Abstract
Intronic polyadenylation (IpA) usually leads to changes in the coding region of an mRNA, and its implication in diseases has been recognized, although at its very beginning status. Conveniently and accurately identifying IpA is of great importance for further evaluating its biological significance. Here, we developed IPAFinder, a bioinformatic method for the de novo identification of intronic poly(A) sites and their dynamic changes from standard RNA-seq data. Applying IPAFinder to 256 pan-cancer tumor/normal pairs across six tumor types, we discovered 490 recurrent dynamically changed IpA events, some of which are novel and derived from cancer-associated genes such as TSC1, SPERD2, and CCND2. Furthermore, IPAFinder revealed that IpA could be regulated by factors related to splicing and m6A modification. In summary, IPAFinder enables the global discovery and characterization of biologically regulated IpA with standard RNA-seq data and should reveal the biological significance of IpA in various processes.
Alternative polyadenylation (APA) of mRNA is a widespread phenomenon in diverse species, serving as an important contributor to transcriptome diversity (Elkon et al. 2013; Tian and Manley 2017). There are different types of APA based on the location of the polyadenylation (pA) site in an mRNA, such as 3′ untranslated region APA (UTR-APA), coding region APA (CR-APA), and intronic polyadenylation (IpA) (Tian and Manley 2017). UTR-APA changes the length of 3′ UTR, thereby altering RNA stability, translation efficiency, RNA localization, or even protein localization (Elkon et al. 2013; Berkovits and Mayr 2015; Tian and Manley 2017). Both CR-APA and IpA introduce a premature termination signal and lead to changes in either the coding sequence or the 3′ UTR of the corresponding mRNA (Tian et al. 2007). Although UTR-APA is widespread and involved in diverse biological processes (Mayr and Bartel 2009; Chen et al. 2018), CR-APA and IpA are less prevalent and their biological functions are not well understood. Recent studies have begun to highlight the biological significance of IpA. For example, IpA diversifies immune cell proteomes via loss of the C-terminal domain (Singh et al. 2018). Cancer cells use aberrant intronic pA sites more frequently than normal cells, and the partial loss of function of tumor suppressor genes (TSGs) caused by IpA can contribute crucially to tumorigenesis (Lee et al. 2018). Intronic polyadenylation of Pdgfra produces a truncated protein that inhibits PDGF signaling and protects mice from fibrosis (Mueller et al. 2016). CDK12 suppresses IpA as a mode of regulating DNA repair genes, which is conserved in human tumors that contain loss-of-function CDK12 mutations (Dubbury et al. 2018). These lines of evidence suggest that genome-wide IpA regulation may play a previously underestimated role in diverse biological processes and pathological conditions.
Conveniently and accurately identifying genome-wide IpA is of great importance for further evaluating its biological significance and regulatory mechanism. Although direct 3′-end sequencing of mRNA has provided invaluable insight into the global landscape of APA including IpA (Shepard et al. 2011; Hoque et al. 2013; Ni et al. 2013), it has not yet been widely adopted as a routine study strategy, and consequently the availability of such data is currently limited. Conversely, standard RNA-seq has been used in a variety of physiological and pathological conditions, and the amount of related data has increased exponentially in the last decade. Some methods such as DaPars (Xia et al. 2014) and QAPA (Ha et al. 2018) that use RNA-seq data to identify UTR-APA have also been established. However, there is a strong need for a bioinformatic method to de novo identify IpA and its dynamic changes using standard RNA-seq data.
To meet this demand, we developed a novel bioinformatic algorithm called IPAFinder to identify intronic pA sites and directly infer dynamically changed IpA events by comparative analysis on standard RNA-seq data from different conditions.
Results
IPAFinder identifies dynamically changed IpA events
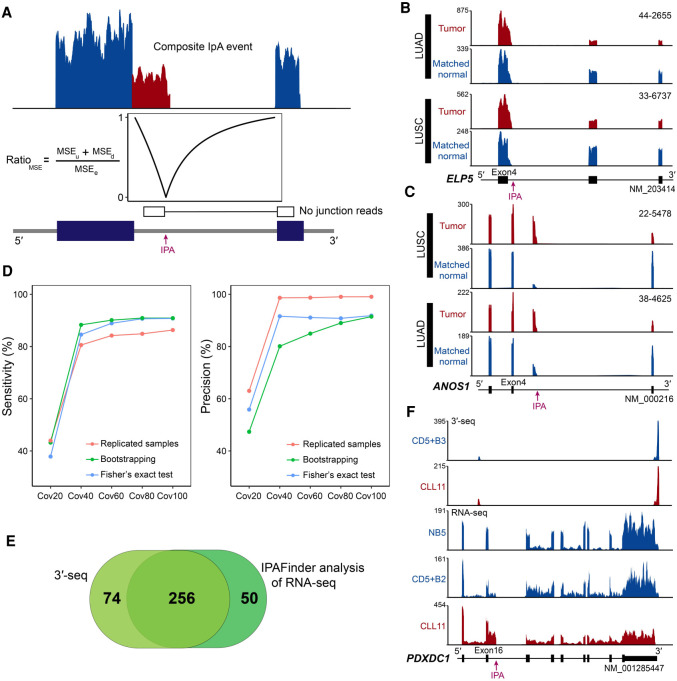
IPAFinder performs de novo identification and quantification of IpA events, without the need for any prior poly(A) site annotation. IpA events are usually classified into two groups: composite terminal exon IpA (or composite IpA) and skipped terminal exon IpA (or skipped IpA) (Supplemental Fig. S1A; Tian et al. 2007). Assuming that there is an intronic poly(A) site (IPA site) used in a given intron, IPAFinder models the normalized RNA-seq read coverage profiles at single-nucleotide resolution and identifies the drop in coverage to infer the potential IPA site, as reflected by the lowest ratio of the sum of mean squared error (MSE) in the upstream and downstream segments split by breakpoint and the MSE computed for the entire intron region (RatioMSE) (Fig. 1A; for details, see Methods). Such a strategy could detect composite IpA. To detect skipped IpA (or splicing-coupled IpA), IPAFinder recognizes cryptic 3′ splice sites by junction-spanning reads and concatenates the preceding exon to the potential terminal exon (Supplemental Fig. S1B). IPAFinder can also exclude alternative splicing events such as alternative 5′ splice site and cryptic exon activation using junction-spanning reads to remove potential false-positive events when identifying IpA. Finally, the difference in IpA usage between different conditions can be quantified as changes in the intronic poly(A) site usage index (ΔIPUI), which can identify dynamically changed IpA events. For example, IPAFinder identified ELP5 with an increased usage of a composite IPA site in two lung cancer types compared with that in matched normal tissues (Fig. 1B). In addition, ANOS1 showed dynamic changes of splicing-coupled IpA in lung cancers (Fig. 1C). The canonical polyadenylation signal (PAS) AAUAAA exists upstream of both IPA sites (Supplemental Fig. S2), which supports the authenticity of these intronic pA sites identified by IPAFinder.
Figure 1.
Overview of the IPAFinder algorithm and evaluation of its performance. (A) Schematic diagram of IPAFinder in detecting composite terminal exon IpA event. Intronic poly(A) site is determined based on the expected drop in read coverage downstream from the predicted poly(A) site. Alternative splicing events are excluded by recognizing junction-spanning reads. (B,C) Two examples of IPAFinder-identified dynamically changed IpA events from TCGA RNA-seq data. IpA usage of the ELP5 gene (B, composite IPA site) and ANOS1 gene (C, skipped IPA site) is increased in both lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) compared with that in matched normal tissues. Sample IDs are shown at the top right corner of the corresponding RNA-seq density plot. IPA site is indicated by a red arrow. (D) Performance of IPAFinder in detecting differentially used IPA sites in terms of sensitivity and precision. The number of TPs, FPs, and predefined true differentially used IPA sites (Ps) are used to calculate sensitivity (TP/P) and precision (TP/[TP + FP]). For each coverage level, we repeated 10 times to calculate the mean value of sensitivity and precision. For samples without replicates, two methods, including a bootstrapping-based method and Fisher's exact test–based method, were assessed. (E) Venn diagram comparison of the number of recurrent up-regulated IpA events identified by IPAFinder and those by 3′-seq using the same data from CLL and immune cell samples. (F) An example of dynamically changed IpA events (PDXDC1) between CLL samples and normal B cells detected by IPAFinder, which was absent in 3′-seq.
