Abstract
The human progesterone receptor (PR) exists as two functionally distinct isoforms, hPRA and hPRB. hPRB functions as a transcriptional activator in most cell and promoter contexts, while hPRA is transcriptionally inactive and functions as a strong ligand-dependent transdominant repressor of steroid hormone receptor transcriptional activity. Although the precise mechanism of hPRA-mediated transrepression is not fully understood, an inhibitory domain (ID) within human PR, which is necessary for transrepression by hPRA, has been identified. Interestingly, although ID is present within both hPR isoforms, it is functionally active only in the context of hPRA, suggesting that the two receptors adopt distinct conformations within the cell which allow hPRA to interact with a set of cofactors that are different from those recognized by hPRB. In support of this hypothesis, we identified, using phage display technology, hPRA-selective peptides which differentially modulate hPRA and hPRB transcriptional activity. Furthermore, using a combination of in vitro and in vivo methodologies, we demonstrate that the two receptors exhibit different cofactor interactions. Specifically, it was determined that hPRA has a higher affinity for the corepressor SMRT than hPRB and that this interaction is facilitated by ID. Interestingly, inhibition of SMRT activity, by either a dominant negative mutant (C'SMRT) or histone deacetylase inhibitors, reverses hPRA-mediated transrepression but does not convert hPRA to a transcriptional activator. Together, these data indicate that the ability of hPRA to transrepress steroid hormone receptor transcriptional activity and its inability to activate progesterone-responsive promoters occur by distinct mechanisms. To this effect, we observed that hPRA, unlike hPRB, was unable to efficiently recruit the transcriptional coactivators GRIP1 and SRC-1 upon agonist binding. Thus, although both receptors contain sequences within their ligand-binding domains known to be required for coactivator binding, the ability of PR to interact with cofactors in a productive manner is regulated by sequences contained within the amino terminus of the receptors. We propose, therefore, that hPRA is transcriptionally inactive due to its inability to efficiently recruit coactivators. Furthermore, our experiments indicate that hPRA interacts efficiently with the corepressor SMRT and that this activity permits it to function as a transdominant repressor.
The progesterone receptor (PR) is a ligand-activated transcription factor that belongs to the nuclear receptor superfamily of transcription factors (16). In the absence of hormone, the transcriptionally inactive receptor remains associated with a large complex of heat shock proteins in the nuclei of target cells (52). Upon hormone binding, the receptor dissociates from the heat shock protein complex, dimerizes, and binds to progesterone-responsive elements (PREs) within the regulatory regions of target genes (4, 36). When bound to DNA, the PR dimer contacts components of the general transcription machinery, either directly (28) or indirectly via cofactors such as coactivators and corepressors (21, 45, 51, 59), and either positively or negatively modulates target gene transcription.
Adding to the complexity of its signal transduction pathway is the fact that PR exists in humans as two isoforms, hPRA (94 kDa) and hPRB (114 kDa) (33). hPRA is a truncated form of hPRB, lacking the B upstream sequence (amino acids [aa] 1 to 164). The two isoforms are transcribed from a single gene by alternate initiation of transcription from two distinct promoters (20, 30). While the two forms of PR have similar DNA- and ligand-binding affinities (11), they have opposite transcriptional activities (9, 37, 56, 58, 61). In most contexts, hPRB functions as an activator of progesterone-responsive genes, while hPRA is transcriptionally inactive (56, 58). In addition, hPRA also functions as a strong transdominant repressor of hPRB (58) and human estrogen receptor (hER) transcriptional activity in the presence of both PR agonists and antagonists (18, 38, 58, 61).
Although the precise mechanism underlying the differential activities of the two human PR isoforms is not fully understood, recent structure-function studies of the two receptor isoforms suggest that hPRB contains three specific activation functions (AF-1, -2, and -3) whereas hPRA contains only two. AF-1, located within the amino terminus, and AF-2, in the carboxyl terminus, are common to both hPRA and hPRB. The third putative activation function, AF-3, is located within the B upstream sequence, a region which is absent in hPRA (47). We believe that AF-3 contributes to hPRB transcriptional activity by suppressing the activity of an inhibitory domain (ID) contained within sequences common to hPRA and hPRB. In support of this view, Giangrande et al. identified within the first 140 aa of hPRA an ID which has been shown to prevent hPRA from functioning as a transcriptional activator and permits this receptor isoform to function as a transdominant repressor of heterologous steroid receptor transcriptional activity (18). Deletion of the N-terminal 140 aa (ID) from hPRA results in a receptor mutant that is functionally indistinguishable from hPRB (18). Furthermore, Hovland et al. have shown that sequences within hPRA which contain an ID inhibit both AF-1 and AF-2 but not AF-3 (25). Cumulatively, these results support the hypothesis that hPRA, like hPRB, contains all of the sequences necessary for proper transcriptional activation; however, hPRA is transcriptionally inactive because in the absence of AF-3, ID prevents AF-1 and/or AF-2 from activating transcription. Thus, it seems that the role of AF-3 is to override the inhibitory function of ID, thereby allowing hPRB to activate transcription (18, 25).
The presence of an ID within hPR, whose function is masked in hPRB but not in hPRA, suggests that the distinct functions of the two receptors may be due to the ability of these proteins to adopt different conformations within the cell. This hypothesis is further supported by our recent studies which show that the amino termini of hPRB and hPRA interact differently with the carboxyl terminus of the human PR ligand-binding domain (hLBD) (54). Specifically, it was shown that the amino terminus of hPRB, but not that of hPRA, interacts efficiently with its hLBD both in vivo and in vitro in an agonist-dependent manner. Thus, the differential interaction between the carboxyl and amino termini of hPRB and hPRA may contribute to different cofactor interactions, which in turn may result in differences in the transcriptional activities of the two human PR isoforms. A potential mechanistic basis for these differential effects was revealed recently with the demonstration that the residue Ser294, which lies within the ID, is preferentially phosphorylated in the context of hPRB as opposed to hPRA (12).
To investigate the potential role(s) of differential cofactor interactions, we examined the ability of hPRA and hPRB to associate with different coactivators and corepressors and assessed the effect of these interactions on the receptors' transcriptional activity. We also investigated whether any of these factors could be implicated in hPRA-mediated transrepression of hER transcriptional activity. From these analyses, we found that antagonist-bound hPRA has a higher affinity for the corepressor SMRT than does antagonist-bound hPRB. The physiological significance of this interaction was demonstrated using a dominant negative variant of SMRT, C'SMRT, to partially reverse hPRA transrepression of hER-mediated transcriptional activity. Furthermore, using both in vivo and in vitro methodologies, we found that unlike hPRB, hPRA did not associate efficiently with the coactivators SRC-1 and GRIP1. Thus, differential cofactor association appears to explain why hPRA and hPRB manifest distinct transcriptional activities in target cells.
