Abstract
Background
Although there are several severity predictors for COVID-19, none are specific. Serum levels of phenylalanine were recently associated with increased inflammation, higher SOFA scores, ICU admission, and mortality rates among non-COVID-19 patients. Here, we investigated the relationship between phenylalanine and inflammatory markers in adults with COVID-19.
Methods
We assessed adults with COVID-19 at hospital admission for clinical and laboratory data. Nuclear magnetic resonance spectroscopy measured serum levels of phenylalanine and other amino acids of its metabolomic pathway. Flow Cytometry measured serum levels of IL-2, IL-4, IL-6, Il-10, TNF-α, and IFN-γ. Linear regression models adjusted for potential confounders assessed the relationship between serum levels of phenylalanine and inflammatory cytokines.
Results
Phenylalanine and tyrosine were significantly lower in mild disease as compared to moderate and severe groups. Linear regression models showed that phenylalanine is independently and positively associated with disease severity regardless of the cytokine analyzed and after adjustment for potential confounders. In addition, mild cases showed consistently lower serum phenylalanine levels within the first ten days from disease onset to hospital admission.
Conclusions
Phenylalanine is a marker of disease severity. This association is independent of the time between the onset of symptoms and the magnitude of the inflammatory state.
Keywords: COVID-19, SARS-CoV-2, Cytokines, Phenylalanine, Tyrosine
1. Background
COVID-19 exhibits a spectrum of manifestations ranging from asymptomatic or mild flu-like symptoms to severe pneumonia with acute respiratory distress syndrome and death [1]. The degrees of increasing severity reflect different clinical and laboratory findings, response to therapy, and clinical evolution [2]. In this regard, the literature points out that about 50 markers (including demographic, patient history, physical examination, laboratory, radiological data, and a high Sequential Organ Failure Assessment -SOFA score) can provide valuable prognostic information about mortality disease severity [3]. However, most of these markers are not specific and share similarities with other conditions. Moreover, they do not explain why some patients progress to severe states or the determinants of the speed at which it occurs. Thus, there is a need to investigate alternative predictors of disease severity and death.
Approximately 20% of COVID-19 patients develop a severe inflammatory-mediated hypoxemic disease that resembles a cytokine-storm syndrome characterized by high levels of pro-inflammatory cytokines such as interleukin 1 (IL-1), IL-6, and TNF-α [4]. Accordingly, we and others have shown that serum IL-6 and IL-10 levels are significantly associated with disease severity and progression [5], [6], [7], [8]. Recently, a few studies have shown the incidence of clinically significant metabolic changes that may interfere with disease severity and prognosis [9], [10], [11].
Metabolomic studies can provide a portrait of the physiological state of an organism responding to pathogenic challenges and treatment [12]. To this point, only a few studies have used mass spectrometry metabolomics to identify COVID-19-induced alterations in circulating metabolites. A metabolic phenotyping study of blood plasma coupled with cytokine/chemokine analysis from 59 patients with COVID-19 demonstrated significant elevations of nine plasma metabolites, including quinolinic acid, glutamic acid, nicotinic acid, aspartic acid, neopterin, kynurenine, phenylalanine, 3-hydroxykynurenine, and taurine [13]. Moreover, the correlation between metabolite changes and concentrations of inflammatory cytokines and chemokines points to an immunometabolic disorder [13]. Other studies have focused on the correlation of metabolic parameters with a clinical presentation [14], circulating IL-6 levels [15], or sex-specific metabolic changes [16], [17]. However, the limited sample size is the major limitation of these studies, and also, none took into account possible confounding factors.
Nevertheless, Wannemacher et al. [18] provided significant advances in understanding the pattern of change in individual plasma-free amino acids during infectious processes. According to their findings, inflammation and infection often lead to increased levels of some serum amino acids, especially in serum phenylalanine and in the phenylalanine-tyrosine ratio [19]. It occurs due to an accelerated release from skeletal muscle to meet the rates of glucose turnover and oxidation in the infected host [18]. However, there is a lack of studies investigating the relationship between phenylalanine levels and inflammatory markers to identify potential determinants of disease severity and progression. Thus, we investigated this relationship in adult patients with COVID-19, considering disease severity and the main markers of the phenylalanine metabolic pathway.
2. Methods
2.1. Study design, participants and setting
We conducted a cross-sectional study at the University Hospital of the Federal University of São Carlos (HU-UFSCar). From July to December 2020, patients included if they were diagnosed with COVID-19 according to the WHO guidelines and had <11 days from the illness onset to hospital admission. We excluded subjects having more than 11 days after symptom onset to hospital admission because this is the period above the 75th percentile for viral clearance. Thus, we aimed to reduce the chance of secondary interferences on the pattern of inflammatory cytokines beyond the relationship between SARS-CoV-2 infection and immune response. The study was conducted according to the guidelines laid down in the Declaration of Helsinki and all procedures involving research study participants were approved by the UFSCar’s Research Ethics Committee (Number: 30184220.8.0000.5504). All subjects provided written informed consent.
