Abstract
To evaluate how age and Apolipoprotein E-ε4 (APOE4) status interact with APOE-independent polygenic risk score (PRSnon-APOE), we estimated PRSnon-APOE in superagers (age ≥ 90 years, N=346), 89− controls (age 60–89, N=2,930) and Alzheimer’s Disease (AD) cases (N=1,760). Employing superagers, we see a nearly five times greater odds ratio (OR) for AD comparing the top PRSnon-APOE decile to the lowest decile (OR=4.82, P=2.5×10−6), which is twice the OR as using 89 controls (OR=2.38, P=4.6×10−9). Thus PRSnon-APOE is correlated with age, which in turn is associated with APOE. Further exploring these relationships, we find that PRSnon-APOE modifies age-at-onset among APOE4 carriers, but not among non-carriers. More specifically, PRSnon-APOE in the top decile predicts an age-at-onset five years earlier compared to the lowest decile (70.1 vs 75.0 years; t-test P=2.4×10−5) among APOE4 carriers. This disproportionally large PRSnon-APOE among younger APOE4-positive cases is reflected in a significant statistical interaction between APOE4 status and age-at-onset (β=−0.02, P=4.8×10−3) as a predictor of PRSnon-APOE. Thus, the known AD risk variants are particularly detrimental in young APOE4 carriers.
Keywords: Alzheimer’s disease, Polygenic risk score, Superager, Genetic risks, Risk interactions, APOE
1. Introduction
Alzheimer’s disease (AD) has an estimated heritability of 59–79% (Gatz et al., 2006) and a decades long pre-clinical stage, during which AD-specific pathological changes develop without clinical signs of cognitive decline (Bateman et al., 2012; Jack et al., 2010). Low-cost non-invasive testing would ideally identify at-risk individuals in the pre-clinical stage to enable preventive intervention. To this end, genetic risk factors have been proposed as useful predictors. APOE is a major risk gene for AD with dose-dependent risk effects. Compared to non-carriers of the APOE-ε4 allele (APOE4), heterozygous carriers of APOE4 have a three- to fourfold and homozygous carriers up to a 14-fold increased odds of developing AD (Farrer et al., 1997; Genin et al., 2011), whereas the APOE-ε2 allele (APOE2) is protective (odds ratio, OR=0.6) (Bertram et al., 2008; Bickeböller et al., 1997; Corder et al., 1994; Genin et al., 2011; Kunkle et al., 2019a). The effects of APOE variants have been found to be age-dependent (Rasmussen et al., 2018). In addition to the APOE gene, genome-wide association studies (GWAS) and meta-analysis of GWAS have identified 39 risk loci in subjects with European ancestry (Jansen et al., 2019; Kunkle et al., 2019b; Lambert et al., 2013b; Marioni et al., 2018). Desikan et al. constructed a polygenic hazard score (PHS) by combining AD-associated single nucleotide polymorphisms (SNPs) from GWAS from the International Genomics of Alzheimer’s Project (IGAP) and disease incidence estimates from the United States population (Desikan et al., 2017). This PHS was reported to be predictive for age-at-onset (AAO) and useful to identify individuals at greatest risk for developing AD at a given age. However, the interactions between APOE-independent genetic risks, age and APOE have not been evaluated. Similar to PHS, individual genetic risk can be aggregated into a polygenic risk score (PRS) by summing the number of risk alleles, each weighted by its effect size derived from independent GWAS summary statistics (Euesden et al., 2015; Khera et al., 2018; Purcell et al., 2009). Both PRS and PHS have been applied to AD and showed similar results (Leonenko et al., 2019) although a conflicting study (Fan et al., 2020) reported that PHS is superior for predicting age at onset and other age-related phenotypes, particularly when stratified by sex. PRS, rather than PHS is broadly used and may be considered the current state of the art for genetic risk prediction. In addition, because PRS quantifies genetic risk in individuals independently of age, PRS can also be used to explore the interactions between genetics and other causes of disease, including age. PRS has been reported to be useful to identify individuals at high risk for common diseases such as coronary artery disease (Khera et al., 2018). For AD, the predictive power of PRS derived from the GWAS meta-analysis (Lambert et al., 2013a) reached an area under the curve (AUC) of 0.78 in clinically-defined AD when genetic risk factors were combined with demographic factors, and 0.84 in pathologically-confirmed AD (Escott-Price et al., 2017; Escott-Price et al., 2015).
The predictive performance of PRS was shown to depend on the prevalence of the disease in a specific population (Gibson et al., 2019), which is meaningful for AD, given that prevalence of AD increases exponentially with age (Mayeux and Stern, 2012). Furthermore, it is important to note that APOE is not only a major constituent for predicting AD (Desikan et al., 2017; Escott-Price et al., 2015), but is also strongly associated with age itself (Deelen et al., 2019; Sebastiani et al., 2019) as well as AAO of AD (Olarte et al., 2006). Therefore, it is important to investigate whether and how APOE-independent PRS (PRSnon-APOE) depends on age and APOE, both for the interpretation of AD PRS and to understand how genetic risk is distributed in case-control AD genetics studies.
To address this question, we stratified healthy controls into superagers aged 90 years or older without dementia (90+; range 90–109 years) and subjects aged 89 or younger (89−; range 60–89 years). To understand the role of age with regard to PRSnon-APOE, we compared these two control cohorts to AD cases with a broad range of AAO (range 60–99 years). We further investigated whether PRSnon-APOE is correlated with AAO in cases and age of controls and whether the APOE4 risk allele would modify such a correlation.
2. Methods
2.1. Participants
To ensure that there is no overlap with the samples included in the 2013 IGAP meta-analysis (Lambert et al., 2013b), we selected 13 independent cohorts consisting of a total of 1,733 cases and 3,121 controls from the Alzheimer’s Disease Genetics Consortium (ADGC) (Supplementary Methods). Additionally, we included an independent sample of 182 subjects with Ashkenazi ancestry from the Litwin Zucker Alzheimer’s Research Center at the Feinstein Institutes for Medical Research (LZ). The sample ascertainment and assessment methods were described previously (Adelson et al., 2019; Freudenberg-Hua et al., 2016; Koppel et al., 2018). A visual overview of data preparation and analysis workflow is in Supplementary Figure 1.
Age was defined using ADGC criteria. For ADGC cases, age is defined as AAO as reported by the study (n=1,574). If AAO is not reported, we estimated AAO by subtracting ten years from age at death (AAD, n=102) and for those without AAD, age at last examination (AAE) is used (n=57). AAO estimation has been used in previous studies using the same cohorts (Desikan et al., 2017; Jun et al., 2010; Naj et al., 2014). To ensure that estimated AAO does not introduce biases, we further performed sensitivity analysis by removing individuals with estimated AAO from the dataset. For the ADGC controls, AAE is used (n=2,995) and AAD is used for a small proportion of controls (n=126). For the LZ cohort, age is reported as age at enrollment for cases (n=27), and as AAE for controls (89−: n=35, 90+: n=120). The total sample utilized in this study includes 1,760 AD cases (60–99 years), 2,930 89− controls, and 346 90+ controls. Sample and genotype data quality control procedures and principal component analysis are described in Supplementary Methods. The demographics of the sample set following merging and QC are shown in Supplementary Table 1. The distribution of APOE genotypes among cases and controls is described in Supplementary Table 2.
2.2. Polygenic Risk Score Estimation
A PRS is a sum of genetic variants, weighted by variant effect sizes for a given trait. The effect sizes are estimated from independent GWAS summary statistics. PRSnon-APOE is calculated at various P-value thresholds (PT) based on GWAS summary statistics (Lambert et al., 2013c) excluding the APOE region (Supplementary Methods). We separately optimized PRSnon-APOE using two control sets – ADGC controls without age restriction and 90+ controls (Supplementary Figure 2) – which resulted in identical optimal PT for the most predictive PRS at PT = 1×10−5. Details of PRS calculation are in Supplementary Methods.
