Abstract
In many bacteria, the reactions of proline catabolism are catalyzed by the bifunctional enzyme known as proline utilization A (PutA). PutA catalyzes the two-step oxidation of L-proline to L-glutamate using distinct proline dehydrogenase (PRODH) and L-glutamate-γ-semialdehyde dehydrogenase (GSALDH) active sites, which are separated by over 40 Å and connected by a complex tunnel system. The tunnel system consists of a main tunnel that connects the two active sites and functions in substrate channeling, plus six ancillary tunnels whose functions are unknown. Here we used tunnel-blocking mutagenesis to probe the role of a dynamic ancillary tunnel (tunnel 2a) whose shape is modulated by ligand binding to the proline dehydrogenase active site. The 1.90 Å resolution crystal structure of Geobacter sulfurreducens PutA variant A206W verified that the side chain of Trp206 cleanly blocks tunnel 2a without perturbing the surrounding structure. Steady-state kinetic measurements indicate the mutation impaired PRODH activity without affecting the GSALDH activity. Single-turnover experiments corroborated a severe impairment of PRODH activity with flavin reduction decreased by nearly 600-fold in A206W relative to wild-type. Substrate channeling is also significantly impacted as A206W exhibited a 3000-fold lower catalytic efficiency in coupled PRODH-GSALDH activity assays, which measure NADH formation as a function of proline. The structure suggests that Trp206 inhibits binding of the substrate L-proline by preventing the formation of a conserved glutamate-arginine ion pair and closure of the PRODH active site. Our data are consistent with tunnel 2a serving as an open space through which the glutamate of the ion pair travels during the opening and closing of the active site in response to binding L-proline. These results confirm the essentiality of the conserved ion pair in binding L-proline and support the hypothesis that the ion pair functions as a gate that controls access to the PRODH active site.
Keywords: X-ray crystallography, proline utilization A, proline dehydrogenase, proline catabolism, protein tunnels
Graphical Abstract
1. Introduction
Proline utilization A (PutA) is a bifunctional enzyme that catalyzes both steps of proline catabolism using spatially-separated active sites and a substrate-channeling mechanism (Fig. 1) [1]. The proline dehydrogenase (PRODH) active site catalyzes the FAD-dependent oxidation of proline to Δ1-pyrroline-5-carboxylate (P5C). P5C, or perhaps its hydrolysis product L-glutamate-γ-semialdehyde (GSAL), is channeled over 40 Å to the GSAL dehydrogenase (GSALDH, a.k.a. P5CDH) active site, which catalyzes the NAD+-dependent oxidation of GSAL to glutamate.
Fig. 1.
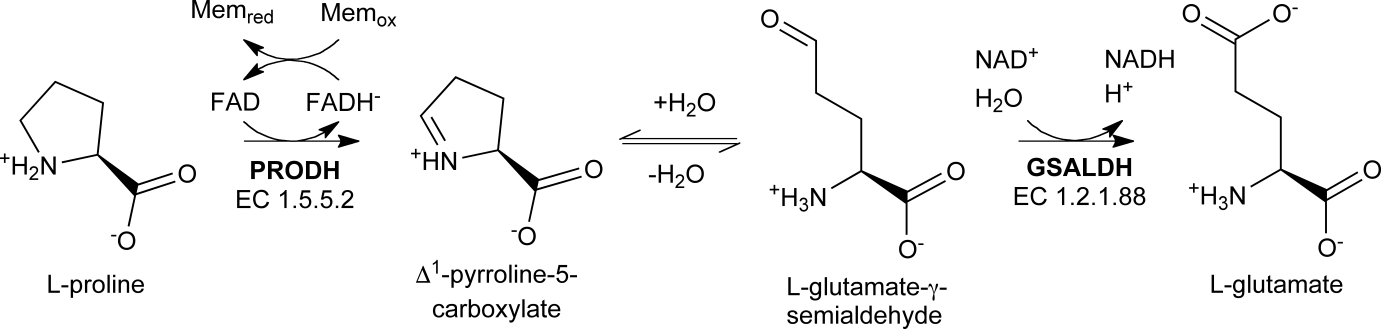
Reactions catalyzed by PutA.
Substrate channeling is a defining feature of PutA. All PutAs studied to date exhibit kinetic hallmarks of substrate channeling, including the absence of a lag phase in the coupled PRODH-GSALDH reaction and protection of the intermediate P5C/GSAL [2–7]. Also, in one well-studied PutA, the channeling step of the coupled PRODH-GSALDH reaction was shown to be both rate-limiting and hysteretic [4]. Hysteresis in this case refers to the novel dependence of the rate constant for the channeling step on the number of enzyme turnovers; the rate is slowest during the first turnover and increases 40-fold in subsequent turnovers. Hysteresis implies that the channeling step is activated, although the mechanistic details of activation are unknown. In summary, substrate channeling is a hallmark of the catalytic mechanism of PutA.
Crystal structures of PutAs have revealed a conserved tunnel system, which is thought to be the structural basis of substrate channeling (Fig. 2) [2, 5–7]. The system consists of a main tunnel that connects the two active sites, as well as six additional tunnels that connect the main tunnel to the protein surface. The function of the main tunnel is obvious – it is the conduit through which the intermediate P5C/GSAL diffuses from the PRODH site to the GSALDH site. This functional assignment is consistent with a mutagenesis study showing that blocking the main tunnel via site-directed mutagenesis to Trp impaired the coupled activity of PutA [8]. In contrast, the functions of the offshoots from the main tunnel have not been investigated; however, their conservation among PutAs with different oligomeric structures and low sequence identities implies functional importance.
Fig. 2.
