Abstract
We have recently identified the Raf kinase inhibitor protein (RKIP) as a physiological endogenous inhibitor of the Raf-1/MEK/extracellular signal-regulated kinase (ERK) pathway. RKIP interfered with MEK phosphorylation and activation by Raf-1, resulting in the suppression of both Raf-1-induced transformation and AP-1-dependent transcription. Here we report the molecular mechanism of RKIP's inhibitory function. RKIP can form ternary complexes with Raf-1, MEK, and ERK. However, whereas MEK and ERK can simultaneously associate with RKIP, Raf-1 binding to RKIP and that of MEK are mutually exclusive. RKIP is able to dissociate a Raf-1–MEK complex and behaves as a competitive inhibitor of MEK phosphorylation. Mapping of the binding domains showed that MEK and Raf-1 bind to overlapping sites in RKIP, whereas MEK and RKIP associate with different domains in Raf-1, and Raf-1 and RKIP bind to different sites in MEK. Both the Raf-1 and the MEK binding sites in RKIP need to be destroyed in order to relieve RKIP-mediated suppression of the Raf-1/MEK/ERK pathway, indicating that binding of either Raf-1 or MEK is sufficient for inhibition. The properties of RKIP reveal the specific sequestration of interacting components as a novel motif in the cell's repertoire for the regulation of signaling pathways.
In metazoans, the Ras/Raf-1/MEK/extracellular signal-regulated kinase (ERK) module is a ubiquitously expressed signaling pathway that conveys mitogenic and differentiation signals from the cell membrane to the nucleus (6). This kinase cascade appears to be spatially organized in a signaling complex nucleated by Ras proteins (15). The small G protein Ras is activated by many growth factor receptors and binds the Raf-1 kinase with high affinity when activated. This induces the recruitment of Raf-1 from the cytosol to the cell membrane and its subsequent activation by mechanisms which remain incompletely understood (16). Activated Raf-1 then phosphorylates and activates MEK, a kinase that in turn phosphorylates and activates ERK, the prototypic mitogen-activated protein kinase (MAPK) (13). Activated ERKs can translocate to the nucleus and regulate gene expression by the phosphorylation of transcription factors (19).
Studies with yeasts have revealed the important role of scaffolding proteins which assemble the components of MAPK pathways and thereby ensure that the signal transfer is efficient and specific (5). Mammalian homologues of such scaffolding proteins have been postulated, but despite extensive efforts, only a few candidates have been identified. These include JIP-1, a scaffolding protein for the stress-activated MAPKs/JNKs (24), as well as Ksr, a protein kinase identified in genetic screens (4), which could have a similar function in the ERK pathway. Ksr binds to Raf-1, MEK, and ERK, but as both activation and inhibition by Ksr were observed, the physiological role of Ksr remains enigmatic (3, 10, 14, 23, 25, 27). Since scaffolding proteins are expected to function in a stoichiometric manner, these discrepancies may have arisen from situations of nonstoichiometric expression levels (20) but also could reflect additional regulatory properties of Ksr. These observations suggest that the Raf-1/MEK/ERK pathway is subject to an additional level of regulation exerted by associated proteins. This hypothesis was further confirmed by the cloning of MP-1, a MEK-1-binding protein that specifically enhances the activation of ERK-1 (21).
Using the yeast two-hybrid system, we recently identified a protein which binds to Raf-1, MEK, and ERK in vitro and in vivo (26). This protein was dubbed the Raf kinase inhibitor protein (RKIP) because it interfered with the activation of the Raf→MEK→ERK signaling pathway in vitro and in vivo. RKIP overexpression suppressed the ERK pathway and, as a consequence, interfered with Raf-1-induced transformation and AP-1-dependent transcription, whereas the downregulation of RKIP had the opposite effect. Genetic evidence indicated that RKIP functions at the Raf-1/MEK interface, because it suppressed signaling by activated Raf-1 mutants but not by activated MEK alleles. Here we describe the molecular mechanism of how RKIP works to inhibit the ERK pathway.
MATERIALS AND METHODS
Plasmids and protein expression.
RKIP expression plasmids have been previously described (26). Deletion mutants of pCMV5-HA-RKIP (26) for expression in mammalian cells were generated by PCR. To construct FLAG-tagged Raf-1, the Raf-1 cDNA was PCR amplified for in-frame cloning into pCMV2-FLAG. For expression in Escherichia coli, deletion mutants were made as follows. GNX, which contains the BXB cDNA cloned into pGEX-KG (7), was cut with HindIII and other restriction enzymes (see Fig. 5a). HindIII cuts downstream of the BXB cDNA and upstream of stop codons in all three reading frames. After blunt ending with T4 polymerase, the plasmids were religated. The same strategy was used to make glutathione S-transferase (GST)–RKIP deletion mutants. MEK-1 deletion mutants were generated by PCR and cloned into pRSETA, resulting in the addition of an N-terminal six-His tag. Proteins were expressed and purified as described previously (7, 26). Activated Raf-1 was purified from Sf-9 insect cells coinfected with GST–Raf-1 plus RasV12 and Lck as previously described (18). GST–MEK-1–Raf-1 complexes were produced in Sf-9 insect cells and purified by adsorption to glutathione Sepharose, as described previously (18).
