sites created in‐house pipelines to perform MRI segmentation on site‐specific data specifically. Fracasso et al. (2016) developed a custom workflow (used, e.g., in Bergmann et al., 2019) which we also adopt for labelling GM and WM. The placement of the GM/CSF boundary is based on the location of the 75% quantile of the variability of T1w partial volume estimates across cortical depth, while GM/WM boundary from a combination of AFNI‐3dSeg (Cox, 1996) and a clustering procedure (see Section 2.3.2 for more details).
As an attempt to develop a site‐independent approach, Bazin et al. (2014) presented a computational framework for whole brain segmentation at 7T, specifically optimised for MP2RAGE sequences. The authors develop a rich atlas of brain structures, on which they combine a statistical and a geometrical model. The method, which includes a non‐trivial pre‐processing chain for skull stripping and dura estimation, achieves whole brain segmentation and cortical extraction, all within a computation time below 6 hr. Despite these efforts, the most existing solutions, including Bazin et al. (2014) and Nighres by Huntenburg et al. (2018), still generate a variety of segmentation errors that needs to be manually addressed, as reported in Gulban et al. (2018).
All aforementioned solutions—including FreeSurfer v6, FreeSurfer v7, or even BrainVoyager (Goebel, 2012)—can work well, but require multiple pre‐processing and parameters' tweaking steps. The effect is a huge inter‐ and intra‐user variability in the output analysis, as shown for example in Botvinik‐Nezer et al. (2020), in which authors asked 70 independent teams to analyse the same data set, testing the same nine ex‐ante hypotheses; results showed how every single team choses a different workflow for data analysis, leading to sizeable variations in the results of hypothesis tests. In this respect, having a validated tool that works in a fully automatic fashion, which produces systematic outputs—errors included—would be desirable to minimise variability, thus boosting reproducibility and facilitating comparability of downstream research.
2.1.1. Other DL methods for MRI segmentation
Recent advances in DL offer a novel way to improve and automate complex tasks that up until now could only be performed by professionals (Yu et al., 2018). In particular, the advanced classification and segmentation capabilities ensured by DL methods have impacted several medical imaging domains (Litjens et al., 2017; Hamidinekoo et al., 2018). Various segmentation algorithms, which exploit the generalisation capabilities of DL and convolutional neural networks (CNNs) on unseen data, made possible a drastic improvement in the performance with respect to other traditional, mostly atlas‐based, segmentation tools.
To the best of our knowledge, no DL architectures have been directly applied on 7T data for segmentation purposes yet. The only attempt made by Bahrami et al. (2016) to use CNN in this field, aimed at reconstructing 7T‐like images from 3T MRI data. Specifically, from the 3T image intensities and the segmentation labels of 3T patches, the CNN learns a mapping function so as to generate the corresponding 7T‐like image, with quality similar to the ground‐truth 7T MRI.
Restricting the scope to 3T data only, recent DL‐based methods such as QuickNat (Roy et al., 2019), MeshNet (Fedorov et al., 2017; McClure et al., 2019), NeuroNet (Rajchl et al., 2018), DeepNAT (Wachinger et al., 2018) and FastSurfer (Henschel et al., 2020) have been the most effective solutions among those which proposed to obtain a whole brain segmentation. However, a common trait of all the aforementioned methods is that none of them fully exploit the 3D spatial nature of MRI data, thus making segmentation accuracy sub‐optimal. In fact, such solutions partition the brain into 3D sub‐volumes (DeepNAT, MeshNet and NeuroNet) or 2D patches (QuickNAT, FastSurfer), which are processed independently and only eventually reassembled; as recently shown in Reina et al. (2020), the use of “tiling” introduces small but relevant differences during inference that can negatively affect the overall quality of the segmentation result. For example in MRI segmentation, tiling entails a loss of global contextual information, such as the absolute and relative positions of different brain structures, which negatively impacts the segmentation outcome.
The DL model CEREBRUM (Bontempi et al., 2020) is the first attempt to fully exploit the 3D nature of MRI 3T data, taking advantage of both global and local spatial information. This 3D approach adopts an end‐to‐end encoding/decoding fully convolutional structure. Trained in a weakly supervised fashion with 900 whole brain volumes segmented with FreeSurfer v6 (Fischl, 2012), CEREBRUM learns to segment out‐of‐the‐scanner brain volumes, with neither atlas‐registration, pre‐processing, nor filtering, in just ~5–10 s on a desktop GPU.
2.2. Scenarios of application
CEREBRUM‐7T is a segmentation tool useful for researchers working in almost any condition of data availability. The first scenario accounts for the availability of a large brain MRI 7T database (e.g., scans >50). Under this hypothesis, the code we provide allows for training the CEREBRUM‐7T model from scratch, by exploiting inaccurate labelling produced from automatic tools (e.g., Freesurfer, BrainVoyager, etc.).
In a second scenario, only a few brain MRI 7T scans from a different site are available (e.g., 20 < scans < 40). The model trained from scratch using a limited number of samples could lack generalisation abilities. We therefore provide a CEREBRUM‐7T model (pre‐trained on Glasgow data), and a fine‐tuning procedure (and code) to specialise it on a new site data. The term fine‐tuning refers to the procedure of adjusting weights of a network, trained on a large data set, and specialising them to optimally work on a different, usually smaller, data set. Considering that the new data set is in general not drastically different in context to the larger one, as in our case, the pre‐trained model will already have learnt features that are relevant to the specialised classification problem.
The last investigated scenario explores the condition of extreme data scarcity (MRI scans <10, from a different site), which is partially compensated by the presence of excellent segmentation masks obtained via manual procedures. Also in this case we provide a CEREBRUM‐7T pre‐trained model and the data augmentation procedures needed to fine‐tune the network.
The three data sets described in Table 1—one for each of the investigated scenarios—include a large one for training the model from scratch (Glasgow data set, 142 scans, 120 for training); a reduced size data set (Amsterdam Ultra‐high field adult lifespan database [AHEAD] data set, Alkemade et al., 2020; 105 scans, 20 for training) with automatically obtained segmentations; an extremely tiny set (Schneider et al., 2019; 4 scans, 3 for training), with manually segmented masks.
TABLE 1.
Data sets details
| Parameter | Glasgow data | Alkemade et al. (2020) | Schneider et al. (2019) | |||
|---|---|---|---|---|---|---|
| Sequence used | T1w MP2RAGE | |||||
| Field strength | 7 tesla | |||||
| Voxel size |
|
|
|
|||
| Original volume sizes |
|
|
|
|||
| Training volume sizes |
|
|||||
| Training | vol. (+1.1k vol. augmented offline) | vol. (+300 vol. augmented offline) | vol. (+90 vol. augmented offline) | |||
| Validation | volumes | volumes | (original) volumes | |||
| Testing | volumes | volumes | volumes | |||
| Testing (Turing test) | vol. areas | n.a. | n.a. | |||
Neck cropping.
2.3. Scenario 1: Training from scratch with a large data set
In this scenario we train CEREBRUM‐7T model from scratch on a large data set which has been inaccurately labelled via an automated pipeline.
2.3.1. Glasgow data: Acquisition and split
The database consists of 142 out‐of‐the‐scanner volumes obtained with a MP2RAGE sequence at isotropic resolution, using a Siemens 7T Terra Magnetom MRI scanner with 32‐channel head coil. All volumes were collected, as reconstructed DICOM images, at the Imaging Centre of Excellence (ICE) at the Queen Elizabeth University Hospital, Glasgow (UK). The columns of Figure 3 show some selected slices of the out‐of‐the‐scanner T1w, the segmentation resulting from FreeSurfer v6, the one from Fracasso et al. (2016), the one from Huntenburg et al. (2018), the reference iGT, the CEREBRUM‐7T mask and the manual segmentation, respectively.
FIGURE 3.
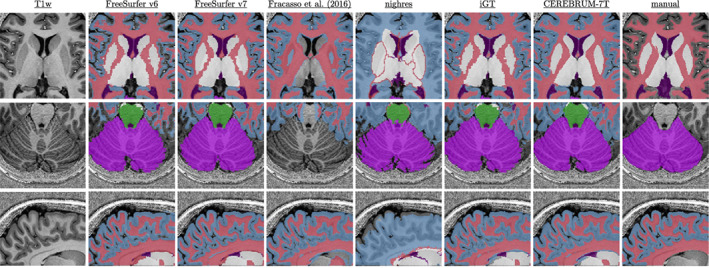
Visual examples of the data set and results. Columns, from left to right, show: T1w scan (left), FreeSurfer v6 and v7 segmentations (Fracasso et al. 2016), nighres, iGT (obtained as described in Section 2.3.2), CEREBRUM‐7T, and manual segmentation. Coloured labels are shown in overlay only when returned by the specific method
Out of the total volumes, are used for training, for validation and for testing (3 of which used in a Turing test). The only pre‐processing applied is the neck cropping using the INV2 scan obtained during acquisition. Data set details are shown in Table 1.
