Abstract
Although the health benefits of exercise in adults with obesity are well described, the direct effects of exercise on adipose tissue that may lead to improved metabolic health are poorly understood. The primary aims of this study were to perform an unbiased analysis of the subcutaneous abdominal adipose tissue transcriptomic response to acute exercise in adults with obesity, and to compare the effects of moderate-intensity continuous exercise versus high-intensity interval exercise on this response. Twenty-nine adults with obesity performed a session of either high-intensity interval exercise (HI; 10 × 1 min at 90%HRpeak, 1 min recovery between intervals; n = 14) or moderate-intensity continuous exercise (MI; 45 min at 70%HRpeak; n = 15). Groups were well matched for BMI (HI 33 ± 3 vs. MI 33 ± 4 kg/m2), sex (HI: 9 women vs. MI: 10 women), and age (HI: 32 ± 6 vs. MI: 29 ± 5). Subcutaneous adipose tissue was collected before and 1 h after the session of HI or MI, and samples were processed for RNA sequencing. Gene set enrichment analysis revealed 7 of 21 gene sets enriched postexercise overlapped between HI and MI. Interestingly, both HI and MI upregulated gene sets involved in inflammation (IL6-JAK-STAT3 signaling, allograft rejection, TNFα signaling via NFκB, and inflammatory response; FDR q value < 0.25). Exercise also downregulated adipogenic and oxidative metabolism gene sets in both groups. Overall, these data suggest genes involved in subcutaneous adipose tissue metabolism and inflammation may be an important part of the initial response after a session of exercise.
NEW & NOTEWORTHY This study compared the effects of a single session of high-intensity interval exercise versus moderate-intensity continuous exercise on transcriptional changes in subcutaneous abdominal adipose tissue collected from adults with obesity. Our novel findings indicate exercise upregulated inflammation-related gene sets, while it downregulated metabolism-related gene sets – after both high-intensity and moderate-intensity exercise. These data suggest exercise can alter the adipose tissue transcriptome 1 h after exercise in ways that may impact inflammation and metabolism.
Keywords: acute exercise, adipose tissue, HIIT, obesity, transcriptomics
INTRODUCTION
In obesity, adipose tissue is often associated with chronic, low-grade inflammation, which in turn has been linked to systemic insulin resistance and elevated cardiovascular disease risk (1, 2). Participation in regular exercise is known to reduce cardiometabolic disease risk (3), but the direct impact of exercise on adipose tissue is poorly understood, especially in humans. Although there is growing interest in the potential for exercise-induced browning of white adipose tissue by the release of myokines such as irisin (4), studies in humans have not found clear evidence for adipose tissue “browning” after exercise training (5, 6). There are limited data concerning mitochondrial biogenesis in human white adipose tissue after exercise training, with some suggestion of reduced or unchanged mitochondrial capacity/content (7–9). Furthermore, we recently reported a decrease in adipose tissue progenitor content 12 h after acute exercise in adults with obesity (10), whereas others have reported decreases in adipogenic signaling after acute exercise in mice (11). Overall, targeted approaches to identify the metabolic pathways in adipose tissue that are altered by exercise have not yet provided a clear understanding of the pathways that might be responsible for the local metabolic adaptations to exercise in humans.
Adipose tissue immune cell composition is also plastic and has been shown to change in a matter of days of exposure to a high-fat diet (12). Most work assessing the immune response in the hours after exercise has measured circulating immune cells (13–15), with no clear consensus on whether acute exercise is pro or anti-inflammatory (16). MacPherson et al. (17) reported a decrease in proinflammatory macrophage content 2 h after acute exercise in inguinal white adipose tissue from high-fat-fed mice; but work in young, lean male subjects has demonstrated a broad increase in monocyte/macrophage signature genes 4 h after exercise (18). The effects of acute exercise on inflammatory signaling in adipose tissue from adults with obesity remain unresolved.
Adaptations to exercise typically accrue in response to repeated exposure to transient exercise-induced transcriptional responses after each exercise session (19). Therefore, identifying the signals that occur in the hours after exercise is an important step toward understanding mechanisms for chronic adaptations to exercise training, and whether this can be manipulated to induce favorable adipose tissue remodeling. Subcutaneous adipose tissue is responsible for the vast majority (80%–90%) of fatty acids released into the systemic circulation at rest and during exercise (20, 21). In contrast, visceral adipose tissue has been found to contribute <10% to the systemic fatty acid pool (20). Importantly, the intensity of the exercise stimulus often influences the magnitude of key metabolic adaptations to exercise (22). For example, classic work has established that exercise-induced muscle mitochondrial biogenesis is most robust in response to high-intensity exercise (23, 24). In addition, cellular stress, blood flow, substrate mobilization, and fuel selection during/after exercise are also greatly impacted by the intensity, duration, and energy expenditure of the exercise session (25–27). Therefore, the objectives of this study were twofold: 1) to perform an unbiased analysis of the subcutaneous adipose tissue transcriptomic response to a session of exercise in adults with obesity, and 2) to compare the effects of high-intensity interval exercise with more conventional moderate-intensity continuous exercise. We hypothesized that acute exercise would increase the expression of factors involved in regulating fat metabolism, while reducing the expression of proinflammatory genes within adipose tissue; and this effect would be more robust in response to high-intensity exercise. Expanding our understanding about the adipose tissue transcriptional response to exercise can help uncover new mechanisms for improving metabolic health in obesity.
METHODS
Subjects
Subjects participating in this study also participated in a related study from our laboratory examining the metabolic responses to 3-mo exercise training (28). All data presented here are novel and do not overlap with our previous study (28). The current study was done at the midway point of this 3-mo exercise training study to assess the transcriptomic response to exercise in “habitual” exercisers, of two distinct training protocols. This prolonged run-in exercise training period was an important part of our goal to assess responses to exercise in people who exercise regularly, and was selected for its applicability to real-life exercise prescription. Twenty-nine men and women with obesity, who were nondiabetic and weight stable (±3 kg for ≥ 6 mo) participated in this study. This sample size was based on power calculations from work by MacPherson et al. (17) indicating a predicted sample size of 11 (per group) is required to address a typical mean difference in gene expression of 0.5-fold with a SD of 0.5. Therefore, to ensure sufficient subjects for this study, we enrolled 14–15 participants per group and used 12 subjects per group for mRNA sequencing (based on sample quality control testing). Subjects completed a medical history questionnaire, physical examination, and resting 12-lead electrocardiogram before entering the study and were excluded if they were taking medications that affected lipid or glucose metabolism, had electrocardiogram abnormalities, or were pregnant/smokers. The study protocol was approved by the University of Michigan Institutional Review Board, and all subjects provided written, informed consent to participate.
