Abstract
Although we develop influenza immunity from an early age, it is insufficient to prevent future infection with antigenically novel strains. One proposed way to generate long-term protective immunity against a broad range of influenza virus strains is to boost responses to the conserved epitopes on the hemagglutinin, the major surface glycoprotein on the influenza virus. Influenza-specific humoral immunity comprises a large fraction of the overall immune memory in humans, and it has been long recognized that preexisting immunity to influenza shapes the response to subsequent influenza infections and vaccinations. However, the mechanisms by which preexisting immunity modulates the response to influenza vaccination are still not completely understood. Using a set of mathematical models, we explore several hypotheses that may contribute to diminished boosting of antibodies to conserved epitopes after repeated vaccinations.
With the exception of severely immunocompromised individuals and highly isolated populations on remote islands, no one is immunologically naive to influenza. Even infants acquire immunity transferred to them from their mothers, which can affect the immune response to vaccination or infection (Albrecht et al. 1977; Halasa et al. 2008; Voysey et al. 2017; Willis et al. 2020). In unvaccinated populations it is estimated that the first influenza infection occurs within the first 5 years of life (Bodewes et al. 2011). Influenza immunity is therefore established early in life and generates long-lasting immunity to the infecting strain. For instance, elderly individuals were largely protected from infection by the 2009 pandemic H1N1 strain (Centers for Disease and Prevention 2009), and it has been suggested that this is due to increased cross-reactive antibody titers elicited by antigenically similar H1N1 strains that circulated more than 60 years earlier (Hancock et al. 2009; Skountzou et al. 2010). Thus, immunity to influenza can last several decades. However, we can be re-infected with influenza multiple times as the virus constantly mutates and reassorts to escape preestablished immunity. In fact, it has been estimated that unvaccinated individuals are infected with a new strain of influenza every 3–7 yr (Kucharski et al. 2015).
There are two ways in which circulating influenza strains change over time. Antigenic drift arises from the gradual accumulation of mutations in the hemagglutinin (HA) and neuraminidase (NA) glycoproteins that are the main antigens on the viral surface. Antigenically drifted viral strains can evade antibodies elicited following infection with previous strains, thus allowing the drifted virus to spread in the population. Antigenic drift is responsible for seasonal outbreaks of influenza and requires periodic reformulation of the vaccine. Antigenic shifts largely arise from reassortment of zoonotic influenza strains. Antigenic shifts can cause more severe pandemics such as the 1957 and 1968 pandemics of H2N2 and H3N2 subtypes. Although there is significant sequence diversity between strains, there are some constraints on the variation of the HA protein. In particular, the mutations that cause antigenic drift are concentrated in the “head” of HA (the portion furthest from the membrane), and there are more conserved regions on the “stem” of the HA molecule that can elicit cross-protective antibodies (Doud and Bloom 2016; Neu et al. 2016).
The effect of preestablished immunity on the immune response to influenza infection or vaccination has been a conundrum that has riddled the influenza field since its inception. Studies conducted in the 1950s demonstrated that there was an effect of preexisting immunity on subsequent immune responses to infection or vaccination (Francis 1955, 1960). To complicate matters, preestablished influenza immunity varies based on personal history of influenza exposure, which is highly variable, particularly between different age groups (Linderman et al. 2014; Gostic et al. 2019). Although the sequential exposure to antigenically distinct strains constantly shapes the antibody repertoire in a given individual, we might expect boosting of responses to conserved epitopes with each new influenza infection. In animal model systems, an increase in the level of antibodies to the conserved region of the stem of HA has been shown to provide heterosubtypic protection (Wei et al. 2010; Kanekiyo et al. 2013; Krammer et al. 2013, 2014; Margine et al. 2013; Impagliazzo et al. 2015; Yassine et al. 2015). Similarly, there is evidence in humans suggesting that antibodies elicited by responses to virus subtypes in a given clade can provide limited protection to other viruses in the same clade (but not necessarily to viruses in a different clade) (Gostic et al. 2016).
One longstanding question is why repeated influenza vaccinations and infections do not boost cross-reactive antibody titers to sufficient levels to prevent infection. Here, we review our understanding of human immune responses to influenza vaccination and how preexisting immunity affects the response to vaccination. We discuss the data on the magnitude of antibody responses to different epitopes of influenza and how these are boosted by vaccination. We then examine how simple mathematical models can be used to understand the factors contributing to the effect that preexisting immunity has on the boosting of humoral immune responses by vaccination.
