Abstract
Long terminal repeat (LTR) retrotransposons are widely distributed across the human genome. They have accumulated through retroviral integration into germline DNA and are latent genetic modules. Active LTR promoters are observed in germline cells; however, little is known about the mechanisms underlying their active transcription in somatic tissues. Here, by integrating our previous transcriptome data set with publicly available data sets, we show that the LTR families MLT2A1 and MLT2A2 are primarily expressed in human four-cell and eight-cell embryos and are also activated in some adult somatic tissues, particularly pineal gland. Three MLT2A elements function as the promoters and first exons of the protein-coding genes ABCE1, COL5A1, and GALNT13 specifically in the pineal gland of humans but not in that of macaques, suggesting that the exaptation of these LTRs as promoters occurred during recent primate evolution. This analysis provides insight into the possible transition from germline insertion to somatic expression of LTR retrotransposons.
Retrotransposons (LTRs, LINEs, and SINEs) are genetic elements that account for >40% of the human genome (International Human Genome Sequencing Consortium 2001). Retrotransposons are an essential source of regulatory sequences (Rebollo et al. 2012; Chuong et al. 2017) and contribute to primate-specific gene regulation (Jacques et al. 2013; Trizzino et al. 2017). LTR retrotransposons might have the greatest potential to be adapted as regulatory elements because of their unique structure (Thompson et al. 2016). The full-length elements typically consist of two or three genes encoding viral proteins flanked by two identical LTRs. Homologous recombination facilitated by LTRs leads to the elimination of internal open reading frames (Mager and Goodchild 1989; Belshaw et al. 2007) and yields solitary LTRs containing regulatory elements, such as promoters, transcription factor binding sites, and splice sites.
Relics of LTR retrotransposons, including solitary LTRs, originate from exogenous retroviruses that were repeatedly integrated into germline genomes during evolution, enabling transmission to subsequent generations (Belshaw et al. 2004). Human germline cells express multiple distinct LTR families, such as human endogenous retrovirus L and H (HERV-L and HERV-H) (Göke et al. 2015). During embryonic genome activation (EGA), HERV-L elements are activated by the homeobox transcription factor DUX4 (Hendrickson et al. 2017). DUX4 also activates retrotransposons, thereby creating new promoters and fusion transcripts in facioscapulohumeral muscular dystrophy (Young et al. 2013; Mitsuhashi et al. 2021). The function of HERV-L derived transcripts in early embryos remains unknown, whereas a long noncoding RNA derived from HERV-H may be involved in the maintenance of pluripotency of embryonic stem cells and inner cell mass (Lu et al. 2014). Some LTR elements may have been recruited as functional components in germline development (Fort et al. 2014; Durruthy-Durruthy et al. 2016; Zhang et al. 2019).
Because LTRs originate from germline integration events, it is somewhat surprising that LTRs are also transcribed in various somatic tissues (Faulkner et al. 2009). Although transcription from LTRs tends to be less active in somatic tissues than in embryonic tissues, some LTR elements act as primary promoters in somatic tissues (Cohen et al. 2009). However, the evolutionary mechanisms underlying the transition from germline insertion to somatic expression of LTR retrotransposons remain unclear.
Using single-cell-tagged reverse transcription (STRT), we have profiled the expression levels and transcription start sites (TSSs) of maternal and embryonic transcripts in more than 300 human oocytes, zygotes, and four- and eight-cell blastomeres (Töhönen et al. 2015).
Here, we integrate our previous transcriptome data set with public resources that cover stages from early embryos to somatic tissues across human, macaque, and marmoset to define LTR families most transcriptionally active in the early stages of embryonic development, and we explore the transcriptional activity of these LTR families in a wide range of somatic cells.
Results
MLT2A elements are transcribed in four-cell embryos
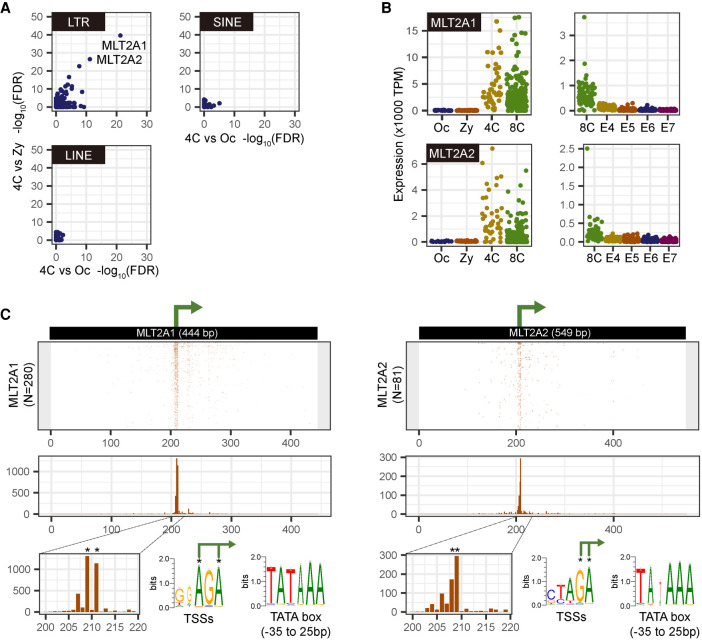
Using our STRT data set (Töhönen et al. 2015) and another single-cell RNA-seq data set (Supplemental Fig. S1A; Petropoulos et al. 2016), we explored repetitive elements that are activated at the beginning of EGA. The LTR families MLT2A1 and MLT2A2 were up-regulated with the lowest false discovery rate (FDR) at the four-cell stage compared with oocytes and zygotes (Fig. 1A; Supplemental Fig. S1B). This observation is consistent with the earlier finding of the MLT2A1 activation in the cleavage stage (Hendrickson et al. 2017). MLT2A1 and MLT2A2 are closely related HERV-L retrotransposons; each family has more than 3000 copies in the human genome. The total expression of all MLT2A copies was markedly increased from zygote to the four-cell stage, followed by a rapid decrease after the eight-cell stage (Fig. 1B; Supplemental Fig. S1C), similar to that of other embryonic genes such as ZSCAN4 (Supplemental Fig. S1D). Most transcription occurred from MLT2A copies longer than 200 bp (“long” elements), whereas truncated elements (200 bp or less; “short” elements) were rarely transcribed (Supplemental Fig. S1E,F). Therefore, in this study, we analyzed only transcripts from long MLT2A1 elements (N = 2416; median length, 413 bp [range, 200–572 bp]) and MLT2A2 (N = 3069; median length, 517 bp [range, 200–636 bp]). Coordinates of all the elements and their expression values are provided in Supplemental Tables S1 and S2.