Cardiorespiratory Fitness Testing
After enrolling in the study, we assessed cardiorespiratory fitness by measuring peak oxygen consumption (V̇o2peak) using an incremental exercise test to exhaustion on a cycle ergometer (Corvival, Lode, the Netherlands). The test began with a 4-min warm-up, followed by an increase in resistance every minute (beginning at 40 W and increasing in 20 W increments) until volitional fatigue. This exercise test was also used to establish heart rate targets during the experimental exercise sessions.
Treatment Assignment and Exercise Training
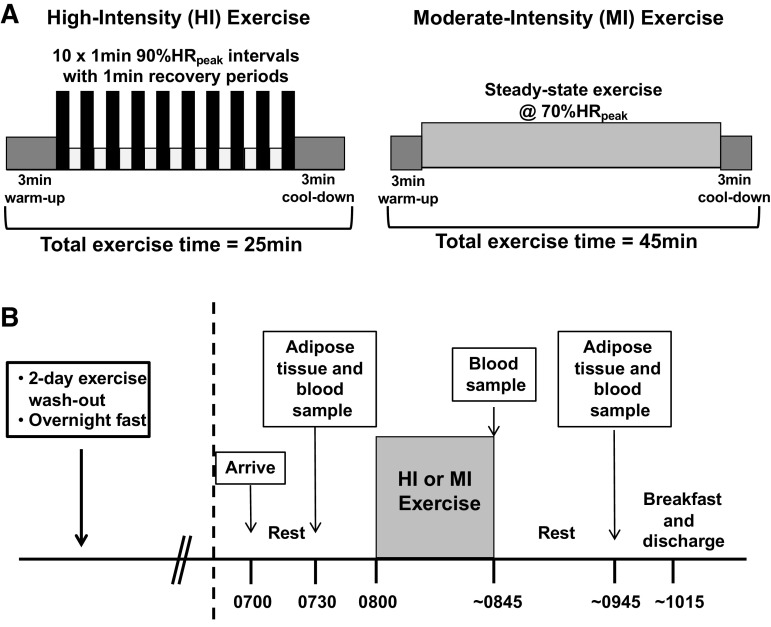
Subjects were assigned to either a high-intensity (HI) or moderate-intensity (MI) exercise group in a counter balanced manner. Groups were matched for sex, BMI, and V̇o2peak. Subjects in the HI group performed 10×1-min intervals at 90% HRpeak, with 1 min of active recovery between intervals. The HI exercise session also included a 3-min warmup and cooldown before and after the interval protocol, so the total exercise session lasted 25 min (Fig. 1A). Subjects in the MI group performed steady-state exercise at 70% HRpeak for 45 min (Fig. 1A). Exercise intensity was monitored by telemetry heart rate devices (Polar, Finland), and recorded after every 1-min interval (HI) or every 5 min (MI) to achieve the desired target heart rate when these values were averaged across the exercise session. To understand the signals induced by these distinct training protocols in habitual exercisers, the experimental trial was conducted after subjects trained for 6 wk (4 sessions/wk). To apply these findings to real-life long-term exercise guidelines, it was important for us to study the response to acute exercise in participants who had followed a well-controlled (HI or MI) training protocol. Participants exercised on their modality of choice throughout training (treadmill, cycle ergometer, rowing ergometer), and performed the experimental exercise session on this same exercise mode. The distribution of modalities was largely balanced between groups (i.e., cycle ergometer: HI = 9, MI = 8; treadmill: HI = 5, MI = 5; rowing ergometer: HI = 1, MI = 0), and one MI subject completed half of their exercise training sessions on a cycle ergometer and half on a treadmill. The experimental trial was conducted 2 days after subjects’ last exercise training session.
Figure 1.
Experimental design. A: subjects completed a single bout of MI (45 min at ∼70% HRpeak) or HI (10×1 min at ∼90% HRpeak with 1 min recovery in 25 min total). B: sample collection was performed 2 days after the last regular exercise training session, following a 12-h overnight fast. HI, high-intensity exercise; MI, moderate-intensity exercise.
Experimental Protocol
The experimental trial is outlined in Fig. 1B. Subjects reported to the laboratory between 0700 and 1000 h, after an overnight fast. Participants then rested for 20–30 min before the preexercise (Pre) blood and subcutaneous abdominal adipose tissue samples were collected by needle aspiration as described previously (29). In brief, ∼200 mg adipose tissue was collected by aspiration with a 16-G needle following administration of local anesthetic (2% lidocaine with no epinephrine). The sample was thoroughly rinsed of blood with saline, frozen in liquid nitrogen, and stored at −80°C until analysis. After this preexercise sample collection, subjects performed the supervised exercise session, consistent with their group assignment (HI or MI). Importantly, HI and MI differed by intensity (90% vs. 70% HRpeak), duration (25 vs. 45 min), and total energy expended (∼150 vs. ∼250 kcal) during exercise. Immediately postexercise (IPEX), another blood sample was taken, and subjects rested quietly in the laboratory for 1 h before postexercise (1hPost) blood and fat samples were collected from the opposite side of the umbilicus to the Pre biopsy.
Analytical Procedures
Adipose tissue samples were partitioned for RNA sequencing as described below. RNA was isolated from ∼80 mg tissue using a Qiagen RNeasy Plus Universal Mini kit (Cat. No. 73404), with Turbo DNase treatment (Cat. No. AM1907) after RNA isolation.
RNA-sequencing.
Library preparation and sequencing were performed by the University of Michigan Advanced Genomics Core. The Lexogen QuantSeq 3′ mRNA library prep (Cat. No. 015) was used on 100 ng total RNA to generate fragment lengths of ∼100 nucleotides. Single-end sequencing was performed on the NovaSeq 6000 with 100 cycles and ∼10 million reads per sample. Data were analyzed in collaboration with the University of Michigan Bioinformatics Core. Data passed FastQC analysis and samples had ∼80% alignment to the reference genome. Differential expression (DE) analysis was performed on log-CPM values calculated using Voom, which were further normalized by the trimmed mean of M-values technique through edgeR. The Limma software package of Bioconductor was used for DE analysis. Gene set enrichment analysis (GSEA; Broad Institute) was used to identify gene sets from the Hallmark database (Liberzon) that were up or downregulated for the same comparisons. This was established by normalized enrichment scores (NES), which reflect the degree to which the expression of genes from a given gene set is collectively increased or decreased 1 h after exercise (1hPost) compared with all genes detected in the experiment. Leading edge analysis (30) was used to identify the key genes driving these gene set differences. RNA sequencing source data are available at https://doi.org/10.17632/2ytjynczcb.2.
Blood measurements.