HIGH PREEXISTING IMMUNITY TO INFLUENZA IN HUMANS
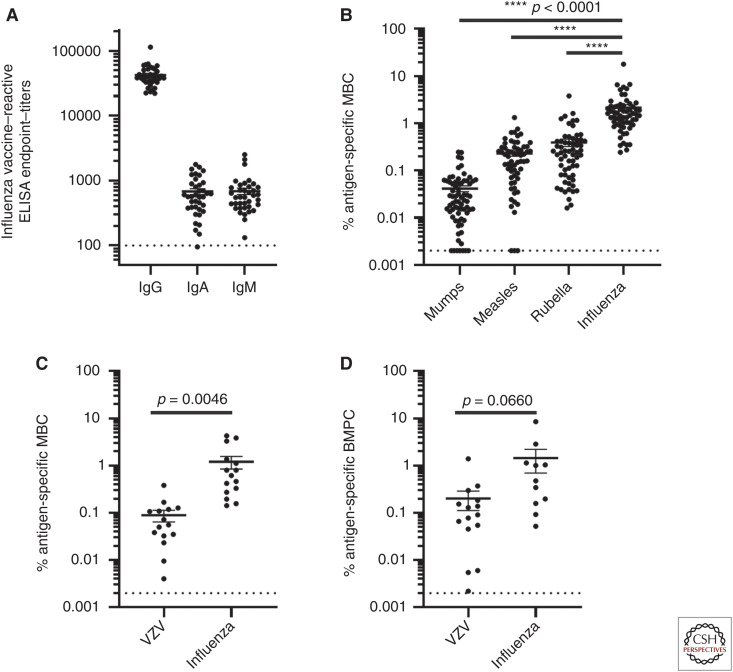
Healthy adults accumulate high levels of immunity to influenza viruses via infections and vaccinations. For instance, influenza vaccine–reactive IgG, IgA, and IgM antibodies are readily measurable by enzyme-linked immunosorbent assay (ELISA) in the peripheral blood of healthy adults (Fig. 1A). Influenza vaccine–specific memory B cells (MBCs) account for ∼1% of IgG MBCs in the blood. This is significantly higher than is found for other common vaccine antigens such as mumps, measles, rubella, or varicella-zoster virus (VZV) (Fig. 1B,C; Rasheed et al. 2019; Eberhardt et al. 2020). Additionally, influenza vaccine–specific plasma cells account for ∼1% of total IgG plasma cells in the bone marrow (Fig. 1D; Davis et al. 2020; Eberhardt et al. 2020). This may be an underestimate for at least two reasons: there may be additional influenza-reactive MBCs and long-lived plasma cells to other influenza antigens not included in these this analyses, and we are not detecting the responses to prior strains if they are not cross-reactive with the current vaccine strains.
Figure 1.
Adults have high levels of preexisting immunity to influenza. Influenza vaccine–reactive IgG, IgA, and IgM antibody titers were measured in healthy human adults by ELISA (n = 38) (A). Frequency of antigen-specific memory B cells specific for mumps, measles, rubella, and influenza (B), or of VZV and influenza (C) were measured by stimulation of PBMCs and quantification by ELISpot as previously described (Rasheed et al. 2019; Eberhardt et al. 2020). VZV and influenza-specific IgG bone marrow plasma cells were quantified by ELISpot and plotted as the percentage of total IgG secreting bone marrow plasma cells (D).
Because of the continuous mutation and reassortment of circulating influenza strains, novel epitopes arise over time, predominantly in the head of HA, and these changes allow the virus to escape despite preexisting immunity. Thus, to improve vaccine effectiveness, we need to better understand how preexisting immunity impacts the ability to boost cross-reactive epitopes and generate antibodies to new epitopes of the currently circulating virus.
MEMORY RECALL RESPONSES AFTER VACCINATION
It has been previously described that there is a transient increase in plasmablast numbers in the peripheral blood at around day 7 postvaccination or postinfection (Wrammert et al. 2011; Li et al. 2012; Ellebedy et al. 2016). This cell population has been shown to be largely influenza antigen–specific and somatically mutated, suggesting that it is derived from a memory population that is rapidly activated upon reexposure to cross-reactive antigen. After primary vaccination or infection with a highly divergent influenza strain such as the 2009 pandemic H1N1 strain or a 2005 H5N1 strain, the majority of these plasmablasts are specific for the more highly conserved HA stem region (Wrammert et al. 2011; Li et al. 2012; Ellebedy et al. 2014). This is likely a result of the high precursor frequency of cross-reactive HA stem specific MBCs and a low frequency of B cells to the head of the new influenza HA (because of the low sequence homology between HA head of novel pandemic and seasonal influenza strains). Increased precursor frequency of expanded MBC clones compared to naive B cells gives memory B cells an advantage during a recall response. Additionally, because of previous somatic hypermutation, MBC B-cell receptors are also expected to have higher affinity for antigen than naive B cells (Eisen et al. 1969). Selective recall of cross-reactive MBCs rather than exclusive activation of naive B cells likely contributes to the observation of “original antigenic sin” (OAS) (Francis 1955, 1960) in which the immune response is skewed toward cross-reactive epitopes that may have a lower affinity for the antigenically drifted strain.
PREEXISTING ANTIBODY TITERS AFFECT THE MAGNITUDE OF RESPONSE
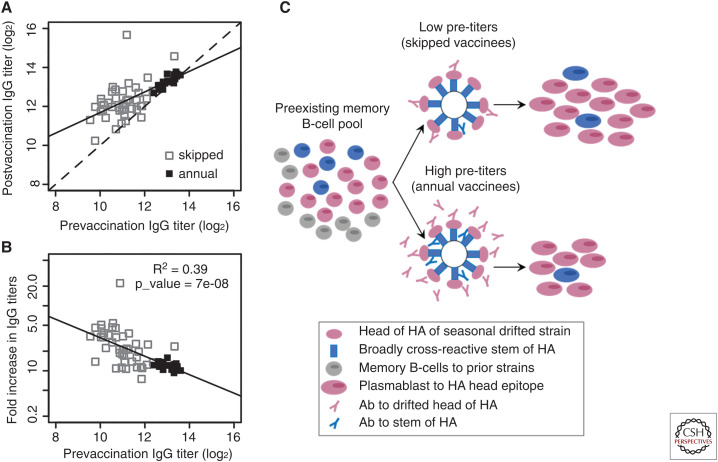
Despite the advantages of memory recall after immunization, there is also evidence for interference of the immune response by preexisting memory. Typically, individuals with high preexisting cross-reactive antibody titers have a significantly reduced peak in circulating antigen-specific plasmablast numbers after influenza vaccination (Ellebedy et al. 2020). Individuals with higher preexisting antibody titers also have a reduced boost in antibody titers after vaccination (Ellebedy et al. 2014, 2020). Therefore, individual immune responses may be modulated by an individual's preexisting antibody titer, and as antibody titers to influenza wane over time, the extent of modulation may change over time. This became evident from immune responses to influenza vaccination in individuals who are vaccinated annually compared to individuals who have skipped influenza vaccination for at least 3 years. Figure 2 shows the response to seasonal influenza vaccination in these two groups of people. Figure 2A shows the relationship between the pre- and postvaccination antibody titers. Prior to vaccination the antibody titers are significantly lower in individuals who have skipped getting an influenza vaccination for at least 3 years (open squares) compared to individuals who have received an influenza vaccination the previous year (filled squares). If antibody titers remain unchanged after vaccination, the data points would fall on the line with slope = 1 (dashed line). An increase in antibody titers after vaccination would position the points above this line. If preexisting antibody titers have no effect on boosting, we would expect the points to line up parallel to the dashed line. However, the best fit (solid line) has slope less than one (least squares; slope = 0.45; 95% CI = [0.37;0.68]), showing that individuals with lower preexisting antibody titers have significantly higher boost, as can be seen from the increased distance above the dashed line. Figure 2B shows the corresponding relationship between the prevaccination antibody titer and magnitude of the boost, defined as fold increase in antibody titer, for responses in the “skipped” (open squares) and “annual” (filled squares) groups. The fold increase (i.e., fold boost) of the antibody response reduces with an increase in the corresponding prevaccination antibody titer. Figure 2C shows how MBC frequency and preexisting antibodies may modulate these responses. With low levels of preexisting immunity, the reactivation of cross-reactive memory B cells becomes a dominant factor defining the response, whereas with high levels of preexisting immunity, the negative feedback from the cross-reactive antibody may contribute to reduce the overall response.