MATERIALS AND METHODS
Biochemicals.
DNA restriction and modification enzymes were obtained from Promega (Madison, Wis.), Boehringer Mannheim, or New England Biolabs (Beverly, Mass.). PCR reagents were obtained from Perkin-Elmer or Promega. 17-β-Estradiol and trichostatin A (TSA) were purchased from Sigma (St. Louis, Mo.). R5020 (promegestone) was purchased from NEN Life Science Products. RU486 was a gift from Ligand Pharmaceuticals (San Diego, Calif.). ZK98299 was a gift from Schering Pharmaceuticals (Berlin, Germany). Secondary antibodies, Hybond-C Extra (nitrocellulose) transfer membrane, and developing film were obtained from Amersham (Arlington Heights, Ill.). The polyclonal antibody raised against hPRA was a gift from Nancy Weigel (Baylor College of Medicine, Houston, Tex.). The monoclonal antibody raised against glutathione S-transferase (GST) was purchased from Sigma.
Plasmids.
pRST7-ERα and SV40-hPRB were provided by Ligand Pharmaceuticals (13); the expression vectors pBKC-hPRA and pBKC-hPRB were reported elsewhere (16); pBKC-Rev-TUP1 and pBKC-βgal have been previously described (35, 60).
The mammalian two-hybrid plasmid pCMX-Gal4-C'SMRT was a gift from J. D. Chen (University of Massachusetts, Worcester); Gal4N-RIP13ΔN4 was provided by D. D. Moore (Baylor College of Medicine); plasmids pM-hPRA (32) and pBKC-DBD (18) have been described previously. pM, containing the yeast Gal4 DNA-binding domain (Gal4DBD), was purchased from Clontech (Palo Alto, Calif.). pM-GRIP1(NR) was constructed as follows. A PCR-generated fragment from pCMV.HA/GRIP1 (provided by M. Stallcup, University of Southern California, Los Angeles) was subcloned into pM previously digested with EcoRI and BamHI. The sequences of the oligonucleotides for PCR are 5′-GGGGAATTCCACAGCCGGCTGCATGACAGC (forward) and 5′-CGCGGATCCTTCCGGTAAACCAATATC (reverse). pM-SRC-1(NR) was constructed by digesting pM with EcoRI and BamHI and subsequent subcloning of a PCR-generated fragment from pCMX-SRC-1 (provided by B. O'Malley, Baylor College of Medicine). The sequences of the oligonucleotides used to generate the PCR product are 5′-CCGGAATTCCCGGGAGACAGTAAATACTCT (forward) and 5′-CGCGGATCCCAGGTTTGGAGTTGATCT (reverse). All PCR-based cloning was verified by sequencing to assess the fidelity of the resulting constructs. The mammalian two-hybrid plasmids pVP16 and pVP16-T were purchased from Clontech; the VP16 fusion constructs pVP16-ERα, pVP16-GR, pVP16-hPRA, and pVP16-hPRB were provided by Ligand Pharmaceuticals. pVP16-ΔhPRA was constructed by digesting the ΔhPRA fragment from pBKC-ΔhPRA (18) with EcoRI and BamHI and subsequent cloning into pVP16 previously digested with EcoRI and BamHI.
The reporter 5XGal4-TATA-LUC was a gift from X.-F. Wang (Duke University Medical Center, Durham, N.C.). 2XPRE-TK-LUC contains two copies of a consensus PRE upstream of the thymidine kinase promoter; 3XERE-TATA-LUC contains three copies of vitellogenin estrogen-responsive element (ERE) cloned into pGL2-TATA-Inr (Stratagene, La Jolla, Calif.).
The GST fusion plasmid pGEX2TK-C'SMRT was provided by J. D. Chen; pGEX-5X-1 was obtained from Pharmacia Biotech (Uppsala, Sweden); pGEX.1-GRIP1 was provided by M. Stallcup. The GST fusion plasmid pGEX-5X-1-SRC-1(NR) was constructed as follows. The SRC-1(NR) fragment was digested from pM-SRC-1(NR) with EcoRI and SalI and subcloned into pGEX-5X-1 previously digested with EcoRI and SalI. pT7-hPRA and pT7-hPRB for in vitro translating hPRA and hPRB, respectively, and baculovirus-purified hPRA and hPRB proteins were prepared as previously described (54). Baculovirus-expressed ERα was provided by PanVera Corporation (Madison, Wis.).
Cloning of peptides identified by phage display into a mammalian expression vector.
pMsx has been previously described (10, 43). pM-LX-H10, pM-LX-E5, and pM-LX-E10 were constructed by digesting the mBax plasmids, isolated using a focused phage peptide library (10), with XhoI and XbaI to release the fragments encoding the peptide sequences. The fragments were then ligated into pMsx previously digested with SalI and XbaI. The sequences of the Gal4DBD-peptide fusion proteins were verified by Perkin-Elmer dye terminator cycle sequencing.
Mammalian transfection and luciferase assays.
HeLa or HepG2 cells were maintained in modified Eagle's medium containing 10% fetal calf serum (Life Technologies, Grand Island, N.Y.). The cells were plated in 24-well plates (coated with 0.1% gelatin for HepG2 cells) 24 h prior to Lipofectin-mediated transfection as described previously (42). Cells were transfected using a total of 3 μg of DNA per well. After 3 to 5 h of incubation with a DNA-Lipofectin mixture, the cells were washed and incubated with phenol red-free medium supplemented with 10% charcoal-stripped fetal calf serum and the appropriate ligand and/or TSA treatment for 24 h. Luciferase and β-galactosidase assays were performed as described previously (42).
GST pull-downs.
Baculovirus-purified hPRA and hPRB or [35S]methionine-labeled hPRA and hPRB, synthesized using a coupled in vitro transcription and translation system as specified by the manufacturer (Promega), were incubated for 24 h at 4°C with either GST-Sepharose, GST-C'SMRT-Sepharose, GST-GRIP1-Sepharose, or GST–SRC-1(NR)–Sepharose, in NETN-A buffer (25 mM NaCl, 20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40) containing 1 μM appropriate ligand. Following incubation, the beads were washed with NENT-B buffer (50 mM NaCl, 20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40), and bound proteins were eluted in sample buffer and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The recombinant GST fusion proteins used for the in vitro pull-down experiments were produced in Escherichia coli BL21. E. coli was transformed with either pGEX2TA-C'SMRT, pGEX.1-GRIP1, pGEX-5X-1-SRC-1(NR), or pGEX-5X-1, grown to an A600 of 2.0, and induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h. The cells were then harvested, lysed by sonication, and incubated with glutathione-Sepharose beads (Pharmacia Biotech) in phosphate-buffered saline containing 0.1% Tween 20 (PBST). The beads were subsequently washed, resuspended in PBST, and used for the in vitro interaction studies.
