Abstract
Chromosomal rearrangements of the NTRK genes generate kinase fusions that are targetable oncogenic drivers in diverse adult and pediatric malignancies. Despite robust clinical response to targeted NTRK inhibition, the emergence of therapeutic resistance poses a formidable clinical challenge. Here we report the characterization of an ETV6-NTRK3 fusion–driven pediatric glioma that progressed through NTRK-targeted treatments with entrectinib and selitrectinib. Genetic analysis of multifocal recurrent/resistant lesions identified a previously uncharacterized NTRK3 p.G623A and a known p.G623E resistance mutation, in addition to other alterations of potential pathogenic impact. Functional studies using heterologous reconstitution model systems and patient-derived tumor cell lines establish that NTRK3G623A and NTRK3G623E mutated kinases exhibit reduced sensitivity to entrectinib and selitrectinib, as well as other NTRK inhibitors tested herein. In summary, this genetic analysis of multifocal recurrent/resistant glioma driven by ETV6-NTRK3 fusion captured a cross section of resistance-associated alterations that, based on in vitro analysis, likely contributed to resistance to targeted therapy and disease progression.
Keywords: neoplasm of the central nervous system
INTRODUCTION
Gene fusion events involving the neurotrophin receptors TRKA, TRKB, and TRKC, encoded by NTRK1, NTRK2, and NTRK3, respectively, are oncogenic drivers in a diversity of adult and pediatric tumor types (Cocco et al. 2018). The neurotrophin receptor family plays a role in nervous system development, maturation, and maintenance through regulation of key intracellular signaling pathways (Huang and Reichardt 2003). Across organ systems, tumors driven by NTRK gene fusions exhibit clinically meaningful responses to targeted NTRK inhibition (Doebele et al. 2020; Hong et al. 2020), leading to approval of larotrectinib and entrectinib for the treatment of NTRK-rearranged cancers by the United States Food and Drug Administration (FDA). Primary central nervous system (CNS) tumors driven by NTRK fusions are increasingly recognized, with cases showing a remarkable range of histological patterns, clinical demographics, radiologic appearances, and co-occurring molecular alterations (Torre et al. 2020). Examples of primary CNS tumors responding to NTRK inhibition are reported, making this a promising molecular alteration for precision oncology strategies (Ziegler et al. 2018).
Despite robust clinical responses to targeted kinase inhibitors, the durability of therapeutic effect is frequently hampered by the emergence of acquired resistance. Elucidating the mechanisms of resistance to targeted kinase inhibitors (TKIs) in order to identify effective second-line therapeutics is thus an important area of investigations in the field. To date, reported mechanisms of clinical resistance to larotrectinib and entrectinib include kinase-intrinsic alterations that negatively affect inhibitor affinity for the kinase (Russo et al. 2016; Drilon et al. 2018a; Somwar et al. 2020) and bypass pathway alterations that compensate for loss of NTRK signaling. Previously reported larotrectinib- and entrectinib-resistant NTRK kinase domain (KD) mutations include substitutions of the glycine residue in the exposed solvent front of the kinase domain (solvent front: NTRK1G595R and NTRK3G623R) or substitutions of the glycine residue preceding the critical aspartic acid (D)-phenyalanine (F)-glycine(G) motif in the activation loop of the kinase (xDFG: NTRK1G667C, NTRK1G667S, and NTRK3G696A) (Drilon et al. 2016a, 2018a; Russo et al. 2016; Somwar et al. 2020). Kinase-extrinsic or bypass alterations are less extensively studied; however, MAPK pathway alterations were reported as a TKI bypass mechanism in non-CNS tumors (Cocco et al. 2019).
Rational drug development strategies to mitigate kinase-intrinsic resistance led to second-generation NTRK inhibitors (NTRKi) including selitrectinib (LOXO-195), repotrectinib (TPX-0005), and taletrectinib (Ds-6051b) (Drilon et al. 2017, 2018b; Katayama et al. 2019). Second-generation inhibitors reportedly possess preclinical efficacy against some resistant mutations (Drilon et al. 2017, 2018b; Katayama et al. 2019), including the solvent front substitutions G595R and G623R in the setting of TPM3-NTRK1 and ETV6-NTRK3 fusions, respectively. Notably, selitrectinib response was observed in a pediatric patient with recurrent infantile fibrosarcoma that harbored the ETV6-NTRK3G623R mutation (Drilon et al. 2017), and repotrectinib response was reported in an ETV6-NTRK3G623E mutant mammary analog secretory carcinoma that progressed on entrectinib (Drilon et al. 2018b).
Here, we report a case of a pediatric patient with a glioma driven by ETV6-NTRK3 fusion who developed therapeutic resistance to entrectinib and subsequently progressed through treatment intervention with a second-generation NTRK inhibitor, selitrectinib. Genetic analysis of the resistant tumors revealed NTRK3G623A and NTRK3G623E resistant mutations. The functional impact of these kinase domain substitutions was characterized through in vitro studies, establishing these variants as contributors to NTRK targeted therapy resistance. This report demonstrates that similar resistance mechanisms for systemic NTRK fusion–driven tumors are relevant in primary CNS tumors with NTRK gene rearrangements, and testing for resistance-associated alterations could be beneficial to guide management of resistant/recurrent NTRK-related CNS neoplasia.