Figure 2.
Boosting of antibodies after influenza vaccination. Influenza vaccine antigen binding antibody titers were measured by ELISA before and 14 d postvaccination in individuals who had been vaccinated the previous year (annual), and those who had skipped influenza vaccination for at least 3 years (skipped). (A) The relationship between the pre- and postvaccination antibody titers. (B) The relationship between fold increase in antibody titers and prevaccination titers. (C) A schematic outlining how preexisting MBCs and antibody titers may affect the recall response after seasonal influenza vaccination.
BOOSTING OF RESPONSES TO DIFFERENT EPITOPES
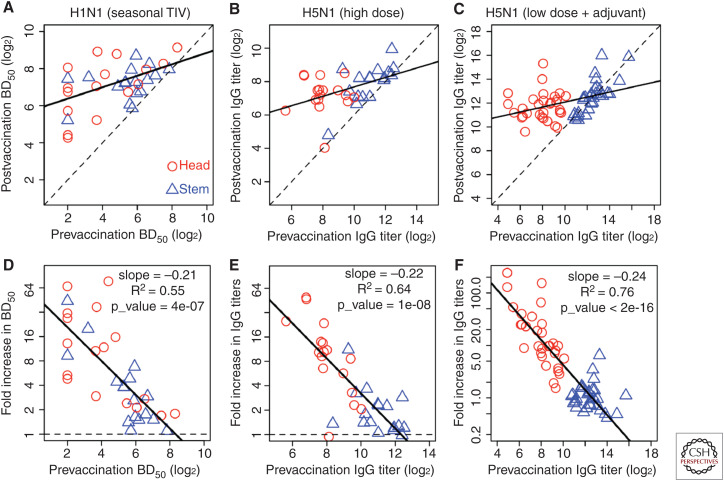
The immune responses to different epitopes following vaccination may be affected by the balance of preexisting memory B cells and preexisting antibody titers to these epitopes. For instance, after a primary immunization with a novel H5N1 strain, the specificity of antibodies elicited by vaccination is predominantly skewed to the more broadly cross-reactive stem specific epitopes. This is a result of the higher number of preexisting stem-specific MBCs elicited by prior exposure to H1N1 that share cross-reactive stem epitopes, and the lack of preexisting memory to the novel H5 head domain. However, after secondary exposure to the H5N1 strain by booster vaccination, the immune response is highly skewed toward head-specific antibody responses, whereas stem-specific antibody titers remain largely unchanged. One potential mechanism for the dominance of HA head- versus stem-specific antibodies after secondary H5N1 vaccination is that high stem-specific antibody titers interfere with the memory recall response. Figure 3 plots the antibody response to head and stem after immunization with seasonal trivalent influenza vaccine (TIV) (Fig. 3A,D), high dose of novel H5N1 strain (Fig. 3B,E), and low dose of H5N1 strain with added adjuvant (Fig. 3C,F). Similar to the data for skipped versus annual vaccinees (Fig. 2), the fold boost of the antibody response to the HA head or stem is negatively correlated with prevaccination antibody titers to these epitopes. In the case of vaccination with seasonal TIV (Fig. 3A), the prevaccination titers of antibodies to both the HA stem and the HA head of H1N1 overlap. In this case, in which preexisting HA stem and head reactive antibody titers are similar, the fold boost in antibody titers is similar regardless of epitope specificity and correlates with the prevaccination titers (Fig. 3D). In the case of secondary vaccination with the novel H5N1 strain (Fig. 3B,C), there were higher levels of antibodies to the stem than the head of HA (and no individuals with very low titers to the stem). In this case, the boost in antibody titers was skewed toward the HA head domain, which exhibited significantly lower prevaccination titers.
Figure 3.
Antibody responses to the head and stem of hemagglutinin (HA). Boosting of HA head- and stem-specific antibodies following vaccination with inactivated H1N1 (from seasonal trivalent influenza vaccine [TIV], [A,D], high-dose H5N1 [ B,E], and low-dose H5N1 with adjuvant [C,F]). For H5N1 we use the data from the boost vaccination (see also Fig. 6). (A–C) Plot of post- versus prevaccination antibody titers. If antibody titers remain unchanged after vaccination, the data points would fall on the line with slope = 1 (dashed line). The best fit line (solid line, slope < 1) shows that there is more boosting when the initial titers are low. (D–F) Prevaccination immunity reduces the boost and the relationship is approximately linear on log-log plot. (Panels created with data from Ellebedy et al. 2014, 2020.)
