Abstract
The treatment of immunotherapy relapsed cutaneous melanoma constitutes a challenge in both research and clinical practice fields given the lack of effective therapeutic options. Homologous recombination deficiency (HRD) has been identified in several solid cancers including cutaneous melanoma. However, the utility of medications targeting HRD cancer cells is an uncharted territory in melanoma. Moreover, preclinical evidence suggests a synergistic role of combining immune checkpoint blockade (ICB) with drugs targeting HRD cancer cells such as PARP inhibitors. Here, we present a case study of a patient with immunotherapy relapsed melanoma who was found to have detected HRD and was treated with nivolumab (ICB) and olaparib (PARP inhibitors).
Keywords: neoplasm of the skin
INTRODUCTION
Immune checkpoint blockade (ICB) has revolutionized the management of malignant cutaneous melanoma leading to significantly improved clinical outcomes. The long-term follow-up studies in patients with metastatic melanoma treated with ICB have demonstrated prolonged and durable responses. For example, the 5-yr survival data with combined nivolumab (PD-1 inhibitor) and ipilimumab (CTLA-4 inhibitor) in the CheckMate-067 study has shown a median overall survival (OS) of >60 mo in patients with metastatic melanoma (Larkin et al. 2019). Similarly, the 5-yr follow-up in the KEYNOTE-001 trial of patients with metastatic melanoma who were treated with pembrolizumab (PD-1 inhibitor) showed a median OS of 38.6 mo in treatment-naive patients (Hamid et al. 2019). Despite these remarkable results, a significant number of patients will eventually stop responding to ICB and develop progressive disease. This has generated an unmet need for novel effective treatment options for patients who develop acquired resistance to immunotherapy. Moreover, primary resistance to immunotherapy is another challenge that hinders the efficacy of ICB in melanoma patients.
Homologous recombination deficiency (HRD) refers to the intracellular state in which cancer cells acquire defects in DNA-damage repair mechanisms. Mutations in the DNA-damage response (DDR) genes as well as other factors like epigenetic alterations could lead to vulnerabilities in DNA repair mechanisms, which lead to HRD status in tumor cells (Pilié et al. 2019). The poly(ADP-ribose) polymerase (PARP) inhibitors are a class of medication that target nuclear proteins involved in the repair of DNA single-stranded and double-stranded breakage. These medications have demonstrated clinical efficacy in ovarian, breast, and pancreatic cancers with defects in the homologous recombination pathway due to BRCA1/2 mutations (Pilié et al. 2019).
In regards to melanoma, Heeke et al. investigated HRD frequency by next-generation sequencing (NGS) in multiple tumors and found that mutations in the DDR genes are commonly prevalent in melanoma (18.1%, n = 670) (Heeke et al. 2018). Similarly, analysis of The Cancer Genome Atlas (TCGA) found a high number of mutations in different DDR pathways in melanoma including mutations in the homologous-recombination, nucleotide-excision-repair, and mismatch-repair genes (Knijnenburg et al. 2018). In addition, there is mounting preclinical evidence of a synergistic role with combination ICB and PARP inhibitors to improve antitumor immunity (Li et al. 2019).
This raises the question of whether combination ICB and PARP inhibitors have efficacy in melanoma with HRD status after progression on immunotherapy.
We previously reported radiological response and mutation clearance after treatment with nivolumab and olaparib in a patient with immunotherapy relapsed melanoma who had a high HRD score due to a germline mutation in CHEK2 and several somatic mutations in DDR genes (Khaddour et al. 2021). Here, we present our new observation in a 42-yr-old male with metastatic melanoma who had evidence of borderline high HRD score and was treated with nivolumab and olaparib after progression on prior ICB (nivolumab). Unlike the previous report, our current patient had a high HRD score despite the absence of mutations in DDR genes and had near-complete response. Liquid biopsy demonstrated mutations cleared after treatment with nivolumab and olaparib.