Figure 1.
Activation of MLT2A1 and MLT2A2 copies in four-cell stage human embryos. (A) Scatter plots of −log10(FDR) values for differential expression between four-cell embryo and oocyte (x-axis) and between four-cell embryo and zygote (y-axis) by using single-map reads; the plots, including multimap reads, are shown in Supplemental Figure S1B. Each dot represents a family of LTR, LINE, or SINE. (B) Normalized expression values (tags per million mapped tags [TPM]) of MLT2A1 and MLT2A2 in each single-cell: (Oc) oocyte; (Zy) zygote; (4C) four-cell embryo; (8C) eight-cell embryo; (E4 to E7) embryo on day 4 to day 7. STRT data from oocyte to eight-cell embryos are shown in the left panels, and RNA-seq data from eight-cell embryo to embryo on day 7 are shown in the right panels. (C) Location of transcription start sites (TSSs) on the active MLT2A1 and MLT2A2 elements. The middle panel shows the frequency of TSSs at each position. Asterisks indicate two most frequent positions.
We identified 280 copies of MLT2A1 and 81 copies of MLT2A2 that are expressed in at least five different cells in the four- or eight-cell stage, and we defined these 361 copies as active elements. They were transcribed from nearly identical positions (Fig. 1C), corresponding to positions 205–215 in the consensus sequences of MLT2A1 (total length, 444 bp) and MLT2A2 (total length, 549 bp) defined in Repbase (Jurka et al. 2005). The TSSs were enriched within AG-rich regions in the middle of the MLT2A copies; TATA boxes were found upstream of the TSSs (Fig. 1C). These results indicate that more than 300 MLT2A copies retain promoters functional in four- or eight-cell blastomeres.
MLT2A loci become accessible in two-cell embryos
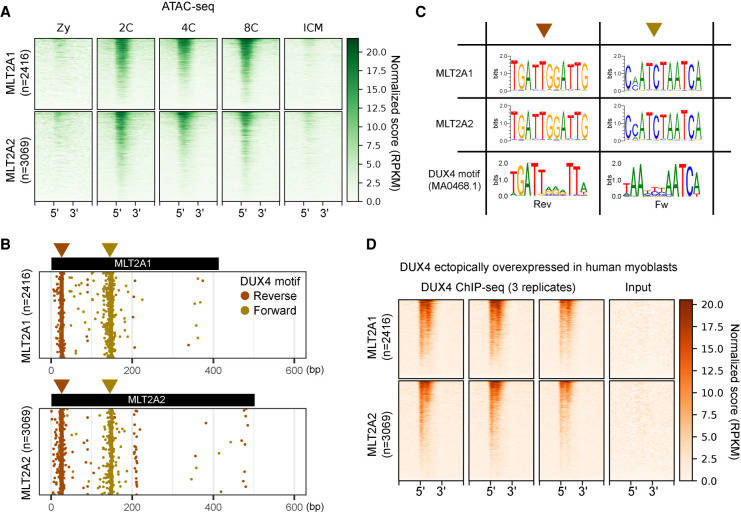
To explore the mechanism of MLT2A activation in four-cell blastomeres, we used a publicly available ATAC-seq data set for human preimplantation embryos (Supplemental Fig. S2A; Wu et al. 2018). We found that the chromatin in thousands of long MLT2A elements with flanking regions, either transcriptionally active or not, becomes accessible during the transition from zygote to the two-cell stage (Fig. 2A; Supplemental Fig. S2B). Searches for the DUX4 binding motif in all long MLT2A elements revealed that both MLT2A families contained two DUX4 motif-like sequences in forward or reverse orientation upstream of the promoter (Fig. 2B). The vast majority of active elements had one or two DUX4 motifs (Supplemental Fig. S2C). The DUX4 binding motif was highly similar to the nucleotide patterns enriched in the two MLT2A families (Fig. 2C), suggesting that DUX4 might be responsible for initiating transcription of MLT2A elements. We analyzed the public ChIP-seq and RNA-seq data sets (Geng et al. 2012; Eidahl et al. 2016) for human myoblasts overexpressing DUX4 (Supplemental Fig. S2A). The ChIP-seq data revealed that DUX4 bound to two motifs in MLT2A elements (Fig. 2D; Supplemental Fig. S2D), and the RNA-seq data revealed that MLT2A elements were up-regulated upon DUX4 overexpression (Supplemental Fig. S2E). However, the loss of DUX has only a minor effect on the mouse EGA transcriptome (Chen and Zhang 2019), suggesting the presence of additional regulators.
Figure 2.
Chromatin states of MLT2A1 and MLT2A2 elements. (A) Chromatin states of all MLT2A elements and flanking regions (±1 kb) in five stages: (Zy) zygote; (2C) two-cell embryo; (4C) four-cell embryo; (8C) eight-cell embryo; (ICM) inner cell mass. All elements are scaled to the same size, with 5′ and 3′ denoting their ends. The elements are sorted from the top to the bottom according to ATAC-seq signal intensity. (B) Distribution of DUX4 binding motifs (MA0468.1) in all MLT2A elements; the motifs were identified by using MAST software. Motifs in the reverse and forward orientations are denoted as brown and gold circles, respectively. (C) Sequence logos of DUX4 motif-enriched regions in MLT2A1 and MLT2A2 and logos of the original DUX4 motif in the reverse (Rev) and forward (Fw) orientations. (D) DUX4 binding states in all MLT2A elements and flanking regions (±1 kb) for three replicates of ChIP-seq samples and one input sample.
In addition to MLT2A1 and MLT2A2, the human genome harbors 10 other MLT2 families. But unlike the A1 and A2 families, they were not expressed in a family-wide manner in early embryos (Supplemental Fig. S2F). This is possibly explained by our finding that DUX4 binding motifs and sites are rarely found in these families (Supplemental Fig. S2G–I).