Plasma was separated from blood samples collected in ice-cold EDTA tubes, and serum was separated by collecting blood samples in silica tubes left to clot for 30 min at room temperature before centrifugation. All samples were centrifuged at 2,000 g for 15 min at 4°C and stored at −80°C until analysis. Plasma glucose (Thermo Fisher Scientific, Waltham, MA), nonesterified fatty acid (Wako Chemicals USA, Richmond, VA), triacylglyceride (Triacylglyceride reagent; Sigma Aldrich), and total- and high-density lipoprotein (HDL; cholesterol E and HDL-cholesterol E; Wako Chemicals) concentrations were measured using commercially available colorimetric assay kits. Serum insulin concentration was measured by IMMULITE 1000 chemiluminescent assay (Siemens, Flanders, NJ). Epinephrine and norepinephrine (Abnova, Taipei City, Taiwan; KA1877), IL6 (R&D Cat. No. HS600C), and cortisol (R&D Cat. No. KGE008B) were measured by ELISA. Some norepinephrine values were undetectable (below the lowest standard) at pre and 1hPost, so statistics were run on the available data with n = 6–7 Pre, n = 13–14 IPEX, and n = 9 1hPost. Glucose, insulin, and blood lipid profile were measured only at Pre to establish basal metabolic profiles were similar between groups. Catecholamines, cortisol, IL6, and nonesterified fatty acids were measured Pre, IPEX, and 1hPost to characterize the exercise stimuli.
Statistics
Student’s t tests were used for baseline anthropometric comparisons of HI versus MI. Linear mixed models (group vs. time) were used for testing IL6, cortisol, NEFA, epinephrine, and norepinephrine concentrations. IL6, cortisol, NEFA, epinephrine, and norepinephrine concentrations were log-transformed to achieve normality before analysis. Sidak’s multiple comparisons test was performed when an interaction effect was observed. RNA sequencing data were analyzed in collaboration with the University of Michigan Bioinformatics Core. DE of RNA-sequencing data was fit to a linear model per Smyth (31), and P values of <0.05 after multiple comparisons correction by false discovery rate (FDR) per Benjamini (32) were considered significant. For GSEA, gene sets found to be enriched with a false discovery rate (FDR) <0.25 were considered significant.
RESULTS
Subject Characteristics
A total of 29 subjects completed the study: 15 HI (9 women and 6 men) and 14 MI (10 women and 4 men). The groups were matched a priori for age, weight, BMI, and V̇o2peak (Table 1). There was no difference between HI and MI in fasting plasma concentrations of glucose, insulin, triglycerides, and cholesterol (Table 1).
Table 1.
Physical characteristics of subjects
| HI | MI | P value | |
|---|---|---|---|
| Age, yr | 32 ± 6 | 29 ± 5 | 0.13 |
| Body mass, kg | 97.2 ± 13.7 | 96.5 ± 11.4 | 0.89 |
| BMI, kg/m2 | 33.2 ± 2.9 | 33.4 ± 3.6 | 0.85 |
| Fasting glucose, mg/dL | 84 ± 9 | 86 ± 9 | 0.46 |
| Fasting insulin, uIU/mL | 18 ± 12 | 13 ± 6 | 0.29 |
| Triglycerides, mg/dL | 104 ± 62 | 114 ± 61 | 0.65 |
| Total cholesterol, mg/dL | 149 ± 30 | 155 ± 33 | 0.61 |
| HDL cholesterol, mg/dL | 34 ± 7 | 34 ± 8 | 0.95 |
| LDL cholesterol, mg/dL | 116 ± 33 | 122 ± 37 | 0.64 |
Data are means ± SD. BMI, body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein. n = 15 HI (9 F, 6 M), 14 MI (10 F, 4 M) subjects per group.
Exercise Response to HI versus MI
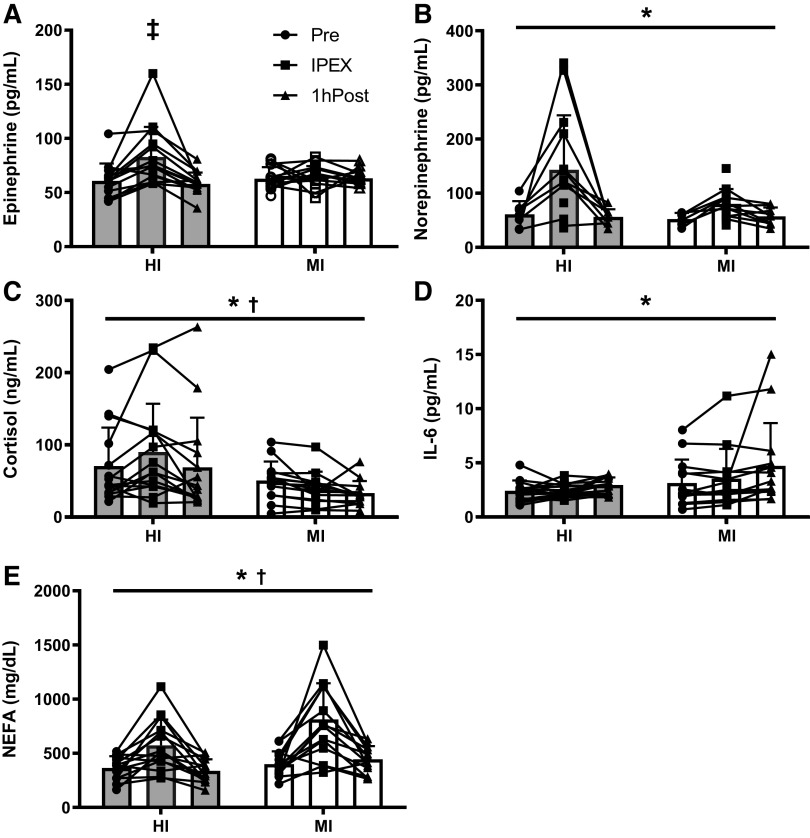
During the experimental exercise session, MI subjects exercised at 71 ± 1% of their HRpeak (132 ± 7 beats/min) for 45 min, whereas HI subjects exercised at 90 ± 2% of their HRpeak (163 ± 8 beats/min) during the 10 × 1 min high-intensity intervals. There was an effect of acute exercise on plasma concentrations of epinephrine, norepinephrine, cortisol, IL6, and NEFA (main effect for time, all P ≤ 0.01; Fig. 2). There was a group × time interaction in the plasma epinephrine response (P < 0.01), such that the increase in plasma epinephrine concentration IPEX was only significant in HI (multiple comparison P < 0.01 compared with Pre and 1hPost). Plasma cortisol concentrations were greater in HI versus MI, whereas plasma NEFA concentration was greater in MI versus HI (main effect for group, both P = 0.03; Fig. 2).
Figure 2.
Exercise response of circulating factors. A: epinephrine concentration – significantly greater IPEX vs. Pre and 1hPost (P < 0.01); interaction (P < 0.01) – main effect for time (P < 0.01). n = 12–14. B: plasma norepinephrine concentration – main effect for time (P < 0.01). n = 6–14. C: cortisol concentration – main effect for time (P = 0.01) and main effect for group (P = 0.03). n = 13–15 subjects per group. D: plasma IL-6 concentration – main effect for time (P < 0.01). n = 13–15. E: plasma NEFA concentration – main effect for time (P < 0.01) and main effect for group (P = 0.03). n = 13–15. Data are means ± SD. *P < 0.05 for time effect; †P < 0.05 for group effect. ‡P < 0.05 for Sidak’s multiple comparison test vs. both Pre and 1hPost HI. Full n breakdown by sex is in Supplemental Table S1 (see https://doi.org/10.17632/2ytjynczcb.2). IPEX, immediate postexercise; NEFA, nonesterified fatty acids.
