Abstract
Novel coronavirus disease 2019 (COVID-19) is a global pandemic caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), which can be transmitted from person to person. As of September 21, 2021, over 228 million cases were diagnosed as COVID-19 infection in more than 200 countries and regions worldwide. The death toll is more than 4.69 million and the mortality rate has reached about 2.05% as it has gradually become a global plague, and the numbers are growing. Therefore, it is important to gain a deeper understanding of the genome and protein characteristics, clinical diagnostics, pathogenic mechanisms, and the development of antiviral drugs and vaccines against the novel coronavirus to deal with the COVID-19 pandemic. The traditional biology technologies are limited for COVID-19-related studies to understand the pandemic happening. Bioinformatics is the application of computational methods and analytical tools in the field of biological research which has obvious advantages in predicting the structure, product, function, and evolution of unknown genes and proteins, and in screening drugs and vaccines from a large amount of sequence information. Here, we comprehensively summarized several of the most important methods and applications relating to COVID-19 based on currently available reports of bioinformatics technologies, focusing on future research for overcoming the virus pandemic. Based on the next-generation sequencing (NGS) and third-generation sequencing (TGS) technology, not only virus can be detected, but also high quality SARS-CoV-2 genome could be obtained quickly. The emergence of data of genome sequences, variants, haplotypes of SARS-CoV-2 help us to understand genome and protein structure, variant calling, mutation, and other biological characteristics. After sequencing alignment and phylogenetic analysis, the bat may be the natural host of the novel coronavirus. Single-cell RNA sequencing provide abundant resource for discovering the mechanism of immune response induced by COVID-19. As an entry receptor, angiotensin-converting enzyme 2 (ACE2) can be used as a potential drug target to treat COVID-19. Molecular dynamics simulation, molecular docking and artificial intelligence (AI) technology of bioinformatics methods based on drug databases for SARS-CoV-2 can accelerate the development of drugs. Meanwhile, computational approaches are helpful to identify suitable vaccines to prevent COVID-19 infection through reverse vaccinology, Immunoinformatics and structural vaccinology.
Keywords: COVID-19, SARS-CoV-2, Application, Bioinformatics technology
Graphical Abstract
Bioinformatics is the application of computational methods and analytical tools in the field of SARS-CoV-2 research which has obvious advantages in predicting the structure, product, function, and evolution of unknown genes and proteins, and in screening drugs and vaccines from a large amount of sequence information.
1. Introduction
Since December 2019, several cases of acute respiratory tract infection have been confirmed in Wuhan, Hubei province, China (C. Huang et al., 2020; Zhu et al., 2020). The large-scale pandemic of confirmed cases jumps unpredictably, spreads rapidly, and poses a severe threat to public health (Mahase, 2020; J.T. Wu et al., 2020). The World Health Organization (WHO) has declared the outbreak an "International Public Health Emergency" on March 11, 2020. The newly identified β-coronavirus (β-CoVs) was named severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) by the Coronavirus Study Group (CSG) of the International Committee. The disease caused by SARS-CoV-2 was suspected to be an infectious disease and officially designated coronavirus disease 2019 (COVID-19) by WHO (Zhu et al., 2020). According to the current data, SARS-CoV-2 is easily transmissible among people through respiratory droplets and survives in the air for about 2 h, remaining highly infectiousness (Y. Liu et al., 2020; Munster et al., 2020; wei Lu et al., 2020; H. Zhang et al., 2020). The incubation period after infection is usually 4–8 days (Chen et al., 2020a; C. Huang et al., 2020; D. Wang et al., 2020). All ages are susceptible to SARS-CoV-2, however, older patients and those who smoke experience severe illness with comorbidities (Chen et al., 2020b; J. Zhang et al., 2020). Importantly, the likelihood of local transmission tends to increase due to cases, including human-to-human transmission in the asymptomatic infection period (Rothe et al., 2020, Yuan et al., 2006).
The two highly pathogenic β-CoVs, which are known to cause severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), are zoonotic pathogens that mainly infect the lower respiratory tract and result in fatal pneumonia in humans, and cause a serious threat to human health (Channappanavar and Perlman, 2017, Luk et al., 2019, Ramadan and Shaib, 2019; F. Wu et al., 2020). The overall mortality rate reported by WHO is 9.6% for SARS and 34.4% for MERS (Munster et al., 2020, World Health Organization, 2020). In comparison, SARS-CoV-2 has a mortality rate of 2.05% in the world, and elder patients with chronic diseases and intensive care unit (ICU) patients have reached mortality rates of 17–38% (Chen et al., 2020a; D. Wang et al., 2020). Global Real Time COVID-19 outbreak map from WHO (https://covid19.who.int/) showing the confirmed cases, deaths and vaccine doses have been administered in different countries and regions worldwide. So far, there is no effective treatment for human coronavirus (CoVs) infection and no drugs are recommended. However, a large number of studies have made progress in the characterization of CoVs, and the therapies and vaccines targeting the virus are being studied extensively. Traditional biology technologies of virus research can not obtain such timely and large-scale data, and has other shortcomings. Bioinformatics technology has made great contributions to the rapid and in-depth study of SARS and MERS and will also bring a new dawn to the study of SARS-CoV-2 (Woo et al., 2010).
Sequencing diagnosis is still the main method in the diagnosis of the novel coronavirus and plays an important role in virus functional research. In this scientific and technological battle, the analysis of sequencing the virus data by bioinformatics tools provides an important basis for the discovery of SARS-CoV-2. Databases of SARS-CoV-2 provide genome and protein structure quality evaluation, variant calling, and interactive visualization and other biological characteristics. Sequence alignment among different species help us to understand virus origin and evolutionary characteristics. The rapid, feasible, accurate, and sensitive point-of-care testing (POCT) approaches for detecting SARS-CoV-2 in different situation are urgently developed. By bioinformatics analysis of single-cell RNA sequencing of SARS-CoV-2 RNA, the features of epithelial and immune cell types were revealed. In addition, it can provide predictive and screening high-throughput computational support to accelerate pandemic treatment and prevention, such as specific drugs, and the development of vaccines. Small molecule drugs, screened from drug databases for SARS-CoV-2 through high-performance computing approaches, such as molecular dynamics simulation, molecular docking and artificial intelligence technology, can competitively bind to the functional sites of viral proteins, which prevents the viral protein from binding to its true substrate, thereby inhibiting viral activity or the receptors induce the host to generate immune responses and produce neutralizing antibodies against novel coronavirus (Yin et al., 2020).
Although we know comparatively little about SARS-CoV-2, it is a highly pathogenic human pathogen, and possibly a zoonotic agent. Challenges will continue to be confronted in several key areas, therefore, in this review, evidence of the virus RNA genome sequence by bioinformatics methods is instrumental in the characteristic, origin, evolution, detection, pathogenesis and function of SARS-CoV-2, and the development of antiviral drugs and vaccines, to provide data for public health decision-making.
2. The bioinformatics technologies relating to COVID-19
The following sections provide several of the most important methods and applications of bioinformatics technologies in combating the COVID-19 based on currently available reports and the descriptive workflow is shown in Fig. 1.
Fig. 1.
The descriptive workflow of the most important methods and applications of bioinformatics technologies on COVID-19.