RESULTS
Case Presentation
A 3-yr-old female presented to an emergency department with ∼2 mo of morning emesis, becoming intractable and associated with headaches in recent weeks. Neuroimaging revealed extensive solid and cystic signal abnormality along the ventricular margins and extending into the left parietal lobe white matter (Fig. 1A,B), along with bilateral cerebellopontine angle masses and foci of abnormal enhancement in the spine. An excisional biopsy of the left parietal region tumor revealed an unusual biphasic spindled and epithelioid neoplasm that showed variable immunoreactivity for glial markers glial fibrillary acidic protein (GFAP) and Olig-2 (Fig. 1H–K). Mitotic figures were present at up to two mitotic figures per 10 high-power fields, whereas necrosis and microvascular proliferation were not identified. A clinical next-generation sequencing study on the Children's Hospital of Los Angeles OncoKids panel identified a fusion between ETV6 and NTRK3 predicted to generate fusion protein composed of exons 1–4 of ETV6 and exons 14–20 of NTRK3, a region that includes most of the NTRK3/TRKC kinase domain (Fig. 1L). No pathogenic mutations were identified in other assessed genes (Hiemenz et al. 2018). Copy-number profiling by single-nucleotide polymorphism microarray showed focal homozygous deletion of 9p21, encompassing the CDKN2A/B tumor suppressor gene, and numerous whole-chromosome losses (Supplemental Fig. S1A). The patient was initially treated with carboplatin and vincristine per Children's Oncology Group protocol A9952 regimen A for 4 wk, and then transitioned to entrectinib (300 mg daily) because of early clinical and radiologic progression. Dramatic clinical and radiographic improvement was seen after 8 wk of therapy (Fig. 1C,D). Entrectinib response persisted for a total of 24 wk, when treatment was discontinued because of tumor progression (Fig. 1E). The patient transitioned to a second-generation NTRK inhibitor, selitrectinib (LOXO-195), with early radiographic recurrence/progression after 8 wk of treatment (Fig. 1F). Subsequent therapies included a combination regimen of larotrectinib, irinotecan, bevacizumab, and temozolomide and focal palliative radiation therapy to midbrain and hypothalamic lesions. Radiographic monitoring of late disease showed numerous cortically based and periventricular nodular lesions and extensive tumor encasement of the spinal cord and spinal nerve roots (Fig. 1G). The patient succumbed to disease while receiving hospice care 14 mo after initial diagnosis. An autopsy limited to the brain and spinal cord was performed.
Figure 1.
Clinical history and histopathological findings of the case. (A–G) Timeline showing the treatment course of the patient with corresponding radiographs indicating location and progression of tumors. Initial pretreatment T2 FLAIR axial and coronal MR images (A,B) demonstrate T2 hyperintense nodular tissue along the ependymal surfaces of the lateral ventricles, greater on the left than the right. T1 post-contrast image shows evidence of progression on carboplatin plus vincristine therapy (C). Following initial entrectinib therapy there is marked reduction in the volume of tumor on axial T2 FLAIR imaging (D). Post-gadolinium axial T1 images (E) reveal disease progression with new enhancing nodular tissue in the sylvian fissure. The image in panel F also shows new T2 FLAIR nodularity at the ventricular margin. Cervical spine sagittal T1 post-gadolinium imaging (G) revealed new leptomeningeal tumor along the dorsal surface of the cord. (H–K) Histology and immunophenotype of the diagnostic excisional biopsy at clinical presentation. Hematoxylin and eosin (H&E)-stained sections showed a predominantly epithelioid neoplasm with spindled and myxoid areas and delicate fibrillary processes (H; scale bar, 100 microns). Most of the tumor cells showed abundant eosinophilic cytoplasm, moderately pleomorphic nuclei, and conspicuous nucleoli (I; scale bar, 20 microns). Immunoreactivity for glial fibrillary acidic protein (GFAP; J) and Olig-2 (K; scale bar, 100 microns applies to J and K). (L) Domain organization for ETV6, NTRK3, and ETV6-NTRK3 fusion. Exons 1–4 of ETV6 and exons 14–20 of NTRK3 form the final fusion product. The helix-loop-helix (HLH) of ETV6 and the tyrosine kinase domain (TKD) of NTRK3 are maintained in the ETV6-NTRK3 fusion. (M–P) H&E stained sections of postmortem tumor samples. Nodules of cerebral hemispheric disease showed a densely cellular, markedly pleomorphic glioma with brisk mitotic activity and focal necrosis (Nec), and limited infiltration of adjacent brain cortex (C) and white matter (WM) (M–N; scale bars, 500 microns). On higher magnification, frequent mitotic figures (arrowheads) were noted, with tumor cells showing high nuclear to cytoplasmic ratios and vesicular chromatin (N; scale bar, 50 microns). A section of spinal leptomeningeal disease showed limited infiltration of the spinal cord (SC) parenchyma, and a coating of pleomorphic tumor encasing the spinal nerve roots (NR) (P; scale bar, 100 microns).