Effects of Exercise on Adipose Tissue Gene Expression
Untargeted analysis of the effects of exercise on adipose tissue gene expression.
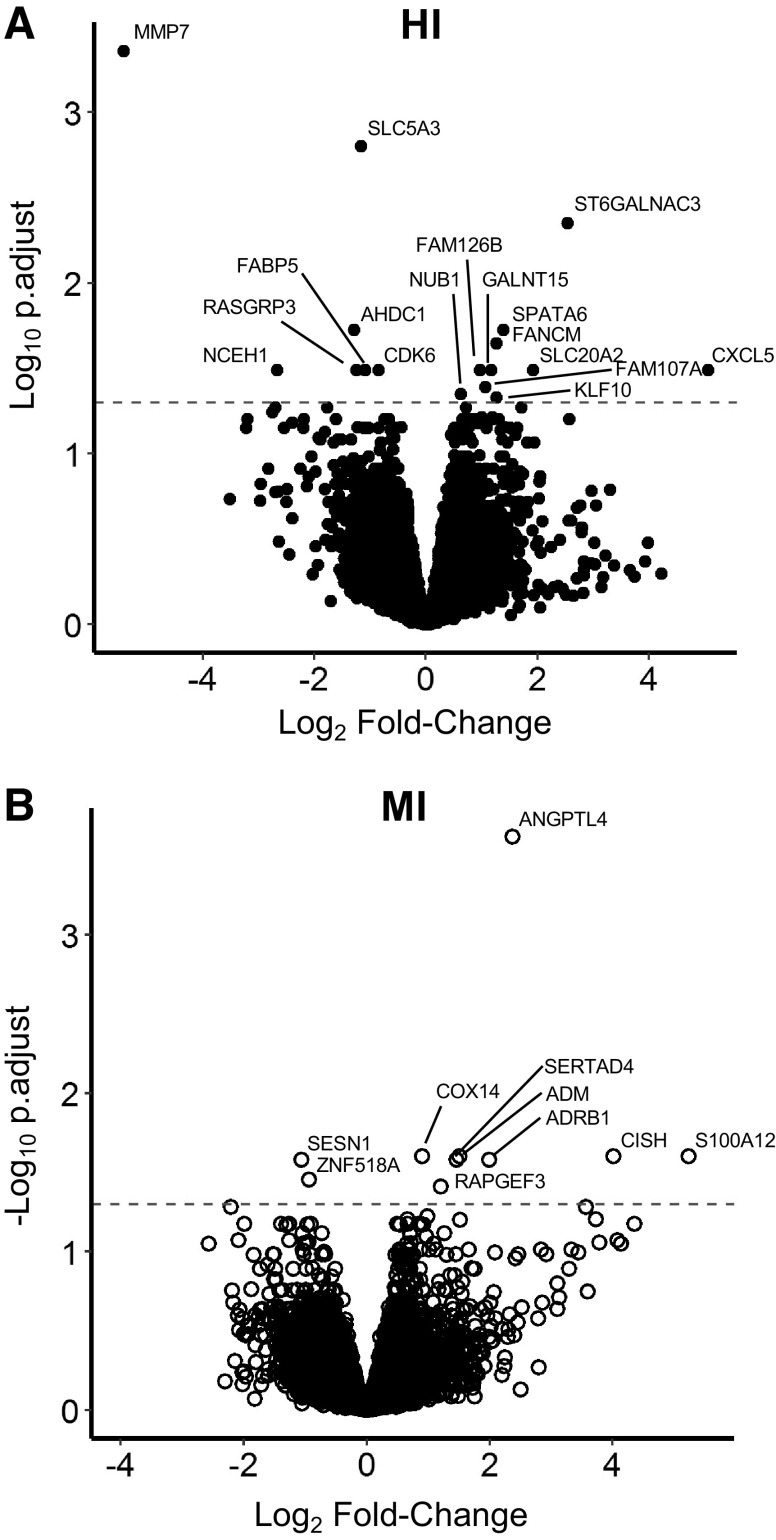
Using QuantSeq RNA sequencing, 12,077 total genes were detected in this experiment. We found 17 DE genes in adipose tissue collected 1 h after HI compared with before exercise (10 upregulated, 7 downregulated – Table 2, Fig. 3A), whereas in MI, there were 10 DE genes (8 upregulated, 2 downregulated – Table 2, Fig. 3B). Surprisingly, there was no overlap in DE genes between HI and MI – the full list of DE genes is provided in Table 2.
Table 2.
Differentially expressed genes in HI and MI
| HI |
MI |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Upregulated |
Upregulated |
||||||||
| Expression |
Expression |
||||||||
| Gene | Pre | Post | logFC | Adj. P | Gene | Pre | Post | logFC | Adj. P |
| ST6GALNAC3 | 7.2 | 12.1 | 0.76 | 0.00 | ANGPTL4 | 159.2 | 254.5 | 0.71 | 0.00 |
| SPATA6 | 16.5 | 21.6 | 0.42 | 0.02 | CISH | 13.6 | 31.8 | 1.21 | 0.03 |
| FANCM | 16.0 | 20.0 | 0.38 | 0.02 | S100A12 | 2.4 | 9.0 | 1.58 | 0.03 |
| CXCL5 | 6.1 | 40.4 | 1.52 | 0.03 | ADM | 151.9 | 202.6 | 0.45 | 0.03 |
| SLC20A2 | 5.1 | 7.5 | 0.58 | 0.03 | COX14 | 161.1 | 196.5 | 0.27 | 0.03 |
| FAM126B | 70.8 | 90.4 | 0.29 | 0.03 | SERTAD4 | 18.0 | 24.3 | 0.44 | 0.03 |
| GALNT15 | 122.4 | 148.2 | 0.35 | 0.03 | ADRB1 | 32.4 | 48.9 | 0.60 | 0.03 |
| FAM107A | 118.3 | 138.2 | 0.32 | 0.04 | RAPGEF3 | 28.9 | 37.6 | 0.36 | 0.04 |
| NUB1 | 107.8 | 121.3 | 0.19 | 0.04 | |||||
| KLF10 | 36.4 | 45.1 | 0.38 | 0.05 | |||||
| Downregulated |
Downregulated |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Expression |
Expression |
||||||||
| Gene | Pre | Post | logFC | Adj. P | Gene | Pre | Post | logFC | Adj. P |
| MMP7 | 7.2 | 1.9 | −1.63 | 0.00 | SESN1 | 60.1 | 47.4 | −0.32 | 0.03 |
| SLC5A3 | 355.2 | 267.0 | −0.35 | 0.00 | ZNF518A | 65.0 | 53.6 | −0.28 | 0.04 |
| AHDC1 | 30.0 | 22.6 | −0.39 | 0.02 | |||||
| NCEH1 | 28.5 | 14.2 | −0.80 | 0.03 | |||||
| FABP5 | 112.8 | 85.7 | −0.33 | 0.03 | |||||
| CDK6 | 96.4 | 80.0 | −0.26 | 0.03 | |||||
| RASGRP3 | 24.8 | 19.0 | −0.38 | 0.03 | |||||
Genes are divided by upregulated (1hPost > pre; top) and downregulated (1hPost < pre; bottom). Expression represents mean normalized logCPM per group. adj. P, adjusted P value for false discovery rate; logFC, log2(fold-change). n = 12 subjects/group (HI: 7 F, 4 M; MI: 9 F, 3 M).
