Abstract
Auxin represents one of the most potent and most versatile hormonal signals in the plant kingdom. Built on a simple core of only a few dedicated components, the auxin signaling system plays important roles for diverse aspects of plant development, physiology, and defense. Key to the diversity of context-dependent functional outputs generated by cells in response to this small molecule are gene duplication events and sub-functionalization of signaling components on the one hand, and a deep embedding of the auxin signaling system into complex regulatory networks on the other hand. Together, these evolutionary innovations provide the mechanisms to allow each cell to display a highly specific auxin response that suits its individual requirements. In this review, we discuss the regulatory networks connecting auxin with a large number of diverse pathways at all relevant levels of the signaling system ranging from biosynthesis to transcriptional response.
As sessile organisms, plants are exposed to a continuously changing environment, which is shaped by a plethora of abiotic and biotic parameters. To cope with these challenges, plants have evolved regulatory networks that allow them to constantly adapt their growth and developmental program to the current conditions. Given the decentralized nature of cellular decision making in plants, these networks need to fulfill a surprisingly large number of sometimes conflicting functions. First, they need to be able to integrate a substantial amount of information into a stable cellular program. For example, a plant may encounter a windy and cold, but very bright, day in spring with the soil moist and nutrients available in abundance. In this hypothetical environment, a plant finds excellent conditions for growth on the one hand with water and nutrient availability near the optimum, as well as enough light to drive a maximum of photosynthesis. On the other hand, UV irradiation, low temperatures, and wind impose substantial stress on the individual. To complicate matters for finding the optimal developmental solution, the opposing inputs mostly affect different parts of the plant, requiring tight communication between roots, leaves, and stems. Here, the plant encounters a second challenge, namely, the system-wide coupling of cell behavior. Whereas in the above example uniform cell behavior across the entire plant likely is beneficial, there are cases, such as localized attack by herbivores or partial shading, where strictly local responses are required. A third challenge for the discussed regulatory networks is that they need to be deeply embedded into the developmental and metabolic programs, both for environmental sensing and for modulating growth and physiology. Last, the decision making has to be aware of temporal components to allow the plant to prepare for certain conditions expected in the future, based on previous exposures.
In this review, we will focus on regulatory networks that have evolved around the auxin signaling system, which plays key roles in diverse aspects of plant development, physiology, and defense. The most abundant and best-characterized auxin, indole acetic acid (IAA), is a small metabolite with similarity to tryptophan and able to move throughout the plant body in the phloem or by active and highly polar cell–cell transport (Okada et al. 1991; Gälweiler et al. 1998; Müller et al. 1998; Palme and Gälweiler 1999; Reinhardt et al. 2003; Petrášek et al. 2006; Tanaka et al. 2006; Adamowski and Friml 2015). The core auxin perception and signaling system is relatively simple, consisting of only three dedicated intracellular components: receptor (Dharmasiri et al. 2005; Kepinski and Leyser 2005), transcriptional executor (Ulmasov et al. 1997a; Hardtke and Berleth 1998), and inhibitor protein (Ulmasov et al. 1997b; Rouse et al. 1998; Hamann et al. 2002), which are coupled to the proteasome system (Gray et al. 2001). However, the pathway has evolved by gene duplication, and in many plants all components are represented by multiple copies with diverse expression patterns and biochemical properties (Vernoux et al. 2011; Weijers and Wagner 2016; Kato et al. 2020), giving rise to an enormous range in direct cellular responses to auxin through combinatorial logic. Importantly, the medium- and long-term auxin response is further shaped by intricate connections of this core auxin-sensing pathway with a dazzling array of signaling systems through transcriptional cross regulation, protein modification, or chromatin remodeling.
CONTROL OF AUXIN HOMEOSTASIS AND TRANSPORT
Many of the biological effects of auxin appear highly localized, such as organ initiation or tropic responses, and it follows that the hormone must be able to act with high spatiotemporal specificity. Indeed, it has turned out that local accumulation of active auxin is tightly regulated at multiple levels including biosynthesis, transport, as well as conjugation and degradation of this small metabolite. Since genetic analysis had suggested that auxin transport would suffice to create local effects, localized auxin biosynthesis, inactivation, or sensing was thought to play only a minor role until recently (Cheng et al. 2006, 2007; Stepanova et al. 2008; Tao et al. 2008; Brumos et al. 2018). However, with the elucidation of auxin anabolic and catabolic pathways and the expression patterns of the underlying genes, it became clear that this level of control is more important than previously recognized. Not only do many auxin metabolic genes exhibit highly specific spatiotemporal expression patterns, they also respond dynamically to a variety of stimuli, lending strong support to the importance of complex gene regulatory networks controlling auxin homeostasis.
Auxin (IAA) biosynthesis can follow both tryptophan (Trp)-dependent and -independent pathways, but currently only the Trp-dependent TAA-YUCCA pathway is well understood at the genetic and mechanistic level (Zhao 2018). To produce IAA from Trp, two sequential steps need to be carried out. The first step is catalyzed by enzymes encoded by members of the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) gene family (Tao et al. 2008), whereas the second and rate-limiting step relies on flavin-containing monooxygenases encoded by the YUCCA (YUC) gene family (Zhao et al. 2001; Won et al. 2011). YUC genes show highly specific expression patterns and higher order yucca mutants show severe development defects (Cheng et al. 2006), lending support to the idea that levels of local auxin synthesis via the transcriptional regulation of YUCs is essential for the correct execution of developmental programs (Eklund et al. 2010; Cui et al. 2013; Martínez-Fernández et al. 2014; Tang et al. 2017; Zhou et al. 2018; Blakeslee et al. 2019; Cao et al. 2019). An interesting example is the induction of YUC expression by the STYLISH (STY) transcription factor (TF) during fruit development (Eklund et al. 2010). STY expression is dependent on the activity of the AP2 TF DORNROESCHEN-LIKE (Eklund et al. 2011), which itself has been shown to act downstream of auxin signals (Cole et al. 2009), hinting at intricate feedforward connections. Similarly, in the root, auxin biosynthesis is controlled by the brassinosteroid (BR) signaling pathway via TAA and YUC genes in a cell-type-specific fashion, which in turn controls meristem development (Vragović et al. 2015). A subset of YUC genes have also been shown to be regulated at the epigenetic level. TFL2/LHP1 (Rizzardi et al. 2011), the HP1 homolog in plants, localizes to promoters of YUC genes and is required for their transcriptional activation. This interaction is functionally relevant, since in tfl2/lhp1 mutants, the activity of the auxin output reporter DR5 is substantially reduced. In addition to these developmental inputs, altered YUC gene expression is also involved in the response to external stimuli, such as light (Li et al. 2012; Zhou et al. 2018), temperature (Sun et al. 2012), and biotic stress (Liu et al. 2016). For example, the growth responses downstream of shade perception and increase in ambient temperature are tightly connected and require direct binding of PHYTOCHROME INTERACTING FACTORS (PIFs) such as PIF4 and PIF7 to the promoters of YUC genes to increase local auxin biosynthesis (Hornitschek et al. 2012; Li et al. 2012; Hersch et al. 2014).
While the TAA-dependent step in the pathway does not appear to be rate limiting, taa mutants still exhibit substantial reduction in the levels of free IAA, along with obvious phenotypes (Stepanova et al. 2008). In line with the hypothesis that regulation of TAA genes is the primary mode of action to modulate auxin levels in response to environmental stimuli, TAA transcription has been shown to be under control of the phytohormones ethylene (Stepanova et al. 2008) and cytokinin (CK) (Yan et al. 2017), as well as light quality (Tao et al. 2008), high temperature (Franklin et al. 2011), or gravity (Cui et al. 2013). In addition to the transcriptional level, local auxin biosynthesis via TAA1 can also be regulated at the protein level by the switch of its enzymatic activity via T101 phosphorylation (Wang et al. 2020).
