Abstract
Sclerosing epithelioid fibrosarcoma (SEF) is a rare and aggressive soft-tissue sarcoma thought to originate in fibroblasts of the tissues comprising tendons, ligaments, and muscles. Minimally responsive to conventional cytotoxic chemotherapies, >50% of SEF patients experience local recurrence and/or metastatic disease. SEF is most commonly discovered in middle-aged and elderly adults, but also rarely in children. A common gene fusion occurring between the EWSR1 and CREB3L1 genes has been observed in 80%–90% of SEF cases. We describe here the youngest SEF patient reported to date (a 3-yr-old Caucasian male) who presented with numerous bony and lung metastases. Additionally, we perform a comprehensive literature review of all SEF-related articles published since the disease was first characterized. Finally, we describe the generation of an SEF primary cell line, the first such culture to be reported. The patient described here experienced persistent disease progression despite aggressive treatment including multiple resections, radiotherapy, and numerous chemotherapies and targeted therapeutics. Untreated and locally recurrent tumor and metastatic tissue were sequenced by whole-genome, whole-exome, and deep-transcriptome next-generation sequencing with comparison to a patient-matched normal blood sample. Consistent across all sequencing analyses was the disease-defining EWSR1–CREB3L1 fusion as a single feature consensus. We provide an analysis of our genomic findings and discuss potential therapeutic strategies for SEF.
Keywords: renal sarcoma
INTRODUCTION
Sclerosing epithelioid fibrosarcoma (SEF) is a rare and aggressive soft-tissue sarcoma, first characterized in 1995 by Dr. Meis-Kindblom et al. (1995) who described the disease as a variant of low-grade fibromyxoid sarcoma (LGFMS). More recently, hybrid tumors have been described that share morphologic and gene fusion variants of both SEF and LGFMS (Arbajian et al. 2017). Despite these shared characteristics, SEF is much more aggressive with a relatively high incidence of local recurrence (>50% of cases), distant metastatic spread (40%–80% of cases), and mortality (25%–57%) (Meis-Kindblom et al. 1995; Antonescu et al. 2001). Because of the rarity of SEF, little information about effective treatment regimens exists. Depending on staging, most patients either undergo surgery alone or also receive systemic chemotherapy and radiotherapy to affected sites. Rates of response to traditional chemotherapy are low, and even those receiving aggressive therapy fare poorly (Chew et al. 2018), especially those with metastatic disease at presentation.
The lack of specific phenotypic differentiation and immunohistochemical signature as well as morphologic similarity of SEF to LGFMS and hybrid-SEF/LGFMS tumors makes the pathologic diagnosis challenging. The aggressive nature of SEF makes accurate identification critical so that patients may receive appropriate care. Microscopically, pure SEF is characterized by small round or ovoid epithelioid fibroblast-like cells with pale or clear cytoplasm arranged in strands, cords, nests, and sheets, embedded in a densely sclerotic and hyalinized collagenous stroma (Meis-Kindblom et al. 1995; Antonescu et al. 2001). Genetically, classic SEF is characterized by fusion rearrangements, the most common being EWSR1–CREB3L1, and rarely FUS–CREB3L1 or ESWR1–CREB3L2/3 (Wang et al. 2012b; Arbajian et al. 2017; Dewaele et al. 2017). We note that EWSR1 rearrangement may suggest a diagnosis of Ewing sarcoma (EWS), yet the morphologic and immunohistochemical profile of SEF differs from that of EWS. SEF typically demonstrates positive staining for CD24, MUC4, and vimentin and displays fibroblastic features (Meis-Kindblom et al. 1995; Antonescu et al. 2001; Jiao et al. 2002; Doyle et al. 2012). SEF has no obvious sex predilection and most commonly affects middle-aged and elderly adults but, rarely, children. Here, we describe the case of a child with SEF, the youngest so far reported in the literature to our knowledge. This report follows a partial case report of this patient's histomorphology (Kurtz et al. 2021) but extends that report by presenting here a comprehensive molecular analysis, the first description of a SEF cell culture and review of the pediatric and nonpediatric disease features.
RESULTS
Clinical Presentation and Family History
A 3-yr-old boy presented to the emergency room (ER) with left leg pain. Plain radiographs of the left lower extremity showed a proximal left fibular complex cystic lesion with poorly defined margins, thought to represent a bone cyst. However, follow-up magnetic resonance imaging (MRI) 3 wk later demonstrated T1 hypointense and T2 hyperintense areas within the cyst, raising concern for malignancy. A computed tomography (CT) guided biopsy was performed, which showed sheets of mononuclear cells and an abundance of multinucleated giant cells upon microscopic examination, without evidence of malignancy.
Three months after symptom onset, the child suffered a fall and presented to the ER with pain in the contralateral (right) leg, leading to further imaging that showed a right distal femur pathologic fracture through another, larger bone cyst. Given these two cystic lesions, the patient underwent a skeletal survey that highlighted additional bony lesions in the lower extremities, a single lesion in the right sixth rib and right proximal humerus, and a soft-tissue density in the abdomen. A subsequent CT of the neck, chest, abdomen, and pelvis showed a large mass (10.5 × 6.9 cm) arising from the superior pole of the left kidney and numerous metastatic nodules in both lungs, the largest of which was 1.8 cm in maximal diameter.
An open renal biopsy yielded pink and gray soft tissue that microscopically showed tumor cells arranged in small cohesive nests in some areas and sheets of cells in other areas. Immunostaining of the biopsy specimen revealed the malignant cells to be diffusely positive for vimentin and focally positive for epithelial membrane antigen (EMA) and cyclin D1 staining was observed in scattered clusters of tumor cells with a variable degree of nuclear staining. The cell membrane stained weakly for CD99 and the nuclei stained positively for BRG1 (SMARCA4) and INI-1 (SMARCB1), and negative for SMA, Melan-A, HMB-95, and S-100 protein. A diagnosis of clear cell sarcoma of the kidney (CCSK) was considered and samples were sent to a renal pathologist for review, following which CCSK was ruled out. Fluorescence in situ hybridization (FISH) break-apart probes were negative for CIC rearrangement but revealed a 3′ telomeric deletion in the EWSR1 gene. Bilateral bone marrow biopsies from the anterior iliac crests were negative for malignancy. Vincristine, doxorubicin, and cyclophosphamide chemotherapy was started while pending a final diagnosis given the tumor burden and concerns for disease progression based on symptoms.
Six weeks after the renal biopsy, a next-generation panel genetic sequencing report from FoundationMedicine identified an EWSR1/CREB3L1 fusion (Table 1), NOTCH1 splice site variant (5167 + 1G > A—sub clonal), and a TMEM30A mutation (Y134fs*1); microsatellite status (MS) was stable and a low tumor mutational burden (2 mutations/Mb) was documented. In light of the EWSR1–CREB3L1 fusion, the diagnosis of SEF was made, and treatment was changed to doxorubicin and ifosfamide, followed by radiation to the right femur, left fibula metastasis, and the left kidney (37.5/37.5/45 Gy, 15 fractions) tumor per Children's Oncology Group protocol ARST0332 Arm D (Spunt et al. 2020). Seven months from symptom onset, the patient underwent curettage surgery of the right femur lesion.
Table 1.
EWSR1–CREB3L1 fusion variant table
| Gene 1 | Gene 2 | Position 1 | Position 2 | Exon #1 | Transcript #1 | Exon #2 | Transcript #2 | Fusion sequence | Variant type | Predicted effect | Platform |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EWSR1 | CREB3L1 | Chr 22: 29292562 | Chr 11: 46311373 | 11 | NM_013986 NM_005243 NM_001163285 NM_001163286 | 6 | NG_033264 | CAGGACCCATGG ATGAAGGACCAGA TCTTGATCTAG___ GCCCACCTGTAGA TCCAGATGAAGACTC TGACAACAGTGCAA TTTATGTACAAGGAT TAAATGACAGTGTGA CTC…|…GCCCAGG CTGGAGTGCAGTGGC TCAATCTCGGCTCACT GAAACATCTGCCTCCC AG___AAATTACAGG GGACATCAGGGCCACT GCTCCTGACAGA GGAGGA | Intergenic- intergenic fusion | In-frame fusion | WGS/RNA |
Two weeks post–curettage surgery, the patient underwent a left radical nephroureterectomy. The histopathology of the residual tumor consisted of nests and cords of epithelioid cells set within a background of densely hyalinized sclerotic matrix. The lesional cells were small to medium sized and contained pale eosinophilic cytoplasm. The nuclei were bland and round or angulated and showed a small nucleolus. Cytoplasmic MUC4 immunostaining was intense in the cytoplasm of the tumor cells.
Three months later (eight months from start of therapy) and after five cycles (21 d each) of doxorubicin and ifosfamide, the patient underwent bilateral thoracotomy with complete wedge resection of eight lung lesions. All lesions showed similar morphological features: well-defined lesions with pushing borders, a bland nodular and densely hyalinized fibrous appearance, and nests and cords of small epithelioid cells (Fig. 1A,B). Some areas displayed a more cellular spindled appearance with mild pleomorphic cytologic features consisting of nuclear enlargement and mild hyperchromasia. Immunohistochemical staining was positive for CD24 (Fig. 1C) and negative for cleaved NOTCH1 (Fig. 2D). Staining was positive for JAGGED1 (upstream of NOTCH) and weakly positive for HEY1 (downstream from NOTCH) (Fig. 1E,F). One month later, the patient underwent resection of a left fibula lesion.
Figure 1.