Figure 3.
Differentially expressed transcripts within HI and MI. A: adjusted P value vs. log2(fold-change) for transcripts from Pre-HI vs. 1hPost-HI. Positive fold-change represents an upregulation 1hPost. B: adjusted P value vs. log2(fold-change) for transcripts from Pre-MI vs. 1hPost-MI. Points above the dashed lines were DE at an FDR-adjusted P < 0.05. n = 12 subjects per group (HI: 7 F, 4 M; MI: 9 F, 3 M). DE, differentially expressed; FDR, false discovery rate; HI, high-intensity exercise; MI, moderate-intensity exercise.
Gene set enrichment analysis.
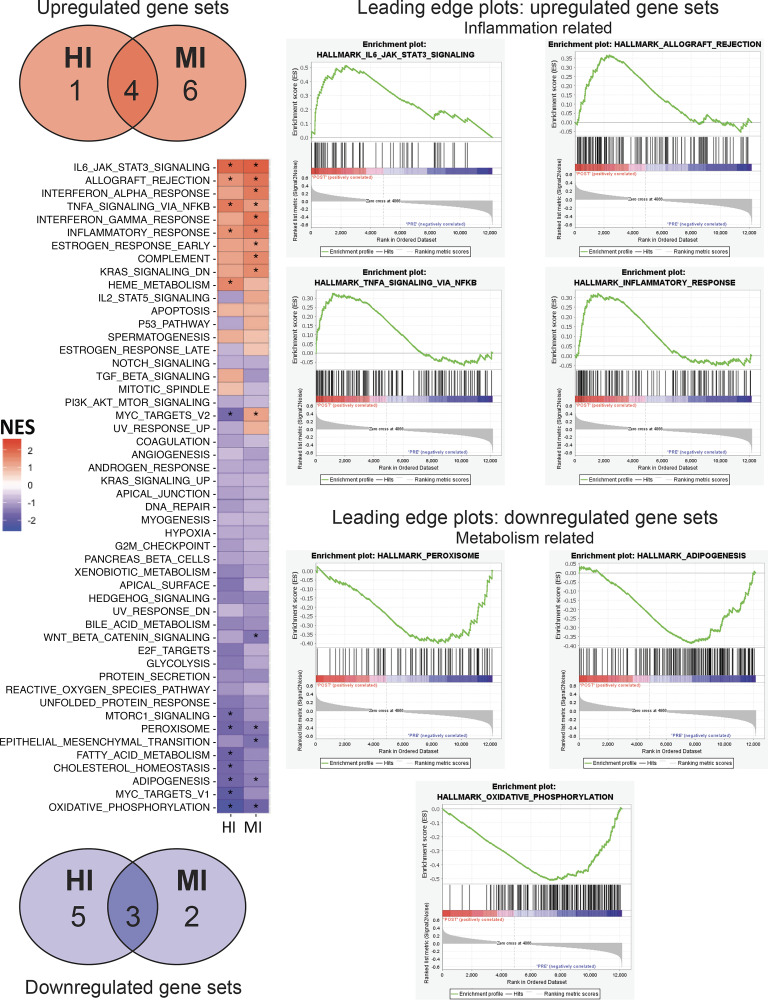
Given the low number and magnitude of DE genes, we used GSEA for its power to identify gene set-level changes in cases where individual gene expression changes may not be robust (33). We identified 13 pathways that were affected by HI (5 increased, 8 decreased), and 15 pathways affected by MI (10 increased, 5 decreased; Fig. 4 and Table 3). We found that seven of these gene sets changed in the same direction between HI and MI (4 upregulated and 3 downregulated; Fig. 4). Individual enrichment plots for these seven gene sets are shown in Fig. 4. Interestingly, the four pathways that were upregulated after exercise were all related to immune function (IL6-JAK-STAT3 signaling, allograft rejection, TNFα signaling via NFκB, and inflammatory response), whereas the three downregulated pathways were broadly related to metabolism (oxidative phosphorylation, adipogenesis, and peroxisome). Leading edge analysis is shown in Supplemental Table S2 (see https://doi.org/10.17632/2ytjynczcb.2).
Figure 4.
Gene set enrichment analysis. Left: heat map depicting the NES for gene sets from the Hallmark database. *FDR q val < 0.25. Venn diagrams presenting common gene set enrichment between HI and MI. Gene sets that were enriched 1hPost are shown in red (top); gene sets that were enriched Pre (i.e. downregulated) are shown in blue (bottom). Right: Seven pathways falling into immune or metabolic function were common between all comparisons; enrichment plots for these common immune (top) and metabolism (bottom) pathways. n = 12 subjects/group (HI: 7F, 4M; MI: 9F, 3M). FDR, false discovery rate; HI, high-intensity exercise; MI, moderate-intensity exercise; NES, normalized enrichment score.
Table 3.