2.1. Genome and protein structure in databases
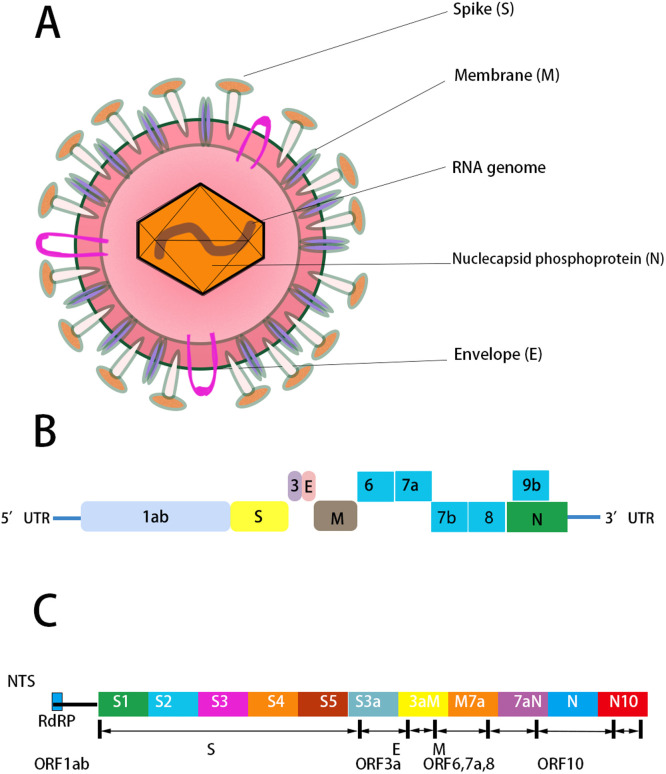
According to the next-generation sequencing (NGS) and third-generation sequencing (TGS) technology, significantly updated and publicly available databases of SARS-CoV-2 offer genome and protein structure quality evaluation, variant calling, and interactive visualization. According to the Global Initiative on Sharing Avian Influenza Data (GISAID, https://www.gisaid.org) (Shu and McCauley, 2017), National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov) (O’Leary et al., 2016), National Microbiology Data Center (NMDC, https://www.nmdc.cn) (Shi et al., 2019), China National GeneBank (CNGB, https://www.cngb.org) (Xiao et al., 2019), China National Center for Bioinformation (CNCB, https://bigd.big.ac.cn/ncov) and several other sources, SARS-CoV-2 genome sequences resources worldwide and their related metadata have been published ( Table 1) (Song et al., 2020; W.M. Zhao et al., 2020). After obtaining nucleic acid sequence, comprehensive bioinformatics analysis demonstrated that COVID-19 was caused by the novel coronavirus SARS-CoV-2 which belongs to lineage B β-CoVs and is a positive single-stranded RNA virus with a genome length of about 29.9 kb, with non-coding regions at both ends, and non-structural protein-coding regions and structural protein-coding regions in the middle (Chan et al., 2020; Roujian Lu et al., 2020; Zhu et al., 2020). SARS-CoV and MERS-CoV have positive-sense RNA genomes of 27.9 kb and 30.1 kb, respectively (De Wit et al., 2016). The complete RNA genome of SARS-CoV-2 includes 29,870 nucleotides and also contains a variable number of open reading frames (ORFs) (Chen et al., 2020a; Y. Liu et al., 2020; Munster et al., 2020; D. Wang et al., 2020; wei Lu et al., 2020; H. Zhang et al., 2020). Excluding the poly (A) tail (GenBank no. MN908947), the ORFs encode 9860 amino acids with five typical ORFs on a common coding strand and in the order of 5′ UTR-replicase polyprotein (orf1a/b, 7096-aa) – Spike glycoprotein (S, 1273-aa) – Envelope protein (E,75-aa) – Membrane protein (M, 222-aa) – Nucleocapsid protein (N, 419-aa)- 3′ UTR, in which the four essential protein-coding regions S, E, M, and N encode structural proteins ( Fig. 2A, B) (Chan et al., 2020). In addition, there are several accessory genes that influence the host innate immune response, such as ORF3b and ORF8. ORF3b encodes a completely novel short protein without an exact function. The new ORF8 probably encodes a secreted protein formed by an alpha-helix, followed by beta sheets that contain six strands that have no known functional domain or motif similarity (Chan et al., 2020). Two-thirds of viral RNA, mainly located in the coding region of non-structural proteins, including the ORF1a and ORF1b genes, translates two replicase polyproteins (pp1a and pp1ab) and are proteolytically cleaved into 16 non-structural proteins (NSP), namely NSP1–16, which are indispensable for viral transcription and replication (Chan et al., 2020; F. Wu et al., 2020; Zhou et al., 2020).
Table 1.
Databases of genome and protein structure.
| Type | Data center |
|---|---|
| Genome sequence | Global Initiative on Sharing Avian Influenza Data (GISAID, https://www.gisaid.org) |
| Genome sequence | National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov) |
| Genome sequence | National Microbiology Data Center (NMDC, https://www.nmdc.cn) |
| Genome sequence | China National GeneBank (CNGB, https://www.cngb.org) |
| Genome sequence | China National Center for Bioinformation (CNCB, https://bigd.big.ac.cn/ncov) |
| Protein sequence/function | Universal Protein (UniProt, https://www.uniprot.org) |
| Protein sequence/structure | Research Collaboratory for Structural Bioinformatics Protein Data Bank Database (RCSB PDB, https://www.rcsb.org) |
Fig. 2.
Structure of SARS-CoV-2. (A) Schematic representation of the structure of SARS-CoV-2. It has four structural proteins, S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the single-strand, positive-sense RNA genome, and the S, E, and M proteins together create the viral envelope. (B) SARS-CoV-2 genome comprises a 5′ untranslated region (5′ UTR) including 5′ leader sequence, open reading frame (ORF) 1a/b, envelope, membrane and nucleoprotein, accessory proteins such as orf 3, 6,7a, 7b, 8, and 9b, and 3′ untranslated region (3′ UTR) in sequence. (C) SARS-CoV-2 structure is based upon amplification targets of the NTS method. NTS detected 12 fragments including ORF1ab and virulence factor-encoding regions (M. Wang et al., 2020).
There are two main methods for predicting the virus structure through high-performance computing and bioinformatics. The first method is to clarify the virus structure through gene sequencing results. To ensure the accuracy of the method, the results require statistical verification. Among them, AlphaFold (Senior et al., 2020) de novo simulation is a deep two-dimensional dilated convolutional residual network, which predicts the protein structure or directs homologous modeling based on similar proteins in the sequence. The higher the sequence similarity, the higher is the reliability of structural simulation, common software include Swiss-Model (https://swissmodel.expasy.org/) (Waterhouse et al., 2018), Robetta (http://robetta.bakerlab.org/) (Kim et al., 2004) I-TASSER suite (Yang et al., 2014), and so on. Novel coronavirus structure can be predicted from the SARS virus structure, so de novo simulations using artificial intelligence programs are seldom needed. The second method to discover the structure of the virus is to reconstruct its three-dimensional structure through imaging techniques. In this process, high-performance computing, mainly using RELION software (Scheres, 2012), can accelerate the reconstruction of the three-dimensional structure. AlphaFold and RELION is recommended to calculate the virus protein structure, but sometimes computing power of AlphaFold may lead to over fitting. The coronavirus molecular weight is large and, therefore, cryo-electron microscopy (Cryo-EM) combined with bioinformatics technologies can accelerate the reconstruction of coronavirus.
Currently, SARS-CoV-2 protein sequences and functional information were described in Universal Protein (UniProt, https://www.uniprot.org) and the world’s first three-dimensional structure of the new coronavirus protein is available in the Research Collaboratory for Structural Bioinformatics Protein Data Bank Database (RCSB PDB, https://www.rcsb.org, PDB ID: 6LU7) (Jin et al., 2020). The functional polypeptides are released from the polyproteins (pp1a and pp1ab) by extensive proteolytic processing, predominantly by a 33.8-kDa main protease (Mpro), which is also referred to as the 3 C-like protease. Mpro digests the polyproteins at no less than 11 conserved sites, starting with the autolytic cleavage of this enzyme itself from pp1a and pp1ab (Hegyi and Ziebuhr, 2002). The important role of Mpro in the viral life cycle, together with few closely related homologs in humans, identifies it as a key drug target against novel coronavirus (Pillaiyar et al., 2016). Through a high-resolution complex structure, academician Chen's team found that the antibody 4A8 has strong virus neutralization ability and can significantly inhibit the activity of the virus (Chi et al., 2020).
Bioinformatics technology has become a powerful tool for identifying, analyzing, and monitoring the novel coronavirus, and has laid an important foundation for controlling the spread of the virus. The rapid acquirement of the genome sequence and protein structure of SARS-CoV-2 by bioinformatics technology has opened a fast channel for targeted drug screening and vaccine development.
2.2. Origin, similarity, and evolution
The origin and evolution of SARS-CoV-2 remain largely unclear. Several studies have suggested that bats are known to be hosts for more than 30 CoVs (Wong et al., 2019), and could be the potential natural reservoir of SARS-CoV-2 (Giovanetti et al., 2020, Paraskevis et al., 2020, Wong et al., 2019). On May 1, 2020, WHO stated that many scientists had studied the gene sequence of COVID-19 and were convinced that it originated from nature. However, there is no definitive evidence of the origin or intermediate host of the novel virus (Li et al., 2020, Phan et al., 2020).