Genomic Profiling of Recurrent/Disseminated Disease after NTRK Inhibitor Treatment
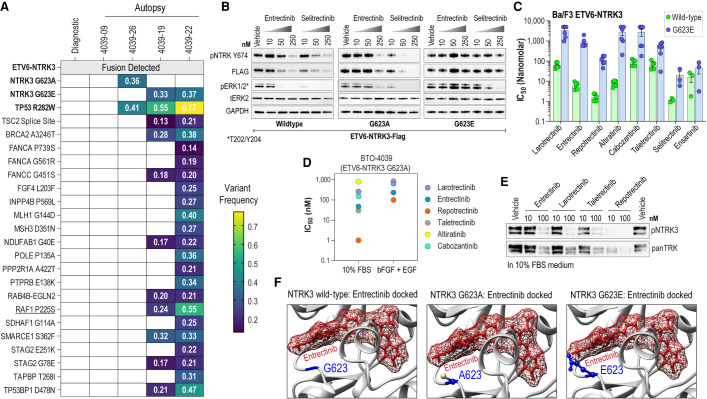
On gross examination of the brain, innumerable nodules of recurrent/disseminated tumor were identified along the ventricular walls. The thalamus, hypothalamus, midbrain, and pons were enlarged and firm. The spinal cord appeared enlarged, because of diffuse coating of the cord and nerve roots by confluent disseminated tumor. Three representative nodular lesions from the cerebral hemispheres and one region of spinal leptomeningeal disease were sampled and showed a pleomorphic, mitotically active glioma with necrosis and parenchymal infiltration along perivascular spaces (Fig. 1M–P). The samples were submitted for GeneTrails, a next-generation sequencing panel that covers mutations and small insertions/deletions in 222 cancer-related genes as well as fusion coverage for 24 genes including NTRK1/2/3. Gene coverage for DNA sequencing and RNA fusion panel is presented in Supplemental Tables 1 and 2, respectively, and sequencing coverage for RNA fusion panel is in Supplemental Table 3. Extracted nucleic acids from the original clinical sample were resequenced on the same panel to help identify mutations associated with recurrent/progressive disease. The pathogenic ETV6-NTRK3 fusion was detected in all four samples (Table 1; Fig. 2A). The sample from disseminated spinal leptomeningeal disease revealed an acquired uncharacterized NTRK3 p.G623A kinase domain mutation (Table 1). Two cerebral lesion showed a NTRK3 p.G623E mutation, known to be associated with NTRK inhibitor resistance, along with multiple acquired mutations with a preponderance of G:C to A:T transitions compatible with sequelae of alkylating agent chemotherapy (Table 1; Fig. 2A; Supplemental Fig. S2; Drilon et al. 2018b). The samples with NTRK3 resistance mutations all showed a shared, acquired, pathogenic mutation TP53 p.R282W, a well-known inactivating mutation impacting p53 protein function (ClinVar ID: 12364). Intriguingly, the final cerebral hemispheric focus with confirmed ETV6-NTRK3 rearrangement showed neither NTRK3 resistance mutation nor TP53 mutation, even though it was histologically similar to the other foci of disease with resistance mutations, with marked pleomorphism and mitotic activity. The complete genetic findings are listed in Supplemental Table 4, and summarized schematically along with variant allele frequencies in Figure 2A.
Table 1.
ETV6-NTRK3 variant table
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | Variant allele frequency | Total reads | Sample ID |
|---|---|---|---|---|---|---|---|---|
| ETV6-NTRK3 | 12 | t(12;15)(p13;q25) (hg19 Chr 12: 12006495 : Chr 15: 88576276) | n/a | ETV6-NTRK3 fusion | Oncogenic, sensitivity to TRK inhibitors. | n/a | 2317 | Diagnostic |
| ETV6-NTRK3 | 12 | t(12;15)(p13;q25) (hg19 Chr 12: 12006495 : Chr 15: 88576276) | n/a | ETV6-NTRK3 fusion | Oncogenic sensitivity to TRK inhibitors | n/a | 1125 | 4093-09 |
| ETV6-NTRK3 | 12 | t(12;15)(p13;q25) (hg19 Chr 12: 12006495 : Chr 15: 88576276) | n/a | ETV6-NTRK3 fusion | Oncogenic sensitivity to TRK inhibitors | n/a | 4439 | 4093-19 |
| ETV6-NTRK3 | 12 | t(12;15)(p13;q25) (hg19 Chr 12: 12006495 : Chr 15: 88576276) | n/a | ETV6-NTRK3 fusion | Oncogenic sensitivity to TRK inhibitors | n/a | 6114 | 4093-22 |
| ETV6-NTRK3 | 12 | t(12;15)(p13;q25) (hg19 Chr 12: 12006495 : Chr 15: 88576276) | n/a | ETV6-NTRK3 fusion | Oncogenic sensitivity to TRK inhibitors | n/a | 9065 | 4093-26 |
| NTRK3 | 15 | c.1868G > C | p. G623A | Substitution | Entrectinib resistance | 36% | 215 | 4093-26 |
| NTRK3 | 15 | c.1868G > A | p. G623E | Substitution | Cabozantinib, entrectinib, larotrectinib, repotrectinib, and selitrectinib resistance | 33% | 269 | 4093-19 |
| NTRK3 | 15 | c.1868G > A | p. G623E | Substitution | Cabozantinib, entrectinib, larotrectinib, repotrectinib, and selitrectinib resistance | 37% | 230 | 4093-22 |
Figure 2.
Genetic analysis of tumors and in vitro characterization of identified NTRK3 mutations. (A) Variant allele frequency of identified mutations from GeneTrails next-generation sequencing of diagnostic and autopsy samples. (B) Immunoblot analysis of phosphorylated NTRK3, flag-tagged NTRK3, and phosphorylated and total ERK from transfected HEK293T17 cell lysates after 4 h of treatment with TKIs as indicated. (C) Bar graph of cell proliferation IC50 values for each of the indicated TKIs against Ba/F3 cells expressing wild-type ETV6-NTRK3 (green) and ETV6-NTRK3G623E (blue). (D) Scatter plot of cell proliferation IC50 values for indicated TKIs against patient cell line BTO-4039 expressing ETV6-NTRK3G623A cultured in 10% FBS media or neural stem cell media supplemented with bFGF and EGF. (E) Immunoblot analysis of phosphorylated NTRK3 and total NTRK3 from patient cell line BTO-4039 after 5 h of treatment with TKIs as indicated. (F) Predicted binding poses for entrectinib with NTRK3WT, NTRK3G623A, and NTRK3G623E.