Acting downstream of auxin biosynthesis, IAA conjugation and degradation are also embedded into the regulatory networks underlying dynamic local auxin homeostasis. IAA conjugation to glutamic acid (IAA-Glu) or other amino acids, is the first step toward auxin degradation. The conjugating process is catalyzed by the Gretchen Hagen 3 (GH3) enzyme family, which are IAA-amido synthetases and first identified as auxin rapid-induced transcripts from soybean (Hagen and Guilfoyle 1985; Wright et al. 1987; Hagen et al. 1991). The feedback loop between auxin-induced increase in GH3 mRNA accumulation and the conversion of IAA to inactive IAA-Glu by GH3 enzymatic activity provides a reset of local auxin levels. Importantly, the activation of GH3 expression is also employed by other pathways, such as by CK signaling. In the root, the CK output TF ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) stimulates GH3.17, which transforms IAA into IAA-Glu in the lateral root cap, creating an auxin minimum defining the transition zone between cell division and cell differentiation in root meristem (Di Mambro et al. 2017, 2019). Unlike IAA-Asp or IAA-Glu, other IAA-amino acid conjugates can serve as a storage of IAA, such as IAA-Ala, and IAA-Leu, which can be hydrolyzed back to free IAA (Woodward and Bartel 2005). However, the oxidization of IAA into 2-oxoindole-3-acetic acid (OxIAA) is chemically irreversible, leading to the inactivation of IAA (Peer et al. 2013). The regulation of IAA oxidization was first revealed in a dao mutant (dioxygenase for auxin oxidation) in rice, which encodes a 2-oxoglutarate-dependent-Fe (II) dioxygenase, oxidizing free IAA into OxIAA (Zhao et al. 2013). Main defects of dao mutants in flowers and seeds highlight importance of auxin concentration in local contexts. Both GH3 and DAO genes are induced by auxin, and inactivate auxin in a concentration-dependent manner, which serves as a dynamic feedback to maintain optimal auxin levels (Mellor et al. 2016). In addition to regulatory conjugation of IAA, the abundance of its precursor IPA is also controlled by the formation of IPA-glucose (IPA-Glc), a reaction mediated by an UDP-glycosyltransferase (UGT76F1). This mechanism contributes significantly to auxin homeostasis via competing with YUC enzymes for the shared substrate IPA (Chen et al. 2020). Again, these steps are tightly integrated into the regulatory landscape and it has been shown that the environmental integrator PIF4 can bind to the promoter of both YUC and UGT76F1 (Chen et al. 2020), suggesting a switch-like activity to tune growth to current conditions.
Whereas synthesis and breakdown only recently have emerged as important players in setting up local gradients of auxin, transport processes have attracted attention for many decades. Directed cell-to-cell transport of auxin has been demonstrated to lie at the heart of a plethora of developmental and adaptive processes, including tissue patterning, body polarity, organ specification, and tropisms. In line with the massive influence on many aspects of plant life, auxin transport is dynamically regulated both at the organismal, as well as the tissue level. One fundamental property influencing this process is the acidic environment found in the cell wall, the extracellular matrix of plants (Wolf et al. 2012), which dictates that auxin is partially protonated. This part of the auxin pool can passively diffuse across the plasma membrane (PM), while the rest is actively transported into cells by the influx carrier AUX1/LAX (Vanneste and Friml 2009; Swarup and Péret 2012) and the nitrate transporter NRT1.1 (Krouk et al. 2010).
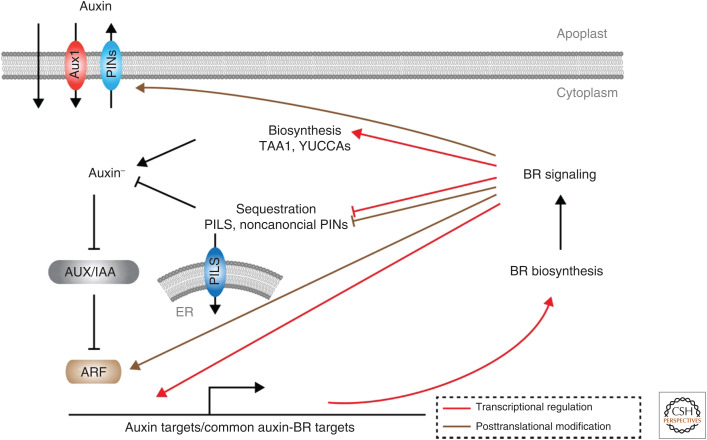
The molecules responsible for the proton gradient between apoplast and cytoplasm are PM-localized H+-ATPase proton pumps. Interestingly, the transport capacity of the pumps can be regulated by auxin itself and environmental cues, such as light (Kinoshita and Shimazaki 1999; Takahashi et al. 2012) and the mechanism for modulating H+-ATPase activity is through protein modification. Both auxin and blue light activate pump activity via SMALL AUXIN UP RNA (SAUR) proteins (McClure and Guilfoyle 1987; Hagen and Guilfoyle 2002) that in turn inhibit type 2C protein phosphatase D (PP2C-D) enzymes, which usually inhibit H+-ATPases (Leyser et al. 1996; Kinoshita and Shimazaki 1999; Takahashi et al. 2012; Spartz et al. 2014; Fendrych et al. 2016; Ren et al. 2018). The transcriptional regulation of SAUR proteins indeed represents a key component of the rapid auxin-induced changes in elongation growth that initiated auxin research more than 150 years ago (Darwin 1880). The SAUR H+-ATPase module also has been shown to couple apoplast pH regulation with other phytohormones or environmental cues. For instance, BR signaling is able to induce phosphorylation of H+-ATPase in hypocotyl elongation either through transcriptional activation of SAURs (Minami et al. 2019) or through a fast BR-regulated PM protein–protein interaction between the BR signaling receptor BRI1 and the H+-ATPase (Fig. 1; Caesar et al. 2011; Miao et al. 2018). In addition, an alternative pathway based on a ligand-receptor system, involving RAPID ALKALINIZATION FACTOR (RALF) and the receptor-like kinase FERONIA, is able to elicit a transient apoplast alkalinization via inhibitory phosphorylation of H+-ATPases (Haruta et al. 2014).
Figure 1.
Regulatory network connecting auxin and brassinosteroid signaling. (ER) Endoplasmic reticulum, (BR) brassinosteroid.
Because deprotonated IAA is essentially incapable of crossing the PM, auxin influx carriers AUXIN1/LIKE-AUX1 (AUX1/LAX) are required for its influx in cells (Swarup and Péret 2012). AUX1, the founder of the AUX1/LAX family, is an amino acid permease-like protein (Bennett et al. 1996) that functions as a H+/IAA− symporter (Yang et al. 2006). AUX/LAX family members in Arabidopsis are all proven functional auxin transporters (Péret et al. 2012) and are involved in a large variety of auxin-mediated developmental processes, such as AUX1 in root gravitropism, root hair development, and phyllotaxy (Bennett et al. 1996; Marchant et al. 1999, 2002); LAX2 in vascular development and quiescent center (QC) maintenance (Péret et al. 2012; Zhang et al. 2013); AUX1 and LAX3 redundant in lateral root initiation and apical hook formation (Hobbie and Estelle 1995; Vandenbussche et al. 2010; Péret et al. 2012); and AUX1, LAX1, and LAX2 redundant in leaf patterning and embryogenesis (Bainbridge et al. 2008; Robert et al. 2015). Specific expression patterns of those family members suggest that these genes are part of an intricate regulatory network and linked to developmental programs; however, most of the underlying mechanistic details still remain to be elucidated. Two examples of transcriptional control of AUX1/LAX genes show the prominent connection to hormone signaling: CK treatment promotes the binding of B-type ARR 1 and 12 TFs to the promoter of LAX2 and causes QC cell division (Zhang et al. 2013). Once again highlighting the significance of feedback regulation in the auxin network, LAX3 expression has shown to be activated by direct promoter binding of the nuclear auxin signaling module ARF7-LBD29 (Porco et al. 2016).
Once inside the cell, auxin is largely present as ionic IAA– and therefore cannot exit the cell by diffusion. Hence, directional transport of auxin depends on two classes of auxin efflux carrier proteins, namely, the ATP-binding cassette of the B subfamily (ABCB) (Geisler et al. 2017) and PIN-FORMED (PIN) families (Okada et al. 1991; Gälweiler et al. 1998; Wisniewska et al. 2006; Adamowski and Friml 2015). A third class of transporters, PIN-like proteins (PILS) (Barbez et al. 2012), together with noncanonical PIN proteins (Bennett et al. 2014), regulate the intracellular auxin homeostasis by importing the hormone to the endoplasmic reticulum (ER). Since auxin mostly acts in the nucleus, this mechanism helps to create local response minima by exploiting the ER as auxin sink (Mravec et al. 2009; Barbez et al. 2012; Béziat et al. 2017; Feraru et al. 2019; Sun et al. 2020).
As the dominant directional auxin efflux carriers, PIN protein families are polarly localized at the PM and PIN localization predicts auxin flux direction (Petrášek et al. 2006; Vanneste and Friml 2009). Analysis of expression patterns and mutant phenotypes of PIN genes demonstrated that PINs are the major determinants of local auxin accumulation (Vieten et al. 2007). Polar localization of PINs on the PM is dependent on controlled exo- and endocytosis (Geldner et al. 2001; Paciorek et al. 2005; Kleine-Vehn et al. 2011) and posttranslational modification represents a major regulatory step in this context (Friml et al. 2004; Abas et al. 2006; Michniewicz et al. 2007; Kleine-Vehn et al. 2009; Leitner et al. 2012; Marhavý et al. 2014; Zourelidou et al. 2014; Weller et al. 2017; Grones et al. 2018; Marhava et al. 2018; Hajný et al. 2020).