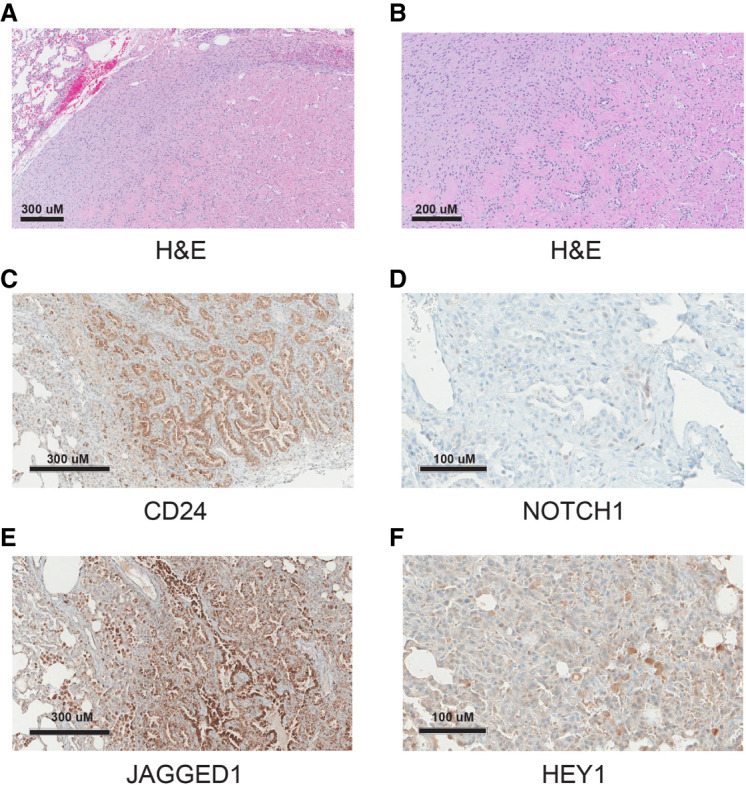
(A) Morphological diagnostic immunochemistry image with hematoxylin and eosin (H&E) staining of the right posterior upper lung nodule biopsy. Circumscribed tumor adjacent to normal tissue (top left corner) showing spindle to sclerotic tumor. The round spaces are thought to be entrapped/residual alveolar spaces. Scale bar, 300 µM. (B) H&E close-up showing spindle to sclerotic tumor. Scale bar, 200 µM. (C) Immunohistochemistry (IHC) staining for CD24. Scale bar, 300 µM. (D) IHC staining for cleaved NOTCH1. Scale bar, 100 µM. (E) IHC staining for JAGGED1 (upstream of NOTCH). Scale bar, 300 µM. (F) IHC staining for HEY1 (downstream from NOTCH). Scale bar, 100 µM.
Figure 2.
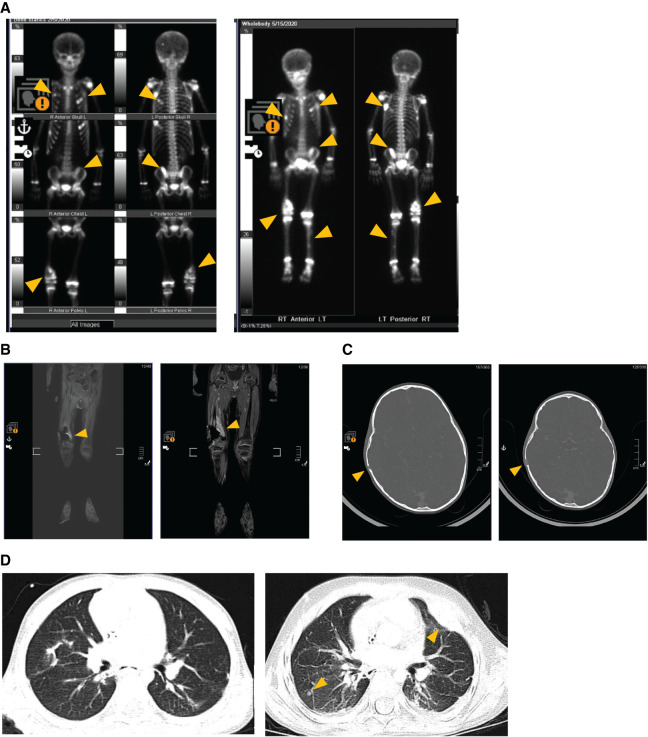
Radiology images showing progression of lung and numerous skeletal metastases over a 16 wk period while enrolled on clinical trial ADVL1615 (pevonedistat, irinotecan, and temozolomide). Left panel images represent metastatic sites at the time of trial enrollment. Right panel images show disease progression at all sites after four treatment cycles. (A) Progression of rib, pelvic, femur, and tibia metastases. (B) Progression of the right femur metastasis. (C) Growth in the skull lesion. (D) Progression of the lung metastases.
A surveillance CT scan (chest, abdomen, and pelvis) 4 wk later showed new metastatic involvement of multiple bones of the skull, ribs, and extremities. The patient was given palliative radiation therapy to selected lesions and started on pazopanib, which had demonstrated activity in other soft-tissue sarcomas.
Four months from first recurrence and 1 month after completing radiation, CT scans of the chest showed a new nodular density in the right upper lobe, and pazopanib was discontinued. Ahead of planned resection of the lung nodule and following appropriate informed consent discussions, the patient was enrolled on an IRB-approved institutional registry study designed to identify targetable mutations in recurrent pediatric tumors via RNA sequencing and pathway analysis. RNA sequencing of the lung metastasis showed high expression of CDK1 and NOTCH3. Chemotherapy was changed to trabectedin based on this agent's activity in other translocation-related sarcomas. Further disease progression after three cycles of trabectedin prompted treatment with gemcitabine and docetaxel, another regime known to be active in many soft-tissue sarcomas. A subsequent positron emission tomography (PET) scan showed continued progression in bony sites (Fig. 2).
At this point the patient was enrolled on a phase I Children's Oncology Group clinical trial (ADVL1615) and completed four treatment cycles of pevonedistat (a NEDD8 activating enzyme inhibitor) with irinotecan and temozolomide. Disease progression in several bony lesions was observed, and treatment was discontinued. Persistent postchemotherapy nausea, vomiting, headaches, and retro-orbital pain led to a follow-up MRI that identified new leptomeningeal spread of disease. The patient then underwent 5 d of palliative whole-brain radiation. Following radiation, the patient started treatment with compassionate use I-131 omburtamab, a radio-labeled antibody against B78H9/CD276 that is widely expressed on most sarcomas with some benefit in patients with central nervous system (CNS) neuroblastoma (Kramer et al. 2017) and administered intraventricularly through an Ommaya reservoir. The patient tolerated two infusions of I-131 omburtamab; however, roughly 6 wk after the second infusion, the child passed away from progression of CNS disease.
Literature Review
A comprehensive literature search led to 59 papers published between 1995 and 2021 describing 230 cases of pure SEF (Meis-Kindblom et al. 1995; Reid et al. 1996; Christensen et al. 1997; Eyden et al. 1998; Gisselsson et al. 1998; Bilsky et al. 2000; Donner et al. 2000; Antonescu et al. 2001; Arya et al. 2001; Boudová et al. 2001; Hanson et al. 2001; Jiao et al. 2002; Hindermann and Katenkamp 2003; Chow et al. 2004; Ogose et al. 2004; Battiata and Casler 2005; Choi et al. 2007; Folk et al. 2007; Sassi et al. 2008; Smith et al. 2008; Kanno et al. 2009; Tomimaru et al. 2009; Grunewald et al. 2010; Tsuchido et al. 2010; Elkins and Wakely 2011; Rekhi et al. 2011; Leisibach et al. 2012; Wang et al. 2012b; Bai et al. 2013; Kilaikode et al. 2013; Monarca et al. 2013; Puerta Roldán et al. 2013; Stockman et al. 2014; Wojcik et al. 2014; Argani et al. 2015; Ertoy Baydar et al. 2015; Ohlmann et al. 2015; Righi et al. 2015; Bell et al. 2016; Xu et al. 2016; Arbajian et al. 2017; Carvalho et al. 2017; Dewaele et al. 2017; Imada et al. 2017; Patterson et al. 2017; Dong et al. 2018; Laliberte et al. 2018; Popli et al. 2018; Zhang and Chou 2018; Perez et al. 2019; Shenoy et al. 2019; Zhang et al. 2019; Ding et al. 2020; Kao et al. 2020; Kosemehmetoglu et al. 2020; Xia et al. 2020; Warmke and Meis 2021). Patient ages ranged from 9 to 87 yr. Primary and metastatic sites are shown in Figure 3. Primary tumor locations were reported most commonly in the lower limb or limb girdle (28.3%) and the trunk (18.7%). Other primary tumor sites included the brain, head and neck, soft tissue, lung or pleura, pancreas, kidney, bone, upper limb or limb girdle, and liver. The most common site of metastasis was the lung or pleura (42.1%). Interestingly, although rare as a primary site, the second most common metastatic site was bone (28.6%). Other sites of metastasis were rare but included the trunk, upper limb or limb girdle, lower limb or limb girdle, pancreas, lymph nodes, brain, heart, liver, kidney, and soft tissues.
Figure 3.
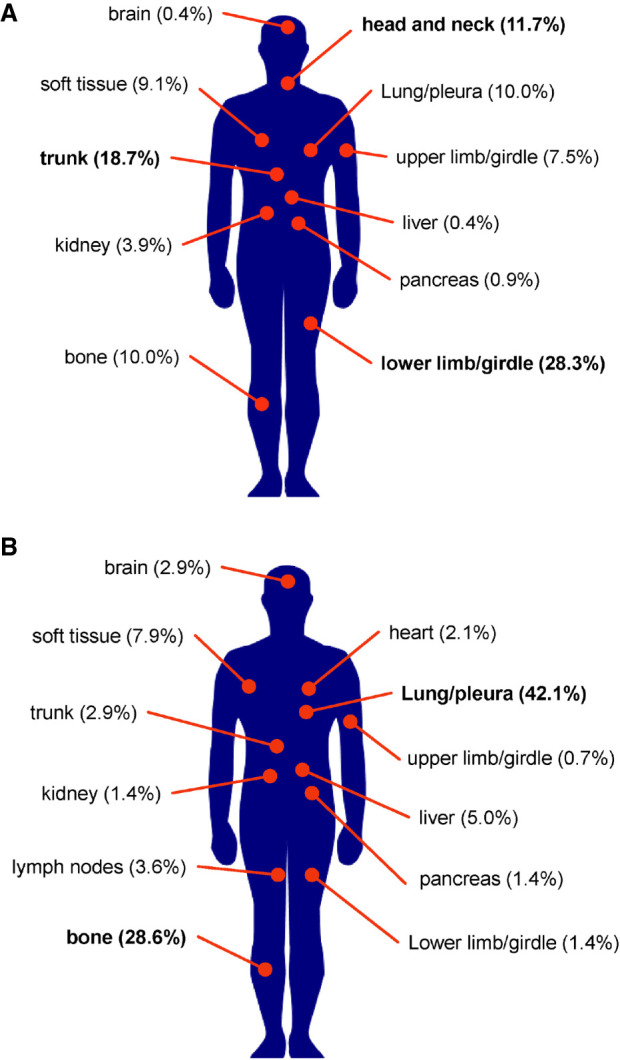
Locations of primary and metastatic SEF tumors. Altogether, 230 cases reported in the literature from 1995 to 2021 were reviewed. (A) Eleven primary tumor sites were reported, the most common being the lower limb or limb girdle (28.3%) and the trunk (18.7%). (B) Eighty-eight patients reported metastasis to 12 sites, the most common being the lung (42.1%) and bone (28.6%).