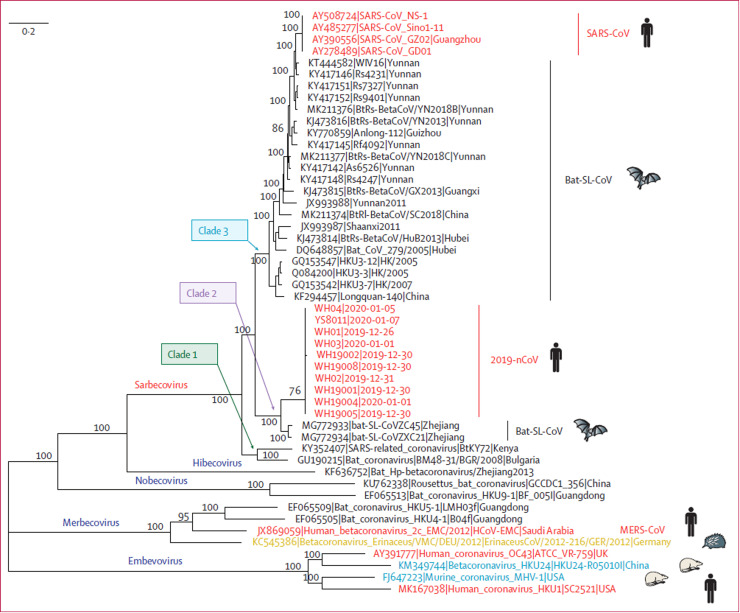
The virus genome sequencing results, sequencing alignment and phylogenetic analysis with bioinformatics tool MUSCLE (3.8.31) (Edgar, 2004) demonstrated that SARS-CoV-2 had a 96.3% sequence identity to BatCoV RaTG13, which originated from Yunnan, China in 2013. This was the closest relative to two bat-derived SARS-like CoVs, bat-SL-CoVZC45 and bat-SL-CoVZXC21, sharing 88% nucleotide similarity, originating from Zhoushan, China, in 2018, whereas it shared 79.5% identity to SARS-CoV and 50% identity to MERS-CoV ( Fig. 3) (Roujian Lu et al., 2020; Paraskevis et al., 2020; Zhou et al., 2020). The results suggest that bat CoV and human SARS-CoV-2 might come from a common ancestor, although bats are not sold in the seafood market, in Wuhan, China (F. Wu et al., 2020). Therefore, the natural host of novel coronavirus SARS-CoV-2 may be bats, but the origin and intermediate host is not yet conclusive. Protein sequences alignment and phylogenetic studies (Y. Zhao et al., 2020) observed that the residues of the receptor were similar in many species, which provides more possibilities of alternative intermediate hosts, such as turtles, pangolins, minks and snakes ( Table 2) (Ghorbani et al., 2021, Ji et al., 2020, Li et al., 2020; Z. Liu et al., 2020; Xiao et al., 2020). A recent study suggests that pangolin has the probability to be an intermediate host of SARS-CoV-2 but may not be the only one. Through the genomic sequence similarity analysis of SARS-CoV-2 obtained from different patients, its homology is more than 99.9%. Therefore, it is reasonable to consider that the SARS-CoV-2 strain originates from one source and can be detected relatively quickly (Roujian Lu et al., 2020; Zhou et al., 2020).
Fig. 3.
Phylogenetic analysis of full-length genomes of SARS-CoV-2 and representative viruses of the genus Betacoronavirus. 2019-nCoV=SARS-CoV-2 = severe acute respiratory syndrome coronavirus type 2. MERS-CoV = Middle East respiratory syndrome coronavirus. SARS-CoV = severe acute respiratory syndrome coronavirus (Roujian Lu et al., 2020).
Table 2.
Comprehensive analysis of the host of SARS-CoV-2.
| Host | Description |
|---|---|
| Bat | Primary host maybe bat. SARS-CoV-2 had a 96.3% sequence identity to BatCoV RaTG13, which originated from Yunnan, China in 2013. SARS-CoV-2 had a 88% identity to bat-SL-CoVZC45 and bat-SL-CoVZXC21 originating from Zhoushan, China, in 2018, whereas it shared 79.5% identity to SARS-CoV and 50% identity to MERS-CoV. |
| Turtle | Turtle maybe Intermediate host. The interaction between the key amino acids of S protein RBD and ACE2 indicated that, turtles (Chrysemys picta bellii, Chelonia mydas, and Pelodiscus sinensis) may act as the potential intermediate hosts. |
| Pangolin | Pangolin maybe Intermediate host. The isolation of a coronavirus from pangolins that is closely related to SARS-CoV-2. |
| Mink | Mink maybe Intermediate host. A high rate of variation within SARS-CoV-2 mink isolates implies that mink populations were infected before human populations. |
| Snake | Snake maybe Intermediate host. SARS-CoV-2 has most similar codon usage bias with snake. |
The S gene of the SARS-CoV-2 has less than 75% sequence identity to those of the bat SARS-like CoVs (SL-CoVZXC21 and ZC45) and human SARS-CoV (Zhou et al., 2020). Similarly, the spike glycoprotein encoded by the S genes of the SARS-CoV-2 was longer than that of the SARS-CoV (Zhou et al., 2020). The spike protein, composed of the S1 and S2 domain, is crucial in determining host tropism and transmission capacity through the mediation of receptor-binding and membrane fusion (Roujian Lu et al., 2020). Among these, the S2 subunit of the SARS-CoV-2 is highly conserved and has a 99% identity with that of SARS-CoV (Roujian Lu et al., 2020). The receptor-binding domain is usually located in the C-terminal domain of S1 to directly associate with the human receptor. Although the S1 domain of the SARS-CoV-2 only has approximately 70% identity with SARS-CoV, homology modeling revealed that the SARS-CoV-2 had a similar receptor-binding domain structure to that of SARS-CoV (Roujian Lu et al., 2020). Notably, Zhou et al. found that SARS-CoV-2, just like SARS-CoV, may use angiotensin-Converting Enzyme 2 (ACE2) as an entry receptor in ACE2-expressing cells, the majority of which are type II alveolar cells (AT2) in human lung (Edgar, 2004; Y. Zhao et al., 2020; Ziegler et al., 2020). The similarities and differences of the S protein of β coronavirus are shown in Table 3. Therefore, high-throughput bioinformatics technology is needed for large-scale screening to monitor whether ACE2 targeted drugs are effective in the treatment of SARS-CoV-2, and individual mutations need to be constantly observed to determine whether they have an effect on drug activity.
Table 3.
Similarities and differences in the S protein of β coronavirus.
| Type | Different sequence structures | Receptors |
|---|---|---|
| SARS-CoV-2 | There are 12 short insertion sequences in the N-terminal domain of S protein, including (PRRA)CCT CGG CGG GCA | ACE2 |
| SARS CoV | S1 domain contains 14 unique amino acids | ACE2 |
| BAT-CoV- RaTG13 | S protein contains short insertion sequences in the N-terminal domain | ACE2 |
| MERS-CoV | The S1 domain RBD is different from SARS-CoV, which contains a core structure and a subsidiary subregion that functions as RBM | DPP4(CD26) |
| HCoV-HKU | Three identical S1 domains form an interwoven cap at the top of the S2 stem | unknown |
2.3. SARS-CoV-2 detection
At this time, diagnostic testing for SARS-CoV-2 can be conducted only at Centers for Disease Control and Prevention (CDC, https://www.cdc.gov/coronavirus/2019-nCoV/guidelines-clinical-specimens.html). Due to the unpredictable emergence of the SARS-CoV-2 virus, the range of diagnostic tools that can be performed in clinical practice is quite limited. To date, SARS-CoV-2 has been detected in human clinical specimens by real-time reverse transcription-polymerase chain reaction (qRT-PCR), next-generation sequencing, third-generation sequencing, RT-loop-mediated isothermal amplification (RT-LAMP), LAMP-Seq, antibody test, pulmonary CT, etc. (W.E. Huang et al., 2020; Lan et al., 2020; Renfei Lu et al., 2020; She et al., 2020). More rapid, feasible, accurate, and sensitive point-of-care testing (POCT) approaches for the diagnosis of COVID-19 in patients infected by SARS-CoV-2 are urgently needed. The comparison of detection methods of SARS-CoV-2 are shown below and in Table 4.
Table 4.
Comparison of detection methods of SARS-CoV-2.
| Methods | Advantages | Disadvantages |
|---|---|---|
| qRT-PCR | Gold standard, high specific, high rapid, economic, the most robust, and widely applied technology | Low sensitivity, speed is limited, require sophisticated equipment and exhibit false-negative rates |
| NGS | High accurate, high throughout, sequencing for high-quality genome | Equipment is expensive, operator expertise, and a turnaround time of > 24 h |
| NTS | Target amplification and long-read, High rapid, high accurate, higher sensitivity than standard qRT-PCR and sequencing for high-quality genome | Equipment is expensive, and more expensive than qRT-PCR |
| RT-LAMP | High sensitivity, simple operation, fast reaction speed (within 50 min), less dependent on complex equipment, and can detect of viruses, various pathogens, and even viral variants | The limitation of the RT-LAMP assay was calculated to be 118.6 copies per reaction. |
| LAMP-Seq | Low cost, high rapid (within 40 min) and highly sensitive protocol, population-scale testing | Without clinical certification |
| Antibody | Traditional mature technology | Lower sensitivity, and antibody production takes a long time |
| CT | Easy to operate and helpful in diagnosis | Asymptomatic infection cannot be diagnosed |
Nucleic acid detection is the golden standard for COVID-19 diagnosis. From the testing methods conducted to detect the causative virus, qRT-PCR is specific, rapid, economic, the most robust, and widely applied technology for qualitative and quantitative diagnosis (Corman et al., 2020, Li et al., 2019a, Wan et al., 2016). However, the qRT-PCR requires sophisticated equipment and exhibits high false-negative rates, low sensitivity, with only 30–50% positive detection ratio, and cannot identify suspected infection, or close contact with confirmed cases, thereby resulting in patients mistakenly considered uninfected or cured and impeding pandemic containment (Chu et al., 2020, Lan et al., 2020, Lo and Chiu, 2020). LAMP is a high sensitivity technology with a simple operation, fast reaction speed (within 50 min), is less dependent on complex equipment, and has been widely used for the development of POCT techniques for the detection of viruses, various pathogens, and even viral variants (Renfei Lu et al., 2020; Notomi et al., 2000; Thai et al., 2004). Later, the mismatch-tolerant version of the LAMP method has higher sensitivity and faster reaction speed than the traditional method (Li et al., 2019b, Zhou et al., 2019). The limitation of the RT-LAMP assay was calculated to be 118.6 copies per reaction.