Directional auxin transport represents an important module for the spatiotemporal regulation of cell behavior. Using this module, auxin feedbacks are integrated with developmental and environmental signals. For example, PIN gene expression and subsequent subcellular transport protein polarization is enhanced by auxin, which generates a powerful positive feedback loop that plays important roles in many developmental processes, such as phyllotaxis (Bhatia et al. 2016), leaf serrations (Bilsborough et al. 2011), canalization (Sauer et al. 2006; Hajný et al. 2020), lateral root initiation (Chen et al. 2015; Wang et al. 2015), and meristem maintenance (Krogan et al. 2016). This mechanism is based on the transcriptional activation of auxin pathway genes, including PINs (Paponov et al. 2008), which is dependent on auxin nuclear signaling and auxin concentration (Vieten et al. 2005). At the molecular level, ARFs can bind to the promoters of PIN genes in a variety of tissues (Chen et al. 2015; Krogan et al. 2016; Simonini et al. 2017) and are responsible for the reinforcement of auxin signaling and auxin transport (Sauer et al. 2006; Bilsborough et al. 2011; Chen et al. 2015; Bhatia et al. 2016; Krogan et al. 2016; Weijers 2016). For example, a small number of MP/ARF5-positive cells at the periphery of the shoot apical meristem (SAM) promote polarization of PIN1 protein in neighboring cells toward the MP/ARF5-expressing cells, thereby further enhancing auxin influx into the founder cells, which eventually will give rise to a new organ primordium (Bhatia et al. 2016). Similarly, during the early steps of early lateral root formation, ARF7 activates PIN3 expression via direct binding to its promoter. Interestingly, ARF7 also activates expression of the MYB TF FOUR LIPS (FLP), which further enhances PIN3 expression. This feedforward regulation helps sustain PIN3 expression and likely makes the system more robust against initial auxin fluctuations (Chen et al. 2015). Another prominent example is the Arabidopsis fruit, where highly specific auxin accumulation patterns are achieved by intricate spatiotemporal control of PIN gene activity and PIN protein localization. First, auxin signaling output is switched from a bilateral to a radial pattern by the direct promotion of PIN1 and PIN3 expression through bHLH TFs of the HECATE clade, which likely act together with the SPATULA (SPT) bHLH factor (Schuster et al. 2015). Importantly, at the fruit tip, auxin transporters need to switch to an apolar localization to allow for accumulation of auxin in these cells and this transition is dependent on transcriptional repression of PINOID (PID) by the INDIHISCENT (IND)/SPT module (Moubayidin and Østergaard 2014). A similar process is required to pattern the valve margins to support seed dispersal. Here, IND directly represses PID and activates the related WAG2 kinase to promote PIN3 delocalization, leading to a local minimum of auxin accumulation, which in turn is the signal for valve margin development (Sorefan et al. 2009). Another important example is the differential deployment of the auxin-PIN1-CUC2 module during leaf development in Arabidopsis thaliana and Cardamine hirsuta, which provides the substrate for leaf shape variations during evolution (Kierzkowski et al. 2019).
Several other hormone pathways are integrated with auxin signaling at the level of the auxin transport machinery. For example, numerous studies have shown that CK signaling can interfere with polar auxin transport through direct transcriptional control of PINs (Pernisová et al. 2009; Ruzicka et al. 2009; Bishopp et al. 2011a,b; Xie et al. 2018), which has in some cases been shown to be mediated directly through B-ARR TFs. In addition, CK has been shown to interfere with auxin signaling–mediated reinforcement of PIN expression through modulating the expression of SHY2/IAA3 (Dello Ioio et al. 2008). CK signaling can also influence PIN activity through posttranslational regulation (Marhavý et al. 2011, 2014; Zhang et al. 2011; Waldie and Leyser 2018), for example, by triggering depletion of PINs from specific polar membrane domains, thus enhancing polarity (Marhavý et al. 2011, 2014). A particularly intriguing example is on display during lateral root formation. Auxin signaling is involved in all stages of lateral root development, which is initiated by a local auxin response maximum in xylem pole pericycle cells near the root tip (Fukaki et al. 2002, 2005; Wilmoth et al. 2005; De Smet et al. 2007, 2010; Dubrovsky et al. 2008; Moreno-Risueno et al. 2010; Xuan et al. 2016). During further steps, auxin flux continues to be regulated in a complex and dynamic manner. For example, auxin itself induces expression of the auxin influx carriers AUX1 and LAX3 in the cells overlaying lateral root primordia. This, in turn, leads to auxin-mediated expression of cell wall remodeling enzymes, which helps to separate cells in the tissues that have to be crossed by the lateral root before emergence (Swarup et al. 2008). CK, on the other hand, negatively affects lateral root initiation and regulates expression and polarity of PIN proteins (Laplaze et al. 2007; Ruzicka et al. 2009; Marhavý et al. 2011, 2014; Bielach et al. 2012). Interestingly, CK produced in and around lateral root primordia forms an inhibitory field preventing the formation of further primordia and thus is important for the regulation of lateral root spacing (Chang et al. 2015). Besides CK, BRs (Retzer et al. 2019), strigolactones (Crawford et al. 2010; Shinohara et al. 2013), gibberellic acid (Willige et al. 2011; Löfke et al. 2013), and other phytohormones significantly influence polar auxin transport (reviewed in Semeradova et al. 2020).
CONTROL OF AUXIN SIGNALING
While the core auxin signaling system is simple and only consists of three basic modules, namely, receptor, inhibitor, and transcriptional output regulator, these elements have been multiplied by gene duplication events and co-opted in a multitude of regulatory programs. In Arabidopsis, there are six TIR1/AFB-like receptors, 29 Aux/IAA signaling inhibitors, and 23 ARF transcriptional regulators providing an extensive network for functional specification. Supporting the idea that diverse combinations of signaling components will result in context-dependent outputs, all members of the system display highly specific spatiotemporal expression patterns (Rademacher et al. 2011; Vernoux et al. 2011; Weijers and Wagner 2016; Kato et al. 2020). Recently, it was shown that activator ARF expression patterns are driven by a combination of constitutively open chromatin at their loci and the interaction of specific transcriptional repressors with the regulatory regions of individual ARF-encoding genes (Truskina et al. 2021). Importantly, because many WRKY regulators were found among the repressors, these findings suggest strong connectivity of activator ARFs to transcriptional inputs relaying environmental information. In addition, key components of developmental circuits, such as WUS, CUC2, and KNAT1, were found to bind to ARF promoters, supporting the view that ARFs may serve as central nodes integrating exogenous and endogenous signals.
Another important aspect is that many components, most prominently the Aux/IAA auxin-response inhibitors, are transcriptionally activated by auxin (Abel et al. 1995), providing a potent feedback regulation in the network. Importantly, these interactions are mediated by ARF binding to promoters of Aux/IAA genes, which therefore have served as models to elucidate auxin output regulation (Krogan et al. 2014). One prominent example for a developmentally controlled input is the posttranscriptional control of ARF3 mRNA accumulation by trans-acting siRNAs (ta-siRNA). Here, several gene-regulatory layers are superimposed to result in spatiotemporal refinement of ARF3 and ARF4 activity. First, the TAS3 mRNA is transcribed, which is then targeted by miR390 for phased processing into ta-siRNAs, which in turn target ARF3 and ARF4 transcripts for degradation. This intricate regulatory regime is required for a number of developmental contexts, including leaf development, lateral root growth, or developmental timing (Allen et al. 2005; Adenot et al. 2006; Fahlgren et al. 2006; Garcia et al. 2006; Marin et al. 2010).
In addition to their specific expression patterns, auxin signaling components exhibit divergent biochemical functions, which greatly contribute to the complexity of the resulting regulatory network. Most notably, ARF TFs, responsible for the transcriptional output of the system, show considerable sequence divergence and can functionally be grouped in those that have the potential to activate transcription (activator ARFs) and those that do not (repressor ARFs) (Mutte et al. 2018). The sequence divergence has also brought about specific DNA-binding behaviors and preferences for orientation and spacing of DNA-binding motifs, which together specify ARF-dependent gene regulatory output (Boer et al. 2014; Freire-Rios et al. 2020; Lanctot and Nemhauser 2020).
In addition to the transcriptional and biochemical level, specificity is also provided by the protein–protein interaction preferences between ARFs and Aux/IAAs, which in turn dictate the transcriptional output of the pathway (Vernoux et al. 2011; Weijers and Wagner 2016). As a rule of thumb, activator ARFs tend to show interactions with a large number of Aux/IAAs, whereas repressor ARFs do not (Vernoux et al. 2011). In addition to the complex dimerization patterns observed within the ARF-Aux/IAA universe, ARFs also directly interact with a number of other transcriptional regulators from diverse families, such as MYB, MADS-box, KANADI, RGA (GRAS), and bHLH (Shin et al. 2007; Varaud et al. 2011; Kelley et al. 2012; Smaczniak et al. 2012; Oh et al. 2014; José Ripoll et al. 2015). Importantly, MP interacts with the SWI/SNF chromatin-remodeling factors BRAHMA (BRM) and SPLAYED (SYD), which act to open chromatin at target loci. These positive regulators of gene expression compete with the binding of Aux/IAA proteins, which in turn interact with proteins of the TOPLESS family of transcriptional corepressors to repress target expression via histone deacetylation (Long et al. 2006; Szemenyei et al. 2008; Wu et al. 2015). Because auxin sensing triggers the degradation of Aux/IAAs, this represents a major switch for downstream gene activity. Another example for an inhibitory ARF interactor is the bHLH TF HECATE1, the activity of which seems to counteract the capacity of MP to drive auxin-dependent transcription in the periphery of the SAM (Gaillochet et al. 2017).