MLT2A elements supply promoters and splicing sites in primate embryos
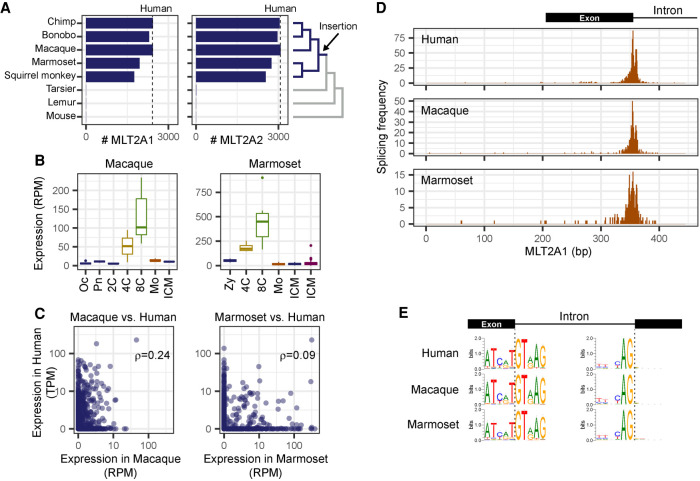
We examined whether the embryonic activation of MLT2A is observed in other primates. There are about 2000–3000 copies of MLT2A in the genomes of simian primates, including rhesus macaque (Macaca mulatta) and common marmoset (Callithrix jacchus), but not in the genomes of other primates or mouse (Fig. 3A). This indicates that both MLT2A1 and MLT2A2 were inserted in the genome of the common ancestor of simian primates about 45–65 million years ago. To examine the expression of MLT2A families in early embryos of simian primates, we used publicly available transcriptome data sets of macaque (Wang et al. 2017) and marmoset (Supplemental Fig. S3A; Boroviak et al. 2018). As in human embryos, MLT2A families were activated in the four-cell stage, and quickly down-regulated before the morula stage (Fig. 3B; Supplemental Fig. S3B). The DUX4 binding motifs in the MLT2A elements were distributed in a similar way to those in humans (Supplemental Fig. S3C). On the other hand, the expression patterns of individual MLT2A elements differed among primates. Spearman's correlation coefficients between human and corresponding macaque or marmoset elements ranged from 0.08 to 0.24 (Fig. 3C; Supplemental Fig. S3D). Taken together, our results suggest that the family-wide activation of MLT2A in four-cell blastomeres, but not the expression patterns of individual elements, is conserved among simian primates.
Figure 3.
Activation of MLT2A1 and MLT2A2 elements in primate embryos. (A) Numbers of MLT2A1 and MLT2A2 elements in seven primate genomes and the mouse genome. Numbers of human MLT2A elements are denoted by dashed lines. The phylogenetic tree is shown on the right side. (B) Box plots of normalized expression values (reads per million mapped reads [RPM]) of MLT2A1 elements: (Oc) oocyte; (Pn) pronuclei; (2C, 4C, and 8C) two-cell, four-cell, and eight-cell embryos, respectively; (Mo) morula; (ICM) inner cell mass. (C) Normalized expression values of individual MLT2A1 elements in macaque versus human and marmoset versus human embryos. The numbers of elements robustly transcribed in two species (more than 5 TPM and 5 RPM) are seven for macaque versus human and 13 for marmoset versus human: (ρ) Spearman's correlation coefficients. (D) Frequency of splicing events in MLT2A1 elements of human, macaque, and marmoset. (E) Sequence logos of splice sites in MLT2A1 elements found in four-cell and eight-cell embryos of human, macaque, and marmoset. The left side shows donor sites inside MLT2A1, and the right side shows acceptor sites outside MLT2A1.
We cloned RNAs transcribed from an MLT2A1 element on Chromosome 1 and an MLT2A2 element on Chromosome 9 in a human eight-cell embryo and sequenced them. All nine transcripts obtained were spliced within the MLT2A element (European Nucleotide Archive [ENA; https://www.ebi.ac.uk/ena/browser/home] accession numbers LR694124–126 for MLT2A1 and LR694118–123 for MLT2A2). To globally determine the splice patterns of activated MLT2A elements, we examined spliced reads in the RNA-seq data sets used above. Splice sites were enriched around nucleotide positions 350 in MLT2A1 (Fig. 3D) and 350 and 450 in MLT2A2 (Supplemental Fig. S3E). DNA sequence analysis of splice sites identified canonical dinucleotides GT for donor sites within MLT2A elements and AG for acceptor sites (Fig. 3E; Supplemental Fig. S3F). We identified 1504 robust splice acceptor sites supported by at least three different samples. About 97% of the acceptor sites were located in unannotated regions or noncoding exons, whereas 25 human, 4 macaque, and 18 marmoset MLT2A elements were connected to protein-coding genes (Supplemental Fig. S3G). Only one gene, SH3BGRL, was detected in all three primates (Supplemental Fig. S3H), most likely because of the low conservation of the expression values of individual MLT2A elements.
Multiple copies of MLT2A are transcribed in pineal gland and amniotic membrane
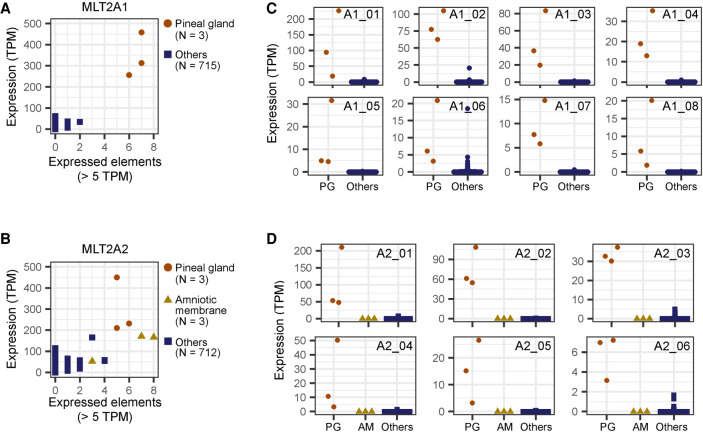
LTR retrotransposons show tissue-specific or stage-specific activity (Faulkner et al. 2009; Hashimoto et al. 2015), consistent with the stage-specific embryonic expression pattern of MLT2A elements. To check whether MLT2A families are reactivated in any somatic cells, we analyzed total expression of MLT2A elements in 178 tissues and 540 primary cells in data from the FANTOM5 consortium (Supplemental Table S3; The FANTOM Consortium and the RIKEN PMI and CLST [DGT] 2014). Among all samples, the triplicate samples of pineal glands derived from three independent donors showed the highest total expression levels of MLT2A1 and MLT2A2 (Fig. 4A,B, y-axes). Most expression was attributed to a small number of these elements (Fig. 4A,B, x-axes), and >92% of MLT2A elements remained silenced in each of the three pineal glands. We identified eight MLT2A1 and six MLT2A2 elements that were robustly (mean TPM >5) and specifically expressed in the three pineal glands (Fig. 4C,D). Triplicate amniotic membrane samples also showed high levels of total MLT2A2 expression relative to the other samples; seven robustly expressed MLT2A2 elements differed from those expressed in pineal gland (Supplemental Fig. S4). Thus, a small number of MLT2A elements are actively transcribed in somatic tissues, such as pineal gland and amniotic membrane.