Solvent Front NTRK3 G623A/E Mutation Confers Broad Resistance to NTRK Inhibitors
To determine the functional significance of the NTRK3G623A and NTRK3G623E variants as contributors to TKI resistance, these amino acid substitutions were engineered into an ETV6-NTRK3-FLAG construct and their impact on NTRK autophosphorylation and signaling effector pathway activation with or without NTRKi treatment was examined. Immunoblotting of cell lysates shows entrectinib and selitrectinib inhibit ETV6-NTRK3 autophosphorylation and downstream activation of MAPKs, ERK1/2. Compared to ETV6-NTRK3WT, ETV6-NTRK3G623A confers partial resistance to entrectinib that is particularly evident in the lack of inhibition of MAPK signaling (ERK1/2 phosphorylation) at a 50 nM dose (Fig. 2B). ETV6-NTRK3G623E is completely resistant to entrectinib in these assays, with no inhibition of autophosphorylation or MAPK signaling noted even at 250 nM treatment and is less sensitive to selitrectinib as compared to ETV6-NTRK3WT.
To further explore the TKI resistance profile of NTRK3G623E mutation, we used the Ba/F3 heterologous model system (Warmuth et al. 2007; Davare et al. 2015; Drilon et al. 2016b; Somwar et al. 2020). Stably transformed Ba/F3 ETV6-NTRK3WT and ETV6-NTRK3G623E cell lines were used for dose–response cell viability assays with a panel of NTRKi. Among these, altiratinib and cabozantinib are Type II inhibitors binding the DFG-out conformation, whereas the others are Type I inhibitors binding the DFG-in conformation. ETV6-NTRK3WT and ETV6-NTRK3G623E harbor comparable transformative propensity in IL-3 withdrawal assays (Supplemental Fig. S1B). The NTRK3G623E mutation has a significant impact on the concentration required to inhibit cell growth by 50% (IC50) for larotrectinib (53-fold increase), entrectinib (148-fold increase), repotrectinib (71-fold increase), altiratinib (344-fold increase), and cabozantinib (32-fold increase). The inhibitory efficacy of taletrectinib, selitrectinib, and ensartinib is also blunted by the NTRK3G623E mutation with approximately eightfold, ∼17-fold, and approximately threefold increases in IC50s, respectively, as compared to the wild-type (Fig. 2C; Supplemental Fig. S3).
Mutations of RAS/RAF/MEK pathway genes that constitutively activate the MAPK pathway were reported as a recurrent mechanism of resistance to NTRKi in NTRK fusion–positive cancers (Cocco et al. 2019). Given this, we were intrigued by our identification of a RAF1P225S variant of unknown significance present in two of the autopsy tumor samples (Fig. 2A). RAF kinases transmit signal from RAS proteins to downstream MAPK proteins. Of note, RAF1P225S resides in a loop connecting the RAF1 RAS-binding and kinase domains (Supplemental Fig. S4A). Another nearby mutation in this loop region, RAF1P261S, is well-established as a gain-of-function mutation in cancer (Noeparast et al. 2019) and as a germline mutation implicated in RASopathies including Noonan syndrome and LEOPARD syndrome (Pandit et al. 2007). We examined the functional impact of RAF1P225S relative to RAF1WT and known activating RAF1P261S as a positive control. Transfection of these constructs into HEK293A cells establish that the RAF1P225S variant is a functionally neutral substitution with activity comparable to RAF1WT as compared to the robust activation of phospho-ERK1/2 by RAF1P261S, supporting that RAF1P225S was not a contributing factor to NTRKi resistance noted in these tumor samples (Supplemental Fig. S4B).
A patient-derived cell line was established from one of the autopsy tissue samples (BTO-4039-26, Supplemental Fig. S1C). Sanger sequencing of cDNA confirmed the expression of ETV6-NTRK3 fusion product and showed the presence of the ETV6-NTRK3G623A mutation (Supplemental Fig. S1D). BTO-4039-26 was cultured in neural stem cell media containing supplementation with recombinant human epidermal growth factor (rh-EGF) and basic fibroblast growth factor (rh-bFGF) and also adapted to grow in DMEM with 10% FBS (D10-FBS). Dose–response cell viability assays of cells cultured in D10-FBS show higher sensitivity to inhibitors as compared with culture in neural stem cell media (Fig. 2D; Supplemental Fig. S5), suggesting that in the presence of growth factors, the tumor would show an even more limited response to targeted inhibitors (Fig. 2D; Supplemental Fig. S5). Immunoblotting of cell lysates prepared from BTO-4039-26 following treatment with select NTRKi shows that at the higher concentration (100 nM) tested, the inhibitors exhibit partial efficacy to block ETV6-NTRK3 autophosphorylation (Fig. 2E). These data are consistent with the partial resistance of ETV6-NTRK3G623A seen in the transient transfection experiments.
To gain insight on the structural impact of these G623A and G623E substitutions on entrectinib binding, we docked entrectinib in the NTRK3 kinase domain using a previously generated computational homology model7 (Fig. 2F; Supplemental Fig. S6). Docking studies suggest that steric hindrance generated from the larger side chains of alanine, and particularly of glutamic acid, as compared to the native glycine at the 623 position, likely interferes with entrectinib binding. This offers a structural rationale for entrectinib resistance of NTRK3G623A and NTRK3G623E.