2.3.2. Generation of the iGT
Similarly to most approaches employing DL frameworks for brain MRI segmentation (Roy et al., 2019; McClure et al., 2019; Fedorov et al., 2017; Rajchl et al., 2018; Bontempi et al., 2020), we also adopt an almost fully automatic procedure for labelling, since the prohibitive time cost required to produce a manual annotation on such large data set. Such a decision is also driven by the consideration that, despite the use of an iGT, in already documented cases the trained models proved to perform the same (Rajchl et al., 2018), or even better (Bontempi et al., 2020; Roy et al., 2019), than the iGT used for training.
Differently from Bontempi et al. (2020), it is not possible to use either FreeSurfer v6 (Fischl, 2012) nor FreeSurfer v7, as unique sources for the generation of the iGT. Both tools perform similarly on structures such as brainstem, basal ganglia, cerebellum and ventricles (see, e.g., Figure 11). However, the quality of WM and GM segmentation masks obtained with FreeSurfer v6 is not acceptable (as also hinted in Figure 3, second column), even considering inaccurate supervision for learning. With respect to FreeSurfer v7, even if it is far superior than v6 when it comes to segment WM and GM, it still produces some systematic failures in segmenting specific structures, for example, the temporal lobes, as we show in Figure 13 and in the Supporting Information (Figure S8).
For these reasons, we design a custom pipeline for the iGT generation process (overview in Figure 4) with performance on GM and WM comparable to those offered by FreeSurfer v7, which is fully automatic, free from unpredictable errors, does not require parameter tweaking, and it is more site‐independent and faster than FreeSurfer (in any version). The pipeline accounts for two main branches: the upper one deals with WM and GM segmentation, while the lower one isolates other brain structures such as cerebellum, ventricles, brainstem and basal ganglia. The two processing branches are combined afterwards, when a manual correction step is also carried out to reduce major errors.
FIGURE 4.
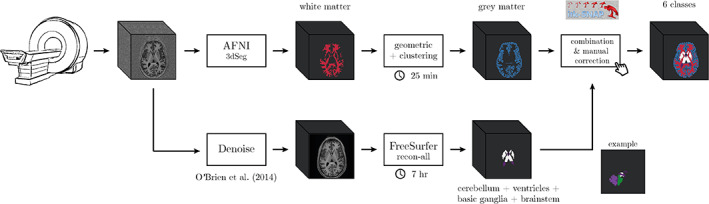
Processing pipeline used to generate the iGT starting from the reconstructed T1w
In the upper branch, the WM mask is obtained using a combination of AFNI‐3dSeg (Cox, 1996) followed by geometric and clustering methods as in Fracasso et al. (2016). Specifically T1w images are co‐registered to an atlas (Desikan et al., 2006) and a brain mask is overlaid to the T1w images to remove the cerebellum and subcortical structures. The T1w images are then separated in six different parts along the posterior to anterior direction to improve intensity homogeneity. Each part is afterwards separately processed by the 3dSeg function in AFNI, to isolate WM. The WM masks obtained from each part are summed together resulting in whole brain WM mask (see Fracasso et al. (2016) for further details).
The GM segmentation exploits such whole brain WM segmentation and an atlas co‐registered to the T1w images (Desikan et al., 2006). Next, a distance map from the WM/GM boundary to the pial surface is built computing the Euclidean distance of each voxel from the WM/GM border. Negative distances are assigned inside WM and positive distances are assigned from WM borders onward. Each region of interest (ROI) in the atlas by Desikan et al. (2006) is selected iteratively. For each ROI the coordinates are divided into four separate subparts using k‐means clustering. For each subpart, voxels within −2 and 7 mm (Euclidean distance) from the WM/GM border are selected and their T1w intensity stored for further analysis. For each cluster 10 bins between −2 and 7 mm are obtained—with each bin containing of the data. For each of them, a partial volume estimate, defined as the SD of T1w intensity as well as the average Euclidean distance for the same bin, is computed. A linear model is then fit between the average Euclidean distance of each bin and the corresponding partial volume estimate. The slope of the linear model can be either positive or negative: if there is a positive slope, the quantile of the SD values is computed among the 10 bins; if, on the other hand, the slope is negative, the quantile is computed. The Euclidean distance of the 25%/75% quantile corresponds to a drop or rise in T1w variability and is considered as the transition between GM and CSF. To improve the obtained GM segmentation, the WM and GM masks are fed to the Cortical Reconstruction using Implicit Surface Evolution algorithm in the Nighres software package (Huntenburg et al., 2018).
Despite the method ensures a robust result in segmenting GM/WM boundary, no cerebellum, ventricles and basal ganglia areas are computed. To address such lack of GT structures, in the lower branch of the pipeline (Figure 4), we use FreeSurfer v6 (Fischl, 2012), which first requires to denoise the T1w volume (O'Brien et al., 2014), to add the following labels: cerebellum, ventricles, brainstem and basal ganglia. The performance of FreeSurfer v6 on these structures is comparable to that offered by FreeSurfer v7 (as shown also in Figure 9), which was just released at the moment of iGT creation. When necessary, a final step of manual correction is carried out to reduce major errors (especially on cerebellum classified as GM) using ITK‐SNAP (Yushkevich et al., 2006).
The z‐scoring procedure, applied to normalise the data before CEREBRUM‐7T training, is obtained using the mean and SD volumes computed on the entire Glasgow data set (shown in Figure 5).
FIGURE 5.
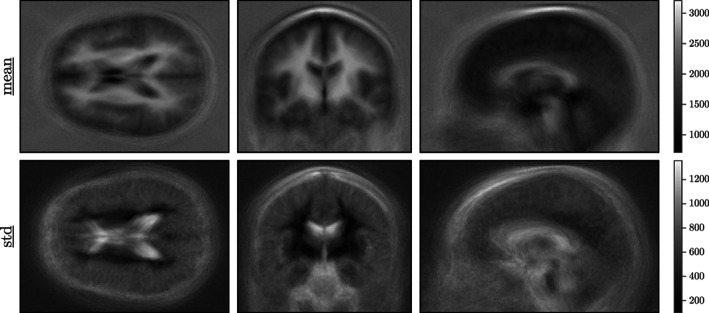
Mean and SD volumes of the database used to z‐score the data. Denoised mean/standard volumes are found in Supporting Information (Figure S5)
2.3.3. Data augmentation
The data corpus used for our experiments is one of the biggest 7T brain MRI publicly and freely available data sets. Yet, given the complexity of the DL architecture, that is, the number of learnable parameters, there are not enough training samples to deliver an off‐the‐shelf model. Therefore, we decide to adopt two customised data augmentation strategies: offline and online data augmentation, as shown in Figure 6.
FIGURE 6.
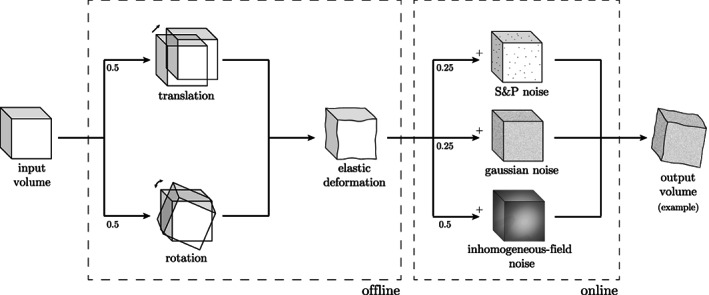
Data augmentation procedure. Offline (with respect to the training procedure), geometric augmentation is performed with rotation or translation and elastic deformation (Çiçek et al., 2016). Online, voxel intensity changes are applied: salt and pepper noise Gaussian noise or inhomogeneous‐field noise An example of data augmentation is shown in the output volume, where one rotation and one elastic deformation are followed by an additional Gaussian noise
Offline augmentation, too computationally demanding to be performed in training‐time, consists in the application of small random shifts voxels) or rotations , on all three axes) and elastic deformations. This ensures an augmentation factor of 10 of the training set.
Online data augmentation, performed during training, comprises variations on voxel intensities only: Gaussian, salt and pepper and inhomogeneous‐field noise. In MRI, and especially in Ultra‐High Field (UHF), the inhomogeneity in the magnetic field produces an almost linear shift in the voxel intensity distributions for different areas in the 3D space (Sled et al., 1998). In other words, the same anatomical structure has different voxel intensities in different areas, for example, GM in frontal and occipital lobes. One of the main limitations of segmentation methods that heavily rely on intensity values is the inability to correctly classify the same class having different local distributions, even if inhomogeneity correction methods are applied as pre‐processing. To increase our model invariance, we introduce, as an additional data augmentation strategy, a synthetic inhomogeneous field noise. We start by pre‐computing a 3D multivariate normal distribution, with zero‐means and twice the dimension (for each axis, i.e., the volume) of the original volume. For each training volume, we randomly sample from the 3D multivariate normal distribution a noise volume as big as the former volume. The so‐generated noise volume is then summed to the anatomical MRI, adding further variability to the volume intensities and simulating distortions along different directions. In Figure 7, a sketch of the method, when applied on a 2D slice example, is shown (see Supporting Information for further examples on 1D and 3D cases—Section 3).