Genomic Analyses
The patient described in this report had multiple sequencing analyses, allowing for comparisons between different metastatic sites and across the timeline of disease progression (Table 2). Tissues resected from the primary kidney tumor, a lung metastasis, a bone metastasis, and a blood-matched patient normal sample were DNA- and RNA-sequenced and analyzed. Whole-genome, whole-exome, and deep-transcriptome sequencing was performed. This sequencing data set was analyzed for both pathogenic germline alterations and actionable (druggable) targets based on the presence of somatic missense mutations altering the function of the original gene, evidence of copy-number gain, and level of gene expression (Table 3; Supplemental Tables S1–S5). Circos plot representations of genomic alterations discovered within each data set are provided in Supplemental Figures S1–S4. Detailed fusion calling data is provided in Supplemental Tables S6 and S7.
Table 2.
Location and type of tissue used for various sequencing analyses
| Sample type | Post Dx (months) | Sequencing type | MS status | TMB |
|---|---|---|---|---|
| Primary kidney biopsy | 0 | DNA WES | Stable | 39 Muts |
| Primary kidney resection | 5 | DNA WES, WGS, RNA, NM DNA | Stable | 39 Muts |
| Femur metastasis | 11 | DNA WES | Unstable | 31 Muts |
| Lung metastasis | 13 | DNA WES, WGS, RNA, NM DNA | Stable | 30 Muts |
| Normal blood | N/A | DNA WGS | N/A | N/A |
Microsatellite instability status is a measure of genetic hypermutability due to impaired DNA mismatch repair. Tumor mutation burden is a measure of total mutations found in the DNA of cancer cells.
(WES) Whole-exome sequencing, (WGS) whole-genome sequencing, (NM) matched normal, (MS) microsatellite instability status, (TMB) tumor mutation burden, (Muts) mutations, (N/A) not available.
Table 3.
Compiled common sequencing results from all samples
| Gene (# of analyses) | Variant type | Transcript ID | HGVS DNA | HGVS protein | PMID reference | CNV (log2) | Median TPM | Samples |
|---|---|---|---|---|---|---|---|---|
| EWSR1–CREB3L1 (7) | Fusion | Kidney Bx, primary kidney, lung relapse | ||||||
| CREB3L1 (4) | ENST00000527342.1 | Loss, gain | 181.3 | Primary kidney, lung relapse | ||||
| EWSR1 (4) | ENST00000331029.11 | c.*1093T > A | Loss, gain | 252.7 | Primary kidney, lung relapse | |||
| CD24 (3) | ENST00000606017.1 | 290 | Primary kidney, lung relapse | |||||
| BCR (2) | ENST00000305877.12 | c.3012 + 2059delG | Loss | 43.7 | Primary kidney, lung relapse | |||
| CARS (2) | ENST00000278224.13 | Loss | 23.7 | Primary kidney, lung relapse | ||||
| EP300 (2) | ENST00000263253.8 | c.95-8921delT | Loss | 128.3 | Primary kidney, lung relapse | |||
| EXT2 (2) | ENST00000343631.3 | Loss | 47.6 | Primary kidney, Lung relapse | ||||
| FANCF (2) | ENST00000327470.5 | Loss | 2.5 | Primary kidney, lung relapse | ||||
| GON4L (2) | Substitution—missense | ENST00000271883.9 | c.4615C > G | Q1539E | 116.3 | Primary kidney, lung relapse | ||
| HCAR3 (2) | Substitution—missense | ENST00000528880.2 | c.1127C > G | A376G | 0.65 | Primary kidney, lung relapse | ||
| HRAS (2) | ENST00000311189.7 | Loss | 34.5 | Primary kidney, lung relapse | ||||
| IGL (2) | Loss | Primary kidney, lung relapse | ||||||
| IRF1 (2) | ENST00000245414.8 | V175A | Kidney Bx, primary kidney | |||||
| LMO1 (2) | ENST00000335790.7 | Loss | 0 | Primary kidney, lung relapse | ||||
| LMO2 (2) | ENST00000290246.10 | Loss | 15.58 | Primary kidney, lung relapse | ||||
| MKL1 (2) | Substitution—missense | ENST00000355630.7 | c.902G > A | Loss | 39.26 | Primary kidney, lung relapse | ||
| MLL2 (2) | P2210L | Kidney Bx, primary kidney | ||||||
| MYH9 (2) | ENST00000216181.10 | c.*5992G > A | Loss | 636.5 | Primary kidney, lung relapse | |||
| MYOD1 (2) | ENST00000250003.3 | Loss | 0 | Primary kidney, lung relapse | ||||
| NF2 (2) | ENST00000334961.11 | c.199-7820G > A | Loss | 45.47 | Primary kidney, Lung relapse | |||
| NOTCH1 (2) | Splice site (subclonal) | ENST00000277541.7 | c. 5167 + 1G > A | 21798893 | 16.48 | Kidney Bx, lung relapse | ||
| NTRK1 (2) | ENST00000358660.3 | P171S | Kidney Bx, primary kidney | |||||
| NUP98 (2) | ENST00000324932.11 | Loss | 43.1 | Primary kidney, lung relapse | ||||
| PATZ1 (2) | ENST00000215919.3 | Loss | 25.89 | Primary kidney, lung relapse | ||||
| PDGFB (2) | ENST00000331163.10 | Loss | 20.85 | Primary kidney, Lung relapse | ||||
| PLCG2 (2) | ENST00000359376.7 | K1019E | Kidney Bx, primary kidney | |||||
| PRDM1 (2) | ENST00000369089.3 | K532Q | Kidney Bx, primary kidney | |||||
| ROS1 (2) | ENST00000368507.7 | R2269* | 25186949 | Kidney Bx, primary kidney | ||||
| SEPT5 (2) | ENST00000383045.7 | Loss | 102.5 | Primary kidney, lung relapse | ||||
| SIGLEC10 (2) | Substitution—missense | ENST00000339313.9 | c.430C > A | Q144K | 26925973 | 25.95 | Primary kidney, lung relapse | |
| SPARC (2) | ENST00000231061.8 | Gain | 20457 | Primary kidney, lung relapse | ||||
| TMEM30A (2) | Insertion—frameshift | ENST00000230461.10 | c.401dupA | Y134fs | 30.46 | Kidney Bx, primary kidney | ||
| WT1 (2) | ENST00000332351.7 | c.1002-8172dupT | Loss | 0.89 | Primary kidney, lung relapse |
Of the 256 total genetic events observed, 33 were found to be common to multiple samples. The human reference genome used was GRCh38.
(HGVS) Human Genome Variation Society, (PMID) PubMed ID, (CNV) copy-number variation, (TPM) transcripts per million, (Bx) biopsy.
The most common/recurrent genetic change was the EWSR1–CREB3L1 fusion. Studies by Mertens et al. (Arbajian et al. 2017) suggest that this fusion alone can dramatically alter the normal fibroblast transcriptional program, up-regulating CD24 and CD276 among other genes. Indeed, CD24 and CD276 were clearly up-regulated across this patient's sequencing samples. The average CD276 transcripts per million (TPM) was 178, compared to a normal CD276 TPM average expression of 19.9 (as determined by the Genome Tissue Expression [GTEx] project). Average CD24 TPM across this patient's sequencing samples was 170. For comparison, 12 epithelioid sarcoma patient samples and cell lines were probed for CD24 expression levels, resulting in an average CD24 TPM level of 34.7 and a median TPM level of 3.7 (data not shown). SIGLEC10 was of interest as a known cell–cell interactor for CD24 (Barkal et al. 2019), as well as NOTCH1 whose signaling pathway has been observed to be activated when CD24 is expressed (Tang et al. 2018). The TP53 tumor suppressor gene was mutated in the femur metastasis sample, and TP53-axis genes MDM2, MDM4, and CDKN2A (ARF) were overexpressed in the primary tumor and lung metastasis compared to healthy comparison tissue.
Primary Cell Culture
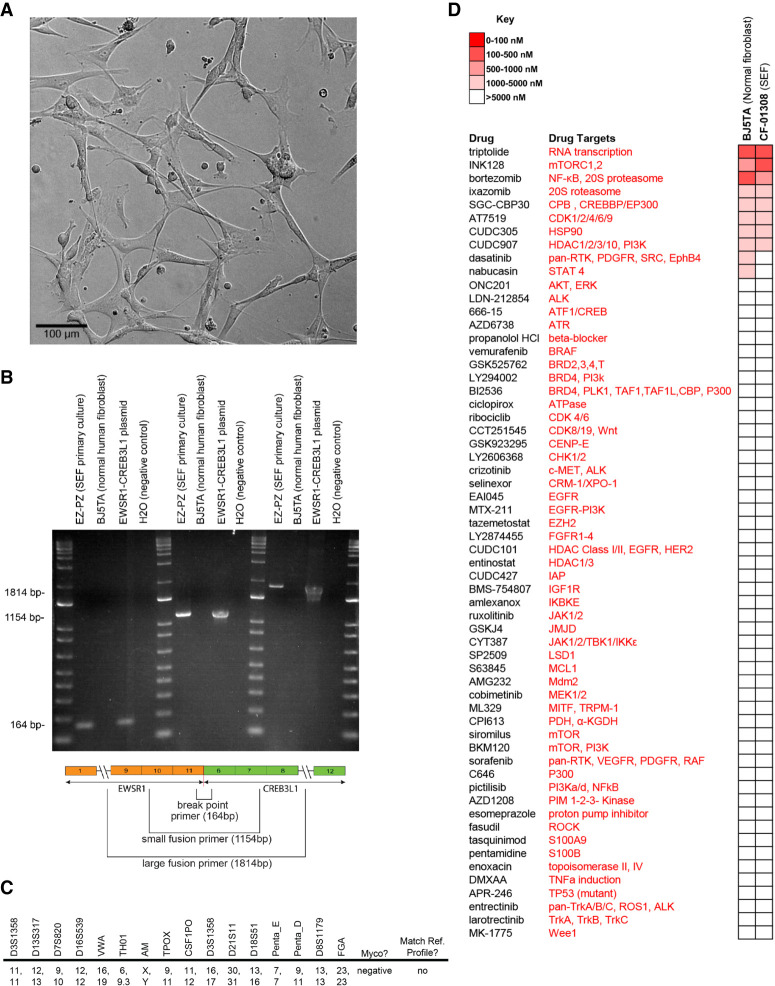
Postmortem tumor tissue samples were collected by rapid autopsy. A small section of tumor tissue from the left lung was minced by hand and digested with a Miltenyi Gentle Macs tumor dissociation system. Resultant cultures were maintained in Gibco's AmnioMAX C-100 complete medium, a specialized media developed for the short-term culture of human amniotic fluid cells. Cell cultures retained a fibroblastic morphology displaying elongated (spindle-shaped) processes extending out from the ends of cell bodies as well as flattened and oval nuclei (Fig. 4A). To distinguish this cell culture from normal fibroblasts, we performed polymerase chain reaction (PCR) analysis to detect the EWSR1–CREB3L1 gene fusion (Fig. 4B), short tandem repeat (STR) (Fig. 4C), and drug screen validations (Fig. 4D).