Figure 4.
Specific expression of MLT2A elements in pineal gland. (A,B) Scatter plots of 718 human samples including three pineal glands denoted by brown circles. The x-axis indicates the numbers of expressed elements, and the y-axis indicates normalized expression values, measured by CAGE (tags per million mapped tags [TPM]). (C) Normalized expression values of individual MLT2A1 elements that are actively transcribed in pineal gland. Expression values are shown separately for three pineal glands (PG) and 715 other samples. (D) Normalized expression values of individual MLT2A2 elements.
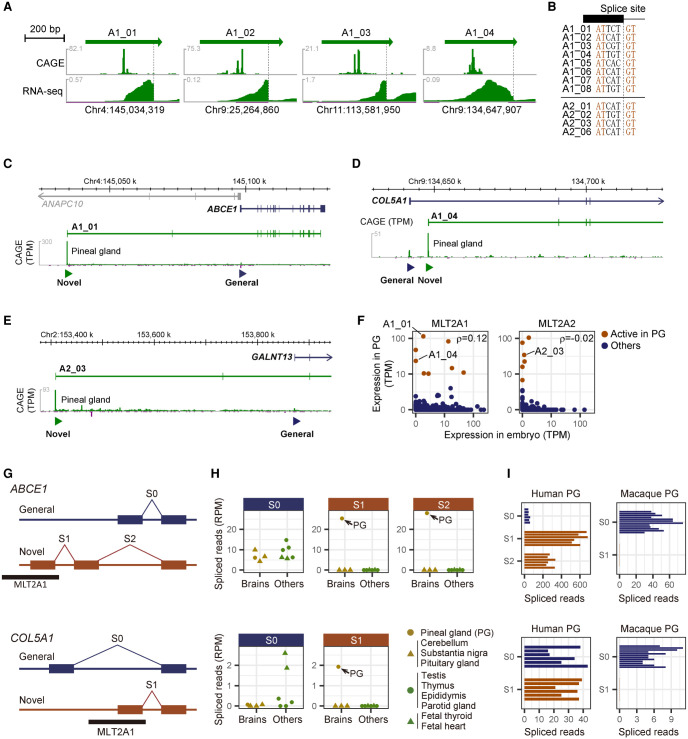
MLT2A elements supply novel promoters for ABCE1, COL5A1, and GALNT13 genes
We then asked whether the transcription of MLT2A elements plays a role in the pineal gland. We used a publicly available RNA-seq data set for human pineal glands derived from six donors (NCBI Gene Expression Omnibus [GEO; https://www.ncbi.nlm.nih.gov/geo/] accession number GSE100472) to investigate transcripts that initiate from the 14 robustly expressed MLT2A elements. In 12 cases, we found multiexon transcripts, where the first exon was entirely contained within MLT2A elements (Fig. 5A; Supplemental Fig. S5A). The promoter architecture and splice sites in the pineal gland was similar to those in embryos (Fig. 5B).
Figure 5.
Pineal gland–specific novel promoters and splice junctions derived from MLT2A elements. (A) Expression patterns of individual MLT2A1 elements actively transcribed in pineal gland. CAGE captures the 5′ ends of the transcripts, whereas RNA-seq captures entire exons. Spliced sites are denoted by dashed lines. (B) DNA sequences of splice sites in the MLT2A1 and MLT2A2 elements. (C–E). Novel promoters used in pineal glands for ABCE1 (C), COL5A1 (D), and GALNT13 (E) genes. The novel promoters overlapped with MLT2A elements are denoted by green triangles. The transcript models (ENST00000296577.8 for ABCE1, ENST00000371817.7 for COL5A1, and ENST00000392825.7 for GALNT13) are from GENCODE. Expression signals of CAGE (tags per million mapped tags [TPM]) are shown in green peaks. (F) Normalized mean expression values (TPM) of individual MLT2A1 and MLT2A2 elements in early embryo (N = 225, all at four-cell or eight-cell stage) and pineal gland (PG, N = 3). The 14 elements actively transcribed in pineal gland are shown in brown. (G) Schematic representation of splice sites of ABCE1 and COL5A1. Novel splice junctions derived from MLT2A1 elements are denoted as S1 and S2, whereas common splice junctions are denoted as S0. (H) Normalized splice counts (spliced reads per million mapped reads [RPM]) in four human brain (gold) and six non-brain (green) tissues for ABCE1 (upper) and COL5A1 (lower). Pineal glands (PG) are denoted by gold circles. (I) Normalized splice counts in human and macaque pineal gland for ABCE1 (upper) and COL5A1 (lower). The human samples are from six different donors (GSE100472), and the macaque samples are from 12 different individuals (GSE78165).
None of the first exons in the 12 MLT2A elements is annotated as a part of protein-coding genes in GENCODE release 32 (Frankish et al. 2019). However, three MLT2A-derived transcripts showed a coding probability of nearly 1.0 (100%) according to the Coding Potential Assessment Tool (CPAT) (Supplemental Fig. S5B; Wang et al. 2013). Indeed, these three transcripts overlapped with protein-coding genes ABCE1, COL5A1, and GALNT13. In these transcripts, a novel exon transcribed from the associated MLT2A element was used as the first exon instead of the common first exon (Fig. 5C–E). Replacement of the first exon did not affect the amino acid sequences of ABCE1 or GALNT13 because their start codons were in the second exon, and the new first exons contained no in-frame start codons. On the other hand, COL5A1 proteins encoded by transcripts from MLT2A1 would be expected to be shorter because of the loss of a start codon in the first exon.
We explored whether the same splicing events occurred in four- and eight-cell embryos. The three MLT2A elements connected to protein-coding genes (A1_01, A1_04, and A2_03) were not expressed or were expressed at low levels in early embryos (Fig. 5F), and no spliced reads between MLT2A elements and coding exons were detected (Supplemental Fig. S3H). No strong correlation was found between expression values in embryo versus pineal gland and amniotic membrane (Fig. 5F; Supplemental Fig. S5C), suggesting that MLT2A elements are activated differently in embryo and these somatic tissues.