Gene set enrichment analysis for samples collected Pre vs. 1hPost exercise
| Gene Set Name | Size | ES | NES | FDR q val. | |
|---|---|---|---|---|---|
| HI: pre vs. post | HALLMARK_IL6_JAK_STAT3_SIGNALING | 63 | 0.44 | 1.97 | 0.001 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 158 | 0.33 | 1.72 | 0.014 | |
| HALLMARK_HEME_METABOLISM | 153 | 0.30 | 1.57 | 0.035 | |
| HALLMARK_INFLAMMATORY_RESPONSE | 134 | 0.28 | 1.44 | 0.075 | |
| HALLMARK_ALLOGRAFT_REJECTION | 140 | 0.27 | 1.41 | 0.074 | |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | 180 | −0.51 | −2.27 | 0.000 | |
| HALLMARK_MYC_TARGETS_V1 | 190 | −0.46 | −2.04 | 0.000 | |
| HALLMARK_FATTY_ACID_METABOLISM | 132 | −0.42 | −1.78 | 0.011 | |
| HALLMARK_ADIPOGENESIS | 184 | −0.38 | −1.69 | 0.056 | |
| HALLMARK_PEROXISOME | 81 | −0.43 | −1.69 | 0.057 | |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 62 | −0.44 | −1.69 | 0.059 | |
| HALLMARK_MYC_TARGETS_V2 | 50 | −0.45 | −1.64 | 0.102 | |
| HALLMARK_MTORC1_SIGNALING | 185 | −0.36 | −1.61 | 0.144 | |
| MI: pre vs. post | HALLMARK_IL6_JAK_STAT3_SIGNALING | 63 | 0.47 | 2.07 | 0.000 |
| HALLMARK_ALLOGRAFT_REJECTION | 140 | 0.37 | 1.81 | 0.005 | |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 173 | 0.34 | 1.75 | 0.006 | |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 89 | 0.38 | 1.73 | 0.005 | |
| HALLMARK_INFLAMMATORY_RESPONSE | 134 | 0.34 | 1.66 | 0.011 | |
| HALLMARK_COMPLEMENT | 154 | 0.31 | 1.56 | 0.020 | |
| HALLMARK_KRAS_SIGNALING_DN | 63 | 0.35 | 1.53 | 0.022 | |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 158 | 0.26 | 1.30 | 0.130 | |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 141 | 0.26 | 1.28 | 0.129 | |
| HALLMARK_MYC_TARGETS_V2 | 50 | 0.30 | 1.22 | 0.194 | |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | 180 | −0.39 | −1.76 | 0.022 | |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 163 | −0.35 | −1.55 | 0.120 | |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 36 | −0.44 | −1.49 | 0.151 | |
| HALLMARK_PEROXISOME | 81 | −0.37 | −1.46 | 0.160 | |
| HALLMARK_ADIPOGENESIS | 184 | −0.31 | −1.38 | 0.247 |
Only pathways with FDR q val. < 0.25 are shown. Size represents the number of genes in the listed gene set. A negative normalized enrichment score (NES) represents enrichment in the precondition. ES, enrichment score; FDR q val., false-discovery rate-adjusted significance score; NES, normalized enrichment score. n = 12 subjects/group (HI: 7 F, 4 M; MI: 9 F, 3 M).
DISCUSSION
A key finding from this study was that acute exercise alters the adipose tissue transcriptome 1 h after exercise. In addition, despite HI and MI representing very distinct exercise stimuli, we found many similar gene sets were altered 1 h after HI and MI. Specifically, pathways related to inflammation were upregulated after both HI and MI exercise, whereas pathways related to adipogenesis and oxidative metabolism were downregulated. These data suggest that gene set-level increases in inflammatory signaling pathways are likely an important component of the early response in adipose tissue after a session of exercise, whereas reductions in metabolic pathways may reflect downregulation during the early phase recovery in response to exercise.
The intensity of exercise is known to have profound impact on the metabolic response during the exercise session (26, 34) and for at least the few hours afterwards (35). Furthermore, there is now a large body of literature suggesting that high-intensity interval training (HIIT) can induce similar adaptations to more conventional moderate-intensity continuous training (MICT) despite a lower time commitment and estimated overall energy expenditure. The high-intensity exercise stimulus is often credited for the robust adaptive responses to HIIT, but other factors associated with the HIIT protocol such as the inherent pulsatile nature may also contribute to the adaptive response (22). Much of the previous work examining the effects of HIIT versus MICT focused on whole body responses and changes within skeletal muscle (34), with very little information available regarding effects on signaling responses within adipose tissue. In our study, the greater adrenergic/cortisol levels in HI indicated that we did indeed achieve distinct metabolic stimuli between the two exercise protocols. Interestingly, the group effect for a higher cortisol concentration in HI versus MI appeared to be largely a consequence of an elevated baseline cortisol concentration in HI, perhaps resulting from the few weeks of HI training before the acute exercise session in this study, or a heightened anticipatory stress response in the HI group (36). However, despite robust differences in exercise intensity, as well as differences in exercise duration and estimated energy expenditure between HI and MI, the gene set responses demonstrated overlap in many key responses, such as IL6-JAK-STAT3 signaling and oxidative phosphorylation. Interestingly, all gene sets that were upregulated after exercise were inflammation-related, whereas most of the gene sets that were downregulated were related to metabolic control, in both HI and MI.
Our finding that acute exercise upregulated gene sets involved in inflammation just 1 h after exercise suggests that inflammatory signaling may be an important early phase postexercise response within human subcutaneous abdominal adipose tissue. This is consistent with recent work in lean participants reporting that the top upregulated gene ontology (GO) pathways in subcutaneous adipose tissue collected 4 h after exercise were also inflammation-related (18). Among the inflammatory gene sets that were upregulated in this cohort, the IL6-JAK-STAT3 signaling pathway had the greatest normalized enrichment score (NES), indicating this pathway exhibited the most robust collective upregulation of genes within the overall transcriptome. This is consistent with the work by MacPherson et al. (17), who reported increased IL6-signaling in inguinal adipose tissue samples collected 2 h after exercise from high-fat diet mice (17), and provides some evidence that exercise could trigger proinflammatory signaling through canonical IL6 signaling pathways including STAT3 and SOCS3. However, we did not have sufficient samples from this study to assess these targeted signaling pathways, so an important follow-up to these findings will be to assess activation of key inflammatory signaling pathways that were identified through the gene set enrichment analysis in this study (i.e., phosphorylated ERK, phosphorylated STAT3, NFκB localization). Importantly, it is unclear from this whole tissue analysis what cell population is responsible for the exercise-induced inflammatory signaling. Therefore, it will also be important for future studies to explore the signaling response to acute exercise in isolated adipocytes and stromal cells to elucidate the regulation of what appears to be a broad and complex response. As the magnitude of inflammatory gene induction in these samples is quite low, measures in isolated cell populations might also provide improved resolution to elucidate this response.
Our GSEA finding that the adipogenic gene set was downregulated after a session of exercise is consistent with previous work (11, 37). This reduction in adipogenic signaling 1 h after exercise may reflect the switch from fatty acid storage to release that accompanies acute exercise. Adipogenesis is regulated by a complex transcriptional programming, which can be altered by environmental cues in vitro, but whose in vivo regulation in humans remains unclear. There is some overlap between genes in the adipogenic gene set and the oxidative metabolism gene set that both decreased after exercise, with both gene sets including many mitochondrial and fat metabolism genes whose reductions may be transient, as subcutaneous adipose tissue oxidative capacity and mitochondrial biogenesis have been reported by some to remain stable with chronic exercise training in human subjects (5, 7, 8). However, it is hard to speculate on the signal for decreased oxidative metabolism gene transcription without knowing the primary cell types responsible for this metabolic signature. This again underscores the importance of assessing the transcriptional response to exercise in isolated cells in future studies, to determine which cell populations contribute to the exercise-induced decrease in adipogenic and oxidative phosphorylation gene programs. Furthermore, a thorough assessment of the time course of changes in gene expression and protein level responses in the individual cell populations within adipose tissue (e.g., progenitor cells and adipocytes) will be required to more fully elucidate the mechanisms for exercise-induced metabolic adaptations in adipose tissue.