Figure 4.
SEF cell culture EZ-PZ. (A) Brightfield confocal image of SEF cell culture EZ-PZ at 10× magnification. Scale bar, 100 µM. (B) Polymerase chain reaction (PCR) indicating the presence of a EWSR1–CREB3L1 gene fusion in the EZ-PZ cell culture. BJ5TA is a normal human fibroblast cell culture that shows an absence of the EWSR1–CREB3L1 fusion. Three primer sets captured the fusion breakpoint, the majority and the entirety of the gene fusion. (C) Baseline short tandem repeat (STR) analysis of the cell culture. Results did not match to any known cell lines. The cell culture was free of mycoplasma. (D) Drug screen results from the EZ-PZ and BJ5TA cell cultures. Heatmap indicates IC50 values and shows a general increase in resistance and sensitivity of the SEF cell culture to TORC 1/2 inhibition.
DISCUSSION
SEF lacks a specific phenotypic differentiation and immunohistochemical signature and shares clinical and morphological features with several differential cancer types, challenging diagnosis in the absence of genetic data. SEF most commonly occurs in the lower limb or limb girdle of middle-aged or elderly adults, yet also presents in a wide range of alternative anatomical sites and across a wide range of ages (Meis-Kindblom et al. 1995; Antonescu et al. 2001; Wang et al. 2012b; Kao et al. 2020). The case presented here represents a primary SEF tumor of the kidney in conjunction with multiple bony metastases discovered in a very young patient. This case emphasizes the importance of considering an SEF diagnosis for patients of all ages and tumor locations.
Multiple sequencing analyses allowed for a complete modeling of the genomic landscape of this particular SEF patient. Given the patient's young age, one could hypothesize that the genetic abnormalities presented here are most likely disease-related and less likely to be from potentially confounding factors often observed in adults, such as lifestyle or long-term exposure to carcinogens. Common to all analyses was the EWSR1–CREB3L1 fusion. A list of genes that were abnormal across different sequencing analyses is shown in Table 3.
CD24
The CD24 gene is primarily expressed by immune cells and has been shown to be overexpressed in a variety of human cancers including SEF, but not on human erythrocytes. Arbajian et al. (2017) showed that increased expression of CD24 in SEF is a direct result of the EWSR1–CREB3L1 gene fusion event. As expected, CD24 gene expression was detected in this patient's transcriptome sequencing. CD24 is a highly glycosylated GPI-anchored cell surface molecule that resembles cell surface mucin (Kristiansen et al. 2004). CD24 has been associated with poor prognosis (Lee et al. 2009), is a cancer stem cell marker (Ortiz-Montero et al. 2018), and has been shown to regulate tumor cell proliferation, migration, and invasion (Baumann et al. 2005). CD24 has been suggested as a therapeutic target in several cancer types (Salnikov et al. 2013).
CD24 has multiple binding partners including P-selectin and Siglecs. In ovarian and breast cancers, the CD24-P-selectin interaction has been shown to support the rolling of tumor cells on endothelial cells and the adhesion of tumor cells to platelets and mesothelium (Aigner et al. 1998; Carroll et al. 2018), suggesting a role in metastasis. Recently, CD24 was shown to interact with SIGLEC10 on tumor associated macrophages to provide a “don't eat me” signal, leading to a lack of clearance by the immune system (Barkal et al. 2019). Barkal et al. suggested that antibodies that disrupt the interaction between CD24 and SIGLEC10 have therapeutic potential. A variety of antibody treatments have been studied to treat human disease, including the use of naked antibodies, bispecific antibodies, and conjugated antibodies. Besides antibody treatment, Sagiv et al. (2006) showed that overexpressed CD24 can be down-regulated to normal levels after short- and long-term exposure to the selective COX2 inhibitor, celecoxib (Sagiv et al. 2006).
NOTCH1
The NOTCH signaling pathway is dysregulated in the majority of human cancers. In previous studies, CD24 expression was shown to accompany NOTCH signaling pathway activation (Tang et al. 2018), and NOTCH1 inhibition has been shown to decrease CD24 transcription in esophageal adenocarcinoma (Wang et al. 2014b). In the patient presented in this report, a NOTCH1 splice site variant (c.5167 + 1G > A) was observed in the primary kidney and metastatic lung tumors, possibly linked to the observed CD24 overexpression. The c.5167 + 1G > A variant is an intronic substitution listed as pathogenic (FATHMM prediction score = 0.99) in the Catalog of Somatic Mutations in Cancer (COSMIC) database. NOTCH1 signaling has been implicated in a wide range of oncogenic activities including tumor growth, survival, and metastasis and has been shown to be important for cancer stem cell self-renewal, differentiation, proliferation, survival, and migration (Wang et al. 2012a; Venkatesh et al. 2018). In this patient's lung metastasis and primary kidney samples, numerous NOTCH1 target genes were found to be overexpressed compared to normal organ-specific tissue collected within the GTEx project, most notably CyclinD1 (CCND1), which was 45-fold overexpressed in the lung metastasis and 85-fold overexpressed in the primary kidney tumor (Table 4). Cyclin D1 is a regulator of CDK4/6, which in turn regulates cell cycle transition from G1 to S phase. Overexpression of Cyclin D1 leads to rapid cellular growth (Qie and Diehl 2016). NOTCH signaling has been shown to positively regulate MTOR pathway activity, and several NOTCH target genes within the PI3K–AKT-mTOR pathway were overexpressed in this patient's tumor samples (PIK3R1, PIK3R2, AKT1, and MTOR). As previously noted, SEF tumors display a propensity to metastasize, oftentimes progressing to widely metastatic disease and resulting in poor patient outcomes. NOTCH1 target gene HEY1 has been shown to play a fundamental role in tumor vasculature development and angiogenesis (Wang et al. 2014a), and was 53-fold overexpressed in this patient's kidney tumor (data not shown) compared to normal GTEx kidney tissue. No overexpression was observed in the lung metastasis sample. HEY1 paralog genes HEY2 and HES1 were overexpressed in both kidney and lung samples. Within the endothelium, cross-talk between NOTCH and VEGF was shown to be necessary for productive tumor angiogenesis (Gu et al. 2012), and interruption of this interaction resulted in decreased tumor perfusion and enhanced tumor growth inhibition in animal models (Miles et al. 2014). Multiple targets within the NOTCH signaling pathway are considered druggable with small molecule inhibitors, including inhibitors of NOTCH itself, CDK4/6, PI3K, and mTOR. Clinical trials targeting NOTCH are currently underway using γ secretase inhibitors, NOTCH receptor antibody treatment, and NOTCH transcription complex inhibitors (Moore et al. 2020).
Table 4.
NOTCH1 target gene expression levels reported as transcripts per million (TPM)
| Gene | Lung tumor | Lung normal | Fold change (lung) | Kidney tumor | Kidney normal | Fold change (kidney) |
|---|---|---|---|---|---|---|
| HEY2 | 68.24 | 5.4 | 12.64 | 66.93 | 2.287 | 29.27 |
| HES1 | 236.15 | 72.71 | 3.25 | 440.28 | 66.71 | 6.6 |
| CCND1 | 2855.08 | 63.51 | 44.95 | 1951.83 | 23.08 | 84.57 |
| GATA3 | 12.1 | 6.22 | 1.95 | 99.06 | 22.15 | 4.47 |
| MYC | 179.66 | 55.01 | 3.27 | 51.42 | 7.855 | 6.55 |
| PIK3R1 | 217.46 | 44.53 | 4.88 | 165.19 | 9.379 | 17.61 |
| PIK3R2 | 95.57 | 22.27 | 4.29 | 107.66 | 20.85 | 5.16 |
| AKT1 | 393.74 | 80.7 | 4.88 | 461.23 | 27.57 | 16.73 |
| MTOR | 27.99 | 15.39 | 1.82 | 49.52 | 8.864 | 5.59 |
| BCL2 | 40.65 | 6.354 | 6.4 | 55.54 | 4.632 | 11.99 |
| TCF3 | 96.49 | 24.83 | 3.89 | 177.55 | 13.24 | 13.41 |
| NOTCH3 | 268.34 | 79.77 | 3.36 | 155.75 | 18.13 | 8.59 |
| EP300 | 119.55 | 33.78 | 3.54 | 137.1 | 11.53 | 11.89 |
Analysis was performed by Omics Data Automation and Children's Cancer Therapy Development Institute. Primary kidney and lung metastasis TPM levels were compared to normal kidney and lung tissue TPM's (GTEx), respectively.
TP53
The TP53 tumor suppressor gene has been shown to be involved in a variety of human cancers (Hollstein et al. 1994; Levine 1997). TP53 gene axis members include MDM2, MDM4, and CDKN2A (ARF) genes among others (Ozenne et al. 2010). MDM2 overexpression has been observed previously in SEF patients (Jiao et al. 2002). In the patient described in this report, TP53 dysregulation and MDM2 overexpression were observed in the primary tumor and multiple metastatic samples (Table 5). Two of the TP53 mutations (p.Pro301fs and p.Pro72Arg) observed in this patient's femur metastasis sample have been characterized as pathogenic (Malapelle et al. 2017) and associated with a poor drug response (Kim et al. 2009; Khrunin et al. 2010), respectively. When compared to GTEx normal tissue, the lung metastasis sample showed a four- to fivefold greater MDM2 expression and the kidney tumor showed a 15-fold greater expression. MDM2 and TP53 form an autoregulatory feedback loop where TP53 regulates MDM2 transcription and MDM2 regulates TP53 activity (Wu et al. 1993). MDM2 ubiquitinates TP53 for proteasomal degradation and works as a “brake” against normal TP53 tumor suppressor activities (Picksley and Lane 1993; Haupt et al. 1997; Honda et al. 1997). Increased MDM2 levels therefore result in decreased wild-type TP53 tumor suppressive activities. Several MDM2 inhibitors have advanced to clinical trials and could be considered as a potential therapeutic approach for SEF patients with MDM2 overexpression.
Table 5.