2.2. Study assessments
We assessed all patients at admission for demographic and clinical data, chronic comorbidities and illness severity. Patients were categorized as mild, moderate, and severe according to the recommendations set forth by the WHO’s COVID-19 Clinical management living guidance [20]. Within the first 12 h of admission, venous blood was sampled to analyze inflammatory cytokines and metabolomics.
2.3. Systemic markers of inflammation
Samples were analyzed through flow cytometry using BD Accuri C6 (BD Biosciences, Franklin Lakes, NJ, USA), and serum cytokines (IL-2, IL-4, IL-6, IL-10, IFN-γ, and TNF-α) were measured with cytometric bead array human inflammation kit (BD™ CBA Human Th1/Th2 Cytokine Kit, BD Biosciences, San Diego, CA, USA). The procedure was conducted following the manufacturers' instructions. Data were analyzed using FlowJo software (FlowJo LLC, Ashland, OR, USA).
2.4. Serum sample preparation for NMR measurements
An aliquoted blood serum sample was stored at −80 °C until measured. The delivered frozen serum sample was thawed to room temperature. After that, 500 μL of blood serum was added to the filter (Amicon Ultra – 3 kDa) and it was centrifuged at 14,000 g for 30 min at 4 °C. Then, 150 μL of filtered serum was transferred to a 5 mm NMR tube (Wilmad Standard Series 5 mm, Sigma-Aldrich) containing 60 μL of phosphate buffer [(monobasic sodium phosphate, NaH2PO4 H20, 137.99 g·mol−1; dibasic sodium phosphate, Na2HPO3, 141.96 g·mol−1), TMSP-d4 (3-(trimethylsilyl)-2,2′,3,3′-tetradeuteropropionic acid sodium salt) at 0.5 mmol·L−1 (internal reference)] and 390 μL of deionized water (99.9%; Cambridge Isotope Laboratories Inc.) for immediate NMR acquisition.
All the filters were washed with 500 μL of ultra-pure water, followed by centrifugation at 14,000 g for 5 min at 4 °C. This process was repeated five times. After the fifth wash, spinning (filter reverse and rotation at 7500 g for 60 s) was performed to eliminate any residue of ultra-pure water.
2.5. Spectrum acquisition and metabolite quantification
All the NMR measurements were acquired in a 14.1 Tesla Bruker spectrometer (600 MHz for hydrogen frequency), equipped with a 5 mm TXI cryoprobe using temperature 343 K, TSP-d 4 as an internal reference, solvent deionized water, and the inverse detection probe head except for 13C (the broadband direct observe).
For the 1H spectrum with HDO presaturation signal using continuous-wave the parameters were, acquisition time (AQ = 5.45 s), sweep width (SWH = 12019 Hz), relaxation delay (d1 = 3 s), the 90° pulse time (p1 = 9.125 µs) and number of scans (ns = 128). After spectrum acquisition, baseline corrections, identification, and quantification of metabolites present in the samples were conducted using Suite 8.6 Chenomx software (Chenomx Inc., Edmonton, AB, Canada) by the TMSP-d4 signal (known concentration = 0,5 mmol.L−1) as an internal reference to quantify other metabolites. All spectra were processed with 0.3 Hz line broadening (lb) to attenuate the noise in the spectral signals.
2.6. Statistical analysis
Continuous data are presented as median [1st, 3rd quartile] according to the Shapiro-Wilk test. Categorical variables are presented as counts (percentages). Comparisons between groups were performed using Kruskal-Wallis test followed by Dunn’s Post Hoc test with Benjamini-Hochberg p-value correction for continuous variables, and Pearson’s Chi-squared test with Yates’ continuity correction for categorical variables. Multivariate linear regression models were used to investigate the relationship between the circulating levels of phenylalanine and major inflammatory cytokines. Phenylalanine and cytokine measurements were transformed (log10) in order for variables to meet the assumptions of statistical tests. Clinical characteristics and disease severity were selected for the multivariate analysis. All analyses were conducted using R version 4.0.3 (The R Foundation for Statistical Computing, Vienna, Austria) in R-Studio 1.3.1093 (RStudio Inc., Boston, USA).
3. Results
3.1. Baseline characteristics of study participants
We included a total of 166 subjects in this study. The overall median age was 60 years old, ranging from 22 to 99 years. About a half were males (53.6%), with a median Charlson Comorbidity Index (CCI) of 2, and about a fourth (24.7%) of them presented high comorbidity (index ≥ 5). Almost a half were hypertensive (47%), while most were not diabetic (69.9%), nor had cardiovascular diseases (84.9%). Table 1 summarizes baseline demographic and clinical characteristics for each severity group. Patients with mild disease were significantly different from those with moderate and severe conditions as regard to sex, age, CCI, and NEWS2 score on admission. Besides the expected differences in NEWS2 score on admission, there were no significant differences between the moderate and severe groups.
Table 1.