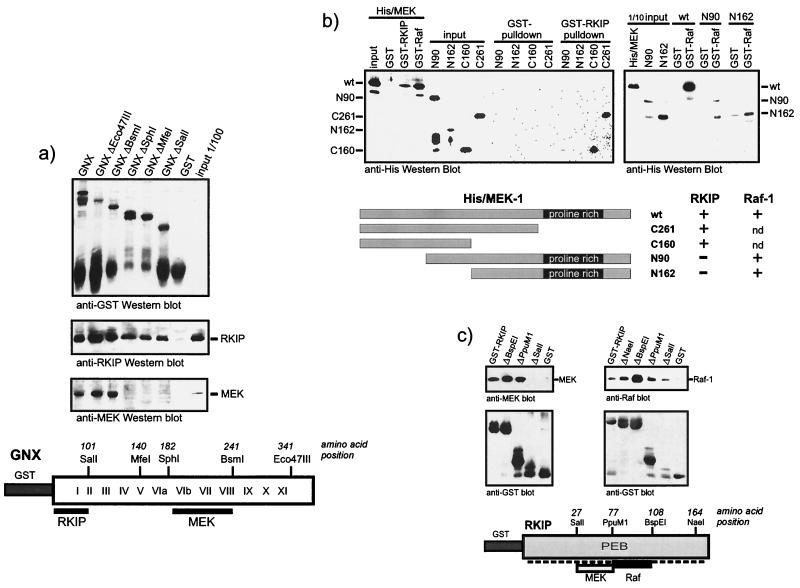
FIG. 5.
Analysis of binding domains. (a) RKIP and MEK bind to different domains of the Raf-1 kinase. GST-tagged BXB, GNX, and the indicated deletion mutants were expressed in E. coli, immobilized on glutathione Sepharose beads, and incubated with purified RKIP or MEK-1. Proteins were visualized by Western blotting. The diagram illustrates the GNX regions deduced to be required for binding. Roman numerals refer to the kinase subdomains as defined by Hanks and Quinn (8). (b) RKIP and Raf-1 bind to different domains of MEK-1. Purified six-His-tagged MEK-1 deletion mutants were tested for binding to GST-RKIP beads (left panel) and GST–Raf-1 beads (right panel). His/MEK-1 proteins were detected by Western blotting with anti-His antibodies. The lower panel shows a schematic summary. nd, not done. (c) Analysis of Raf-1 and MEK binding sites in RKIP. GST-RKIP deletion mutants were tested for binding of MEK-1 and Raf-1. PEB, phosphatidylethanolamine binding motif.
In vitro binding assays.
Typically, binding reactions between purified recombinant proteins were done in phosphate-buffered saline (PBS) containing 10% bovine serum as a nonspecific competitor. Consistent results were obtained with 0.5 or 5% bovine serum albumin. After incubation for 1 to 5 h at 4°C, the samples were washed four times with PBS, resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and blotted. Pulldown assays with the His/MEK-1 deletion mutants were performed by incubating 1 μg of soluble His/MEK-1 proteins with 1 μg of GST or GST fusion proteins immobilized on glutathione Sepharose beads in 0.5 ml of buffer containing 20 mM Tris-HCl (pH 7.4), 0.2 mM EDTA, 0.1 M NaCl, and 1 mM dithiothreitol. The beads were washed twice with the same buffer containing 0.1% NP-40, resolved by SDS-polyacrylamide gel electrophoresis, and immunoblotted with anti-His tag antibody (Qiagen). Since full-length Raf-1 cannot be expressed in E. coli in an active form, Sf-9 insect cells infected with a Raf-1 baculovirus were used. Lysates were prepared by freeze-thawing Sf-9 cells in PBS or by lysis in TBST (20 mM Tris HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, and 1% Triton X-100) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 1 μg of leupeptin/ml). Detergent-free lysis improved the recovery of complexes in the binding reactions but gave qualitatively the same results as Triton X-100 lysates. Lysates were clarified by centrifugation at 23,000 × g for 10 min, and the supernatants were used for the binding reactions. The blots were developed using chemiluminescence. Phosphorylated His/MEK-1 for use in RKIP binding assays (see Fig. 4c) was obtained by incubation with GST–Raf-1 immobilized on glutathione Sepharose in the presence of 20 μM ATP and 0.5 μCi of [γ-32P]ATP for 45 min. The GST–Raf-1 beads were removed by centrifugation. The supernatant was diluted fivefold with PBS and incubated with GST or GST-RKIP beads. To reduce nonspecific binding, the beads were preabsorbed with 10% serum or 2% bovine serum albumin for at least 2 h. Typically, 0.5 to 2 μg of His/MEK-1 per binding reaction was used. Phosphorylated ERK was made in a similar fashion with the following modifications. The GST portion of GST-ERK2 was removed by thrombin cleavage. GST-MEK was activated by GST–Raf-1 as described above except that only cold ATP was used. After 30 min, ERK2 and 0.5 μCi of [γ-32P]ATP were added and incubated for a further 15 min. The reaction was diluted fivefold with PBS, and 20 μl of glutathione Sepharose beads was added to assure the removal of all GST-tagged proteins. The supernatant was used for the binding reactions. For some experiments, activated ERK purchased from New England Biolabs was used with consistent results.