TP53 axis sequencing results showing mutations, copy-number variations, and expression levels of TP53 axis genes TP53, MDM2/4, and CDKN2A (ARF)
| Gene | Chr | Variant | Transcript ID | dbSNP ID | ClinVar ID | HGVS | HGVS protein | CNV (log2) | TPM | TMB (Muts) | MS | Sample type | Post Dx (months) | Data source | Analysis | Data type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDKN2A (ARF) | 9 | ENST00000304494.9 | 0.0176 | 10 | 39 | S | Primary kidney | 5 | NYG | ODA | WGS, RNA, NM | |||||
| MDM2 | 12 | ENST00000258148.11 | 0.1897 | 117 | 39 | S | Primary kidney | 5 | NYG | ODA | WGS, RNA, NM | |||||
| MDM4 | 1 | ENST00000367179.7 | 0.3419 | 166 | 39 | S | Primary kidney | 5 | NYG | ODA | WGS, RNA, NM | |||||
| TP53 | 17 | ENST00000269305.8 | 0.2565 | 121 | 39 | S | Primary kidney | 5 | NYG | ODA | WGS, RNA, NM | |||||
| CDKN2A (ARF) | 9 | Frameshift | ENST00000498124.1 | c.460delA | p.Met154fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| CDKN2A (ARF) | 9 | Frameshift | ENST00000304494.9 | c.64_65delCG | p.Arg22fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| CDKN2A (ARF) | 9 | Frameshift | ENST00000530628.2 | c.115delG | p.Ala39fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000258148.11 | c.730_731insT | p.Glu244fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000258149.9 | c.895_896insT | p.Glu299fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000299252.8 | c.367_368insT | p.Glu123fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000350057.9 | c.802_803insT | p.Glu268fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000360430.6 | c.292_293insT | p.Glu98fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000393415.7 | c.736_737insT | p.Glu246fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000258148.11 | c.1147delC | p.Asn384fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000258149.9 | c.1312delC | p.Asn439fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000299252.8 | c.784delC | p.Asn263fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000348801.6 | c.616delC | p.Asn207fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000350057.9 | c.1219delC | p.Asn408fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000360430.6 | c.709delC | p.Asn238fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000393410.5 | c.550delC | p.Asn185fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000393413.7 | c.475delC | p.Asn160fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM2 | 12 | Frameshift | ENST00000517852.5 | c.211delC | p.Asn72fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000367180.5 | c.90dupA | p.Leu31fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000367179.7 | c.377delT | p.Leu126fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000367182.7 | c.722delT | p.Leu241fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000614459.4 | c.428delT | p.Leu143fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000367179.7 | c.588dupA | p.Phe197fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000367182.7 | c.933dupA | p.Phe312fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000444261.1 | c.267dupA | p.Phe90fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000454264.6 | c.783dupA | p.Phe262fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000612738.4 | c.264dupA | p.Phe89fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| MDM4 | 1 | Frameshift | ENST00000614459.4 | c.639dupA | p.Phe214fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Missense | ENST00000269305.8 | rs1042522 | 12351 | c.215C > G | p.Pro72Arg | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||
| TP53 | 17 | Missense | ENST00000610292.4 | rs1042522 | 12351 | c.98C > G | p.Pro33Arg | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||
| TP53 | 17 | Frameshift | ENST00000269305.8 | c.140delC | p.Pro47fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Frameshift | ENST00000610292.4 | c.23delC | p.Pro8fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Frameshift | ENST00000269305.8 | rs876660726 | 233908 | c.902delC | p.Pro301fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||
| TP53 | 17 | Frameshift | ENST00000504290.5 | rs876660726 | 233908 | c.506delC | p.Pro169fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||
| TP53 | 17 | Frameshift | ENST00000610292.4 | rs876660726 | 233908 | c.785delC | p.Pro262fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||
| TP53 | 17 | Frameshift | ENST00000610623.4 | rs876660726 | 233908 | c.425delC | p.Pro142fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||
| TP53 | 17 | Frameshift | ENST00000615910.4 | rs876660726 | 233908 | c.869delC | p.Pro290fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||
| TP53 | 17 | Frameshift | ENST00000269305.8 | c.636dupT | p.Arg213fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Frameshift | ENST00000504290.5 | c.240dupT | p.Arg81fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Frameshift | ENST00000514944.5 | c.357dupT | p.Arg120fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Frameshift | ENST00000610292.4 | c.519dupT | p.Arg174fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Frameshift | ENST00000615910.4 | c.603dupT | p.Arg202fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| TP53 | 17 | Frameshift | ENST00000618944.4 | c.159dupT | p.Arg54fs | 31 | US | Femur relapse | 11 | MDA | ODA | WES | ||||
| CDKN2A (ARF) | 9 | ENST00000304494.9 | -0.0481 | 14 | 30 | S | Lung relapse | 13 | NYG | ODA | WGS, RNA, NM | |||||
| CDKN2A (ARF) | 9 | ENST00000304494.9 | 13 | Lung relapse | 13 | TH | ODA | RNA | ||||||||
| MDM2 | 12 | ENST00000258148.11 | 0.129 | 144 | 30 | S | Lung relapse | 13 | NYG | ODA | WGS, RNA, NM | |||||
| MDM2 | 12 | ENST00000258148.11 | 99 | Lung relapse | 13 | TH | ODA | RNA | ||||||||
| MDM4 | 1 | ENST00000367179.7 | 0.1851 | 131 | 30 | S | Lung relapse | 13 | NYG | ODA | WGS, RNA, NM | |||||
| MDM4 | 1 | ENST00000367179.7 | 24 | Lung relapse | 13 | TH | ODA | RNA | ||||||||
| TP53 | 17 | ENST00000269305.8 | 0.1395 | 111 | 30 | S | Lung relapse | 13 | NYG | ODA | WGS, RNA, NM | |||||
| TP53 | 17 | ENST00000269305.8 | 35 | Lung relapse | 13 | TH | ODA | RNA |
The human reference genome used was GRCh38.
(NYG) New York Genome Center, (FO) Foundation One, (TH) Treehouse Childhood Cancer Initiative, (ODA) Omics Data Automation, (MS) microsatellite instability status, (S) stable, (US) unstable.
A CD24-TP53 gene axis has recently been established in prostate cancer, in which CD24 expression is associated with aggressive and metastatic disease. Interestingly, mutant KRAS-induced up-regulation of CD24 enhances prostate cancer bone metastasis (Weng et al. 2019). We note that the second most common observed metastatic site in SEF is bone, creating the intriguing hypothesis that a similar biologic mechanism underlies the propensity for bone metastasis in SEF. Studies by Wang et al. (2015) have demonstrated that CD24 can inhibit CDKN2A (ARF) binding to NPM, leading to a decrease in CDKN2A (ARF), which therefore increases MDM2 levels and decreases activity of TP53 and its downstream gene target, P21. These researchers also observed a higher rate of TP53 mutation associated with increased CD24 mRNA expression and suggest the possibility of restoring TP53 tumor-suppressor function while simultaneously disabling mutant TP53 oncogenic activity. Three out of the four most common TP53 mutants (p53R273H, p53V143A, and p53R280T) in CD24 siRNA-silenced cells were almost equally efficient at suppressing colony formation as wild-type TP53. In contrast, TP53 mutants that retained CD24 expression failed to reduce colony formation (Wang et al. 2015). Further studies by Zhang et al. (2016) confirmed the restoration of TP53 tumor-suppressive activities in CD24-silenced TP53 mutants and demonstrated a role for CD24 in tumor progression and metastasis by showing an abnormal nuclear accumulation of TP53 mutants in CD24-expressing prostate cancer cells.
Copy-Number Variation
Increased copy-number variation has been observed previously in SEF and has been proposed to explain SEF's aggressive nature as compared to the closely related LGFMS, which has a better prognosis and has been observed to display only minimal copy-number variation (Arbajian et al. 2017). The primary kidney tumor sample in this report strongly suggests loss of 10q, 11p and Chromosome 22 (Supplemental Fig. S1), concurrent with previous observations within larger cohorts of SEF patients (Arbajian et al. 2017).
Cell Culture
Tumor tissue collected at autopsy resulted in a primary cell culture with fibroblastic features (Fig. 4A), which was named “EZ-PZ.” The resultant EZ-PZ cell culture continued to grow robustly until passage 20 before growth rates declined. To distinguish EZ-PZ from normal fibroblasts, PCR, STR, and drug screen analyses were performed. PCR results showed clear bands with three different primer sets measuring (1) the fusion breakpoint, (2) a portion of the fusion, and (3) the entire fusion, indicating that the SEF primary culture EZ–PZ contains the disease-defining EWSR1–CREB3L1 gene fusion, whereas the fusion is absent from normal human fibroblast BJ5TA cells (Fig. 4B). The fusion sequence and primers used for PCR amplification are provided in Supplemental Table S8. STR results show no cross-contamination between EZ-PZ and known cell lines and establish a baseline for future testing and cell culture validation (Fig. 4C). Drug screens showed activity against EZ-PZ in eight out of 60 compounds tested, with IC50 concentrations of <5 µM (Fig. 4D). Interestingly, EZ-PZ was sensitive to INK128 (IC50 = 0.45 µM), a novel TORC1/2 inhibitor, yet was not sensitive (IC50 = >10 µM) to the other MTOR inhibitors within the screen (sirolimus and BKM120). We speculate that INK128's observed potency is due to dual targeting of both TORC1 and TORC2, and that targeting TORC1 only or MTOR + PI3K is insufficient to reduce EZ–PZ's cell viability. Three compounds within the drug screen (bortezomib, dasatinib, and nabucasin) demonstrated activity against the normal human fibroblast cell line BJ5TA but showed reduced or no activity against EZ–PZ. We suggest that the SEF cell culture generally displays a drug-resistant phenotype, an observation consistent with this patient's clinical response and that is supported by the lack of activity observed in 52 of 60 oncogenic inhibitors used within the screen.
As this case report demonstrates, SEF is an aggressive tumor that is oftentimes resistant to conventional chemotherapy and targeted therapies. SEF can occur across a wide variety of sites and age groups. SEF is a fusion-driven cancer, the most common fusion event being the EWSR1–CREB3L1 gene rearrangement. CD24, MDM2, and NOTCH1 represent potential therapeutic targets for SEF, although further study of these targets in a SEF-specific context is warranted. The EZ–PZ primary cell culture that resulted from this patient's tumor tissue should provide a study model to perform functional assays, which will shed light upon the efficacy of the proposed genetically determined therapeutic targets.
METHODS
Immunohistochemistry
Immunohistochemistry was performed with the following antibodies and dilutions: anti-CD24 antibody (1:100) (Abcam ab199140), cleaved Notch1 (1:400) (Cell Signaling Technology 4147), anti-JAGGED1 (1:100) (Santa Cruz Biotechnology sc-390177), and anti-HEY1 (1:150) (Abcam ab235173).
Genetic Sequencing
Genetic sequencing was performed at New York Genome Center, Foundation One Medicine, MD Anderson Cancer Center, and Covance by Lab Corp. DNA sequencing was performed with at least 100× coverage, paired-end reads. DNA sequencing was performed on an Ilumina HiSeq X or DNBseq. RNA sequencing was performed on an Illumina HiSeq 2500. Variant and sequencing coverage tables are presented in Supplemental Tables S4 and 5.