FIGURE 7.
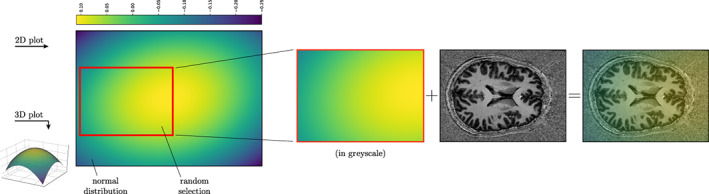
Cartoon to describe the inhomogeneous‐field noise for the 2D case. After pre‐computing a multivariate normal distribution, we randomly extract a noise sample as big as the sample to augment, for every training batch. The extracted patch is then summed to the original sample
2.3.4. Glasgow manual segmentation subset
A portion of the considered data set has been manually annotated by one of the author from the University of Glasgow, who accumulated several years of experience in neuroscience and brain MRI segmentation and reviewed by a radiologist with 20 years of experience. In particular, volumes from three subjects have been randomly selected from the 7T MRI data set, and for each of them, eight regions have been selected and labelled—that is, early visual cortex (EVC), high‐level visual areas (HVC), motor cortex (MCX), cerebellum (CER), hippocampus (HIP), early auditory cortex (EAC), brainstem (BST) and basal ganglia (BGA). Such regions have been chosen among the most commonly brain areas investigated with functional MRI. Since each of the sub‐volumes of interest includes adjacent slices of dimension , the manually labelled data set accounts for a total number of voxels ().
2.3.5. DL model
Similarly to Bontempi et al. (2020), the model architecture is designed to deal with the dimensionality of the training data (i.e., voxels) at once. As shown in Figure 2, the model is a deep encoder/decoder network with three layers, with one, two and three 3D convolutional blocks in the first, second and third levels, respectively .
Since the network is fed with a whole volume as an input, each convolutional block (kernel size , processes the whole brain structure. The full volume helps the model to learn both local and global structures and spatial features (e.g., the absolute and relative positions of different brain components), which are then propagated to subsequent blocks. Dimensionality reduction is achieved using strided convolutions instead of max‐pooling, which contributes to learning the best down‐sampling strategy. A dimensionality reduction (of Factor 4 on each dimension) is computed after the first layer, to explore more abstract spatial features. Eventually, the adoption of both tensor sum and skip connections, instead of concatenation, helps in containing the dimension of the parameter space to .
Training, which takes ~24 hr, is performed on a multi‐GPU machine equipped with GeForce® GTX Ti, on which different parts of the model are distributed. 5 During training, we optimise the categorical cross‐entropy function using Adam (Kingma and Ba, 2014) with a learning rate of and , using dropout on second and third level) and without batch normalisation (Ioffe and Szegedy, 2015), achieving convergence after epochs. The code is written in TensorFlow and Keras.
2.4. Scenario 2: Fine‐tuning with few automatic segmentations
In this scenario, we simulate the condition in which only few brain MRI 7T scans are available, and it is therefore not possible to train the network model from scratch. We provide the CEREBRUM‐7T model pre‐trained on Glasgow data, which is further specialised by fine‐tuning on a smaller data set from a different site, which has been automatically labelled.
2.4.1. Fine‐tuning procedure
Since data acquired from different sites usually significantly differ in statistical distribution—the so called distribution shift (Quionero‐Candela et al., 2009)—simply using the already trained DL on Glasgow data would be ineffective, as the learnt data statistics would not be sufficient to carry out the task on data from different sites. Therefore we present a fine‐tuning procedure which enables to extend the previously trained model on other data sets with fewer scans, also collected in different sites.
All steps of the fine‐tuning procedure are detailed in Figure 8: data preparation (Step 1) includes operations of rotation and cropping and requires the computation of the training labels (either by automatic tools or manually) and the mean/std volumes of the new data set. Afterwards, geometric data augmentation, for both the anatomical and the labelled volumes, is performed offline (Step 2). Step 3 describes the “warming‐up” of the model, in which the new layer weights are learnt without compromising the frozen layer features obtained during the training . Finally, Step 4 is responsible for the fine‐tuning of the entire network .
FIGURE 8.
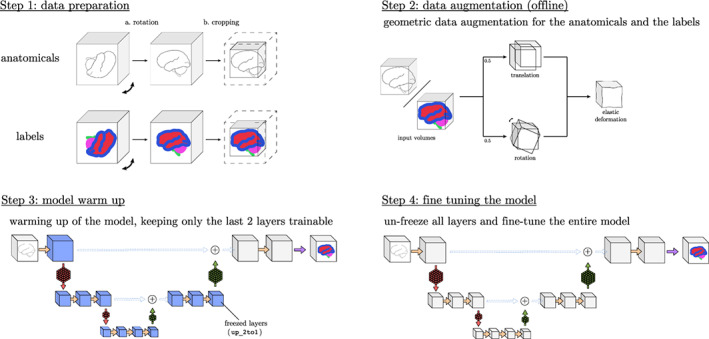
Steps to fine tune the method to a new sequence or site
2.4.2. AHEAD data: Preparation and split
In this scenario, we fine‐tune the model (pre‐trained on Glasgow data) by using only 20 volumes from the AHEAD (Alkemade et al., 2020). The full database consists of 105 7T whole‐brain MRI scans, including both male and female subjects (age range 18–80 years). In order to mimic one real scenario, the labels used for fine‐tuning are obtained by one of the most recent tool openly available on the market: FreeSurfer v07, which is also useful to completely disentangle the model from Fracasso et al. (2016) which was used for training.
We then select 20 volumes, making sure that their segmentation masks is accurate (FreeSurfer v07 presents frequent errors, as discussed above), and we augment every volume times. Validation is performed on 4 volumes, while the remaining 81 are used as a testing set.
2.5. Scenario 3: Fine‐tuning with very few manual segmentations
In this third scenario, we simulate a condition of extreme 7T data scarcity. With the same fine‐tuning procedure used in the previous scenario, we specialise the pre‐trained CEREBRUM‐7T model with only four manually labelled volumes belonging to Schneider et al. (2019).
2.5.1. Schneider et al. (2019) Data set: Preparation and split
Such data set contains sub‐millimetre 7T MRI images of the human brain, which are thought for the supervised training of algorithms to perform tissue class segmentation. In particular it includes pre‐processed MRI images (co‐registered + bias corrected) and corresponding ground truth labels: WM, GM, CSF, ventricles, subcortical, vessels and sagittal sinus. For our purposes we exploit only the MP2RAGE subset, which exhibits accurate manual segmentation masks based on four subjects.
Since the data set contains only four volumes, we exploit a cross‐validation strategy, using three volumes for training and one for testing. However, since the method needs a larger training set, we apply a stronger augmentation procedure, concatenating different volume manipulation strategies: translation, rotation and morphing. Doing so, we create 30 volumes for each training sample (for a total of 90). Due to technical limitations, we decided to fine‐tune the model on six classes only: WM, GM, CSF, ventricles, subcortical and vessels. To ease the task, we also apply a brain mask to the volume, cropping outside the skull.
3. RESULTS
3.1. Experiments on Scenario 1: Training from scratch on Glasgow data
To evaluate CEREBRUM‐7T, in Section 3.1.1 we first provide a quantitative assessment of the obtained segmentation, with and without data augmentation, with respect to the inaccurate labelling obtained by other state‐of‐the‐art methods (iGT). Then, in Section 3.1.2, we evaluate if CEREBRUM‐7T actually outperforms in quality the inaccurate labelling it was trained on. To do so, we present the outcome of the Turing test carried out on a data portion by experienced neuroscientists who were asked to subjectively evaluate the best segmentations among CEREBRUM‐7T, the iGT and a manual segmentation, which serves as a gold standard. In Section 3.1.3, we use manually segmented masks as a reference to rank CEREBRUM‐7T among state‐of‐the‐art methods including FreeSurfer v6, FreeSurfer v7, Fracasso et al. 2016, Huntenburg et al. 2018, and the iGT pipeline. Finally in Section 3.1.4, we show some soft masks associated with regions segmented by CEREBRUM‐7T.
3.1.1. Contribution of data augmentation: CEREBRUM‐7T versus iGT
In order to evaluate the effect of the data augmentation strategy, CEREBRUM‐7T architecture is compared in two variants, with and without data augmentation, against the iGT. The two models are trained by minimising the same loss and using the same learning rate.
Performance is assessed by three metrics adopted by the MICCAI MRBrainS18 challenge, which are among the most popular ones used in the context of segmentation (Taha and Hanbury, 2015): the dice coefficient (DC), a similarity measure which accounts for the overlap between segmentation masks; the Hausdorff Distance computed on its 95th percentile (HD95), which evaluates the mutual proximity between segmentation contours (Huttenlocher et al., 1993); the volumetric similarity (VS) as in (Cárdenes et al., 2009), a non‐overlap‐based metric which considers the similarity between volumes.