Affinity measurements and enzyme-linked immunosorbent assay (ELISA).
Aliquots (0.02 μg) of baculovirus-expressed full-length hPRA or hPRB were diluted in 100 μl of 100 mM NaHCO3 (pH 8.5) containing 1 μM RU486, added to the wells of a 96-well Immunolon 4 plate (Dynex Technologies, Inc.), and incubated at 4°C for 24 h. An equal amount of bovine serum albumin (BSA) was used as a negative control target. The wells were then blocked with 150 μl of 5% milk plus 0.1% BSA in 100 mM NaHCO3 (pH 8.5) for 1 h at room temperature (RT). Excess protein was removed with five washes of NENT-B buffer. Increasing concentrations (0 to 12 μg) of bacterially purified GST-C'SMRT diluted in 100 μl of NENT-A containing 1 μM RU486 were added to the immobilized proteins in each well and incubated at 4°C for 24 h. The unbound protein was removed by washing five times as mentioned above. Then 100 μl of a 1:1,000 solution of anti-GST antibody was added to each well and incubated at RT for 1 h, after which the wells were washed to remove excess antibody; 100 μl of a 1:5,000 solution of horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibody was then added to the wells and incubated at RT for 1 h, followed by five washes of NENT-B. The response was detected by incubation for 30 min at RT in ABTS [2′,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid)] plus 0.05% H2O2, and the color change was measured at an optical density of 405 nm (OD405).
Purification of recombinant hPRA and hPRB.
Full-length His6-tagged human PRA and PRB were expressed in the baculovirus insect cell system and purified by affinity chromatography with nickel affinity resins (Ni-nitrilotriacetic acid; Qiagen, Chatsworth, Calif.) as previously described (7). PR was bound to R5020 during expression in Sf9 insect cells and was approximately 90% pure based on silver staining of the gel used for SDS-PAGE.
Isolation of hPRA-selective peptides.
Affinity selection of phage display peptide libraries was performed to identify peptides that could interact specifically with hPRA bound to R5020. Specifically, 0.2 μg of baculovirus expressed full-length hPRA bound to R5020 was diluted in 100 μl of 100 mM NaHCO3 (pH 8.5) containing 1 μM R5020, added to the wells of a 96-well Immunolon 4 plate (Dynex Technologies), and incubated at 4°C for 24 h. An equal amount of BSA was added to the adjacent well as control target. The wells were then blocked as mentioned above. Excess protein was removed with five washes of NENT-B buffer; 25 μl of phage peptide library (with >1010 page particles) diluted in 125 μl of PBST plus 1 μM R5020 and 0.1% BSA was added to the wells, and the plate was sealed and incubated for 8 h at RT. Nonbinding phage were removed by washing with PBST. The bound phage was eluted with 100 μl of prewarmed (50°C) 50 mM glycine-HCl (pH 2.0) followed by 100 μl of 100 mM ethanolamine (pH 11.0). The first eluent was neutralized by adding 200 μl of 200 mM Na2HPO4 (pH 8.5) and combined with the second eluent. Phage eluted from the targets were amplified in E. coli DH5αF′ cells for 5 h, and the supernatant containing amplified phage was collected for use in subsequent rounds of panning. A total of three rounds of panning were performed. Enrichment of hPRA-binding phage was confirmed by ELISA as described above. Individual phage were plaque purified after the second panning, and the peptide sequences were deduced by DNA sequencing.
RESULTS
hPRA and hPRB repress steroid hormone receptor transcriptional activity by distinct mechanisms.
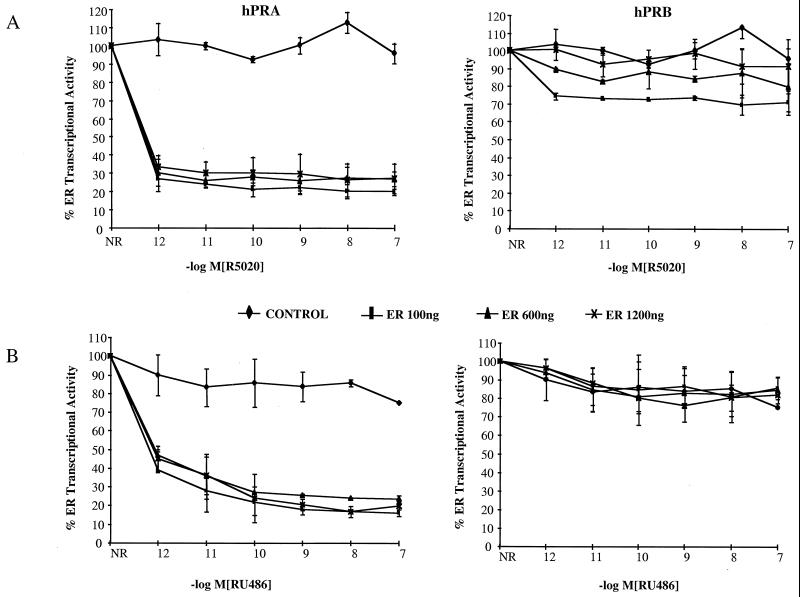
Previously, we have shown that hPRB is a transcriptional activator while hPRA functions predominantly as a repressor of progesterone-responsive promoters (18, 58, 61). The functional differences between hPRA and hPRB became even more obvious when we assessed the impact of hPRA and hPRB on ERα-mediated transcription. This was done by comparing the abilities of hPRA and hPRB to inhibit estrogen activity in cells expressing different levels of ERα. Specifically, HeLa cells were transfected with a 3XERE reporter construct and expression vector for hPRA or hPRB, along with increasing concentrations of an ERα expression vector. Transcriptional activity was measured following the addition of increasing concentrations of either the agonist R5020 (Fig. 1A) or the antagonist RU486 (Fig. 1B). As expected, given our previous findings (61), we noted that increasing the expression level of ERα did not relieve inhibition of ERα transcriptional activity by hPRA in the presence of R5020 or RU486. This indicates that hPRA-mediated repression of ERα activity occurs in a manner which appears to be independent of ERα expression level. In contrast, increasing ERα expression levels completely reversed inhibition of ERα transcriptional activity by R5020-bound hPRB (Fig. 1A). In addition, the observation that hPRB is a weak repressor of ERα activity only when bound to agonist (Fig. 1A), not antagonist (Fig. 1B), suggests that hPRB probably competes for a common coactivator required for maximal ERα activity. Together, these data, along with our previous observations, suggest that hPRB, but not hPRA, competes directly with a cofactor required for ERα transcriptional activity.