Genomic and Transcriptomic Analysis
Genetic sequencing analysis was performed in the following manner: whole-exome and whole-genome sequencing data were analyzed for the presence of somatic point mutations, somatic functional and structural mutations, potential germline mutations, polynucleotide insertions and deletions, and gene copy-number variation. Somatic mutations, variations, and indels were called using Genome Analysis Toolkit (GATK) Version 4.0 with strict calling criterion (Tumor logarithm of odd [TLOD)] scores >6.3). Gene copy-number variations were identified using SAMtools and VarScan2 quantified as a log ratio of tumor copy to normal copy using the GRCh38 human reference genome. RNA sequencing data were analyzed for gene expression and gene fusion events. Transcriptome data were aligned to STAR-derived human transcriptome from GRCh38 human reference genome. Normalized gene expression was quantified using RSEM. NOTCH target gene expression analysis was performed by Omics Data Automation and Children's Cancer Therapy Development Institute.
Literature Review
To determine common site locations of primary and metastatic SEF tumors, search terms “sclerosing epithelioid fibrosarcoma,” “CD24,” “EWSR1,” and “EWSR1–CREB3L1” were entered into PubMed and Google search engines. Papers that did not define SEF primary location were excluded. Of the included studies, those that mentioned metastatic disease were used to determine metastatic site locations.
Clinical Presentation
Medical history was gathered from patient medical records and discussions with the patient's family and medical oncologist. Clinical molecular diagnostic whole-genome and transcriptome sequencing was performed at the New York Genome Center as previously described (Wrzeszczynski et al. 2018).
Cell Culture Generation and Validation
Autopsy tumor samples were collected with informed consent (study # ccTDI-IRB-1). Tumor tissue was minced by hand and processed in a GentleMacs dissociator with the GentleMacs tumor dissociation kit (130-093-235 and 130-095-929, Miltenyi Biotec GmbH) according to manufacturer's protocol. Resultant cultures were maintained in AmnioMAX C-100 complete medium (Gibco, Thermo Fisher Scientific). All cell cultures were maintained in a humidified incubator at 37°C supplemented with 5% CO2. STR validation and mycoplasma testing was performed on all cell cultures. STR analysis was performed with the Promega PowerPlex16HS Assay: 15 Autosomal Loci, X/Y, with a random match probability of approximately 1 in 1.83 × 1017.
For PCR testing, DNA was collected with the PureLink Genomic DNA mini kit (Invitrogen K182002). Dye-based qPCR was performed with DreamTaq Green PCR master mix (Thermo Scientific K1081). An amount of 25 µL of master mix, forward and reverse primers, nuclease-free water, and 2–4 µL of DNA for each sample was made up on ice and then loaded onto a GeneAmp PCR System 9700 (Applied Biosystems) and run for 35 cycles with an annealing temperature of 60°C. Samples were then loaded into a 2% agarose gel and placed on an electrophoresis apparatus for 90 min at 155 V. To determine resultant band size, a 1 Kb Plus DNA ladder (Invitrogen 10787018) was loaded onto the gel with the samples. After electrophoresis, the gel was placed in ethidium bromide (ETBR) and placed on a rocker for 10 min at medium speed, then washed with TAE buffer for 5 min. Bands were visualized with a Model SA-1000 (red) Personal Gel Imaging System SR737 (Alpha Innotech).
Chemical screens were conducted using an investigator-selected 60-agent drug screen. Each agent was tested in triplicate at four dosage points. Initial drug stocks were diluted in appropriate solvents, then plated onto Nunc 384-Well Polystyrene White Microplates (Thermo Fisher Scientific 164610) with DMSO and media-only controls. Two thousand cells/well were added to the drug plates with a Multi-Flo liquid dispenser (BioTek), then placed in a humidified incubator for 72 h at 37°C supplemented with 5% CO2. Cell viability (ATP) was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega G7573) following manufacturer's protocol. Output was read with a Synergy HT plate reader (BioTek).
ADDITIONAL INFORMATION
Data Deposition and Access
Sequencing data within this paper is deposited in the European Genome-phenome Archive (EGA; https://ega-archive.org/). The study accession number is EGAS00001005214; the data set accession number is EGAD00001007515.
Ethics Statement
All human tissue samples were acquired through the Cancer Registry for Familial and Sporadic Tumors (CuRe-FAST) tumor banking program. All patients enrolled in CuRe-FAST provided written informed consent. All aspects of the study were reviewed and approved by the Children's Cancer Therapy Development Institute (cc-TDI) Institutional Review Board (Advarra, study # cc-TDI-IRB-1).
Author Contributions
A.D.W., R.P., L.M., A.W., and C.K. wrote the manuscript; S.L.S. edited the manuscript for intellectual content; K.L. and O.V. provided genetic sequencing data; H.G., K.O.W., and V.J. provided sequencing data and performed genomic analysis; M.L. and G.S. performed genomic analysis; E.B. and A.W. provided tissue samples; J.C., K.A.C., and A.W. provided patient information and case history; E.R.R. performed histopathology; and all authors approved the final version of the manuscript.
Funding
Funding for this project was provided by an anonymous private foundation and the Rucker Collier Foundation (https://www.ruckercollierfoundation.com).
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Aigner S, Ramos CL, Hafezi-Moghadam A, Lawrence MB, Friederichs J, Altevogt P, Ley K. 1998. CD24 mediates rolling of breast carcinoma cells on P-selectin. FASEB J 12: 1241–1251. 10.1096/fasebj.12.12.1241 [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Rosenblum MK, Pereira P, Nascimento AG, Woodruff JM. 2001. Sclerosing epithelioid fibrosarcoma: a study of 16 cases and confirmation of a clinicopathologically distinct tumor. Am J Surg Pathol 25: 699–709. 10.1097/00000478-200106000-00001 [DOI] [PubMed] [Google Scholar]
- Arbajian E, Puls F, Antonescu CR, Amary F, Sciot R, Debiec-Rychter M, Sumathi VP, Järås M, Magnusson L, Nilsson J, et al. 2017. In-depth genetic analysis of sclerosing epithelioid fibrosarcoma reveals recurrent genomic alterations and potential treatment targets. Clin Cancer Res 23: 7426–7434. 10.1158/1078-0432.CCR-17-1856 [DOI] [PubMed] [Google Scholar]
- Argani P, Lewin JR, Edmonds P, Netto GJ, Prieto-Granada C, Zhang L, Jungbluth AA, Antonescu CR. 2015. Primary renal sclerosing epithelioid fibrosarcoma: report of 2 cases with EWSR1-CREB3L1 gene fusion. Am J Surg Pathol 39: 365–373. 10.1097/PAS.0000000000000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya M, Garcia-Montes F, Patel HR, Emberton M, Mundy AR. 2001. A rare tumour in the pelvis presenting with lower urinary symptoms: ‘sclerosing epithelioid fibrosarcoma’. Eur J Surg Oncol 27: 121–122. 10.1053/ejso.2000.1023 [DOI] [PubMed] [Google Scholar]
- Bai S, Jhala N, Adsay NV, Wei S. 2013. Sclerosing epithelioid fibrosarcoma of the pancreas. Ann Diagn Pathol 17: 214–216. 10.1016/j.anndiagpath.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, Krishnan V, Hatakeyama J, Dorigo O, Barkal LJ, et al. 2019. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572: 392–396. 10.1038/s41586-019-1456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiata AP, Casler J. 2005. Sclerosing Epithelioid fibrosarcoma: a case report. Ann Otol Rhinol Laryngol 114: 87–89. 10.1177/000348940511400201 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, Yagita H, Sleeman JP. 2005. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res 65: 10783–10793. 10.1158/0008-5472.CAN-05-0619 [DOI] [PubMed] [Google Scholar]
- Bell MD, Brimo F, Jung S, Aprikian AG. 2016. Sclerosing epithelioid fibrosarcoma metastasizing to the penile shaft. Can Urol Assoc J 10: E398–E400. 10.5489/cuaj.3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsky MH, Schefler AC, Sandberg DI, Dunkel IJ, Rosenblum MK. 2000. Sclerosing epithelioid fibrosarcomas involving the neuraxis: report of three cases. Neurosurgery 47: 956–959. 10.1097/00006123-200010000-00031 [DOI] [PubMed] [Google Scholar]
- Boudová L, Michal M, Kinkor Z, Bencík V, Husek K, Mukensnabl P, Rousarová M. 2001. Sclerosing epithelioid fibrosarcoma. Cesk Patol 37: 158–162. [PubMed] [Google Scholar]
- Carroll MJ, Fogg KC, Patel HA, Krause HB, Mancha AS, Patankar MS, Weisman PS, Barroilhet L, Kreeger PK. 2018. Alternatively-activated macrophages upregulate mesothelial expression of P-selectin to enhance adhesion of ovarian cancer cells. Cancer Res 78: 3560–3573. 10.1158/0008-5472.CAN-17-3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho E, Bonomi DO, Pinho AJM, Salles PGO, Vieira HC. 2017. Primary sclerosing epithelioid fibrosarcoma of the pleura. J Bras Pneumol 43: 490–491. 10.1590/s1806-37562017000000198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew W, Benson C, Thway K, Hayes A, Miah A, Zaidi S, Lee ATJ, Messiou C, Fisher C, van der Graaf WT, et al. 2018. Clinical Characteristics and efficacy of chemotherapy in sclerosing epithelioid fibrosarcoma. Med Oncol 35: 138. 10.1007/s12032-018-1192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HY, Kwon NS, Lee SJ, Lee DK, Park K, Lee CW, Jo MK. 2007. Sclerosing epithelioid fibrosarcoma of the kidney. Korean J Urol 48: 986–989. 10.4111/kju.2007.48.9.986 [DOI] [Google Scholar]