The quantitative comparison is outlined in Figure 9 where average results for DC, HD95 and VS obtained on the test volumes are shown class‐wise (i.e., on GM, WM, ventricles, cerebellum, brainstem and basal ganglia). Independently from the observed metric, it is evident the beneficial effect of applying data augmentation strategies.
FIGURE 9.
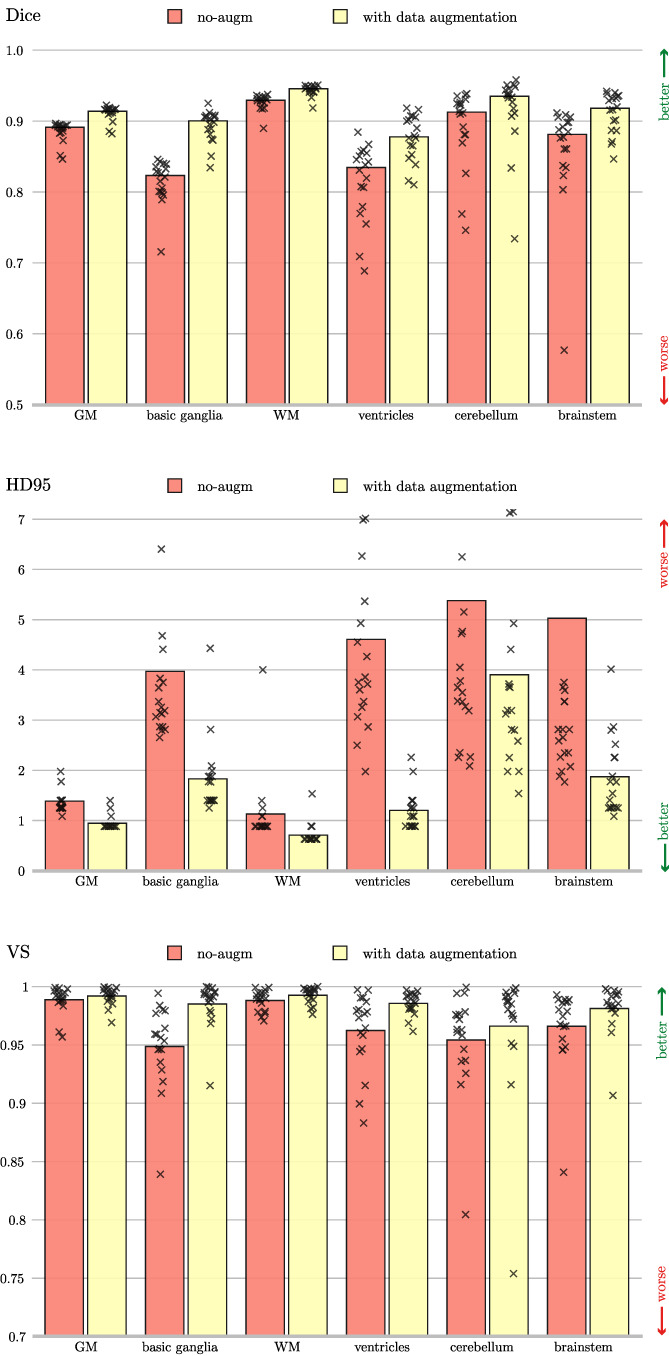
Dice coefficient, 95th percentile Hausdorff distance, and volumetric similarity computed using the inaccurate ground truth (iGT) segmentation as a reference. The data augmented model (yellow), and the model trained without data augmentation (red) are compared. The height of the bar indicates the mean across all the test subjects, while every mark is a tested volume
3.1.2. Turing test: CEREBRUM‐7T versus iGT versus manual segmentation
From the quantitative assessment presented in Section 3.1.1 emerges that there is a relative difference in performance between CEREBRUM‐7T architectures and the iGT used for training. Nevertheless, since performance are measured with respect to an iGT, CEREBRUM‐7T segmentations might be superior to those provided by the inaccurate labelling, as in Bontempi et al. (2020) and Roy et al. (2019), as we also suggest in Figure 3 where CEREBRUM‐7T masks appear more accurate than iGT masks.
To test this hypothesis, we design a Turing test in which seven expert neuroscientists (different from those who generated the manual segmentation) are asked to choose the most accurate results among three provided ones: the mask produced by CEREBRUM‐7T, the iGT and the manual segmentation (intended as gold standard).
If systematically proven, the superiority of CEREBRUM‐7T against the iGT would confirm the validity of the weakly supervised learning approach, resulting in a learnt model with generalisation capability over its training set obtained with state‐of‐the‐art methods. Furthermore, a human expert evaluation, compared to a purely numerical measure, has the advantage to account for the grade of severity of every single segmentation error, giving important feedback on the suitability of the segmentation for the application (Taha and Hanbury, 2015).
The survey participants are presented with a set of randomly arranged slices taken from the manually annotated data set: they are either axial, sagittal, or coronal views from the eight selected areas of interests (see Section 2.3.4 for details) segmented with the three compared methods (CEREBRUM‐7T, iGT and manual segmentation). For each presented couple of segmentation results, the expert is asked to choose the best one between the two, or to skip to the next slice set if unsure. Each participant inspects all eight areas of interest, for each of the three test volumes. To better compare results in a volumetric fashion, it is also possible for the participant to browse among neighbouring slices (two slices before and two after) and interactively change the mask opacity to more easily check for an exact anatomical overlap. A snapshot of the survey interface, coded with PsychoPy (Peirce et al., 2019), is provided in the Supporting Information (Section 1).
The aggregated results of the Turing test are shown in Figure 10a, while results split per different brain areas are given in Figure 10b. In both figures, it is evident that participants judged the segmentation masks generated by CEREBRUM‐7T as more accurate with respect to those of the iGT.
FIGURE 10.
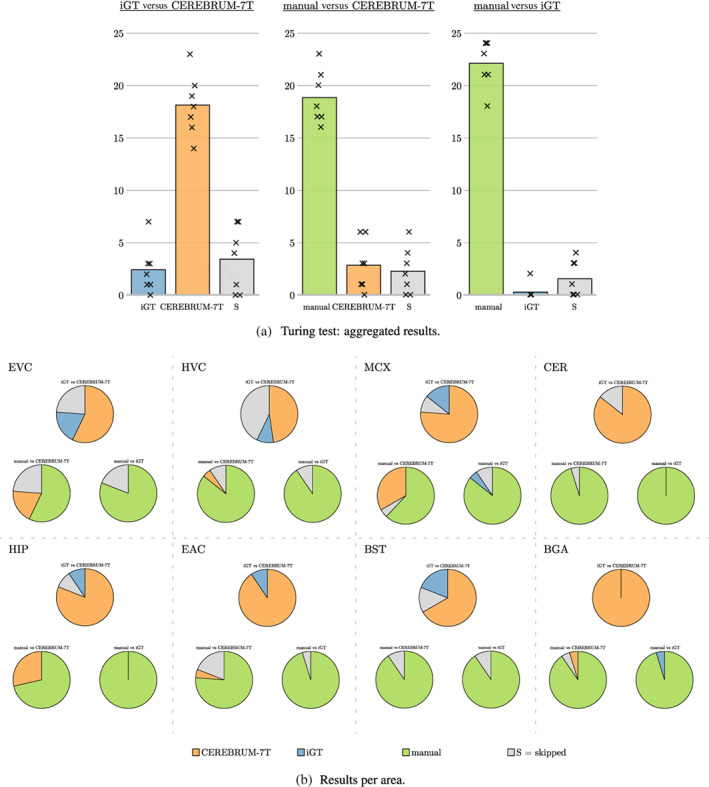
Results of the Turing test. (a) The three subplots show the three comparisons questioned during the survey (manual vs. CEREBRUM‐7T, manual vs. iGT, iGT vs. CEREBRUM‐7T), since segmentations masks were presented in couples. iGT votes are displayed in blue, CEREBRUM‐7T in orange, while skipped responses (S), meaning participants could not choose between the two segmentations, are displayed in grey. The height of bars indicate the means across subjects (i.e., how many times a selection was made, where max. is volumes areas = 24); every mark x is a participant. (b) Results are split per area of interest: early visual cortex (EVC), high‐level visual areas (HVC), motor cortex (MCX), cerebellum (CER), hippocampus (HIP), early auditory cortex (EAC), brainstem (BST), and basal ganglia (BGA)
3.1.3. Quantitative ranking: State‐of‐the‐art methods versus manual segmentation
On the same data set of voxels used in the Turing test (three volumes, eight selected areas per volume) we also perform a purely numerical evaluation, based on DC. Considering manual annotations as a reference, Figure 11 shows that the quality of segmentation labels produced by CEREBRUM‐7T is above other state‐of‐the‐art labelling methods including the iGT, labels obtained by FreeSurfer v6 and Freesurfer v7, by Fracasso et al. (2016) (on applicable classes only, i.e., GM and WM), and those from Huntenburg et al. (2018).
FIGURE 11.