2.3. Statistical Analysis
Statistical analysis was performed using the R software version 3.6.3 (R Core Team, 2019). Linear or logistic regression models included covariates for sex and the top ten principal components (PCs), as well as the variables of interest. All plots and statistical tests (except for those in Figure 1a) used Z-standardized PRS. To facilitate comparison with previous findings on polygenic risks and survival, we plotted a Kaplan-Meier survival curve with the primary event being AAO of AD and we calculated hazard ratio using the R packages “survival” and “survminer”.
Figure 1. PRSnon-APOE Distribution and Effect Sizes in the AD Case and Control Groups (Residualized).
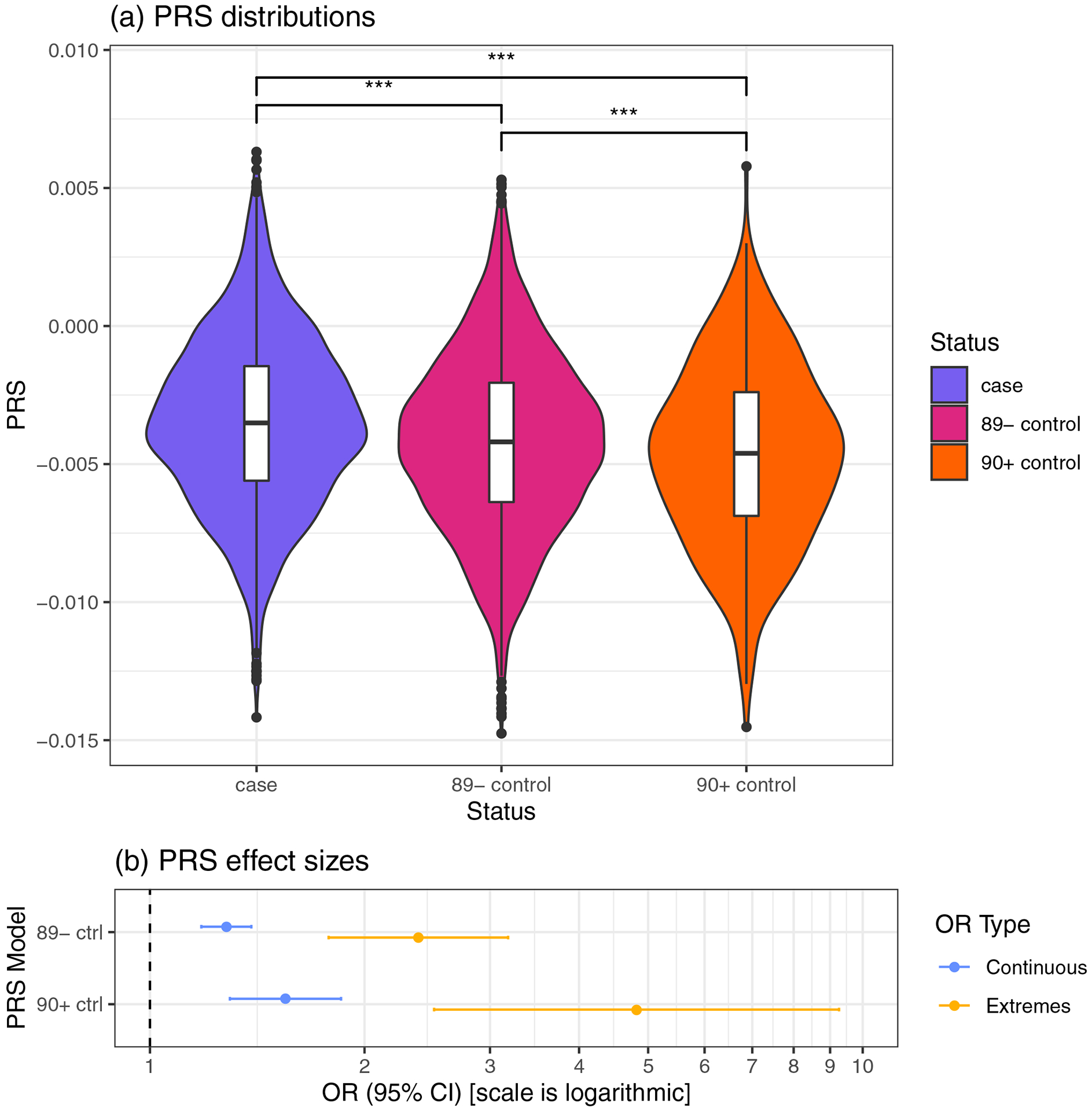
(a), Violin plots comparing PRSnon-APOE in AD cases (case), controls age 60–89 years (89−), and superager controls age ≥ 90 years (90+). Pairwise comparison Bonferroni P-values: *** highly significant (P < 0.001). (b), PRS effects on AD risk. Continuous OR (blue): odds ratio of having AD for a one SD increase in PRSnon-APOE. Extremes OR (orange): odds ratio of having AD with PRSnon-APOE in the top decile compared to having PRSnon-APOE in the lowest decile.
Abbreviations: 90+ ctrl, controls ≥ 90 years old were used in analysis; 89− ctrl, controls age 60–89; AD, Alzheimer’s disease; CI, confidence interval; OR, odds ratio; PRSnon-APOE, APOE-independent polygenic risk score.
To compare PRS distributions across cases, 89− controls, and 90+ controls, we performed a one-way ANOVA and pairwise Student’s t-tests. In order to ensure that observed differences were not due to ethnicity or sex, we tested the residuals of a linear model with sex and ten PCs as predictors and PRS as the response variable.
We evaluated the association between PRS and AD status in two datasets: 1) cases and 89− controls; and 2) cases and superager (90+) controls. To determine the difference of odds ratio (OR) for AD between the extremes of PRS, we selected the bottom and top deciles of the PRS distribution and performed logistic regressions between PRS decile and AD status (extremes model). To determine the effect of a one standard deviation (SD) increase in PRS, we performed logistic regressions with PRS – z-standardized across all samples – as the predictor and AD status as the response (continuous model). The effect size of this continuous model is useful because it does not depend on the extremeness of PRS quantization (e.g. top and bottom 10% vs. 5%).
It has been previously shown that polygenic risk predicts AAO of AD. In this study we further investigate how age and APOE4 status modify the effects of PRS. Therefore, we modeled the association between age, APOE genotype (i.e., 2/2, 2/3, 3/4, or 4/4, dummied with APOE3/3 as the contrast variable), and PRS using linear regression, with covariates for confounding variables as before. To determine if these effects are independent of AD status, we performed an additional linear regression controlling for AD status. We also tested for collinearity by calculating the variance inflation factor (VIF) with the “car” package.
To test the interaction between age and APOE genotype on PRS, we stratified by AD case-control status, categorized samples as APOE4 carriers and non-carriers, and then tested for interactions. To improve interpretability by reducing the number of interactions tested, we collapsed the APOE genotypes into binary categories – an APOE4 carrier was defined as having APOE3/4 or 4/4 (excluding 2/4), and a non-carrier as APOE2/2, 2/3 or 3/3. We first performed linear regressions separately in cases and controls, with APOE4 carrier status and age as predictors and PRS as the response. We then performed a second linear regression stratified by AD status, adding the interaction between APOE4 carrier status and age as a predictor.
Finally, to test for the effect of these interactions on case-control status, we performed logistic regressions for the relationship between AD case-control status, APOE4 carrier status, age, and PRS, with and without interactions and confirmed independence of predictors with VIF.