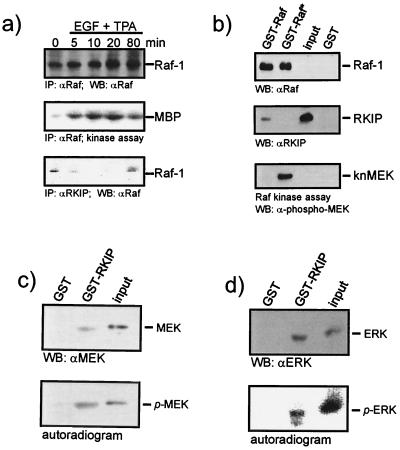
FIG. 4.
Analysis of RKIP binding to activated Raf-1, MEK, and ERK. (a) Mitogen activation of Raf-1 decreases its association with RKIP. COS-1 cells were transiently transfected with Raf-1 and RKIP expression vectors. Serum-starved cells were treated with epidermal growth factor (EGF) (20 ng/ml) plus TPA (100 ng/ml) for the times indicated. Raf-1 immunoprecipitates were analyzed for kinase activity, and RKIP immunoprecipitates were examined for Raf-1. IP, immunoprecipitation; WB, Western blot. (b) Purified RKIP produced in E. coli was tested for binding to GST-Raf and activated (*) GST-Raf beads. GST-Raf proteins were produced in Sf-9 cells and activated by coexpression of RasV12 and Lck. An aliquot of the GST-Raf beads was examined for phosphorylation of kinase-negative MEK (knMEK). (c and d) MEK and ERK proteins were phosphorylated in the presence of [γ-32P]ATP and tested for binding to GST-RKIP beads. Binding of phosphorylated proteins was detected by autoradiography. Binding of total protein was visualized by Western blotting (WB). The contribution of phosphoproteins to the Western blot signal is minimal, because they represent less than 10% of the total protein.
Kinase assays.
For the enzyme kinetic analysis, activated Raf-1 was prepared from Sf-9 cells coinfected with GST–Raf-1 and Ras plus Lck in Sf-9 cells, as previously described (7). Kinase reactions were carried out in 30 μl of Raf kinase buffer (7) supplemented with 10 μM ATP and 2.5 μCi of [γ-32P]ATP using GST–MEK-1 as substrate. Reaction mixtures were incubated 20 min at 25°C and resolved on SDS–10% acrylamide gels; MEK-1 phosphorylation was then quantitated using a Fuji phosphorimager. MEK-1 autophosphorylation was subtracted, and the data were analyzed with the SigmaPlot software. The kinase activity of Raf-1 immunoprecipitates (see Fig. 4a) was measured as described previously (1). The phosphorylation of negative His/MEK-1 was detected (see Fig. 4b) with a phosphospecific MEK antiserum (New England Biolabs).
Reporter gene assays.
AP-1 luciferase assays and microinjection experiments with affinity-purified RKIP antiserum and TRE-lacZ reporter plasmids were carried out as previously described (26).
RESULTS
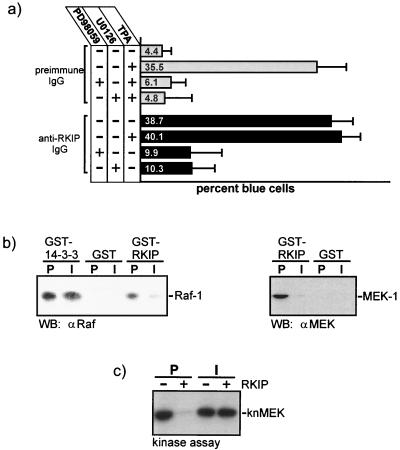
The microinjection of anti-RKIP antibodies raised against the full-length RKIP protein efficiently activated an AP-1-dependent reporter gene. This induction was due to the activation of MEK, since it could be suppressed by two structurally different MEK inhibitors, U0126 and PD98059 (Fig. 1a). This showed that the expression of the reporter gene is controlled by the ERK pathway and supports our previous conclusion that RKIP inhibits this pathway by downregulating the activation of MEK by Raf-1 (26). The induction of the reporter gene could be completely prevented by coinjection of an RKIP expression vector (26), indicating that the RKIP antibodies specifically neutralized RKIP function. These antibodies are therefore useful tools for investigating the molecular mechanism by which RKIP works. The RKIP antiserum interfered with the binding of Raf-1 and MEK to RKIP (Fig. 1b). This effect was specific, as (i) the corresponding preimmune serum had no effect and (ii) the RKIP antibodies did not prevent the binding of Raf-1 to 14-3-3. Furthermore, the RKIP antibodies reversed the inhibitory effect of RKIP on MEK phosphorylation by Raf-1 (Fig. 1c). These results indicated that the inhibitory effect of RKIP on MEK activation by Raf-1 depends on RKIP binding to Raf-1 and/or to MEK.