Baseline characteristics of enrolled subjects.
| Variable | Overall (N = 166) |
Mild (n = 21) |
Moderate (n = 84) |
Severe (n = 61) |
p |
|---|---|---|---|---|---|
| Male sex | 89 (53.6) | 5 (23.8) | 46 (54.8) | 38 (62.3) | 0.009ab |
| Age, years | 60 [44, 74] | 38 [35, 49] | 61 [47, 73] | 65 [48, 79] | < 0.001ab |
| 65 + years | 65 (39.2) | 3 (14.3) | 31 (36.9) | 31 (50.8) | 0.01b |
| Comorbidities | |||||
| Hypertension | 78 (47) | 6 (28.6) | 43 (51.2) | 29 (47.5) | 0.1 |
| Diabetes mellitus | 50 (30.1) | 3 (14.3) | 29 (34.5) | 18 (29.5) | 0.1 |
| Cardiovascular diseases | 25 (15.1) | 2 (9.5) | 12 (14.3) | 11 (18) | 0.6 |
| Charlson Comorbidity Index | 2 [0, 4] | 0 [0, 0] | 2 [0, 4] | 3 [1,5] | 0.001ab |
| Time from symptom onset to hospital admission, days | 7 [4,10] | 7 [6,10] | 7 [4,10] | 7 [4,10] | 0.8 |
| NEWS2 at hospital admission | 4 [2,5] | 1 [1,1] | 3 [2,4] | 5 [5,6] | < 0.001abc |
Continuous data are presented as median [1st, 3rd quartile]. Categorical variables are presented as counts (percentages). Post-hoc significant (p < 0.05) pairwise comparisons: a Mild vs Moderate, b Mild vs Severe, c Moderate vs Severe.
3.2. Phenylalanine and other laboratory parameters according to disease severity
Baseline serum levels of phenylalanine and other laboratory parameters were evaluated at hospital admission and stratified according to the disease severity (see Table 2 ). Post hoc tests indicated that all groups were significantly different from each other, except for serum levels of IL-4, TNF-α, IFN-γ, histidine, ornithine, and creatine phosphokinase. Phenylalanine was significantly lower in mild disease as compared to moderate and severe groups (both p adjusted < 0.001). Tyrosine was also lower in mild disease (both p adjusted = 0.03). Tyrosine-related analyzes can be found in the Supplementary Material. Moreover, subjects with severe disease presented a lower median estimated glomerular filtration rate when compared to mild and moderate groups (p adjusted = 0.05 and 0.04, respectively).
Table 2.
Phenylalanine and other laboratory parameters at hospital admission, according to the severity groups.
| Variable | Overall (N = 166) |
Mild (n = 21) |
Moderate (n = 84) |
Severe (n = 61) |
p |
|---|---|---|---|---|---|
| Lymphocyte count (×109/L) | 1.16 [0.72, 1.58] | 2.17[1.55, 2.65] | 1.17 [0.35, 1.52] | 0.8 [0.52, 1.42] | <0.001abc |
| Lactate dehydrogenase (U/L) | 276 [215, 387] | 185 [167, 265] | 247 [205, 350] | 336 [270, 486] | <0.001abc |
| C-reactive protein (mg/dL) | 6.5 [1.1, 13.6] | 0.6 [0.1, 0.6] | 5.1 [1.1, 11.6] | 11.6 [5, 18.5] | <0.001abc |
| IL-2 (pg/dL) | 14.1 [12.9, 14.7] | 12.2 [11.5, 13.6] | 14.1 [13.2, 14.8] | 14.4 [13.6, 14.9] | <0.001ab |
| IL-4 (pg/dL) | 20 [19.1, 21] | 19.4 [18.6, 20.5] | 19.9 [18.8, 21] | 20.3 [19.4, 21.1] | 0.1 |
| IL-6 (pg/dL) | 35.3 [15.8, 94.1] | 20.1 [18.8, 22.1] | 34.4 [23.5, 70.6] | 59.7 [31.9, 140.4] | <0.001abc |
| IL-10 (pg/dL) | 40.4 [36.9, 48.5] | 36.1 [34.8, 37.6] | 39.2 [36.7, 46.6] | 45.5 [40, 66.6] | <0.001abc |
| TNF-α (pg/dL) | 13.1 [12.7, 14.5] | 13.1 [12.6, 14.7] | 13.1 [12.7, 14.1] | 13.3 [12.9, 14.6] | 0.5 |