MODELING OF RESPONSES TO THE HEAD AND STEM OF HA
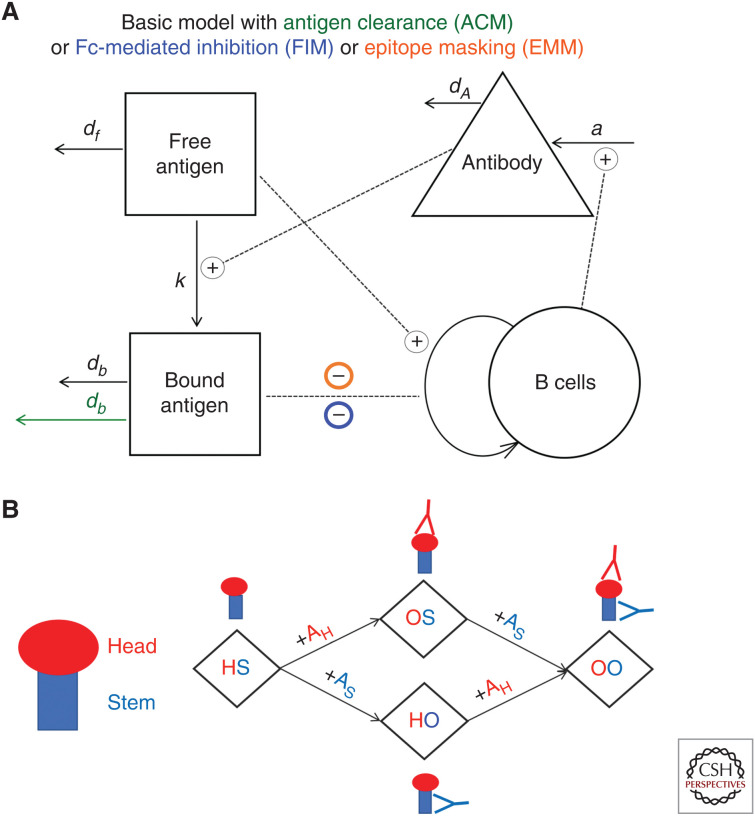
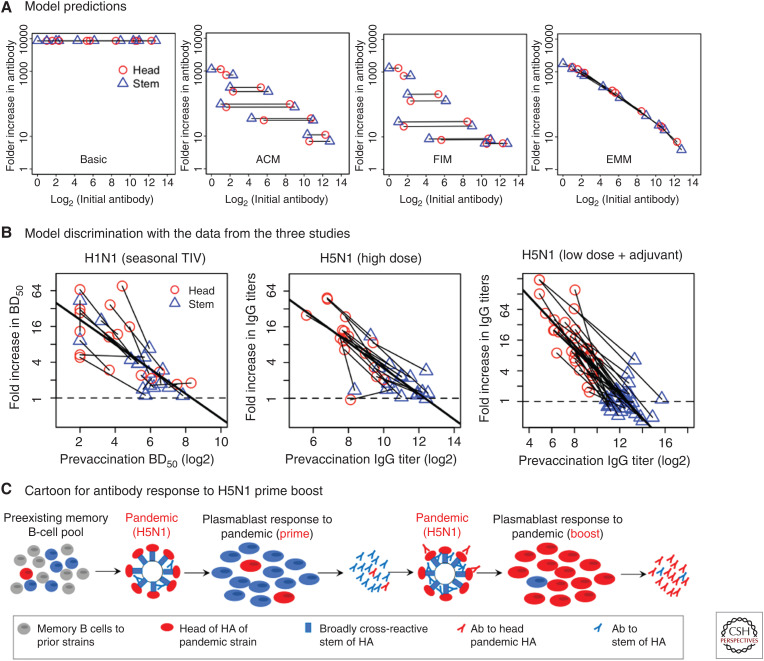
Mathematical models of the humoral immune response can be used to explore different hypotheses for how preexisting memory B cells and antibodies affect the boosting of responses to different epitopes following immunization. Figure 4 shows a previously described simple two-epitope model that focuses on epitopes of the head and stem of HA and their interactions with B cells and antibodies specific for these epitopes (Zarnitsyna et al. 2016). Figure 4A shows a schematic for a basic model of humoral immune response (“basic” model) and its three modifications reflecting three additional mechanisms (color-coded). In the basic model, antigen-specific B cells are stimulated by the antigen, undergo clonal expansion, and produce antibody. Stimulation of specific B cells continues until all the antigen has decayed. In the basic model, the fold increase in antibody titers is independent of prevaccination titers. There are several ways in which preexisting antibodies could reduce recall responses (Zarnitsyna et al. 2016). The antigen clearance model (ACM) hypothesizes that preexisting antibodies cause faster clearance of the antigen reducing the time for B-cell interaction with it and, thus, leading to less B-cell expansion and less generated antibody. The Fc receptor–mediated inhibition model (FIM) hypothesizes that preexisting antibodies form inhibitory immune complexes that reduce the activation of antigen-specific B cells by cross-linking B-cell receptor bound to the antigenic epitope and B-cell FcγRIIB bound to the Fc portion of the antibody in the immune complex (Nimmerjahn and Ravetch 2010). The epitope masking model (EMM) hypothesizes that preexisting antibodies and B cells specific for the same epitope would compete for binding to that epitope on the antigen, and it inhibits the proliferation of epitope-specific B cells. This happens as a result of physical constraints associated with the size of antibodies and does not affect binding and proliferation of B cells specific for other epitopes that are spatially distant. In contrast to the ACM model, in which antigen is removed more quickly by binding to antibody, in the EMM model, production of antibody is down-regulated despite the continued presence of antigen.