Evaluation of IPAFinder using simulated RNA-seq data and experimental 3′-seq data
To assess the performance of IPAFinder, we first used simulated RNA-seq data to test whether IPAFinder could accurately infer intronic poly(A) sites. We created simulated data of 5000 genes, of which 500 had a composite IPA site, 500 had a skipped IPA site, 500 had a retained intron, 500 had an alternative 5′ splice site, and the others were negative controls. IPAFinder could recover ∼80% and ∼90% IPA sites at a sequencing coverage of 40× and 60×, respectively (Supplemental Fig. S3A). Furthermore, IPAFinder could exclude the interference of alternative splicing events such as alternative 5′ splice site and intron retention (Supplemental Fig. S3B). Next, we evaluated the ability of IPAFinder to detect dynamically changed IpA events. We simulated 3000 genes with different coverage in two conditions. Among these genes, 500 had increasing usage of IPA sites (ΔIPUI > 0.1), 500 had decreasing usage of IPA sites (ΔIPUI < −0.1), and 2000 served as negative controls (|ΔIPUI| < 0.1). If a predicted differentially used IPA site is within 100 bp of a predefined differentially used IPA site, then the prediction is considered as a true positive (TP); otherwise, it is considered a false positive (FP). For replicated samples, IPAFinder could recover ∼80% differentially used IPA sites at a sequencing coverage of 40× with a high precision (Fig. 1D). The performance of IPAFinder improved with the increase of sequencing depth (Fig. 1D). For samples without replicates, we used two methods including a bootstrapping-based method and Fisher's exact test–based method to statistically assess the significance of difference for each IpA event between two conditions. As shown, these two methods had comparable sensitivity, but the Fisher's exact test–based method had better precision (Fig. 1D). These results indicate that IPAFinder can infer and quantify IPA sites across a very broad range of RNA-seq coverage levels.
Next, we compared IPAFinder-identified IpA events with those found by 3′-seq. We analyzed 3′-seq and standard RNA-seq data of normal and malignant B cells from patients with chronic lymphocytic leukemia (CLL) (Lee et al. 2018). In their original analysis, the investigators identified 330 recurrent up-regulated IpA events through 3′-seq analysis, followed by validation with standard RNA-seq. Inversely, we first predicted recurrent up-regulated IpA events by applying IPAFinder to RNA-seq data, and then used 3′-seq data to validate the results. IPAFinder inferred 306 recurrent up-regulated IpA events in malignant B cells compared with those in normal ones, ∼84% (256) of which were also found by the original analysis (Fig. 1E; Supplemental Fig. S4A). A heatmap of these 256 IpA events also showed an overall increase of IPA site usage in CLL samples, as reflected by lower MSE ratios and higher IPUI values (Supplemental Fig. S4B), consistent with the results reported by 3′-seq (Lee et al. 2018). These data support the overall agreement between the IPAFinder results based on RNA-seq and those based on 3′-seq. We then undertook a close inspection of those IpA events not overlapping between IPAFinder and the original study (Lee et al. 2018). For IpA events specifically identified by IPAFinder, we found that some genes did have up-regulated IpA usage in malignant B cells, as exemplified by PDXDC1 (Fig. 1F). The presence of a noncanonical poly(A) signal AAUACA, a PAS variant ranking eighth among 18 known PASs (Gruber et al. 2016; Ha et al. 2018), was observed upstream of the predicted intronic pA site (Supplemental Fig. S5A,B). In addition, a clear drop in RNA-seq coverage at the pA site, which has been used for IpA validation (Singh et al. 2018), was observed in PDXDC1. The reduced usage of downstream exons of the intronic pA site in PDXDC1 was also detected in both CLL samples (Fig. 1F) and other cancer types such as LUAD and LUSC (Supplemental Fig. S5C–E). These lines of evidence combined to support the existence and potential regulation of an IpA event in PDXDC1. For IpA events specifically identified by 3′-seq, we found example genes, such as SPTBN1, which had significantly up-regulated IpA usage detected by 3′-seq but did not have significantly higher coverage in the upstream region of the intronic pA site compared with that in the downstream region (Supplemental Fig. S4C). As such, IPAFinder could not detect it easily. Based on these results, we conclude that IPAFinder can reliably detect dynamic changes of IpA events between different conditions using standard RNA-seq resources.
IPAFinder identifies the global landscape of dynamic IpA across tumor types
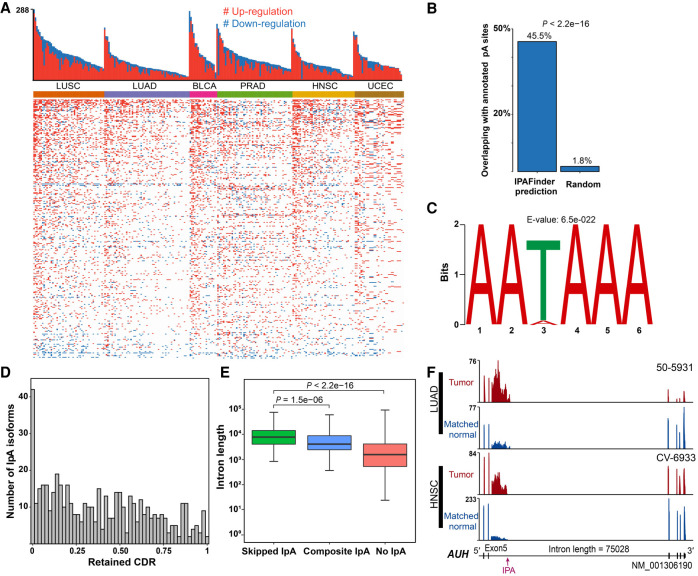
A previous study showed that IpA could inactivate tumor suppressor genes via the up-regulation of truncated mRNAs and proteins and thus contribute to tumorigenesis in CLL (Lee et al. 2018). However, it remains unclear how common IpA-mediated up-regulation of truncated mRNAs is in cancers. To examine whether it also occurs in other cancer types, we use The Cancer Genome Atlas (TCGA) database (which archives thousands of RNA-seq data derived from multiple cancer types but lacks the 3′ end sequencing data) for IpA analysis. We focused our analysis on six tumor types—namely, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), head and neck squamous cell carcinoma (HNSC), prostate adenocarcinoma (PRAD), uterine corpus endometrioid carcinoma (UCEC), and bladder urothelial carcinoma (BLCA), each of which has at least 19 tumor/normal pairs (Supplemental Table S1). We identified 130–285 genes with significantly and recurrently (occurrence rate > 10%) changed IpA events for each tumor type and found a total of 490 nonredundant IpA events across the six tumor types (Fig. 2A; Supplemental Fig. S6A,B; Supplemental Data S1). Furthermore, 56% of the 490 dynamically changed IpA events occurred in at least two tumor types (Supplemental Fig. S6C), which indicates the presence of mechanisms potentially acting in concert in IpA regulation across tumor types. Consistent with the phenomenon in CLL, global up-regulation of intronic polyadenylation was also prevalent in all six cancer types (Fig. 2A). For IPAFinder-identified intronic pA sites, 45.5% are within 50 nt of the annotated ones compiled from RefSeq, UCSC, Ensembl, and PolyASite 2.0 (Fig. 2B; Herrmann et al. 2020). There is an ∼25-fold enrichment of annotated pA sites in IPAFinder predictions compared with the level in random controls. Enrichment of the canonical poly(A) signal AATAAA in the ±50 bp flanking sequences of IPAFinder-identified intronic pA sites (Fig. 2C) further supported the reliability of our method in discovering IpA in the pan-cancer data sets (Bailey et al. 2009).
Figure 2.
IPAFinder reveals the global landscape of IpA events across six TCGA tumor types. (A) IPAFinder discovers prevalent up-regulation of IpA events across six tumor types. The upper histogram shows the number of dynamically changed IpA events per tumor. The lower heatmap shows IpA events (rows) undergoing up-regulation (red) or down-regulation (blue) in each of the 256 tumors (columns) compared with the levels in matched normal tissues across six tumor types. (B) Bar plots showing the percentages of IPAFinder-predicted breakpoints (left) and the randomly selected positions (right) that overlap with annotated intronic pA sites (RefSeq, UCSC, Ensembl, PolyASite 2.0). The P-value was calculated by t-test using 100× bootstrapping of data. (C) MEME identifies the enrichment of the canonical poly(A) signal AATAAA in the ±50 bp region around IPAFinder-inferred IPA sites. The corresponding genomic sequences (from human reference sequence hg38) serve as input. (D) The distribution of the retained coding region fraction (resulting from IpA usage) of the annotated longest coding region (CDR). (E) Box plot for lengths of introns with skipped IpA, composite IpA, and introns without IpA. The P-value was calculated based on a two-sided Wilcoxon rank-sum test. (F) AUH as an example to display skipped IPA sites with increased usage in two types of cancer (LUAD and HNSC) located in an extremely long intron. Sample IDs are shown at the top right corner of the corresponding RNA-seq density plot.