3. Results
3.1. Superagers have the lowest PRSnon-APOE
PRSnon-APOE is different between cases, 89− controls, and 90+ controls after residualizing for sex and ancestry PCs (ANOVA P=1×10−19) (Figure 1a). AD subjects have significantly higher PRSnon-APOE compared to both 89− controls (Bonferroni adj. post-hoc t-test, P=5.5×10−4) and 90+ controls (P=3.6×10−20). The 89− controls have significantly higher PRSnon-APOE than 90+ controls (P=5.1×10−14). This difference shows the dependence of PRSnon-APOE on age among healthy controls. To further evaluate this age dependence, we assessed the effect of PRSnon-APOE on AD risk by comparing cases with 89− controls and then with 90+ controls (Figure 1b). We first calculated “extremes” ORs between subjects in the bottom and top deciles of PRSnon-APOE. Comparing 90+ controls and AD cases, subjects having PRSnon-APOE in the top decile are nearly five times more likely to have AD than subjects with PRSnon-APOE in the lowest decile (OR=4.82, CI: 2.50–9.26, P=2.5×10−6). This effect size is double the odds ratio using 89− controls (OR=2.38, CI: 1.78–3.18, P=4.6×10−9). This difference remains present with the alternative approach of “continuous” ORs measuring the effects of a one SD increase in PRSnon-APOE, albeit less pronounced (90+ controls: OR=1.55, CI: 1.29–1.85, P=1.8×10−6; 89-controls: OR=1.28, CI: 1.18–1.39, P=2.1×10−9). The higher effect sizes of PRSnon-APOE observed when comparing with 90+ controls are consistent with the effect sizes seen for the APOE4 alleles and genotypes (Supplementary Table 3).
3.2. The negative association between age and PRSnon-APOE interacts with APOE genotype in cases
The particularly low PRSnon-APOE observed in superagers led us to elucidate the relationship between PRS and age. Biologically, PRS can affect age of inclusion or onset in AD studies by modifying both AD risk and AAO. Here, we investigated age as a predictor for PRS to explore how non-APOE genetic predictors change with age in case-control studies, and how APOE genotype affects those changes. Age is negatively correlated with PRSnon-APOE among all subjects (Supplementary Table 4). In a linear regression including sex, APOE and ten PCs, age is a significant predictor of PRSnon-APOE (P=2.7×10−5, β=−0.007). This effect remains significant when accounting for case-control status (P=1.6×10−4, β=−0.006) (Supplementary Table 4). No APOE genotype is directly associated with PRSnon-APOE.
We next stratified the subjects nto APOE4 carriers and non-carriers to investigate whether APOE4 carrier status modifies the correlation between age and PRSnon-APOE. We find that this correlation depends on both case-control status and APOE4 status (Figure 2a). Interestingly, the negative correlation between age and PRSnon-APOE depends on APOE4 carrier status in cases, but not in controls (Figure 2b–c, Supplementary Table 5). More specifically, in AD cases, AAO is correlated with PRSnon-APOE (P=0.029, β=−0.007) and APOE4 carrier status is not associated with PRSnon-APOE (P=0.58). However, when the interaction between AAO and APOE (P=4.8×10−3, β=−0.02) is accounted for, APOE4 status becomes a significant predictor of PRSnon-APOE (P=6.1×10−3, β=1.29), whereas age is no longer a significant predictor (P=0.72). Thus, the inverse correlation between AAO and PRSnon-APOE among cases is primarily driven by the relationship between PRSnon-APOE on AAO among APOE4 carriers (Figure 2b). Consequently, among AD cases who are APOE4 carriers, those with PRSnon-APOE in the top decile have an AAO five years earlier than those in the bottom decile (70.1 vs. 75.0 years; 95% CI of difference in means: −7.2 – −2.7; P=2.4×10−5). This difference in AAO remains significant after removing APOE4/4 homozygous cases (71.1 vs. 76.7 years; P=2.1×10−5). A sensitivity analysis removing cases with estimated AAO further shows that these interaction results are robust (Supplementary Table 6).
Figure 2. Regression Plots Visualizing Interactions Between PRS, APOE4, and Age, in Cases and Controls.
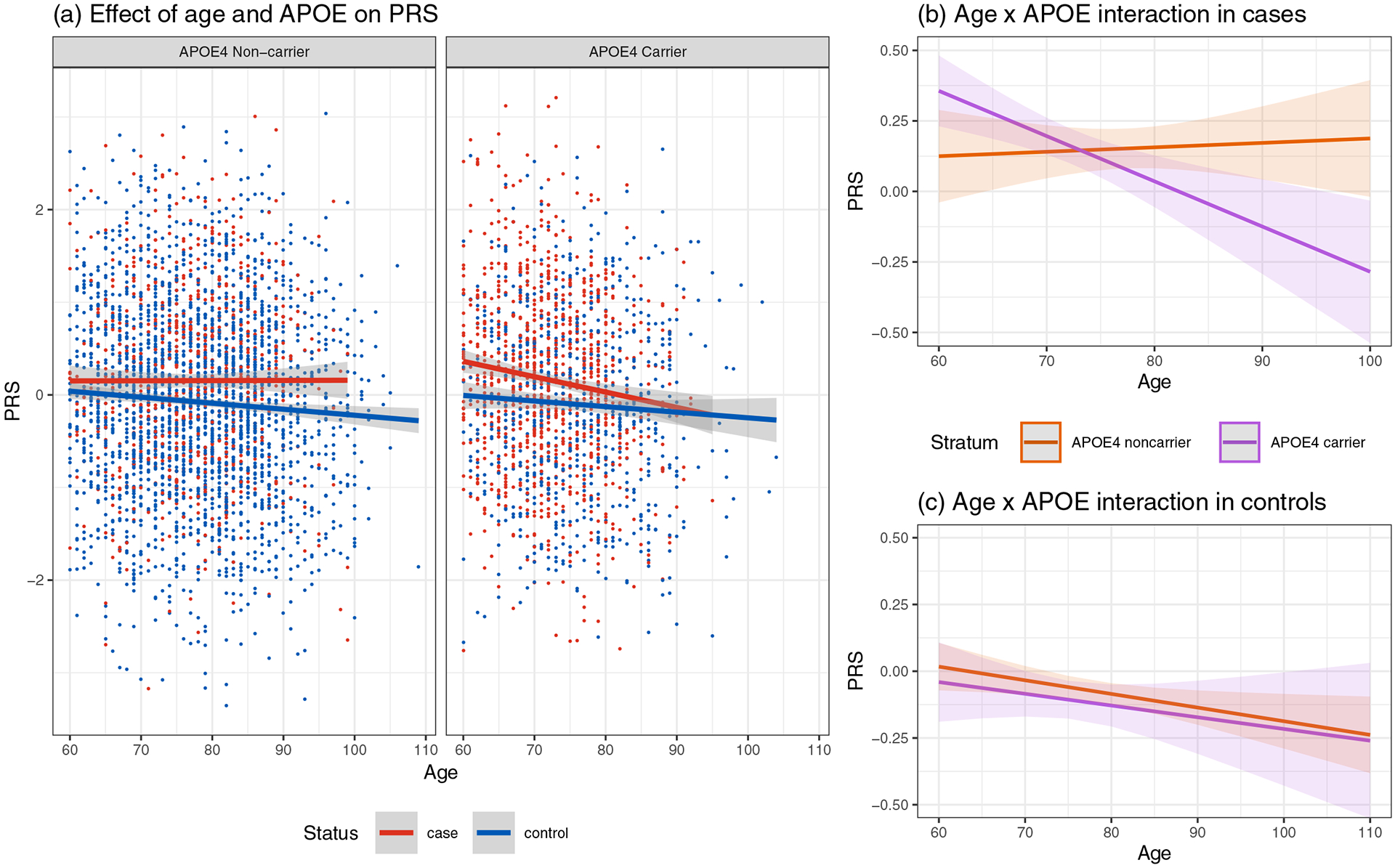
(a) Scatter plots and regression models showing the dependence of PRSnon-APOE on age in AD cases (red) and controls (blue). In AD cases age is defined as age-at-onset. The shaded areas show the 95% confidence intervals. PRSnon-APOE in cases and controls over age is shown separately among APOE4 non-carriers and carriers. (b) Age x APOE interaction in AD cases. (c) Age x APOE interaction in controls, which is negatively associated with PRS for both APOE4 carriers (purple) and non-carriers (orange). Abbreviations: AD, Alzheimer’s disease; APOE4, Apolipoprotein E-ε4 allele; PRSnon-APOE, APOE-independent polygenic risk score.