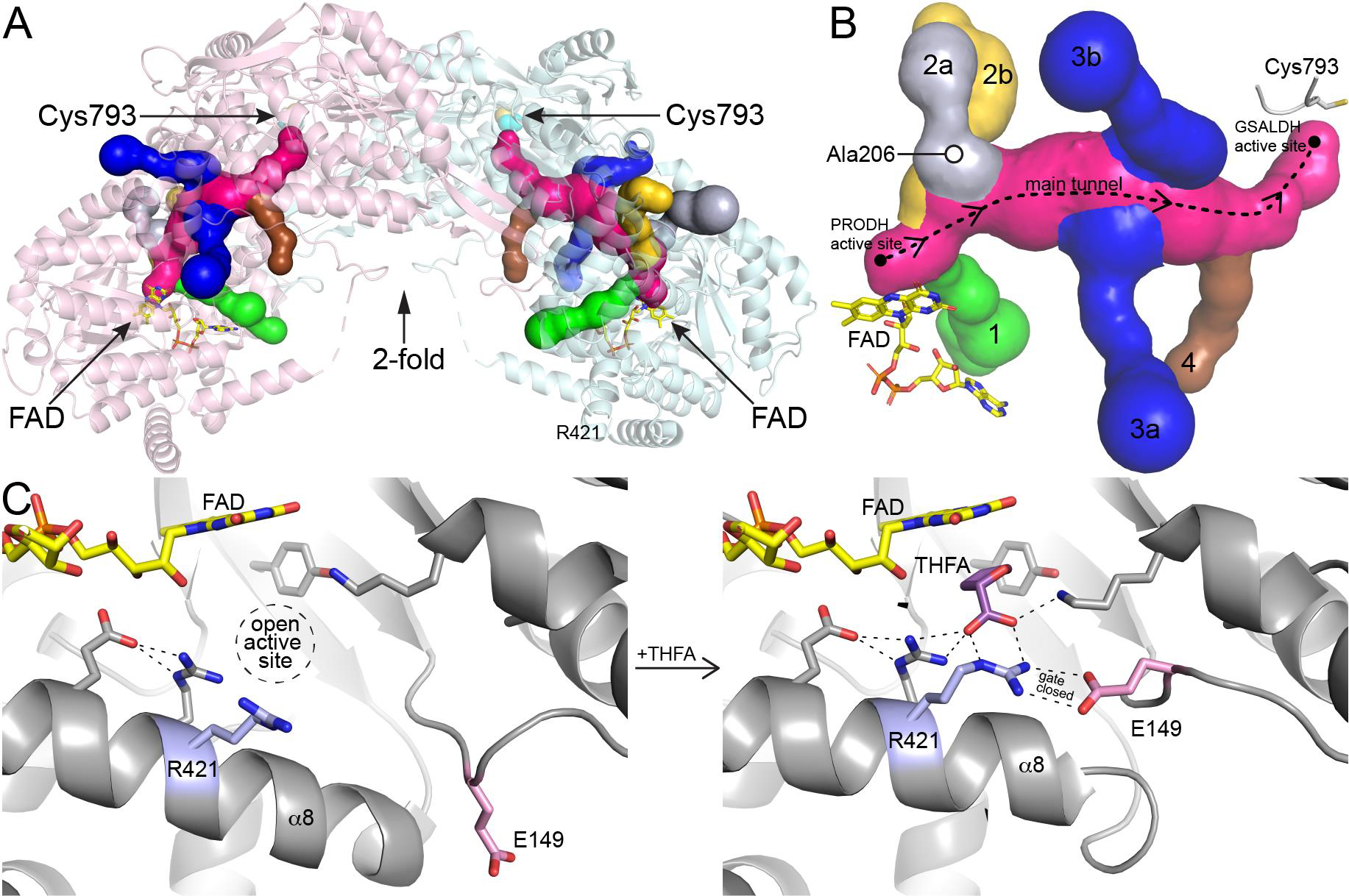
Tunnel system of PutA and dynamics of the PRODH site. (A) Ribbon drawing of a dimer of the resting state of Geobacter sulfurreducens PutA (GsPutA, PDB ID 4NM9) with the tunnel system represented as surfaces. The two protomers are colored pink and cyan. (B) Close-up view of the tunnel system. The dashed curve shows the direction of channeling of P5C/GSAL through the main tunnel. The six tributary tunnels, which connect the main tunnel to the protein surface, are colored green (tunnel 1), silver (2a), yellow (2b), blue (3a, 3b), and brown (4). (C) Conformational changes in the PRODH site associated with the binding of the proline analog THFA. Residues of the conserved ion pair are colored pink (Glu149) and blue (Arg421). Left: resting enzyme with no ligand in the PRODH site (PDB ID 4NM9). Right: complexed with the proline analog THFA (PDB ID 4NMA).
Herein we probe the importance of ancillary tunnel 2a and its involvement in controlling access to the PRODH site of PutA. Tunnel 2a connects to the main tunnel near the PRODH active site, approximately 14 Å from the FAD and 34 Å from catalytic Cys793 in the GSALDH site (Fig. 2B). Tunnel 2a was chosen for this study because it appeared from previous structural studies be functionally important, yet its precise role in catalysis was unknown. In particular, tunnel 2a connects the PRODH active site to the bulk medium, suggesting it may be an entrance for the substrate proline. Furthermore, tunnel 2a is unique among the PutA tunnels in that changes shape in response to ligand binding to the PRODH site, suggesting that its dynamics may be functionally relevant [5]. The tunnel is present in the resting state of the enzyme, before the binding of the substrate L-proline (as in Fig. 2, PDB ID 4NM9); however, the tunnel collapses upon the binding of the proline analog, S-(−)-tetrahydro-2-furoic acid (THFA, PDB ID 4NMA). Collapse of tunnel 2a is accompanied by the closure of the active site and the formation of an ion pair between conserved glutamate and arginine residues, the latter stabilizing the carboxylate of the proline analog (Fig. 2C). These observations have led to the hypothesis that the ion pair functions as an active site gate, which opens to allow the binding of L-proline, closes during hydride transfer to the FAD, and reopens to release the product P5C into the main tunnel [2, 5, 9].
To investigate the importance of tunnel 2a and the ion pair gate, we installed Trp at residue 206 of PutA from Geobacter sulfurreducens PutA (GsPutA) in place of Ala with the goal of blocking tunnel 2a. Ala206 is located on the wall of tunnel 2a, near its connection to the main tunnel (Fig. 2B). The crystal structure of A206W shows that the mutation achieved the intended blockage without perturbing the surrounding structure. Steady-state and single-turnover kinetic measurements show that blockage of the tunnel severely impairs the PRODH activity of PutA, resulting in 3000-lower catalytic efficiency in the coupled PRODH-GSALDH reaction. These results suggest that tunnel 2a functions as a passageway through which the glutamate of the ion pair gate travels during the opening and closing of the active site and demonstrate the essentiality of gating of the PRODH site for PRODH function.
2. Materials and Methods
2.1. Protein production
All experiments were performed on PutA from Geobacter sulfurreducens PCA (GsPutA, 1004 residues, UniProt Q746X3). Mutagenesis was carried out on the wild-type GsPutA gene in the pNIC28-Bsa4 protein expression vector using the Quik-change II XL Site-Directed Mutagenesis Kit (Agilent). The presence of the mutation was verified by Sanger sequencing prior to further analysis and subsequently confirmed with X-ray crystallography.