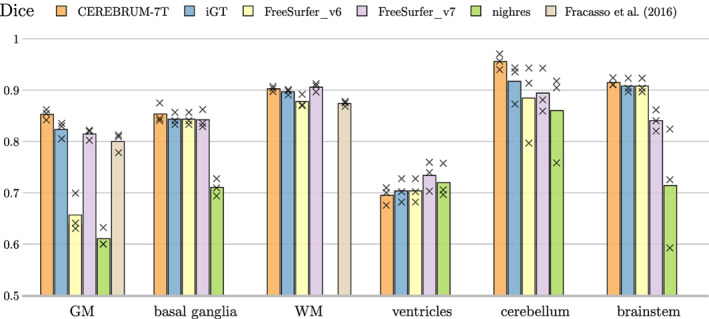
Using manual annotation as a reference, comparison of Dice coefficient between our method (CEREBRUM‐7T), the iGT used for training, FreeSurfer v6, FreeSurfer v7, and the segmentation tools in Fracasso et al. (2016) (only GM/WM) and Nighres by Huntenburg et al. (2018). Every mark is a tested volume from the manually annotated testing set. Nighres result (green bar) is missing for WM since the Dice coefficient is below 0.5
3.1.4. Probability maps
To appreciate the quality of CEREBRUM‐7T output on Glasgow data, in Figure 12 we show the segmentation inferred by the model before thresholding. In testing, the model outputs both the probability maps and the thresholded segmentation mask by default. Since in such probability maps each voxel intensity is associated with the probability of belonging to the most likely class, the reader can inspect the almost total absence of voxel activation outside the correct areas.
FIGURE 12.
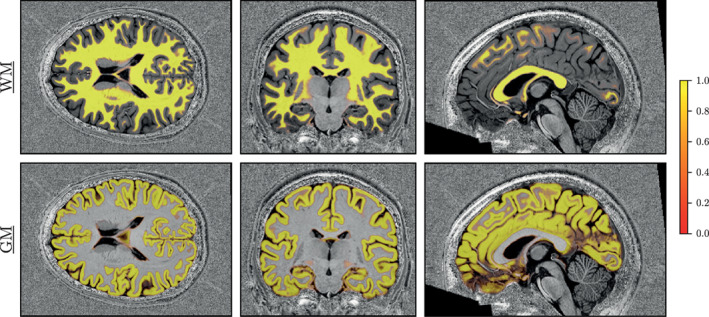
Soft segmentation maps (i.e., probability maps) of a testing volume of Glasgow data for WM and GM classes. The model produces maps with very consistent probabilities, giving additional flexibility than a hard thresholded map. Remaining classes are shown in Supporting Information (Figure S6)
3.2. Experiments on Scenario 2: Fine‐tuning on AHEAD data set
In Figure 13, we show the results of a qualitative comparison (on slices from five different subjects of the AHEAD testing set) using different tools. In particular, the reader can inspect and appreciate the different level of smoothness achieved on the segmentation masks produced by FreeSurfer v7 (top row), CEREBRUM‐T7 off‐the‐shelf (middle row) and CEREBRUM‐T7 fine‐tuned on only 20 volumes (bottom row).
FIGURE 13.
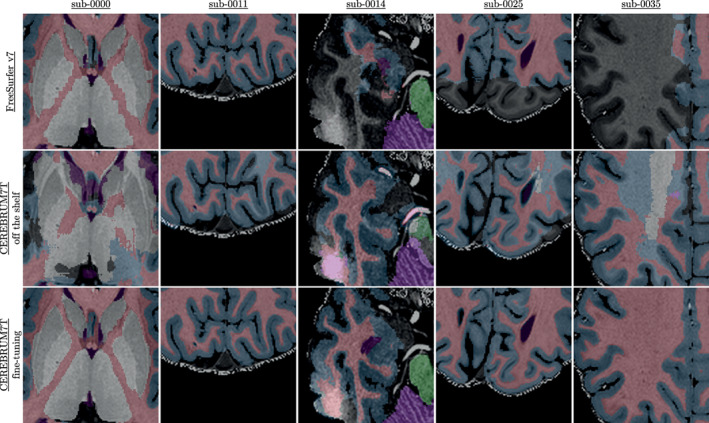
Fine‐tuning results on AHEAD data: comparison between FreeSurfer v7, plain CEREBRUM‐T7 (trained on Glasgow data and tested on AHEAD data), and CEREBRUM‐T7 fine‐tuned for AHEAD data. More results in Supporting Information (Figure S8). Animated GIF on the project website. AHEAD, Amsterdam Ultra‐high field adult lifespan database
3.2.1. Mesh reconstruction on AHEAD data
With CEREBRUM‐7T, it is fast to produce 3D high‐quality models useful for the neuroscientific and biomedical communities. For example, in fMRI studies, researchers need first to isolate specific brain structures (e.g., GM) in order to analyse the spatio‐temporal patterns of activity happening within it. As such, we show in Figure 14 a view on four reconstructed meshes (WM/GM boundary and outer GM boundary) obtained from a testing volume of the independent data set AHEAD, processed by FreeSurfer V7 (left) and CEREBRUM‐7T (right), respectively.
FIGURE 14.
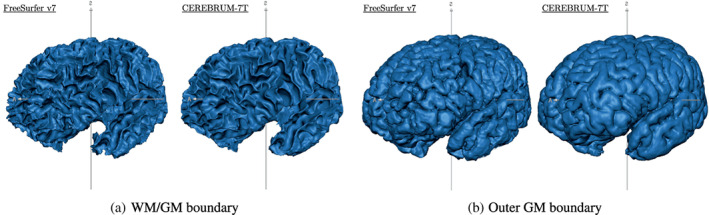
Reconstructed meshes of (a) WM/GM boundary and (b) outer GM boundary of a testing volume of the independent data set AHEAD—sub. 88—for FreeSurfer V7 (left) and CEREBRUM‐7T (right). A light smoothing operation is performed on both meshes (50 iterations—BrainVoyager, Brain Innovation; Goebel, 2012)—no manual corrections performed. We added unsmoothed meshes on Supporting Information (Figure S9). More results (animated GIF) on the website page. AHEAD, Amsterdam Ultra‐high field adult lifespan database; GM, grey matter; WM, white matter
3.3. Experiments on Scenario 3: Fine‐tuning on Schneider et al. (2019) data set
Table 2 shows very accurate segmentation performance obtained by fine‐tuning CEREBRUM‐7T model with only three volumes. Results are obtained in cross‐validation for all four volumes in the data set. Other visual results can be inspected on the project website.
TABLE 2.
Dice coefficient computed on the four brain volumes of Schneider et al. (2019) data
| Subject | Sub‐001 | Sub‐013 | Sub‐014 | Sub‐019 | ||||
| Dice coeff. (tot) |
|
|
|
|
4. DISCUSSION
In this work we present a CNN‐based segmentation algorithm for 7T MRI brain data, which starting from a single MRI sequence (T1w), produces a 3D segmentation mask in only few seconds. Similarly to CEREBRUM, also CEREBRUM‐7T processes the whole brain volume as one, avoiding the drawbacks of the tiling process (Reina et al., 2020), thus preserving both global and local contexts. This partially resembles what happens during manual segmentation: first, the expert looks at the brain volume from afar to identify where different brain structures are located (global clues). Once a coarse segmentation is apparent, the expert begins to segment voxel by voxel at the pixel level, focusing only on a specific area (local processing). For a human, both of these two levels (or scales) of information are needed to perform the segmentation. CEREBRUM‐7T preserves such two‐scale analysis: global features are obtained by analysing the volume at once, without partitioning. The full‐resolution processing of the first layer enables to perform a maximum resolution analysis. A table reporting the receptive fields for each convolutional block of CEREBRUM‐7T can be found in the Supporting Information (Table 1).
Classical automatic (pre‐DL) segmentation tools, instead, emulate these two steps using atlases to gain global clues and, for most of them, gradient methods for the local processing. For what concerns DL segmentation methods based on tiling, they conceptually lack in the gain of global clues. Furthermore, limitations in memory size of accelerator cards, prevented so far large medical volumes from being processed as a whole: thanks to the reduction of network layers we applied on the model architecture, it was possible to make the exploitation of global spatial information computationally tractable. In fact increasing the depth of a CNN does not always allows the model to capture richer structures and yielding better performance. On the contrary, as highlighted by works such as Perone et al. (2018), in some cases the low‐level features extracted by the network prove to be the most important ones—even if the task is complex. Maintaining a small number of layers allow us to analyse the volume at full resolution and at once, gaining both global and local scale: this brings in a sense our DL model closer to an atlas, with respect to any other previous approach, since it finally learns a‐priori probabilities for every voxel.
Since the lack of a universally accepted labelling system, when deciding for the widely adopted labelling strategy and metrics proposed by the MICCAI Society—one of the most prestigious societies dedicated to the practice in the field of medical image computing—we chose to maximise research reproducibility and the possibility to compare our method with state‐of‐the‐art literature. GM, WM, CSF, ventricles, cerebellum, brainstem and basal ganglia were chosen as labels, while results were evaluated in terms of the dice similarity coefficient, the 95th Hausdorff distance and the VS coefficient. Choosing for a different labelling system or metrics would have hampered research comparison and reproducibility. In addition to this, despite the fact that software tools like Freesurfer or MAGeTbrain return a higher number of structures, it is often the case that, depending on the final application, having too many labels is not always useful and re‐clustering is often needed. Moreover, it is in general true that the higher the number of labels, the less accurate the available segmentation. In short, the MICCAI suggested labelling system constitutes a good compromise between flexibility and research reproducibility.