| IFN-γ (pg/dL) | 14.8 [13.9, 17.1] | 13.9 [13.4, 17] | 14.9 [13.9, 17] | 15.1 [14, 18.3] | 0.1 |
| IL-6/IL-10 ratio | 0.8 [0.6, 1.7] | 0.6 [0.5, 0.6] | 0.8 [0.6, 1.4] | 1.1 [0.7, 2.7] | <0.001abc |
| IL-6/IFN-γ ratio | 2.2 [1.5. 4.5] | 1.4 [1.1, 1.6] | 2.1 [1.5, 4.3] | 3.5 [2.0, 8.4] | <0.001abc |
| TNF-α/IL-10 ratio | 3 [2.7, 3.5] | 2.8 [2.5, 2.9] | 2.9 [2.7, 3.4] | 3.3 [2.9, 4.1] | <0.001abc |
| Phenylalanine (mg/dL) | 1.51 [1.18, 2.08] | 1 [0.85, 1.35] | 1.5 [1.14, 2.23] | 1.71 [1.43, 2.17] | <0.001ab |
| Histidine (mg/dL) | 0.84 [0.36, 1.07] | 0.68 [0.22, 1.03] | 0.9 [0.49, 1.11] | 0.81 [0.35, 1.03] | 0.2 |
| Ornithine (mg/dL) | 0.65 [0.48, 0.88] | 0.59 [0.48, 0.75] | 0.64 [0.47, 0.86] | 0.68 [0.5, 0.92] | 0.4 |
| Tyrosine (mg/dL) | 1.28 [0.94, 1.60] | 1.01 [0.9, 1.3] | 1.29 [1, 1.71] | 1.36 [1, 1.61] | 0.04ab |
| Creatinine (mg/dL) | 0.9 [0.7, 1.3] | 0.9 [0.8, 1] | 0.8 [0.7, 1.12] | 1 [0.8, 1.6] | 0.02c |
| Estimated Glomerular Filtration Rate, eGFR (mL/min/1.73 m2) | 88 [54, 105] | 95 [83, 108] | 91 [66, 109] | 75 [39, 171] | 0.02bc |
| Creatine phosphokinase (mg/dL) | 87 [49, 201] | 72 [48, 114] | 86 [51, 140] | 100 [47, 264] | 0.2 |
Continuous data are presented as median [1st, 3rd quartile]. Categorical variables are presented as counts (percentages). Post-hoc significant (p < 0.05) pairwise comparisons: a Mild vs Moderate, b Mild vs Severe, c Moderate vs Severe.
3.3. Relationship between serum phenylalanine levels and inflammatory markers
The relationship between serum levels of phenylalanine and major inflammatory cytokines (IL-2, IL-6, and IL-10) was evaluated using linear regression analysis adjusted for sex, age, comorbidity (CCI), and disease severity groups. Table 3 shows that serum phenylalanine levels are independently and positively associated only with disease severity regardless of the cytokine analyzed.
Table 3.
Linear regression models for the association between serum levels of phenylalanine and inflammatory markers, adjusted for sex, age, comorbidity (CCI), and disease severity groups.
| Dependent variable: Phenylalanine (log mg/dL) |
||||||
|---|---|---|---|---|---|---|
| Independent variables | Model 1 |
Model 2 |
Model 3 |
|||
| Standardized β Coefficient (95% CI) |
p | Standardized β Coefficient (95% CI) |
p | Standardized β Coefficient (95% CI) |
p | |
| IL-2 (log pg/mL) | 0.06 (−0.09 to 0.21) | 0.4 | – | – | – | – |
| IL-6 (log pg/mL) | – | – | 0.13 (−0.02 to 0.28) | 0.08 | – | – |
| IL-10 (log pg/mL) | – | – | – | – | 0.09 (−0.06 to 0.24) | 0.2 |
| Male sex | 0.07 (−0.23 to 0.39) | 0.1 | 0.09 (−0.21 to 0.4) | 0.5 | 0.08 (−0.22 to 0.39) | 0.5 |
| Age, years | 0.12 (−0.15 to 0.39) | 0.3 | 0.11 (−0.6 to 0.38) | 0.4 | 0.11 (−0.16 to 0.38) | 0.4 |
| Charlson Comorbidity Index | −0.03 (−0.3 to 0.24) | 0.8 | −0.01 (−0.28 to 0.25) | 0.9 | −0.01 (0.28 to 0.25) | 0.9 |
| Disease severity | ||||||
| Mild | Reference | – | Reference | – | Reference | – |
| Moderate | 0.73 (0.23–1.22) | 0.004 | 0.70 (0.22–1.19) | 0.005 | 0.70 (0.21–1.19) | 0.005 |
| Severe | 0.91 (0.39–1.43) | <0.001 | 0.85 (0.34–1.37) | 0.001 | 0.86 (0.35–1.38) | 0.001 |
95% CI, 95% Confidence Interval.