Wild-type and mutant protein expression constructs were transformed into BL21-AI expression cells (Thermo Fisher Scientific) for protein expression. A 10 mL starter culture grown overnight at 37°C from a single transformant was used to inoculate 1 L of terrific broth. Cultures were grown at 37°C and protein expression was induced at 18°C with 0.5 mM IPTG. The following day, cells were pelleted, frozen, and stored at −80°C for downstream applications.
For purification, cell pellets were resuspended in 50 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole, 10% (v/v) glycerol, and 1% (v/v) Tween-20 and lysed by sonication. Cell-debris and unbroken cells were pelleted at 16,000 rpm for 1 h. The supernatant was then loaded by gravity onto a Ni-NTA column and washed with 40 bed volumes of 50 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole, and 10% (v/v) glycerol. The His-tagged protein was eluted with six bed volumes of 50 mM HEPES pH 7.5, 500 mM NaCl, 250 mM imidazole, and 10% (v/v) glycerol. The His-tag was cleaved from the purified protein using Tobacco Etch Virus protease. The His-tag cleavage reaction was carried out by incubation for 2 h at 28°C, followed by an overnight dialysis step against 50 mM HEPES pH 7.5, 50 mM NaCl, 0.5 mM Tris(carboxyethyl)phosphine, and 5% (v/v) glycerol. The cleaved His-tag was removed by passing the protein solution over Ni-NTA. The flow-through from this step was concentrated and further purified by size-exclusion chromatography on Superdex 200 10–30, in the presence of 50 mM HEPES pH 7.5, 50 mM NaCl, 0.5 mM Tris(carboxyethyl)phosphine, and 5% (v/v) glycerol. Fractions containing the protein were pooled and concentrated. Concentrated protein was aliquoted in PCR strip tubes and flash-frozen in liquid nitrogen.
2.2. Crystal structure determination
Crystals of A206W were grown in sitting drops at 20 °C using an enzyme stock solution of ~6 mg/mL and the drops formed by mixing equal volumes of the enzyme and reservoir solutions. Although we had determined the structure of wild-type GsPutA previously [5], crystal screening trials were repeated to identify a more robust crystallization condition. These experiments revealed Hampton Index condition E9, which consists of 0.05 M ammonium sulfate, 0.05 M BIS-TRIS pH 6.5, and 30% (v/v) pentaerythritol ethoxylate (15/4 EO/OH). Crystals of the mutant variant were grown with slight variations of this condition as the reservoir solution, aided by microseeding. The crystals were prepared for low temperature data collection by soaking them in the reservoir solution supplemented with 15–20% (v/v) ethylene glycol and then flash-cooling in liquid nitrogen.
Diffraction data were collected in shutterless mode at the Advanced Light Source beamline 4.2.2 using an RDI CMOS-8M detector. The data were indexed, integrated, and scaled with XDS [10]. Intensities were merged and converted to amplitudes with AIMLESS [11]. The space group is P212121, with a dimer in the asymmetric unit; the dimer has been confirmed by small-angle X-ray scattering as the oligomer formed in solution [5]. We note this is one of the crystal forms we reported previously despite a different crystallization condition.
Interactive model building and restrained refinement were performed with COOT [12, 13] and PHENIX [14], respectively. A deposited structure of GsPutA in the resting (open) state (PDB ID 4NM9) was used to initiate refinement. The structure was validated using MolProbity, the PDB validation server, and polder omit maps [15–17].
The tunnel system in GsPutA was analyzed with MOLEonline using the default parameters [18]. The main tunnel was calculated using a starting point in the PRODH active site near the si face of the FAD (defined by residues 308 and 385) and an end point in the GSALDH active site (defined by catalytic Cys793). The tributary tunnels were identified using the aforementioned starting point and no end point.
2.3. Steady-state kinetic measurements.
The coupled PRODH-GSALDH activities of wild-type GsPutA and A206W were measured by monitoring NADH production (340 nm) at 23 °C in 96-well plates using a BioTek microplate spectrophotometer. The assay included 0.5 μM enzyme, 0.1 mM menadione, 0.2 mM NAD+ and varying concentration of L-proline (0 – 200 mM) in a buffer containing 50 mM sodium phosphate pH 7.5, 50 mM NaCl, and 5% (v/v) glycerol. The PRODH activities of wild-type GsPutA and A206W were measured using a dye-coupled oxidoreductase assay, in which L-proline is the variable substrate (0 – 200 mM), 2,6-dichlorophenolindophenol is the terminal electron acceptor (monitored at 600 nm), and phenazine methosulfate is a secondary electron acceptor that mediates electron transfer from the flavoenzyme to the terminal electron acceptor [19]. The GSALDH activities of wild-type GsPutA and A206W were measured by monitoring NADH production at 340 nm with L-P5C as the variable substrate (0 – 3.5 mM) and NAD+ fixed at a saturating concentration as described previously [5]. D,L-P5C was synthesized from D,L-5-hydroxylysine-HCl according to the method of Williams and Frank [20].
2.4. Transient-state kinetic measurements
Stopped-flow kinetics experiments were carried out as previously described with slight modifications [21]. GsPutA wild-type and the A206W variant were purified as described above, except that the size exclusion chromatography step was omitted. Prior to initializing the experiments, enzyme, reaction buffer A (50 mM sodium phosphate pH 7.5, 50 mM NaCl, and 5% (v/v) glycerol), and reaction buffer B (50 mM sodium phosphate pH 7.5, 50 mM NaCl, 5% (v/v) glycerol, 80 mM L-proline, and 0.4 mM NAD+) were degassed via 30 consecutive cycles of vacuum followed by flushing with oxygen-scrubbed nitrogen. To remove additional dissolved molecular oxygen, protocatechuate 3,4-dioxygenase and protocatechuic acid were added to each solution to a final concentration of 0.05 U/mL and 100 μM, respectively, in an anaerobic chamber under a nitrogen atmosphere. Reaction materials were then transferred to anaerobic syringes in the chamber and capped until use.
All stopped-flow experiments were carried out using a Hi-Tech Scientific SF-61DX2 stopped-flow instrument equipped with a photodiode array detector and KinetAsyst software. Prior to performing stopped-flow experiments, the stopped-flow mixing devices were washed with deoxygenated water, followed by incubation in deoxygenated buffer A or B for 30 minutes. All stopped-flow experiments were performed at 25°C.