FIG. 1.
RKIP inhibits the ERK pathway by preventing MEK activation. (a) Rat-1 cells were microinjected with a TRE-LacZ reporter plasmid and affinity-purified RKIP antibodies or preimmune immunoglobulin G (IgG) and treated as indicated. The MEK inhibitors PD98059 and U0126 were administered 1 h before microinjection of TPA (100 ng/ml). (b) RKIP antibodies prevent binding of RKIP to Raf-1 or MEK. GST, GST-RKIP, or GST–14-3-3 beads were incubated with saturating amounts of RKIP antibodies (I) or the corresponding preimmune serum (P) and tested for binding of Raf-1 or MEK-1. WB, Western blot. (c) The phosphorylation of kinase-negative MEK-1 (knMEK) by activated Raf-1 was examined in the presence (+) or absence (−) of 10 μM purified RKIP. RKIP was preincubated with RKIP antibodies or the corresponding preimmune serum for 1 h.
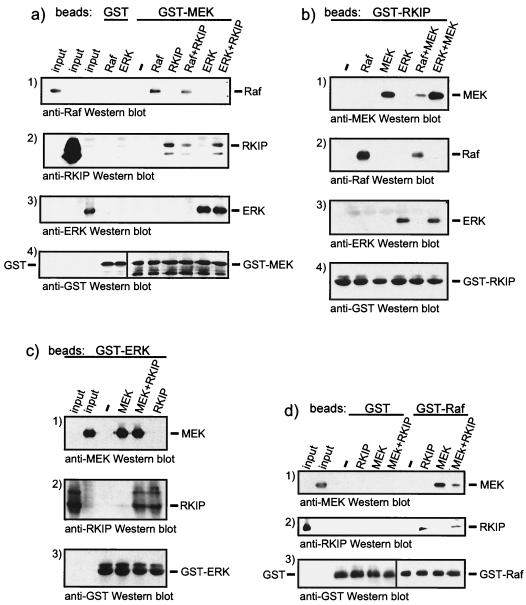
Therefore, we analyzed the role of RKIP in the formation of ternary protein complexes with Raf, MEK, and ERK in more detail (Fig. 2). Immobilized GST-MEK could bind Raf-1, ERK, and RKIP (Fig. 2a). However, while GST-MEK could bind both ERK and RKIP simultaneously (Fig. 2a, panels 2 and 3), Raf-1 and RKIP seemed to compete for binding (Fig. 2a, panels 1 and 2). Consistent results were obtained when GST-RKIP, GST-ERK, or GST–Raf-1 beads were used for binding assays. RKIP decreased the binding of Raf-1 to GST-MEK beads (Fig. 2a, panel 1) and the binding of MEK to GST–Raf-1 beads (Fig. 2d, panel 1). In both cases, RKIP competed for binding. In contrast, RKIP did not interfere with the association of ERK with GST-MEK beads (Fig. 2a, panel 3) or of MEK with GST-ERK beads (Fig. 2c, panel 1). When GST-RKIP beads were used as bait, Raf and MEK mutually diminished their binding to GST-RKIP (Fig. 2b, panels 1 and 2), whereas MEK and ERK mixed together bound with an efficiency similar to that of each individual protein alone (Fig. 2b, panels 1 and 3). In summary, these experiments demonstrated that MEK and ERK can bind to RKIP at the same time but the binding of Raf-1 to RKIP and that of MEK are mutually exclusive. Further, these data suggest that the binding of Raf-1 or MEK to RKIP may compete with their binding to each other and thus interfere with the formation of Raf-1–MEK complexes.
FIG. 2.
Analysis of the composition of RKIP protein complexes. (a) GST-MEK beads were incubated with RKIP, Raf, and MEK in the indicated combinations. GST-RKIP beads (b), GST-ERK beads (c), or GST-Raf-1 beads (d) were incubated with recombinant purified proteins as indicated. Incubations were done as described in Materials and Methods, and associated proteins were visualized by Western blotting.