4.1. Fully trained model
From the inspection of results in Figure 9, we observe that the architecture with data augmentation outperforms the baseline solution on every class, independently from the observed metric. This is especially noticeable for HD95, where the difference in the average score between the two configurations (computed on all test volumes) is proportionally more prominent than for other metrics. This might be due either to a larger variability (which might affect the reliability of the measure), or to the fact that, since HD95 accounts for differences in segmentation contours, the beneficial effects given by offline data augmentation (i.e., shifts, rotations and morphing) reflects on an increased accuracy of the segmentation borders. Such interpretation is supported by the observation that smaller brain structures, such as ventricles, brainstem and, where the identification of segmentation boundaries is most critical, are the ones that benefit the most from such augmentation. In summary, results in Figure 9 point out how much the applied data augmentation strategy helps segmentation, significantly improving results from Bontempi et al. (2020).
As for the aggregated results of the Turing test shown in Figure 10a, the direct comparison between CEREBRUM‐7T and the iGT shows that survey participants clearly favoured our proposed solution. Moreover, when both CEREBRUM‐7T and the iGT are compared against manual segmentation, CEREBRUM‐7T obtains more favourable results than iGT. This is confirmed also when results are split per different brain areas, as in Figure 10b: in the comparison against manual, the iGT is almost never chosen, while in selected areas (i.e., EVC, MCX, HIP) CEREBRUM‐7T becomes competitive also against the gold standard offered by manual segmentation. Although the gold standard is built by a single neuroscientist (on three subjects), manual segmentation is here intended only as a reference: even considering multiple annotators (and also in case of low level of inter‐agreements between them) manual segmentation would remain anyway by far the best, 6 not impacting on the comparison between CEREBRUM‐7T and the iGT used for training the model itself.
The manually segmented masks are used as a reference also for quantitatively comparing CEREBRUM‐7T, the iGT, FreeSurfer v6, Freesurfer v7, Fracasso et al. (2016), Huntenburg et al. (2018) on each of the six brain categories. Results shown in Figure 11 confirm that CEREBRUM‐7T returns the most accurate segmentation on all brain structures against all other state‐of‐the‐art methods.
Eventually, the advantages of a fully 3D segmentation method are visible in the soft masks shown in Figure 12. Since most voxels are associated with significant probability of belonging to their correct brain class, such maps highlight the ability of the model to make use of both global and local spatial cues. Furthermore, the almost total absence of spurious activations, confirms the high level of confidence achieved by the model.
4.2. Fine‐tuning
Fine‐tuning experiments are important because they directly tackle one of the main limitation of DL: the need for large training set. Although in such scenarios it is pretty straightforward to apply safer strategies—like decomposing the volumes in slices and apply a slice‐based method—it is, however, important to prove the portability of the trained model and the fine‐tuning procedure, with the objective of releasing an effective and efficient tool applicable on data from new sites.
Looking for example at Figure 13, although we use only 20 automatically segmented volumes for fine‐tuning CEREBRUM‐T7 (Scenario 2), the improvements in the results with respect to using CEREBRUM‐T7 off‐the‐shelf are notable, also with respect to FreeSurfer v7. In particular, in the comparison versus Freesurfer 7, it is evident that CEREBRUM‐7T produces smoother masks. Whereas FreeSurfer v7, which has been improved for UHF data, is able to segment multiple areas (e.g., GM/WM boundary), the inhomogeneity of the scan still affects its ability to correctly select all regions (e.g., the parietal and temporal lobes), often producing holes in the segmentation masks (see the project website for more results). This is also the main reasons why we did not exploit FreeSurfer v7 for the creation of the iGT.
The advantages delivered by a fully 3D segmentation are also visible in the reconstructed meshes built on AHEAD data and shown in Figure 14. By operating as a true 3D structure model, CEREBRUM‐7T ensures globally smoother and more coherent surfaces across slices with respect to 2D methods, both manual and automatic. Commonly adopted editing tools, such as ITK‐SNAP (Yushkevich et al., 2006) or ilastik (Berg et al., 2019), usually display three synchronised 2D orthogonal views onto which the operator draws the contour of the structures. The extraction of a continuous 3D surface from the collection of 2D contours, as well as from 3D tiles, is a nontrivial post‐processing task, where bumps in the reconstructed 3D surface are often inevitable due to inter‐slice inconsistencies in segmentation.
Results obtained in the third scenario show that, with only three (although accurately segmented) volumes employed for fine‐tuning, the predicted labels are very accurate (see Table 2). To further comment results, we need to distinguish two different cases. If we consider classes which are already known by the CEREBRUM‐7T model (i.e., the seven MICCAI labels) such as GM, WM and ventricles—or a combination of previous classes—such as CSF or subcortical (which is a combination of basal ganglia, brainstem and cerebellum), the model takes advantage of the previous learning (on Glasgow data) and simply transfers/applies the knowledge on the new data set, producing accurate results. Conversely, when inferencing on new classes never seen before, like vessels, on which the model has not a prior knowledge, segmentation results are qualitatively lower. 7 However, if a researcher is interested in adding a set of different labels not currently handled by CEREBRUM‐7T, by the provided code it is possible to train (from scratch or via fine‐tuning) a model, using new labels provided by FreeSurfer as iGT or by manual segmentation masks, as similarly done for the seven MICCAI labels. The performance obtained on the seven MICCAI labels, despite their variety in size and morphology, indicates that potential results even on new structures will likely outperform the labels generated by other existing tools. For example, we tested CEREBRUM‐7T ability to segment other small structures, such as blood vessels (see Figure S7 of Supporting Information and the project website for this and other examples).
4.3. Error analysis
Errors made by CEREBRUM‐7T are usually of two kinds: systematic errors and casual ones. While not so much can be said about casual errors (see Figure S8 of Supporting Information for few examples), since they are unpredictable by definition, we observe that systematic errors derive from incorrect training labels. If the model is consistently fed with the same type of errors during training, it will eventually learn to replicate them. Figure 15 shows some systematic errors derived from inaccurate training labels. The first row highlights part of the temporal lobe of sub‐005, with different segmentation masks, FreeSurfer v7, Fracasso et al. (2016), and our method, while the second row reports the segmentation of the basal ganglia area of sub‐013 (all data can be found in EBRAINS Knowledge Graph). The red circles point out the errors made by each method. As it is possible to see, both Fracasso et al. (2016) and CEREBRUM‐7T make mistakes in the same areas, like confusing vessels as GM (second row). Fracasso et al. (2016) also segments as GM the inner boundary close to basal ganglia (second row); this is only partially made by CEREBRUM‐7T, since the basal ganglia and the ventricles labels help to solve the problem masking the incorrect labels. In the first row, it is possible to notice how FreeSurfer v7 performs poorly in the temporal lobe; this is due to the strong inhomogeneity values within every scan, which would require an expert intervention and more time. These errors are consistent and occur in almost every scan (as noticeable in the results on the independent data set AHEAD, visible at the project website). While some types of errors, such as the inner GM, can be easily overcome using more training samples, few others are easily removable performing manual correction post CEREBRUM‐7T. The type of error made by FreeSurfer v7 in the temporal lobe may require a few hours to be manually corrected. These errors are the fundamental reason behind our choice to use Fracasso et al. (2016) to compose the training masks, alongside with random errors made by the default setting of FreeSurfer v7, as shown in Figure 13, which could have required a large amount of time in pre‐processing and parameter tweaking by experts.
FIGURE 15.
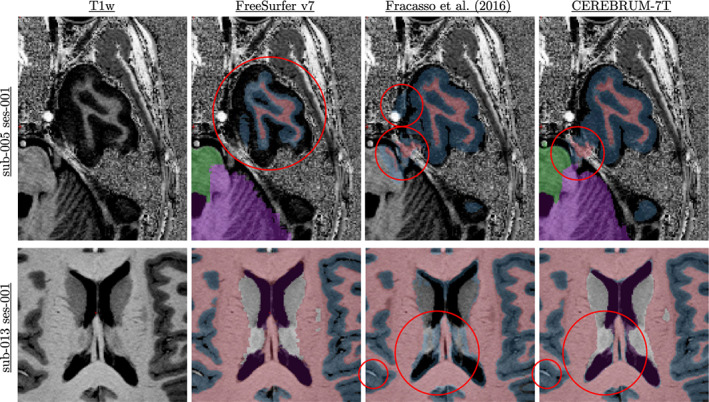
Systematic errors made by different methods. The first row highlights part of the temporal lobe of sub‐005, with different segmentation masks, FreeSurfer v7 (Fracasso et al. 2016), and our method, while the second row reports the segmentations of the basal ganglia area of sub‐013. The red circles point out the errors made by each method
4.4. Limitations
Although the inference can be easily done in a few seconds on a normal CPU, full training is much more time consuming (24 hr) and needs dedicated hardware (i.e., GPUs 1,080 Ti or RTX8000). Furthermore, as described above, although the quality of obtained results is superior to labels used for training (in the condition of inaccurate GT), such model is not able to overcome systematic errors.