3. Results
3.1. Crystal structure of A206W
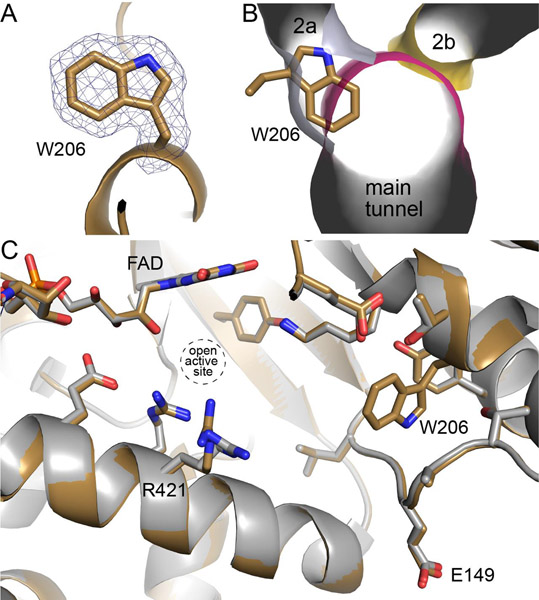
X-ray crystallography was used to determine whether the mutation had the intended impact, as well as any other unexpected effects on the structure. The structure of A206W was determined at 1.90 Å resolution (Table 1). The electron density maps verified the mutation and clearly defined the conformation of Trp206 (Fig. 3A). As intended, the side chain of Trp206 has inserted into tunnel 2a, as shown by overlaying the tunnel system of the wild-type enzyme resting state onto the structure of A206W (Fig. 3B). Part of the indole ring also pierces the main tunnel. By occupying vacant space in the wild-type enzyme, the bulky side chain of Trp206 was accommodated without any significant conformational changes to the surrounding structure (Fig. 3C). The only perceptible difference is a slight deflection of Arg421. Thus, structure of A206W is essentially identical to the resting enzyme except that tunnel 2a is blocked.
Table 1.
X-ray Diffraction Data Collection and Refinement Statistics
| A206W | |
|---|---|
| Space group | P212121 |
| Unit cell parameters (Å) | a = 95.42, b = 151.38, c = 175.55 |
| Wavelength (Å) | 1.000 |
| Resolution (Å)a | 58.52 – 1.90 (1.93 – 1.90) |
| Observationsa | 1441588 (64876) |
| Unique reflectionsa | 199837 (9787) |
| Rmerge(I)a | 0.141 (1.002) |
| Rmeas(I)a | 0.152 (1.088) |
| Rpim(I)a | 0.057 (0.421) |
| Mean I/σa | 13.4 (2.0) |
| Mean CC1/2a | 0.997 (0.762) |
| Completeness (%)a | 100 (100) |
| Multiplicitya | 7.2 (6.6) |
| No. protein residues | 1959 |
| No. of atoms | |
| Protein | 15361 |
| FAD | 106 |
| Water | 1462 |
| R work a | 0.1688 (0.2922) |
| R free a,b | 0.1986 (0.3391) |
| RMSD bonds (Å) | 0.006 |
| RMSD angles (°) | 0.795 |
| Ramachandran plotc | |
| Favored (%) | 98.20 |
| Outliers (%) | 0.00 |
| Clashscore (PR)c | 1.6 (100) |
| MolProbity score (PR)c | 0.98 (100) |
| Average B-factor (Å2) | |
| Protein | 20.8 |
| FAD | 15.9 |
| Water | 27.2 |
| Coordinate error (Å)d | 0.21 |
| PDB ID | 7NA0 |
Values for the outer resolution shell of data are given in parenthesis.
5% test set.
From MolProbity. The percentile ranks (PR) for Clashscore and MolProbity score are given in parentheses.
Maximum likelihood-based coordinate error estimate reported by phenix.refine.
Fig. 3.
Structure of A206W. (A) Electron density for Trp206 (polder omit, 4σ). (B) Overlay of A206W onto the tunnel system of the open conformation of the wild-type GsPutA, emphasizing how Trp206 blocks tunnel 2a. (C) Comparison of A206W (sand) with the open conformation of wild-type GsPutA (gray, PDB ID 4NM9).
3.2. Steady-state kinetic analysis
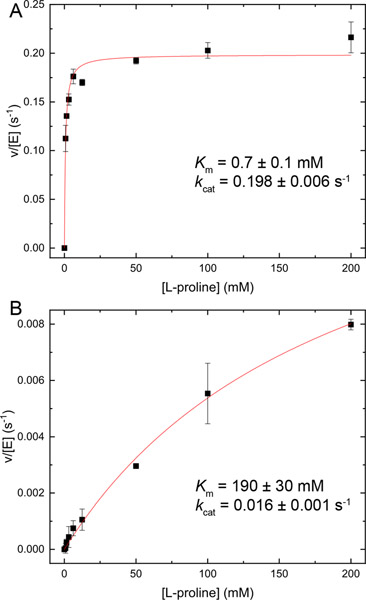
The coupled PRODH-GSALDH activity of A206W was measured using an assay that monitors NADH production as a function of L-proline concentration in the presence of an electron acceptor for the FAD (menadione). GsPutA shows hyperbolic dependence on L-proline concentration characterized by the kinetic parameters Km of 0.7 mM, kcat of 0.2 s−1, and catalytic efficiency (kcat/Km) of 286 M−1s−1 (Fig. 4A). The mutation of Ala206 to Trp severely diminished the coupled activity. A206W could not be saturated with L-proline (Fig. 4B). The kinetic parameters estimated for A206W are Km of 190 mM and kcat of 0.016 s−1. Thus, the mutation decreased the catalytic efficiently of the coupled reaction by a factor of 3000 to approximately 0.1 M−1s−1.
Fig. 4.
Coupled PRODH-GSALDH activities of (A) wild-type GsPutA and (B) A206W. Note the vertical scales of the two panels differ.
The individual PRODH and GSALDH activities of the A206W variant were measured to identify the source of the severely diminished coupled PRODH-GSALDH activity. A206W exhibited very low PRODH activity in a dye-coupled assay and we were not able to determine kinetic constants. In contrast, the GSALDH activity of A206W was robust and characterized by a Km for L-P5C of 65.0 ± 18.3 μM and kcat of 6.0 ± 0.2 s−1, which are within a factor of two of the wild-type values (30.0 ± 13.0 μM, 4.7 ± 0.2 s−1). The catalytic efficiency of the GSALDH activity of A206W is 92 ± 30 mM−1 s−1 compared to 156 ± 75 mM−1 s−1 for GsPutA, indicating near-normal GSALDH activity. These results suggest that the low coupled PRODH-GSALDH activity of A206W is due to a defect in PRODH activity.