RESULTS
Clinical Presentation
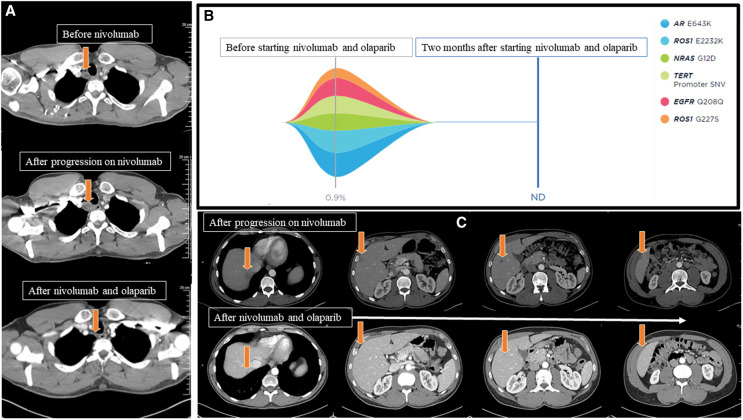
A 42-yr-old male was diagnosed with cutaneous melanoma in February 2017 (cT3aN2CM0: Stage IIIC), which was treated with wide surgical excision followed by adjuvant therapy with ipilimumab. He had grade IV colitis after the second cycle of ipilimumab, which required prolonged hospitalization and permanent discontinuation of cancer treatment. He continued surveillance without any evidence of recurrence of melanoma until September 2020 when computed tomography (CT) showed a 1.3-cm right paratracheal lymph node without presence of any other lesions (Fig. 1A). Biopsy was performed and pathology showed metastatic melanoma. NGS revealed BRAF-wild type. The patient started nivolumab (480 mg IV, every 4 wk). Repeat imaging after two cycles showed increase in the size of the paratracheal node, a new lung lesion, and multiple new hypodense hepatic lesions consistent with disease progression (Fig. 1A,C). HRD testing on the sequencing data from a Tempus xT test showed genome-wide loss of heterozygosity (LOH) at 32.9%. Based on this LOH score, the patient started off-label combination nivolumab (480 mg IV, every 4 wk) and olaparib (300 mg PO, twice daily). After 2 mo, CT demonstrated interval response and decrease in size of the paratracheal node, resolution of a left lower lobe pulmonary nodule, and near-complete/complete resolution of multiple hepatic hypoattenuating lesions (Fig. 1A,C). Longitudinal circulating tumor DNA (ctDNA) assessment revealed clearance of all previously detected mutations (Fig. 1B).
Figure 1.
(A) Cross-sectional computed tomography of the chest: The upper image demonstrates right paratracheal lesion without compressive features prior to starting nivolumab monotherapy, the middle image demonstrates an increase in the size of the paratracheal lesion after two cycles of nivolumab, and the lower image demonstrates near-complete resolution of the right paratracheal lymph node after 2 mo of combined treatment with nivolumab and olaparib (arrows). (B) Tumor response map using Guardant 360 showing several somatic mutations detected on liquid biopsy (gray solid line) prior to starting treatment with nivolumab and olaparib. All detected somatic mutations were cleared 2 mo after initiating nivolumab and olaparib (blue line). (C) Cross-sectional computed tomography of the abdomen demonstrating (upper images) new hepatic lesions prior to starting nivolumab and olaparib, which were not present before starting nivolumab monotherapy. The lower images show near-complete and complete resolution of multiple hepatic lesions after 2 mo of combined treatment with nivolumab and olaparib (arrows). (ND) Not detected, (SNV) single-nucleotide variant.
The patient developed grade III hepatitis before his third cycle of treatment, which required discontinuation of cancer therapy and treatment with steroids with complete resolution of the immune therapy–related adverse event. Repeat CT after 2 mo of treatment discontinuation demonstrated no lesions in the liver and stable disease in the lungs.