Figure 4.
Schematic for mathematical models to explain reduced boosting of antibody responses. (A) Schematic for the basic one-epitope model and its three modifications incorporating three different mechanisms: enhanced antibody-bound antigen clearance (ACM, in green), FcγR-mediated inhibition (FIM, in blue), and epitope masking (EMM, in orange). (B) The one-epitope model is extended to a two-epitope model by including one epitope on the head of HA (H) and one epitope on the stem of HA (S). Binding of head-specific antibody (AH) and stem-specific antibody (AS) defines four possible states of the antigen: free antigen with no antibody bound (HS), antigen bound by antibody to the head (OS), antigen bound by antibody to the stem (HO), and antigen bound by both antibodies (OO).
When considering a simple two-epitope model with responses to the head and stem of HA (Fig. 4B), the antigen could be in four states, and the states are defined by antibody binding to the head and/or stem. The free antigen is named HS, whereas OS, HO, and OO represent antigen with antibodies bound to H (head epitope), S (stem epitope), or both epitopes, respectively.
The three mechanisms incorporated in the ACM, FIM, and EMM models could each explain the reduction in the antibody fold boost due to preexisting antibodies, but we found that the two-epitope models make different predictions if we compare how responses to the head and stem of HA for each individual depend on prevaccination titers to head and stem (Fig. 5A; Zarnitsyna et al. 2016). For each individual, we assume different initial levels of antibody to the head (red circles) and to the stem (blue triangles), and we connect the predicted responses to the head and stem in a given individual after vaccination with a line. The boosting results for 10 different initial conditions mixing high and low initial antibody titers for the head and stem are shown.
Figure 5.
Model discrimination. (A) Model predictions for the basic model and its modifications incorporating three different mechanisms: antigen clearance (ACM), Fc-receptor mediated inhibition of B-cell activation (FIM), and epitope masking (EMM). Using a two-epitope model, we predict how different levels of prevaccination head- and stem-specific antibodies affect the boosting of antibody titers to head and stem of HA. Ten different sets of various prevaccination head- and stem- antibody titers (imitating 10 different individuals) were used for prediction of the postvaccination fold boost in antibody titers. Responses to the head (red circle) and stem (blue triangle) in each individual are connected by a line. (B) The corresponding plots from three experimental studies. (C) Schematic illustrating how prevaccination memory B cells and antibody affect the responses to prime and boost vaccinations with novel pandemic H5N1. High prevaccination frequency of stem-specific memory B cells defines the primary response with a strong recall of stem-specific antibodies. However, increased titers of stem-specific antibodies after the first H5N1 vaccination suppress further boosting of stem-specific antibodies following the secondary vaccination. Instead, the newly generated antibodies after the second vaccination are predominantly head-specific.
The basic model predicts that boosting will be independent of the level of preexisting immunity and, therefore, all lines are horizontal and at the same height. The ACM model predicts that antigen clearance equally affects the responses to head and stem as binding of antibody to any epitope will remove the entire antigen. Thus, the lines are parallel, and their heights are determined by the total preexisting antibody titers in each individual. In the FIM model, binding of antibody–antigen complexes (through the Fc portion of antibody) to FcγRIIB on a B cell leads to inhibition of B-cell activation regardless of its epitope specificity. The FIM model predicts that preexisting antibody to either epitope similarly affects generation of the responses to both head and stem of HA and, thus, the lines for each individual are horizontal. The EMM model proposes that preexisting immunity only affects the response to the antibody-bound epitope, and the head- and stem-specific responses can be boosted by different amounts depending on their prevaccination antibody titers. More preexisting antibody to a given epitope would result in more masking and less boosting of the response to that epitope, and the EMM model predicts that the lines connecting responses to head and stem for all individuals fall along a diagonal with a negative slope.
We now compare the models’ predictions with data from three vaccine studies in which individuals were vaccinated with HAs from H1N1 and H5N1 (data in Fig. 3). The experimental data were reanalyzed to generate a plot similar to that in Figure 5A by connecting antibody responses to the head and stem of HA with a line for each individual. For all three vaccination studies, the lines connecting responses to head and stem in each individual (thin lines) and an average line using all the data (thick line) are shown in Figure 5B. We find that the average of the slopes from the thin individual's lines (not shown) to be significantly different from zero. This suggests that responses to head and stem are boosted to different degrees in each individual and, thus, it is inconsistent with the ACM and FIM models in which inhibition of the head- and stem-specific responses is predicted to be similar. On the contrary, the averaged slope is not significantly different from the slope of fit to all data (thick line) supporting the EMM model.
The EMM model assumes that responses to the head and stem of HA are generated independently, and preexisting titers to the head would not affect boosting of antibody to the stem (and vice versa). We would like to note that in all three considered vaccination studies the initial level of antibody to the novel head was relatively low. In this case, we do not expect much interference between the antibodies and B cells binding to the epitopes on the head with boosting of the responses to the epitopes on the stem of HA, and we found no significant correlation between the initial titer of head-specific antibody and fold boost in stem-specific antibody in these studies. However, this assumption may not be true in the case of high preexisting head-specific antibody, and contributions of the different mechanisms described by the ACM, FIM, and EMM models on recall responses in different scenarios, for example, after repeated seasonal vaccination or infection, has yet to be determined.