- Chow LT, Lui YH, Kumta SM, Allen PW. 2004. Primary sclerosing epithelioid fibrosarcoma of the sacrum: a case report and review of the literature. J Clin Pathol 57: 90–94. 10.1136/jcp.57.1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DR, Ramsamooj R, Gilbert TJ. 1997. Sclerosing epithelioid fibrosarcoma: short T2 on MR imaging. Skeletal Radiol 26: 619–621. 10.1007/s002560050297 [DOI] [PubMed] [Google Scholar]
- Dewaele B, Libbrecht L, Levy G, Brichard B, Vanspauwen V, Sciot R, Debiec-Rychter M. 2017. A novel EWS-CREB3L3 gene fusion in a mesenteric sclerosing epithelioid fibrosarcoma. Genes Chromosomes Cancer 56: 695–699. 10.1002/gcc.22474 [DOI] [PubMed] [Google Scholar]
- Ding Q, Shao X, Liu X, Fu Y, Huang F, Wang C. 2020. Case report: radiological features of sclerosing epithelioid fibrosarcoma in the right fibula. Front Oncol 10: 603127. 10.3389/fonc.2020.603127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Quade BJ, Dal Cin P, Jo VY. 2018. Expanding the spectrum of translocations in sclerosing epithelioid fibrosarcoma: a new case with EWSR1-CREB3L3 fusion. Genes Chromosomes Cancer 57: 675–677. 10.1002/gcc.22677 [DOI] [PubMed] [Google Scholar]
- Donner LR, Clawson K, Dobin SM. 2000. Sclerosing epithelioid fibrosarcoma: a cytogenetic, immunohistochemical, and ultrastructural study of an unusual histological variant. Cancer Genet Cytogenet 119: 127–131. 10.1016/S0165-4608(99)00225-3 [DOI] [PubMed] [Google Scholar]
- Doyle LA, Wang WL, Dal Cin P, Lopez-Terrada D, Mertens F, Lazar AJ, Fletcher CD, Hornick JL. 2012. MUC4 is a sensitive and extremely useful marker for sclerosing epithelioid fibrosarcoma: association with FUS gene rearrangement. Am J Surg Pathol 36: 1444–1451. 10.1097/PAS.0b013e3182562bf8 [DOI] [PubMed] [Google Scholar]
- Elkins CT, Wakely PE JR. 2011. Sclerosing epithelioid fibrosarcoma of the oral cavity. Head Neck Pathol 5: 428–431. 10.1007/s12105-011-0277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertoy Baydar D, Kosemehmetoglu K, Aydin O, Bridge JA, Buyukeren B, Aki FT. 2015. Primary sclerosing epithelioid fibrosarcoma of kidney with variant histomorphologic features: report of 2 cases and review of the literature. Diagn Pathol 10: 186. 10.1186/s13000-015-0420-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyden BP, Manson C, Banerjee SS, Roberts IS, Harris M. 1998. Sclerosing epithelioid fibrosarcoma: a study of five cases emphasizing diagnostic criteria. Histopathology 33: 354–360. 10.1046/j.1365-2559.1998.00530.x [DOI] [PubMed] [Google Scholar]
- Folk GS, Williams SB, Foss RB, Fanburg-Smith JC. 2007. Oral and maxillofacial sclerosing epithelioid fibrosarcoma: report of five cases. Head Neck Pathol 1: 13–20. 10.1007/s12105-007-0002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Andreasson P, Meis-Kindblom JM, Kindblom LG, Mertens F, Mandahl N. 1998. Amplification of 12q13 and 12q15 sequences in a sclerosing epithelioid fibrosarcoma. Cancer Genet Cytogenet 107: 102–106. 10.1016/S0165-4608(98)00098-3 [DOI] [PubMed] [Google Scholar]
- Grunewald TG, von Luettichau I, Weirich G, Wawer A, Behrends U, Prodinger PM, Jundt G, Bielack SS, Gradinger R, Burdach S. 2010. Sclerosing epithelioid fibrosarcoma of the bone: a case report of high resistance to chemotherapy and a survey of the literature. Sarcoma 2010: 431627. 10.1155/2010/431627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JW, Rizzo P, Pannuti A, Golde T, Osborne B, Miele L. 2012. Notch signals in the endothelium and cancer “stem-like” cells: opportunities for cancer therapy. Vasc Cell 4: 7. 10.1186/2045-824X-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson IM, Pearson JM, Eyden BP, Slawik S, Harris M. 2001. Evidence of nerve sheath differentiation and high grade morphology in sclerosing epithelioid fibrosarcoma. J Clin Pathol 54: 721–723. 10.1136/jcp.54.9.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299. 10.1038/387296a0 [DOI] [PubMed] [Google Scholar]
- Hindermann W, Katenkamp D. 2003. Sclerosing epithelioid fibrosarcoma. Pathologe 24: 103–108. 10.1007/s00292-002-0569-6 [DOI] [PubMed] [Google Scholar]
- Hollstein M, Rice K, Greenblatt MS, Soussi T, Fuchs R, Sørlie T, Hovig E, Smith-Sørensen B, Montesano R, Harris CC. 1994. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res 22: 3551–3555. [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27. 10.1016/S0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- Imada H, Yamaguchi T, Iida S, Ueda Y. 2017. A case of sclerosing epithelioid fibrosarcoma with EWSR1 translocation. AJSP Rev Rep 22: 171–174. [Google Scholar]
- Jiao YF, Nakamura S, Sugai T, Uesugi N, Habano W, Ogata M, Fujioka T. 2002. Overexpression of MDM2 in a sclerosing epithelioid fibrosarcoma: genetic, immunohistochemical and ultrastructural study of a case. Pathol Int 52: 135–140. 10.1046/j.1440-1827.2002.01326.x [DOI] [PubMed] [Google Scholar]
- Kanno A, Hatori M, Hosaka M, Kishimoto KN, Watanuki M, Watanabe M, Itoi E. 2009. Multiple bone metastasis of sclerosing epithelioid fibrosarcoma 12 years after initial surgery-increasing Ki-67 labeling index. Sarcoma 2009: 953750. 10.1155/2009/953750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YC, Lee JC, Zhang L, Sung YS, Swanson D, Hsieh TH, Liu YR, Agaram NP, Huang HY, Dickson BC, et al. 2020. Recurrent YAP1 and KMT2A gene rearrangements in a subset of MUC4-negative sclerosing epithelioid fibrosarcoma. Am J Surg Pathol 44: 368–377. 10.1097/PAS.0000000000001382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrunin AV, Moisseev A, Gorbunova V, Limborska S. 2010. Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer patients. Pharmacogenomics J 10: 54–61. 10.1038/tpj.2009.45 [DOI] [PubMed] [Google Scholar]
- Kilaikode S, Kuril S, Sedrak A, Sadanandan S. 2013. Sclerosing epithelioid fibrosarcoma of the parietal bone and adjacent meninges in an adolescent: a case report. World J Oncol 4: 255–257. 10.4021/wjon742w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Sohn SK, Chae YS, Song HS, Kwon KY, Do YR, Kim MK, Lee KH, Hyun MS, Lee WS, et al. 2009. TP53 codon 72 polymorphism associated with prognosis in patients with advanced gastric cancer treated with paclitaxel and cisplatin. Cancer Chemother Pharmacol 64: 355–360. 10.1007/s00280-008-0879-3 [DOI] [PubMed] [Google Scholar]
- Kosemehmetoglu K, Ardic F, Kilpatrick SE, Aydingoz U, Sumathi VP, Michal M. 2020. Sclerosing epithelioid fibrosarcoma of bone: morphological, immunophenotypical, and molecular findings of 9 cases. Virchows Arch 478: 767–777. 10.1007/s00428-020-02953-y [DOI] [PubMed] [Google Scholar]
- Kramer K, Kushner BH, Modak S, Pandit-Taskar N, Tomlinson U, Wolden SL, Zanzonico P, John HL, Haque S, Souweidane MM, et al. 2017. A curative approach to central nervous system metastases of neuroblastoma. J Clin Oncol 35: 10545. 10.1200/JCO.2017.35.15_suppl.10545 [DOI] [Google Scholar]
- Kristiansen G, Sammar M, Altevogt P. 2004. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol 35: 255–262. 10.1023/B:HIJO.0000032357.16261.c5 [DOI] [PubMed] [Google Scholar]
- Kurtz JL, Tan SY, Hazard FK. 2021. Sclerosing epithelioid fibrosarcoma of the kidney: first reported case in a young child. Pediatr Dev Pathol 24: 148–153. 10.1177/1093526620977738 [DOI] [PubMed] [Google Scholar]
- Laliberte C, Leong IT, Holmes H, Monteiro EA, O'Sullivan B, Dickson BC. 2018. Sclerosing epithelioid fibrosarcoma of the jaw: late recurrence from a low grade fibromyxoid sarcoma. Head Neck Pathol 12: 619–622. 10.1007/s12105-017-0879-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim SH, Lee ES, Kim YS. 2009. CD24 overexpression in cancer development and progression: a meta-analysis. Oncol Rep 22: 1149–1156. [DOI] [PubMed] [Google Scholar]
- Leisibach P, Weder W, Soltermann A, Jungraithmayr W. 2012. Primary sclerosing epithelioid fibrosarcoma of the lung in a patient with Lynch syndrome. Lung 190: 691–695. 10.1007/s00408-012-9401-0 [DOI] [PubMed] [Google Scholar]
- Levine AJ. 1997. p53, the cellular gatekeeper for growth and division. Cell 88: 323–331. 10.1016/S0092-8674(00)81871-1 [DOI] [PubMed] [Google Scholar]
- Malapelle U, Morra F, Ilardi G, Visconti R, Merolla F, Cerrato A, Napolitano V, Monaco R, Guggino G, Monaco G, et al. 2017. USP7 inhibitors, downregulating CCDC6, sensitize lung neuroendocrine cancer cells to PARP-inhibitor drugs. Lung Cancer 107: 41–49. 10.1016/j.lungcan.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Meis-Kindblom JM, Kindblom LG, Enzinger FM. 1995. Sclerosing epithelioid fibrosarcoma. A variant of fibrosarcoma simulating carcinoma. Am J Surg Pathol 19: 979–993. 10.1097/00000478-199509000-00001 [DOI] [PubMed] [Google Scholar]
- Miles KM, Seshadri M, Ciamporcero E, Adelaiye R, Gillard B, Sotomayor P, Attwood K, Shen L, Conroy D, Kuhnert F, et al. 2014. Dll4 blockade potentiates the anti-tumor effects of VEGF inhibition in renal cell carcinoma patient-derived xenografts. PLoS One 9: e112371. 10.1371/journal.pone.0112371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monarca C, Fino P, Rizzo MI, Palmieri A, Tarallo M, Scuderi N. 2013. The hundredth case of sclerosing epithelioid fibrosarcoma (SEF). Ann Ital Chir 84: 315–318. [PubMed] [Google Scholar]
- Moore G, Annett S, McClements L, Robson T. 2020. Top notch targeting strategies in cancer: a detailed overview of recent insights and current perspectives. Cells 9: 1503. 10.3390/cells9061503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogose A, Kawashima H, Umezu H, Hotta T, Gu W, Yamagiwa H, Ito T, Tohyama T, Nishijima H, Endo N. 2004. Sclerosing epithelioid fibrosarcoma with der(10)t(10;17)(p11;q11). Cancer Genet Cytogenet 152: 136–140. 10.1016/j.cancergencyto.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Ohlmann CH, Brecht IB, Junker K, van der Zee JA, Nistor A, Bohle RM, Stöckle M, Metzler M, Hartmann A, Agaimy A. 2015. Sclerosing epithelioid fibrosarcoma of the kidney: clinicopathologic and molecular study of a rare neoplasm at a novel location. Ann Diagn Pathol 19: 221–225. 10.1016/j.anndiagpath.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Ortiz-Montero P, Liu-Bordes WY, Londoño-Vallejo A, Vernot JP. 2018. CD24 expression and stem-associated features define tumor cell heterogeneity and tumorigenic capacities in a model of carcinogenesis. Cancer Manag Res 10: 5767–5784. 10.2147/CMAR.S176654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenne P, Eymin B, Brambilla E, Gazzeri S. 2010. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer 127: 2239–2247. 10.1002/ijc.25511 [DOI] [PubMed] [Google Scholar]