There are several limitations to consider in interpreting these data. As this ancillary study was carried out using an untargeted approach to identify novel signaling pathways induced in adipose tissue by acute exercise, validation using similar unbiased approaches in a different cohort will provide important corroboration of the gene set findings identified here. These findings are particularly relevant for adults with obesity, and this was a novel aspect of this study. Future work should expand on our understanding of the transcriptomic response to exercise in adults with metabolic disease, especially those who might have more advanced tissue inflammation. It is possible that the proinflammatory signaling observed here would be enhanced in participants with more advanced metabolic disease due to increased immune cell content in the tissue. Sampling at several time points after exercise would also provide important information regarding the time course of transcriptional responses in adipose tissue after exercise. It is possible the very small number of DE genes after exercise was a consequence of the timing of our sample collection (i.e., 1 h after exercise). Fabre et al. (18) recently identified ∼350 DE genes in lean, trained subjects when measured 4 h after exercise. Our decision to collect adipose tissue samples 1 h after exercise was based on our interest in the early signals induced in adipose tissue after exercise. The use of GSEA allowed us to identify that this response, though mild at the individual gene level, is consistent at the gene set level. Furthermore, because our subjects completed a few weeks of either HI or MI exercise training before the exercise session in this study, we recognize this prior exposure to HI or MI training may have also had some impact on our outcomes. Therefore, our findings here should be interpreted as a reflection of the acute adipose tissue transcriptomic responses to MI and HI in adults with obesity who exercise regularly, which is likely different from the responses to exercise in individuals unaccustomed to MI or HI exercise. The analyses in this study were not designed to explore sex differences in the adipose tissue response to exercise but reveal pathways that are up/downregulated with both sexes pooled, providing insight to consistent exercise-induced signals that are triggered in both sexes – which is important for practical exercise prescription. Finally, although the central focus of this study was to expand our understanding of exercise-induced adaptations to abdominal subcutaneous adipose tissue that could improve its storage capacity, thereby reducing ectopic lipid deposition and inflammation in visceral adipose tissue during periods of weight gain, there are well-known regional differences among adipose tissue depots. Therefore, the findings reported here may not translate to the response in visceral adipose tissue, or even gluteal/femoral subcutaneous adipose tissue.
In summary, our key findings indicate that 1 h after a session of exercise, there was an upregulation of gene sets involved in inflammation, and a downregulation of gene sets related to oxidative metabolism and adipogenesis in adipose tissue from adults with obesity. These data suggest that an upregulation in inflammatory pathways is an important part of the immediate response to regular acute HI and MI exercise. This provides new evidence suggesting immune signaling in adipose tissue from adults with obesity could be a key early trigger for the purported long-term adipose tissue adaptations to exercise training such as reduced proinflammatory macrophage content (17). Finally, our findings indicate that low-volume, high-intensity exercise evokes many similar adipose tissue transcriptomic responses to more conventional moderate-intensity exercise [consistent with most standard exercise guidelines (38)], despite meaningful differences in energy expenditure and time commitment between these two exercise protocols.
SUPPLEMENTAL DATA
Supplemental Tables S1 and S2: https://doi.org/10.17632/2ytjynczcb.2.
GRANTS
This project was supported by the National Institutes of Health (DK077966, T32DK007245, and F32DK117522) and the Canadian Institutes of Health Research (338735 and 146190).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.C.L., B.J.R., J.B.G., and J.F.H. conceived and designed research; A.C.L., M.W.S., E.M.K., N.M.T., T.C.B., C.A., and P.V. performed experiments; A.C.L., M.W.S., E.M.K., N.M.T., B.J.R., T.C.B., and P.V. analyzed data; A.C.L., M.W.S., E.M.K., N.M.T., B.J.R., T.C.B., J.B.G., C.A., P.V., and J.F.H. interpreted results of experiments; A.C.L. and M.W.S. prepared figures; A.C.L. and J.F.H. drafted manuscript; A.C.L., M.W.S., E.M.K., N.M.T., B.J.R., T.C.B., J.B.G., C.A., P.V., and J.F.H. edited and revised manuscript; A.C.L., M.W.S., E.M.K., N.M.T., B.J.R., T.C.B., J.B.G., C.A., P.V., and J.F.H. approved final version of manuscript.
ACKNOWLEDGMENTS
We are grateful to Suzette Howton for subject recruitment and coordination and Thomas Rode for technical assistance. RNA sequencing was performed by the University of Michigan Advanced Genomics Core and analysis was performed in collaboration with the University of Michigan Bioinformatics Core.
REFERENCES
- 1.Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest 127: 1–4, 2017. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lumeng CN, Deyoung SM, Saltiel AR. Macrophages block insulin action in adipocytes by altering expression of signaling and glucose transport proteins. Am J Physiol Endocrinol Metab 292: E166–E174, 2007. doi: 10.1152/ajpendo.00284.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tremmel M, Gerdtham UG, Nilsson P, Saha S. Economic burden of obesity: a systematic literature review. Int J Environ Res Public Health 14: 435, 2017. doi: 10.3390/ijerph14040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boström P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Boström EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Højlund K, Gygi SP, Spiegelman BM. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481: 463–468, 2012. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stinkens R, Brouwers B, Jocken JW, Blaak EE, Teunissen-Beekman KF, Hesselink MK, van Baak MA, Schrauwen P, Goossens GH. Exercise training-induced effects on the abdominal subcutaneous adipose tissue phenotype in humans with obesity. J Appl Physiol (1985) 125: 1585–1593, 2018. doi: 10.1152/japplphysiol.00496.2018. [DOI] [PubMed] [Google Scholar]
- 6.Tsiloulis T, Carey AL, Bayliss J, Canny B, Meex RCR, Watt MJ. No evidence of white adipocyte browning after endurance exercise training in obese men. Int J Obes (Lond) 42: 721–727, 2018. doi: 10.1038/ijo.2017.295. [DOI] [PubMed] [Google Scholar]
- 7.Camera DM, Anderson MJ, Hawley JA, Carey AL. Short-term endurance training does not alter the oxidative capacity of human subcutaneous adipose tissue. Eur J Appl Physiol 109: 307–316, 2010. doi: 10.1007/s00421-010-1356-3. [DOI] [PubMed] [Google Scholar]