Having decided to process the whole volume at once, which required to maintain a model with low level of complexity, it was not possible to include network elements which are very popular in recent DL architectures (e.g., dense layers). As another downside of the choice of processing the whole brain volume at once, it was not possible to increase the batch size to a value greater than one, due to the technological constraints of GPU memory. We chose to analyse the volume in its entirety, instead of exploiting the advantages that the increase of the batch size could carry. With the rapid increase of hardware capabilities, we are confident to be able soon to manage more recent architecture elements and larger batch sizes.
Being developed on proprietary data, and especially inserted in a study pipeline mostly focused on WM/GM analysis, we performed neck cropping on data. Such reduction in the size of the input volume allowed for an increase in the filter number, increasing model capability. To provide another example of this, additionally showing the flexibility of the method, we crop both T1w and iGT on the visual cortex and we retrain the model on GM and WM classes only. Segmentation results are presented in the Supporting Information (see Section 5).
As another main limitation, which is currently under investigation, we cite the need of fine‐tuning in case of new data sets coming from different sites. Despite this phase requires manual intervention, however, for each newly given 7T data set, fine‐tuning is performed just once and does not depend on the operator, nor on the used data set, does not require intervention on single volumes, and once trained, the model works without any additional step, in just few seconds. This makes our solution more generalizable and more leaning towards reproducibility than other common software suites, which heavily suffer from high variability within and between operators. Besides this, on a longer‐term vision, the fine‐tuning phase can be fully engineered too and integrated with the other main modules in a software suite, which is again more suitable for clinical studies (e.g., as in Isensee et al. (2021).
Lastly, our choice to perform segmentation only on one sequence (i.e., T1w) was made in order to limit the scanning time, which is a constraint often imposed for reducing the patient discomfort. This choice also avoids the need for sequence alignment, and the reduction of distortion and morphing which are typical of each sequence. However, for whoever would like to develop a segmentation method combining multiple sequences, the code provided can be easily extended to other sequences without adding much more complexity to the model.
5. CONCLUSIONS
In this work, we design and test CEREBRUM‐7T, an optimised end‐to‐end DL architecture that allows the segmentation of a whole MRI brain volume acquired at 7T at once. The speed of computation, which could be decisive in clinical situations where turnaround time is important for timely decision‐making, and the quality of obtained results (i.e., above the labelling used for training), make CEREBRUM‐7T one of the most advantageous fully automatic solutions for 7T MRI brain segmentation among the few currently available. Furthermore, as shown above, by following a simple fine‐tuning procedure, any researcher in the field is able to use CEREBRUM‐7T to segment brain data from different MRI sites. In order to allow other researchers to replicate and build upon CEREBRUM‐7T findings, we make code, 7T data, and other materials (including iGT and Turing test) available to readers. 8
AUTHOR CONTRIBUTIONS
Michele Svanera: Conceptualisation, methodology, software, validation, formal analysis, investigation, data curation, writing‐review and editing, visualisation. Sergio Benini: Conceptualisation, writing‐original draft, writing‐review and editing, supervision, project administration. Dennis Bontempi: Conceptualisation, methodology, software, writing‐original draft, writing‐review and editing. Lars Muckli: Resources, funding acquisition.
Supporting information
Data S1 Supporting Information
ACKNOWLEDGMENTS
This project has received funding from the European Union Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 and 945539 (Human Brain Project SGA2 and SGA3). We thank Dr Alessio Fracasso for assistance with the inaccurate ground truth creation, the Muckli lab for the Turing test and neuroimaging knowledge, Dr Mattia Savardi for very useful technical comments, Dr Alberto Signoroni for fruitful discussion on learning strategies, Dr Lucy Petro for comments improving the manuscript and Mrs Frances Crabbe for useful feedbacks on manual segmentation.
Svanera, M. , Benini, S. , Bontempi, D. , & Muckli, L. (2021). CEREBRUM‐7T: Fast and Fully Volumetric Brain Segmentation of 7 Tesla MR Volumes. Human Brain Mapping, 42(17), 5563–5580. 10.1002/hbm.25636
[Correction added on 07 October 2021, after first online publication: Supplementary material has been updated.]
Funding information European Union Horizon 2020 Framework Programme for Research and Innovation
Endnotes
OpenNeuro: https://openneuro.org/
EBRAINS: https://search.kg.ebrains.eu/
With the term “out‐of‐scanner” we refer to the reconstructed data saved in DICOM 2D images.
Data are openly available under EBRAINS knowledge graph (http://doi.org/10.25493/RF12‐09N).
Other graphic cards are also suitable for the purpose, such as RTX 8000 or RTX 3090.
In order to let the reviewer verify the quality of the produced maks, we have also released the segmentations on the openNeuro project (https://openneuro.org/datasets/ds003642/).
Visual results on these structures can be inspected on the project website.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in EBRAINS Knowledge Graph at http://doi.org/10.25493/RF12‐09N, reference number RF12‐09N. We additionally released a dedicated website with the code, docker and singularity containers, data link and several visual results at https://rocknroll87q.github.io/cerebrum7t/.
REFERENCES
- Alkemade, A. , Mulder, M. J. , Groot, J. M. , Isaacs, B. R. , van Berendonk, N. , Lute, N. , … Forstmann, B. U. (2020). The Amsterdam ultra‐high field adult lifespan database (ahead): A freely available multimodal 7 tesla submillimeter magnetic resonance imaging database. NeuroImage, 221, 117200. [DOI] [PubMed] [Google Scholar]
- Archila‐Meléndez, M. E. , Valente, G. , Correia, J. M. , Rouhl, R. P. W. , van Kranen‐Mastenbroek, V. H. , & Jansma, B. M. (2018). Sensorimotor representation of speech perception. Cross‐decoding of place of articulation features during selective attention to syllables in 7t fmri. eNeuro, 5(2). https://www.eneuro.org/content/5/2/ENEURO.0252-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami, K. , Shi, F. , Rekik, I. , & Shen, D. (2016). Convolutional neural network for reconstruction of 7t‐like images from 3t mri using appearance and anatomical features. In Carneiro G., Mateus D., Peter L., Bradley A., Tavares J. M. R. S., Belagiannis V., et al. (Eds.), Deep learning and data labeling for medical applications (pp. 39–47). Cham: Springer International Publishing. [Google Scholar]
- Bazin, P.‐L. , Weiss, M. , Dinse, J. , Schaefer, A. , Trampel, R. , & Turner, R. (2014). A computational framework for ultra‐high resolution cortical segmentation at 7 tesla. NeuroImage, 93, 201–209. [DOI] [PubMed] [Google Scholar]
- Berg, S. , Kutra, D. , Kroeger, T. , Straehle, C. N. , Kausler, B. X. , Haubold, C. , … Kreshuk, A. (2019). ilastik: Interactive machine learning for (bio)image analysis. Nature Methods, 16, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Bergmann, J. , Morgan, A. T. & Muckli, L. (2019). Two distinct feedback codes in v1 for “real” and “imaginary” internal experiences. bioRxiv.