FIG. 1.
hPRA and hPRB repress SHR transcriptional activity by distinct mechanisms. HeLa cells were transiently transfected with 1.3 μg of 3XERE-TATA-LUC, 50 ng of pBKC-βgal, either 103 ng of pBKC-hPRA or 112 ng of pBKC-hPRB, and increasing concentrations of pRST7-ERα (100 ng, 600 ng, or 1,200 ng) or 100 ng of pBKC-TUP1 (CONTROL) plus 1,200 ng of pRST7-ERα. Variable amounts of pBSII-KS were used for a total of 3 μg of DNA. Transcriptional activity of the 3XERE-TATA-LUC reporter was measured 24 h after the addition of 10−8 M 17-β-estradiol alone or in the presence of increasing concentrations of R5020 (A) or RU486 (B). A control was done in the absence of ligands (not shown). The data are presented as percent activation where 100% represents a measure of 17-β-estradiol dependent transactivation by hER in the absence of hPRA or hPRB (n = 2). The average coefficient of variation at each point was <15%. NR, no progestins.
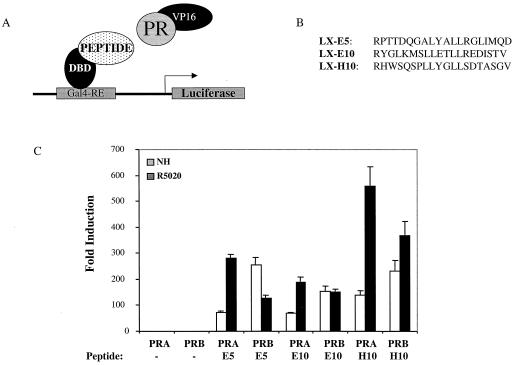
hPRA-selective peptides differentially modulate hPRA and hPRB transcriptional activity.
The finding that hPRA, but not hPRB, functioned as a transdominant inhibitor of hER signaling and the observation that hPRA was a weak activator of progesterone-responsive genes suggested that these two PR isoforms may display different cofactor preferences. We reasoned, therefore, that it might be possible to identify peptides which could competitively block PR-cofactor interactions and that when expressed in target cells these peptides would have different effects on hPRA and hPRB signaling. To date, all of the coactivators which interact with and modulate PR transcriptional activity have been shown to contain a canonical LXXLL motif. Therefore, we screened a phage display library which was created in the format X7LXXLLX7 for peptides which interacted with agonist (R5020)-activated hPRA. The peptides implicated from these screens were then subcloned into mammalian expression vectors to assess their ability to interact with either hPRA or hPRB in a mammalian two-hybrid assay (Fig. 2). Specifically, we transiently transfected HepG2 cells with a 5XGal4 reporter construct and expression vectors for either VP16-hPRA or VP16-hPRB, along with constructs expressing the receptor-interacting peptides fused onto the Gal4DBD (Fig. 2A and B). Transcriptional activity was measured in the absence or in the presence of the agonist R5020. Interestingly, we observed that while all peptides tested were capable of interacting with both receptors, subtle binding preferences for either hPRA or hPRB were observed (Fig. 2C). These data suggest that although hPRA and hPRB may adopt slightly different conformations within target cells, the coactivator binding pocket required for LXXLL binding is available on both receptors.
FIG. 2.
Interaction of hPRA-selective peptides with hPRA and hPRB in vivo. (A) Schematic of the mammalian two-hybrid assay. (B) Sequences of peptides which were fused to the Gal4DBD and used in the mammalian two-hybrid assay. These peptides were isolated by affinity selection from an X7LXXLLX7 phage display library using R5020-activated hPRA. (C) HepG2 cells were transiently transfected with 1,000 ng of 5XGal4-TATA-LUC reporter, 200 ng of pBKC-βgal, either 400 ng of VP16-hPRA, VP16-hPRB, or VP16 alone, and either 400 ng of a vector encoding Gal4DBD alone or Gal4DBD fused to a peptide; 1,000 ng of pBSII-KS was used to bring the total amount of DNA per triplicate up to 3 μg. At 24 h after transfection, cells were treated with no hormone or with 10−7 M R5020 for 24 h. Transcriptional activity was assayed on the 5XGal4-TATA-LUC reporter and represents an indirect measure of the binding of the fusion proteins. Transfections were normalized for efficiency using an internal β-galactosidase control plasmid (pBKC-βgal). The data are represented as fold induction over the no-peptide response for each condition tested, which was set to 1.0. Each data point represents the average of triplicate determinations of the transcriptional activity under the given experimental conditions (n = 3).
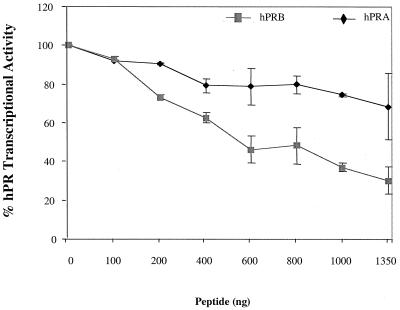
To investigate the effects of these peptides on hPRA and hPRB function, we tested the effect of expressing the LX-H10 peptide on the ability of these two receptors to activate transcription. We reasoned that if hPRA and hPRB activated target gene transcription in the same manner, overexpression of peptides which bound to the major coactivator pocket, present on both receptors, should inhibit their positive transcriptional activity. To examine this possibility, we transfected HeLa cells with a 3XPRE reporter construct along with vectors expressing either hPRA or hPRB and increasing amounts of a plasmid expressing the LX-H10 peptide. Transcriptional activity was measured following the addition of R5020 (Fig. 3). Surprisingly, we observed that although LX-H10 binds to both receptors, it differentially affects their transcriptional activity. Specifically, we found that expression of the LX-H10 peptide inhibited hPRB transcriptional activity by approximately 70% but had only a modest effect on hPRA activity (Fig. 3). A similar result was obtained with other LXXLL peptides (data not shown). We conclude from these experiments that the mechanisms by which hPRA and hPRB activate transcription are not the same and that the AF-2 coactivator binding pocket does not seem to be required for hPRA transcriptional activity. Furthermore, these data suggest that hPRA and hPRB may interact with different subsets of coactivators within target cells.