As mentioned above, intronic polyadenylation tends to disrupt the coding region of an mRNA at different degrees depending on the location at which it occurs. We thus evaluated the impact of IpA on gene expression by computing the fraction of retained coding regions for each IpA isoform relative to the full-length annotated coding regions. An overrepresentation of IpA isoforms that lose the majority of their coding regions was observed (Fig. 2D). The remaining IpA isoforms showed a relatively uniform distribution along the coding region (Fig. 2D). We found that introns with splicing-coupled IpA events were longer than those with composite IpA or no IpA events (Fig. 2E), consistent with the previous finding that a large intron size is a determining factor for IpA events (Tian et al. 2007). A typical example is the AUH gene with a splicing-coupled IpA event occurring in an extremely long intron (Fig. 2F). These results indicate that IPAFinder can reveal the overall landscape of IpA in six cancer types.
Cancer-associated genes are regulated by intronic polyadenylation
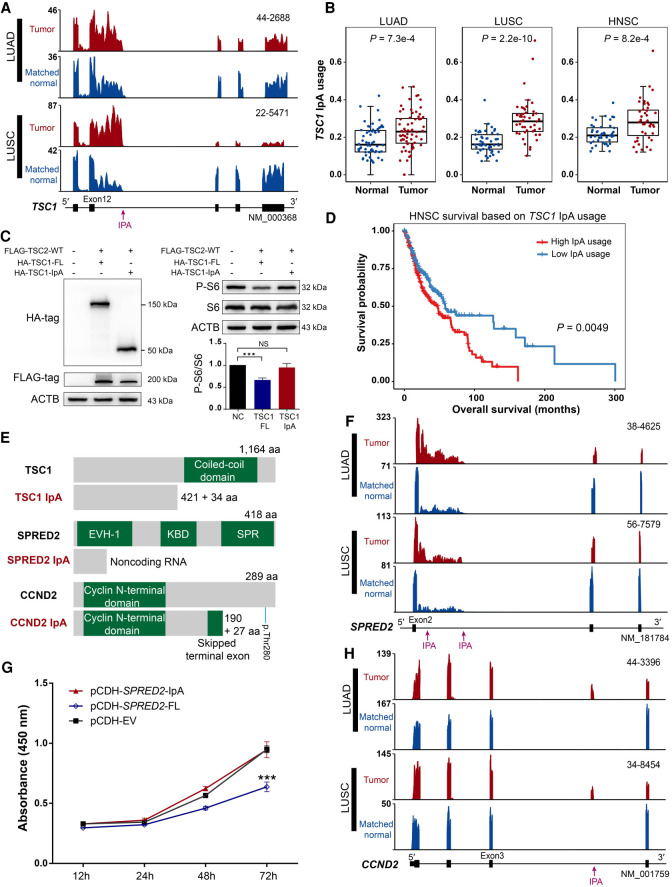
To evaluate the relevance of dysregulated IpA events during cancer development, we performed domain analysis and survival analysis on IpA-generated truncated proteins. We found that IpA events occurring at different positions of coding regions all had the possibility of generating truncated proteins with important functional impacts. The first example is TSC1, which is required to interact with and stabilize TSC2 as the TSC1-TSC2 complex, and the GAP domain on TSC2 hydrolyzes Rheb-GTP to Rheb-GDP, thereby inhibiting the activation of mTOR kinase (Garami et al. 2003; Inoki et al. 2003; Chong-Kopera et al. 2006). We found that TSC1 IpA was significantly up-regulated in LUAD, LUSC, and HNSC compared with that in normal tissues (Fig. 3A,B). The IpA isoform of TSC1 was predicted to generate a truncated protein that loses the C-terminal coiled-coil domain (Fig. 3E), which is necessary for heterodimerizing with TSC2 (van Slegtenhorst et al. 1998; Yang et al. 2021). Thus, the IpA-derived truncated protein may fail to form a functional TSC1/2 complex and lead to the aberrant activation of mTOR kinase. To test this possibility, we transiently cotransfected human HEK293T cells with full-length or IpA-truncated HA-tagged TSC1 and FLAG-tagged TSC2. Then, we performed immunoblot analysis to assess the phosphorylation of S6, an indicator of mTOR kinase activation. Compared with full-length TSC1-expressing cells, IpA-truncated TSC1 failed to inhibit the phosphorylation of S6 (Fig. 3C), which indicates that the truncated protein templated by the IpA isoform of TSC1 has impaired function compared with its full-length version. Furthermore, HNSC patients with higher IpA usage in TSC1 were found to be associated with worse survival (Fig. 3D), which was consistent with previous studies indicating that Tsc1 inactivation could promote tumor progression in mice (Kladney et al. 2010; Sun et al. 2015). The second example is the SPRED2 gene. SPRED2 inhibits the MAP kinase pathway by suppressing the phosphorylation and activation of RAF, in which both EVH-1 and SPR domains are essential (Wakioka et al. 2001; Nobuhisa et al. 2004). SPRED2 increased the usage of intronic poly(A) sites in multiple cancers (LUAD, LUSC, and HNSC) and thus produced more truncated transcripts with low coding potential (resulting in noncoding RNA as predicted by three different algorithms) (Fig. 3E,F; Supplemental Figs. S7A, S8). Experimental validation showed that the truncated transcript of SPRED2 did not produce any protein in both HEK293T and HeLa cells (Supplemental Fig. S7B). Consistent with this, overexpression of the IpA isoform of SPRED2 failed to inhibit cell proliferation, whereas the full-length version of SPRED2 did so in the human lung cancer cell line NCI-H520 (Fig. 3G). HNSC patients with higher IpA usage in SPRED2 were also associated with worse survival (Supplemental Fig. S8C). The third example is CCND2, which encodes cyclin D2. Cyclin D2 has been widely implicated in cell-cycle transition and cellular transformation, and its overexpression is highly correlated with poor prognosis in various cancers (Takano et al. 1999, 2000). IPAFinder identified that CCND2 frequently used IpA to produce a new protein isoform in LUAD, LUSC, and HNSC (Fig. 3H; Supplemental Fig. S9A–C). This IpA isoform loses the 3′ UTR miRNA repression sites (Mayr and Bartel 2009; Yang et al. 2020) as well as the important residue Thr280 (Fig. 3E), which can be phosphorylated by GSK3B and render cyclin D2 susceptible to ubiquitin–proteasome-mediated degradation (Mirzaa et al. 2014). Thus, increased IpA usage in CCND2 likely causes resistance to protein degradation, which results in CCND2 accumulating to promote cell-cycle progression. In line with this, the presence of CCND2 IpA was associated with worse survival in LUSC (Supplemental Fig. S9D). These three examples show that IPAFinder can identify functional IpA regulation in cancer-related genes.
Figure 3.
IpA generates truncated proteins with important functional impacts. (A) RNA-seq density plots showing that TSC1 has increased IpA usage in lung cancers. Numbers on the y-axis indicate RNA-seq read coverage. Sample IDs are shown at the top right corner of the corresponding RNA-seq density plot. (B) Box plots showing that TSC1 has significantly higher IpA usage in LUAD, LUSC, and HNSC tumors. The P-value was calculated based on a two-sided Wilcoxon rank-sum test. (C) Immunoblot analysis of S6 phosphorylation in HEK293T cells with overexpression of the IpA and full-length (FL) isoform of TSC1. Successful expression of HA-tagged IpA and full-length isoforms of TSC1 was confirmed by western blot using anti-HA and anti-FLAG antibodies (left). Both total S6 protein and its phosphorylated form (P-S6) were also quantified by western blot (right). ACTB was used as an internal control. Negative control (NC) means a sample without FLAG-TSC2-WT, HA-TSC1-FL, and HA-TSC1-IpA. (***) P-value < 0.001, t-test. (D) Kaplan–Meier curves of overall survival for two HNSC tumor groups stratified by the IpA usage of TSC1. The P-value was calculated using the log-rank test. (E) Diagrams showing the domain information of full-length and IpA-generated truncated proteins, with known domains shown in green. The numbers of retained and novel amino acids (aa) and amino acids of full-length proteins are given. The position of the important residue Thr280 of CCND2 is denoted by a short blue line (p.Thr280). (F) RNA-seq density plots showing that SPRED2 has increased IpA usage in lung cancers. (G) CCK-8 assay showing that IpA-truncated SPRED2 fails to inhibit cell proliferation in NCI-H520 cells. (***) P-value < 0.001, t-test. (H) RNA-seq density plots showing that CCND2 has increased IpA usage in lung cancers.