Next-generation and third-generation sequencing are widely applied for mutation identification, pathogen identification and monitoring virus evolution (Metsky et al., 2017, Wilson et al., 2014) including SARS-CoV, but require expensive equipment, operator expertise, and a turnaround time of > 24 h, making them unsuitable for the current crisis. To effectively interrupt the transmission of SARS-CoV-2, promising POCT detection methods are needed. And to break the cycle of isolation and spread, LAMP-Seq and nanopore target sequencing (NTS) are capable of population-scale processing.
Because traditional PCR detection needs a professional laboratory, its speed is greatly limited. LAMP-Seq, a low cost, rapid (within 40 min) and highly sensitive protocol, allows for population-scale testing using massively parallel RT-LAMP by using sequence-specific barcodes. It can pool processing, parallelize deep sequencing, and perform standard computing analysis to identify positive individuals (Schmid-Burgk et al., 2020; Y. Zhang et al., 2020). Most importantly, this method, in which the false negative and false positive rates are lower than 0.2%, is expected to scale hundreds of thousands of samples per day per sequencing facility (Schmid-Burgk et al., 2020). Although LAMP, DNA barcode and high-throughput sequencing technology are not new, their combination provides a new large-scale diagnostic idea for potentially larger outbreaks.
On January 31, 2020, The New England Journal of Medicine reported the first patient diagnosed with COVID-19 in the United States and indicated nanopore sequencing as part of the sequencing toolkit. Subsequently, Belgium, China, Canada, and more than 16 countries used the ARTIC protocol and nanopore sequencing to complete the sequencing of the novel coronavirus in about 7 h (Fauver et al., 2020; J. Lu et al., 2020). Combining the advantage of target amplification and long-read real-time nanopore sequencing, the NTS to detect SARS-CoV-2 and other respiratory viruses was established simultaneously within 6–10 h, with higher sensitivity than standard qPCR (Taiaroa et al., 2020). For the virulence regions, NTS designed 11 fragments of 600–950 bp as targets, completely covering the 9115 bp region, and 26 primers were used to develop the SARS-CoV-2 primer panel (Fig. 2C) (M. Wang et al., 2020). For more testing samples, NTS could rapidly detect many positive samples within 10 min (for quick detection) and 4 h (for final evaluation), showing good concordance with qPCR results. Parallel testing with approved qPCR of SARS-CoV-2 and NTS using a variety of nucleic acid samples from suspected COVID-19 cases confirmed that NTS identified more infected patients as positive, and could also monitor mutations in the nucleic acid sequence or in other respiratory viruses. NTS is a rapid, accurate, and comprehensive detection method, which makes it suitable for the diagnosis of contemporary COVID-19 to reduce mortality. In addition, this approach has the potential to be further extended to diagnose more viruses or pathogens (Fauver et al., 2020; M. Wang et al., 2020).
Oxford Nanopore MinION and GridION were used for NTS. MinION, the smallest Oxford Nanopore sequencer, is smaller than a cell phone and can be used in various environments with low equipment cost. Base-calling and the quality assessment of MinION sequencing data use Guppy (v. 3.1.5) software, and GridION uses MinKNOW (v. 3.6.5). The reads of each sample are mapped against the virus genome reference database using BLASTn (v. 2.9.0+) (Altschul et al., 1990). Sequence correction of nanopore sequencing data was performed using medaka (v. 0.10.5) (Oxford Nanopore Technologies, 2018). The ARTIC bioinformatics workflow is used to obtain consistent sequence, SNPs, and mutation data through published SARS-CoV-2 sequence information. Variants were identified in infected patients by single nucleotide mutations at seven sites by NTS and qPCR. Silent variants exist that have not been characterized by existing whole-genome sequencing methods (M. Wang et al., 2020). Finally, for data analysis, bioinformatics analysis may also be introduced for high-quality genome, variants, and high-throughput detection (Charalampous et al., 2019, Greninger et al., 2015).
At the time of writing, qRT-PCR has become the gold standard method for diagnosis and clinical decision-making of antiviral therapy. While, next-generation and third-generation sequencing based on bioinformatics technology were established to conduct virus detection and genome assembly and large-scale sequencing analysis of COVID-19 in patient samples. Combining the detection of pathogenic organisms in respiratory specimens in clinical virology laboratories with bioinformatics analysis of the downstream data of viruses will help to gain insight into the mutation, spread, evolution, pathogenesis of the virus, and drug and vaccine development.
2.4. A single-cell atlas revealed COVID-19 immune features
Single-cell RNA sequencing (scRNA-seq) is another important technology which provides a comprehensive cell atlas comprehensive to identify that different peripheral immune subtype changes are associated with distinct clinical features of COVID-19. By bioinformatics analysis, SARS-CoV-2 RNA was found in a variety of epithelial and immune cell types, accompanied by significant changes in the transcriptome of virus positive cells. Monocytes and lymphocytes do not express large amounts of pro-inflammatory cytokines in peripheral blood (Wilk et al., 2020). While, upregulation of S100A8/A9 of peripheral megakaryocytes and monocytes mainly causes cytokine storm in severe patients (Ren et al., 2021). Collectively, single-cell RNA sequencing results represent a rich resource for understanding the complex multicellular immune response induced by COVID-19 and developing effective therapeutic directions for COVID-19.
2.5. Medicine screening for COVID-19
At present, bioinformatics based on high-performance computing is the standard of scientific research tools and will certainly be used to accelerate the development of specific drugs. In general, it is necessary for modern pharmaceuticals to understand the structure of the virus, and then evaluate the small protein molecules that can bind to and inactivate the virus. ACE2 as an entry receptor, potential drug target, plays an important role in the discovery and optimization of inhibitors blocking the entry of cells to treat disease. Small molecule drugs can competitively bind to the functional sites of viral proteins, which prevents the viral protein from binding to its true substrate, thereby inhibiting viral activity (Abd El-Aziz and Stockand, 2020). Drug databases for SARS-CoV-2 (D3Targets-2019-nCoV, CoViLigands, CORDITE, DockCov2, DrugBank, TCMD), accelerating molecular dynamics simulation (vsREMD), molecular docking (AutoDock, D3Docking, D3Similarity, CDOCKER), and artificial intelligence (AI) technology (DNN, MolAICal) will assist in drug design and screening through high-throughput computing with bioinformatics analysis.
Drug databases for SARS-CoV-2 ( Table 5). D3Targets-2019-nCoV, a molecular docking-based webserver, is accessible freely to public (https://www.d3pharma.com/D3Targets-2019-nCoV/index.php) (Shi et al., 2020), which currently contains 42 viral proteins with 69 conformations for predicting potential target protein for active compounds and for drugs screening against targets that may have potential inhibitory effects on SARS-CoV-2 viral RNA polymerase. CoViLigands is an anti-coronavirus active compounds database which collected from previous publications at https://www.d3pharma.com/D3Targets-2019-nCoV/CoViLigands/2019-nCoV.php, which is updated regularly and available free of charge (Yang et al., 2021). CORDITE (CORona Drug InTEractions database, https://cordite.mathematik.uni-marburg.de) is accessible web server and integrates all available publications about potential drugs for SARS-CoV-2 (Martin et al., 2020). DockCoV2 a predicting the binding affinity of drugs with the SARS-CoV-2 virus encodes proteins database which contains 3109 drugs. DockCoV2 is available at https://covirus.cc/drugs/. Seven target structural proteins involved in viral entry and replication process, including 3 CLpro (PDB ID: 6LU7), PLpro (PDB ID: 6WX4), RdRp (PDB ID: 7BV2), spike receptor-binding domain (RBD) (PDB ID: 6M0J), N protein (PDB ID: 6M3M) and ACE2 (PDB ID: 1R42) were retrieved from Protein Data Bank (Chen et al., 2021). Drugbank database (https://www.drugbank.ca) contains comprehensive bioinformatics and chemoinformatics resources, and provides detailed drug, drug target information and its mechanism, including pharmacochemistry, pharmacology, pharmacokinetics, and drug-drug interaction information. Traditional Chinese Medicines Database (TCMD, 2009) provides traditional knowledge and modern science to understand traditional Chinese medicines (Traditional Chinese Medicine Database (TCMD), 2009).