FIG. 3.
hPRA-interacting peptides differentially modulate hPRA and hPRB transcriptional activities. HeLa cells were transiently transfected with 1,500 ng of 3XPRE-TATA-LUC reporter, 50 ng of pBKC-βgal, either 50 ng of pBKC-hPRA or 50 ng of pBKC-hPRB, and increasing amounts of pM-LX-H10 (from 0 to 1,350 ng). Various amounts of pM vector were used for a total of 3,000 ng of DNA per triplicate. Transcriptional activity was assayed following the addition of 10−7 M R5020. Transfections were normalized for efficiency as mentioned above. The data are represented as percent hPR transcriptional activity where 100% represents hPR transcriptional activity in the absence of peptide. Each data point represents the average of triplicate determinations from two separate experiments.
The ID present within PR facilitates the interaction of hPRA with the corepressor SMRT.
In addition to results of our peptide studies, data from other studies suggest that the opposing activities of the two isoforms of human PR may be due to the ability of the two receptors to interact with different cofactors within the cell (Fig. 1 and 3; references 18 and 25). To determine whether hPRA and hPRB bind to different cofactors, we assessed, using both in vivo and in vitro binding assays, whether hPRA and hPRB could interact with several corepressors and coactivators which have been shown to be important for PR signaling.
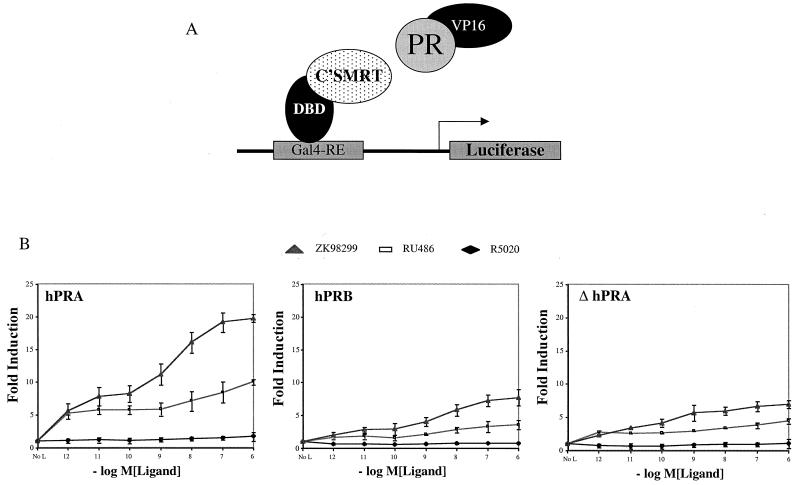
We first examined whether there is a difference between the abilities of hPRA and hPRB to interact with corepressors. This was done by performing a series of in vivo and in vitro binding studies to assess the abilities of hPRA and hPRB to interact with SMRT in the presence of different ligands. The ability of hPRA to interact in vivo with SMRT was tested using a mammalian two-hybrid system (60). Specifically, we evaluated whether full-length hPRA or hPRB, fused to the heterologous VP16 acidic activation domain, could interact with the nuclear receptor-interacting domains (NR boxes) of SMRT (C'SMRT; aa 981 to 1495) fused to the Gal4DBD (Fig. 4A). The interaction between the two isoforms of PR and C'SMRT was assayed by measuring the ability of VP16-hPRA or VP16-hPRB fusions to activate transcription from a Gal4-responsive reporter plasmid (5XGal4-TATA-LUC) with increasing concentrations of different PR ligands. Consistent with our previous report, hPRB interacted with C'SMRT (Fig. 4B) in the presence of RU486 and ZK98299 but not in the presence of the agonist R5020 or in the absence of ligand, and the interaction between hPRB and SMRT was stronger with the class II antagonist ZK98299 (60). Like hPRB, hPRA interacted with C'SMRT when bound to antagonists but not agonists. Interestingly, the interaction of hPRA with C'SMRT was ∼5-fold stronger than that of hPRB with C'SMRT. The specificity of this interaction was assessed by showing that there was no difference in the ability of hPRA and hPRB to interact with the NR box of NCoR (data not shown). These results indicate that both hPRA and hPRB associate with SMRT in the presence of PR antagonists and that antagonist-bound hPRA interacts more efficiently with the corepressor SMRT than antagonist-bound hPRB. Interestingly, ΔhPRA, the deletion mutant of hPRA lacking the ID (18), does not interact with C'SMRT as efficiently as the full-length receptor (∼5-fold) (Fig. 4B). These observations suggest that in the context of hPRA, ID facilitates binding to SMRT and that this function of ID is repressed in hPRB. The VP16-ID fusion alone does not interact with Gal4-C'SMRT (data not shown), suggesting that ID is not sufficient for the interaction of hPRA with SMRT. The differences in the interactions of the various VP16 fusion proteins were not due to differences in protein expression since all VP16 fusion constructs were shown to express at similar levels by Western immunoblot analysis (data not shown).
FIG. 4.
The ID facilitates hPRA's interaction with the corepressor SMRT. (A) Schematic of the mammalian two-hybrid assay. The receptor-interacting domain of SMRT (C'SMRT; aa 981 to 1495) was fused to the Gal4DBD (aa 1 to 147). hPRA, hPRB, or ΔhPRA was fused onto VP16 (VP16 acidic activation domain; aa 411 to 455). The fusion constructs were cotransfected into HeLa cells along with a reporter plasmid containing five copies of a Gal4-responsive element (Gal4-RE) upstream of the luciferase gene. (B) HeLa cells were transiently transfected with 0.5 μg of 5XGal4-TATA-LUC, 50 ng of pBKC-βgal, 1 μg of pCMX-Gal4-C'SMRT (Gal4-C'SMRT), 1 μg of either pVP16-hPRB, pVP16-hPRA, or pVP16-ΔhPRA, and 0.45 μg of pBSII-KS. Transcriptional activity was assayed on the 5XGal4-TATA-LUC reporter and represents an indirect measure of the binding of the fusion proteins. Transcriptional activity was measured following the addition of increasing concentrations of R5020, RU486, or ZK98299. Transfections were normalized for efficiency using an internal β-galactosidase control plasmid (pBKC-βgal). The data are represented as fold induction over the no-hormone (No L) response, which was set to 1.0. Each data point represents the average of triplicate determinations of the transcriptional activity under the given experimental conditions (n = 2).