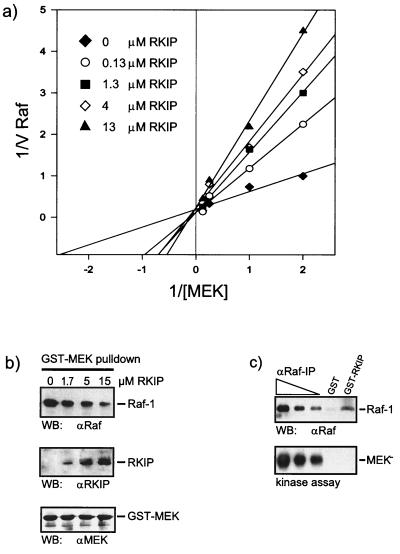
This possibility was tested. The analysis of the kinetics of MEK phosphorylation by Raf-1 revealed that RKIP diminished the Km but not the Vmax of the reaction, indicating a competitive type of inhibition (Fig. 3a). Control proteins, such as GST and 14-3-3, had no effect, and since RKIP is not a Raf-1 substrate, it did not compete for phosphorylation (data not shown). We have previously shown that the association of Raf-1 and MEK is required for efficient MEK phosphorylation and activation (12). Therefore, we tested whether RKIP could disturb the physical interaction between Raf-1 and MEK. For this purpose we coexpressed Raf-1 and GST–MEK-1 in Sf-9 insect cells and purified the GST–MEK-1–Raf-1 complex by adsorption to glutathione Sepharose beads. The Raf-1–GST–MEK complex was incubated with increasing amounts of purified RKIP. After a washing, the composition of the complex was examined by Western blotting (Fig. 3b). The addition of RKIP resulted in RKIP binding and a concomitant displacement of Raf-1 from the GST-MEK beads, thus confirming a competitive mode of inhibition. These data suggest that at least two populations of Raf-1 can be distinguished, one that is associated with MEK and competent for MEK phosphorylation and another that is bound to RKIP and disabled for MEK phosphorylation. To test this prediction, Raf-1 was produced in Sf-9 insect cells and recovered either by affinity adsorption to GST-RKIP beads or by immunoprecipitation with Raf antibodies from serial dilutions of the same lysate (Fig. 3c). When assayed for MEK phosphorylation, the kinase activity of RKIP-associated Raf-1 was severely impaired compared to an equivalent amount of immunoprecipitated Raf-1. These results confirmed the hypothesis that Raf-1 bound to RKIP is inactive as MEK kinase.
FIG. 3.
RKIP inhibits Raf-1 by a competitive mechanism. (a) Lineweaver-Burk plot of Raf-1 inhibition by RKIP. Activated GST–Raf-1 was used to phosphorylate GST–MEK-1 in the presence of increasing amounts of RKIP, as indicated. Phosphorylation was quantified with a Fuji phosphorimager. The data shown are the averages of three independent experiments. (b) RKIP disrupts the Raf-1–MEK complex. GST-MEK and Raf-1 were coexpressed in Sf-9 cells. The GST-MEK–Raf-1 complex was purified by adsorption to glutathione Sepharose beads, washed, and resuspended in PBS. Purified RKIP was added at the concentrations indicated. After 1 h at 4°C, the GST-MEK beads were washed three times with PBS and examined for associated proteins by Western blotting (WB) with the indicated antisera. (c) Raf-1 bound to RKIP does not phosphorylate MEK. A lysate of Sf-9 cells expressing activated Raf-1 was incubated with 5 μg of GST or GST-RKIP beads. Serial dilutions of the same lysate were immunoprecipitated with the anti-Raf serum crafVI. After three washes with PBS, the pellets were resuspended in kinase buffer and incubated with 100 μM ATP and kinase-negative MEK as substrate. MEK phosphorylation was visualized by immunoblotting with a phospho-MEK-specific antiserum. Raf-1 was stained with crafVI.
These results also suggested that only the fraction of Raf-1 which is not bound to RKIP is available for activation. Therefore, we examined whether Raf-1 dissociates from RKIP during activation. For this purpose, RKIP and Raf-1 were coexpressed in COS-1 cells (Fig. 4a). Raf-1 coprecipitated with RKIP in quiescent cells. Stimulation of the cells with tetradecanoyl phorbol acetate (TPA) plus epidermal growth factor caused an increase in Raf-1 kinase activity which correlated with a decrease of RKIP association. At later time points, as Raf-1 catalytic activity declined, the levels of Raf-1 coprecipitating with RKIP increased again. To investigate whether the changes in RKIP association are related to the activation status of Raf-1, the binding of purified RKIP to inactive and activated GST–Raf-1 beads was determined (Fig. 4b). Activated GST–Raf-1 was produced in Sf-9 insect cells coinfected with RasV12 and Lck, which results in a robust activation of the catalytic activity. GST–Raf-1 proteins were purified by adsorption to glutathione Sepharose beads and incubated with recombinant RKIP produced in E. coli. Less RKIP bound to activated GST–Raf-1, indicating that Raf-1 activation weakens the affinity towards RKIP. This finding, however, did not seem to depend on the kinase activity of Raf-1 per se. Kinase-negative Raf-1 mutants, such as RafK375W (11) or RafS621A (17), as well as activated Raf-1 mutants, such as RafS259D (17) or the isolated kinase domain BXB, bound to RKIP at levels comparable to that of the wild-type Raf-1 (reference 26 and data not shown). We also tested whether activation affected the binding of MEK and ERK to RKIP. Purified MEK and ERK were phosphorylated in vitro with recombinant Raf-1 or Raf-1 plus MEK, respectively, and incubated with GST or GST-RKIP beads. The binding reaction products were washed, separated on SDS gels, and immunoblotted with the appropriate antisera. We did not observe any differences in binding between activated and nonactivated forms (data not shown). However, since only small fractions of MEK and ERK become phosphorylated (1), we also carried out the phosphorylation in the presence of [γ-32P]ATP in order to avoid misinterpretation due to low phosphorylation efficiencies (Fig. 4c and d). The blots were autoradiographed to detect phosphorylated MEK and ERK and were subsequently stained with the cognate antisera to visualize total protein bound. Under these conditions, binding of phosphorylated MEK and ERK to RKIP was evident.