- Berron, D. , Vieweg, P. , Hochkeppler, A. , Pluta, J. , Ding, S.‐L. , Maass, A. , … Wisse, L. (2017). A protocol for manual segmentation of medial temporal lobe subregions in 7tesla mri. NeuroImage: Clinical, 15, 466–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontempi, D. , Benini, S. , Signoroni, A. , Svanera, M. , & Muckli, L. (2020). Cerebrum: A fast and fully‐volumetric convolutional encoder‐decoder for weakly‐supervised segmentation of brain structures from out‐of‐the‐scanner mri. Medical Image Analysis, 62, 101688. [DOI] [PubMed] [Google Scholar]
- Botvinik‐Nezer, R. , Holzmeister, F. , Camerer, C. F. , Dreber, A. , Huber, J. , Johannesson, M. , … Schonberg, T. (2020). Variability in the analysis of a single neuroimaging dataset by many teams. Nature, 582, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenes, R. , de Luis‐García, R. , & Bach‐Cuadra, M. (2009). A multidimensional segmentation evaluation for medical image data. Computer Methods and Programs in Biomedicine, 96(2), 108–124. [DOI] [PubMed] [Google Scholar]
- Çiçek, Ö. , Abdulkadir, A. , Lienkamp, S. S. , Brox, T. , & Ronneberger, O. (2016). 3D U‐net: Learning dense volumetric segmentation from sparse annotation. In Ourselin S., Joskowicz L., Sabuncu M. R., Unal G., & Wells W. (Eds.), Medical image computing and computer‐assisted intervention—MICCAI 2016 (pp. 424–432). Cham: Springer International Publishing. [Google Scholar]
- Clarke, W. T. , Mougin, O. , Driver, I. D. , Rua, C. , Morgan, A. T. , Asghar, M. , … Bowtell, R. (2020). Multi‐site harmonization of 7 tesla mri neuroimaging protocols. NeuroImage, 206, 116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R. W. (1996). Afni: Software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research, 29(3), 162–173. [DOI] [PubMed] [Google Scholar]
- Desikan, R. S. , Ségonne, F. , Fischl, B. , Quinn, B. T. , Dickerson, B. C. , Blacker, D. , … Killiany, R. J. (2006). An automated labeling system for subdividing the human cerebral cortex on mri scans into gyral based regions of interest. NeuroImage, 31(3), 968–980. [DOI] [PubMed] [Google Scholar]
- Duyn, J. H. (2012). The future of ultra‐high field mri and fmri for study of the human brain. NeuroImage, 62(2), 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva, A. , Robicquet, A. , Ramsundar, B. , Kuleshov, V. , DePristo, M. , Chou, K. , … Dean, J. (2019). A guide to deep learning in healthcare. Nature Medicine, 25(1), 24–29. [DOI] [PubMed] [Google Scholar]
- Fedorov, A. , Johnson, J. , Damaraju, E. , Ozerin, A. , Calhoun, V. , and Plis, S. (2017). End‐to‐end learning of brain tissue segmentation from imperfect labeling. 2017 International Joint Conference on Neural Networks (IJCNN), pages 3785–3792. https://ieeexplore.ieee.org/document/7966333. [Google Scholar]
- Fischl, B. (2012). FreeSurfer. NeuroImage, 62(2), 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracasso, A. , van Veluw, S. J. , Visser, F. , Luijten, P. R. , Spliet, W. , Zwanenburg, J. J. , … Petridou, N. (2016). Lines of baillarger in vivo and ex vivo: Myelin contrast across lamina at 7t mri and histology. NeuroImage, 133, 163–175. [DOI] [PubMed] [Google Scholar]
- Goebel, R. (2012). BrainVoyager—Past, present, future. NeuroImage, 62(2), 748–756. [DOI] [PubMed] [Google Scholar]
- Gulban, O. F. , Schneider, M. , Marquardt, I. , Haast, R. A. , & De Martino, F. (2018). A scalable method to improve gray matter segmentation at ultra high field mri. PLoS One, 13(6), e0198335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidinekoo, A. , Denton, E. , Rampun, A. , Honnor, K. , & Zwiggelaar, R. (2018). Deep learning in mammography and breast histology, an overview and future trends. Medical Image Analysis, 47, 45–67. [DOI] [PubMed] [Google Scholar]
- Henschel, L. , Conjeti, S. , Estrada, S. , Diers, K. , Fischl, B. , & Reuter, M. (2020). Fastsurfer—A fast and accurate deep learning based neuroimaging pipeline. NeuroImage, 219, 117012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntenburg, J. M. , Steele, C. J. , & Bazin, P.‐L. (2018). Nighres: Processing tools for high‐resolution neuroimaging. GigaScience, 7(7), giy082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher, D. P. , Klanderman, G. A. , & Rucklidge, W. J. (1993). Comparing images using the hausdorff distance. IEEE Transactions on Pattern Analysis and Machine Intelligence, 15(9), 850–863. [Google Scholar]
- Ioffe, S. , & Szegedy, C. (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. CoRR, 37, 448–456. [Google Scholar]
- Isensee, F. , Jaeger, P. F. , Kohl, S. A. , Petersen, J. , & Maier‐Hein, K. H. (2021). nnu‐net: A self‐configuring method for deep learning‐based biomedical image segmentation. Nature Methods, 18(2), 203–211. [DOI] [PubMed] [Google Scholar]
- Kingma, D. P. and Ba, J. (2014). Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
- Koizumi, A. , Zhan, M. , Ban, H. , Kida, I. , De Martino, F. , Vaessen, M. J. , … Amano, K. (2019). Threat anticipation in pulvinar and in superficial layers of primary visual cortex (v1). Evidence from layer‐specific ultra‐high field 7t fmri. eNeuro, 6(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens, G. , Kooi, T. , Bejnordi, B. E. , Setio, A. A. A. , Ciompi, F. , Ghafoorian, M. , … Sanchez, C. I. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60–88. [DOI] [PubMed] [Google Scholar]
- Lu, B. , Li, H.‐X. , Chang, Z.‐K. , Li, L. , Chen, N.‐X. , Zhu, Z.‐C. , Zhou, H.‐X. , Li, X.‐Y. , Wang, Y.‐W. , Cui, S.‐X. , et al. (2021). A practical alzheimer disease classifier via brain imaging‐based deep learning on 85,721 samples. bioRxiv, 2020–2008 pp.
- McClure, P. , Rho, N. , Lee, J. A. , Kaczmarzyk, J. R. , Zheng, C. Y. , Ghosh, S. S. , … Pereira, F. (2019). Knowing what you know in brain segmentation using bayesian deep neural networks. Frontiers in Neuroinformatics, 13, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrik, A. M. , Vincken, K. L. , Kuijf, H. J. , Breeuwer, M. , Bouvy, W. H. , de Bresser, J. , … Viergever, M. A. (2015). MRBrainS challenge: Online evaluation framework for brain image segmentation in 3T MRI scans. Computational Intelligence and Neuroscience, 2015, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, K. R. , Kober, T. , Hagmann, P. , Maeder, P. , Marques, J. , Lazeyras, F. , … Roche, A. (2014). Robust t1‐weighted structural brain imaging and morphometry at 7t using mp2rage. PLoS One, 9(6), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce, J. , Gray, J. R. , Simpson, S. , MacAskill, M. , Höchenberger, R. , Sogo, H. , … Lindeløv, J. K. (2019). PsychoPy2: Experiments in behavior made easy. Behavior Research Methods, 51(1), 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perone, C. S. , Calabrese, E. , & Cohen‐Adad, J. (2018). Spinal cord gray matter segmentation using deep dilated convolutions. Scientific Reports, 8(1), 5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quionero‐Candela, J. , Sugiyama, M. , Schwaighofer, A. , & Lawrence, N. D. (2009). Dataset shift in machine learning. Cambridge, MA: The MIT Press. [Google Scholar]
- Rajchl, M. , Pawlowski, N. , Rueckert, D. , Matthews, P. M. , and Glocker, B. (2018). NeuroNet: Fast and robust reproduction of multiple brain image segmentation pipelines. arXiv:1806.04224.
- Reina, G. A. , Panchumarthy, R. , Thakur, S. P. , Bastidas, A. , & Bakas, S. (2020). Systematic evaluation of image tiling adverse effects on deep learning semantic segmentation. Frontiers in Neuroscience, 14, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A. G. , Conjeti, S. , Navab, N. , Wachinger, C. , & ADNI . (2019). QuickNAT: A fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage, 186, 713–727. [DOI] [PubMed] [Google Scholar]
- Schneider, M. , Gulban, F. O. , and Goebel, R. (2019). Data set for sub‐millimetre MRI tissue class segmentation.
- Sled, J. G. , Zijdenbos, A. P. , & Evans, A. C. (1998). A nonparametric method for automatic correction of intensity nonuniformity in mri data. IEEE Transactions on Medical Imaging, 17(1), 87–97. [DOI] [PubMed] [Google Scholar]
- Taha, A. A. , & Hanbury, A. (2015). Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Medical Imaging, 15(1), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen, D. C. , Smith, S. M. , Barch, D. M. , Behrens, T. E. , Yacoub, E. , & Ugurbil, K. (2013). The WU‐Minn human connectome project: An overview. NeuroImage, 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachinger, C. , Reuter, M. , & Klein, T. (2018). DeepNAT: Deep convolutional neural network for segmenting neuroanatomy. NeuroImage, 170, 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger, E. , Mårtensson, J. , Noack, H. , Bodammer, N. C. , Kühn, S. , Schaefer, S. , … Lövdén, M. (2014). Comparing manual and automatic segmentation of hippocampal volumes: Reliability and validity issues in younger and older brains: Comparing manual and automatic segmentation of Hc volumes. Human Brain Mapping, 35(8), 4236–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, K.‐H. , Beam, A. L. , & Kohane, I. S. (2018). Artificial intelligence in healthcare. Nature Biomedical Engineering, 2(10), 719–731. [DOI] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Piven, J. , Cody Hazlett, H. , Gimpel Smith, R. , Ho, S. , Gee, J. C. , & Gerig, G. (2006). User‐guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage, 31(3), 1116–1128. [DOI] [PubMed] [Google Scholar]
- Zhan, M. , Goebel, R. , & de Gelder, B. (2018). Ventral and dorsal pathways relate differently to visual awareness of body postures under continuous flash suppression. eNeuro, 5(1), ENEURO.0285–ENEU17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Supporting Information
Data Availability Statement
The data that support the findings of this study are openly available in EBRAINS Knowledge Graph at http://doi.org/10.25493/RF12‐09N, reference number RF12‐09N. We additionally released a dedicated website with the code, docker and singularity containers, data link and several visual results at https://rocknroll87q.github.io/cerebrum7t/.