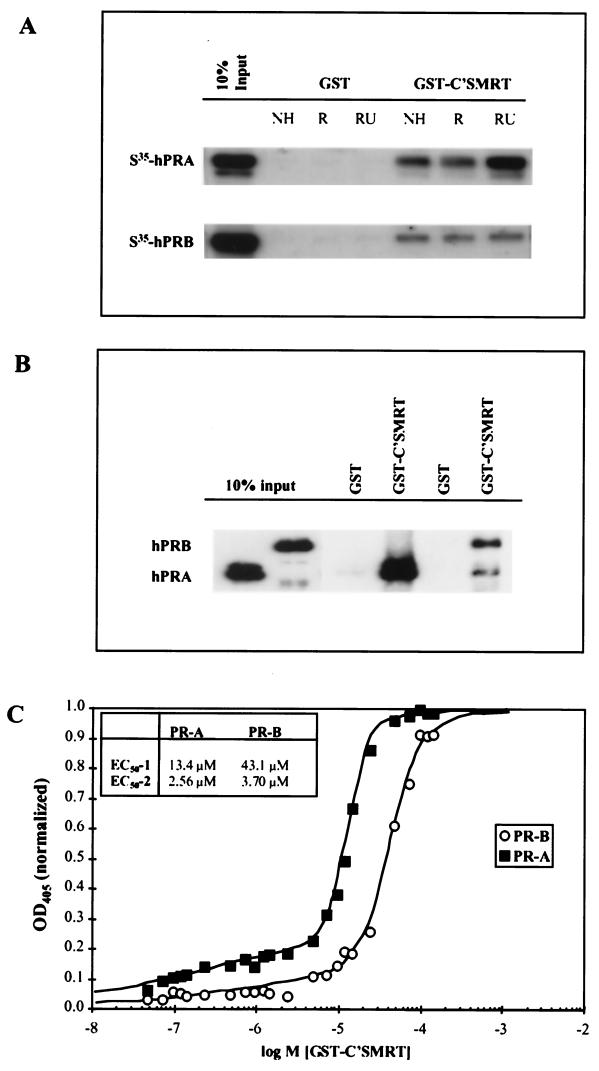
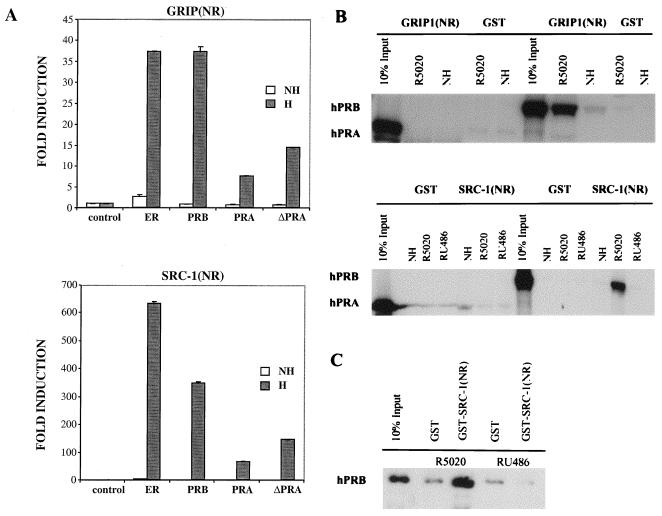
The differential interaction of hPRA and hPRB with C'SMRT was also assessed by in vitro binding analysis (Fig. 5A). Specifically, the ability of 35S-labeled hPRA or 35S-labeled hPRB to interact with either bacterially expressed GST alone or a GST-C'SMRT fusion protein was assessed. These studies revealed a specific, robust interaction between hPRA and C'SMRT in the presence of RU486. As previously reported, hPRB also interacts with C'SMRT, albeit in a ligand-independent manner (60). In agreement with the mammalian two-hybrid assay, in vitro-translated ΔhPRA did not interact efficiently with GST-C'SMRT under any ligand treatment condition (data not shown). In conclusion, these in vitro data correlate with the mammalian two-hybrid data shown in Fig. 4B.
FIG. 5.
hPRA has a higher affinity for the corepressor SMRT than hPRB. For GST pull-down assays, the fusion protein GST-C'SMRT, containing the carboxyl terminus of SMRT fused to GST, was immobilized on glutathione beads and incubated at 4°C for 24 h with either in vitro-translated hPRA or hPRB, in the presence of either vehicle (NH), R5020, or RU486 (A) or baculovirus-purified receptors bound to RU486 (B). An equimolar amount of GST was used as a negative control for each condition tested. Following incubation at 4°C, the unbound proteins were removed with five washes of NENT-B buffer. The bound receptors were subsequently visualized by Western analysis using a polyclonal antibody against PR. (C) Equal amounts of BSA and either baculovirus-purified hPRA or hPRB bound to RU486 were immobilized onto 96-well plates and incubated in the presence of increasing concentrations of bacterially purified GST-C'SMRT. Following incubation at 4°C for 24 h, the unbound fusion protein was removed by washing five times with NENT-B buffer. The amount of bound GST-C'SMRT was determined by ELISA. The response was measured at 405 nm after 30 min of incubation with ABTS plus 0.05% H2O2. The OD405 readings for hPRA and hPRB were normalized by subtracting those obtained with the BSA control and subsequently setting the highest reading value to OD405 = 1. The data were fitted to a two-site binding curve, and the values for each curve are reported. Hill-1 and Hill-2 for hPRA = 3.66 and 0.33, respectively; Hill-1 and Hill-2 for hPRB = 2.49 and 0.43 respectively. ymax-1 and ymax-2 for hPRA = 0.272 and 0.098 respectively; ymax-1 and ymax-2 for hPRB = 0.274 and 0.052, respectively.
To determine whether the interaction of hPRA and hPRB with C'SMRT was direct, we repeated the GST pull-down experiment using RU486-activated hPRA and hPRB which had been purified from baculovirus (Fig. 5B). Not surprisingly, a greater amount of hPRA than hPRB bound to GST-C'SMRT. Together these data suggest that both hPRA and hPRB bind to C'SMRT directly and that the interaction of hPRA with C'SMRT is stronger than that of hPRB with C'SMRT.
hPRA has a higher affinity for C'SMRT than hPRB.
To determine whether hPRA has a higher affinity for C'SMRT than hPRB, we used an ELISA to quantitate the binding of the receptors to C'SMRT (Fig. 5C). Equal amounts of baculovirus-purified hPRA and hPRB were immobilized onto 96-well plates and incubated with increasing concentrations of GST-C'SMRT. Any unbound fusion protein was washed away, and the amount of GST-C'SMRT bound was determined using an antibody against GST. Our binding data best fit a two-site binding curve, suggesting that the receptors contact C'SMRT at multiple points. The first binding site has 50% effective concentrations (EC50) for hPRA and hPRB of 13.4 and 43.1 μM, respectively (Fig. 5C, EC50-1) suggesting that hPRA has ∼3-fold-higher affinity for C'SMRT at this site than hPRB. This finding further supports our in vivo binding data (Fig. 4B). In addition to site 1, hPRA and hPRB contact C'SMRT on a second site which has lower capacity but higher binding affinity than site 1 (Fig. 5C, EC50-2). Determination of whether this is in fact the case requires further experimentation.