Recent studies have revealed that ARF6 forms a trimeric complex with PIF4 and the BR responsive TF BZR1 to integrate multiple cues into regulation of cell elongation. DNA binding of the three TFs to their common target loci is inhibited by DELLA proteins, thus GA signaling is required to release inhibition (Oh et al. 2014). BR and auxin signaling are connected at multiple levels in addition to convergence on target genes (Nemhauser et al. 2004). For example, auxin transcriptionally regulates BR biosynthesis and signaling (Chung et al. 2011; Yoshimitsu et al. 2011; Sakamoto et al. 2013; Zhang et al. 2015), whereas a BR input is wired into auxin signaling through interaction of the GSK3-like kinase BIN2, a BR signaling component, and ARFs (Fig. 1). BIN2 phosphorylates the (repressive) B-class ARF2, which decreases DNA binding of the TF, and thus enhances auxin response (Vert et al. 2008). BIN2 can also phosphorylate ARF7/19 to promote DNA binding by disturbing interaction with IAAs, thus enhancing auxin response. However, in this case, BIN2 does not seem to provide BR input but acts downstream of the signaling peptide TDIF (CLE41/44) and its receptor PXY (Cho et al. 2014).
It has recently emerged that posttranslational modification of ARFs seems to be a more general mechanism to mediate integration of inputs form other pathways. For example, ARF2 can also be phosphorylated in response to potassium stress (Zhao et al. 2016), whereas root branching toward water involves ARF7 SUMOylation. Here, SUMO negatively affects DNA binding activity and enhances interaction with IAA3, thus attenuating auxin signaling (Orosa-Puente et al. 2018).
CONTROL OF AUXIN OUTPUT
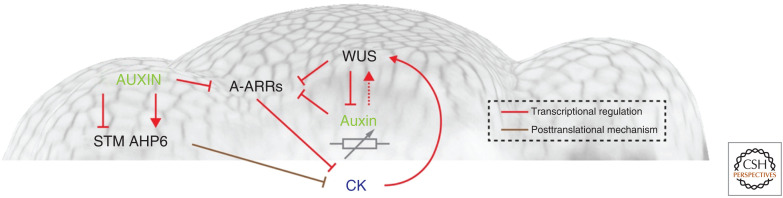
For auxin to be able to play so many divergent roles, the output of the signaling pathway needs to be tightly integrated with other regulatory programs. Importantly, since many activities are highly local, this also requires that the competence for specific auxin responses is limited to few cells by independent repression of downstream genes. Plant stem cell systems, or meristems, represent prominent examples for this scenario, since auxin plays essential, but highly specific roles in subsets of meristematic cells. In the root meristem, auxin accumulates maximally in niche and stem cells and is required for the maintenance of both populations (Sabatini et al. 1999; Friml et al. 2002; Vanneste and Friml 2009); however, adjacent cells that are also exposed to high levels of auxin do not exhibit stem cell fate. In the shoot meristem, high auxin signaling output is excluded from niche and stem cells, but required for inducing differentiation in adjacent cells (Vernoux et al. 2011; Luo et al. 2018; Ma et al. 2019). And finally, auxin provides a locally restricted signal for the development of wood from the cambium, despite a substantially broader accumulation in this tissue (Brackmann et al. 2018). All of these cases have in common that auxin signaling interacts with regulatory modules controlled by WUSCHEL-RELATED HOMEOBOX (WOX) TFs. WUSCHEL (WUS) is a master regulator in shoot apical stem cells (Mayer et al. 1998; Leibfried et al. 2005; Busch et al. 2010; Yadav et al. 2013; Ma et al. 2019), whose family members play conserved functions in stem cell induction, organization, and maintenance, such as WOX5 in root QC cells (Sarkar et al. 2007), WOX4 in cambium stem cells (Brackmann et al. 2018), and root cambium organizer cells (Smetana et al. 2019), as well as other WOXs in embryonic stem cells (WOX2) (Zhang et al. 2017), leaf mediolateral axis cells (WOX1 and WOX3/PRS) (Nakata et al. 2012; Guan et al. 2017; Zhang et al. 2020), and de novo organogenesis (WOX11/12) (Liu et al. 2014). In the stem cambium, WOX4 promotes stem cell fate and is activated by ARF3/4, while it is repressed by ARF5/MP (Brackmann et al. 2018). These results showed that auxin not only is required to position the stem cells, but at the same time to define the fate of one of the derived cell lineages. In the SAM, auxin acts mainly as a local signal to trigger differentiation in the periphery; however, SAM treatments even with high doses and in the absence of auxin transport do not lead to stem cell termination (Reinhardt et al. 2003; Guenot et al. 2012). This robustness is conferred by the local activity of WUS in niche and stem cells by acting as a global repressor of auxin-dependent transcriptional activation (Ma et al. 2019). Interestingly, even in the shoot, stem cells require a basal level of auxin signaling, and it turns out that WUS acts as an auxin response rheostat via the promotion of histone deacetylation (Ma et al. 2019). The dependence of stem cells on low auxin signaling may be connected to the cross talk of auxin with CK (Fig. 2). In the SAM, auxin acts to repress the expression of type-A ARRs, negative feedback regulators of CK via direct binding of MP to ARR promoters, thereby synergizing with the activity of WUS (Leibfried et al. 2005; Zhao et al. 2010). Breaking this interaction may lead to reduced local CK signaling and hence a termination of the stem cell pool. Similarly, during embryonic stem cell initiation, WOX2 represses auxin signaling and in parallel activates CK, which shields the stem cells from differentiation (Zhang et al. 2017). Later, during the initiation of the root meristem, auxin induced expression of type-A ARRs, which in turn limits CK output in the hypophysis to the cells that will give rise to the QC (Müller and Sheen 2008). Along these lines, auxin signaling is required to restrict WOX5 expression to the QC of the mature root meristem, probably through microRNA MIR169-targeted ARF10 and ARF16 auxin-response factors (Ding and Friml 2010). Importantly, WOX5 expression and QC function depend on PLETHORA (PLT) proteins, AP2 TFs that play important roles in setting the balance between cell proliferation and differentiation in roots and shoots (Aida et al. 2004; Galinha et al. 2007; Cruz-Ramírez et al. 2013; Mähönen et al. 2014; Shimotohno et al. 2018). PLT expression, in turn, depends on auxin, and PLT proteins promote auxin biosynthesis and signaling, forming a positive feedback loop (Santuari et al. 2016). PLT proteins form a relatively stable and inert gradient that is generally aligned with the auxin-response gradient in the root meristem (Sabatini et al. 2003; Billou et al. 2005; Grieneisen et al. 2007), but uncouples developmental patterning from rapid auxin fluctuations (Mähönen et al. 2014), for example, those observed during growth adjustments and tropic responses (Shih et al. 2015; Fendrych et al. 2018; Harmer and Brooks 2018). This auxin-PLT module is antagonized by CK signaling to accomplish robust zonation of the longitudinal root meristem (Di Mambro et al. 2017; Salvi et al. 2020).
Figure 2.
Regulatory network connecting auxin and cytokinin signaling in the shoot apical meristem (SAM). (CK) Cytokinin.
Several other regulators interact with auxin to coordinate developmental decisions. At the shoot apex, differentiation is repressed by the class I KNOX homeobox TF SHOOTMERISTEMLESS (STM), whose combined activity with WUS is required for stem cell fate (Lenhard et al. 2002). To allow differentiation at the periphery, STM expression is turned off in response to local PIN1-mediated auxin accumulation (Heisler et al. 2005) via histone deacetylation-mediated transcriptional silencing (Chung et al. 2019). This epigenetic reprogramming of the STM locus is achieved directly by ARF3/ARF4 and indirectly by MP through the intermediary regulator FILAMENTOUS FLOWER (FIL) (Chung et al. 2019). Interestingly, STM at the same time receives positive regulatory inputs by mechanical signals (Landrein et al. 2015), providing an important integration point, which likely serves to properly position organ primordia at the shoot apex. In parallel to this system, auxin signaling at the SAM interacts with an elaborate prepattern setup by the polarity factors KANADI (KAN) and REVOLUTA (REV), one of the HD-ZIP III TFs. While REV is expressed throughout a wide central domain, but excluded from the periphery due to the activity of the mobile miR165/166, KAN accumulation is restricted to the very periphery. Interestingly, both factors repress organ formation, directing maximal PIN1 expression and auxin response to the boundary of their expression domains (Caggiano et al. 2017). This not only defines a competence zone for differentiation, but also prepatterns emerging organ primordia, because the cells in the specification field inherit the REV/KAN expression domains, which in turn will instruct dorsoventral patterning of leaves.