Table 5.
Drug databases for SARS-CoV-2.
| Databases | Websites/Resources |
|---|---|
| D3Targets-2019-nCoV | https://www.d3pharma.com/D3Targets-2019-nCoV/index.php |
| CoViLigands | https://www.d3pharma.com/D3Targets-2019-nCoV/CoViLigands/2019-nCoV.php |
| CORDITE | https://cordite.mathematik.uni-marburg.de |
| DockCov2 | https://covirus.cc/drugs/ |
| DrugBank | https://www.drugbank.ca |
| TCMD | Software |
Molecular dynamics (MD) simulation is a method that simulates experimental conditions and can display the microscopic evolution of the system from the atomic level (J. Wang et al., 2020). After the vsREMD simulation, the homology model of the novel coronavirus Mpro was constructed and N3 could fit inside the substrate-binding pocket, which could target the current novel coronavirus Mpro. Kinetic studies by balancing the binding constant Ki (designated as K2 / K1) and the inactivation rate constant K3 formed by covalent bonds also showed that N3 has a strong inhibitory effect on the novel coronavirus Mpro (Jin et al., 2020). Moreover, Joshi et al. conducted phylogenetic and sequence similarity network analysis on the main drug target, Mpro, and identified molecules that were strongly active against SARS-CoV-2 Mpro, such as δ-viniferin, myricitrin, Taiwanhomoflavone A, lactucopicrin 15-oxalate, nympholide A, afzelin, biorobin, hesperidin, and phyllaemblicin B, which also proved that these molecules have a powerful combination to other potential targets of SARS-CoV-2 infection, such as the viral receptor human angiotensin-converting enzyme 2 (HACE-2) and RNA-dependent RNA polymerase (RdRp) (Joshi et al., 2020). In addition, Chen et al. prepared a three-dimensional model of COVID-19 virus 3 C-like protease (3 CLpro, Mpro) and used the three-dimensional model computer to virtually screen 16 candidate drugs. Among them, the antiviral drugs ledipasvir and velpatasvir are effective as therapeutic drugs against novel coronaviruses with minimal side effects, such as fatigue and headache (Jo et al., 2020, Kang et al., 2020; Z. Wang et al., 2020).
Molecular docking calculations are based on receptor active site regions to search for whether ligands interact with receptors and the optimal binding mode between them. The targets of SARS-CoV-2 were chosen Mpro, 3 CLpro, and RdRp and ligands were obtained from the DrugBank and D3Targets-2019-nCoV database. AutoDock (MGLTools, version 1.5.6) simulates and calculates each small molecule in a small molecule library, and observes the energy released through binding with proteins from various angles. The more stable the binding, the more likely this small molecule will be developed into a drug (Morris et al., 2009). D3Docking is a structure-based module embedded in the D3Targets-2019-nCoV web server, which utilized molecular docking to explore drug targets for drugs or active compounds observed from previous clinical studies and identify compounds against potential drug targets via protein structure based virtual screening (Shi et al., 2020). D3Similarity is a complementary approach to docking-based methods for the SARS-CoV-2 target prediction, virtual screening and target identification of potential coronavirus antivirals according to two-dimensional (2D)/three-dimensional (3D) molecular similarity, which lays the foundation for the screening of new COVID-19 drugs (Jin et al., 2020). For example, Remdisvir is an RdRp inhibitor, which is reported to effectively inhibit SARS-CoV and SARS-CoV-2 in D3Docking and clinical practice. Recent research reported that a batch of old and traditional Chinese medicines that could be effective in the therapy of pneumonia were discovered by using high-performance computing and bioinformatics methods (Ma et al., 2020; Z. Wang et al., 2020). A strategy of combining virtual screening and enzymatic testing can be used. It focuses on drug screening for marketed drugs, as well as self-establishing a database of highly mature medicinal compounds and ingredients of medicinal plant-derived compounds, which are rapidly distributed. Ma et al. (2020) established an optimized docking model and made a high-throughput virtual screening based on molecular docking technology. They obtained the crystal structures of Mpro and papain-like protein (PLP) from the PDB database and homology model and used CDOCKER program to conduct virtual screening of the TCMD, 11294 potential active ingredients were obtained by Mpro and PLP inhibitor screening. Rhubarb screened by Mpro and pollen screened by PLP both contain Maxing Shigan Decoction and Xuanbai Chengqi Decoction, which can be used for pandemic virus obstruction of the lung. The old drugs that are active natural products in traditional Chinese medicines and may have therapeutic effects on SARS-CoV-2 have been discovered, which greatly shortens the development time (Beck et al., 2020, Jin et al., 2020).
2.6. Artificial intelligence (AI)
Deep Neural Network (DNN) algorithm of AI technology is an efficient way to screen the potential drugs for the treatment of COVID-19 by using two different learning databases in which one is composed of compounds which have been reported or confirmed to have activity against SARS-CoV, SARS-CoV-2, human immunodeficiency virus, influenza virus, and the other contains the known 3C-like protease inhibitors (Ke et al., 2020). After selection, 13 drugs (bedaquiline, brequinar, celecoxib, clofazimine, conivaptan, gemcitabine, tolcapone, vismodegib, boceprevir, chloroquine, homoharringtonine, tilorone, and salinomycin) may prevent virus replication and indicate the potential to develop antiviral drugs (Ke et al., 2020). Deep learning is another approach of AI that applied into medicine and 2D/3D ligand design. MolAICal software supplies for 3D drug design of protein targets by artificial intelligence and classical algorithm (Bai et al., 2020).
At present, the SARS-CoV-2 infectious disease outbreak is tending to expand. Screening and developing of therapeutic drugs for viral diseases have become the focus and difficulty of world scientific research. Old drugs and traditional Chinese medicines of anti-SARS-CoV-2 are screened by many kinds of molecular dynamics simulation, molecular docking and artificial intelligence technology with bioinformatics which are considered to be more effective and to short the development time. After screening, ledipasvir, velpatasvir and remdisvir are effective as therapeutic drugs against SARS-CoV-2. As well, these molecules, δ-viniferin, myricitrin, Taiwanhomoflavone A, lactucopicrin 15-oxalate, nympholide A, afzelin, biorobin, hesperidin, and phyllaemblicin B, have a powerful combination to other potential targets of SARS-CoV-2 infection. In addition, the bedaquiline, brequinar, celecoxib, clofazimine, conivaptan, gemcitabine, tolcapone, vismodegib, boceprevir, chloroquine, homoharringtonine, tilorone, and salinomycin are the potential antiviral drugs. And, one of the traditional Chinese medicines named Rhubarb, can be used for pandemic virus obstruction of the lung. However, new drugs also need to be designed and still need a large number of clinical trials.
2.7. Vaccine for COVID-19
Vaccination is the most effective means of preventing and controlling infectious diseases. Since the COVID-19 pandemic, researchers around the world have been accelerating the development of an effective candidate vaccine to combat this viral disease. At present, several applications of bioinformatics to vaccine research can develop the effectiveness and safety of the vaccines through reverse vaccinology, Immunoinformatics and structural vaccinology (Chukwudozie et al., 2021a).
Reverse vaccinology (RV) is a promising approach that aims to find novel vaccine candidates through genome or proteome of pathogens. Systematic sequencing revealed that many isolates have deletions in the core genomes and may have a significant impact on viral structure or function. RV relies on bioinformatics methods to identify target antigens according to genomic information. The open reading frame (ORF) of organism genome was identified by VaxiJen server to determine various antigens and physicochemical properties related to antigen epitopes, reducing the time on the antigen selection process. Using the existing SARS-COV-2 genome sequence information, few researchers have used RV in the design of novel COVID-19 vaccine. In addition, it is also applied to develop a multi-epitope chimeric vaccine against SARS-COV-2 (Enayatkhani et al., 2021, Ong et al., 2020).
Immunoinformatics approach involves the application of computational methods to epitope-based peptide vaccine design. After getting the SARS-COV-2 genome and 3D protein structure, the most antigenic polyprotein and inhibitor-binding sites were predicted using various bioinformatics tools, which can generate a specific immune response. In order to predict the immunogenic targets from the viral genome sequences, computational tools such as TEpredict, CTLPred, NetMHC, and Epitopemap were applied. Immunoinformatics tools can help researchers develop potentially suitable vaccine candidates by satisfactorily understanding the human immune response to organisms in a short time (Delany et al., 2013, María et al., 2017).