3.3. Transient-state kinetic analysis
Rapid-reaction kinetic measurements employing single turnover conditions were also performed (anaerobic, no external electron acceptor for the reduced flavin). Enzyme, proline (40 mM after mixing), and NAD+ (0.2 mM after mixing) were rapidly mixed, and the absorbance spectra of the FAD cofactor and NADH product were recorded using a stopped-flow instrument.
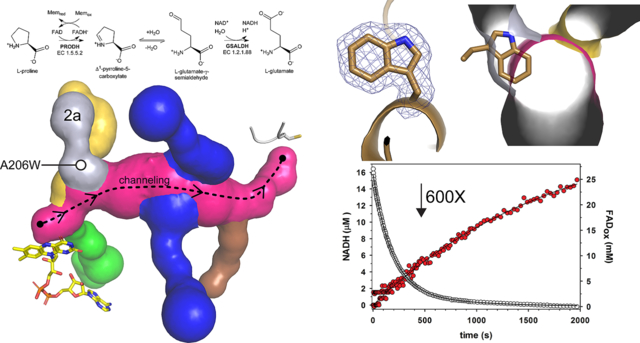
Analysis of the wild-type enzyme revealed rapid redution of the FAD by proline and the production of NADH, as expected (Fig. 5A). Flavin reduction was modeled using a double-exponential decay model. The rate of flavin reduction under these conditions is 3.6 (± 0.2) s−1 for the major phase followed by a slower rate of 0.2 (± 0.014) s−1 for the minor phase (19% amplitude). NADH production was modeled with a single-exponential fit to the first phase yielding a rate of 0.38 (± 0.04) s−1 followed by a slow linear rate (0.02 ± 0.001 μM s−1). Note the production of NADH occurs without a lag phase, consistent with substrate channeling.
Fig. 5.
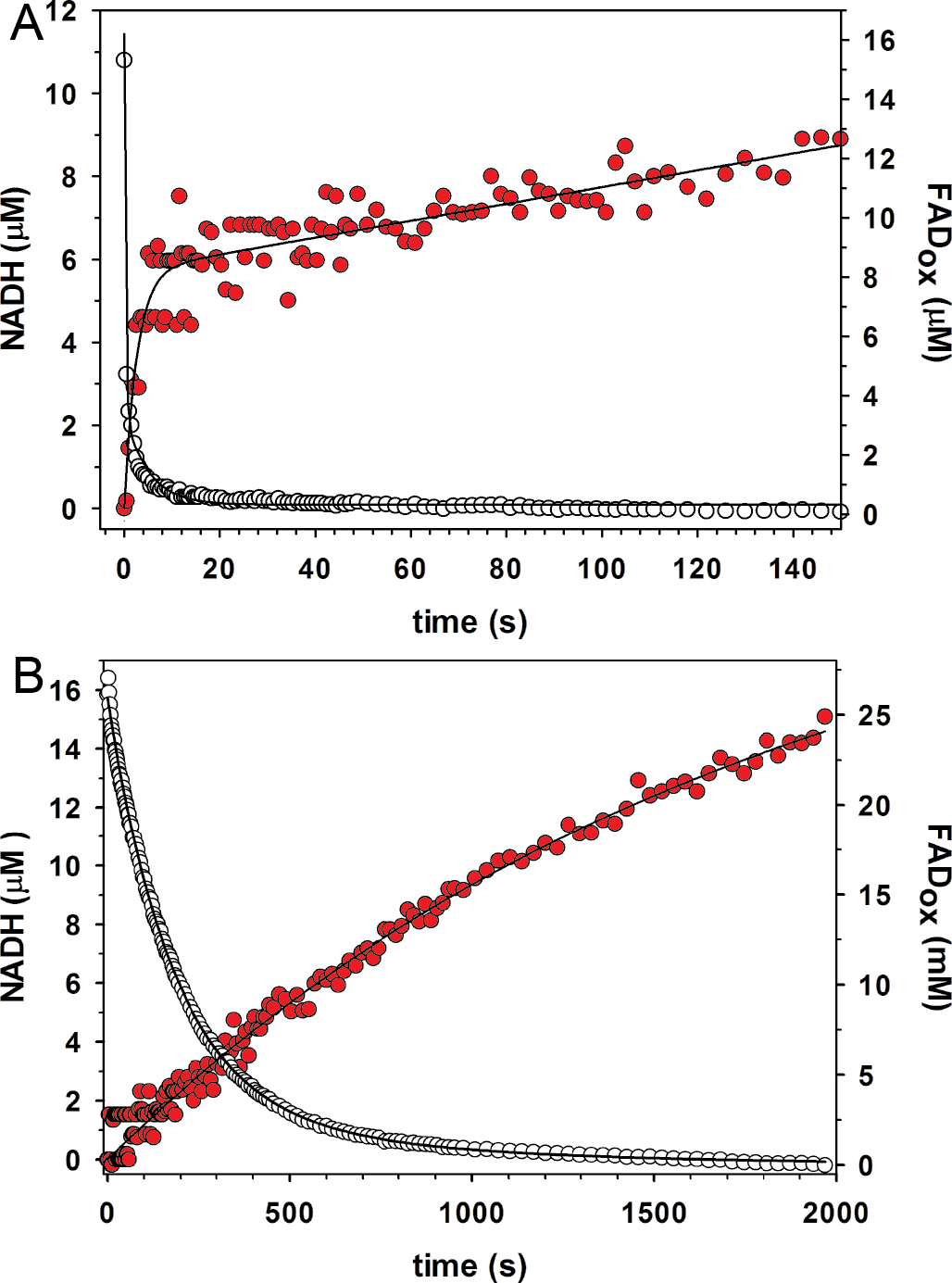
Rapid-reaction kinetic data acquired under single turnover conditions for (A) wild-type GsPutA and (B) A206W. The open circles show FAD reduction monitored at 450 nm. The red filled circles show NADH production monitored at 340 nm. Fits to the data are shown.
Single-turnover analysis of A206W revealed a severe defect in PRODH function, consistent with the steady-state kinetic data (Fig. 5B). The estimated rate of flavin reduction using a double-exponential fit is 0.006 s−1 (87% amplitude for the major phase), which is a factor of ~600 times slower than wild-type. The rate of NADH production for A206W is 0.0006 (± 0.00003) s−1 (single-exponential fit), which is also ~600 times lower than wild-type. Thus, the single turnover experiments reveal flavin reduction as a major defect in the coupled activity of A206W thereby limiting delivery of P5C/GSAL into the main tunnel and NADH production at the GSALDH site.