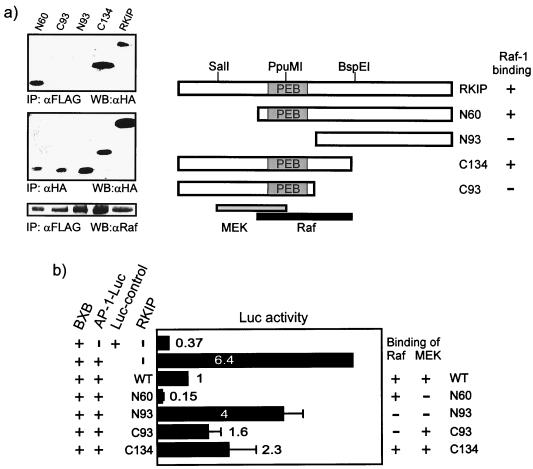
These data were consistent with Raf-1 being the main regulatory target of RKIP. To further examine the molecular basis for the observed competitive mode of RKIP inhibition, we mapped the domains in the Raf-1 kinase domain, BXB, which are necessary for RKIP and MEK binding (Fig. 5a). BXB deletion mutants were expressed as GST fusion proteins in E. coli and were examined for binding to purified RKIP or MEK in vitro. Surprisingly, the required binding domains were different. Raf-1 kinase subdomains VIb to VIII were essential for MEK binding, whereas RKIP bound to subdomains I and II. The latter region contains the ATP binding site, but RKIP did not compete for ATP (data not shown). Likewise, RKIP and Raf-1 bound to different domains in MEK-1 (Fig. 5b). As previously reported (2), Raf-1 bound to MEK-1 constructs containing the proline-rich region, whereas RKIP bound to the N-terminus of MEK-1. Thus, RKIP's ability to dissociate Raf-MEK complexes does not seem to involve a direct competition for the same binding sites. Rather, it must be due to an allosteric reduction of the binding affinity induced by RKIP or to mutual steric hindrance that excludes simultaneous binding of RKIP and Raf to MEK or of RKIP and MEK to Raf-1, respectively. When we mapped the binding sites of Raf-1 and MEK-1 to RKIP (Fig. 5c), the RKIP domain required for MEK binding could be clearly located, while Raf-1 interacted with multiple domains in RKIP. Notably, removal of the RKIP carboxy terminus up to the BspEI site enhanced Raf-1 association, whereas further deletion up to the PpuMI site decreased Raf-1 binding again. These data suggest that the interaction between Raf-1 and RKIP is complex, involving a main site of binding to amino acids 77 to 108 in the BspEI-PpuMI fragment, as well as minor contacts with several other domains. The partial overlap between the MEK and Raf-1 binding sites, however, is consistent with the observation that RKIP cannot bind Raf-1 and MEK simultaneously (Fig. 2).
In summary, all these data suggested that RKIP mutants that are defective for Raf-1 binding should also be compromised as inhibitors of the ERK pathway. To examine this possibility, we generated RKIP deletion mutants suitable for expression in mammalian cells. The analysis of Raf-1 binding to the RKIP deletion mutants in mammalian cells was consistent with the in vitro mapping of the main Raf-1 binding site to amino acids 77 to 108 (Fig. 6a). The N93 and the C93 RKIP mutants, which both disrupt this domain, failed to coimmunoprecipitate with Raf-1. However, C93 RKIP still contains the MEK binding domain. When tested for suppression of Raf-mediated AP-1 induction, only N93 RKIP showed a clear decrease in inhibitory activity (Fig. 6b). Since N93 RKIP is the only mutant that lacks both the Raf-1 and MEK interaction domains, we conclude that either Raf-1 or MEK binding is sufficient for suppression of the ERK pathway.
FIG. 6.
RKIP binding to Raf-1 or MEK is sufficient for inhibition. (a) Coimmunoprecipitation of RKIP deletion mutants with Raf-1. FLAG–Raf-1 and hemagglutinin (HA)-RKIP or HA-RKIP deletion mutants were coexpressed in COS cells. Lysates were immunoprecipitated (IP) with anti-FLAG antibodies, and associated HA-RKIP proteins were detected by Western blotting (WB) with anti-HA antibodies. PEB, phosphatidylethanolamine binding motif. (b) The effect of RKIP deletion mutants on Raf-induced AP-1 reporter gene expression. HA-RKIP mutants were cotransfected with the Raf-1 kinase domain, BXB, and an AP-1–luciferase plasmid.