Structural vaccinology is a reasonable method to develop effective vaccines by joining epitopes and immunogenic domains of the SARS-COV-2 proteins. The conformational characteristics of viral epitopes can make them become good candidate antigens (María et al., 2017). Structural properties (structural stability of peptides, solvent exposure, hydrophobicity and codon optimization) are used to map antigen epitopes to detect conformational features that may affect immunogenicity. In a word, structural vaccine technologies have been used to predict candidate vaccines for SARS-COV-2 via molecular docking, dynamics simulations, and homology modeling. These techniques are helpful to identify effective vaccination for the prevention of COVID-19 infection (Chukwudozie et al., 2021b).
Although many bioinformatics tools and algorithms have been used to develop safe candidate vaccines for COVID-19, there are still some limitations in fully integrating this prediction method in the design and development of traditional vaccines. Compared with most other DNA viruses with stable genomes such as smallpox virus, novel coronavirus has a distinctive feature and is more prone to gene mutations because it is an RNA virus, which is likely to change the nature of the virus itself and also causes a great reduction in the effectiveness of the vaccine. One of the main deficiencies of bioinformatics in finding effective vaccines against COVID-19 is that it can not solve the long-standing problem of selecting a suitable animal model to test vaccine candidates. In addition, reverse vaccinology can only target proteins and identify linear and discontinuous epitopes, which may lead to some predicted epitopes hidden in viral proteins and not easy to detect in vivo (Silva-Arrieta et al., 2020). Also, for the immunoinformatics screening, the pleomorphism of MHC class I molecules may prevent the precision of vaccines designing (Greenbaum et al., 2011). These may be the reasons for the failure of candidate vaccine screening. Ultimately, it is required to solve these limitations to improve the safety and effectiveness of vaccine candidates against SARS-COV-2 or other novel viruses.
Modern vaccines research is inseparable from bioinformatics technology. As of September 22, 2021, there are 13 different vaccines have been administered by researchers around the world and more than 5.87 million people have been vaccinated. For instance, the Pfizer/BioNtech Comirnaty vaccine was listed for WHO Emergency Use Listing (EUL) on December 31, 2020. The COVID-19 vaccine of Sinopharm was successfully launched in EUL on May 7, 2021 which is produced by Beijing Bio-Institute of Biological Products Co Ltd, subsidiary of China National Biotec Group (CNBG). Although the efficacy of several COVID-19 vaccines is amazingly high, it has not been reached 100% yet. The SARS-COV-2 continues to mutate, but the current viral mutations have not caused a great impact on vaccine development and disease diagnosis and treatment. The protective effectiveness of the two doses BNT162b2 and ChAdOx1 vaccines against B.1.617.2 (delta) variant of SARS-COV-2 were 88% and 67%, respectively (Lopez Bernal et al., 2021). No matter how the virus mutates, the more we know about its variants, the better the vaccines that will be developed by combining bioinformatics methods and biological technologies. The pandemic is not over yet, and whether it will return in the future remains unknown. Even if the pandemic ends suddenly, we should continue to develop the most promising vaccine candidates.
3. Conclusion
We reviewed and systematically analyzed the current studies on the characteristics of SARS-CoV-2 based on bioinformatics methods as follows. Firstly, a large number of computer resources have been used to sequence the genome and variants of the novel coronavirus, and the sequencing results have been compared to find the source of the virus and the maximum probability of host transmission combining bioinformatics methods and other technologies. At present, the sequence resources on SARS-CoV-2 are evolving very rapidly. The simple sequence alignment of viruses consumes little resources and can be done using online databases. However, to accelerate complex and detailed research, high-performance computing environments are necessary. High quality SARS-CoV-2 genome was assembled by the NGS and TGS technology. After sequencing alignment and phylogenetic analysis, evolutionary characteristics indicated that the bat may be the natural host of the novel coronavirus. However, the origin and intermediate host of the virus has not been fully determined, so the sequence alignment and mutations of individuals need to be constantly continued. By comparing different virus detection methods, they have different advantages in different aspects. qRT-PCR is the gold standard approach for virus detection and auxiliary clinical diagnosis. Other SARS-CoV2 detection methods have different advantages and provide comprehensive support.
Secondly, so far, the protein three-dimensional structure and RNA sequence characteristics of SARS-CoV-2 will help us to understand the structural basis and functional characteristics of coronavirus entering the target cells. ACE2, as an entry receptor, provides the entry point for SARS-CoV2 to infect human cells, acts as a drug target to treat COVID-19 patients. Bioinformatics analysis of single-cell RNA sequencing provide abundant resource for discovering the mechanism of immune response induced by COVID-19. The up-regulation of S100A8/A9 on megakaryocytes and monocytes in severe patients mainly causes cytokine storm. A large number of drug databases provide the basis and possibility for drug research of SARS-CoV-2. Molecular dynamics simulation, molecular docking and artificial intelligence technology of high-throughput computing and bioinformatics methods are applied to accelerate and support the design and screening of auxiliary drugs. Several effective drugs for the treatment of SARS-CoV-2 were screened, such as ledipasvir, velpatasvir and remdisvir and so on. In addition, computational approaches can screen effective vaccines against SARS-COV-2 in a short time through reverse vaccinology, Immunoinformatics and structural vaccinology. However, whether these antiviral drugs have a definite effect on the treatment of patients with COVID-19 and whether vaccines can protect against disease, still requires solid data from clinical trials. Bioinformatics tools will continue to play a key and complementary role in all stages of the COVID-19 pandemic.
Ethics approval and consent to participate
Not applicable.
CRediT authorship contribution statement
Lifei Ma: Conceptualization, Investigation, Methodology, Data curation, Resources, Software, Writing – review & editing, Funding acquisition, Supervision, Visualization. Huiyang Li: Conceptualization, Investigation, Methodology, Data curation, Resources, Software, Writing – review & editing, Visualization. Jinping Lan: Resources, Writing – review & editing, Visualization. Xiuqing Hao: Resources, Writing – review & editing. Huiying Liu: Writing – review & editing. Xiaoman Wang: Conceptualization, Resources, Methodology, Writing -– review & editing, Supervision. Yong Huang: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Science and Technology Research Project of Hebei Higher Education Institutions (QN2021012, ZD2021007), the Youth Funding Project of Hebei North University (QN2020024) and the Innovation and Entrepreneurship Training Program for College Students in Hebei Province (S202110092029, and xj202154), China.