Genomic Analysis
An NGS panel of 596 genes identified mutations in TERT (c.-124C > T, VAF 73.1%) , PIK3CA (p.H1047R, VAF 24.1%), NRAS (p.G12D, VAF 55.5%), TET2 (p.Q1445*, VAF 37.5%), SETD2 (p.K916fs, VAF 17.8%), and CDKN2A copy-number loss, in addition to microsatellite stable (MSS) and tumor mutation burden (TMB) of 27.4 m/MB (Tempus xT). Also, testing of HRD (Tempus xT) revealed genome-wide LOH of 32.9% nominally below the 33% threshold required for the HRD status to be detected in cancer types other than breast, ovarian, and pancreatic (defined by commercial vendor). Liquid biopsy analysis of ctDNA in December 2020 demonstrated pathogenic mutations and variants of unknown significance (VUSs) (Table 1). These mutations included NRAS (G12D) (VAF 0.6%), TERT (promoter SNV) (VAF 0.6%), and detected additional somatic mutations in AR (E643K) (VAF 0.9%), ROS1 (E2232K VAF 0.8% and G227S VAF 0.3%), and EGFR (Q208Q) (VAF 0.6%) as shown in Fig. 1B (Guardant360). The follow-up liquid biopsy using Guardant360 showed clearance of detected somatic mutations and VUSs after 1 and 2 mo of treatment with nivolumab and olaparib. Of note, Guardant360 panel does not capture mutations in TET2 and SETD2.
Table 1.
Mutations and variants identified by circulating free tumor DNA sequencing of melanoma
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP/dbVar ID | Genotype |
|---|---|---|---|---|---|---|---|
| NRAS | Chr 1:115258747 | c.35G > A | G12D | Pathogenic | Substitution | rs121913237 | Heterozygous |
| TERT | Chr 5:1295228 | c.124C > T | Promoter-124C > T | Pathogenic | Promoter/gene regulation | rs1242535815 | Heterozygous |
| ROS1 | Chr 6:117622176 | c.6694G > A | E2232K | VUS | Substitution | rs1275344272 | Heterozygous |
| ROS1 | Chr 6:117718178 | c.679G > A | G227S | VUS | Substitution | Not available | Heterozygous |
| AR | Chr X:66931285 | c.1927G > A | E643K | VUS | Substitution | rs2076093229 | Heterozygous |
(VUS) Variant of unknown significance.
DISCUSSION: COMBINATION CHECKPOINT INHIBITORS AND PARP INHIBITORS IN THE TREATMENT OF MELANOMA WITH DETECTED HRD
Response to olaparib (PARP inhibitor) in melanoma patients has been previously reported when used as monotherapy and in combination with ICB (Khaddour et al. 2021; Lau et al. 2021). The previously reported patients had high LOH scores of 28.4% and 26.5%. Interestingly in the current report, there were no detected mutations in DDR genes unlike the previous reports that identified somatic mutations in DDR genes such as PALB2, BRCA2, ATRX, and germline mutation in CHEK2. Notably, mechanisms other than DDR gene mutations, such as epigenetic alterations like DNA hypermethylation, could contribute to HRDness in cancer cells (Moschetta et al. 2016; Sahnane et al. 2020). Therefore, our observation is intriguing as it highlights the potential detection of HRD status without the presence of genetic alterations in DDR genes as well as the need for establishing an LOH threshold specific to melanoma. Olaparib in combination with bevacizumab is currently approved for HRD-positive advanced ovarian cancer based on significant improvement in progression-free survival (PFS; 37.2 mo vs. 17.7 mo with bevacizumab alone) (Ray-Coquard et al. 2019).
PARP inhibitors can enhance sensitivity in the tumor microenvironment to ICB through a variety of mechanisms including increased chromosomal aberrations due to deficiency in DNA repair pathways, and modulation of immune response in the tumor microenvironment (Teo et al. 2018; Chabanon et al. 2019). In addition, the presence of HRD in cancer cells as well as the use of PARP inhibitor–induced synthetic lethality could lead to neoantigen formation that may resensitize tumor cells to ICB (Kakoti et al. 2020).