The dominant negative variant of SMRT, C'SMRT, can partially reverse hPRA-mediated transrepression of hER transcriptional activity.
Recently, we showed that removal of ID from the A isoform of PR causes hPRA to lose its ability to transrepress heterologous steroid hormone receptor transcriptional activity (18). This observation, together with the mammalian two-hybrid data (Fig. 4B), suggests that ID, in the context of hPRA, allows human PR to acquire a conformation that is optimal for corepressor binding and/or ID is one of the corepressor binding sites present in PR. These observations suggest that the ability of hPRA to transrepress ERα-mediated activity may involve the corepressor SMRT.
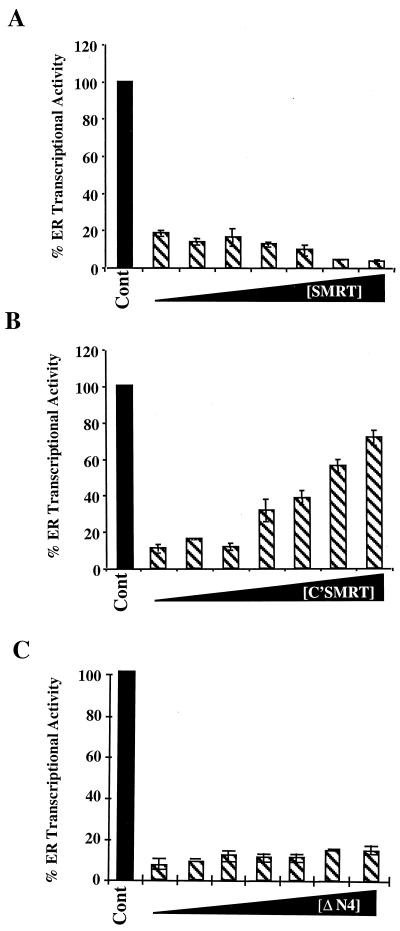
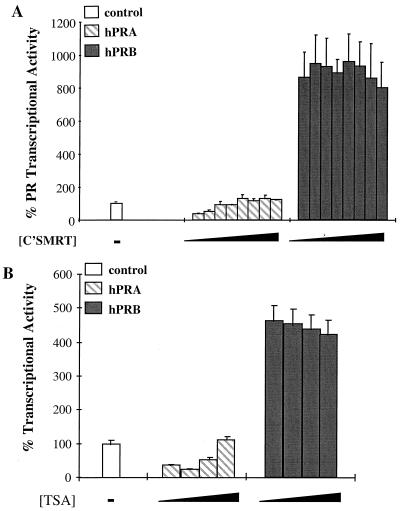
To test whether SMRT is involved in hPRA-mediated transrepression of hER transcriptional activity, we studied the effect of overexpressing the dominant negative C'SMRT on hPRA-mediated transrepression of hER transcriptional activity. This was accomplished by transiently transfecting HeLa cells with expression vectors for hER and hPRA and the 3XERE-TATA-LUC reporter construct in the presence of increasing amounts of either Gal4-SMRT, Gal4-C'SMRT, or Gal4-ΔN4 (Fig. 6). In this experiment, increasing amounts of Gal4-ΔN4 had no effect on hPRA-mediated transrepression in the presence of 10−7 M RU486 (Fig. 6C). Interestingly, C'SMRT reversed hPRA-mediated transrepression of ERα activity in a dose-dependent manner (from 15 to 77% ERα transcriptional activity). Increasing amounts of Gal4-SMRT further enhanced the ability of hPRA to repress ERα transcriptional activity (from 20 to 5% ERα transcriptional activity) (Fig. 6A), directly implicating SMRT in hPRA-mediated transrepression. Increasing amounts of Gal4-C'SMRT had no effect on estradiol-mediated ERα transcriptional activity in the absence of hPRA (data not shown).
FIG. 6.
C'SMRT can partially reverse hPRA-mediated repression of hER transcriptional activity. HeLa cells were transiently transfected with 1 μg of 3XERE-TATA-LUC, 50 ng of pBKC-βgal, 450 ng of pRST7-ERα, 300 ng of pBKC-hPRA, and increasing concentrations (ranging from 0 to 1.2 μg) of either Gal4-SMRT (A), Gal4-C'SMRT (B), or ΔN4, used as a control (C). Various amounts of pBKC-DBD were added to balance the amount of input Gal4DBD. Transcriptional activity of the 3XERE-TATA-LUC reporter was measured 24 h after the addition of 10−7 M 17-β-estradiol and 10−7 M RU486. A control was done in the absence of ligands (not shown). The data are presented as percent activation where 100% represents a measure of 17-β-estradiol-dependent transactivation by hER in the absence of RU486 (Cont). The average coefficient of variation at each point was <12% (n = 2).
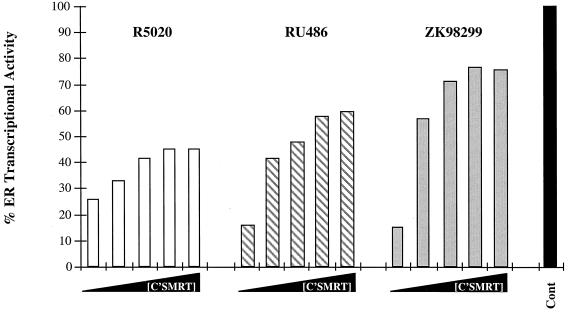
To test the effects of various PR ligands on the ability of C'SMRT to reverse hPRA-mediated transrepression, we transfected HeLa cells as described above and induced them with 10−7 M estradiol alone or with either 10−7 M R5020, 10−7 M RU486, or 10−7 M ZK98299 (Fig. 7). In the presence of R5020, C'SMRT reversed hPRA-mediated transrepression of ERα activity in a dose-dependent manner (from 25 to 46% ERα transcriptional activity). Reversal of hPRA-mediated transrepression by C'SMRT in the presence of the class I antagonist RU486 was from 15 to 62% of ERα transcriptional activity. Interestingly, in the presence of class II antagonist ZK98299, C'SMRT reversed hPRA-mediated transrepression of hER activity from 16% to about 80% (Fig. 7). These results suggest that C'SMRT is better at reversing hPRA-mediated transrepression when the receptor is bound to antagonists rather than to agonists. This observation correlates with the mammalian two-hybrid data that indicate that the interaction of hPRA with Gal4-C'SMRT is greater in the presence of antiprogestins (Fig. 4B).