Intronic polyadenylation can be influenced by factors related to splicing and m6A modification
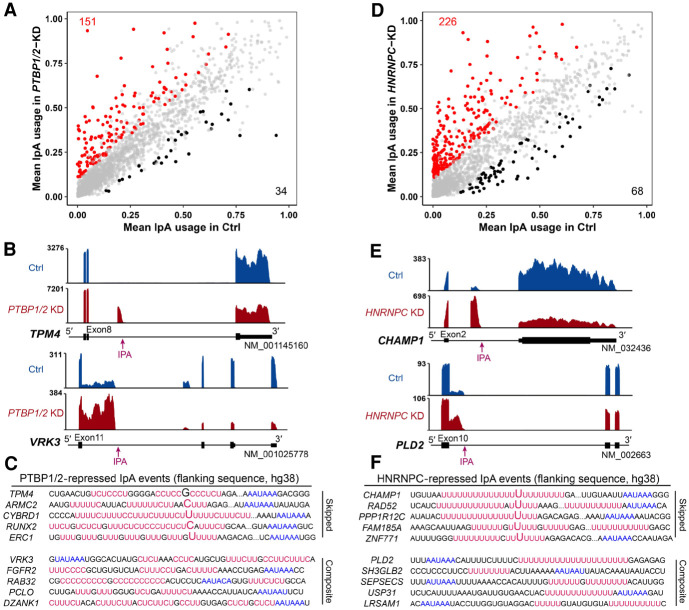
The fidelity of RNA splicing is regulated by an orchestration of splicing enhancers and repressors, and multiple RNA-binding proteins (RBPs) can protect the transcriptome from the aberrant exonization of transposable elements (Zarnack et al. 2013; Attig et al. 2018) and modulate cleavage and polyadenylation at poly(A) sites where they bind (Licatalosi et al. 2008; Hilgers et al. 2012). Applying IPAFinder to published RNA-seq data sets generated from concurrent knockdown of PTBP1 and PTBP2 (Gueroussov et al. 2015), two splicing factors preferentially binding to CU repeats (Oberstrass et al. 2005), we found that PTBP1/2 deficiency resulted in many more up-regulated IpA events than down-regulated ones, consistent with the findings of PTBP1/2 in repressing cryptic exons in the original study (Fig. 4A,B). Sequence analysis confirmed the presence of adjacent CU microsatellites around these activating poly(A) sites (Fig. 4C), which suggests the direct binding of PTBP1/2. We also tested another RBP heterogeneous nuclear ribonucleoprotein C (HNRNPC), which has been reported to modulate the processing of pre-mRNA 3′-ends (Gruber et al. 2016). Applying IPAFinder to RNA-seq data sets of HEK293T cells obtained upon the knockdown of this protein (Liu et al. 2015), we found that the loss of HNRNPC also led to the widespread up-regulation of IpA events, and the majority (72.1%) were skipped IpA events (Fig. 4D,E). Sequence analysis of the major IpA isoforms showed that the density of (U)5 tracts, reported to be the binding site for HNRNPC (König et al. 2010), was markedly higher around cryptic 3′ splice sites whose usage increased upon HNRNPC knockdown compared with those without apparent changes in usage (Fig. 4F; Supplemental Fig. S10). Experimental validation supported the up-regulation of IpA events upon knockdown of these RBPs (Supplemental Fig. S11). Altogether, these results show that PTBP1/2 and HNRNPC can protect pre-mRNAs from premature cleavage and polyadenylation by inhibiting the usage of IPA sites.
Figure 4.
PTBP1/2 and HNRNPC inhibit the usage of IPA sites. (A) Scatterplot of IPUI value reflecting the relative IpA usage before and after concurrent knockdown of PTBP1 and PTBP2 (PTBP1/2-KD) in HEK293 cells. Red and blue dots represent genes with increased and decreased IpA usage upon knockdown of PTBP1/2, respectively. (B) Representative examples of IpA events repressed by PTBP1/2. Both the skipped IpA event (top) and the composite IpA event (bottom) are shown. (C) Sequences flanking example IPA sites repressed by PTBP1/2 have CU repeats (red) and poly(A) signals (blue). Five genes with a skipped terminal exon (top) and five genes with a composite terminal exon (bottom) are shown. The first bases of skipped terminal exons are denoted by enlarged characters. (D) Scatterplot of IPUI value reflecting the relative IpA usage before and after knockdown of HNRNPC (HNRNPC-KD) in HEK293T cells. Red and blue dots represent genes with increased and decreased IpA usage in HNRNPC-KD cells, respectively. (E) Representative examples of HNRNPC-repressed IpA events. (F) Sequences flanking example IPA sites repressed by HNRNPC have (U)5 tracts (red) and poly(A) signals (blue).
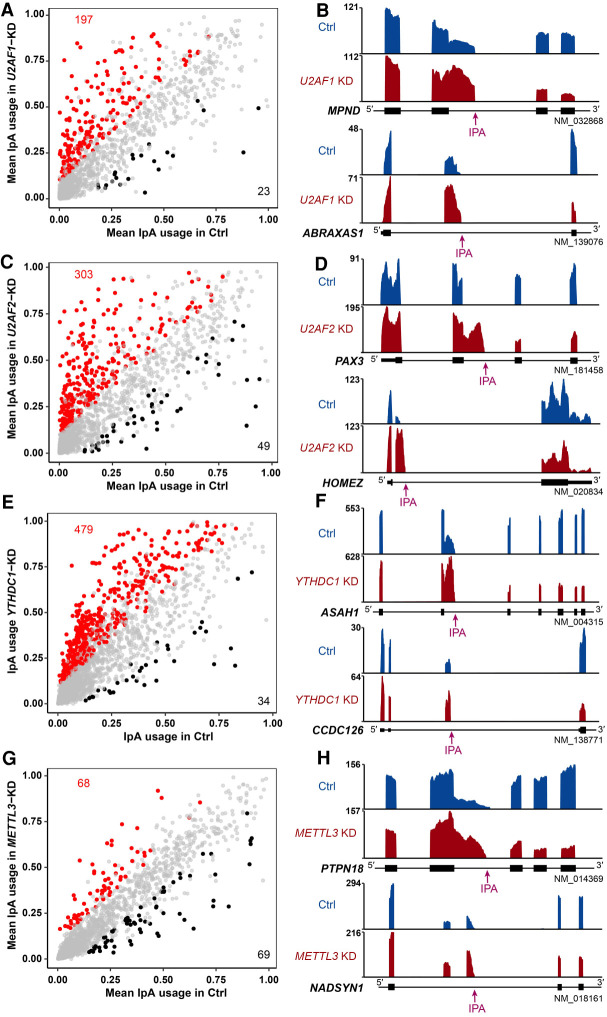
To explore other factors regulating intronic polyadenylation, we next applied IPAFinder to RNA-seq data derived from samples with the knockdown of relevant RBPs. U2AF1 and U2AF2 are two auxiliary factors for U2 small nuclear RNA, which bind to the AG dinucleotide and polypyrimidine tract of the 3′ splice site, respectively, to facilitate splice site recognition (Zamore et al. 1992; Wu et al. 1999). Multiple studies have reported links between splicing and 3′-end processing (Kyburz et al. 2006; Millevoi et al. 2006). Thus, we explored whether these two splicing factors could impact intronic pA site usage by analyzing our custom-made RNA-seq data derived from human foreskin fibroblasts (HFFs) with the knockdown of U2AF1 or U2AF2. We found that depletion of U2AF1 or U2AF2 globally increased the usage of intronic pA sites (Fig. 5A–D), consistent with a previous analysis of 3′-seq data showing that the knockdown of U2af2 in mouse C2C12 myoblast cells led to the overall up-regulation of IpA events (Li et al. 2015). For skipped IPA sites with increased usage upon the knockdown of U2AF1 or U2AF2, the splicing strength of their cryptic 3′ splice sites was significantly weaker than that of downstream 3′ splice sites (Supplemental Fig. S12A). Furthermore, intronic pA sites with increased usage showed considerable overlap between U2AF1-KD and U2AF2-KD samples (Supplemental Fig. S12B,C), suggesting the potential coordination of these two factors in regulating IpA. In line with the similarity in IpA level changes between U2AF1-KD and U2AF2-KD samples, we also found that both U2AF1-KD and U2AF2-KD could lead to senescence-associated phenotypes at the cellular level (Supplemental Fig. S13; Yao et al. 2020). Although the causal relationship between IpA and senescence deserves further investigation, we did reveal that U2AF1 and U2AF2 have a genome-wide effect on intronic poly(A) site selection.