- Patterson JW, Tchernev G, Chokoeva AA, Wick MR. 2017. Sclerosing epithelioid fibrosarcoma. Wien Med Wochenschr 167: 120–123. 10.1007/s10354-016-0509-3 [DOI] [PubMed] [Google Scholar]
- Perez D, Fullmer JM, Naous R. 2019. A rare case of TLE1-positive sclerosing epithelioid fibrosarcoma expanding the differential diagnosis of TLE1-positive tumors: a case report. AME Case Rep 3: 6. 10.21037/acr.2019.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley SM, Lane DP. 1993. The p53-mdm2 autoregulatory feedback loop: a paradigm for the regulation of growth control by p53? Bioessays 15: 689–690. 10.1002/bies.950151008 [DOI] [PubMed] [Google Scholar]
- Popli A, Mahajan R, Rustagi T, Gupta S, Verma V, Gupta H. 2018. Sclerosing epithelioid fibrosarcoma of the coccyx: a case report and review of literature. Cureus 10: e2407. 10.7759/cureus.2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta Roldán P, Rodríguez Rodríguez R, Bagué Rossell S, de Juan Delago M, Molet Teixidó J. 2013. Sclerosing epithelioid fibrosarcoma of the paravertebral column. Case report and literature review. Neurocirugia (Astur) 24: 178–182. 10.1016/j.neucir.2012.09.002 [DOI] [PubMed] [Google Scholar]
- Qie S, Diehl JA. 2016. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl) 94: 1313–1326. 10.1007/s00109-016-1475-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid R, Barrett A, Hamblen DL. 1996. Sclerosing epithelioid fibrosarcoma. Histopathology 28: 451–455. 10.1046/j.1365-2559.1996.342383.x [DOI] [PubMed] [Google Scholar]
- Rekhi B, Folpe AL, Deshmukh M, Jambhekar NA. 2011. Sclerosing epithelioid fibrosarcoma—a report of two cases with cytogenetic analysis of FUS gene rearrangement by FISH technique. Pathol Oncol Res 17: 145–148. 10.1007/s12253-010-9277-3 [DOI] [PubMed] [Google Scholar]
- Righi A, Gambarotti M, Manfrini M, Benini S, Gamberi G, Cocchi S, Casadei R, Picci P, Vanel D, Dei Tos AP. 2015. Sclerosing epithelioid fibrosarcoma of the thigh: report of two cases with synchronous bone metastases. Virchows Arch 467: 339–344. 10.1007/s00428-015-1810-z [DOI] [PubMed] [Google Scholar]
- Sagiv E, Memeo L, Karin A, Kazanov D, Jacob-Hirsch J, Mansukhani M, Rechavi G, Hibshoosh H, Arber N. 2006. CD24 is a new oncogene, early at the multistep process of colorectal cancer carcinogenesis. Gastroenterology 131: 630–639. 10.1053/j.gastro.2006.04.028 [DOI] [PubMed] [Google Scholar]
- Salnikov AV, Bretz NP, Perne C, Hazin J, Keller S, Fogel M, Herr I, Schlange T, Moldenhauer G, Altevogt P. 2013. Antibody targeting of CD24 efficiently retards growth and influences cytokine milieu in experimental carcinomas. Br J Cancer 108: 1449–1459. 10.1038/bjc.2013.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi SH, Dhouib R, Ben Dhaou S, Mrad K, Driss M, Abbes I, Arbi H, Haourai H, Ben Romdhane K. 2008. Sclerosing epithelioid fibrosarcoma. A case report. Rev Chir Orthop Reparatrice Appar Mot 94: 92–95. 10.1016/j.rco.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Shenoy A, Surrey L, Jain P, Foster J, Straka J, Resnick A, Waanders A, Luo M, Li M, Kazahaya K, et al. 2019. Sclerosing epithelioid fibrosarcoma of the bone with rare EWSR1-CREB3L3 translocation driving upregulation of the PI3K/mTOR signaling pathway. Pediatr Dev Pathol 22: 594–598. 10.1177/1093526619864230 [DOI] [PubMed] [Google Scholar]
- Smith PJ, Almeida B, Krajacevic J, Taylor B. 2008. Sclerosing epithelioid fibrosarcoma as a rare cause of ascites in a young man: a case report. J Med Case Rep 2: 248. 10.1186/1752-1947-2-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spunt SL, Million L, Chi YY, Anderson J, Tian J, Hibbitts E, Coffin C, McCarville MB, Randall RL, Parham DM, et al. 2020. A risk-based treatment strategy for non-rhabdomyosarcoma soft-tissue sarcomas in patients younger than 30 years (ARST0332): a Children's Oncology Group prospective study. Lancet Oncol 21: 145–161. 10.1016/S1470-2045(19)30672-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockman DL, Ali SM, He J, Ross JS, Meis JM. 2014. Sclerosing epithelioid fibrosarcoma presenting as intraabdominal sarcomatosis with a novel EWSR1-CREB3L1 gene fusion. Hum Pathol 45: 2173–2178. 10.1016/j.humpath.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Tang MR, Guo JY, Wang D, Xu N. 2018. Identification of CD24 as a marker for tumorigenesis of melanoma. Onco Targets Ther 11: 3401–3406. 10.2147/OTT.S157043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimaru Y, Nagano H, Marubashi S, Kobayashi S, Eguchi H, Takeda Y, Tanemura M, Kitagawa T, Umeshita K, Hashimoto N, et al. 2009. Sclerosing epithelioid fibrosarcoma of the liver infiltrating the inferior vena cava. World J Gastroenterol 15: 4204–4208. 10.3748/wjg.15.4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchido K, Yamada M, Satou T, Otsuki Y, Shimizu S, Kobayashi H. 2010. Cytology of sclerosing epithelioid fibrosarcoma in pleural effusion. Diagn Cytopathol 38: 748–753. 10.1002/dc.21310 [DOI] [PubMed] [Google Scholar]
- Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, Kuppanna G, Kallappa CG, Basalingappa KM. 2018. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig 5: 5. 10.21037/sci.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sullenger BA, Rich JN. 2012a. Notch signaling in cancer stem cells. Adv Exp Med Biol 727: 174–185. 10.1007/978-1-4614-0899-4_13 [DOI] [PubMed] [Google Scholar]
- Wang WL, Evans HL, Meis JM, Liegl-Atzwanger B, Bovee JV, Goldblum JR, Billings SD, Rubin BP, López-Terrada D, Lazar AJ. 2012b. FUS rearrangements are rare in ‘pure’ sclerosing epithelioid fibrosarcoma. Mod Pathol 25: 846–853. 10.1038/modpathol.2011.214 [DOI] [PubMed] [Google Scholar]
- Wang X, He Z, Xia T, Li X, Liang D, Lin X, Wen H, Lan K. 2014a. Latency-associated nuclear antigen of Kaposi sarcoma–associated herpesvirus promotes angiogenesis through targeting notch signaling effector Hey1. Cancer Res 74: 2026–2037. 10.1158/0008-5472.CAN-13-1467 [DOI] [PubMed] [Google Scholar]
- Wang Z, da Silva TG, Jin K, Han X, Ranganathan P, Zhu X, Sanchez-Mejias A, Bai F, Li B, Fei DL, et al. 2014b. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res 74: 6364–6374. 10.1158/0008-5472.CAN-14-2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P, Sakabe K, Zheng X, Wu W, Zhang P, et al. 2015. Intracellular CD24 disrupts the ARF–NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun 6: 5909. 10.1038/ncomms6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke LM, Meis JM. 2021. Sclerosing epithelioid fibrosarcoma: a distinct sarcoma with aggressive features. Am J Surg Pathol 45: 317–328. 10.1097/PAS.0000000000001559 [DOI] [PubMed] [Google Scholar]
- Weng CC, Ding PY, Liu YH, Hawse JR, Subramaniam M, Wu CC, Lin YC, Chen CY, Hung WC, Cheng KH. 2019. Mutant Kras-induced upregulation of CD24 enhances prostate cancer stemness and bone metastasis. Oncogene 38: 2005–2019. 10.1038/s41388-018-0575-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik JB, Bellizzi AM, Dal Cin P, Bredella MA, Fletcher CD, Hornicek FJ, Deshpande V, Hornick JL, Nielsen GP. 2014. Primary sclerosing epithelioid fibrosarcoma of bone: analysis of a series. Am J Surg Pathol 38: 1538–1544. 10.1097/PAS.0000000000000265 [DOI] [PubMed] [Google Scholar]
- Wrzeszczynski KO, Felice V, Abhyankar A, Kozon L, Geiger H, Manaa D, London F, Robinson D, Fang X, Lin D, et al. 2018. Analytical validation of clinical whole-genome and transcriptome sequencing of patient-derived tumors for reporting targetable variants in cancer. J Mol Diagn 20: 822–835. 10.1016/j.jmoldx.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 7: 1126–1132. 10.1101/gad.7.7a.1126 [DOI] [PubMed] [Google Scholar]
- Xia W, Yang Y, Huang Y. 2020. Imaging features of sclerosing epithelioid fibrosarcoma of the pancreas: a case report. Front Oncol 10: 901. 10.3389/fonc.2020.00901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang J, Zhang M, Li B. 2016. Skull sclerosing epithelioid fibrosarcoma: a case report and review of the literature. Oncol Lett 11: 3417–3420. 10.3892/ol.2016.4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chou D. 2018. Spinal sclerosing epithelioid fibrosarcoma at the cervicothoracic junction. World Neurosurg 114: 155–160. 10.1016/j.wneu.2018.03.105 [DOI] [PubMed] [Google Scholar]
- Zhang W, Yi B, Wang C, Chen D, Bae S, Wei S, Guo RJ, Lu C, Nguyen LL, Yang WH, et al. 2016. Silencing of CD24 enhances the PRIMA-1-induced restoration of mutant p53 in prostate cancer cells. Clin Cancer Res 22: 2545–2554. 10.1158/1078-0432.CCR-15-1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang Y, Guo L, Zhang H, Yu Y, Li D, Xu L, Duan X, Liu Y, Li T. 2019. Primary sclerosing epithelioid fibrosarcoma of chest wall in a patient with breast cancer: a case report. Int J Clin Exp Pathol 12: 365–371. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data within this paper is deposited in the European Genome-phenome Archive (EGA; https://ega-archive.org/). The study accession number is EGAS00001005214; the data set accession number is EGAD00001007515.