References
- Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2) - an update on the status. Infect. Genet. Evol. 2020:83. doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bai Q., Tan S., Xu T., Liu H., Huang J., Yao X. MolAICal: a soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalampous T., Kay G.L., Richardson H., Aydin A., Baldan R., Jeanes C., Rae D., Grundy S., Turner D.J., Wain J., Leggett R.M., Livermore D.M., O’Grady J. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 2019;37:783–792. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and Clinical Characteristics of 99 Cases of 2019-Novel Coronavirus (2019-nCoV) Pneumonia in Wuhan. China SSRN Electron. J. 2020 doi: 10.2139/ssrn.3523861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.F., Chang Y.C., Hsiao Y., Lee K.H., Hsiao Y.C., Lin Y.H., Tu Y.C.E., Huang H.C., Chen C.Y., Juan H.F. DockCoV2: a drug database against SARS-CoV-2. Nucleic Acids Res. 2021;49:D1152–D1159. doi: 10.1093/nar/gkaa861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, X., Yan, R., Zhang, Jun, Zhang, G., Zhang, Y., Hao, M., Zhang, Z., Fan, P., Dong, Y., Yang, Y., Chen, Z., Guo, Y., Zhang, Jinlong, Li, Y., Song, X., Chen, Y., Xia, L., Fu, L., Hou, L., Xu, J., Yu, C., Li, J., Zhou, Q., Chen, W., 2020. A potent neutralizing human antibody reveals the N-terminal domain of the Spike protein of SARS-CoV-2 as a site of vulnerability. 〈 10.1101/2020.05.08.083964〉. [DOI] [PMC free article] [PubMed]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwudozie O.S., Duru V.C., Ndiribe C.C., Aborode A.T., Oyebanji V.O., Emikpe B.O. The relevance of bioinformatics applications in the discovery of vaccine candidates and potential drugs for COVID-19 treatment. Bioinform. Biol. Insights. 2021:15. doi: 10.1177/11779322211002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwudozie O.S., Gray C.M., Fagbayi T.A., Chukwuanukwu R.C., Oyebanji V.O., Bankole T.T., Adewole R.A., Daniel E.M. Immuno-informatics design of a multimeric epitope peptide based vaccine targeting SARS-CoV-2 spike glycoprotein. PLoS One. 2021:16. doi: 10.1371/journal.pone.0248061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I., Rappuoli R., Seib K.L. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb. Perspect. Med. 2013:3. doi: 10.1101/cshperspect.a012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Gouklani H., Davoodian P., Ahmadi N., Einakian M.A., Karmostaji A., Ahmadi K. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J. Biomol. Struct. Dyn. 2021;39:2857–2872. doi: 10.1080/07391102.2020.1756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., Vogels C.B.F., Brito A.F., Alpert T., Muyombwe A., Razeq J., Downing R., Cheemarla N.R., Wyllie A.L., Kalinich C.C., Ott I.M., Quick J., Loman N.J., Neugebauer K.M., Greninger A.L., Jerome K.R., Roychoudhury P., Xie H., Shrestha L., Huang M.L., Pitzer V.E., Iwasaki A., Omer S.B., Khan K., Bogoch I.I., Martinello R.A., Foxman E.F., Landry M.L., Neher R.A., Ko A.I., Grubaugh N.D. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181:990–996. doi: 10.1016/j.cell.2020.04.021. (990--996.e5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani A., Samarfard S., Eskandarzade N., Afsharifar A., Eskandari M.H., Niazi A., Izadpanah K., Karbanowicz T.P. Comparative phylogenetic analysis of SARS-CoV-2 spike protein-possibility effect on virus spillover. Brief. Bioinform. 2021;22 doi: 10.1093/bib/bbab144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanetti M., Benvenuto D., Angeletti S., Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J. Med. Virol. 2020;92:518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V., Stryke D., Bouquet J., Somasekar S., Linnen J.M., Dodd R., Mulembakani P., Schneider B.S., Muyembe-Tamfum J.J., Stramer S.L., Chiu C.Y. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015:7. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang Xinglou, You T., Liu Xiaoce, Yang Xiuna, Bai F., Liu H., Liu Xiang, Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Ziliang, Zhou Zhechong, Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y.Y., Peng T.T., Yeh T.K., Huang W.Z., Chang S.E., Wu S.H., Hung H.C., Hsu T.A., Lee S.J., Song J.S., Lin W.H., Chiang T.J., Lin J.H., Sytwu H.K., Chen C.T. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020;43:355–362. doi: 10.1016/j.bj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004:32. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA - J. Am. Med. Assoc. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. New Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/nejmoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wan Z., Hu Y., Zhou Y., Chen Q., Zhang C. A mismatch-tolerant RT-quantitative PCR: Application to broad-spectrum detection of respiratory syncytial virus. Biotechniques. 2019;66:225–230. doi: 10.2144/btn-2018-0184. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhou Y., Ma Y., Xu R., Jin X., Zhang C. A mismatch-tolerant RT-LAMP method for molecular diagnosis of highly variable viruses. Bio-Protoc. 2019:9. doi: 10.21769/bioprotoc.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Yingle, Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K. fai, Kan H., Fu Q., Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.M.D., Chiu R.W.K. Racing towards the development of diagnostics for a novel coronavirus (2019-nCoV) Clin. Chem. 2020;66:503–504. doi: 10.1093/clinchem/hvaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C.N.J., Amirthalingam G., Edmunds M., Zambon M., Brown K.E., Hopkins S., Chand M., Ramsay M. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. New Engl. J. Med. 2021;385:585–594. doi: 10.1056/nejmoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., Plessis, L. du, Liu, Z., Hill, V., Kang, M., Lin, H., Sun, J., Francois, S., Kraemer, M.U.G., Faria, N.R., McCrone, J.T., Peng, J., Xiong, Q., Yuan, R., Zeng, L., Zhou, P., Liang, C., Yi, L., Liu, J., Xiao, J., Hu, J., Liu, T., Ma, W., Li, W., Su, J., Zheng, H., Peng, B., Fang, S., Su, W., Li, K., Sun, R., Bai, R., Tang, X., Liang, M., Quick, J., Song, T., Rambaut, A., Loman, N., Raghwani, J., Pybus, O., Ke, C., 2020. Genomic epidemiology of SARS-CoV-2 in Guangdong Province, China. medRxiv 2020.04.01.20047076. 〈 10.1101/2020.04.01.20047076〉. [DOI] [PMC free article] [PubMed]
- Lu Renfei, Wu X., Wan Z., Li Y., Zuo L., Qin J., Jin X., Zhang C. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 2020;35:344–347. doi: 10.1007/s12250-020-00218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Roujian, Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk H.K.H., Li X., Fung J., Lau S.K.P., Woo P.C.Y. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect. Genet. Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Huo X.Q., Chen X., Zhu W.X., Yao M.C., Qiao Y.J., Zhang Y.L. Study on screening potential traditional Chinese medicines against 2019-nCoV based on Mpro and PLP. Zhongguo Zhong Yao Za Zhi. 2020;45:1219–1224. doi: 10.19540/j.cnki.cjcmm.20200216.401. [DOI] [PubMed] [Google Scholar]
- Mahase E. China coronavirus: what do we know so far? BMJ. 2020;368:m308. doi: 10.1136/bmj.m308. [DOI] [PubMed] [Google Scholar]
- María R.R., Arturo C.J., Alicia J.A., Paulina M.G., Gerardo A.O. The impact of bioinformatics on vaccine design and development. Vaccines. 2017;38:1966–1972. doi: 10.5772/intechopen.69273. [DOI] [Google Scholar]
- Martin R., Löchel H.F., Welzel M., Hattab G., Hauschild A.C., Heider D. CORDITE: the curated CORona drug InTERactions database for SARS-CoV-2. iScience. 2020:23. doi: 10.1016/j.isci.2020.101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsky H.C., Matranga C.B., Wohl S., Schaffner S.F., Freije C.A., Winnicki S.M., West K., Qu J., Baniecki M.L., Gladden-Young A., Lin A.E., Tomkins-Tinch C.H., Ye S.H., Park D.J., Luo C.Y., Barnes K.G., Shah R.R., Chak B., Barbosa-Lima G., Delatorre E., Vieira Y.R., Paul L.M., Tan A.L., Barcellona C.M., Porcelli M.C., Vasquez C., Cannons A.C., Cone M.R., Hogan K.N., Kopp E.W., Anzinger J.J., Garcia K.F., Parham L.A., Ramírez R.M.G., Montoya M.C.M., Rojas D.P., Brown C.M., Hennigan S., Sabina B., Scotland S., Gangavarapu K., Grubaugh N.D., Oliveira G., Robles-Sikisaka R., Rambaut A., Gehrke L., Smole S., Halloran M.E., Villar L., Mattar S., Lorenzana I., Cerbino-Neto J., Valim C., Degrave W., Bozza P.T., Gnirke A., Andersen K.G., Isern S., Michael S.F., Bozza F.A., Souza T.M.L., Bosch I., Yozwiak N.L., Macinnis B.L., Sabeti P.C. Zika virus evolution and spread in the Americas. Nature. 2017;546:411–415. doi: 10.1038/nature22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Ruth H., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China — key questions for impact assessment. New Engl. J. Med. 2020;382:692–694. doi: 10.1056/nejmp2000929. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000:28. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C.M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V.S., Kodali V.K., Li W., Maglott D., Masterson P., McGarvey K.M., Murphy M.R., O’Neill K., Pujar S., Rangwala S.H., Rausch D., Riddick L.D., Schoch C., Shkeda A., Storz S.S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R.E., Vatsan A.R., Wallin C., Webb D., Wu W., Landrum M.J., Kimchi A., Tatusova T., DiCuccio M., Kitts P., Murphy T.D., Pruitt K.D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E., Wong M.U., Huffman A., He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front. Immunol. 2020;11:1581. doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford Nanopore Technologies, 2018. Medaka: Sequence correction provided by ONT Research. GitHub. 〈https://github.com/nanoporetech/medaka〉.