MLT2A promoters for human pineal gland are not activated in macaque
To evaluate the transcriptional balance between the common and novel promoters, we used publicly available RNA-seq data for four brain and six non-brain human tissues (Supplemental Fig. S5D, DDBJ Sequence Read Archive [DRA; https://www.ddbj.nig.ac.jp/dra/index-e.html] accession number DRA000991); Hon et al. 2017). We estimated the alternative exon usage on the basis of the numbers of spliced reads that spanned exon–exon junctions; the common exon junctions were denoted as S0, and novel ones as S1 and S2 (Fig. 5G; Supplemental Fig. S5E). The novel exon junctions were specifically used in pineal gland, whereas the common ones were typically used in multiple tissues including pineal gland (Fig. 5H; Supplemental Fig. S5F). In pineal gland, the expression levels of novel exons were the same as, or greater than, those of the common exons, indicating that these MLT2A elements act as the main promoters for ABCE1, COL5A1, and GALNT13.
These three MLT2A elements are conserved among most of the primate genomes according to the phastCons track in the UCSC Genome Browser (Lee et al. 2020). To explore whether these elements are transcribed similarly in macaque and human pineal glands, we used public RNA-seq data for pineal gland extracted from 12 adult rhesus macaques (Supplemental Fig. S5D; Backlund et al. 2017). We detected S0 in nearly all samples but did not identify any spliced reads that would support junctions corresponding to human S1 (Fig. 5I; Supplemental Fig. S5G). The finding that ABCE1, COL5A1, and GALNT13 are transcribed from common promoters rather than MLT2A promoters in macaque pineal gland suggests that these MLT2A elements might have evolved differently in primate lineages. The coordinates of the exon–exon junctions are listed in Supplemental Figure S5H.
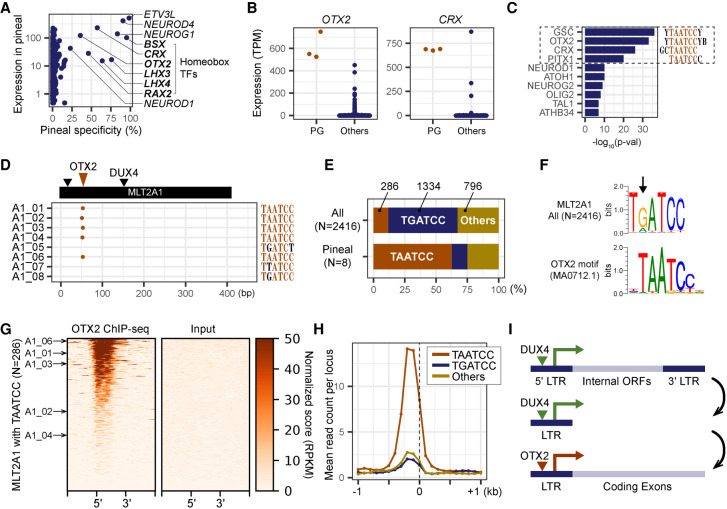
OTX2 is a potential regulator of MLT2A1 elements in pineal gland
Finally, we explored the mechanism of MLT2A element activation in pineal gland. We analyzed single- and multimap reads from DUX4 loci, including DUX4L1 to DUX4L8, and detected no expression in pineal gland. To check whether the MLT2A elements are activated by different transcription factors specific to pineal gland, we evaluated the expression values of 1665 transcription factors in 718 samples of human primary cells using the FANTOM5 expression atlas. We identified several transcription factor genes, including the homeobox genes BSX, CRX, OTX2, LHX3, LHX4, and RAX4, that are predominantly expressed in pineal gland (Fig. 6A,B). We also searched for transcription factor binding motifs enriched in promoter regions of the genes that are specifically expressed in pineal gland. We identified a TAATCC motif commonly recognized by some homeobox transcription factors (Fig. 6C). Among them OTX2 and CRX are reportedly key regulators in murine pineal glands (Rohde et al. 2019). According to Liu et al. (2019), MLT2A1 elements contain the OTX2 binding motif. We detected this motif (MA0712.1) in five of the eight MLT2A1 elements expressed in pineal gland (Fig. 6D). The TAATCC motif was located around nucleotide position 50 between two DUX4-binding sites. However, the most frequent sequence at this position in MLT2A1 elements is not TAATCC (11.8%) but TGATCC (55.2%) (Fig. 6E,F). Thus, the presence of TAATCC at this position might be important for activation by OTX2 in pineal gland.
Figure 6.
OTX2 as a potential regulator of MLT2A elements in pineal gland. (A) Scatter plot of the levels of pineal gland expression of 1665 human transcription factors. The x-axis indicates the specificity of pineal gland expression (pineal gland expression as a percentage of the total expression of all samples). Gene names of the top 10 transcription factors (displaying the highest pineal specificity) are listed. (B) Normalized expression values of OTX2 and CRX genes in three pineal glands (PG) and 715 other samples. (C) The top 10 transcription factors significantly enriched in promoter regions of pineal-specific genes identified by using HOMER. (D) Distribution of OTX2 binding motifs (MA0712.1) in eight MLT2A1 elements; the motifs were identified by using MAST software. (E) The percentages of MLT2A elements that have TAATCC, TGATCC, or other hexamer sequences. (F) Sequence logos of OTX2 motif-enriched regions in MLT2A1 and the original OTX2 motif. (G) OTX2 binding states in 286 MLT2A elements that have the TAATCC sequence and flanking regions (±1 kb) for ChIP-seq and input samples. The pineal-active five MLT2A elements are denoted by arrows. (H) Normalized ChIP-seq read counts on MLT2A1 elements that have TAATCC (N = 286), TGATCC (N = 1334), or other (N = 796) sequences. (I) Schematic representation of the transition of LTR retrotransposons from full-length to the solo-LTR fused to a protein-coding gene.
To examine whether OTX2 binds to MLT2A1 elements, we used a publicly available OTX2 ChIP-seq data set for a human medulloblastoma cell line, D283 (GEO accession number GSE92582) (Boulay et al. 2017). We found that MLT2A1 elements with TAATCC (N = 286) have clear OTX2-binding signals (Fig. 6G), unlike those with TGATCC (N = 1334) and others (N = 796) (Fig. 6H; Supplemental Fig. S6A). Phylogenetic analysis indicated that the pineal-active MLT2A1 elements with TAATCC are not a single lineage (Supplemental Fig. S6B). These results suggest that OTX2 might activate MLT2A1 elements that happened to have the TAATCC motif in the course of evolution (Fig. 6I). Because hundreds of MLT2A1 elements with TAATCC motifs in human and macaque genomes were not expressed in pineal gland, the presence of TAATCC does not necessarily indicate the expression by OTX2 in pineal gland. Conversely, because few MLT2A2 elements have the TAATCC motif and ChIP-seq signals (Supplemental Fig. S6C,D), other transcription factors probably activate MLT2A2 elements in pineal gland.