Finally, a recent study replicated previous findings of the common prevalence of HR-DDR gene mutations in melanoma and fond a frequency of 21.4% (18 of 84 patient samples). This study also showed that PARP inhibitors could have efficacy in melanoma cell lines harboring mutations in the HR-DDR genes, which suggests a possible role for PARP inhibitors monotherapy in cutaneous melanoma (Kim et al. 2021). However, the role of PARP inhibitors in metastatic melanoma with HRD status requires further validation both in vitro and in vivo and in well-designed prospective clinical trials with larger cohorts. Several questions remain to be answered as to whether the addition of ICB to PARP inhibitors has any added benefit to monotherapy with PARP inhibitors. Another challenge is the absence of a unified test of HRDness in tumor specimens and the lack of insight on a reliable cutoff point that could potentially help select patients who could benefit from therapy targeting the homologous recombination repair (HRR) pathway. Finally, we acknowledge the limitation in our observation given that it is a single case study, the absence of a comparative arm, and the possibility that the observed response could have been a delayed effect of ICB. In addition, it is important to mention the pseudoprogression phenomenon that results from tumor infiltration with immune cells after treatment with ICB and could be misinterpreted on imaging as true progression (Nishino et al. 2019). Our patient developed multiple new scattered hepatic lesions after starting nivolumab, which makes pseudoprogression unlikely but cannot be completely excluded per iRECIST criteria (Seymour et al. 2017).
In conclusion, we describe a case of a patient with immunotherapy relapsed melanoma with detected HRD in the absence of mutations in DDR genes who was treated with nivolumab and olaparib. The patient had radiological response and mutation clearance by NGS 2 mo after initiation of therapy. Before the third cycle, the patient developed grade III hepatitis during treatment, which required discontinuation of therapy. This observation suggests the need for further investigation to assess the safety and efficacy of ICB and PARP inhibitor combination in immunotherapy relapsed melanoma with detected HRD. Currently, several clinical trials are ongoing to evaluate the efficacy of PARP inhibitors and ICB in HRD melanoma (NCT04633902 and NCT04187833).
METHODS
The patient's tumor tissue was analyzed with a Tempus xT gene panel using NGS for mutation testing (Beaubier et al. 2019). Longitudinal analysis of ctDNA was performed on the patient's peripheral blood using Guardant360 CDx before starting nivolumab and olaparib and at 2 mo (Lanman et al. 2015).
ADDITIONAL INFORMATION
Data Deposition and Access
Sequencing data from the TEMPUS xT gene panel and the ctDNA assay from the Guardant360 CDx assay (Guardant Health, Inc.) were not made available for public distribution. The variants were submitted to ClinVar (https://www.ncbi.nlm.nif.gov/clinvar/) and can be found under accession numbers SCV000503722.1 (for NRAS), SCV001976515 (for TERT), SCV001976502 and SCV001976503 (for ROS1), and SCV001976504 (for AR).
Ethics Statement
The corresponding author obtained informed consent to publish information and/or images from the patient.
Acknowledgments
The authors thank Greg Call for reviewing the paper.
Author Contributions
K.K. and G.A. acquired clinical data and images, performed the literature search, and wrote the report. K.K., G.A., and M.A. reviewed the report and approved the final version.
Competing Interest Statement
The authors have declared no competing interest.