ANTIBODY RESPONSES TO STEM OF HA DURING PRIME AND BOOST H5N1 VACCINATION
One of the remaining questions is why the response to the stem of HA was boosted during the primary vaccination but not the secondary vaccination with H5N1 (Ellebedy et al. 2014, 2020). We use the adjuvanted H5N1 vaccine study as an example as it generated a stronger overall response. Figure 6 shows the fold boost of antibody to the stem of HA versus prevaccination titers for both prime and boost vaccinations with adjuvanted H5N1. Initial antibody titers to the stem before the primary vaccination were lower compared to the initial titers before the secondary vaccination, and this potentially explains the relatively large fold boost during primary vaccination and limited boost during secondary vaccination. This is similar to the response of annual and skipped vaccinees shown in Figure 2 in which individuals who had been more recently vaccinated and had higher preexisting antibody titers had reduced fold boost in antibody titers.
Figure 6.
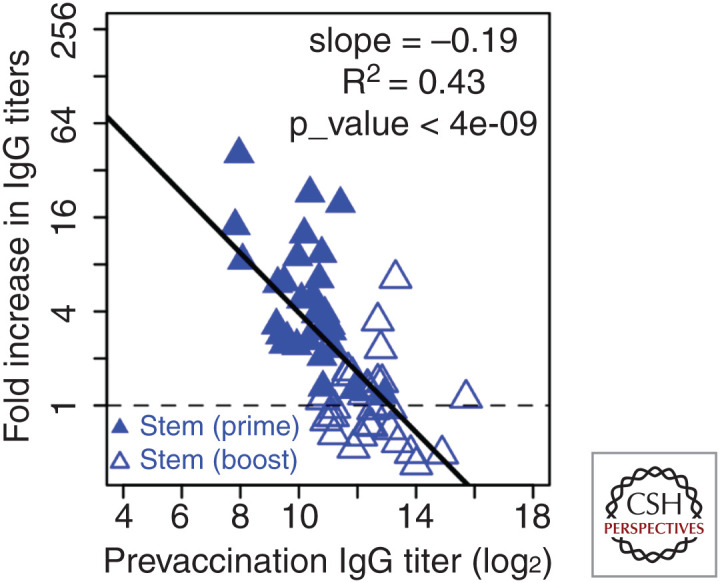
Boosting of antibody response to the stem of HA during prime vaccination with adjuvanted H5N1 vaccine and lack of boosting after the boost vaccination. Filled triangles and open triangles are the responses to the stem of HA after prime and boost vaccinations, respectively. Prevaccination immunity reduces the boost, and the relationship is approximately linear on log-log plot. (Figure created from data in Ellebedy et al. 2020.)
DISCUSSION
Influenza is a pathogen that exhibits extensive strain variation. This enables it to infect individuals multiple times, each time shaping and modulating influenza-specific immune memory. As a result, the level of immunity to influenza in humans is strikingly high, with influenza-specific MBCs commonly comprising at least 1% of total circulating MBCs. This is significantly higher than the frequency of MBCs specific for other commonly used childhood vaccine antigens (Fig. 1). In spite of the high level of acquired immunity, immune responses to the conserved epitopes on the virus are insufficient to prevent new infections.
Here we analyze three vaccine studies in which immunity to influenza has been boosted with novel pandemic strains with the goal of understanding how preexisting immunity modulates the antibody responses to the conserved epitopes on the stem of HA. Preexisting immunity includes both memory B cells and antibodies, and although the increased frequency of memory B cells may benefit the secondary response, negative feedback from antibodies could limit it. We find that individuals with high preexisting cross-reactive antibody titers have a lower-magnitude response to vaccination with a reduced recall response despite high levels of preestablished MBC frequency (Figs. 1 and 2). Using mathematical models we analyzed the contribution of different factors such as MBC frequency, prevaccination antibody titers, increased clearance of antigen bound to antibody in comparison to free antigen, inhibition of B-cell activation through binding of antibody–antigen immune complexes to inhibitory receptors on B cells, and epitope masking on the antigen by antibody to recall responses after vaccination (see models schematic in Fig. 4).
In the case of primary responses to novel influenza strains such as the 2009 pandemic H1N1 strain, or a highly divergent H5N1 strain, there are low preexisting antibody titers and higher preestablished cross-reactive HA stem–specific MBC frequencies in comparison to HA head–specific MBCs. Thus, in response to primary infection or vaccination with these novel strains, there is a strong recall response of cross-reactive HA stem–specific antibodies. Intriguingly, upon secondary exposure to the H5N1 strain, the antibody response is predominantly skewed toward the HA head epitopes rather than the HA stem epitopes (Figs. 3 and 5). We hypothesize that this is due to the high preexisting HA stem–reactive antibody titers at the time of secondary vaccination (Fig. 5C) and explore three different mechanisms that may lead to the reduction of the response using simple mathematical models (Figs. 4 and 5). We found that the epitope masking model most closely recapitulates the data in the scenario of a response to a novel pandemic strain. We note that other considered mechanisms, such as antibody-enhanced antigen clearance and inhibition of B-cell activation by triggering inhibitory receptors on B cells by antigen–antibody complexes, may largely define the recall responses in different scenarios of vaccination and infection. The fact that the antibody response is skewed toward specificities not as prevalent in the antibody repertoire at the time of secondary H5N1 vaccination may be a beneficial outcome of epitope masking as it expands the immune repertoire.