3.4. Serum levels of phenylalanine as a marker of disease severity
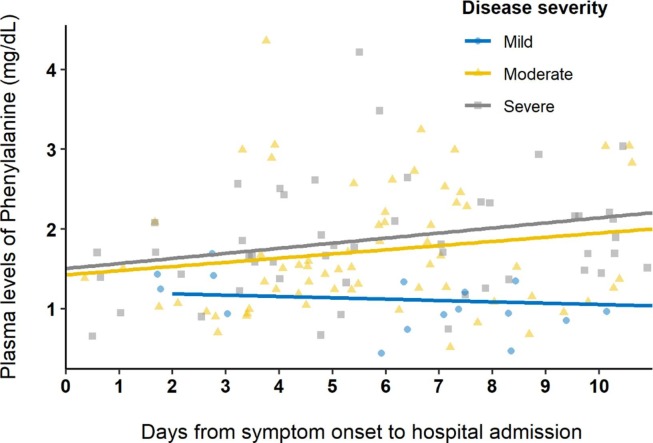
Considering a possible role of phenylalanine as a marker of disease severity independent from major markers of inflammation, we further evaluated the temporal evolution of serum levels of phenylalanine during the first ten days from illness onset to hospital admission. Fig. 1 shows the distribution of serum levels of phenylalanine in each disease severity group as a function of the time from illness onset to hospital admission. In the figure, serum levels of phenylalanine in mild severity have lower and less variable concentrations throughout the time as moderate and severe groups.
Fig. 1.
Distribution of serum levels of phenylalanine according to the time from illness onset to hospital admission and disease severity.
4. Discussion
There is an increasing attempt to answer why some COVID-19 patients progress to severe states and the determinants of the speed at which it occurs. Currently, there are several studies on clinical parameters [21], specific comorbidities [22], [23], biochemical [24] and hematological [25] laboratory parameters, and inflammatory cytokines alterations [26]. More recently, some have brought a new perspective to COVID-19 through the prism of metabolomics [27], [28].
We analyzed the metabolomic and inflammatory profile of adults with COVID-19 according to disease severity. We demonstrated that phenylalanine and tyrosine are markedly higher among subjects with moderate to severe disease. Inflammatory cytokines promote muscle breakdown with the release of phenylalanine for gluconeogenesis to supply the metabolic demand during the infection. Thus, higher phenylalanine levels would be associated with the catabolic state. However, we have shown that phenylalanine maintained an independent association with disease severity even after adjusting for different pro-inflammatory cytokines. This association is independent of the time between the onset of symptoms and the magnitude of the inflammatory state. In particular, these findings point to phenylalanine as a distinct marker of disease severity.
The evidence for phenylalanine as a marker of severity is not new. A study carried out between 2017 and 2018, that is, before the COVID-19 pandemic, evaluated patients with different serious infections (SOFA ≥ 2), analyzing levels of plasma phenylalanine, leucine, C-reactive protein (CRP), and nutritional indices (albumin, pre-albumin, and transferrin) and monitored their outcomes for three months. Phenylalanine was associated with higher mortality and ICU admission rates, SOFA scores, episodes of bacteremia, CRP levels, and lower levels of pre-albumin, transferrin, and leucine. When dividing patients according to phenylalanine levels, participants with higher phenylalanine levels had higher SOFA scores, ICU admission rates, and levels of CRP and leucine [29]. However, this study did not analyze the relationship of phenylalanine levels with inflammatory markers taking into account possible confounding factors in a multivariate model.
Despite that, phenylalanine appears to be a severity marker related to respiratory distress. Recently Xu et al. showed that patients with acute respiratory distress syndrome (ARDS) have significantly different metabolomic profiles than healthy controls. [30] According to their results, phenylalanine, D-phenylalanine, and phenylacetylglutamine levels were higher among non-survivors. Phenylalanine metabolism was the most notably altered pathway among non-survivors and survivors. In vivo animal experiments have also shown that higher phenylalanine levels were associated with severe lung injury and increased mortality from ARDS. Moreover, Chen et al. have recently reported that phenylalanine predicted mortality in critically ill patients with acute heart failure (HF) or acute on chronic HF [31]. In multivariate analysis, phenylalanine levels ≥ 112 μM predicted death over one year, regardless of age, APACHE II and SOFA scores, atrial fibrillation, C-reactive protein, cholesterol, prealbumin, transferrin, and IL-8 and IL-10. Thus, a growing body of evidence points to phenylalanine as a marker of disease severity, especially related to respiratory distress.
The dominant respiratory feature of COVID-19 is the progressive worsening of arterial hypoxemia, eventually leading to ARDS. To this end, the classification of severity proposed by the WHO reflects different clinical and laboratory findings, responses to therapy, and clinical evolution [2], [20]. However, most of these class markers are non-specific and share similarities with other inflammatory and respiratory conditions. Furthermore, as far as we know, no study has demonstrated an independent predictive value of a serum marker on disease severity in patients with COVID-19. Accordingly, previous studies using NMR metabolomic analysis have shown several abnormalities when comparing COVID-19 patients to healthy individuals, including the reduction in some essential amino acids (methionine, isoleucine, histidine, lysine, tyrosine, and glutamine) and increased levels of phenylalanine (81%) and 2-hydroxybutyric acid, suggesting a condition of metabolic stress in COVID-19 [32]. Untargeted analysis of non-critical COVID-19 patients demonstrated that phenylalanine, tyrosine and tryptophan biosynthesis, phenylalanine metabolism, aminoacyl-tRNA degradation, arachidonic acid metabolism, and the tricarboxylic acid (TCA) cycle were the most important metabolic pathways affected by the SARS-CoV-2 infection [33]. A genome-scale metabolic study complemented these findings by revealing a significantly higher phenylalanine flux among COVID-19 deceased patients. It suggested that the usage of phenylalanine residues for the production of viral proteins and their subsequent assembly in viral particles would somehow be related to disease severity [34].