Discussion
We identified the embryonic LTR families MLT2A1 and MLT2A2, which start to be transcribed at the four-cell stage. The family-wide activation in embryos is conserved among simian primates (human, macaque, and marmoset), whereas individual elements are expressed differently. We observed that, after the eight-cell stage, the MLT2A families are repressed, and they generally remain silenced in somatic tissues. However, we discovered that 21 MLT2A elements are expressed in human pineal gland or amniotic membrane; in the former, three of them serve as novel promoters of protein-coding genes. The pineal gland is a neuroendocrine organ responsible for synthesis of the hormone melatonin (Maronde and Stehle 2007), in which homeobox genes play essential roles (Rath et al. 2013). Our results that the pineal homeobox transcription factor OTX2 binds to a subset of MLT2A1 elements suggest that MLT2A elements started to be used or optimized during evolution as parts of genes expressed in pineal gland.
How can embryonic MLT2As create promoters for somatic tissues? Our hypothesis is that a subset of MLT2A elements altered their tissue specificity and gained the ability to be expressed in somatic tissues by tissue-specific local transcription factors (Fig. 6I). First, about 45–65 million years ago, many copies of the MLT2A families were inserted into genomes, most likely by the activity of DUX4 during early embryogenesis. Next, the inserted full-length MLT2A elements were gradually transformed into solo-LTRs, which lost their open reading frames. The formation of solo-LTRs through homologous recombination between 5′ and 3′ LTRs is a unique feature of LTR retrotransposons and might be beneficial to the host genome evolution (Huh et al. 2006; Belshaw et al. 2007). The advantage of the transformation is to fix LTRs as potential regulatory elements without the threat of further genomic damage. Eventually, thousands of MLT2A elements were fixed in the population as a large pool of promoter modules. These elements did not necessarily confer immediate selective advantage to the host, and therefore their sequences were free to accumulate mutations. Later, a small subset of MLT2A elements started to be expressed in somatic tissues by local transcription factors. In pineal gland, three MLT2A elements fused to ABCE1, COL5A1, or GALNT13 as their novel promoters. COL5A1 encodes collagen type V alpha 1 chain; transcription from the MLT2A1 promoter would result in in-frame translation of a shortened COL5A1 protein. COL5A1 is the most commonly mutated gene in classic Ehlers–Danlos syndrome type 1, with hyperelasticity and dystrophic scarring of the skin and joint laxity as hallmarks (Villefranche criteria), as well as muscular hypotonia with variable presentation in some patients (Symoens et al. 2012). Inappropriate activation of DUX4 in facioscapulohumeral muscular dystrophy might contribute to its pathogenesis by activating MLT2A1 and interfering with COL5A1 synthesis.
OTX2 binds the TAATCC motif (Bunt et al. 2012), which was present in ∼12% of MLT2A1 elements. Indeed, the MLT2A1 elements with TAATCC were bound by OTX2, according to the ChIP-seq data of a human medulloblastoma cell line. In pineal gland, OTX2 alone cannot explain the expression of all the MLT2A elements. This study suggests an evolutionary process in which embryonic LTR retrotransposons supply novel promoters to a somatic tissue.
Methods
Differential expression analysis using STRT data
STRT raw sequence data under European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) accession number PRJEB8994 were aligned to the human genome (hg38/GRCh38) by using BWA (Burrows-Wheeler Aligner) version 0.7.15 with “mem -L 5,0” options (Li and Durbin 2009). Uniquely mapped reads (single-map reads) with a minimum mapping quality of 10 were used in this study except for two analyses shown in Supplemental Figure S1C and S2F, denoted as multimap. The reads were counted for each family of LINE, SINE, and LTR retrotransposons according to the definition in RepeatMasker (http://www.repeatmasker.org) by using the intersectBed command in BEDTools version 2.26.0 (Quinlan and Hall 2010). Differentially expressed repeat families were identified by using the Bioconductor package edgeR version 3.14.0 with estimateCommonDisp, estimateTagwiseDisp, and exactTest functions (Robinson et al. 2010). The coordinates were transformed to local coordinates on consensus sequences of MLT2A1 and MLT2A2 on the basis of the pairwise alignment between the individual elements and consensus sequences in Repbase (release 21.04).
Expression and splicing analysis using RNA-seq data
We downloaded publicly available RNA-seq data for human embryos from ArrayExpress (https://www.ebi.ac.uk/arrayexpress) accession number E-MTAB-3929, macaque embryos from NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) accession number GSE86938, marmoset embryos (ArrayExpress accession number E-MTAB-7078), immortalized human myoblasts transfected with plasmids encoding DUX4 (GEO accession number GSE85935), 10 human somatic tissues from DDBJ (https://www.ddbj.nig.ac.jp) accession number DRA000991, human pineal glands (GEO accession number GSE100472), and macaque pineal glands (GEO accession number GSE78165) (Supplemental Figs. S1A, S2A, S3A, and S5D). The raw sequences were aligned to the human (hg38/GRCh38), the macaque (rheMac8), or the marmoset (calJac3) genome, by using STAR with “‐‐outSAMattributes NH HI AS nM NM XS ‐‐outSAMattrIHstart 0 ‐‐outSAMstrandField intronMotif ‐‐outFilterIntronMotifs RemoveNoncanonical” options (Dobin et al. 2013). Genomic coordinates of high-confidence splice junctions reported in “SJ.out.tab” by the STAR aligner were used to evaluate the frequency of splice events within MLT2A elements and to identify exon–exon junctions between MLT2A elements and protein-coding exons. RNA-seq alignments were visualized by using ZENBU (Severin et al. 2014).
In the analysis of pineal gland, transcripts were assembled by using StringTie version 1.3.5 (Pertea et al. 2015) with the “‐‐fr” option. The assembled transcripts were merged into a single file by using StringTie with the “‐‐merge” function. Spliced reads with S0, S1, and S2 exon–exon junctions were counted on the basis of the mapped positions and CIGAR strings reported in BAM files. Spliced reads were considered to be true if the donor and accepter positions of the read exactly matched the coordinates listed in Supplemental Figure S5H.