In contrast, the negative correlation between age and PRSnon-APOE (P=0.013, β=−0.005) does not depend on APOE4 carrier status among controls (Figure 2c, Supplementary Table 5), and APOE is not associated with PRSnon-APOE (P=0.27). Accounting for interactions between age and APOE4 status, the negative association between age and PRSnon-APOE remains significant (P=0.021, β=−0.005) and APOE4 status non-significant (P=0.78). The interaction between APOE4 and age is not significant (P=0.88). It is worthwhile to note that the main effect sizes while account ng for interactions do not have any intuitive interpretation due to the extrapolation of the age variable (Aschard, 2016).
3.3. Interactions between age, PRSnon-APOE, and APOE on AD status
The interactions between AAO and APOE4 status led us to evaluate the effect of the interactions between APOE4 status, age and PRSnon-APOE on AD status using logistic regression (Supplementary Table 7). Without modeling for interactions, age (P=2.4×10−6, β=−0.02), APOE4 carrier status (P=8.6×10−86, β=1.31) and PRSnon-APOE (P=2.7×10−12, β=0.23) are all predictors of AD status. When all interactions between these three predictors are included in the model, the main effects of age (P=0.60) and PRSnon-APOE (P=0.41) are no longer significant, indicating that the effects of age and PRSnon-APOE on AD status are dependent on APOE. APOE4 carrier status remains a significant predictor (P=8.2×10−15, β=4.79). Similarly, the main effects of the interactions between age and APOE (P=1.3×10−8, β=−0.05), between PRSnon-APOE and APOE (P=0.035, β=1.31), and the three-way interaction between age, PRS and APOE (P=0.035, β=−0.02) are all significant predictors for AD status.
The Receiver Operator Characteristics (ROC) are similar to previous reports when APOE SNPs are included (Supplementary Figure 3) and the Kaplan-Meier survival curve for PRSnon-APOE stratified by APOE4 carriers status is similar to that previously reported (Supplementary Figure 4) (Desikan et al., 2017).
4. Discussion
We show that APOE-independent polygenic risk has a disproportionally large effect in younger APOE4 carriers. Among APOE4 carriers, the AAO is on average five years earlier for cases with PRSnon-APOE in the top decile compared to those with PRSnon-APOE in the lowest decile. In contrast, no correlation between PRSnon-APOE and AAO was observed among APOE4 non-carriers. We further observe a negative correlation between age and PRSnon-APOE that is independent of APOE4 carrier status among controls, consistent with the notion that APOE-independent genetic risk increases AD incidence throughout late life. Our findings indicate that younger APOE4 carriers bear greater detrimental effect from currently known AD risk variants, as captured by PRSnon-APOE, whereas superager controls have fewer AD risk variants independent from APOE. Even though the age effect per year is small, it can be considerable in a population of subjects with ages ranging from 60 to over 100 years. This pattern may either be inherent to genetically conferred risk for AD or reflect biases in current PRS estimates as a result of ascertainment of previous GWAS subjects.
Compared with healthy superagers, subjects having PRSnon-APOE in the top decile have nearly five times the odds of developing AD than those in the bottom decile. This large effect size may be caused by depletion of AD risk alleles among superagers, indicating that many typical controls will develop AD later in life. This is consistent with the long pre-clinical stage of 20 to 25 years during which a subject does not display dementia symptoms (Bateman et al., 2012). Our observation also corroborates the reported higher effect sizes of GWAS risk alleles when superagers are used as controls (Tesi et al., 2019). The lifetime risk of AD for healthy females and males at 75 years old is estimated to be 26.2% and 19.9%, respectively (Brookmeyer et al., 2018). As the majority of controls enrolled in AD genetic studies are in their 70s and 80s, a considerable proportion of them are expected to convert to AD at a later age unless they die from other causes. These cryptic preclinical AD cases reduce the power of case-control studies. The larger magnitude of effect sizes for both PRSnon-APOE and APOE when 90+ controls were deployed in our study highlights the importance of inclusion of cognitively healthy superagers in future studies. Increasing the number of superager controls in case-control studies for AD is expected to increase the power to detect additional risk variants as well as protective variants.
The greater PRSnon-APOE among younger APOE4 positive cases, as reflected in the statistical interaction between AAO and APOE4 carrier status, suggests a greater role for the currently known APOE-independent genetic risk in younger APOE4 carriers. Although PRSnon-APOE alone is not useful to clinically predict individual risk for developing AD – perhaps unlike PHS/PRS including APOE (Desikan et al., 2017; Escott-Price et al., 2015) – the interactions we observed have implications for the design of future genetic association studies. In particular, enrollment of APOE4-negative cases irrespective of age may lead to the identification of novel variants, given the absent correlation of current PRSnon-APOE with AAO in this group. Novel non-genetic protective factors may best be found among healthy elderly APOE4 carriers with high PRSnon-APOE based on known GWAS risk loci, whereas novel risk factors may best be found among AD patients who have both low PRSnon-APOE and no APOE4 alleles.
Recently another study, investigating subjects that do not over ap with our study and using different parameters, reported a greater effect of APOE in younger participants (Bellou et al., 2020). Our stratified interaction analysis indicating a larger effect of PRSnon-APOE in younger APOE4 carriers leads us to a novel conclusion about APOE-independent genetic risk and provides a logical explanation for their observation: among all APOE4 cases, those with a younger onset carry the largest PRSnon-APOE.
Our study has several limitat ons, some of which are inherently shared by many large-scale AD genetic studies In order to reach sufficient power, genetic studies of complex diseases require very large sample size which is often achieved by pooling subjects from many studies that were initially independently designed. This approach introduces heterogeneity in diagnostic criteria, recorded demographics, and comorbidities (Jun et al., 2010; Naj et al., 2011). Such heterogeneity typically introduces noise and reduces statistical power for detecting true signals. There are now collaborative efforts by the National Institute on Aging to harmonize genetic, epidemiologic and clinical data for AD. Notwithstanding the importance of clinical details, genetic studies based solely on self-report of parental AD in the UK Biobank have yielded results that are highly correlated with the previously known IGAP loci (Andrews et al., 2020; Marioni et al., 2018). This indicates that genetic studies can generate valid findings without considering or correcting for all clinical details, when large sample sizes are used. In our study, AAO for AD was generally provided by the participating cohorts of ADGC (Desikan et al., 2017; Jun et al., 2010). Our sensitivity analysis shows that removing subjects with an estimated AAO does not substantially affect our overall results. However, a larger study may be required to fully measure the interactions between PRSno-APOE and the number of APOE4 and APOE2 alleles. Additionally, because of differences across ethnicities in PRS applicability (Martin et al., 2019) and APOE effects (Rajabli et al., 2018), studies in non-European populations are necessary to determine whether the same conclusions apply.
5. Conclusion
In summary, we show that current APOE-independent PRS has a particularly detrimental effect in younger APOE4 risk allele carriers, predicting significantly earlier age at onset for AD. APOE-independent PRS is negatively correlated with dementia-free aging. Future larger studies are needed to evaluate whether interactions between PRS, APOE and additional demographic, clinical, and environmental risk factors, as reflected in health data and biomarkers, can improve AD risk prediction.