DISCUSSION
In a previous study (26), we established that RKIP is a physiologically relevant inhibitor of the Raf-1/MEK/ERK pathway. Overexpression of RKIP suppressed signaling through this pathway, whereas downregulation of RKIP enhanced it. RKIP did not inhibit Raf-1 catalytic activity but specifically interfered with the phosphorylation of MEK by Raf-1. Since MEK was not inhibited and activated MEK mutants could rescue ERK activation, we concluded that RKIP blocks the pathway at the Raf-1/MEK interface. Here we describe the molecular mechanism of RKIP's inhibitory function.
According to enzyme kinetic analysis, RKIP acted like a competitive inhibitor of MEK phosphorylation. Since we have previously shown that MEK phosphorylation requires physical interaction with Raf-1 (12), this mode of inhibition can be explained by RKIP's ability to dissociate Raf-1–MEK complexes. This interpretation is supported by the observation that RKIP is a monomer and that artificial oligomerization converts it into an activator, presumably by cross-linking Raf-1 with its substrate MEK (data not shown). Surprisingly, mapping the domains in Raf-1 which are necessary for the binding of RKIP and MEK revealed different sites. The same was true for the binding sites of RKIP and Raf-1 in MEK. Thus, rather than acting as a direct competitor for binding, RKIP must reduce the affinity of Raf-1 and MEK for each other, possibly by inducing a conformational change. A potential precedent for such a mechanism is exemplified by an antibody raised against an amino-terminal peptide of Raf-1. This antibody, whose epitope is approximately 450 amino acids away from the MEK binding site, dramatically reduced the association of Raf-1 with MEK (9). An alternative but not exclusive possibility is that RKIP binding to either Raf or MEK creates a steric obstacle for the association of the other partner. The binding sites of RKIP and MEK in Raf-1 and of RKIP and Raf-1 in MEK, respectively, are both located in the kinase domain. Although the crystal structure of neither Raf-1 nor MEK is known, RKIP binds to a region in Raf-1 and MEK that by comparison with other kinases (22) is expected to be part of the small lobe. The small lobe is in close proximity to the substrate binding domain in the large lobe, where Raf-1 interacts with MEK. Thus, a sterical interference of RKIP with Raf-MEK binding seems conceivable. Either hypothesis is compatible with our observations that although Raf-1 bound to RKIP is disabled as MEK kinase, the presence of either the Raf or the MEK binding domain in RKIP is sufficient for full repression.
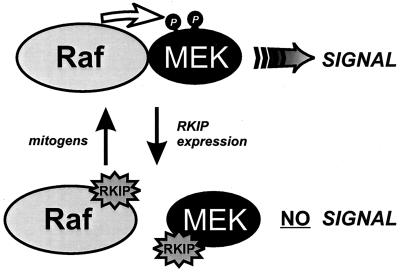
In this context, it is important to note that (as shown by a detailed analysis of ternary RKIP complexes) RKIP can bind either MEK or Raf-1 but not both simultaneously. This can be explained by the partial overlap of their binding domains and predicts the existence of at least two different pools of Raf-1 and MEK, one which is bound to RKIP and one which is not. Only the latter pool is available for transducing signals through the Raf/MEK/ERK cascade (Fig. 7). The size of this pool appears to be determined by the expression levels of RKIP. Since coimmunoprecipitation, pulldown, and immunodepletion experiments apprehend only the steady-state levels of RKIP complexes, it is very difficult to estimate the true size of this pool in the cell under any condition. Our observation that MEK phosphorylation does not compromise its ability to associate with RKIP in vitro suggests that MEK sequestration by RKIP is not limiting. However, since mitogens decrease the association of RKIP with Raf-1, RKIP–Raf-1 complexes seem to provide the main interface for regulation. We have previously shown that RKIP association with Raf-1 decreases concomitant with activation of the ERK pathway during mitogenic stimulation and increases again when ERK activity declines (26). Here we demonstrate that the changes in RKIP association show an inverse correlation with Raf-1 activation (Fig. 4a and b). But Raf-1 catalytic activity per se does not seem to play a major role, since RKIP bound inactive Raf-1 and activated Raf-1 mutants with comparable affinities (data not shown). Therefore, it is rather a mitogen-induced modification of Raf-1 or RKIP, or both, that is responsible for this effect. The nature and role of this modification are currently under investigation. The discovery of RKIP and its inhibitory mechanism adds the selective and regulated disruption of signaling complexes as a new concept to how the cell controls its intricate signaling circuitry.
FIG. 7.