The developing vascular system of the root provides another great example for how auxin signaling is embedded into developmentally regulated transcriptional networks. In a first step, a local auxin maximum established by polar auxin transport confers provascular initial identity to some of the inner cells of the embryo at the early globular stage. MP plays a crucial role in the formation of vascular tissue and activates a suite of downstream TFs termed TARGET OF MONOPTEROS (TMO) (Hardtke and Berleth 1998; Schlereth et al. 2010). One of these, namely, the bHLH regulator TMO7, moves from the basal cell of the embryo proper to the topmost cell of the suspensor to trigger hypophysis development. In contrast, TMO5, another MP target in the embryo, acts cell-autonomously by dimerizing with LONESOME HIGHWAY, but holds the key to setting up the vascular CK signaling domain to promote periclinal cell divisions in both the embryo and the primary root (De Rybel et al. 2013, 2014; Ohashi-Ito et al. 2014). In xylem precursor cells, the TMO5/LHW complex promotes the expression of LONELY GUY4 (LOG4) encoding for a rate-limiting enzyme in CK biosynthesis (De Rybel et al. 2014). Concomitantly, xylem precursor cells are desensitized against the CK they produce through auxin-induced expression of the CK signaling inhibitor AHP6 (Mähönen et al. 2006; Bishopp et al. 2011a; Ohashi-Ito et al. 2014). However, within the xylem axis, AHP6 is only expressed in protoxylem precursor cells, due to the presence of auxin-induced HD-ZIP III TFs in the central xylem cells (Carlsbecker et al. 2010; Bishopp et al. 2011a; Ursache et al. 2014). As these central cells are also largely insensitive toward CK, the existence of another, yet unidentified inhibitory factor besides AHP6 has been postulated. HD-ZIP III TFs also feed back on the auxin response as PHABULSOSA promotes the expression of both MP and the non-auxin-responsive IAA20, indicating an elaborate regulation of auxin signaling strength (Müller et al. 2016). Taken together, high auxin response in xylem precursor cells promotes the biosynthesis of CK and simultaneously cell autonomously interferes with CK signaling. Xylem precursor cell-derived CK thus acts non–cell autonomously in the adjacent procambial domain, where CK signaling promotes formative (periclinal) cell divisions (Mähönen et al. 2000, 2006). In addition, CK in the procambium, which is also derived from phloem transport cells, is required for the expression and subcellular localization of PIN proteins that direct auxin flow toward the xylem axis (Bishopp et al. 2011a,b). This intricate network is further stabilized by a group of DOF TFs called PEARs, which are induced in protophloem sieve-element precursor cells by CK. These factors move to neighboring procambial cells and promote cell division and in addition enhance HD-ZIP III expression, which in turn antagonize PEARs (Miyashima et al. 2019). Interestingly, the bilateral symmetry of the root vasculature, established through the mutually inhibitory interdigitation of auxin and CK signaling, also impacts on tissues outside of the stele (Andersen et al. 2018).
Later in development, during secondary root growth, MP and HD-ZIP III activities synergize to activate a canonical WOX organizer module, in this case built around WOX4. Importantly, vascular stem cell fate is induced non–cell autonomously in the procambial cell adjacent to the WOX4 expressing organizer cell, leading to a direct and indirect induction of xylem and phloem identity, respectively, via a common stem cell (Smetana et al. 2019). This elegant coupling of highly local auxin signaling, CK production, and mobile factors allow for a stepwise elaboration of a complex vascular system from a single inductive cell in the embryo.
Taken together, research in past decades has uncovered that the auxin signaling system is deeply embedded into a multitude of regulatory networks at all levels from biosynthesis to transcriptional output and from the organismal to the subcellular scale. This very embedding appears to be the mechanism allowing one small molecule to play so many and so different roles during plant life.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Abas L, Benjamins R, Malenica N, Paciorek TT, Wiřniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. 2006. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8: 249–256. 10.1038/ncb1369 [DOI] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Theologis A. 1995. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549. 10.1006/jmbi.1995.0454 [DOI] [PubMed] [Google Scholar]
- Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H. 2006. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932. 10.1016/j.cub.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Andersen TG, Naseer S, Ursache R, Wybouw B, Smet W, De Rybel B, Vermeer JEM, Geldner N. 2018. Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature 555: 529–533. 10.1038/nature25976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge K, Guyomarc'h S, Bayer E, Swarup R, Bennett M, Mandel T, Kuhlemeier C. 2008. Auxin influx carriers stabilize phyllotactic patterning. Genes Dev 22: 810–823. 10.1101/gad.462608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, et al. 2012. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122. 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. 1996. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950. 10.1126/science.273.5277.948 [DOI] [PubMed] [Google Scholar]
- Bennett T, Brockington SF, Rothfels C, Graham SW, Stevenson D, Kutchan T, Rolf M, Thomas P, Wong GKS, Leyser O, et al. 2014. Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Mol Biol Evol 31: 2042–2060. 10.1093/molbev/msu147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat C, Barbez E, Feraru MI, Lucyshyn D, Kleine-Vehn J. 2017. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat Plants 3: 17105. 10.1038/nplants.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia N, Bozorg B, Larsson A, Ohno C, Jönsson H, Heisler MG. 2016. Auxin acts through MONOPTEROS to regulate plant cell polarity and pattern phyllotaxis. Curr Biol 26: 3202–3208. 10.1016/j.cub.2016.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielach A, Podlešáková K, Marhavý P, Duclercq J, Cuesta C, Müller B, Grunewald W, Tarkowski P, Benkováa E. 2012. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell 24: 3967–3981. 10.1105/tpc.112.103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billou I, Xu J, Wildwater M, Willemsen V, Paponov I, Frimi J, Heldstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci 108: 3424–3429. 10.1073/pnas.1015162108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. 2011a. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol 21: 917–926. 10.1016/j.cub.2011.04.017 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Lehesranta S, Vatén A, Help H, El-Showk S, Scheres B, Helariutta K, Mähönen AP, Sakakibara H, Helariutta Y. 2011b. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr Biol 21: 927–932. 10.1016/j.cub.2011.04.049 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Spatola Rossi T, Kriechbaumer V. 2019. Auxin biosynthesis: spatial regulation and adaptation to stress. J Exp Bot 70: 5041–5049. 10.1093/jxb/erz283 [DOI] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, Van Den Berg WAM, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, De Vries SC, Solano R, et al. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. 10.1016/j.cell.2013.12.027 [DOI] [PubMed] [Google Scholar]
- Brackmann K, Qi J, Gebert M, Jouannet V, Schlamp T, Grünwald K, Wallner ES, Novikova DD, Levitsky VG, Agustí J, et al. 2018. Spatial specificity of auxin responses coordinates wood formation. Nat Commun 9: 875. 10.1038/s41467-018-03256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN. 2018. Local auxin biosynthesis is a key regulator of plant development. Dev Cell 47: 306–318.e5. 10.1016/j.devcel.2018.09.022 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. 2010. Transcriptional control of a plant stem cell niche. Dev Cell 18: 841–853. 10.1016/j.devcel.2010.03.012 [DOI] [PubMed] [Google Scholar]
- Caesar K, Elgass K, Chen Z, Huppenberger P, Witthöft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K. 2011. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J 66: 528–540. 10.1111/j.1365-313X.2011.04510.x [DOI] [PubMed] [Google Scholar]
- Caggiano MP, Yu X, Bhatia N, Larsson A, Ram H, Ohno CK, Sappl P, Meyerowitz EM, Jönsson H, Heisler MG. 2017. Cell type boundaries organize plant development. eLife 6: e27421. 10.7554/eLife.27421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yang H, Shang C, Ma S, Liu L, Cheng J. 2019. The roles of auxin biosynthesis YUCCA gene family in plants. Int J Mol Sci 20: 8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. 10.1038/nature08977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. 2015. Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. J Exp Bot 66: 4759–4768. 10.1093/jxb/erv252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Liu Y, Maere S, Lee E, Van Isterdael G, Xie Z, Xuan W, Lucas J, Vassileva V, Kitakura S, et al. 2015. A coherent transcriptional feed-forward motif model for mediating auxin-sensitive PIN3 expression during lateral root development. Nat Commun 6: 8821. 10.1038/ncomms9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Huang XX, Zhao SM, Xiao DW, Xiao LT, Tong JH, Wang WS, Li YJ, Ding Z, Hou BK. 2020. IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci 117: 6910–6917. 10.1073/pnas.2000172117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799. 10.1101/gad.1415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2007. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439. 10.1105/tpc.107.053009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. 2014. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol 16: 66–76. 10.1038/ncb2893 [DOI] [PubMed] [Google Scholar]
- Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, Takatsuto S, Tsujimoto M, Kim H, Cho S, et al. 2011. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J 66: 564–578. 10.1111/j.1365-313X.2011.04513.x [DOI] [PubMed] [Google Scholar]