Supplementary Material
Highlights.
APOE-independent polygenic risk is more detrimental in younger APOE4 carriers
Age-at-onset in APOE4+ cases is 5-years earlier with high vs. low polygenic risk
Lower APOE-independent genetic risk in older controls than younger controls
In APOE4 negative patients, age-at-onset is not correlated with polygenic risk
Acknowledgements
Funding: This work was supported by the National Institutes of Health, National Institute on Aging (NIH-NIA) (ADGC and grant numbers U01 AG032984 and RC2 AG036528). Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement awarded by the NIH-NIA (grant number U24 AG21886), were used in this study. The NACC database is funded by the NIH-NIA (grant number U01 AG016976). Data for this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (grant number U24-AG041689-01). In addition, this work was funded by the NIH (grant numbers U01AG052411, U01AG058635, K08AG054727, R01 AG618381, and R01 AG046949), and the Einstein Nathan Shock Center (grant number P30 AG038072). Additional support was provided by the Mildred and Frank Feinberg Family Foundation, the Advancing Women in Science and Medicine Foundation, and the JPB Foundation.
The authors would like to thank the contributors who collected samples used in this study during a period of 30 years, as well as patients and their families, whose help and participation made this work possible. A list of ADGC members and their affiliations are included in the Appendix within the Supplementary Content file.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
AMG has consulted for Eisai, Biogen, Pfizer, AbbVie, Cognition Therapeutics and GSK. She also served on the SAB at Denali Therapeutics from 2015–2018. YFH co-owns stock and stock options of Regeneron Pharmaceuticals. All other authors have no interests to declare. None of the author’s institution has contracts relating to this research through which it or any other organization may stand to gain financially now or in the future.
The work described in our manuscript entitled “Effect of Polygenic Score on Alzheimer’s Disease Risk Depends on Age and APOE Status” has not been published previously. It is not under consideration for publication elsewhere and its publication is approved by all authors and tacitly or explicitly by the responsible authorities where the work was carried out, and that, if accepted, it will not be published elsewhere in the same form, in English or in any other language, including electronically without the written consent of the copyright-holder.
All authors have reviewed the contents of the manuscript being submitted, approve of its contents and validate the accuracy of the data.
References
- Adelson RP, Renton AE, Li W, Barzilai N, Atzmon G, Goate AM, Davies P, Freudenberg-Hua Y, 2019. Empirical design of a variant quality control pipeline for whole genome sequencing data using replicate discordance. Sci Rep 9(1), 16156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SJ, Fulton-Howard B, Goate A, 2020. Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol 19(4), 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, 2016. A perspective on interaction effects in genetic association studies. Genet Epidemiol 40(8), 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, Marcus DS, Cairns NJ, Xie X, Blazey TM, Holtzman DM, Santacruz A, Buckles V, Oliver A, Moulder K, Aisen PS, Ghetti B, Klunk WE, McDade E, Martins RN, Masters CL, Mayeux R, Ringman JM, Rossor MN, Schofield PR, Sperling RA, Salloway S, Morris JC, Network DIA, 2012. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 367(9), 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellou E, Baker E, Leonenko G, Bracher-Smith M, Daunt P, Menzies G, Williams J, Escott-Price V, Initiative, A.s.D.N., 2020. Age-dependent effect of APOE and polygenic component on Alzheimer’s disease. Neurobiol Aging 93, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE, 2008. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet 83(5), 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickeböller H, Campion D, Brice A, Amouyel P, Hannequin D, Didierjean O, Penet C, Martin C, Pérez-Tur J, Michon A, Dubois B, Ledoze F, Thomas-Anterion C, Pasquier F, Puel M, Demonet JF, Moreaud O, Babron MC, Meulien D, Guez D, Chartier-Harlin MC, Frebourg T, Agid Y, Martinez M, Clerget-Darpoux F, 1997. Apolipoprotein E and Alzheimer disease: genotype-specific risks by age and sex. Am J Hum Genet 60(2), 439–446. [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R, Abdalla N, Kawas CH, Corrada MM, 2018. Forecasting the prevalence of preclinical and clinical Alzheimer’s disease in the United States. Alzheimers Dement 14(2), 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Rimmler JB, Locke PA, Conneally PM, Schmader KE, 1994. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7(2), 180–184. [DOI] [PubMed] [Google Scholar]
- Deelen J, Evans DS, Arking DE, Tesi N Nygaard M, Liu X, Wojczynski MK, Biggs ML, van der Spek A, Atzmon G, Ware EB, Sarnowski C, Smith AV, Seppälä I, Cordell HJ, Dose J, Amin N, Arnold AM, Ayers KL, Barzilai N, Becker EJ, Beekman M, Blanché H, Christensen K, Christiansen L, Collerton JC, Cubaynes S, Cummings SR, Davies K, Debrabant B, Deleuze JF, Duncan R, Faul JD, Franceschi C, Galan P, Gudnason V, Harris TB, Huisman M, Hurme MA, Jagger C, Jansen I, Jylhä M, Kähönen M, Karasik D, Kardia SLR, Kingston A, Kirkwood TBL, Launer LJ, Lehtimäki T, Lieb W , Lyytikäinen LP, Martin-Ruiz C, Min J, Nebel A, Newman AB, Nie C, Nohr EA, Orwoll ES, Perls TT, Province MA, Psaty BM, Raitakari OT, Reinders MJT, Robine JM, Rotter JI, Sebastiani P, Smith J, Sørensen TIA, Taylor KD, Uitterlinden AG, van der Flier W, van der Lee SJ, van Duijn CM, van Heemst D, Vaupel JW, Weir D, Ye K, Zeng Y, Zheng W, Holstege H, Kiel DP, Lunetta KL, Slagboom PE, Murabito JM, 2019. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun 10(1), 3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, Thompson WK, Besser L, Kukull WA, Holland D, Chen CH, Brewer JB, Karow DS, Kauppi K, Witoelar A, Karch CM, Bonham LW, Yokoyama JS, Rosen HJ, Miller BL, Dillon WP, Wilson DM, Hess CP, Pericak-Vance M, Haines JL, Farrer LA, Mayeux R, Hardy J, Goate AM, Hyman BT, Schellenberg GD, McEvoy LK, Andreassen OA, Dale AM, 2017. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med 14(3), e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Myers AJ, Huentelman M, Hardy J, 2017. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann Neurol 82(2), 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, Morgan K, Passmore P, Holmes C, Powell J, Brayne C, Gill M, Mead S, Goate A, Cruchaga C, Lambert JC, van Duijn C, Maier W, Ramirez A, Holmans P, Jones L, Hardy J, Seshadri S, Schellenberg GD, Amouyel P, Williams J, GERAD/PERADES, consortia, I., 2015. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 138(Pt 12), 3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF, 2015. PRSice: Polygenic Risk Score software. Bioinformatics 31(9), 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CC, Banks SJ, Thompson WK, Chen CH, McEvoy LK, Tan CH, Kukull W, Bennett DA, Farrer LA, Mayeux R, Schellenberg GD, Andreassen OA, Desikan R, Dale AM, 2020. Sex-dependent autosomal effects on clinical progression of Alzheimer’s disease. Brain 143(7), 2272–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM, 1997. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278(16), 1349–1356. [PubMed] [Google Scholar]