Figure 5.
IPAFinder reveals that intronic polyadenylation can be influenced by factors related to splicing and m6A modification. (A,C,E,G) Scatterplot of IPUI values reflecting the relative IpA usage in cells with knockdown (KD) of U2AF1 (A, in HFF cell), U2AF2 (C, in HFF cell), YTHDC1 (E, in HeLa cell), or METTL3 (G, in HEK293T cell). Red and blue dots represent genes with increased and decreased IpA usage upon knockdown of corresponding genes. (B,D,F,H) Representative RNA-seq density plots for genes with significantly increased IpA usage upon knockdown of U2AF1 (B), U2AF2 (D), YTHDC1 (F), or METTL3 (H). Each knockdown condition has two IpA examples: the composite terminal exon IpA (top) and the skipped terminal exon IpA (bottom).
N6-methyladenosine (m6A) has been identified as the most abundant modification that ubiquitously occurs in eukaryotic mRNAs and affects multiple aspects of mRNA metabolism including alternative splicing (Yang et al. 2018). However, whether factors related to m6A modification affect IpA is unclear. Applying IPAFinder to RNA-seq data derived from HeLa cells deficient in YTHDC1, the only known m6A reader in the nucleus that has been reported to regulate splicing (Xiao et al. 2016), we found that YTHDC1 deficiency led to increased IPA site usage (Fig. 5E). Furthermore, IPA sites with increased usage upon YTHDC1 knockdown had considerable overlap with those upon SRSF3 knockdown (Supplemental Fig. S14), consistent with previous findings that YTHDC1 and SRSF3 interact with each other to regulate mRNA splicing and 3′ UTR length (Xiao et al. 2016; Kasowitz et al. 2018). In addition, analyzing RNA-seq data derived from HEK293T cells deficient in METTL3, a methyltransferase implicated in placing m6A on RNA (Śledź and Jinek 2016), we found that the loss of METTL3 also changed the IPA site usage, but the numbers of up-regulated or down-regulated IpA events were relatively equal (Fig. 5G). Example genes with changed IpA usage upon knockdown of either YTHDC1 or METTL3 are shown (Fig. 5F,H). However, it should be kept in mind that it is currently unclear whether the regulation is directly mediated by m6A modification, which warrants further investigation. These results indicate that IpA events have a complex regulatory system, and IPAFinder is a valuable tool capable of discovering IpA events from RNA-seq data sets of varied sources, which would also facilitate screening for regulators involved in IpA determination.
Comparison of methods for analyzing IpA
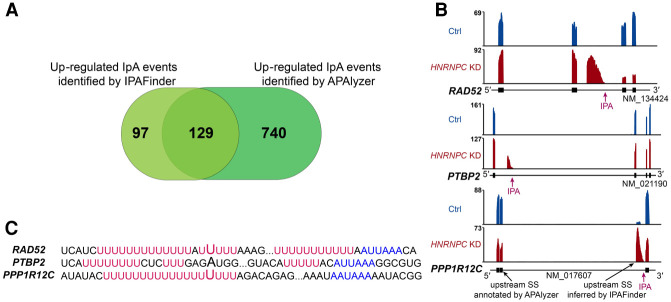
IPAFinder was inspired by DaPars, which identifies the breakpoint that can best explain the localized read-density change to perform de novo identification and quantification of dynamic UTR-APA events using standard RNA-seq (Xia et al. 2014). However, IpA identification is more complicated than UTR-APA and could be interfered with by at least three events, including cryptic exon activation, alternative 5′ splice site, and intron retention. IPAFinder uses BAM format as the input file, which contains splice junction information, and considers most of the interference factors to improve the identification of IpA events (Supplemental Fig. S15). We also compared IPAFinder with APAlyzer, which analyzes intronic polyadenylation by using RNA-seq data based on known poly(A) sites (such as those annotated in the PolyA_DB database) (Wang et al. 2018; Wang and Tian 2020). Applying APAlyzer to RNA-seq data sets obtained upon HNRNPC-KD, we observed widespread usage changes of intronic poly(A) sites according to their suggested cutoff (P-value < 0.05, using t-test for significance analysis) (Supplemental Fig. S16A). Intronic poly(A) sites with increased usage upon HNRNPC-KD analyzed by IPAFinder and APAlyzer had considerable overlap (Fig. 6A). However, IPAFinder could identify dynamic IPA sites that were not annotated by the PolyA_DB database, as exemplified by the IPA sites of RAD52 and PTBP2 in HNRNPC-KD condition (Fig. 6B). IPAFinder could infer upstream splice sites by recognizing junction reads and quantify the usage of corresponding skipped IPA sites accurately, as exemplified by the gene PPP1R12C (Fig. 6B). Sequence analysis showed that (U)5 tracts existed in the flanking region of these three IPA sites (Fig. 6C), which indicated the direct binding of HNRNPC (König et al. 2010). Although IPAFinder and APAlyzer have different criteria for calling differential IpA events, they have a relatively consistent trend in quantifying the usage of IPA sites (Supplemental Fig. S16 B,C). Overall, IPAFinder is a specialized tool for de novo IpA analysis that is distinct from existing methods such as APAlyzer. Different tools have their own strengths and weaknesses (Supplemental Table S2), and users may need to apply multiple programs in their research to obtain comprehensive and complementary results.
Figure 6.
Comparison between IPAFinder and APAlyzer. (A) Venn diagram illustrating the overlap of up-regulated IpA events upon HNRNPC-KD identified by IPAFinder and APAlyzer, respectively. (B) Representative RNA-seq tracks for genes with increased IpA usage inferred by IPAFinder but not APAlyzer. (C) Sequence flanking three IPA sites shown in B have (U)5 tract (red) and poly(A) signal (blue). The first bases of skipped terminal exons are denoted by enlarged characters.
Discussion
In this study, we developed IPAFinder, a method for the de novo identification of intronic poly(A) sites and dynamically changed IpA events from standard RNA-seq data. Multiple lines of evidence support the reliability of IPAFinder. Applying IPAFinder to 256 pan-cancer tumor/normal pairs across six tumor types archived in TCGA revealed 490 recurrently changed IpA events, among which there were genes with novel IpA regulation, such as TSC1, SPERD2, and CCND2. Furthermore, genes harboring dynamic IpA events were found being enriched in TSGs but not in the oncogenes (Supplemental Fig. S17A). In additional tumor samples (without matched normal tissues), we also found well-known TSGs (NF1, PTEN, and CDH1) with increased IPA site usage (Supplemental Fig. S17B). Thus, IPAFinder should help to reveal potential IpA events playing roles in diverse physiological and pathological processes by exploiting the huge amount of standard RNA-seq data.
RNA-seq data tend to have coverage biases that are more predominant in untranslated region, and we can observe this phenomenon in both real and simulated RNA-seq data (Supplemental Fig. S18). Many well-developed methods (such as DaPars, APAtrap, and PAQR) (Xia et al. 2014; Gruber et al. 2018; Ye et al. 2018), which de novo infer internal poly(A) sites in 3′ UTR from standard RNA-seq data, are based on the MSE model, regardless of potential coverage bias in 3′ UTR. It is difficult to detect poly(A) sites from RNA-seq data at single-nucleotide precision, thus some degree of flexibility (e.g., 100 nt for IPAFinder) is used to match predicted poly(A) sites to the annotated ones (Supplemental Fig. S3). Although 3′-end sequencing strategies such as 3′-seq coupled with dedicated bioinformatic pipelines can identify IPA sites and detect changes in IPA site usage between different conditions, they are less extensively used in diverse biological processes than standard RNA-seq. In addition, standard RNA-seq has multiple advantages in detecting intronic pA sites compared with 3′-seq: (1) RNA-seq covers the whole gene body and thus junction reads can be used to distinguish skipped IPA sites from composite IPA sites; and (2) the use of RNA-seq to infer IPA sites can avoid the interference of internal priming according to continuous upstream read coverage for composite IPA sites and junction-spanning reads for skipped IPA sites.