DISCUSSION
Although targeted therapeutics are becoming increasingly available, therapeutic resistance is an inevitable issue. Here, we present a case of primary ETV6-NTRK3 fusion–driven pediatric glioma with progression on multiple targeted NTRK inhibitors. This analysis identified solvent front mutations in the tyrosine kinase domain of NTRK3, similar to previously identified resistance mechanisms in NTRK1 (G595R) and NTRK3 (G623R) in extracranial NTRK fusion–driven neoplasia (Drilon et al. 2016a, 2017, 2018a,b; Russo et al. 2016; Hemming et al. 2020). This case report demonstrates the clinical relevance of these mechanisms of resistance in patients with primary CNS malignancies that, to our knowledge, have yet to be reported. Notably, this case also demonstrates that a next-generation inhibitor such as selitrectinib, which was previously shown to have efficacy in ETV6-NTRK3G623E-driven extracranial tumors, may have limited benefit in primary CNS tumors, potentially because of lower achievable CNS concentration.
Of the mutations found in the autopsy samples, relatively few are definitively pathogenic. TP53R282W identified in three postmortem samples is a known hotspot in human cancers affecting the DNA binding ability of p53 (Supplemental Table 4; Xu et al. 2011; Chang et al. 2016; Gao et al. 2017). The TSC2 splice-site variant identified in two postmortem samples is likely to be pathogenic, as splice variants of TSC2 are reported in a variety of cancers to have potential impact on activity (Cerami et al. 2012; Gao et al. 2013). The majority of the mutations are variants of uncertain significance with unknown impact on protein function or disease pathology. Here we showed that the RAF1P225S is a neutral impact mutation, unlikely to contribute to drug resistance or disease progression. POLEP135A has not been previously reported, but POLEP135S mutation was suggested to be a nonpathogenic passenger mutation (Campbell et al. 2017). From a structural perspective, we postulate that the alanine (A) and serine (S) amino acids have similar bulk and are not charged; thus, it is unlikely that P135A would impact function if P135S does not. Of the remaining mutations, many have been previously identified in patients, but functional impacts have not been elucidated (Cerami et al. 2012; Gao et al. 2013).
Although NTRK3G623A and NTRKG623E were not detected in the original clinical sample, we cannot exclude the possibility of a minor preexisting population of tumor cells harboring these alterations that was below the threshold of detection of these NGS assays. Regardless of whether NTRK3G623A or NTRK3G623E represent a preexisting subclone or de novo acquired mutation, their reduced affinity for NTRKi was most likely permissive to their selection and expansion in the presence of inhibitor, thus contributing to disease progression. The NTRK3G623R solvent front substitution has been shown to be sensitive to selitrectinib, both in model systems and patients (Drilon et al. 2017). Specifically, Drilon et al. (2017) demonstrated that selitrectinib treatment induced rapid tumor response and extended disease control in a larotrectinib-resistant ETV6-NTRK3G623R fusion–positive infantile fibrosarcoma. In contrast to this, the ETV6-NTRK3G623A and ETV6-NTRK3G623E lesions progressed during the 8 wk of selitrectinib treatment (ClinicalTrials.gov identifier: NCT03215511 [43 mg/m2]). Ba/F3 cell model–based studies show that ETV6-NTRK3G623E exhibits ∼18-fold increase in IC50 for selitrectinib. We postulate that achievable CNS concentrations of selitrectinib with the pediatric dosing regimen may have been insufficient to control tumor cells harboring the kinase domain mutant ETV6-NTRK3 fusion protein that required 18-fold higher inhibitor amounts compared to wild-type fusion in our model cell system. It is also possible that the trophic microenvironment of the pediatric brain confers exogenous resistance, dampening the tumor response by activating basal growth or survival pathways in these cells. Indeed the patient derived cell line is highly resistant to the NTRKi when cultured with recombinant human EGF or bFGF, as opposed to generic media with fetal bovine serum (Fig. 2D).
Genetic analyses of multiple postmortem tumors showed variation in mutational landscape across distinct disseminated sites of disease in the CNS. These postmortem samples were collected after progression on entrectinib, selitrectinib, and a combination regimen that included larotrectinib and temozolomide. In the postmortem sample with the highest somatic mutation burden, >95% of them are G:C to A:T transitions, a pattern consistent with temozolomide-associated mutation (Supplemental Fig. S2; Johnson et al. 2014; Degasperi et al. 2020). Interestingly, one of the autopsy samples (4039-09) showed no additional alterations as compared to the original clinical sample. Despite first- and second-generation NTRK inhibitor treatments, this tumor lesion grew and formed a persistent, viable nodule. This suggests several possibilities. Although NTRKi are cytotoxic in NTRK fusion–driven in vitro model systems, their effect in vivo may be cytostatic rather than cytotoxic because of the pharmacokinetics in the CNS and the brain microenvironment. It is also possible that alterations in genes not included in our NGS panel are responsible for therapeutic resistance in 4039-09 tumor.
Based on functional data presented here, we conclude that NTRK3G623E and, to a lesser degree, NTRK3G623A were the causative mechanisms of targeted therapeutic resistance in this tumor. Furthermore, these alterations are found to confer cross-resistance to essentially all current NTRK inhibitors. Solvent front mutations have previously been observed in NTRK fusion–driven cancers, but this case represents the first report of these mutations in a primary CNS tumor. Further studies are needed to continue to assemble the profile of NTRK mutations that exhibit resistance to TRK inhibitors as well as NTRK-extrinsic mechanisms of resistance. Genetic profiling of residual and recurrent disease can shed light on these resistance mechanisms and potentially impact treatment decisions in this challenging and unusual type of glioma.