- Freudenberg-Hua Y, Li W, Abhyankar A, Vacic V, Cortes V, Ben-Avraham D, Koppel J, Greenwald B, Germer S, Darnell RB, Barzilai N, Freudenberg J, Atzmon G, Davies P, T2D-GENES Consortium, 2016. Differential burden of rare protein truncating variants in Alzheimer’s disease patients compared to centenarians. Hum Mol Genet 25(14), 3096–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL, 2006. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63(2), 168–174. [DOI] [PubMed] [Google Scholar]
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, Pasquier F, Dubois B, Tognoni G, Fievet N, Brouwers N, Bettens K, Arosio B Coto E, Del Zompo M, Mateo I, Epelbaum J, Frank-Garcia A, Helisalmi S, Porcellini E, Pilotto A, Forti P, Ferri R, Scarpini E, Siciliano G, Solfrizzi V, Sorbi S, Spalletta G, Valdivieso F, Vepsalainen S, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Hanon O, Piccardi P, Annoni G, Seripa D, Galimberti D, Licastro F, Soininen H, Dartigues JF, Kamboh MI, Van Broeckhoven C, Lambert JC, Amouyel P, Campion D, 2011. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry 16(9), 903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Russ TC , Clarke TK, Howard DM, Hillary RF, Evans KL, Walker RM, Bermingham ML, Morris SW, Campbell A, Hayward C, Murray AD, Porteous DJ, Horvath S, Lu AT, McIntosh AM, Whalley HC, Marioni RE, 2019. A meta-analysis of genome-wide association studies of epigenetic age acceleration. PLoS Genet 15(11), e1008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ, 2010. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 9(1), 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, Sealock J, Karlsson IK, Hägg S, Athanasiu L, Voyle N, Proitsi P, Witoelar A, Stringer S, Aarsland D, Almdahl IS, Andersen F, Bergh S, Bettella F, Bjornsson S, Brækhus A, Bråthen G, de Leeuw C, Desikan RS, Djurovic S, Dumitrescu L, Fladby T, Hohman TJ, Jonsson PV, Kiddle SJ, Rongve A, Saltvedt I, Sando SB, Selbæk G, Shoai M, Skene NG, Snaedal J, Stordal E, Ulstein ID, Wang Y, White LR, Hardy J, Hjerling-Leffler J, Sullivan PF, van der Flier WM, Dobson R, Davis LK, Stefansson H, Stefansson K, Pedersen NL, Ripke S, Andreassen OA, Posthuma D, 2019. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 51(3), 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, Inzelberg R, Kramer P, Rogaeva E, St George-Hyslop P, Cantwell LB, Dombroski BA, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Lunetta KL, Martin ER, Montine TJ, Goate AM, Blacker D, Tsuang DW, Beekly D, Cupples LA, Hakonarson H, Kukull W, Foroud TM, Haines J, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD, Consortium, A.s.D.G., 2010. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol 67(12), 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S, 2018. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50(9), 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel J, Sousa A, Gordon ML, Giliberto L, Christen E Davies P, 2018. Association Between Psychosis in Elderly Patients With Alzheimer Disease and Impaired Social Cognition. JAMA Psychiatry 75(6), 652–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, Frizatti A, Chouraki V, Martin ER, Sleegers K, Badarinarayan N, Jakobsdottir J, Hamilton-Nelson KL, Moreno-Grau S, Olaso R, Raybould R, Chen Y, Kuzma AB, Hiltunen M, Morgan T, Ahmad S, Vardarajan BN, Epelbaum J, Hoffmann P, Boada M, Beecham GW, Garnier JG, Harold D, Fitzpatrick AL, Valladares O, Moutet ML, Gerrish A, Smith AV, Qu L, Bacq D, Denning N, Jian X, Zhao Y, Del Zompo M, Fox NC, Choi SH, Mateo I, Hughes JT, Adams HH, Malamon M, Sanchez-Garcia F, Patel Y, Brody JA, Dombroski BA, Naranjo MCD, Daniilidou M, Eiriksdottir G, Mukherjee S, Wallon D, Uphill J, Aspelund T, Cantwell LB, Garzia F, Galimberti D, Hofer E, Butkiewicz M, Fin B, Scarpini E, Sarnowski C, Bush WS, Meslage S, Kornhuber J, White CC, Song Y, Barber RC, Engelborghs S, Sordon S, Voijnovic D, Adams PM, Vandenberghe R, Mayhaus M, Cupples LA, Albert MS, De Deyn PP, Gu W, Himali JJ, Beekly D, Squassina A, Hartmann AM, Orellana A, Blacker D, Rodriguez-Rodriguez E, Lovestone S, Garcia ME, Doody RS, Munoz-Fernadez C, Sussams R, Lin H, Fairchild TJ, Benito YA, Holmes C, Karamujić-Čomić H, Frosch MP, Thonberg H, Maier W, Roshchupkin G, Ghetti B, Giedraitis V, Kawalia A, Li S, Huebinger RM, Kilander L, Moebus S, Hernández I, Kamboh MI, Brundin R, Turton J, Yang Q, Katz MJ, Concari L, Lord J, Beiser AS, Keene CD, Helisalmi S, Kloszewska I, Kukull WA, Koivisto AM, Lynch A, Tarraga L, Larson EB, Haapasalo A, Lawlor B, Mosley TH, Lipton RB, Solfrizzi V, Gill M, Longstreth WT, Montine TJ, Frisardi V, Diez-Fairen M, Rivadeneira F, Petersen RC, Deramecourt V, Alvarez I, Salani F, Ciaramella A, Boerwinkle E, Reiman EM, Fievet N, Rotter JI, Reisch JS, Hanon O, Cupidi C, Andre Uitterlinden AG, Royall DR, Dufouil C, Maletta RG, de Rojas I, Sano M, Brice A, Cecchetti R, George-Hyslop PS, Ritchie K, Tsolaki M, Tsuang DW, Dubois B, Craig D, Wu CK, Soininen H, Avramidou D, Albin RL, Fratiglioni L, Germanou A, Apostolova LG, Keller L, Koutroumani M, Arnold SE, Panza F, Gkatzima O, Asthana S, Hannequin D, Whitehead P, Atwood CS, Caffarra P, Hampel H, Quintela I, Carracedo Á, Lannfelt L, Rubinsztein DC, Barnes LL, Pasquier F, Frölich L, Barral S, McGuinness B, Beach TG, Johnston JA, Becker JT, Passmore P, Bigio EH, Schott JM, Bird TD, Warren JD, Boeve BF, Lupton MK, Bowen JD, Proitsi P, Boxer A, Powell JF, Burke JR, Kauwe JSK, Burns JM, Mancuso M, Buxbaum JD, Bonuccelli U, Cairns NJ, McQuillin A, Cao C, Livingston G, Carlson CS, Bass NJ, Carlsson CM, Hardy J, Carney RM, Bras J, Carrasquillo MM, Guerreiro R, Allen M, Chui HC, Fisher E, Masullo C, Crocco EA, DeCarli C, Bisceglio G, Dick M, Ma L, Duara R, Graff-Radford NR, Evans DA, Hodges A, Faber KM, Scherer M, Fallon KB, Riemenschneider M, Fardo DW, Heun R, Farlow MR, Kölsch H, Ferris S, Leber