Nevertheless, our results agree with the significant association between higher phenylalanine levels and COVID-19 severity [35]. This association is also in line with reports of low occurrence of severe COVID-19 among patients with phenylketonuria, which has previously been attributed to the lower rates of vitamin D deficiency in these patients as a result of protein substitutes in their diet [36]. One of our strengths was that we demonstrated that this association is independent of the time between the onset of symptoms and the magnitude of the inflammatory state. However, the role of phenylalanine in the viral cycle and the impact of changes in its levels on the worsening of disease severity is still unknown.
Our study has some limitations. It includes its cross-sectional design and single-center nature that avoid predictive causal result inferences. Moreover, the metabolic and inflammatory parameters were recorded once at hospital admission, whereas consecutive measurements would have given a better idea of the dynamics between metabolic and inflammatory states and disease severity. Therefore, there is little information available on variations in phenylalanine levels during hospitalization and respiratory deterioration. However, our study provided a significant picture of the relationship between phenylalanine levels and inflammatory markers in adult patients with COVID-19, considering disease severity and the main features of the phenylalanine metabolic pathway. Considering that serum phenylalanine screening is a well-known blood test for diagnosing phenylketonuria (PKU) in newborns. We believe that future strategies may enable the rapid and practical measurement of phenylalanine levels after extensive validation of phenylalanine as a COVID-19-related biomarker. In this sense, the follow-up of these patients over time will be relevant to assess the diagnostic performance of phenylalanine levels as a marker of disease severity, an object of further analysis.
We showed that phenylalanine is a marker of disease severity. This association is independent of the time between the onset of symptoms and the magnitude of the inflammatory state. These results may help to better understand the metabolic and inflammatory pathways involved in COVID-19 and offer new pathophysiological insights into the interaction between inflammation and metabolism in COVID-19.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo - Brasil (FAPESP) - [grant numbers 2014/50867-3, 2019/16135-9, and 2020/13939-7], and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.108313.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Ramos-Casals M., Brito-Zeron P., Mariette X. Systemic and organ-specific immune-related manifestations of COVID-19. Nat. Rev. Rheumatol. 2021;17(6):315–332. doi: 10.1038/s41584-021-00608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddiqi H.K., Mehra M.R. COVID-19 illness in native and immunosuppressed states: A clinical-therapeutic staging proposal. J. Heart Lung Transplant. 2020;39(5):405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izcovich A., Ragusa M.A., Tortosa F., Lavena Marzio M.A., Agnoletti C., Bengolea A., Ceirano A., Espinosa F., Saavedra E., Sanguine V., Tassara A., Cid C., Catalano H.N., Agarwal A., Foroutan F., Rada G. Prognostic factors for severity and mortality in patients infected with COVID-19: A systematic review. PLoS One. 2020;15(11):e0241955. doi: 10.1371/journal.pone.0241955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D. Blanco-Melo, B.E. Nilsson-Payant, W.C. Liu, S. Uhl, D. Hoagland, R. Moller, T.X. Jordan, K. Oishi, M. Panis, D. Sachs, T.T. Wang, R.E. Schwartz, J.K. Lim, R.A. Albrecht, B.R. tenOever, Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19, Cell 181(5) (2020) 1036–1045 e9. [DOI] [PMC free article] [PubMed]
- 5.Coomes E.A., Haghbayan H. Interleukin-6 in Covid-19: A systematic review and meta-analysis. Rev. Med. Virol. 2020;30(6):1–9. doi: 10.1002/rmv.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon J.S., Kim J.Y., Kim M.C., Park S.Y., Kim B.N., Bae S., Cha H.H., Jung J., Kim M.J., Lee M.J., Choi S.H., Chung J.W., Shin E.C., Kim S.H. Factors of Severity in Patients with COVID-19: Cytokine/Chemokine Concentrations, Viral Load, and Antibody Responses. Am. J. Trop. Med. Hyg. 2020;103(6):2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. Luís Luporini, J.M. de A. Rodolpho, L. Tatsuo Kubota, A. Carolina Baptista Moreno Martin, M.R. Cominetti, F. de Freitas Anibal, H. Pott-Junior, IL-6 and IL-10 are associated with disease severity and a higher comorbidity in adults with COVID-19, Cytokine (2021) 155507. [DOI] [PMC free article] [PubMed]