METHODS
Inhibitors
Selitrectinib (LOXO-195) and repotrectinib (TPX-0005) were purchased from MedChemExpress. Entrectinib (RXDX-101), altiratinib (DCC-2701), cabozantinib (XL184), ensartinib (X-396), larotrectinib (LOXO-101), and taletrectinib (DS-6051b) were purchased from Selleck Chemicals.
Cell Culture
Parental Ba/F3 cells (American Type Culture Collection) were cultured in complete medium (RPMI medium 1640 with 10% fetal bovine serum [FBS], 2 mmol/L L-glutamine, penicillin, streptomycin, and fungizome) supplemented with 2 ng/mL recombinant murine IL-3 and maintained at densities of 0.5 × 106 to 1.5 × 106. HEK293A and HEK293T were cultured in complete medium (DMEM with 10% bovine growth serum, 2 mmol/L l-glutamine, penicillin, streptomycin). All cell lines were grown under standard mammalian cell culture conditions (37°C, 5% CO2).
Ba/F3 Cell Line Generation
The ETV6-NTRK3 mutants were generated using site-directed mutagenesis following manufacturer's protocol (Agilent). Platinum-E cells (Cell Biolabs, Inc.) were transfected with pCX4 ETV6-NTRK3 wild-type or mutant constructs using DNA transfection reagent from Biotool to generate replication incompetent ecotropic retrovirus. Ba/F3 cells were infected with retrovirus encoding wild-type or mutant versions of ETV6-NTRK3. Cells were selected with 2 µg/mL puromycin treatment. Selected Ba/F3 cells were washed three times with IL-3 free complete medium and suspended at a density of 0.5 × 106 per ml. The number of viable cells were counted every other day and expanded as needed to maintain a density of 0.5 × 106 per mL. Cells that grew out after IL-3 withdrawal were maintained in complete medium without IL-3 and used for in vitro assays.
Sanger Sequencing Validation of Endogenous NTRK3 Mutation
RNA was isolated using the Quick-RNA Miniprep kit (Zymo Research). cDNA was synthesized using OneScript Plus cDNA Synthesis kit (Applied Biological Materials Inc.) according to manufacturer's specifications. The NTRK3 kinase domain was polymerase chain reaction (PCR)-amplified using the primers ETV6F2 (5′-CTTCCACCCTGGAAACTCTATAC-3′) and NTRKR (5′-GTCCTCCCACCCTGTAATAATC-3′) and sequenced using ETV6F2. Chromatographs were aligned to identify mutations using Benchling software.
DNA Transfections
HEK293A and HEK293T cells were transiently transfected using TransIT-LT1 transfection reagent (Mirus Bio) to manufacturer's specification.
Patient-Derived Tumor Cell Line
BTO-4039-26 cells were derived from an autopsy specimen. Harvested tissue was washed with sterile saline buffer, minced under aseptic conditions, and transferred to neural stem cell medium (1:1 DMEM/F12 and neural stem cell media [ThermoFisher] with 20 ng/mL EGF, 20 ng/mL FGF, 1 ng/mL LIF, 20 µg/mL heparin, penicillin, streptomycin, 0.025 M HEPES, 1× B27 Supplement without A [ThermoFisher], 1× GlutaMAX [ThermoFisher], 1× Non-Essential Amino Acids Solution [ThermoFisher]). The cells were maintained in neural stem cell media. For adaptation to DMEM + 10% fetal bovine serum medium, 1 × 106 cells were pelleted and without rinsing, transferred to new flask containing 15 mL of DMEM with 10% fetal bovine serum, 2 mmol/L L-glutamine, penicillin, and streptomycin. The cells were fed with 5 mL of DMEM + 10% FBS once a week and cultured for no more than three population doublings in this medium.
Immunoblot Analysis
Transfected HEK293T and patient-derived cell lines were treated with the indicated concentrations of inhibitors for 4 h. Following treatment, cells were pelleted, washed once in ice-cold PBS, and lysed in cell lysis buffer supplemented with 0.25% deoxycholate, 0.05% SDS, and protease and phosphatase inhibitors. Protein concentrations were determined using the Pierce BCA Protein Assay (ThermoFisher Scientific). Protein was extracted with NuPAGE LDS Sample Buffer (4×) (ThermoFisher Scientific) sample buffer, supplemented with β-mercaptoethanol, for 10 min at 75°C. An amount of 10–25 µg of extracted lysate protein was run on 4%–12% Bis-Tris (Invitrogen; ThermoFisher Scientific) and transferred to nitrocellulose membranes (Prometheus). Blots were probed with antibodies specific for phospho-TRK (CST; 4621; 1:1000), pan-TRK (CST; 3266; 1:1000), phospho-p44/42 MAPK (CST; 9101; 1:1000), total ERK2 (Santa Cruz; sc-1647;1:1000), phospho-S6 (CST; 4858; 1:1000), total S6 (CST; 2216; 1:1000), GFP (Origene; TA150032; 1:5000), phospho-RAF1 (CST; 9421; 1:1000), and GAPDH (Origene; OTI2D9; 1:5000). We used the Bio-Rad Chemidoc imaging station or LI-COR Odyssey imaging system and followed manufacturer's recommendations for immunoblot detection with use of HRP-conjugated or IR dye secondary antibodies, respectively.