M, Foroud TM, Heuser I, Galasko DR, Giegling I, Gearing M, Hüll M, Geschwind DH, Gilbert JR, Morris J, Green RC, Mayo K, Growdon JH, Feulner T, Hamilton RL, Harrell LE, Drichel D, Honig LS, Cushion TD, Huentelman MJ , Hollingworth P, Hulette CM, Hyman BT, Marshall R, Jarvik GP, Meggy A, Abner E, Menzies GE, Jin LW, Leonenko G, Real LM, Jun GR, Baldwin CT, Grozeva D , Karydas A, Russo G, Kaye JA, Kim R, Jessen F, Kowall NW, Vellas B, Kramer JH, Vardy E, LaFerla FM, Jöckel KH, Lah JJ, Dichgans M, Leverenz JB, Mann D, Levey AI, Pickering-Brown S, Lieberman AP, Klopp N, Lunetta KL, Wichmann HE, Lyketsos CG, Morgan K, Marson DC, Brown K, Martiniuk F, Medway C, Mash DC, Nöthen MM, Masliah E, Hooper NM, McCormick WC, Daniele A, McCurry SM, Bayer A, McDavid AN, Gallacher J, McKee AC, van den Bussche H, Mesulam M, Brayne C, Miller BL, Riedel-Heller S, Miller CA, Miller JW, Al-Chalabi A, Morris JC, Shaw CE, Myers AJ, Wiltfang J, O’Bryant S, Olichney JM, Alvarez V, Parisi JE, Singleton AB, Paulson HL, Collinge J, Perry WR, Mead S, Peskind E, Cribbs DH, Rossor M, Pierce A, Ryan NS, Poon WW, Nacmias B, Potter H, Sorbi S, Quinn JF, Sacchinelli E, Raj A, Spalletta G, Raskind M Caltagirone C, Bossù P, Orfei MD, Reisberg B, Clarke R, Reitz C, Smith AD, Ringman JM, Warden D, Roberson ED, Wilcock G, Rogaeva E, Bruni AC, Rosen HJ, Gallo M, Rosenberg RN, Ben-Shlomo Y, Sager MA, Mecocci P, Saykin AJ, Pastor P, Cuccaro ML, Vance JM, Schneider JA, Schneider LS, Slifer S, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tang M, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Saba Y, Pilotto A, Bullido MJ, Peters O, Crane PK, Bennett D, Bosco P, Coto E, Boccardi V, De Jager PL, Lleo A, Warner N, Lopez OL, Ingelsson M, Deloukas P, Cruchaga C, Graff C, Gwilliam R, Fornage M, Goate AM, Sanchez-Juan P, Kehoe PG, Amin N, Ertekin-Taner N, Berr C, Debette S, Love S, Launer LJ, Younkin SG, Dartigues JF, Corcoran C, Ikram MA, Dickson DW, Nicolas G, Campion D, Tschanz J, Schmidt H, Hakonarson H, Clarimon J, Munger R, Schmidt R, Farrer LA, Van Broeckhoven C, C O’Donovan M, DeStefano AL, Jones L, Haines JL, Deleuze JF, Owen MJ, Gudnason V, Mayeux R, Escott-Price V, Psaty BM, Ramirez A, Wang LS, Ruiz A, van Duijn CM, Holmans PA, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Lambert JC, Pericak-Vance MA, Alzheimer Disease Genetics Consortium (ADGC), European Alzheimer’s Disease Initiative (EADI), Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE), Genetic and Environmental Risk in AD/Defining Genetic, P.l.a.E.R.f.A.s.D.C.G.P., 2019a. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 51(3), 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, Frizatti A, Chouraki V, Martin ER, Sleegers K, Badarinarayan N, Jakobsdottir J, Hamilton-Nelson KL, Moreno-Grau S, Olaso R, Raybould R, Chen Y, Kuzma AB, Hiltunen M, Morgan T, Ahmad S, Vardarajan BN, Epelbaum J, Hoffmann P, Boada M, Beecham GW, Garnier JG, Harold D, Fitzpatrick AL, Valladares O, Moutet ML, Gerrish A, Smith AV, Qu L, Bacq D, Denning N, Jian X, Zhao Y, Del Zompo M, Fox NC, Choi SH, Mateo I, Hughes JT, Adams HH, Malamon J, Sanchez-Garcia F, Patel Y, Brody JA, Dombroski BA, Naranjo MCD, Daniilidou M, Eiriksdottir G, Mukherjee S, Wallon D, Uphill J, Aspelund T, Cantwell LB, Garzia F, Galimberti D, Hofer E, Butkiewicz M, Fin B, Scarpini E, Sarnowski C, Bush WS, Meslage S, Kornhuber J, White CC, Song Y, Barber RC, Engelborghs S, Sordon S, Voijnovic D, Adams PM, Vandenberghe R, Mayhaus M, Cupples LA, Albert MS, De Deyn PP, Gu W, Himali JJ, Beekly D, Squassina A, Hartmann AM, Orellana A, Blacker D, Rodriguez Rodriguez E, Lovestone S, Garcia ME, Doody RS, Munoz-Fernadez C, Sussams R, Lin H, Fairchild TJ, Benito YA, Holmes C, Karamujić-Čomić H, Frosch MP, Thonberg H, Maier W, Roshchupkin G, Ghetti B, Giedraitis V, Kawalia A, Li S, Huebinger RM, Kilander L, Moebus S, Hernández I, Kamboh MI, Brundin R, Turton J, Yang Q, Katz MJ, Concari L, Lord J, Beiser AS, Keene CD, Helisalmi S, Kloszewska I , Kukull WA, Koivisto AM, Lynch A, Tarraga L, Larson EB, Haapasalo A, Lawlor B , Mosley TH, Lipton RB, Solfrizzi V, Gill M, Longstreth WT, Montine TJ, Frisardi V, Diez-Fairen M, Rivadeneira F, Petersen RC, Deramecourt V, Alvarez I, Salani F, Ciaramella A, Boerwinkle E, Reiman EM, Fievet N, Rotter JI, Reisch JS, Hanon O, Cupidi C, Andre Uitterlinden AG, Royall DR, Dufouil C, Maletta RG, de Rojas I, Sano M, Brice A, Cecchetti R, George-Hyslop PS, Ritchie K, Tsolaki M , Tsuang DW, Dubois B, Craig D, Wu CK, Soininen H, Avramidou D, Albin RL, Fratiglioni L, Germanou A, Apostolova LG, Keller L, Koutroumani M, Arnold SE, Panza F, Gkatzima O, Asthana S, Hannequin D, Whitehead P, Atwood CS , Caffarra P, Hampel H, Quintela I, Carracedo Á, Lannfelt L, Rubinsztein DC, Barnes LL, Pasquier F, Frölich L, Barral S, McGuinness B, Beach TG, Johnston JA, Becker JT, Passmore P, Bigio EH, Schott JM, Bird TD, Warren JD, Boeve BF, Lupton MK, Bowen JD, Proitsi P, Boxer A, Powell JF, Burke JR, Kauwe JSK, Burns JM, Mancuso M, Buxbaum JD, Bonuccelli U, Cairns NJ, McQuillin A, Cao C, Livingston G, Carlson CS, Bass NJ, Carlsson CM, Hardy J, Carney RM, Bras J, Carrasquillo MM, Guerreiro R, Allen M, Chui HC, Fisher E, Masullo C, Crocco EA, DeCarli C, Bisceglio G, Dick M, Ma L, Duara R, Graff-Radford NR, Evans DA, Hodges A, Faber KM, Scherer M, Fallon KB, Riemenschneider M, Fardo DW, Heun R, Farlow MR, Kölsch H, Ferris S, Leber M, Foroud TM, Heuser I, Galasko DR, Giegling I, Gearing M, Hüll M, Geschwind DH, Gilbert JR, Morris J, Green RC, Mayo K, Growdon JH, Feulner T, Hamilton RL, Harrell LE, Drichel D, Honig LS, Cushion TD, Huentelman MJ, Hollingworth P, Hulette CM, Hyman BT, Marshall R, Jarvik GP, Meggy A, Abner E, Menzies GE, Jin LW, Leonenko G, Real LM, Jun GR, Baldwin CT, Grozeva D, Karydas A, Russo G, Kaye JA, Kim R, Jessen F, Kowall NW, Vellas B, Kramer JH, Vardy E, LaFerla FM, Jöckel KH, Lah JJ, Dichgans M, Leverenz JB, Mann D, Levey AI, Pickering-Brown S, Lieberman AP, Klopp N, Lunetta KL, Wichmann HE, Lyketsos CG, Morgan K, Marson DC, Brown K, Martiniuk F, Medway C, Mash DC, Nöthen MM, Masliah E, Hooper NM, McCormick WC, Daniele A, McCurry SM, Bayer A, McDavid AN, Gallacher J, McKee AC, van den Bussche H, Mesulam M, Brayne C, Miller BL, Riedel-Heller S, Miller CA, Miller JW, Al-Chalabi A, Morris JC, Shaw CE, Myers AJ, Wiltfang J, O’Bryant S, Olichney JM, Alvarez V, Parisi JE, Singleton AB, Paulson HL, Collinge J, Perry WR, Mead S, Peskind E, Cribbs DH, Rossor M, Pierce A, Ryan NS, Poon WW, Nacmias B, Potter H, Sorbi S, Quinn JF, Sacchinelli E, Raj A, Spalletta G, Raskind M, Caltagirone C, Bossù P, Orfei MD, Reisberg B, Clarke R, Reitz C, Smith AD, Ringman JM, Warden D, Roberson