- 8.Pott-Junior H., Bittencourt N.Q.P., Chacha S.F.G., Luporini R.L., Cominetti M.R., Anibal F.D.F. Elevations in Liver Transaminases in COVID-19: (How) Are They Related? Front. Med. 2021;8 doi: 10.3389/fmed.2021.705247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y.M., Zheng Y., Yu Y., Wang Y., Huang Q., Qian F., Sun L., Song Z.G., Chen Z., Feng J., An Y., Yang J., Su Z., Sun S., Dai F., Chen Q., Lu Q., Li P., Ling Y., Yang Z., Tang H., Shi L., Jin L., Holmes E.C., Ding C., Zhu T.Y., Zhang Y.Z. Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. EMBO J. 2020;39(24) doi: 10.15252/embj.2020105896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J.W. Song, S.M. Lam, X. Fan, W.J. Cao, S.Y. Wang, H. Tian, G.H. Chua, C. Zhang, F.P. Meng, Z. Xu, J.L. Fu, L. Huang, P. Xia, T. Yang, S. Zhang, B. Li, T.J. Jiang, R. Wang, Z. Wang, M. Shi, J.Y. Zhang, F.S. Wang, G. Shui, Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis, Cell Metab. 32(2) (2020) 188–202 e5. [DOI] [PMC free article] [PubMed]
- 11.Shi D., Yan R., Lv L., Jiang H., Lu Y., Sheng J., Xie J., Wu W., Xia J., Xu K., Gu S., Chen Y., Huang C., Guo J., Du Y., Li L. The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism. 2021;118 doi: 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016;17(7):451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawler N.G., Gray N., Kimhofer T., Boughton B., Gay M., Yang R., Morillon A.C., Chin S.T., Ryan M., Begum S., Bong S.H., Coudert J.D., Edgar D., Raby E., Pettersson S., Richards T., Holmes E., Whiley L., Nicholson J.K. Systemic Perturbations in Amine and Kynurenine Metabolism Associated with Acute SARS-CoV-2 Infection and Inflammatory Cytokine Responses. J. Proteome Res. 2021;20(5):2796–2811. doi: 10.1021/acs.jproteome.1c00052. [DOI] [PubMed] [Google Scholar]
- 14.B. Shen, X. Yi, Y. Sun, X. Bi, J. Du, C. Zhang, S. Quan, F. Zhang, R. Sun, L. Qian, W. Ge, W. Liu, S. Liang, H. Chen, Y. Zhang, J. Li, J. Xu, Z. He, B. Chen, J. Wang, H. Yan, Y. Zheng, D. Wang, J. Zhu, Z. Kong, Z. Kang, X. Liang, X. Ding, G. Ruan, N. Xiang, X. Cai, H. Gao, L. Li, S. Li, Q. Xiao, T. Lu, Y. Zhu, H. Liu, H. Chen, T. Guo, Proteomic and Metabolomic Characterization of COVID-19 Patient Sera, Cell 182(1) (2020) 59–72 e15. [DOI] [PMC free article] [PubMed]
- 15.Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., Hudson K.E., Zimring J.C., Hansen K.C., Hod E.A., Spitalnik S.L., D'Alessandro A. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14) doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Y. Cai, D.J. Kim, T. Takahashi, D.I. Broadhurst, S. Ma, N.J.W. Rattray, A. Casanovas-Massana, B. Israelow, J. Klein, C. Lucas, T. Mao, A.J. Moore, C.M. Muenker, J. Silva, P. Wong, A.J. Ko, S.A. Khan, A. Iwasaki, C.H. Johnson, Kynurenic acid underlies sex-specific immune responses to COVID-19, medRxiv (2020).
- 17.Zheng H., Jin S., Li T., Ying W., Ying B., Chen D., Ning J., Zheng C., Li Y., Li C., Chen C., Li X., Gao H. Metabolomics reveals sex-specific metabolic shifts and predicts the duration from positive to negative in non-severe COVID-19 patients during recovery process. Comput. Struct. Biotechnol. J. 2021;19:1863–1873. doi: 10.1016/j.csbj.2021.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wannemacher R.W., Jr. Key role of various individual amino acids in host response to infection. Am. J. Clin. Nutr. 1977;30(8):1269–1280. doi: 10.1093/ajcn/30.8.1269. [DOI] [PubMed] [Google Scholar]
- 19.Wannemacher R.W., Jr., Klainer A.S., Dinterman R.E., Beisel W.R. The significance and mechanism of an increased serum phenylalanine-tyrosine ratio during infection. Am. J. Clin. Nutr. 1976;29(9):997–1006. doi: 10.1093/ajcn/29.9.997. [DOI] [PubMed] [Google Scholar]
- 20.WHO, COVID-19 clinical management: living guidance, 31 March 2021, World Health Organization, Geneva, Switzerland, 2021, p. 85.