Cell Viability Assays
All inhibitors were prepared as 1 mM stock in DMSO. Inhibitors were distributed into 384-well plates that were preseeded with 25 µL of complete RPMI-10% FBS medium using a D300 Digital Dispenser (Hewlett-Packard). Ba/F3 cells expressing wild-type or mutant ETV6-NTRK3 were seeded at 1000 cells per well in 25 µL onto drug plates using a Multidro Combi Reagent Dispenser (Thermo Scientific). Plates were incubated for 72 h at 37°C. Viability was measured using a water-soluble tetrazolium salt (WST-8) based assay (Bimake) and absorbance (450 nm) was read 1 h after adding reagent using a BioTek Synergy 2 plate reader. Data from drug-treated wells were normalized to Vehicle (0.05% DMSO) treated wells in Microsoft Excel. Each experiment had three replicate wells per condition and the average and SEM were used for curve fit analysis. Plotting and nonlinear regression curve fit analysis for determination of IC50 values was performed using GraphPad Prism software.
Homology Model
Chimera (Pettersen et al. 2004) software was used to generate G623A and G623E substitutions (structural editing) within a previously published (Somwar et al. 2020) homology model of NTRK3 with docked entrectinib.
Mutational Signature Analysis
Signal (https://signal.mutationalsignatures.com/) was used to analyze mutational signatures from next-generation sequencing. Human GRCh37 was used as the reference genome (Degasperi et al. 2020).
Next-Generation Sequencing
Tumor-rich regions of formalin-fixed, paraffin-embedded sections of autopsy tissue were macrodissected, deparaffinized, and total nucleic acid was extracted and purified using the Nucleospin tissue kit (Machery-Nagel). Amplicon-based sequencing libraries (custom QiaSeq panel, QIAGEN, Inc.) covering the protein-coding regions of 222 cancer-related genes (see Supplemental Table 1) were prepared from tumor nucleic acid samples and sequenced on an Illumina NextSeq550. The Knight Diagnostic Laboratories GeneTrails sequencing panel covers 222 cancer-associated genes, with a total sequencing footprint of 0.61 Mb and average read depth of ∼2000. Single-nucleotide variations and small insertions/deletions are identified using FreeBayes and MuTect2 algorithms in a custom analysis pipeline. Copy-number alterations and microsatellite instability at 227 short repeats are also assessed. An RNA-based panel covering breakpoints in 21 genes detects rearrangements with any fusion partner (see Supplemental Table 2). The ETV6-NTRK3 fusion was confirmed in each sample by next-generation sequencing of tumor RNA using a custom amplicon-based library (QiaSeq) run on an Illumina NextSeq550. The panel includes partner-agnostic coverage of common known breakpoints in NTRK3.
ADDITIONAL INFORMATION
Data Availability
Data produced for clinical diagnostic purposes will not be publicly available. Research-related data will be made available without restrictions upon request by contacting the corresponding authors. The variants reported herein were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV001781535–SCV001781538.
Ethics Statement
Studies performed on deidentified human postmortem tissue are not considered “human subjects” research and have been exempted from review by the IRB. Unrestricted consent for use of the tissue is obtained from the next of kin before autopsy.
Acknowledgments
The authors thank Qianyue Yang for her assistance with maintenance of patient-derived cell lines and Dr. Phil Stork (Oregon Health & Science University) for generously sharing the RAF1 constructs. This work was partially supported by funding from Hero-Up for Children's Cancer.
Author Contributions
J.P.N., K.J.N., R.W., Y-J.C., and M.D.W. provided clinical care of the patient and clinical data and/or autopsy tumor specimen or other resources. C.K., J.H., C.Z.B., R.W., M.D.W., and M.A.D. performed experiments and analyzed data. M.A.D. conducted molecular modeling. C.K., J.P.N., M.D.W., and M.A.D. assembled figures. M.D.W. and M.A.D. conceived the project. C.K., T.N., C.Z.B., Y.A., J.J., S.M., C.L.C., C.B., M.D.W., and M.A.D. performed data analysis. C.K., M.D.W., and M.A.D. wrote the manuscript. All authors reviewed and edited the paper.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Theodore Nicolaides
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel KP, et al. 2017. Comprehensive analysis of hypermutation in human cancer. Cell 171: 1042–1056.e1010. 10.1016/j.cell.2017.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: figure 1. Cancer Discov 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, Gao J, Socci ND, Solit DB, Olshen AB, et al. 2016. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 34: 155–163. 10.1038/nbt.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco E, Scaltriti M, Drilon A. 2018. NTRK fusion–positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 15: 731–747. 10.1038/s41571-018-0113-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco E, Schram AM, Kulick A, Misale S, Won HH, Yaeger R, Razavi P, Ptashkin R, Hechtman JF, Toska E, et al. 2019. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat Med 25: 1422–1427. 10.1038/s41591-019-0542-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davare MA, Vellore NA, Wagner JP, Eide CA, Goodman JR, Drilon A, Deininger MW, O'Hare T, Druker BJ. 2015. Structural insight into selectivity and resistance profiles of ROS1 tyrosine kinase inhibitors. Proc Natl Acad Sci 112: E5381–E5390. 10.1073/pnas.1515281112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degasperi A, Amarante TD, Czarnecki J, Shooter S, Zou X, Glodzik D, Morganella S, Nanda AS, Badja C, Koh G, et al. 2020. A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies. Nat Cancer 1: 249–263. 10.1038/s43018-020-0027-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, Blakely CM, Seto T, Cho BC, Tosi D, et al. 2020. Entrectinib in patients with advanced or metastatic NTRK fusion–positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol 21: 271–282. 