- Chung Y, Zhu Y, Wu MF, Simonini S, Kuhn A, Armenta-Medina A, Jin R, Østergaard L, Gillmor CS, Wagner D. 2019. Auxin response factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat Commun 10: 886. 10.1038/s41467-019-08861-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W. 2009. DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136: 1643–1651. 10.1242/dev.032177 [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913. 10.1242/dev.051987 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Wachsman G, Du Y, Arteága-Vázquez M, Zhang H, Benjamins R, Blilou I, Neef AB, Chandler V, et al. 2013. A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol 11: e1001724. 10.1371/journal.pbio.1001724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D, Zhao J, Jing Y, Fan M, Liu J, Wang Z, Xin W, Hu Y. 2013. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet 9: e1003759. 10.1371/journal.pgen.1003759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1880. The power of movements in plants. John Murray, London. [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D. 2013. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 24: 426–437. 10.1016/j.devcel.2012.12.013 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Adibi M, Breda AS, Wendrich JR, Smit ME, Novák O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J, et al. 2014. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345: 1255215. 10.1126/science.1255215 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, dit Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690. 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voß U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci 107: 2705–2710. 10.1073/pnas.0915001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci 114: E7641–E7649. 10.1073/pnas.1705833114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mambro R, Svolacchia N, Dello Ioio R, Pierdonati E, Salvi E, Pedrazzini E, Vitale A, Perilli S, Sozzani R, Benfey PN, et al. 2019. The lateral root cap acts as an auxin sink that controls meristem size. Curr Biol 29: 1199–1205.e4. 10.1016/j.cub.2019.02.022 [DOI] [PubMed] [Google Scholar]
- Ding Z, Friml J. 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci 107: 12046–12051. 10.1073/pnas.1000672107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E, Benková E. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci 105: 8790–8794. 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund D, Ståldal V, Valsecchi I, Cierlik I, Eriksson C, Hiratsu K, Ohme-Takagi M, Sundström JF, Thelander M, Ezcurra I, et al. 2010. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22: 349–363. 10.1105/tpc.108.064816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund D, Cierlik I, Ståldal V, Claes AR, Vestman D, Chandler J, Sundberg E. 2011. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol 157: 2069–2080. 10.1104/pp.111.182253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. 2006. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944. 10.1016/j.cub.2006.03.065 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J. 2016. Tir1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048. 10.7554/eLife.19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Barbez E, Waidmann S, Sun L, Gaidora A, Kleine-Vehn J. 2019. PILS6 is a temperature-sensitive regulator of nuclear auxin input and organ growth in Arabidopsis thaliana. Proc Natl Acad Sci 116: 3893–3898. 10.1073/pnas.1814015116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. 2011. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci 108: 20231–20235. 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Rios A, Tanaka K, Crespo I, Van der Wijk E, Sizentsova Y, Levitsky V, Lindhoud S, Fontana M, Hohlbein J, Roeland Boer D, et al. 2020. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc Natl Acad Sci 117: 24557–24566. 10.1073/pnas.2009554117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. 2002. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673. 10.1016/S0092-8674(02)00656-6 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg G, et al. 2004. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865. 10.1126/science.1100618 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. 2002. Lateral root formation is blocked by a gain-of-function mutation in the solitary-root/IAA14 gene of Arabidopsis. Plant J 29: 153–168. 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. 2005. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44: 382–395. 10.1111/j.1365-313X.2005.02537.x [DOI] [PubMed] [Google Scholar]
- Gaillochet C, Stiehl T, Wenzl C, Ripoll JJ, Bailey-Steinitz LJ, Li L, Pfeiffer A, Miotk A, Hakenjos JP, Forner J, et al. 2017. Control of plant cell fate transitions by transcriptional and hormonal signals. eLife 6: e30135. 10.7554/eLife.30135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. 10.1038/nature06206 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230. 10.1126/science.282.5397.2226 [DOI] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. 2006. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938. 10.1016/j.cub.2006.03.064 [DOI] [PubMed] [Google Scholar]
- Geisler M, Aryal B, Di Donato M, Hao P. 2017. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol 58: 1601–1614. 10.1093/pcp/pcx104 [DOI] [PubMed] [Google Scholar]
- Geldner N, Firml J, Stierhof YD, Jürgens G, Palme K. 2001. Auxin transport inhibitors block PIN1 and vesicle trafficking. Nature 413: 425–428. 10.1038/35096571 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. 10.1038/nature06215 [DOI] [PubMed] [Google Scholar]
- Grones P, Abas M, Hajný J, Jones A, Waidmann S, Kleine-Vehn J, Friml J. 2018. PID/WAG-mediated phosphorylation of the Arabidopsis PIN3 auxin transporter mediates polarity switches during gravitropism. Sci Rep 8: 10279. 10.1038/s41598-018-28188-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C, Wu B, Yu T, Wang Q, Krogan NT, Liu X, Jiao Y. 2017. Spatial auxin signaling controls leaf flattening in Arabidopsis. Curr Biol 27: 2940–2950. 10.1016/j.cub.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenot B, Bayer E, Kierzkowski D, Smith RS, Mandel T, Zádníková P, Benková E, Kuhlemeier C. 2012. Pin1-independent leaf initiation in Arabidopsis. Plant Physiol 159: 1501–1510. 10.1104/pp.112.200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ. 1985. Rapid induction of selective transcription by auxins. Mol Cell Biol 5: 1197–1203. 10.1128/MCB.5.6.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. 2002. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385. 10.1023/A:1015207114117 [DOI] [PubMed] [Google Scholar]
- Hagen G, Martin G, Li Y, Guilfoyle TJ. 1991. Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol Biol 17: 567–579. 10.1007/BF00040658 [DOI] [PubMed] [Google Scholar]
- Hajný J, Prát T, Rydza N, Rodriguez L, Tan S, Verstraeten I, Domjan D, Mazur E, Smakowska-Luzan E, Smet W, et al. 2020. Receptor kinase module targets PIN-dependent auxin transport during canalization. Science 370: 550–557. 10.1126/science.aba3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. 2002. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615. 10.1101/gad.229402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411. 10.1093/emboj/17.5.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Brooks CJ. 2018. Growth-mediated plant movements: hidden in plain sight. Curr Opin Plant Biol 41: 89–94. 10.1016/j.pbi.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911. 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, De Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C. 2014. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc Natl Acad Sci 111: 6515–6520. 10.1073/pnas.1320355111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. 1995. The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J 7: 211–220. 10.1046/j.1365-313X.1995.7020211.x [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711. 10.1111/j.1365-313X.2012.05033.x [DOI] [PubMed] [Google Scholar]
- José Ripoll J, Bailey LJ, Mai QA, Wu SL, Hon CT, Chapman EJ, Ditta GS, Estelle M, Yanofsky MF. 2015. MicroRNA regulation of fruit growth. Nat Plants 1: 1–9. 10.1038/nplants.2015.36 [DOI] [PubMed] [Google Scholar]
- Kato H, Mutte SK, Suzuki H, Crespo I, Das S, Radoeva T, Fontana M, Yoshitake Y, Hainiwa E, van den Berg W, et al. 2020. Design principles of a minimal auxin response system. Nat Plants 6: 473–482. 10.1038/s41477-020-0662-y [DOI] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. 2012. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109. 10.1242/dev.067918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D, Runions A, Vuolo F, Strauss S, Lymbouridou R, Routier-Kierzkowska AL, Wilson-Sánchez D, Jenke H, Galinha C, Mosca G, et al. 2019. A growth-based framework for leaf shape development and diversity. Cell 177: 1405–1418.e17. 10.1016/j.cell.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki KI. 1999. Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558. 10.1093/emboj/18.20.5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J. 2009. PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849. 10.1105/tpc.109.071639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martinière A, Łangowski Ł, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, et al. 2011. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7: 540. 10.1038/msb.2011.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Yin X, Ckurshumova W, Berleth T. 2014. Distinct subclades of Aux/IAA genes are direct targets of ARF5/MP transcriptional regulation. New Phytol 204: 474–483. 10.1111/nph.12994 [DOI] [PubMed] [Google Scholar]
- Krogan NT, Marcos D, Weiner AI, Berleth T. 2016. The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol 212: 42–50. 10.1111/nph.14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937. 10.1016/j.devcel.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Lanctot A, Nemhauser JL. 2020. It's Morphin’ time: how multiple signals converge on ARF transcription factors to direct development. Curr Opin Plant Biol 57: 1–7. 10.1016/j.pbi.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, Laufs P, Takeda S, Aida M, Traas J, et al. 2015. Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. eLife 4: e07811. 10.7554/eLife.07811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900. 10.1105/tpc.107.055863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JPCC, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. 2005. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175. 10.1038/nature04270 [DOI] [PubMed] [Google Scholar]
- Leitner J, Petrášek J, Tomanov K, Retzer K, Pařezová M, Korbei B, Bachmair A, Zažímalová E, Luschnig C. 2012. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci 109: 8322–8327. 10.1073/pnas.1200824109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M, Jürgens G, Laux T. 2002. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129: 3195–3206. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M. 1996. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413. 10.1046/j.1365-313x.1996.10030403.x [DOI] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. 2012. Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L. 2014. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093. 10.1105/tpc.114.122887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Gao S, Tian H, Wu W, Robert HS, Ding Z. 2016. Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet 12: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Zwiewka M, Heilmann I, Van Montagu MCE, Teichmann T, Friml J. 2013. Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc Natl Acad Sci 110: 3627–3632. 10.1073/pnas.1300107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. 10.1126/science.1123841 [DOI] [PubMed] [Google Scholar]