- 21.Li J., Huang D.Q., Zou B., Yang H., Hui W.Z., Rui F., Yee N.T.S., Liu C., Nerurkar S.N., Kai J.C.Y., Teng M.L.P., Li X., Zeng H., Borghi J.A., Henry L., Cheung R., Nguyen M.H. Epidemiology of COVID-19: A systematic review and meta-analysis of clinical characteristics, risk factors, and outcomes. J. Med. Virol. 2021;93(3):1449–1458. doi: 10.1002/jmv.26424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.H. Pott-Jr, M.R. Cominetti, Comorbidities predict 30-day hospital mortality of older adults with COVID-19, Geriatric Nursing in press (2021). [DOI] [PMC free article] [PubMed]
- 23.Fernandez Villalobos N.V., Ott J.J., Klett-Tammen C.J., Bockey A., Vanella P., Krause G., Lange B. Effect modification of the association between comorbidities and severe course of COVID-19 disease by age of study participants: A systematic review and meta-analysis. Syst. Rev. 2021;10(1):194. doi: 10.1186/s13643-021-01732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciaccio M., Agnello L. Biochemical biomarkers alterations in Coronavirus Disease 2019 (COVID-19) Diagnosis (Berl) 2020;7(4):365–372. doi: 10.1515/dx-2020-0057. [DOI] [PubMed] [Google Scholar]
- 25.Delshad M., Tavakolinia N., Pourbagheri-Sigaroodi A., Safaroghli-Azar A., Bagheri N., Bashash D. The contributory role of lymphocyte subsets, pathophysiology of lymphopenia and its implication as prognostic and therapeutic opportunity in COVID-19. Int. Immunopharmacol. 2021;95 doi: 10.1016/j.intimp.2021.107586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darif D., Hammi I., Kihel A., El Idrissi Saik I., Guessous F., Akarid K. The pro-inflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021;153 doi: 10.1016/j.micpath.2021.104799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casari I., Manfredi M., Metharom P., Falasca M. Dissecting lipid metabolism alterations in SARS-CoV-2. Prog. Lipid Res. 2021;82 doi: 10.1016/j.plipres.2021.101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin B., Liu J., Liu Y., Qin X. Progress in understanding COVID-19: Insights from the omics approach. Crit. Rev. Clin. Lab. Sci. 2021;58(4):242–252. doi: 10.1080/10408363.2020.1851167. [DOI] [PubMed] [Google Scholar]
- 29.Huang S.S., Lin J.Y., Chen W.S., Liu M.H., Cheng C.W., Cheng M.L., Wang C.H. Phenylalanine- and leucine-defined metabolic types identify high mortality risk in patients with severe infection. Int. J. Infect. Dis. 2019;85:143–149. doi: 10.1016/j.ijid.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 30.Xu J., Pan T., Qi X., Tan R., Wang X., Liu Z., Tao Z., Qu H., Zhang Y., Chen H., Wang Y., Zhang J., Wang J., Liu J. Increased mortality of acute respiratory distress syndrome was associated with high levels of plasma phenylalanine. Respir. Res. 2020;21(1):99. doi: 10.1186/s12931-020-01364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W.S., Wang C.H., Cheng C.W., Liu M.H., Chu C.M., Wu H.P., Huang P.C., Lin Y.T., Ko T., Chen W.H., Wang H.J., Lee S.C., Liang C.Y. Elevated plasma phenylalanine predicts mortality in critical patients with heart failure. ESC Heart Fail. 2020;7(5):2884–2893. doi: 10.1002/ehf2.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C. Bruzzone, M. Bizkarguenaga, R. Gil-Redondo, T. Diercks, E. Arana, A. Garcia de Vicuna, M. Seco, A. Bosch, A. Palazon, I. San Juan, A. Lain, J. Gil-Martinez, G. Bernardo-Seisdedos, D. Fernandez-Ramos, F. Lopitz-Otsoa, N. Embade, S. Lu, J.M. Mato, O. Millet, SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum, iScience 23(10) (2020) 101645. [DOI] [PMC free article] [PubMed]
- 33.Barberis E., Timo S., Amede E., Vanella V.V., Puricelli C., Cappellano G., Raineri D., Cittone M.G., Rizzi E., Pedrinelli A.R., Vassia V., Casciaro F.G., Priora S., Nerici I., Galbiati A., Hayden E., Falasca M., Vaschetto R., Sainaghi P.P., Dianzani U., Rolla R., Chiocchetti A., Baldanzi G., Marengo E., Manfredi M. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020;21(22) doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mei Q., Wang A.Y., Bryant A., Yang Y., Li M., Wang F., Du S., Kurts C., Wu P., Ma K., Wu L., Chen H., Luo J., Li Y., Hu G., Yuan X., Li J. Survival Factors and Metabolic Pathogenesis in Elderly Patients (>/=65) With COVID-19: A Multi-Center Study. Front. Med. (Lausanne) 2020;7 doi: 10.3389/fmed.2020.595503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.T. Dierckx, J. van Elslande, H. Salmela, B. Decru, E. Wauters, J. Gunst, Y. Van Herck, J. Wauters, B. Stessel, P. Vermeersch, The metabolic fingerprint of COVID-19 severity, medRxiv (2020) 2020.11.09.20228221.
- 36.Rocha J.C., Calhau C., MacDonald A. Reply to Jakovac; Severity of COVID-19 infection in patients with phenylketonuria: Is vitamin D status protective? Am. J. Physiol. Endocrinol. Metab. 2020;318(6):E890–E891. doi: 10.1152/ajpendo.00195.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.