- 8.Larsen S, Danielsen JH, Søndergård SD, Søgaard D, Vigelsoe A, Dybboe R, Skaaby S, Dela F, Helge JW. The effect of high-intensity training on mitochondrial fat oxidation in skeletal muscle and subcutaneous adipose tissue. Scand J Med Sci Sports 25: e59–e69, 2015. doi: 10.1111/sms.12252. [DOI] [PubMed] [Google Scholar]
- 9.Dohlmann TL, Hindsø M, Dela F, Helge JW, Larsen S. High-intensity interval training changes mitochondrial respiratory capacity differently in adipose tissue and skeletal muscle. Physiol Rep 6: e13857–11, 2018. doi: 10.14814/phy2.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludzki AC, Krueger EM, Baldwin TC, Schleh MW, Porsche CE, Ryan BJ, Muir LA, Singer K, Lumeng CN, Horowitz JF. Acute aerobic exercise remodels the adipose tissue progenitor cell phenotype in obese adults. Front Physiol 11: 903, 2020. doi: 10.3389/fphys.2020.00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y, Zhou H, Jin W, Lee HJ. Acute exercise regulates adipogenic gene expression in white adipose tissue. Biol Sport 33: 381–391, 2016. doi: 10.5604/20831862.1224395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muir LA, Kiridena S, Griffin C, DelProposto JB, Geletka L, Martinez-Santibañez G, Zamarron BF, Lucas H, Singer K, O' Rourke RW, Lumeng CN. Frontline science: rapid adipose tissue expansion triggers unique proliferation and lipid accumulation profiles in adipose tissue macrophages. J Leukoc Biol 103: 615–628, 2018. doi: 10.1002/JLB.3HI1017-422R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson E, Durrer C, Simtchouk S, Jung ME, Bourne JE, Voth E, Little JP. Short-term high-intensity interval and moderate-intensity continuous training reduce leukocyte TLR4 in inactive adults at elevated risk of type 2 diabetes. J Appl Physiol (1985) 119: 508–516, 2015. doi: 10.1152/japplphysiol.00334.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slusher AL, Zúñiga TM, Acevedo EO. Maximal exercise alters the inflammatory phenotype and response of mononuclear cells. Med Sci Sports Exerc 50: 675–683, 2018. doi: 10.1249/MSS.0000000000001480. [DOI] [PubMed] [Google Scholar]
- 15.Campbell JP, Riddell NE, Burns VE, Turner M, van Zanten J, Drayson MT, Bosch JA. Acute exercise mobilises CD8+ T lymphocytes exhibiting an effector-memory phenotype. Brain Behav Immun 23: 767–775, 2009. doi: 10.1016/j.bbi.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Peake JM, Neubauer O, Walsh NP, Simpson RJ. Recovery of the immune system after exercise. J Appl Physiol (1985) 122: 1077–1087, 2017. doi: 10.1152/japplphysiol.00622.2016. [DOI] [PubMed] [Google Scholar]
- 17.Macpherson REK, Huber JS, Frendo-Cumbo S, Simpson JA, Wright DC. Adipose tissue insulin action and IL-6 signaling after exercise in obese mice. Med Sci Sports Exerc 47: 2034–2042, 2015. doi: 10.1249/MSS.0000000000000660. [DOI] [PubMed] [Google Scholar]
- 18.Fabre O, Ingerslev LR, Garde C, Donkin I, Simar D, Barrès R. Exercise training alters the genomic response to acute exercise in human adipose tissue. Epigenomics 10: 1033–1050, 2018. doi: 10.2217/epi-2018-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry CGR, Lally J, Holloway GP, Heigenhauser GJF, Bonen A, Spriet LL. Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J Physiol 588: 4795–4810, 2010. doi: 10.1113/jphysiol.2010.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen S, Guo Z, Johnson CM, Hensrud DD, Jensen MD. Splanchnic lipolysis in human obesity. J Clin Invest 113: 1582–1588, 2004. doi: 10.1172/JCI21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arner P, Kriegholm E, Engfeldt P, Bolinder J. Adrenergic regulation of lipolysis in situ at rest and during exercise. J Clin Invest 85: 893–898, 1990. doi: 10.1172/JCI114516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacInnis MJ, Gibala MJ. Physiological adaptations to interval training and the role of exercise intensity. J Physiol 595: 2915–2930, 2017. doi: 10.1113/JP273196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudley GA, Tullson PC, Terjung RL. Influence of mitochondrial content on the sensitivity of respiratory control. J Biol Chem 262: 9109–9114, 1987. doi: 10.1016/S0021-9258(18)48054-4. [DOI] [PubMed] [Google Scholar]
- 24.Holloszy JO. Biochemical adaptations in muscle. J Biol Chem 242: 2278–2282, 1967. doi: 10.1016/S0021-9258(18)96046-1. [DOI] [PubMed] [Google Scholar]
- 25.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 17: 162–184, 2013. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Romijn JA, Coyle EF, Sidossis LS, Zhang XJ, Wolfe RR. Relationship between fatty acid delivery and fatty acid oxidation during strenuous exercise. J Appl Physiol (1985) 79: 1939–1945, 1995. doi: 10.1152/jappl.1995.79.6.1939. [DOI] [PubMed] [Google Scholar]
- 27.Frayn KN, Karpe F. Regulation of human subcutaneous adipose tissue blood flow. Int J Obes (Lond) 38: 1019–1029, 2014. doi: 10.1038/ijo.2013.200. [DOI] [PubMed] [Google Scholar]
- 28.Ryan BJ, Schleh MW, Ahn C, Ludzki AC, Gillen JB, Varshney P, Van Pelt DW, Pitchford LM, Chenevert TL, Gioscia-Ryan RA, Howton SM, Rode T, Hummel SL, Burant CF, Little JP, Horowitz JF. Moderate-intensity exercise and high-intensity interval training affect insulin sensitivity similarly in obese adults. J Clin Endocrinol Metab 105: 1–19, 2020. doi: 10.1210/clinem/dgz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Pelt DW, Guth LM, Horowitz JF. Aerobic exercise elevates markers of angiogenesis and macrophage IL6 gene expression in the subcutaneous adipose tissue of overweight-to-obese adults. J Appl Physiol (1985) 123: 1150–1159, 2017. doi: 10.1152/japplphysiol.00614.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3, 2004. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300, 2007. [Google Scholar]
- 33.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273, 2003. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 34.van Loon LJ, Greenhaff PL, Constantin-Teodosiu D, Saris WH, Wagenmakers AJ. The effects of increasing exercise intensity on muscle fuel utilisation in humans. J Physiol 536: 295–304, 2001. doi: 10.1111/j.1469-7793.2001.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulla NA, Simonsen L, Bülow J. Post-exercise adipose tissue and skeletal muscle lipid metabolism in humans: the effects of exercise intensity. J Physiol 524(Pt 3): 919–928, 2000. doi: 10.1111/j.1469-7793.2000.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Paridon KN, Timmis MA, Nevison CM, Bristow M. The anticipatory stress response to sport competition: a systematic review with meta-analysis of cortisol reactivity. BMJ Open Sport Exerc Med 3: e000261–11, 2017. doi: 10.1136/bmjsem-2017-000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeve D, Millay DP, Seo J, Graff JM. Exercise-induced skeletal muscle adaptations alter the activity of adipose progenitor cells. PLoS One 11: e0152129, 2016. doi: 10.1371/journal.pone.0152129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.U.S. Department of Health and Human Services. Physical Activity Guidelines for Americans (2nd ed). Washington, DC: U.S. DHHS, 2018, p. 1–117. [Google Scholar]