- Luo L, Zeng J, Wu H, Tian Z, Zhao Z. 2018. A molecular framework for auxin-controlled homeostasis of shoot stem cells in Arabidopsis. Mol Plant 11: 899–913. 10.1016/j.molp.2018.04.006 [DOI] [PubMed] [Google Scholar]
- Ma Y, Miotk A, Šutiković Z, Ermakova O, Wenzl C, Medzihradszky A, Gaillochet C, Forner J, Utan G, Brackmann K, et al. 2019. WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat Commun 10: 5093. 10.1038/s41467-019-13074-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. 2000. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943. 10.1101/gad.189200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. 2006. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98. 10.1126/science.1118875 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. 10.1038/nature13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ, Perrot-Rechenmann C, Bennett MJ. 1999. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073. 10.1093/emboj/18.8.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G. 2002. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597. 10.1105/tpc.010354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P, Bassukas AEL, Zourelidou M, Kolb M, Moret B, Fastner A, Schulze WX, Cattaneo P, Hammes UZ, Schwechheimer C, et al. 2018. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558: 297–300. 10.1038/s41586-018-0186-z [DOI] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Pařezová M, Petrášek J, Friml J, Kleine-Vehn J, et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804. 10.1016/j.devcel.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benková E. 2014. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr Biol 24: 1031–1037. 10.1016/j.cub.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. mir390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. 10.1105/tpc.109.072553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Fernández I, Sanchís S, Marini N, Balanzá V, Ballester P, Navarrete-Gómez M, Oliveira AC, Colombo L, Ferrándiz C. 2014. The effect of NGATHA altered activity on auxin signaling pathways within the Arabidopsis gynoecium. Front Plant Sci 5: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. 10.1016/S0092-8674(00)81703-1 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T. 1987. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623. 10.1007/BF00020537 [DOI] [PubMed] [Google Scholar]
- Mellor N, Band LR, Pěňík A, Novák O, Rashed A, Holman T, Wilson MH, Vo U, Bishopp A, King JR, et al. 2016. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci 113: 11022–11027. 10.1073/pnas.1604458113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao R, Wang M, Yuan W, Ren Y, Li Y, Zhang N, Zhang J, Kronzucker HJ, Xu W. 2018. Comparative analysis of Arabidopsis ecotypes reveals a role for brassinosteroids in root hydrotropism. Plant Physiol 176: 2720–2736. 10.1104/pp.17.01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. 2007. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. 10.1016/j.cell.2007.07.033 [DOI] [PubMed] [Google Scholar]
- Minami A, Takahashi K, Inoue SI, Tada Y, Kinoshita T. 2019. Brassinosteroid induces phosphorylation of the plasma membrane H+-ATPase during hypocotyl elongation in Arabidopsis thaliana. Plant Cell Physiol 60: 935–944. 10.1093/pcp/pcz005 [DOI] [PubMed] [Google Scholar]
- Miyashima S, Roszak P, Sevilem I, Toyokura K, Blob B, Heo JO, Mellor N, Help-Rinta-Rahko H, Otero S, Smet W, et al. 2019. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 565: 490–494. 10.1038/s41586-018-0839-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311. 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Østergaard L. 2014. Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr Biol 24: 2743–2748. 10.1016/j.cub.2014.09.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrášek J, Zhang J, Gaykova V, Stierhof YD, et al. 2009. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140. 10.1038/nature08066 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. 10.1038/nature06943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. 1998. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911. 10.1093/emboj/17.23.6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CJ, Valdés AE, Wang G, Ramachandran P, Beste L, Uddenberg D, Carlsbecker A. 2016. PHABULOSA mediates an auxin signaling loop to regulate vascular patterning in Arabidopsis. Plant Physiol 170: 956–970. 10.1104/pp.15.01204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GKS, Weijers D. 2018. Origin and evolution of the nuclear auxin response system. eLife 7: e33399. 10.7554/eLife.33399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K. 2012. Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24: 519–535. 10.1105/tpc.111.092858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: e258. 10.1371/journal.pbio.0020258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. 2014. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. 10.7554/eLife.03031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Saegusa M, Iwamoto K, Oda Y, Katayama H, Kojima M, Sakakibara H, Fukuda H. 2014. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr Biol 24: 2053–2058. 10.1016/j.cub.2014.07.050 [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. 1991. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684. 10.2307/3869249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. 2018. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362: 1407–1410. 10.1126/science.aau3956 [DOI] [PubMed] [Google Scholar]
- Paciorek T, Zažímalová E, Ruthardt N, Petrášek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al. 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256. 10.1038/nature03633 [DOI] [PubMed] [Google Scholar]
- Palme K, Gälweiler L. 1999. PIN-pointing the molecular basis of auxin transport. Curr Opin Plant Biol 2: 375–381. 10.1016/S1369-5266(99)00008-4 [DOI] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JAH, Palme K. 2008. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1: 321–337. 10.1093/mp/ssm021 [DOI] [PubMed] [Google Scholar]
- Peer WA, Cheng Y, Murphy AS. 2013. Evidence of oxidative attenuation of auxin signalling. J Exp Bot 64: 2629–2639. 10.1093/jxb/ert152 [DOI] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, et al. 2012. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885. 10.1105/tpc.112.097766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisová M, Klíma P, Horák J, Válková M, Malbeck J, Souček P, Reichman P, Hoyerová K, Dubová J, Friml J, et al. 2009. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci 106: 3609–3614. 10.1073/pnas.0811539106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wiśniewska J, Tadele Z, Kubeš M, Čovanová M, et al. 2006. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918. 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Porco S, Larrieu A, Du Y, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, et al. 2016. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 143: 3340–3349. 10.1242/dev.136283 [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Möller B, Lokerse AS, Llavata-Peris CI, Van Den Berg W, Weijers D. 2011. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 68: 597–606. 10.1111/j.1365-313X.2011.04710.x [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. 10.1038/nature02081 [DOI] [PubMed] [Google Scholar]
- Ren H, Park MY, Spartz AK, Wong JH, Gray WM. 2018. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genet 14: e1007455. 10.1371/journal.pgen.1007455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzer K, Akhmanova M, Konstantinova N, Malínská K, Leitner J, Petrášek J, Luschnig C. 2019. Brassinosteroid signaling delimits root gravitropism via sorting of the Arabidopsis PIN2 auxin transporter. Nat Commun 10: 5516. 10.1038/s41467-019-13543-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi K, Landberg K, Nilsson L, Ljung K, Sundås-Larsson A. 2011. TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J 65: 897–906. 10.1111/j.1365-313X.2010.04470.x [DOI] [PubMed] [Google Scholar]
- Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J. 2015. Plant embryogenesis requires aux/lax-mediated auxin influx. Development 142: 702–711. 10.1242/dev.115832 [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. 1998. Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373. 10.1126/science.279.5355.1371 [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Šimášková M, Duclercq J, Petrášek J, Zažímalová E, Simon S, Friml J, Van Montagu MCE, Benková E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci 106: 4284–4289. 10.1073/pnas.0900060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. 10.1016/S0092-8674(00)81535-4 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358. 10.1101/gad.252503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S. 2013. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J 73: 676–688. 10.1111/tpj.12071 [DOI] [PubMed] [Google Scholar]
- Salvi E, Rutten JP, Di Mambro R, Polverari L, Licursi V, Negri R, Dello Ioio R, Sabatini S, Ten Tusscher K. 2020. A self-organized PLT/auxin/ARR-B network controls the dynamics of root zonation development in Arabidopsis thaliana. Dev Cell 53: 431–443.e23. 10.1016/j.devcel.2020.04.004 [DOI] [PubMed] [Google Scholar]
- Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L, Gorte M, Prasad K, Bao D, Timmermans-Hereijgers JLPM, et al. 2016. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28: 2937–2951. 10.1105/tpc.16.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. 10.1038/nature05703 [DOI] [PubMed] [Google Scholar]
- Sauer M, Balla J, Luschnig C, Wiśniewska J, Reinöhl V, Friml J, Benková E. 2006. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20: 2902–2911. 10.1101/gad.390806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916. 10.1038/nature08836 [DOI] [PubMed] [Google Scholar]