U1 snRNP can protect pre-mRNAs from drastic premature termination by cleavage and polyadenylation at cryptic polyadenylation signals in introns (Kaida et al. 2010; Berg et al. 2012). Applying IPAFinder to publicly available RNA-seq data derived from HeLa cells upon treatment of U1 Antisense Morpholino Oligonucleotide (AMO) (Oh et al. 2017), which has been shown to pair efficiently with U1 snRNA and thereby functionally inhibit U1 snRNP, we found that U1 AMO treatment globally increased the usage of IPA sites (Supplemental Fig. S19). These data support the ability of IPAFinder in detecting the usage changes of cryptic IPA sites.
We also compared a bootstrapping-based method with a Fisher's exact test–based method by analyzing RNA-seq data set obtained by knockdown of HNRPNC (merge two replicates as one sample). The bootstrapping-based method identified 197 significantly up-regulated IpA events, whereas the Fisher's exact test–based method identified 279 up-regulated IpA events; up-regulated IpA events identified by these two methods had considerable overlap (Supplemental Fig. S20A). Furthermore, we found that the bootstrapping-based method is sensitive to IpA events whose usage difference is relatively large (Supplemental Fig. S20B), as exemplified by IpA events of genes ZCCHC4 and VPS4B (Supplemental Fig. S20C). By comparing with up-regulated IpA events identified by the DEXSeq method on replicated samples (Anders et al. 2012), we found that the Fisher's exact test–based method could identify more up-regulated IpA events supported by the DEXSeq method than the bootstrapping-based method (174 vs. 118). Up-regulated IpA events identified by the Fisher's exact test–based method have higher fraction supported by the DEXSeq method than those identified by the bootstrapping-based method (62.4% vs. 59.9%). Thus, for samples without replicates between two conditions, users could try both these two statistical methods in their research to obtain comprehensive and complementary results.
In conclusion, the IPAFinder method should open up a new avenue for discovering IpA events and changes in their usage in numerous biological processes using standard RNA-seq data. This should help to reveal the functional roles of IpA in diverse conditions.
Methods
IPAFinder algorithm
IPAFinder performs de novo identification and quantification of dynamically changed IpA events between two conditions, regardless of any prior poly(A) site annotation. Assuming that there is an intronic pA site used in a given intron, there will be a significant drop in RNA-seq read coverage because of polyadenylation processing. Thus, IPAFinder models the normalized RNA-seq read coverage at single-nucleotide resolution and progressively segments the intron region into two regions with distinct mean coverage. This enables inference of the potential intronic poly(A) site, where the squared deviation decreases most from the mean coverage of the intron when dividing the segment into two regions compared with considering it as a single segment. IPAFinder separately calculates the MSE of read coverage for upstream (MSEu) and downstream (MSEd) segments split by every point in the intron region and compares the sum of MSEu and MSEd (MSEu+MSEd) with the MSE computed for the entire intron region (MSEe). The ratio of the sum of MSEu and MSEd to MSEe is defined as RatioMSE (Fig. 1A) and, if the lowest value of RatioMSE ≤ 0.5, a cutoff used to infer internal poly(A) site in 3′ UTR by a previous study (Gruber et al. 2018), the corresponding breakpoint is considered as a potential intronic poly(A) site. In addition, the mean coverage in the upstream region of the candidate poly(A) site must be higher than that in the downstream region. Alternative splicing events such as alternative 5′ splice site may also have similar segmentation breakpoints, so we exclude those breakpoints where there are splice sites supported by junction-spanning reads around them. If the given intron has no composite terminal exon IpA event, IPAFinder next searches for whether it has a skipped terminal exon IpA event (Supplemental Fig. S1B). We regard a splice site in an internal intron as a cryptic 3′ splice site if it is supported by more than 10 splice junction reads or >10% of upstream 5′ splice site junction reads. Then, we concatenate the preceding exon to this potential skipped terminal exon and identify the best segmentation breakpoint in the newly formed narrowed intron region, as performed for the composite terminal exon. Alternative splicing events such as cryptic exon activation are also excluded by recognizing junction-spanning reads.
IPAFinder could also detect multiple IPA sites in a single intron. IPAFinder first infers the breakpoint with the lowest RatioMSE in the entire intron region. If there is an alternative intronic poly(A) site in the inferred terminal exon, another drop in RNA-seq read coverage inside the terminal exon will be observed. Similarly, the alternative used pA site allows the best segmentation of the terminal exon into upstream and downstream regions with distinct coverage. Therefore, IPAFinder can infer its location by calculating the ratio of MSE recursively (Supplemental Fig. S21A). The alternative intronic poly(A) sites of SPRED2 are identified in such a strategy (Supplemental Fig. S21B,C).
Once the intronic poly(A) sites have been identified, library size-normalized expression levels and relative usage of IPA sites are calculated. We define the intronic poly(A) site usage index (IPUI) to quantify the relative IpA usage for sample j as follows:
| (1) |
where , , and are the estimated expression levels of IpA isoform, full-length isoform, and constitutive preceding exon located upstream of the IPA site for a given sample (j), respectively. In principle, is equal to the sum of and (Supplemental Fig. S22).
For samples with replicates, to detect differential usage of IpA isoform between two conditions, we examined the difference in relative usage of terminal exon inferred by IPAFinder. We applied DEXSeq to model the read counts of all exons across conditions by negative binomial distribution and tested for the significance of an interaction term between exon and condition (Supplemental Fig. S23; Anders et al. 2012). We defined an IpA isoform to be significantly differentially used if its corresponding terminal exon usage is significantly different between two conditions (FDR-adjusted P-value < 0.05) and the difference of IPA site usage is more than 0.1 (|ΔIPUI| > 0.1).
For samples without replicates, such as paired tumor-normal samples from TCGA, we used Fisher's exact test to infer differential usage of IpA isoform between conditions, which is a similar approach taken by previous methods for detecting 3′ UTR-APA events (Xia et al. 2014; Guvenek and Tian 2018). We defined an IpA isoform to be significantly differentially used if its FDR-adjusted P-value < 0.05 and |ΔIPUI| > 0.1.
We also provided a bootstrapping-based method to statistically assess the significance of difference for each IpA event between two samples without replicates, which is inspired by the significance analysis of alternative polyadenylation (SAAP) method (Li et al. 2015). Briefly, for an IpA event from two comparing samples, IPUI was first calculated and was called observed IPUI. Then we sampled reads based on the assumption that the relative abundance of each IpA isoform was the same in two samples. Sampling was performed m times (default m = 20) to obtain mean and standard deviation of IPUI, which were then used to convert observed IPUI to Z-score. False discovery rate (FDR) and Q-value were calculated by comparing observed Z (Zo) of IPUI and expected Z (Ze) of IPUI for a given Z cutoff value (Zc). The Q-value for an IpA event x is the FDR using the absolute value of its Zo (Zox) as Zc. We used Q-value < 0.05 and |ΔIPUI| > 0.1 to select significantly differential IpA events. We updated our pipeline in GitHub to indicate which option the users should choose for samples with or without replicates.
Data download
All the TCGA RNA-seq BAM files for tumor and matched normal samples were obtained from the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/). Here, we processed LUAD, LUSC, HNSC, UCEC, BLCA, and PRAD cancers. Other publicly available raw sequencing data of 3′-seq and RNA-seq, including those derived from normal immune cells (Singh et al. 2018) and malignant B cells (Lee et al. 2018) from patients with chronic lymphocytic leukemia (CLL) (Gene Expression Omnibus [GEO; https://www.ncbi.nlm.nih.gov/geo/] accession numbers GSE111310 and GSE111793), PTBP1/2 knockdown in HEK293 cells (GEO: GSE69656) (Gueroussov et al. 2015), HNRNPC knockdown and METTL3 knockdown in HEK293T cells (GEO: GSE56010) (Liu et al. 2015), YTHDC1 knockdown and SRSF3 knockdown in HeLa cells (GEO: GSE71095) (Xiao et al. 2016), U2AF1 knockdown in HFF cells (BioProject [https://www.ncbi.nlm.nih.gov/bioproject] accession number PRJNA565612) (Yao et al. 2020), and U1 inhibition in HeLa cells (GEO: GSE135140) (So et al. 2019), were obtained from NCBI.
RNA-seq and 3′-seq data analyses
Among the raw paired-end reads obtained from RNA-seq experiments, low-quality reads were filtered out, followed by alignment to the human reference genome sequence (UCSC hg38 assembly) using STAR (Dobin et al. 2013) with the default settings. Analysis of the 3′-seq data was performed as described previously (Singh et al. 2018). The peaks were assigned to RefSeq-annotated genes (downloaded on January 1, 2020). Isoforms with an expressed level of at least three transcripts per million mapped reads (TPM) and usage of at least 0.1 in at least one sample were used for subsequent analyses. We only analyzed IpA isoforms of protein-coding genes.