ED, Wilcock G, Rogaeva E, Bruni AC, Rosen HJ, Gallo M, Rosenberg RN, Ben-Shlomo Y, Sager MA, Mecocci P, Saykin AJ, Pastor P, Cuccaro ML, Vance JM, Schneider JA, Schneider LS, Slifer S, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tang M, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE Yu L, Saba Y, Pilotto A, Bullido MJ, Peters O, Crane PK, Bennett D, Bosco P, Coto E, Boccardi V, De Jager PL, Lleo A, Warner N, Lopez OL, Ingelsson M, Deloukas P, Cruchaga C, Graff C, Gwilliam R, Fornage M, Goate AM, Sanchez-Juan P, Kehoe PG, Amin N, Ertekin-Taner N, Berr C, Debette S, Love S, Launer LJ, Younkin SG, Dartigues JF, Corcoran C, Ikram MA, Dickson DW, Nicolas G, Campion D, Tschanz J, Schmidt H, Hakonarson H, Clarimon J, Munger R, Schmidt R Farrer LA, Van Broeckhoven C, C O’Donovan M, DeStefano AL, Jones L Haines JL, Deleuze JF, Owen MJ, Gudnason V, Mayeux R, Escott-Price V, Psaty BM, Ramirez A, Wang LS, Ruiz A, van Duijn CM, Holmans PA, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Lambert JC, Pericak-Vance MA, Consortium, A D.G., Initiative, E.A.s.D., Heart, C.f., Consortium, A.R.i.G.E., Genetic, Environmental Risk in AD/Defining Genetic, P.l., Consortium, E.R.f.A.s.D., 2019b. Genetic meta analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 51(3), 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer’s Disease, I., Genetic, Environmental Risk in Alzheimer’s, D., Alzheimer’s Disease Genetic, C., Cohorts for, H., Aging Research in Genomic, E., Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr., Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, 2013a. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh M, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ , Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K , Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, , E.A.s.D.I., Disease, G., Alzheimer’s, E.R.i., Consortium, A.s.D.G., Epidemiology, C.f.H., Genomic, A.R.i., 2013b. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, (EADI), E.A.s.D.I., Dis ase, G.a.E.R.i.A.s., Consortium, A.s.D.G., Epidemiology, C.f.H.a.A.R.i.G., 2013c. Meta analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease Nat Genet 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonenko G, Sims R, Shoai M, Frizzati A, Bossù P, Spalletta G, Fox NC, Williams J, Hardy J, Escott-Price V, consortium G, 2019. Polygenic risk and hazard scores for Alzheimer’s disease prediction. Ann Clin Transl Neurol 6(3), 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Zhang Q , McRae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM , Goate AM, Porteous DJ, Yang J, Evans KL, Deary IJ, Wray NR, Visscher PM 2018. GWAS on family history of Alzheimer’s disease. Transl Psychiatry 8(1), 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kama ani Y, Okada Y, Neale BM, Daly MJ, 2019. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 51(4), 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeux R, Stern Y, 2012. Epidemiology of Alzheimer disease. Cold Spring Harb Perspect Med 2(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43(5), 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Reitz C, Kunkle BW, Perry W, Park YS, Beecham GW, Rajbhandary RA, Hamilton-Nelson KL, Wang LS, Kauwe JS, Huentelman MJ, Myers AJ, Bird TD, Boeve BF, Baldwin CT, Jarvik GP, Crane PK, Rogaeva E, Barmada MM, Demirci FY, Cruchaga C, Kramer PL, Ertekin-Taner N, Hardy J, Graff-Radford NR, Green RC, Larson EB, St George-Hyslop PH, Buxbaum JD, Evans DA, Schneider JA, Lunetta KL, Kamboh MI, Saykin AJ, Reiman EM De Jager PL, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Martin ER, Haines JL, Mayeux RP, Farrer LA, Schellenberg GD, Pericak-Vance MA, Albert MS, Albin RL, Apostolova LG, Arnold SE, Barber R, Barnes LL, Beach TG, Becker JT, Beekly D, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Cantwell LB, Cao C, Carlson CS, Carney RM, Carrasquillo MM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cribbs DH, Crocco EA, DeCarli C, DeKosky ST, Dick M, Dickson D, Duara R, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Kramer JH LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lin CF, Lopez OL, Lyketsos CG, Mack WJ, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Murrell JR, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Peskind E, Petersen RC, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosen HJ, Rosenberg RN, Sano M, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Valladares O, Van Deerlin VM, Van Eldik LJ, Vardarajan BN, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Wishnek S, Woltjer RL, Wright CB, Younkin SG, Yu CE, Yu L, Consortium, A.D.G., 2014. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol 71(11), 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte L, Schupf N, Lee JH, Tang MX, Santana V, Williamson J, Maramreddy P, Tycko B, Mayeux R, 2006. Apolipoprotein E epsilon4 and age at onset of sporadic and familial Alzheimer disease in Caribbean Hispanics. Arch Neurol 63(11), 1586–1590. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P, Consortium IS, 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460(7256), 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2019. R: A language and environment for statistical computing.
- Rajabli F, Feliciano BE, Celis K, Hamilton-Nelson KL, Whitehead PL, Adams LD, Bussies PL, Manrique CP, Rodriguez A, Rodriguez V, Starks T, Byfield GE, Sierra Lopez CB, McCauley JL, Acosta H, Chinea A, Kunkle BW, Reitz C, Farrer LA, Schellenberg GD, Vardarajan BN, Vance JM, Cuccaro ML, Martin ER, Haines JL, Byrd GS, Beecham GW, Pericak-Vance MA, 2018. Ancestral origin of ApoE epsilon4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet 14(12), e1007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KL, Tybjaerg-Hansen A, Nordestgaard BG, Frikke-Schmidt R, 2018. Absolute 10-year risk of dementia by age, sex and APOE genotype: a population-based cohort study. CMAJ 190(35), E1033–E1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Gurinovich A, Nygaard M, Sasaki T, Sweigart B, Bae H, Andersen SL, Villa F, Atzmon G, Christensen K, Arai Y, Barzilai N, Puca A Christiansen L, Hirose N, Perls TT, 2019. APOE Alleles and Extreme Human Longevity. J Gerontol A Biol Sci Med Sci 74(1), 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesi N, van der Lee SJ, Hulsman M, Jansen IE, Stringa N, van Schoor N, Meijers-Heijboer H, Huisman M, Scheltens P, Reinders MJT , van der Flier WM, Holstege H, 2019. Centenarian controls increase variant effect sizes by an average twofold in an extreme case-extreme control analysis of Alzheimer’s disease. Eur J Hum Genet 27(2), 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