- Schuster C, Gaillochet C, Lohmann JU. 2015. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142: 3343–3350. 10.1242/dev.120444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeradova H, Montesinos JC, Benkova E. 2020. All roads lead to auxin: post-translational regulation of auxin transport by multiple hormonal pathways. Plant Commun 1: 100048. 10.1016/j.xplc.2020.100048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HW, Depew CL, Miller ND, Monshausen GB. 2015. The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr Biol 25: 3119–3125. 10.1016/j.cub.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Shimotohno A, Heidstra R, Blilou I, Scheres B. 2018. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev 32: 1085–1100. 10.1101/gad.314096.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. 2007. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453. 10.1105/tpc.107.050963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. 2013. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. 10.1371/journal.pbio.1001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S, Bencivenga S, Trick M, Østergaard L. 2017. Auxin-induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis. Plant Cell 29: 1864–1882. 10.1105/tpc.17.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci 109: 1560–1565. 10.1073/pnas.1112871109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana O, Mäkilä R, Lyu M, Amiryousefi A, Sánchez Rodríguez F, Wu MF, Solé-Gil A, Leal Gavarrón M, Siligato R, Miyashima S, et al. 2019. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565: 485–489. 10.1038/s41586-018-0837-0 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galván-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Østergaard L. 2009. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459: 583–586. 10.1038/nature07875 [DOI] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. 10.1105/tpc.114.126037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. 2012. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. 10.1371/journal.pgen.1002594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Feraru E, Feraru MI, Waidmann S, Wang W, Passaia G, Wang ZY, Wabnik K, Kleine-Vehn J. 2020. PIN-LIKES coordinate brassinosteroid signaling with nuclear auxin input in Arabidopsis thaliana. Curr Biol 30: 1579–1588.e6. 10.1016/j.cub.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Péret B. 2012. AUX/LAX family of auxin influx carriers—an overview. Front Plant Sci 3: 225. 10.3389/fpls.2012.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954. 10.1038/ncb1754 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. 10.1126/science.1151461 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Hayashi KI, Kinoshita T. 2012. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol 159: 632–641. 10.1104/pp.112.196428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J. 2006. Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell Mol Life Sci 63: 2738–2754. 10.1007/s00018-006-6116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LP, Zhou C, Wang SS, Yuan J, Zhang XS, Su YH. 2017. FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol 213: 1740–1754. 10.1111/nph.14313 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. 10.1016/j.cell.2008.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truskina J, Han J, Chrysanthou E, Galvan-Ampudia CS, Lainé S, Brunoud G, Macé J, Bellows S, Legrand J, Bågman AM, et al. 2021. A network of transcriptional repressors modulates auxin responses. Nature 589: 116–119. 10.1038/s41586-020-2940-2 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1997a. ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868. 10.1126/science.276.5320.1865 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997b. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R, Miyashima S, Chen Q, Vatén A, Nakajima K, Carlsbecker A, Zhao Y, Helariutta Y, Dettmer J. 2014. Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 141: 1250–1259. 10.1242/dev.103473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Petrášek J, Žádníková P, Hoyerová K, Pešek B, Raz V, Swarup R, Bennett M, Zažímolavá E, Benkova E, et al. 2010. The auxin influx carriers AUX1 and LAX3 are involved in auxin–ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606. 10.1242/dev.040790 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin: a trigger for change in plant development. Cell 136: 1005–1016. 10.1016/j.cell.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23: 973–983. 10.1105/tpc.110.081653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van Den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. 2008. Integration of auxin and brassinosteroid pathways by auxin response factor 2. Proc Natl Acad Sci 105: 9829–9834. 10.1073/pnas.0803996105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wiśniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. 2005. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531. 10.1242/dev.02027 [DOI] [PubMed] [Google Scholar]
- Vieten A, Sauer M, Brewer PB, Friml J. 2007. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci 12: 160–168. 10.1016/j.tplants.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Vragović K, Sela A, Friedlander-Shani L, Fridman Y, Hacham Y, Holland N, Bartom E, Mockler TC, Savaldi-Goldstein S, Selaa A, et al. 2015. Translatome analyses capture of opposing tissues pecific brassinosteroid signals orchestrating root meristem differentiation. Proc Natl Acad Sci 112: 923–928. 10.1073/pnas.1417947112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldie T, Leyser O. 2018. Cytokinin targets auxin transport to promote shoot branching. Plant Physiol 177: 803–818. 10.1104/pp.17.01691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HZ, Yang KZ, Zou JJ, Zhu LL, Xie ZD, Morita MT, Tasaka M, Friml J, Grotewold E, Beeckman T, et al. 2015. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat Commun 6: 8822. 10.1038/ncomms9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Qin G, Cao M, Chen R, He Y, Yang L, Zeng Z, Yu Y, Gu Y, Xing W, et al. 2020. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nat Commun 11: 679. 10.1038/s41467-020-14395-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D. 2016. Phyllotaxis: a Matthew effect in auxin action. Curr Biol 26: R1233–R1235. 10.1016/j.cub.2016.10.019 [DOI] [PubMed] [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67: 539–574. 10.1146/annurev-arplant-043015-112122 [DOI] [PubMed] [Google Scholar]
- Weller B, Zourelidou M, Frank L, Barbosa ICR, Fastner A, Richter S, Jürgens G, Hammes UZ, Schwechheimer C. 2017. Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc Natl Acad Sci 114: E887–E896. 10.1073/pnas.1614380114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Isono E, Richter R, Zourelidou M, Schwechheimer C. 2011. Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23: 2184–2195. 10.1105/tpc.111.086355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. 2005. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43: 118–130. 10.1111/j.1365-313X.2005.02432.x [DOI] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifartová D, Brewer PB, Růžička K, Blilou L, Rouquié D, Benková E, Scheres B, Friml J. 2006. Polar PIN localization directs auxin flow in plants. Science 312: 883. 10.1126/science.1121356 [DOI] [PubMed] [Google Scholar]
- Wolf S, Hématy K, Höfte H. 2012. Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63: 381–407. 10.1146/annurev-arplant-042811-105449 [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. 2011. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci 108: 18518–18523. 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Ann Bot 95: 707–735. 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RM, Hagen G, Guilfoyle T. 1987. An auxin-induced polypeptide in dicotyledonous plants. Plant Mol Biol 9: 625–634. 10.1007/BF00020538 [DOI] [PubMed] [Google Scholar]
- Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. 2015. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4: e09269. 10.7554/eLife.09269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Chen H, Huang L, O'Neil RC, Shokhirev MN, Ecker JR. 2018. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat Commun 9: 1604. 10.1038/s41467-018-03921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Möller BK, Skorzinski N, Njo MF, et al. 2016. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351: 384–387. 10.1126/science.aad2776 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Ohno C, Heisler M, Girke T, Jönsson H, Reddy GV, Venugopala Reddy G. 2013. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol Syst Biol 9: 654. 10.1038/msb.2013.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Liu X, Ljung K, Li S, Zhao W, Yang F, Wang M, Tao Y. 2017. Type B response regulators act as central integrators in transcriptional control of the auxin biosynthesis enzyme TAA1. Plant Physiol 175: 1438–1454. 10.1104/pp.17.00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. 2006. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127. 10.1016/j.cub.2006.04.029 [DOI] [PubMed] [Google Scholar]
- Yoshimitsu Y, Tanaka K, Fukuda W, Asami T, Yoshida S, Hayashi KI, Kamiya Y, Jikumaru Y, Shigeta T, Nakamura Y, et al. 2011. Transcription of DWARF4 plays a crucial role in auxin-regulated root elongation in addition to brassinosteroid homeostasis in Arabidopsis thaliana. PLoS ONE 6: e23851. 10.1371/journal.pone.0023851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, To JPC, Cheng CY, Eric Schaller G, Kieber JJ. 2011. Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J 68: 1–10. 10.1111/j.1365-313X.2011.04668.x [DOI] [PubMed] [Google Scholar]
- Zhang W, Swarup R, Bennett M, Schaller GE, Kieber JJ. 2013. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr Biol 23: 1979–1989. 10.1016/j.cub.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang S, Xu Y, Yu C, Shen C, Qian Q, Geisler M, Jiang DA, Qi Y. 2015. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ 38: 638–654. 10.1111/pce.12397 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tucker E, Hermann M, Laux T. 2017. A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev Cell 40: 264–277.e4. 10.1016/j.devcel.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Runions A, Mentink RA, Kierzkowski D, Karady M, Hashemi B, Huijser P, Strauss S, Gan X, Ljung K, et al. 2020. A WOX/auxin biosynthesis module controls growth to shape leaf form. Curr Biol 30: 1–12. [DOI] [PubMed] [Google Scholar]
- Zhao Y. 2018. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu Rev Plant Biol 69: 417–435. 10.1146/annurev-arplant-042817-040226 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309. 10.1126/science.291.5502.306 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. 2010. Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092. 10.1038/nature09126 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang Y, Liu X, Zhang X, Liu S, Yu X, Ren Y, Zheng X, Zhou K, Jiang L, et al. 2013. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27: 113–122. 10.1016/j.devcel.2013.09.005 [DOI] [PubMed] [Google Scholar]
- Zhao S, Zhang ML, Ma TL, Wang Y. 2016. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28: 3005–3019. 10.1105/tpc.16.00684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang D, An J, Yin H, Fang S, Chu J, Zhao Y, Li J. 2018. TCP transcription factors regulate shade avoidance via directly mediating the expression of both PHYTOCHROME INTERACTING FACTORs and auxin biosynthetic genes. Plant Physiol 176: 1850–1861. 10.1104/pp.17.01566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Absmanner B, Weller B, Barbosa ICR, Willige BC, Fastner A, Streit V, Port SA, Colcombet J, van Bentem SDLF, et al. 2014. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: e02860. 10.7554/eLife.02860 [DOI] [PMC free article] [PubMed] [Google Scholar]