4. Discussion
Voids are common features of protein structure and appear as tunnels, channels, buried cavities, and surface clefts [22]. Tunnels often connect the actives sites in bifunctional enzymes, providing a conduit for substrate channeling [23]. Tunnels provide access for substrates to buried active sites [24]. Channels in integral membrane transporters provide hydrophilic pathways for solutes through the hydrophobic lipid bilayer. Cavities, clefts, grooves, and pockets provide microenvironments for chemical reactions. The ubiquity of voids has motivated the computational analysis of the empty space in protein structures [22]. Still, the functions of many voids are not obvious.
Tunnels are ubiquitous in enzymes. Tunnels have been identified in numerous enzymes representing all six major enzyme classes (reviewed in [24, 25]). The functions of enzyme tunnels have been interrogated, leading to conceptual frameworks for describing the contributions of tunnels to enzyme catalysis. For example, tunnels between the active sites of bifunctional enzymes serve as conduits for substrate channeling, as discussed here. Functionally, channeling protects reactive intermediates and can enhance reaction kinetics by decreasing the transit time between the active sites [26]. Tunnels for channeling have been characterized structurally and biochemically in the classic example, tryptophan synthase [27–31], as well as carbamoyl phosphate synthase [32], PutA [2, 5–7], and many others (reviewed in [23, 33]). Tunnels are also found in many monofunctional enzymes, especially those in which the active site is buried deep in the protein interior. Active site tunnels provide another layer of substrate specificity, in addition to the specificity afforded by the constellation of amino acids that directly contact the substrate in the Michaelis complex. Tunnels to deeply buried active sites have been referred to as the “keyholes” in an updated version of the classic lock-and-key model of substrate recognition [25]. The size, shape, physiochemical properties, and dynamics of the “keyhole” can contribute to substrate specificity and catalytic efficiency [24].
Here we used site directed mutagenesis to explore the function of a tunnel in PutA. Tunnel-modifying mutagenesis has been used for many purposes, including studying substrate channeling [8, 27–29], identifying pathways for ligand diffusion [34], altering substrate specificity [35], endowing an enzyme with new functions [36], engineering improved enzymes [37], and probing the roles of tunnels in the mechanisms of enzymes [38–42].
The tunnel of interest here is part of a complex tunnel system in PutA (Fig. 2B). Tunnel 2a is one of six tunnels that connect the substrate-channeling tunnel to the outside. Its location on the PRODH half of the main tunnel implies a role in PRODH function. Indeed, blocking tunnel 2a by installing Trp at position 206 severely impaired PRODH activity and resulted in a 3000-fold decrease in the catalytic efficiency of the coupled reaction. The impact on catalytic function is especially notable considering that the site of mutation is 14 Å away from the PRODH active site and might be considered a remote mutation. Thus, tunnel 2a apparently has an essential function.
Comparison of the structure of A206W to the wild-type PRODH Michaelis complex suggests a function for tunnel 2a. The binding of THFA (and presumably L-proline) to the resting enzyme is accompanied by large conformational changes, including the tilting of α8 toward proline site and the remodeling of the Glu149 loop (Fig. 2C). The latter results in movement of Glu149 by 10 Å from the protein surface into the active site to ion pair with Arg421. This ion pair has been observed in all structures of PutAs and monofunctional PRODHs complexed with proline analogs, suggesting functional importance. The A206W structure shows that Trp206 occupies the space reserved for Glu149 when in the ion-pairing conformation (Fig. 6). The structure also suggests that the trajectory taken by Glu149 passes through the space occupied by Trp206, i.e., tunnel 2a. Thus, our data are consistent with tunnel 2a serving as open space needed for the movement of Glu149 during the opening and closing of the active site in response to binding L-proline.
Fig. 6.
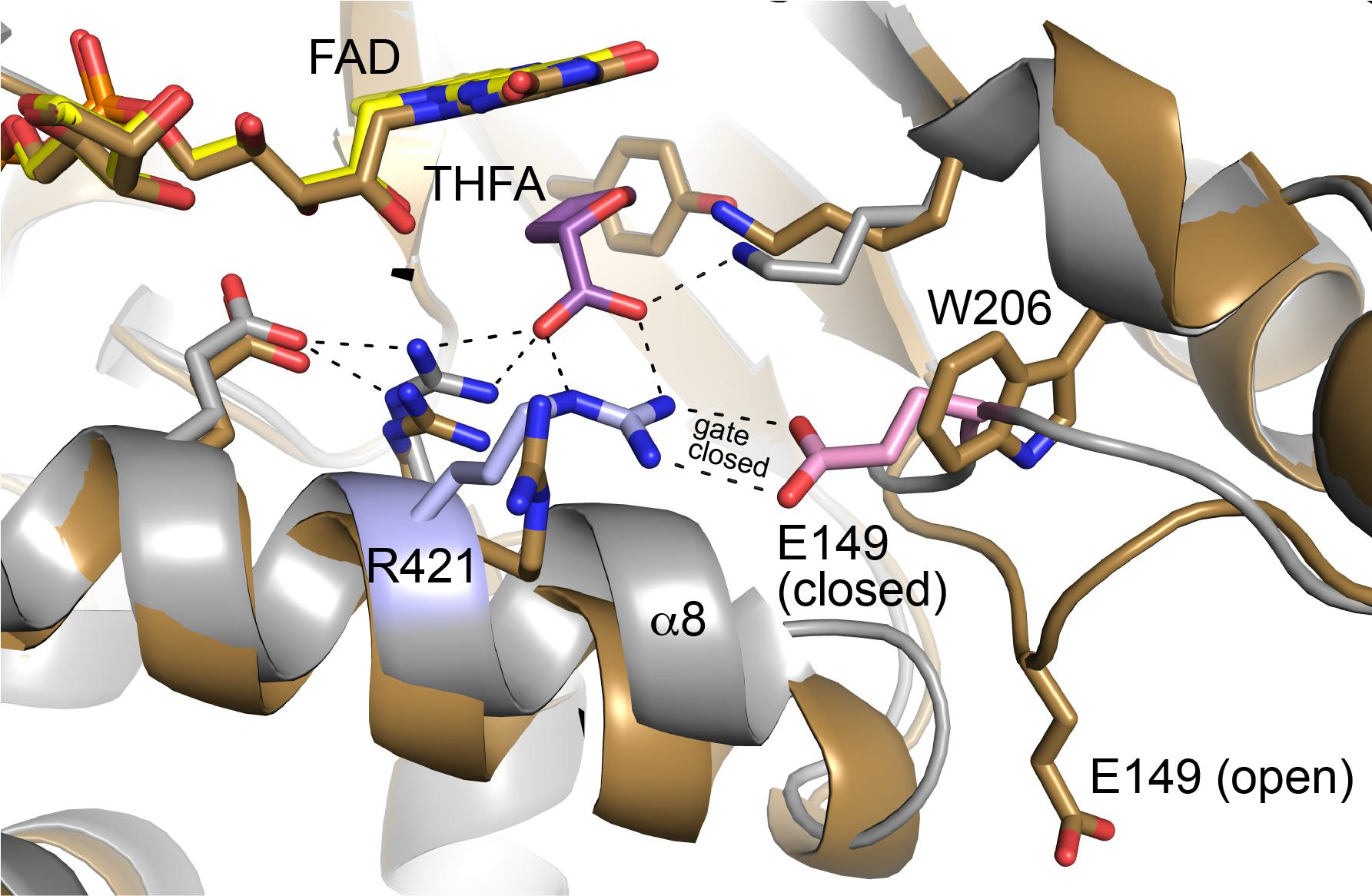
Comparison of A206W (sand) with the closed conformation of wild-type GsPutA complexed with THFA (gray, PDB ID 4NMA). Residues of the conserved ion pair in the closed state are colored pink (Glu149) and blue (Arg421).