10.1016/S1470-2045(19)30691-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Li G, Dogan S, Gounder M, Shen R, Arcila M, Wang L, Hyman DM, Hechtman J, Wei G, et al. 2016a. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann Oncol 27: 920–926. 10.1093/annonc/mdw042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Somwar R, Wagner JP, Vellore NA, Eide CA, Zabriskie MS, Arcila ME, Hechtman JF, Wang L, Smith RS, et al. 2016b. A novel crizotinib-resistant solvent-front mutation responsive to cabozantinib therapy in a patient with ROS1-rearranged lung cancer. Clin Cancer Res 22: 2351–2358. 10.1158/1078-0432.CCR-15-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Nagasubramanian R, Blake JF, Ku N, Tuch BB, Ebata K, Smith S, Lauriault V, Kolakowski GR, Brandhuber BJ, et al. 2017. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion–positive solid tumors. Cancer Discov 7: 963–972. 10.1158/2159-8290.CD-17-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, et al. 2018a. Efficacy of larotrectinib in TRK fusion–positive cancers in adults and children. N Engl J Med 378: 731–739. 10.1056/NEJMoa1714448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Ou SI, Cho BC, Kim DW, Lee J, Lin JJ, Zhu VW, Ahn MJ, Camidge DR, Nguyen J, et al. 2018b. Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent- front mutations. Cancer Discov 8: 1227–1236. 10.1158/2159-8290.CD-18-0484 [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. 10.1126/scisignal.6273er1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Chang MT, Johnsen HC, Gao SP, Sylvester BE, Sumer SO, Zhang H, Solit DB, Taylor BS, Schultz N, et al. 2017. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med 9: 4. 10.1186/s13073-016-0393-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming ML, Nathenson MJ, Lin J-R, Mei S, Du Z, Malik K, Marino-Enriquez A, Jagannathan JP, Sorger PK, Bertagnolli M, et al. 2020. Response and mechanisms of resistance to larotrectinib and selitrectinib in metastatic undifferentiated sarcoma harboring oncogenic fusion of NTRK1. JCO Precis Oncol 4: 79–90. 10.1200/po.19.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemenz MC, Ostrow DG, Busse TM, Buckley J, Maglinte DT, Bootwalla M, Done J, Ji J, Raca G, Ryutov A, et al. 2018. OncoKids: a comprehensive next-generation sequencing panel for pediatric malignancies. J Mol Diagn 20: 765–776. 10.1016/j.jmoldx.2018.06.009 [DOI] [PubMed] [Google Scholar]
- Hong DS, DuBois SG, Kummar S, Farago AF, Albert CM, Rohrberg KS, van Tilburg CM, Nagasubramanian R, Berlin JD, Federman N, et al. 2020. Larotrectinib in patients with TRK fusion–positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol 21: 531–540. 10.1016/S1470-2045(19)30856-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. 2003. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 72: 609–642. 10.1146/annurev.biochem.72.121801.161629 [DOI] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, Mclean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. 2014. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343: 189–193. 10.1126/science.1239947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama R, Gong B, Togashi N, Miyamoto M, Kiga M, Iwasaki S, Kamai Y, Tominaga Y, Takeda Y, Kagoshima Y, et al. 2019. The new-generation selective ROS1/NTRK inhibitor DS-6051b overcomes crizotinib resistant ROS1-G2032R mutation in preclinical models. Nat Commun 10: 3604. 10.1038/s41467-019-11496-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeparast A, Giron P, Noor A, Bahadur Shahi R, De Brakeleer S, Eggermont C, Vandenplas H, Boeckx B, Lambrechts D, De Greve J, et al. 2019. CRAF mutations in lung cancer can be oncogenic and predict sensitivity to combined type II RAF and MEK inhibition. Oncogene 38: 5933–5941. 10.1038/s41388-019-0866-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, et al. 2007. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet 39: 1007–1012. 10.1038/ng2073 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Russo M, Misale S, Wei G, Siravegna G, Crisafulli G, Lazzari L, Corti G, Rospo G, Novara L, Mussolin B, et al. 2016. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov 6: 36–44. 10.1158/2159-8290.CD-15-0940 [DOI] [PubMed] [Google Scholar]
- Somwar R, Hofmann NE, Smith B, Odintsov I, Vojnic M, Linkov I, Tam A, Khodos I, Mattar MS, de Stanchina E, et al. 2020. NTRK kinase domain mutations in cancer variably impact sensitivity to type I and type II inhibitors. Commun Biol 3: 776. 10.1038/s42003-020-01508-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre M, Vasudevaraja V, Serrano J, DeLorenzo M, Malinowski S, Blandin AF, Pages M, Ligon AH, Dong F, Meredith DM, et al. 2020. Molecular and clinicopathologic features of gliomas harboring NTRK fusions. Acta Neuropathol Commun 8: 107. 10.1186/s40478-020-00980-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. 2007. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol 19: 55–60. 10.1097/CCO.0b013e328011a25f [DOI] [PubMed] [Google Scholar]
- Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine J-C, et al. 2011. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 7: 285–295. 10.1038/nchembio.546 [DOI] [PubMed] [Google Scholar]
- Ziegler DS, Wong M, Mayoh C, Kumar A, Tsoli M, Mould E, Tyrrell V, Khuong-Quang DA, Pinese M, Gayevskiy V, et al. 2018. Brief Report: potent clinical and radiological response to larotrectinib in TRK fusion–driven high-grade glioma. Br J Cancer 119: 693–696. 10.1038/s41416-018-0251-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data produced for clinical diagnostic purposes will not be publicly available. Research-related data will be made available without restrictions upon request by contacting the corresponding authors. The variants reported herein were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV001781535–SCV001781538.