Referees
James Hicks
Elin Gray
REFERENCES
- Beaubier N, Tell R, Lau D, Parsons JR, Bush S, Perera J, Sorrells S, Baker T, Chang A, Michuda J, et al. 2019. Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget 10: 2384–2396. 10.18632/oncotarget.26797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabanon RM, Muirhead G, Krastev DB, Adam J, Morel D, Garrido M, Lamb A, Hénon C, Dorvault N, Rouanne M, et al. 2019. PARP inhibition enhances tumor cell–intrinsic immunity in ERCC1-deficient non–small cell lung cancer. J Clin Invest 129: 1211–1228. 10.1172/JCI123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph R, Weber JS, et al. 2019. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann Oncol 30: 582–588. 10.1093/annonc/mdz011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, Baker TM, Marshall JL, Isaacs C. 2018. Prevalence of homologous recombination–related gene mutations across multiple cancer types. JCO Precis Oncol 2018: PO.17.00286. 10.1200/PO.17.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakoti S, Sato H, Laskar S, Yasuhara T, Shibata A. 2020. DNA repair and signaling in immune-related cancer therapy. Front Mol Biosci 7: 205. 10.3389/fmolb.2020.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaddour K, Ansstas M, Visconti J, Ansstas G. 2021. Mutation clearance and complete radiologic resolution of immunotherapy relapsed metastatic melanoma after treatment with nivolumab and olaparib in a patient with homologous recombinant deficiency: any role for PARP inhibitors and checkpoint blockade? Ann Oncol 32: 279–280. 10.1016/j.annonc.2020.10.602 [DOI] [PubMed] [Google Scholar]
- Kim KB, Soroceanu L, de Semir D, Millis SZ, Ross J, Vosoughi E, Dar AA, Nosrati M, Desprez PY, Ice R, et al. 2021. Prevalence of homologous recombination pathway gene mutations in melanoma: rationale for a new targeted therapeutic approach. J Invest Dermatol 141: 2028–2036.e2. 10.1016/j.jid.2021.01.024 [DOI] [PubMed] [Google Scholar]
- Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, et al. 2018. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep 23: 239–254.e6. 10.1016/j.celrep.2018.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, Collisson EA, Divers SG, Hoon DS, Kopetz ES, et al. 2015. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 10: e0140712. 10.1371/journal.pone.0140712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, et al. 2019. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 381: 1535–1546. 10.1056/NEJMoa1910836 [DOI] [PubMed] [Google Scholar]
- Lau B, Menzies AM, Joshua AM. 2021. Ongoing partial response at 6 months to olaparib for metastatic melanoma with somatic PALB2 mutation after failure of immunotherapy: a case report. Ann Oncol 32: 280–282. 10.1016/j.annonc.2020.11.006 [DOI] [PubMed] [Google Scholar]
- Li A, Yi M, Qin S, Chu Q, Luo S, Wu K. 2019. Prospects for combining immune checkpoint blockade with PARP inhibition. J Hematol Oncol 12: 98. 10.1186/s13045-019-0784-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschetta M, George A, Kaye SB, Banerjee S. 2016. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann Oncol 27: 1449–1455. 10.1093/annonc/mdw142 [DOI] [PubMed] [Google Scholar]
- Nishino M, Hatabu H, Hodi FS. 2019. Imaging of cancer immunotherapy: current approaches and future directions. Radiology 290: 9–22. 10.1148/radiol.2018181349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilié PG, Tang C, Mills GB, Yap TA. 2019. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol 16: 81–104. 10.1038/s41571-018-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, Fujiwara K, Vergote I, Colombo N, Mäenpää J, et al. 2019. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 381: 2416–2428. 10.1056/NEJMoa1911361 [DOI] [PubMed] [Google Scholar]
- Sahnane N, Carnevali I, Formenti G, Casarin J, Facchi S, Bombelli R, Di Lauro E, Memoli D, Salvati A, Rizzo F, et al. 2020. BRCA methylation testing identifies a subset of ovarian carcinomas without germline variants that can benefit from PARP inhibitor. Int J Mol Sci 21: 9708. 10.3390/ijms21249708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour L, Bogaerts J, Perrone A, Ford R, Schwartz LH, Mandrekar S, Lin NU, Litière S, Dancey J, Chen A, et al. 2017. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol 18: e143–e152. 10.1016/S1470-2045(17)30074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, Cipolla CK, Bluth MJ, Chaim J, Al-Ahmadie H, et al. 2018. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol 36: 1685–1694. 10.1200/JCO.2017.75.7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data from the TEMPUS xT gene panel and the ctDNA assay from the Guardant360 CDx assay (Guardant Health, Inc.) were not made available for public distribution. The variants were submitted to ClinVar (https://www.ncbi.nlm.nif.gov/clinvar/) and can be found under accession numbers SCV000503722.1 (for NRAS), SCV001976515 (for TERT), SCV001976502 and SCV001976503 (for ROS1), and SCV001976504 (for AR).