Chromatin accessibility and DUX4 binding analysis
We downloaded ATAC-seq data for human embryos (GEO accession number GSE101571), ChIP-seq data of DUX4 for human myoblasts transduced with lentivirus carrying DUX4 (GEO accession number GSE33838), and ChIP-seq data of OTX2 for a human medulloblastoma cell line, D283 (GEO accession number GSE92582). The raw sequences were aligned to the human genome (hg38/GRCh38) by using BWA version 0.7.15 with the “mem” option. Coverage track files were generated by using deepTools2 with “bamCoverage binSize 20 ‐‐normalizeUsing RPKM ‐‐minMapping Quality 10 –ignoreDuplicates” options (Ramírez et al. 2016). Signal scores within 1 kb from individual MLT2 elements in 20-bp bins were calculated by using deepTools2 with “computeMatrix scale-regions -a 1000 -b 1000” options, and the scores were visualized as heatmap and intensity plots with the “plotHeatmap” function.
A position-specific scoring matrix file of the DUX4 binding motif (MA0468.1) was downloaded from footprintDB (Sebastian and Contreras-Moreira 2014). DNA sequences of individual MLT2 elements were extracted from the human genome (hg38/GRCh38) by using fastaFromBed command in BEDTools. DUX4-binding sites in MLT2 sequences were identified by using MAST (Bailey and Gribskov 1998) with the “-hit_list” option. Sequences of DUX4-binding sites in forward and reverse orientations, TATA boxes, TSSs, and splice sites were identified from multiple sequence alignments of MLT2A elements constructed by MAFFT version 7.307 with the “‐‐maxiterate 1000” option (Katoh and Standley 2013), and sequence logos were generated by using WebLogo version 3.6.0 (Crooks et al. 2004).
Phylogenetic analysis
The RepeatMasker tracks of seven primates (genome assemblies: panTro6, panPan2, rheMac8, calJac3, saiBol1, tarSyr2, micMur2) and mouse (mm10) were downloaded from the UCSC Table Browser (Karolchik et al. 2004). Copy numbers of long MLT2A1 and MLT2A2 elements (>200 bp) were counted for each genome on the basis of the RepeatMasker definition of these elements.
The genomic coordinates of macaque and marmoset MLT2A elements were converted to the coordinates in the human genome using liftOver with the “-minMatch=0.5” option. A phylogenetic tree was constructed using the ngphylogeny.fr pipeline with MAFFT alignment, BMGE curation, FastME tree inference, and iTOL visualization (Lemoine et al. 2019).
cDNA cloning
Embryos were collected in Sweden (Dnr 2010/937–31/4 of the Regional Ethics Board in Stockholm), and three cDNA libraries derived from different human eight-cell embryos were prepared (Tang et al. 2010; Töhönen et al. 2015). MLT2A transcripts were PCR amplified from the cDNA libraries by using Phusion High-Fidelity DNA polymerase or HotStarTaq Plus DNA polymerase. Each PCR product was cloned into a pCR4Blunt-TOPO vector (for the product by Phusion polymerase) or a pCRII-dual promoter TOPO vector (for the product by HotStarTaq Plus polymerase) and sequenced using T7 primer at Eurofins Genomics, Germany.
Expression analysis of MLT2A1 and MLT2A2 in 718 samples
We downloaded CAGE data in the ctss.bed format from the FANTOM5 web sites (https://fantom.gsc.riken.jp/5/datafiles/reprocessed/hg38_latest/basic/human.tissue.hCAGE/ and https://fantom.gsc.riken.jp/5/datafiles/reprocessed/hg38_latest/basic/human.primary_cell.hCAGE/). Expression values of individual MLT2A elements were calculated for samples of the 178 tissues and 540 primary cells that had at least 1 million reads. The full sample list is available in Supplemental Table S3.
OTX2 binding analysis
Expression values of 1665 human transcription factors for 718 samples were obtained from the FANTOM5 expression atlas. For each transcription factor, expression specificity in pineal gland was calculated as the total expression of pineal gland samples divided by the total expression of all samples.
Known motif enrichment analysis was performed by using the findMotifs.pl script in HOMER version 4.9.1 (Heinz et al. 2010). The input DNA sequences were extracted from transcription start sites with upstream 150-bp sequences of selected genes. Pineal-specific genes (>50% specificity) were selected for the foreground, and pineal unexpressed genes (0% specificity) were selected for the background from all genes expressed at least 100 TPM in total of all samples.
Data access
Sequences of cDNA clones generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) under accession numbers LR694118–LR694126.
Supplementary Material
Acknowledgments
This work was supported by a Research Grant from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) to the RIKEN Center for Integrative Medical Sciences. K.H. was supported by a Research Grant from Institute for Protein Research, Osaka University. S.K. was supported by Jane and Aatos Erkko Foundation. This study was initiated when J.K. was a Japan Society for the Promotion of Science Fellow at RIKEN Yokohama.
Author contributions: K.H. and S.K. analyzed the data. K.H., J.K., and S.K. wrote and edited the manuscript. E.-M.J., V.T., J.K., and S.K. prepared resources. P.C., J.K., and S.K. supervised research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275354.121.
Competing interest statement
The authors declare no competing interests.