We used a simple two-epitope model with one epitope on the head and one epitope on the stem of HA to make predictions for model discrimination. It has been established that HA has multiple epitopes on the head and fewer epitopes on the stem (Caton et al. 1982; Das et al. 2013). The incorporation of multiple epitopes on the head of HA to the model would require consideration of steric interference. Steric interference arises from the comparable size of the HA and antibody molecules and the close proximity of head epitopes. Steric interference has been previously proposed in the context of neutralization efficiency of epitopes (Ndifon et al. 2009), and it is supported by stoichiometric studies of binding of antibodies to the head of HA on intact virus (Taylor et al. 1987; Taylor and Dimmock 1994). We can introduce a parameter that defines steric interference so that antibody bound to a head epitope not only blocks the binding of further antibody or B-cell receptors to the same epitope, but also inhibits binding to nearby epitopes on the head of HA. We have shown that multiepitope models with additional epitopes on the head of HA, and which considers steric interference between the epitopes on the head, generate dynamics and predictions similar to the two-epitope models shown in Fig. 5 (Zarnitsyna et al. 2016).
In the studies described here, the preexisting head-specific antibodies were low at the time of vaccination as both H1N1 and H5N1 were novel pandemic strains. As there is no correlation between the preexisting antibody titers to the stem and the fold boost of antibody to the head, this suggests that stem-specific antibody titers do not affect the generation of responses to the head of HA (Zarnitsyna et al. 2016). In the case of seasonal drifted strains, we would expect higher levels of preexisting immunity to the head of HA, and it is possible that head specific antibodies may interfere with B-cell binding to the stem because of steric hindrance. Our modeling approach can be easily extended to consider asymmetric steric interference in which head antibodies could suppress the generation of stem antibodies but not vice versa. As the relevant experimental data becomes available, this should allow us to assess the potential role of asymmetric steric interference on boosting of stem-reactive antibody titers during different scenarios of immunization or infection with antigenically drifted strains.
Although the boost in antibody titers in response to vaccination was very low in subjects receiving the seasonal influenza vaccine who had been repeatedly vaccinated including the year prior to the study (Fig. 2), annual vaccination provides significant benefits. For instance, it is important not to overlook the fact that despite the increased magnitude of the response in skipped vaccinees, the postvaccination titers did not surpass that of vaccinees who had been vaccinated in the previous year. Additionally, although the memory recall response seems to be limited in repeat vaccinees with high preexisting cross-reactive antibody titers, this does not preclude the possibility that novel strain specific antibody specificities are elicited by vaccination. As the magnitude of novel antibody specificities derived from naive B cells would likely be of significantly lower titers compared with the high preexisting cross-reactive antibody titers, this may not dramatically increase the overall titers but may still be immunologically relevant in the case of a viral challenge. Although there is a strong negative correlation between preexisting antibody titers and the magnitude of the boost of cross-reactive antibody titers, there may be an expansion of novel antibody specificities in germinal centers. Thus, repeated vaccination may affect and update the overall immune repertoire.
The limited boosting of HA stem–reactive antibodies after secondary H5 vaccination and of antibody titers in repeated vaccinees raises questions regarding the limits of boosting of cross-reactive antibodies. The epitope masking model predicts that increasing the antigen dose may overcome the limited boosting of antibody because of masking of epitopes on the antigen by preexisting antibody (Zarnitsyna et al. 2016). Additional strategies such as inclusion of adjuvants, novel vaccine platforms, or novel immunogens will likely be required for further boosting of broadly cross-reactive antibody titers.
ACKNOWLEDGMENT
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
- Albrecht P, Ennis FA, Saltzman EJ, Krugman S. 1977. Persistence of maternal antibody in infants beyond 12 months: mechanism of measles vaccine failure. J Pediatr 91: 715–718. 10.1016/S0022-3476(77)81021-4 [DOI] [PubMed] [Google Scholar]
- Bodewes R, de Mutsert G, van der Klis FR, Ventresca M, Wilks S, Smith DJ, Koopmans M, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2011. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin Vaccine Immunol 18: 469–476. 10.1128/CVI.00396-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31: 417–427. 10.1016/0092-8674(82)90135-0 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC). 2009. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep 58: 521–524. [PubMed] [Google Scholar]
- Das SR, Hensley SE, Ince WL, Brooke CB, Subba A, Delboy MG, Russ G, Gibbs JS, Bennink JR, Yewdell JW. 2013. Defining influenza A virus hemagglutinin antigenic drift by sequential monoclonal antibody selection. Cell Host Microbe 13: 314–323. 10.1016/j.chom.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CW, Jackson KJL, McCausland MM, Darce J, Chang C, Linderman SL, Chennareddy C, Gerkin R, Brown SJ, Wrammert J, et al. 2020. Influenza vaccine-induced human bone marrow plasma cells decline within a year after vaccination. Science (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud MB, Bloom JD. 2016. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses 8: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt CS, Wieland A, Nasti TH, Grifoni A, Wilson E, Schmid DS, Pulendran B, Sette A, Waller EK, Rouphael N, et al. 2020. Persistence of varicella-zoster virus-specific plasma cells in adult human bone marrow following childhood vaccination. J Virol 94. 10.1128/JVI.02127-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen HN, Little JR, Steiner LA, Simms ES, Gray W. 1969. Degeneracy in the secondary immune response: stimulation of antibody formation by cross-reacting antigens. Isr J Med Sci 5: 338–351. [PubMed] [Google Scholar]
- Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, et al. 2014. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci 111: 13133–13138. 10.1073/pnas.1414070111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy AH, Jackson KJL, Kissick HT, Nakaya HI, Davis CW, Roskin KM, McElroy AK, Oshansky CM, Elbein R, Thomas S, et al. 2016. Defining antigen-specific plasmablast and memory B cell subsets in blood following viral infection and vaccination of humans. Nat Immunol 17: 1226–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy AH, Nachbagauer R, Jackson KJL, Dai YN, Han J, Alsoussi WB, Davis CW, Stadlbauer D, Rouphael N, Chromikova V, et al. 2020. Adjuvanted H5N1 influenza vaccine enhances both cross-reactive memory B cell and strain-specific naive B cell responses in humans. Proc Natl Acad Sci 117: 17957–17964. 10.1073/pnas.1906613117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T Jr. 1955. The current status of the control of influenza. Ann Intern Med 43: 534–538. 10.7326/0003-4819-43-3-534 [DOI] [PubMed] [Google Scholar]
- Francis T. 1960. On the doctrine of original antigenic sin. Proc Am Philos Soc 104: 572–578. [Google Scholar]
- Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. 2016. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354: 722–726. 10.1126/science.aag1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostic KM, Bridge R, Brady S, Viboud C, Worobey M, Lloyd-Smith JO. 2019. Childhood immune imprinting to influenza A shapes birth year-specific risk during seasonal H1N1 and H3N2 epidemics. PLoS Pathog 15: e1008109. 10.1371/journal.ppat.1008109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasa NB, Gerber MA, Chen Q, Wright PF, Edwards KM. 2008. Safety and immunogenicity of trivalent inactivated influenza vaccine in infants. J Infect Dis 197: 1448–1454. 10.1086/587643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, et al. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 361: 1945–1952. 10.1056/NEJMoa0906453 [DOI] [PubMed] [Google Scholar]
- Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349: 1301–1306. 10.1126/science.aac7263 [DOI] [PubMed] [Google Scholar]
- Kanekiyo M, Wei CJ, Yassine HM, McTamney PM, Boyington JC, Whittle JR, Rao SS, Kong WP, Wang L, Nabel GJ. 2013. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499: 102–106. 10.1038/nature12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Pica N, Hai R, Margine I, Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87: 6542–6550. 10.1128/JVI.00641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Margine I, Hai R, Flood A, Hirsh A, Tsvetnitsky V, Chen D, Palese P. 2014. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol 88: 2340–2343. 10.1128/JVI.03183-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski AJ, Lessler J, Read JM, Zhu H, Jiang CQ, Guan Y, Cummings DA, Riley S. 2015. Estimating the life course of influenza A(H3N2) antibody responses from cross-sectional data. PLoS Biol 13: e1002082. 10.1371/journal.pbio.1002082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, et al. 2012. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci 109: 9047–9052. 10.1073/pnas.1118979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, Ellebedy AH, Carter DM, Andrews SF, Zheng NY, et al. 2014. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci 111: 15798–15803. 10.1073/pnas.1409171111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, Runstadler JA, Wilson PC, Albrecht RA, García-Sastre A, et al. 2013. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol 87: 10435–10446. 10.1128/JVI.01715-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndifon W, Wingreen NS, Levin SA. 2009. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc Natl Acad Sci 106: 8701–8706. 10.1073/pnas.0903427106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu KE, Henry Dunand CJ, Wilson PC. 2016. Heads, stalks and everything else: how can antibodies eradicate influenza as a human disease? Curr Opin Immunol 42: 48–55. 10.1016/j.coi.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. 2010. Antibody-mediated modulation of immune responses. Immunol Rev 236: 265–275. 10.1111/j.1600-065X.2010.00910.x [DOI] [PubMed] [Google Scholar]
- Rasheed MAU, Hickman CJ, McGrew M, Sowers SB, Mercader S, Hopkins A, Grimes V, Yu T, Wrammert J, Mulligan MJ, et al. 2019. Decreased humoral immunity to mumps in young adults immunized with MMR vaccine in childhood. Proc Natl Acad Sci 116: 19071–19076. 10.1073/pnas.1905570116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skountzou I, Koutsonanos DG, Kim JH, Powers R, Satyabhama L, Masseoud F, Weldon WC, Martin Mdel P, Mittler RS, Compans R, et al. 2010. Immunity to pre-1950 H1N1 influenza viruses confers cross-protection against the pandemic swine-origin 2009 A (H1N1) influenza virus. J Immunol 185: 1642–1649. 10.4049/jimmunol.1000091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HP, Dimmock NJ. 1994. Competitive binding of neutralizing monoclonal and polyclonal IgG to the HA of influenza A virions in solution: only one IgG molecule is bound per HA trimer regardless of the specificity of the competitor. Virology 205: 360–363. 10.1006/viro.1994.1653 [DOI] [PubMed] [Google Scholar]
- Taylor HP, Armstrong SJ, Dimmock NJ. 1987. Quantitative relationships between an influenza virus and neutralizing antibody. Virology 159: 288–298. 10.1016/0042-6822(87)90466-1 [DOI] [PubMed] [Google Scholar]
- Voysey M, Kelly DF, Fanshawe TR, Sadarangani M, O'Brien KL, Perera R, Pollard AJ. 2017. The influence of maternally derived antibody and infant age at vaccination on infant vaccine responses: an individual participant meta-analysis. JAMA Pediatr 171: 637–646. 10.1001/jamapediatrics.2017.0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CJ, Boyington JC, McTamney PM, Kong WP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang ZY, et al. 2010. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329: 1060–1064. 10.1126/science.1192517 [DOI] [PubMed] [Google Scholar]
- Willis E, Pardi N, Parkhouse K, Mui BL, Tam YK, Weissman D, Hensley SE. 2020. Nucleoside-modified mRNA vaccination partially overcomes maternal antibody inhibition of de novo immune responses in mice. Sci Transl Med 12: eaav5701. 10.1126/scitranslmed.aav5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208: 181–193. 10.1084/jem.20101352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21: 1065–1070. 10.1038/nm.3927 [DOI] [PubMed] [Google Scholar]
- Zarnitsyna VI, Lavine J, Ellebedy A, Ahmed R, Antia R. 2016. Multi-epitope models explain how pre-existing antibodies affect the generation of broadly protective responses to influenza. PLoS Pathog 12: e1005692. 10.1371/journal.ppat.1005692 [DOI] [PMC free article] [PubMed] [Google Scholar]