Benchmarking of IPAFinder using simulated RNA-seq data
We first generated a synthetic RNA-seq data set to assess the performance of IPAFinder to infer intronic poly(A) sites from standard RNA-seq data. To simulate the different coverage levels, baseline coverage for each gene was uniformly sampled between 20× and 80×. An “n×” coverage means that an exonic genomic locus is covered by n reads on average. The usage of IpA isoform or alternative splicing isoform was uniformly sampled from a usage range (40%–60%). We also evaluated the ability of IPAFinder to detect dynamically changed IpA events at different levels of sequencing coverage between two conditions. IPUI values for each gene were randomly sampled until the conditions outlined were met. For samples with replicates, three replicates per condition were generated using negative binomial distribution. The R package polyester was applied to simulate paired-end 100-nt reads from the human genome (hg38) with the default parameters (Frazee et al. 2015). We provided the full-length transcript structure for IPAFinder to infer and quantify IPA sites based on the synthetic RNA-seq data set.
Comparison between IPAFinder-analyzed RNA-seq and custom-analyzed 3′-seq
A total of 330 recurrent CLL-IpA events were obtained from the data sets of Lee et al. (2018). When IPAFinder was applied to RNA-seq data, an IpA isoform was considered as a recurrently up-regulated IpA isoform if it had significant up-regulation in at least three malignant B cell samples (11 samples in total) compared with the level in normal immune cell samples. With this criterion, we obtained 306 recurrently up-regulated IpA events. A lower RatioMSE value means that there is a better coverage segmentation point in the given intron region. Thus, CLL samples with a larger number of CLL-IpA events as reported by the original publication (including CLL4, CLL7, CLL11, and CLL12) have more low RatioMSE values than samples with a smaller number of CLL-IpA events or normal samples (Supplemental Fig. S4B), which suggests that RatioMSE is a rational index for identifying potential intronic poly(A) sites. Furthermore, CLL samples with a larger number of CLL-IpA events have higher IPUI (Supplemental Fig. S4B), consistent with previous results (Lee et al. 2018), which indicates that IPUI is also a rational index for quantifying IpA isoform usage.
Motif frequency analysis
The genomic sequences (from the human reference sequence hg38) of 200 nt upstream of and downstream from the cryptic 3′ splice sites were used for motif analysis. The frequency of HNRNPC binding motif (U)5 tracts was calculated by counting the number of (U)5 motifs (smoothened by ±5 nt centered on the position of interest) along these specified annotation features.
Clinical significance analysis of IpA usage
We obtained clinical information including overall survival time of patients from the GDC data portal (https://portal.gdc.cancer.gov/). A log-rank test and Kaplan–Meier survival analysis were performed to identity the association between intronic pA site usage and overall survival. For the gene TSC1, groups with high and low IpA usage were separately used for a survival plot by splitting the ordered IPUI with an equal number of samples in each group. For the gene CCND2, patients whose CCND2 IpA usage is greater than 0.1 were grouped into patients with CCND2 IpA. All statistical analyses were performed in R (v.3.5.1) (R Core Team 2018).
RT-PCR validation of up-regulated IpA isoforms upon knockdown of RBPs
Endogenous PTBP1 and HNRNPC were knocked down using pLKO.1-puro lentiviral vector-based shRNAs (Sigma-Aldrich). HEK293T cells were transduced in six-well plates using Lipofectamine 2000 (Invitrogen). Virus was produced using the helper plasmids VSVG and gag/pol. After infection over 36 h, the cells were selected with puromycin (2.5 μg/mL) for 2 d and the surviving cells were cultured for two more days and then collected for RT-PCR analysis.
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instruction. RNA was reversely transcribed using the FastKing RT Kit (with gDNase; Tiangen). Twenty microliters cDNA product was diluted fivefold and 2 µL diluted cDNA was used as the template for each semiquantitative RT-PCR reaction. We used a typical reaction containing 500 nM forward and reverse primers for individual isoforms. The PCR reaction products were analyzed by gel electrophoresis. Primers are listed in Supplemental Table S3.
Vector construction
The full-length TSC1, TSC2, and SPRED2 mRNA was amplified from HEK293T cDNA. Plasmids for the expression of full length of HA-TSC1 (ENSG00000165699, 1164aa), FLAG-TSC2 (ENSG00000103197, 1807aa), and HA-SPRED2 (ENSG00000198369, 418aa) were constructed by cloning full-length CDS of TSC1, TSC2, and SPRED2 into the pRK5 vector with either FLAG or HA tag at their N terminus. TSC1 IpA was PCR-amplified from two fragments. Fragment 1 was amplified from HEK293T cDNA and corresponds to amino acids 1–421, whereas fragment 2 was amplified from genomic DNA of HEK293T and corresponds to intronic sequence upstream predicted IPA site.
To construct the pCDH-SPRED2 plasmid, full-length CDS of SPRED2 cloned from HEK293T cDNA was inserted into the pCDH-CMV-MCS-T2Apuro plasmid using EcoRI/BamHI restriction sites. SPRED2 IpA was also PCR-amplified from two fragments. Fragment 1 was amplified from HEK293T cDNA and corresponds to amino acids 1–68, whereas fragment 2 was amplified from genomic DNA and corresponds to intronic sequence upstream predicted IPA site. The integrity of all constructs was confirmed by Sanger sequencing.
Western blotting
Cells were rinsed with PBS and lysed in cold RIPA buffer (25 mM Tris at pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing freshly added Protease and Phosphatase Inhibitor Cocktail, EDTA-free (Thermo Fisher Scientific). Cell lysates were incubated on ice for 10 min, and centrifuged at 14,000g for 15 min at 4°C. The supernatant was collected and the protein concentration was determined by Bicinchoninic Acid (BCA) assay (Beyotime). A total of 20 μg protein per sample was resolved by 10% SDS-PAGE, followed by transfer to a PVDF membrane with pore size 0.2 μm (Millipore) for immunoblotting. Quantification was performed by densitometry using ImageJ software, and ACTB served as internal control.
The following primary antibodies were used: anti-ACTB (proteintech HRP-60008, 1:2000), Anti-phospho-S6 ribosomal protein (Cell Signaling Technology 2215S, 1:2000), Anti-S6 ribosomal protein (Cell Signaling Technology 2217S, 1:2000), anti HA-Tag (ABclonal AE008, 1:2000), and Mouse anti DDDDK-Tag (ABclonal AE005, 1:2000). The secondary antibodies used included HRP Goat Anti-Rabbit IgG (H + L) (ABclonal AS014, 1:5000) and HRP Goat Anti-Mouse IgG (H + L) (ABclonal AS003, 1:5000).
Cell proliferation assay
Cells were counted and seeded in 96-well plates with 2000 cells per well and four replicates for each time point. Cell Counting Kit-8 (CCK-8) reagent (Dojindo) was diluted with medium according to the manufacturer's protocol and then added to each testing well. Then, cells were incubated for another 2 h at 37°C, and then the absorbance of each well was measured at 450 nm by a microplate reader (Bio-Rad).
Data access
The IPAFinder method is freely available at GitHub (https://github.com/ZhaozzReal/IPAFinder) and as Supplemental Code. The U2AF2-KD RNA-seq data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA660570.