Finally, tunnel-blocking mutagenesis provided a way to test the importance of a noncovalent interaction for enzyme function without mutating the residues involved in the interaction. By preventing the movement of Glu149, the A206W mutation indirectly demonstrated the essentiality of a conserved ion pair gate for enzyme function.
5. Databases
Coordinates and structure factor amplitudes has been deposited in the Protein Data Bank under accession code 7NA0.
Highlights.
PutA is a bifunctional enzyme that catalyzes the oxidation of Pro to Glu
PutA has a complex network of tunnels whose functions are poorly understood
Trp mutagenesis is used to block one of the tunnels of PutA
The tunnel is shown to be essential for proline dehydrogenase activity
The tunnel contributes to catalysis by enabling the dynamics of an ion-pair gate
Acknowledgments
We thank J. Nix and for help with remote X-ray diffraction data collection and processing. Beamline 4.2.2 of the Advanced Light Source, a DOE Office of Science User Facility under Contract No. DE-AC02-05CH11231, is supported in part by the ALS-ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01.
Funding
Research reported in this publication was supported by the NIGMS of the National Institutes of Health under award number R01GM065546.
Abbreviations:
- PRODH
proline dehydrogenase
- P5C
Δ1-pyrroline-5-carboxylate
- GSAL
L-glutamate-γ-semialdehyde
- GSALDH
L-glutamate-γ-semialdehyde dehydrogenase
- PutA
proline utilization A
- GsPutA
Geobacter sulfurreducens proline utilization A
- THFA
S-(−)-tetrahydro-2-furoic acid
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Liu LK, Becker DF, Tanner JJ, Structure, function, and mechanism of proline utilization A (PutA), Arch Biochem Biophys 632 (2017) 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Srivastava D, Schuermann JP, White TA, Krishnan N, Sanyal N, Hura GL, Tan A, Henzl MT, Becker DF, Tanner JJ, Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum, Proc. Natl. Acad. Sci. USA 107(7) (2010) 2878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Luo M, Christgen S, Sanyal N, Arentson BW, Becker DF, Tanner JJ, Evidence that the c-terminal domain of a type b puta protein contributes to aldehyde dehydrogenase activity and substrate channeling, Biochemistry 53(35) (2014) 5661–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moxley MA, Sanyal N, Krishnan N, Tanner JJ, Becker DF, Evidence for Hysteretic Substrate Channeling in the Proline Dehydrogenase and Delta1-Pyrroline-5-carboxylate Dehydrogenase Coupled Reaction of Proline Utilization A (PutA), J. Biol. Chem 289(6) (2014) 3639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Singh H, Arentson BW, Becker DF, Tanner JJ, Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site, Proc. Nat. Acad. Sci. USA 111(9) (2014) 3389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, Tanner JJ, Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function, J. Biol. Chem 291(46) (2016) 24065–24075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Korasick DA, Gamage TT, Christgen S, Stiers KM, Beamer LJ, Henzl MT, Becker DF, Tanner JJ, Structure and characterization of a class 3B proline utilization A: Ligand-induced dimerization and importance of the C-terminal domain for catalysis, J Biol Chem 292(23) (2017) 9652–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Arentson BW, Luo M, Pemberton TA, Tanner JJ, Becker DF, Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A, Biochemistry 53(31) (2014) 5150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Luo M, Arentson BW, Srivastava D, Becker DF, Tanner JJ, Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release, Biochemistry 51(50) (2012) 10099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kabsch W, XDS., Acta crystallographica. Section D, Biological crystallography 66(Pt 2) (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].French S, Wilson K, IUCr, On the treatment of negative intensity observations, Acta Crystallographica Section A: Crystal Physics, Diffraction, Theoretical and General Crystallography 34(4) (1978) 517–525. [Google Scholar]
- [12].Emsley P, Cowtan K, IUCr, Coot: model-building tools for molecular graphics, Acta crystallographica. Section D, Biological crystallography 60(12) (2004) 2126–2132. [DOI] [PubMed] [Google Scholar]
- [13].Emsley P, Lohkamp B, Scott WG, Cowtan K, IUCr, Features and development of Coot, Acta crystallographica. Section D, Biological crystallography 66(4) (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, IUCr, Towards automated crystallographic structure refinement with phenix.refine, Acta crystallographica. Section D, Biological crystallography 68(4) (2012) 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, MolProbity: all-atom structure validation for macromolecular crystallography, Acta Crystallogr. D Biol. Crystallogr D66(Pt 1) (2010) 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gore S, Sanz Garcia E, Hendrickx PMS, Gutmanas A, Westbrook JD, Yang H, Feng Z, Baskaran K, Berrisford JM, Hudson BP, Ikegawa Y, Kobayashi N, Lawson CL, Mading S, Mak L, Mukhopadhyay A, Oldfield TJ, Patwardhan A, Peisach E, Sahni G, Sekharan MR, Sen S, Shao C, Smart OS, Ulrich EL, Yamashita R, Quesada M, Young JY, Nakamura H, Markley JL, Berman HM, Burley SK, Velankar S, Kleywegt GJ, Validation of Structures in the Protein Data Bank, Structure 25(12) (2017) 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liebschner D, Afonine PV, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, Polder maps: improving OMIT maps by excluding bulk solvent, Acta Crystallogr D Struct Biol 73(Pt 2) (2017) 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pravda L, Sehnal D, Tousek D, Navratilova V, Bazgier V, Berka K, Svobodova Varekova R, Koca J, Otyepka M, MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update), Nucleic Acids Res 46(W1) (2018) W368–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tanner JJ, Structural Biology of Proline Catabolic Enzymes, Antioxid Redox Signal 30(4) (2019) 650–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Williams I, Frank L, Improved chemical synthesis and enzymatic assay of delta-1-pyrroline-5-carboxylic acid, Anal Biochem 64(1) (1975) 85–97. [DOI] [PubMed] [Google Scholar]