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L.T., Nguyen, Thuong V., Luong Q.C., Nguyen, Thinh V., Nguyen H.T., Le H.Q., Nguyen T.T., Cao T.M., Pham Q.D. Importation and human-to-human transmission of a novel coronavirus in vietnam. New Engl. J. Med. 2020;382:872–874. doi: 10.1056/nejmc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan N., Shaib H. Middle east respiratory syndrome coronavirus (MERS-COV): a review. Germs. 2019;9:35–42. doi: 10.18683/germs.2019.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wen W., Fan X., Hou W., Su Bin, Cai P., Li J., Liu Y., Tang F., Zhang F., Yang Y., He Jiangping, Ma W., He Jingjing, Wang P., Cao Q., Chen F., Chen Y., Cheng X., Deng G., Deng X., Ding W., Feng Y., Gan R., Guo C., Guo W., He S., Jiang C., Liang J., Li Y. min, Lin J., Ling Y., Liu H., Liu J., Liu N., Liu S.Q., Luo M., Ma Q., Song Q., Sun W., Wang G.X., Wang F., Wang Y., Wen X., Wu Q., Xu G., Xie X., Xiong X., Xing X., Xu H., Yin C., Yu D., Yu K., Yuan J., Zhang B., Zhang P., Zhang T., Zhao J., Zhao Peidong, Zhou J., Zhou W., Zhong S., Zhong X., Zhang S., Zhu L., Zhu P., Zou B., Zou J., Zuo Z., Bai F., Huang X., Zhou P., Jiang Q., Huang Z., Bei J.X., Wei L., Bian X.W., Liu X., Cheng T., Li X., Zhao Pingsen, Wang F.S., Wang H., Su Bing, Zhang Zheng, Qu K., Wang X., Chen J., Jin R., Zhang Zemin. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913–19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. New Engl. J. Med. 2020;382:970–971. doi: 10.1056/nejmc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J.L., Schmithausen R.M., Li D., Hollstein R., Ben-Shmuel A., Israeli O., Weiss S., Paran N., Wilbring G., Liebing J., Feldman D., Słabicki M., Lippke B., Sib E., Borrajo J., Strecker J., Reinhardt J., Hoffmann P., Cleary B., Hölzel M., Nöthen M.M., Exner M., Ludwig K.U., Regev A., Zhang F. LAMP-Seq: population-scale COVID-19 diagnostics using combinatorial barcoding. bioRxiv. 2020 doi: 10.1101/2020.04.06.025635. [DOI] [Google Scholar]
- Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W.R., Bridgland A., Penedones H., Petersen S., Simonyan K., Crossan S., Kohli P., Jones D.T., Silver D., Kavukcuoglu K., Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- She J., Jiang J., Ye L., Hu L., Bai C., Song Y. 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin. Transl. Med. 2020;9:19. doi: 10.1186/s40169-020-00271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Qi H., Sun Q., Fan G., Liu S., Wang J., Zhu B., Liu H., Zhao F., Wang X., Hu X., Li W., Liu J., Tian Y., Wu L., Ma J. GcMeta: a global catalogue of metagenomics platform to support the archiving, standardization and analysis of microbiome data. Nucleic Acids Res. 2019;47:D637–D648. doi: 10.1093/nar/gky1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Zhang X., Mu K., Peng C., Zhu Z., Wang X., Yang Y., Xu Z., Zhu W. D3Targets-2019-nCoV: a webserver for predicting drug targets and for multi-target and multi-site based virtual screening against COVID-19. Acta Pharm. Sin. B. 2020;10:1239–1248. doi: 10.1016/j.apsb.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017:22. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Arrieta S., Goulder P.J.R., Brander C. In silico veritas? Potential limitations for SARSCoV-2 vaccine development based on T-cell epitope prediction. PLoS Pathog. 2020:16. doi: 10.1371/journal.ppat.1008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Ma L., Zou D., Tian D., Li C., Zhu J., Chen M., Wang A., Ma Y., Li M., Teng X., Cui Y., Duan G., Zhang M., Jin T., Shi C., Du Z., Zhang Y., Liu C., Li R., Zeng J., Hao L., Jiang S., Chen H., Han D., Xiao J., Zhang Z., Zhao W., Xue Y., Bao Y. The Global Landscape of SARS-CoV-2 Genomes, Variants, and Haplotypes in 2019nCoVR. Genom. Proteom. Bioinform. 2020;18:749–759. doi: 10.1016/j.gpb.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiaroa, G., Rawlinson, D., Featherstone, L., Pitt, M., Caly, L., Druce, J., Purcell, D., Harty, L., Tran, T., Roberts, J., Scott, N., Catton, M., Williamson, D., Coin, L., Duchene, S., 2020. Direct RNA sequencing and early evolution of SARS-CoV-2. 〈 10.1101/2020.03.05.976167〉. [DOI]
- Thai H.T.C., Le M.Q., Vuong C.D., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traditional Chinese Medicine Database (TCMD), 2009. Beijing.
- Wan Z., Zhang Y., He Z., Liu J., Lan K., Hu Y., Zhang C. A melting curve-based multiplex RT-qPCR assay for simultaneous detection of four human coronaviruses. Int. J. Mol. Sci. 2016;17:1880. doi: 10.3390/ijms17111880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Peng C., Yu Y., Chen Z., Xu Z., Cai T., Shao Q., Shi J., Zhu W. Exploring conformational change of adenylate kinase by replica exchange molecular dynamic simulation. Biophys. J. 2020;118:1009–1018. doi: 10.1016/j.bpj.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Fu A., Hu B., Tong Y., Liu R., Gu J., Liu J., Jiang W., Shen G., Zhao W., Men D., Deng Z., Yu L., Li Y., Liu T. Nanopore target sequencing for accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. medRxiv. 2020 doi: 10.1101/2020.03.04.20029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chen X., Lu Y., Chen F., Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci. Trends. 2020;14:64–68. doi: 10.5582/BST.2020.01030. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- wei Lu C., fen Liu X., fang Jia Z. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.R., Naccache S.N., Samayoa E., Biagtan M., Bashir H., Yu G., Salamat S.M., Somasekar S., Federman S., Miller S., Sokolic R., Garabedian E., Candotti F., Buckley R.H., Reed K.D., Meyer T.L., Seroogy C.M., Galloway R., Henderson S.L., Gern J.E., DeRisi J.L., Chiu C.Y. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. New Engl. J. Med. 2014;370:2408–2417. doi: 10.1056/nejmoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.C.P., Li X., Lau S.K.P., Woo P.C.Y. Global epidemiology of bat coronaviruses. Viruses. 2019:11. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1805–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Who.Int; China: 2020. Who Statement regarding Cluster of Pneumonia Cases in Wuhan. [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., Zhang Zhipeng, Shu F., Huang W., Li Y., Zhang Ziding, Chen R.A., Wu Y.J., Peng S.M., Huang M., Xie W.J., Cai Q.H., Hou F.H., Chen W., Xiao L., Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Xiao S.Z., Armit C., Edmunds S., Goodman L., Li P., Tuli M.A., Hunter C.I. Increased interactivity and improvements to the GigaScience database. GigaDB Database. 2019;2019 doi: 10.1093/database/baz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2014;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhu Z., Wang X., Zhang X., Mu K., Shi Y., Peng C., Xu Z., Zhu W. Ligand-based approach for predicting drug targets and for virtual screening against COVID-19. Brief Bioinform. 2021;22:1053–1064. doi: 10.1093/bib/bbaa422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Sciences. 2020 doi: 10.1126/science.abc1560. ((80-. ). 368, 1499–1504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Yun H., Lan W., Wang W., Sullivan S.G., Jia S., Bittles A.H. A climatologic investigation of the SARS-CoV outbreak in Beijing, China. Am. J. Infect. Control. 2006;34:234–236. doi: 10.1016/j.ajic.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Zhixiu, Li Zifu, Cui X., Xiao J., Zhan J., Meng T., Zhou W., Liu J., Xu H. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69:1010–1018. doi: 10.1136/gutjnl-2020-320953. [DOI] [Google Scholar]
- Zhang J., Litvinova M., Liang Y., Wang Y., Wang W., Zhao S., Wu Q., Merler S., Viboud C., Vespignani A., Ajelli M., Yu H. Changes in contact patterns shape the dynamics of the COVID-19 outbreak in China. Science. 2020;368:1481–1486. doi: 10.1126/science.abb8001. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 doi: 10.1101/2020.02.26.20028373. [DOI] [Google Scholar]
- Zhao W.M., Song S.H., Chen M.L., Zou D., Ma L.N., Ma Y.K., Li R.J., Hao L.L., Li C.P., Tian D.M., Tang B.X., Wang Y.Q., Zhu J.W., Chen H.X., Zhang Z., Xue Y.B., Bao Y.M. The 2019 novel coronavirus resource. Yi chuan = Hered. 2020;42:212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X., Lou, Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R., Di, Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wan Z., Yang S., Li Y., Li M., Wang B., Hu Y., Xia X., Jin X., Yu N., Zhang C. A mismatch-tolerant reverse transcription loop-mediated isothermal amplification method and its application on simultaneous detection of all four serotype of dengue viruses. Front. Microbiol. 2019:10. doi: 10.3389/fmicb.2019.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C., Allon S.J., Nyquist S.K., Mbano I., Miao V.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth II M.H., Kazer S., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Tzouanas C.N., Taliaferro F., Guo Z., Wang J.P., Dwyer D.F., Buchheit K.M., Boyce J., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Berger B., Leslie A., Flynn J.L., Fortune S.M., Finberg R.W., Kean L., Garber M., Schmidt A., Lingwood D., Shalek A.K., Ordovas-Montanes J., Lung Biological Network H. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3555145. [DOI] [PMC free article] [PubMed] [Google Scholar]