Model of RKIP function. The activation of MEK by Raf-1 requires physical interaction between Raf-1 and MEK. RKIP binding to either Raf-1 or MEK dissociates Raf-MEK complexes and thereby interrupts MEK activation and downstream signaling. The binding of RKIP to Raf-1 is negatively regulated by mitogens.
ACKNOWLEDGMENTS
We appreciate the critical reading and helpful comments by members of the Beatson laboratories.
This work was supported by the CRC, by grants from the AICR, and by NIH grant R01 GM55435 to J.M.S.
W.K. and J.M.S. contributed equally to the work.
REFERENCES
- 1.Alessi D R, Cohen P, Ashworth A, Cowley S, Leevers S J, Marshall C J. Assay and expression of mitogen-activated protein kinase, MAP kinase kinase, and Raf. Methods Enzymol. 1995;255:279–290. doi: 10.1016/s0076-6879(95)55031-3. [DOI] [PubMed] [Google Scholar]
- 2.Catling A D, Schaeffer H-J, Reuter C W M, Reddy G R, Weber M J. A proline-rich sequence unique to MEK1 and MEK2 is required for Raf binding and regulates MEK function. Mol Cell Biol. 1995;15:5214–5225. doi: 10.1128/mcb.15.10.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denouel-Galy A, Douville E M, Warne P H, Papin C, Laugier D, Calothy G, Downward J, Eychene A. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol. 1998;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- 4.Downward J. KSR: a novel player in the RAS pathway. Cell. 1995;83:831–834. doi: 10.1016/0092-8674(95)90198-1. [DOI] [PubMed] [Google Scholar]
- 5.Elion E A. Routing MAP kinase cascades. Science. 1998;281:1625–1626. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]
- 6.Ferrell J E., Jr MAP kinases in mitogenesis and development. Curr Top Dev Biol. 1996;33:1–60. doi: 10.1016/s0070-2153(08)60336-1. [DOI] [PubMed] [Google Scholar]
- 7.Häfner S, Adler H S, Mischak H, Janosch P, Heidecker G, Wolfman A, Pippig S, Lohse M, Ueffing M, Kolch W. Mechanism of inhibition of Raf-1 by protein kinase A. Mol Cell Biol. 1994;14:6696–6703. doi: 10.1128/mcb.14.10.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanks S K, Quinn A M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structures and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 9.Huang W, Alessandrini A, Crews C M, Erikson R L. Raf-1 forms a stable complex with Mek1 and activates Mek1 by serine phosphorylation. Proc Natl Acad Sci USA. 1993;90:10947–10951. doi: 10.1073/pnas.90.23.10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joneson T, Fulton J A, Volle D J, Chaika O V, Bar-Sagi D, Lewis R E. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J Biol Chem. 1998;273:7743–7748. doi: 10.1074/jbc.273.13.7743. [DOI] [PubMed] [Google Scholar]
- 11.Kolch W, Heidecker G, Lloyd P, Rapp U R. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 12.Kolch W, Philipp A, Mischak H, Dutil E M, Mullen T M, Feramisco J R, Meinkoth J L, Rose D W. Inhibition of Raf-1 signaling by a monoclonal antibody, which interferes with Raf-1 activation and with Mek substrate binding. Oncogene. 1996;13:1305–1314. [PubMed] [Google Scholar]
- 13.Marais R, Marshall C J. Control of the ERK MAP kinase cascade by Ras and Raf. Cancer Surv. 1996;27:101–125. [PubMed] [Google Scholar]
- 14.Michaud N R, Therrien M, Cacace A, Edsall L C, Spiegel S, Rubin G M, Morrison D K. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moodie S A, Willumsen B M, Weber M J, Wolfman A. Complexes of Ras/GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 16.Morrison D K, Cutler R E. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 17.Morrison D K, Heidecker G, Rapp U R, Copeland T D. Identification of the major phosphorylation sites of the Raf-1 kinase. J Biol Chem. 1993;268:17309–17316. [PubMed] [Google Scholar]
- 18.Mueller G, Storz P, Bourteele S, Doeppler H, Pfizenmaier K, Mischak H, Philipp A, Kaiser C, Kolch W. Regulation of Raf-1 kinase by TNF via its second messenger ceramide and cross-talk with mitogenic signalling. EMBO J. 1998;17:732–742. doi: 10.1093/emboj/17.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 20.Rose D W, McCabe G, Feramisco J R, Adler M. Expression of c-fos and AP-1 activity in senescent human fibroblasts is not sufficient for DNA synthesis. J Cell Biol. 1992;119:1405–1411. doi: 10.1083/jcb.119.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaeffer H J, Catling A D, Eblen S T, Collier L S, Krauss A, Weber M J. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 22.Taylor S S, Radzio Andzelm E. Three protein kinase structures define a common motif. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 23.Therrien M, Michaud N R, Rubin G M, Morrison D K. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 24.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 25.Xing H, Kornfeld K, Muslin A J. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 26.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis K D, Rose D W, Mischak H, Sedivy J M, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 27.Yu W, Fantl W J, Harrowe G, Williams L T. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1998;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]