Supplementary Material
Acknowledgments
We thank the Edanz Group China (http://www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript. This work was supported by the National Key Research and Development Program of China (2018YFC1003500), the National Natural Science Foundation of China (91949107, 31771336, 31521003), and the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.271627.120.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Anders S, Reyes A, Huber W. 2012. Detecting differential usage of exons from RNA-seq data. Genome Res 22: 2008–2017. 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attig J, Agostini F, Gooding C, Chakrabarti AM, Singh A, Haberman N, Zagalak JA, Emmett W, Smith CWJ, Luscombe NM, et al. 2018. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell 174: 1067–1081.e17. 10.1016/j.cell.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64. 10.1016/j.cell.2012.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Mayr C. 2015. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522: 363–367. 10.1038/nature14321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lyu G, Han M, Nie H, Shen T, Chen W, Niu Y, Song Y, Li X, Li H, et al. 2018. 3′ UTR lengthening as a novel mechanism in regulating cellular senescence. Genome Res 28: 285–294. 10.1101/gr.224451.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong-Kopera H, Inoki K, Li Y, Zhu T, Garcia-Gonzalo FR, Rosa JL, Guan KL. 2006. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J Biol Chem 281: 8313–8316. 10.1074/jbc.C500451200 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbury SJ, Boutz PL, Sharp PA. 2018. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 564: 141–145. 10.1038/s41586-018-0758-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506. 10.1038/nrg3482 [DOI] [PubMed] [Google Scholar]
- Frazee AC, Jaffe AE, Langmead B, Leek JT. 2015. Polyester: simulating RNA-seq datasets with differential transcript expression. Bioinformatics 31: 2778–2784. 10.1093/bioinformatics/btv272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466. 10.1016/S1097-2765(03)00220-X [DOI] [PubMed] [Google Scholar]
- Gruber AJ, Schmidt R, Gruber AR, Martin G, Ghosh S, Belmadani M, Keller W, Zavolan M. 2016. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res 26: 1145–1159. 10.1101/gr.202432.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AJ, Schmidt R, Ghosh S, Martin G, Gruber AR, van Nimwegen E, Zavolan M. 2018. Discovery of physiological and cancer-related regulators of 3′ UTR processing with KAPAC. Genome Biol 19: 44. 10.1186/s13059-018-1415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueroussov S, Gonatopoulos-Pournatzis T, Irimia M, Raj B, Lin ZY, Gingras AC, Blencowe BJ. 2015. An alternative splicing event amplifies evolutionary differences between vertebrates. Science 349: 868–873. 10.1126/science.aaa8381 [DOI] [PubMed] [Google Scholar]
- Guvenek A, Tian B. 2018. Analysis of alternative cleavage and polyadenylation in mature and differentiating neurons using RNA-seq data. Quant Biol 6: 253–266. 10.1007/s40484-018-0148-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KCH, Blencowe BJ, Morris Q. 2018. QAPA: a new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biol 19: 45. 10.1186/s13059-018-1414-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann CJ, Schmidt R, Kanitz A, Artimo P, Gruber AJ, Zavolan M. 2020. PolyASite 2.0: a consolidated atlas of polyadenylation sites from 3′ end sequencing. Nucleic Acids Res 48: D174–D179. 10.1093/nar/gkz918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Lemke SB, Levine M. 2012. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev 26: 2259–2264. 10.1101/gad.199653.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10: 133–139. 10.1038/nmeth.2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17: 1829–1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. 10.1038/nature09479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ. 2018. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 14: e1007412. 10.1371/journal.pgen.1007412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladney RD, Cardiff RD, Kwiatkowski DJ, Chiang GG, Weber JD, Arbeit JM, Lu ZH. 2010. Tuberous sclerosis complex 1: an epithelial tumor suppressor essential to prevent spontaneous prostate cancer in aged mice. Cancer Res 70: 8937–8947. 10.1158/0008-5472.CAN-10-1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W. 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205. 10.1016/j.molcel.2006.05.037 [DOI] [PubMed] [Google Scholar]
- Lee SH, Singh I, Tisdale S, Abdel-Wahab O, Leslie CS, Mayr C. 2018. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 561: 127–131. 10.1038/s41586-018-0465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, et al. 2015. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet 11: e1005166. 10.1371/journal.pgen.1005166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469. 10.1038/nature07488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. 2015. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518: 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. 2006. An interaction between U2AF 65 and CF Im links the splicing and 3′ end processing machineries. EMBO J 25: 4854–4864. 10.1038/sj.emboj.7601331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa G, Parry DA, Fry AE, Giamanco KA, Schwartzentruber J, Vanstone M, Logan CV, Roberts N, Johnson CA, Singh S, et al. 2014. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genet 46: 510–515. 10.1038/ng.2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AA, van Velthoven CT, Fukumoto KD, Cheung TH, Rando TA. 2016. Intronic polyadenylation of PDGFRα in resident stem cells attenuates muscle fibrosis. Nature 540: 276–279. 10.1038/nature20160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Yang Y, Hafez D, Yang W, Kiesewetter K, Wakabayashi Y, Ohler U, Peng W, Zhu J. 2013. Distinct polyadenylation landscapes of diverse human tissues revealed by a modified PA-seq strategy. BMC Genomics 14: 615. 10.1186/1471-2164-14-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuhisa I, Kato R, Inoue H, Takizawa M, Okita K, Yoshimura A, Taga T. 2004. Spred-2 suppresses aorta-gonad-mesonephros hematopoiesis by inhibiting MAP kinase activation. J Exp Med 199: 737–742. 10.1084/jem.20030830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. 2005. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309: 2054–2057. 10.1126/science.1114066 [DOI] [PubMed] [Google Scholar]
- Oh JM, Di C, Venters CC, Guo J, Arai C, So BR, Pinto AM, Zhang Z, Wan L, Younis I, et al. 2017. U1 snRNP telescripting regulates a size-function-stratified human genome. Nat Struct Mol Biol 24: 993–999. 10.1038/nsmb.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-seq. RNA 17: 761–772. 10.1261/rna.2581711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Lee SH, Sperling AS, Samur MK, Tai YT, Fulciniti M, Munshi NC, Mayr C, Leslie CS. 2018. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat Commun 9: 1716. 10.1038/s41467-018-04112-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledź P, Jinek M. 2016. Structural insights into the molecular mechanism of the m6A writer complex. eLife 5: e18434. 10.7554/eLife.18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Di C, Cai Z, Venters CC, Guo J, Oh JM, Arai C, Dreyfuss G. 2019. A complex of U1 snRNP with cleavage and polyadenylation factors controls telescripting, regulating mRNA transcription in human cells. Mol Cell 76: 590–599.e4. 10.1016/j.molcel.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen S, Liu F, Wu H, McHugh J, Bergin IL, Gupta A, Adams D, Guan JL. 2015. Constitutive activation of mTORC1 in endothelial cells leads to the development and progression of lymphangiosarcoma through VEGF autocrine signaling. Cancer Cell 28: 758–772. 10.1016/j.ccell.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Y, Kato Y, Masuda M, Ohshima Y, Okayasu I. 1999. Cyclin D2, but not cyclin D1, overexpression closely correlates with gastric cancer progression and prognosis. J Pathol 189: 194–200. [DOI] [PubMed] [Google Scholar]
- Takano Y, Kato Y, van Diest PJ, Masuda M, Mitomi H, Okayasu I. 2000. Cyclin D2 overexpression and lack of p27 correlate positively and cyclin E inversely with a poor prognosis in gastric cancer cases. Am J Pathol 156: 585–594. 10.1016/S0002-9440(10)64763-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. 2007. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res 17: 156–165. 10.1101/gr.5532707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, van den Ouweland A, Reuser A, Sampson J, Halley D, van der Sluijs P. 1998. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet 7: 1053–1057. 10.1093/hmg/7.6.1053 [DOI] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. 2001. Spred is a Sprouty-related suppressor of Ras signalling. Nature 412: 647–651. 10.1038/35088082 [DOI] [PubMed] [Google Scholar]
- Wang R, Tian B. 2020. APAlyzer: a bioinformatics package for analysis of alternative polyadenylation isoforms. Bioinformatics 36: 3907–3909. 10.1093/bioinformatics/btaa266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Nambiar R, Zheng D, Tian B. 2018. Polya_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res 46: D315–D319. 10.1093/nar/gkx1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. 10.1038/45590 [DOI] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 5: 5274. 10.1038/ncomms6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. 2016. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 61: 507–519. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hsu PJ, Chen YS, Yang YG. 2018. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 28: 616–624. 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Li L, Connelly JP, Porter SN, Kodali K, Gan H, Park JM, Tacer KF, Tillman H, Peng J, et al. 2020. A cancer-specific ubiquitin ligase drives mRNA alternative polyadenylation by ubiquitinating the mRNA 3′ end processing complex. Mol Cell 77: 1206–1221.e7. 10.1016/j.molcel.2019.12.022 [DOI] [PubMed] [Google Scholar]
- Yang H, Yu Z, Chen X, Li J, Li N, Cheng J, Gao N, Yuan HX, Ye D, Guan KL, et al. 2021. Structural insights into TSC complex assembly and GAP activity on Rheb. Nat Commun 12: 339. 10.1038/s41467-020-20522-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ding D, Li X, Shen T, Fu H, Zhong H, Wei G, Ni T. 2020. Prevalent intron retention fine-tunes gene expression and contributes to cellular senescence. Aging Cell 19: e13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Long Y, Ji G, Li QQ, Wu X. 2018. APAtrap: identification and quantification of alternative polyadenylation sites from RNA-seq data. Bioinformatics 34: 1841–1849. 10.1093/bioinformatics/bty029 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614. 10.1038/355609a0 [DOI] [PubMed] [Google Scholar]
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466. 10.1016/j.cell.2012.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.