- [21].Moxley MA, Becker DF, Rapid reaction kinetics of proline dehydrogenase in the multifunctional proline utilization A protein, Biochemistry 51(1) (2012) 511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brezovsky J, Chovancova E, Gora A, Pavelka A, Biedermannova L, Damborsky J, Software tools for identification, visualization and analysis of protein tunnels and channels, Biotechnol Adv 31(1) (2013) 38–49. [DOI] [PubMed] [Google Scholar]
- [23].Raushel FM, Thoden JB, Holden HM, Enzymes with molecular tunnels, Acc Chem Res 36(7) (2003) 539–48. [DOI] [PubMed] [Google Scholar]
- [24].Kingsley LJ, Lill MA, Substrate tunnels in enzymes: structure-function relationships and computational methodology, Proteins 83(4) (2015) 599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Prokop Z, Gora A, Brezovsky J, Chaloupkova R, Stepankova V, Damborský J, Engineering of protein tunnels: Keyhole-lock-key model for catalysis by the enzymes with active sites, in: Lutz S.a.B., U. T. (Ed.), Protein Engineering Handbook, Wiley-VCH Verlag GmbH & Co.2012, pp. 421–464. [Google Scholar]
- [26].Arentson BW, Sanyal N, Becker DF, Substrate channeling in proline metabolism, Front. Biosci. 17 (2012) 375–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hyde CC, Miles EW, The tryptophan synthase multienzyme complex: exploring structure-function relationships with X-ray crystallography and mutagenesis, Biotechnology (N Y) 8(1) (1990) 27–32. [DOI] [PubMed] [Google Scholar]
- [28].Anderson KS, Kim AY, Quillen JM, Sayers E, Yang XJ, Miles EW, Kinetic characterization of channel impaired mutants of tryptophan synthase, J Biol Chem 270(50) (1995) 29936–44. [DOI] [PubMed] [Google Scholar]
- [29].Schlichting I, Yang XJ, Miles EW, Kim AY, Anderson KS, Structural and kinetic analysis of a channel-impaired mutant of tryptophan synthase, J Biol Chem 269(43) (1994) 26591–3. [PubMed] [Google Scholar]
- [30].Dunn MF, Niks D, Ngo H, Barends TR, Schlichting I, Tryptophan synthase: the workings of a channeling nanomachine, Trends Biochem. Sci. 33(6) (2008) 254–64. [DOI] [PubMed] [Google Scholar]
- [31].Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR, Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium, J. Biol. Chem 263(33) (1988) 17857–71. [PubMed] [Google Scholar]
- [32].Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I, Structure of carbamoyl phosphate synthetase: a journey of 96 A from substrate to product, Biochemistry 36(21) (1997) 6305–16. [DOI] [PubMed] [Google Scholar]
- [33].Huang X, Holden HM, Raushel FM, Channeling of substrates and intermediates in enzyme-catalyzed reactions, Annu. Rev. Biochem 70 (2001) 149–80. [DOI] [PubMed] [Google Scholar]
- [34].Winter MB, Herzik MA Jr., Kuriyan J, Marletta MA, Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain, Proc Natl Acad Sci U S A 108(43) (2011) E881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schmitt J, Brocca S, Schmid RD, Pleiss J, Blocking the tunnel: engineering of Candida rugosa lipase mutants with short chain length specificity, Protein Eng 15(7) (2002) 595–601. [DOI] [PubMed] [Google Scholar]
- [36].Yan X, Wang J, Sun Y, Zhu J, Wu S, Facilitating the Evolution of Esterase Activity from a Promiscuous Enzyme (Mhg) with Catalytic Functions of Amide Hydrolysis and Carboxylic Acid Perhydrolysis by Engineering the Substrate Entrance Tunnel, Appl Environ Microbiol 82(22) (2016) 6748–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lu Z, Li X, Zhang R, Yi L, Ma Y, Zhang G, Tunnel engineering to accelerate product release for better biomass-degrading abilities in lignocellulolytic enzymes, Biotechnol Biofuels 12 (2019) 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Biedermannova L, Prokop Z, Gora A, Chovancova E, Kovacs M, Damborsky J, Wade RC, A single mutation in a tunnel to the active site changes the mechanism and kinetics of product release in haloalkane dehalogenase LinB, J Biol Chem 287(34) (2012) 29062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tan X, Loke HK, Fitch S, Lindahl PA, The tunnel of acetyl-coenzyme a synthase/carbon monoxide dehydrogenase regulates delivery of CO to the active site, J Am Chem Soc 127(16) (2005) 5833–9. [DOI] [PubMed] [Google Scholar]
- [40].Tan X, Volbeda A, Fontecilla-Camps JC, Lindahl PA, Function of the tunnel in acetylcoenzyme A synthase/carbon monoxide dehydrogenase, J Biol Inorg Chem 11(3) (2006) 371–8. [DOI] [PubMed] [Google Scholar]
- [41].Kokkonen P, Slanska M, Dockalova V, Pinto GP, Sanchez-Carnerero EM, Damborsky J, Klan P, Prokop Z, Bednar D, The impact of tunnel mutations on enzymatic catalysis depends on the tunnel-substrate complementarity and the rate-limiting step, Comput Struct Biotechnol J 18 (2020) 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kokkonen P, Beier A, Mazurenko S, Damborsky J, Bednar D, Prokop Z, Substrate inhibition by the blockage of product release and its control by tunnel engineering, RSC Chemical Biology (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]