References
- Backlund PS, Urbanski HF, Doll MA, Hein DW, Bozinoski M, Mason CE, Coon SL, Klein DC. 2017. Daily rhythm in plasma N-acetyltryptamine. J Biol Rhythms 32: 195–211. 10.1177/0748730417700458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54. 10.1093/bioinformatics/14.1.48 [DOI] [PubMed] [Google Scholar]
- Belshaw R, Pereira V, Katzourakis A, Talbot G, Pačes J, Burt A, Tristem M. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci 101: 4894–4899. 10.1073/pnas.0307800101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, Tristem M. 2007. Rate of recombinational deletion among human endogenous retroviruses. J Virol 81: 9437–9442. 10.1128/JVI.02216-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Stirparo GG, Dietmann S, Hernando-Herraez I, Mohammed H, Reik W, Smith A, Sasaki E, Nichols J, Bertone P. 2018. Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 145: dev167833. 10.1242/dev.167833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay G, Awad ME, Riggi N, Archer TC, Iyer S, Boonseng WE, Rossetti NE, Naigles B, Rengarajan S, Volorio A, et al. 2017. OTX2 activity at distal regulatory elements shapes the chromatin landscape of group 3 medulloblastoma. Cancer Discov 7: 288–301. 10.1158/2159-8290.CD-16-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R, Kool M. 2012. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer 131: E21–E32. 10.1002/ijc.26474 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang Y. 2019. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet 51: 947–951. 10.1038/s41588-019-0418-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114. 10.1016/j.gene.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durruthy-Durruthy J, Sebastiano V, Wossidlo M, Cepeda D, Cui J, Grow EJ, Davila J, Mall M, Wong WH, Wysocka J, et al. 2016. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat Genet 48: 44–52. 10.1038/ng.3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl JO, Giesige CR, Domire JS, Wallace LM, Fowler AM, Guckes SM, Garwick-Coppens SE, Labhart P, Harper SQ. 2016. Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum Mol Genet 25: 4577–4589. 10.1093/hmg/ddw287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN PMI and CLST (DGT). 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, et al. 2014. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet 46: 558–566. 10.1038/ng.2965 [DOI] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, VanderMaarel SM, Ruzzo WL, Gentleman RC, Tawil R, et al. 2012. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 22: 38–51. 10.1016/j.devcel.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I. 2015. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16: 135–141. 10.1016/j.stem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Suzuki AM, Dos Santos A, Desterke C, Collino A, Ghisletti S, Braun E, Bonetti A, Fort A, Qin XY, et al. 2015. CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Res 25: 1812–1824. 10.1101/gr.191031.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. 2017. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49: 925–934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. 2017. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543: 199–204. 10.1038/nature21374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JW, Kim DS, Ha HS, Kim TH, Kim W, Kim HS. 2006. Formation of a new solo-LTR of the human endogenous retrovirus H family in human chromosome 21. Mol Cells 22: 360–363. [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Jacques PÉ, Jeyakani J, Bourque G. 2013. The majority of primate-specific regulatory sequences Are derived from transposable elements. PLoS Genet 9: e1003504. 10.1371/journal.pgen.1003504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467. 10.1159/000084979 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinricks AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Barber GP, Casper J, Clawson H, Diekhans M, Gonzalez JN, Hinrichs AS, Lee BT, Nassar LR, Powell CC, et al. 2020. UCSC Genome Browser enters 20th year. Nucleic Acids Res 48: D756–D761. 10.1093/nar/gkz1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, Gascuel O. 2019. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res 47: W260–W265. 10.1093/nar/gkz303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Leng L, Liu C, Lu C, Yuan Y, Wu L, Gong F, Zhang S, Wei X, Wang M, et al. 2019. An integrated chromatin accessibility and transcriptome landscape of human pre-implantation embryos. Nat Commun 10: 364. 10.1038/s41467-018-08244-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sachs F, Ramsay LA, Jacques PÉ, Göke J, Bourque G, Ng HH. 2014. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 21: 423–425. 10.1038/nsmb.2799 [DOI] [PubMed] [Google Scholar]
- Mager DL, Goodchild NL. 1989. Homologous recombination between the LTRs of a human retrovirus-like element causes a 5-kb deletion in two siblings. Am J Hum Genet 45: 848–854. [PMC free article] [PubMed] [Google Scholar]
- Maronde E, Stehle JH. 2007. The mammalian pineal gland: known facts, unknown facets. Trends Endocrinol Metab 18: 142–149. 10.1016/j.tem.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi S, Nakagawa S, Sasaki-Honda M, Sakurai H, Frith MC, Mitsuhashi H. 2021. Nanopore direct RNA sequencing detects DUX4-activated repeats and isoforms in human muscle cells. Hum Mol Genet 30: 552–563. 10.1093/hmg/ddab063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F. 2016. Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165: 1012–1026. 10.1016/j.cell.2016.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MF, Rohde K, Klein DC, Møller M. 2013. Homeobox genes in the rodent pineal gland: roles in development and phenotype maintenance. Neurochem Res 38: 1100–1112. 10.1007/s11064-012-0906-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21–42. 10.1146/annurev-genet-110711-155621 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K, Hertz H, Rath MF. 2019. Homeobox genes in melatonin-producing pinealocytes: Otx2 and Crx act to promote hormone synthesis in the mature rat pineal gland. J Pineal Res 66: e12567. 10.1111/jpi.12567 [DOI] [PubMed] [Google Scholar]
- Sebastian A, Contreras-Moreira B. 2014. footprintDB: a database of transcription factors with annotated cis elements and binding interfaces. Bioinformatics 30: 258–265. 10.1093/bioinformatics/btt663 [DOI] [PubMed] [Google Scholar]
- Severin J, Lizio M, Harshbarger J, Kawaji H, Daub CO, Hayashizaki Y, FANTOM Consortium, Bertin N, Forrest ARR. 2014. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat Biotechnol 32: 217–219. 10.1038/nbt.2840 [DOI] [PubMed] [Google Scholar]
- Symoens S, Syx D, Malfait F, Callewaert B, De Backer J, Vanakker O, Coucke P, De Paepe A. 2012. Comprehensive molecular analysis demonstrates type V collagen mutations in over 90% of patients with classic EDS and allows to refine diagnostic criteria. Hum Mutat 33: 1485–1493. 10.1002/humu.22137 [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. 2010. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 5: 516–535. 10.1038/nprot.2009.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PJ, Macfarlan TS, Lorincz MC. 2016. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell 62: 766–776. 10.1016/j.molcel.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töhönen V, Katayama S, Vesterlund L, Jouhilahti EM, Sheikhi M, Madissoon E, Filippini-Cattaneo G, Jaconi M, Johnsson A, Bürglin TR, et al. 2015. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun 6: 8207. 10.1038/ncomms9207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park YS, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res 27: 1623–1633. 10.1101/gr.218149.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. 2013. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res 41: e74. 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu D, He D, Suo S, Xia X, He X, Han JDJ, Zheng P. 2017. Transcriptome analyses of rhesus monkey preimplantation embryos reveal a reduced capacity for DNA double-strand break repair in primate oocytes and early embryos. Genome Res 27: 567–579. 10.1101/gr.198044.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xu J, Liu B, Yao G, Wang P, Lin Z, Huang B, Wang X, Li T, Shi S, et al. 2018. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557: 256–260. 10.1038/s41586-018-0080-8 [DOI] [PubMed] [Google Scholar]
- Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, Tapscott SJ. 2013. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet 9: e1003947. 10.1371/journal.pgen.1003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu Y, Hu R, Lee AY, et al. 2019. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet 51: 1380–1388. 10.1038/s41588-019-0479-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.