FIG. 7.
The ability of C'SMRT to reverse hPRA-mediated transrepression is ligand dependent. HeLa cells were transiently transfected as for Fig. 4. Transcriptional activity was measured 24 h after the addition of 10−7 M 17-β-estradiol and either 10−7 M R5020, 10−7 M RU486, or 10−7 M ZK98299. A control was done in the absence of ligands (not shown). The data are presented as percent activation where 100% represents a measure of 17-β-estradiol-dependent transactivation by hER in the absence of progestins or antiprogestins (Cont) for each experimental condition. The average coefficient of variation at each point was <12%. The data from a single representative experiment are shown (n = 3).
The deacetylase inhibitor TSA partially reverses hPRA-mediated transrepression of hER transcriptional activity.
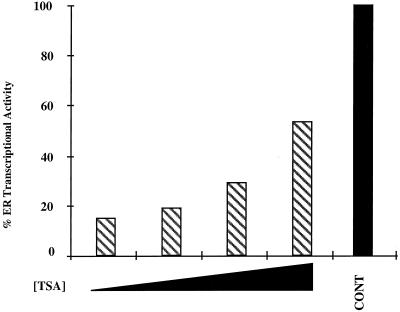
The transcriptional silencers NCoR and SMRT have been shown to exist in a complex with the repressor mSin3 and the histone deacetylase HD-1 (also known as HDAC1), suggesting that corepressors mediate gene repression by acting as bridging factors between the receptor and histone deacetylases, thus recruiting the latter to the receptor-DNA complex (1, 22, 41). To assess whether histone deacetylases play a role in hPRA-mediated transrepression of ERα transcriptional activity, we examined whether the deacetylase inhibitor TSA could reverse hPRA-mediated transrepression (Fig. 8). We transiently transfected HeLa cells with expression constructs for hER and either hPRA or a control plasmid together with the 3XERE-TATA-LUC reporter construct in the presence of 10−7 M estradiol and 10−7 M RU486 alone or together with increasing concentrations of TSA. Estradiol-dependent activation of the 3XERE-TATA promoter in HeLa cells expressing hER together with a control plasmid was not significantly affected by coaddition of increasing concentrations of TSA (data not shown). In this experiment, TSA is capable of partially reversing hPRA-mediated transrepression of ERα activity in a dose-dependent manner. In conclusion, the experiments detailed above suggest that the strong interaction of hPRA with the SMRT corepressor complex might be responsible for the inability of hPRA to activate transcription, as well as its ability to act as a potent transrepressor of heterologous steroid hormone receptor transcriptional activity.
FIG. 8.
The deacetylase inhibitor TSA can partially reverse hPRA-mediated repression of hER transcriptional activity. HeLa cells were transiently transfected with 1.5 μg of 3XERE-TATA-LUC, 50 ng of pBKC-βgal, 500 ng of pRST7-ERα, and either 481 ng of pBKC-hPRA or 467 ng of pBKC-Rev-TUP1 (not shown). Variable amounts of pBSII-KS were used for a total of 3 μg of DNA. Transcriptional activity of the 3XERE-TATA-LUC reporter was measured 24 h after the addition of 10−7 M 17-β-estradiol and 10−7 M RU486, alone or in combination with increasing concentrations of TSA (0, 10−8, 10−7, and 10−6 M). A control was done in the absence of ligands (not shown). The data are presented as percent activation where 100% represents a measure of 17-β-estradiol-dependent transactivation by hER in the absence of RU486 (CONT). The data from one representative experiment are shown (n = 2). The average coefficient of variation at each point was <10%.
Inactivation of the nuclear receptor silencer SMRT does not convert hPRA to a transcriptional activator.
The findings detailed above indicate that hPRA forms a strong association with the corepressor SMRT, implying that hPRA recruits a repressor complex, composed of SMRT and histone deacetylases, to the promoters of target genes, thereby repressing transcription of target genes. Furthermore, the association of hPRA with the SMRT corepressor complex seems to play an important role in transrepression by hPRA (Fig. 6). To test whether a complex of SMRT and histone deacetylases with hPRA was also responsible for the inability of hPRA to activate transcription, we studied the effect of (i) overexpression of C'SMRT (Fig. 9A) and (ii) increasing concentrations of TSA (Fig. 9B) on hPRA-mediated transcription. Specifically, we transfected HeLa cells with an expression vector for hPRA or hPRB, alone or in the presence of increasing concentrations of C'SMRT, together with the 2XPRE-TK-LUC reporter construct, and induced them with 10−7 M R5020 (Fig. 9A). A control transfection, to assess the basal transcriptional activity of the reporter in the absence of receptors, was included, and the value was set to 100%. Clearly, hPRB-mediated transcriptional activity in the presence of ligand was not significantly affected by increasing C'SMRT concentrations. In contrast, while increasing concentrations of C'SMRT completely reversed hPRA-mediated repression of the basal promoter, they failed to activate hPRA transcription beyond the basal level of the reporter (Fig. 9A).
FIG. 9.
Inactivation of the nuclear receptor silencer SMRT does not convert hPRA to a transcriptional activator. (A) HeLa cells were transiently transfected with 1.5 μg of 2XPRE-TK-LUC, 50 ng of pBKC-βgal, either 52 ng of pBKC-hPRB, 48 ng of pBKC-hPRA, or 46 ng of pBKC-RevTUP1, and increasing concentrations (from 0 to 1 μg) of Gal4-C'SMRT. Various amounts of pBKC-DBD were added to balance the amount of input Gal4DBD. pBSK-II was added to normalize the total DNA to 3 μg. The transcriptional activity of these vectors was assayed on a 2XPRE-TK-LUC reporter and measured after the addition of 10−7 M R5020. Transfections were normalized for efficiency as mentioned previously. R5020-mediated transcriptional activity in the presence of increasing concentrations C'SMRT was normalized to the no-ligand control for each concentration of C'SMRT used. Each data point represents the average of triplicate determinations (± standard error of the mean) from two separate experiments (n = 2). The control represents basal reporter activity in the presence of control vector and was set to 100%. (B) HeLa cells were transiently transfected with 1.5 μg of 2XPRE-TK-LUC, 50 ng of pBKC-βgal, either 50 ng of pBKC-hPRA or 48 ng of pBKC-Rev-TUP1, and various amounts of pBSK-II for a total of 3 μg. Transcriptional activity of the constructs was measured following the addition of 10−7 M R5020 alone or in combination with increasing concentrations (0, 10−8, 10−7, and 10−6 M) of the deacetylase inhibitor TSA. Transfections were normalized for efficiency as mentioned above. R5020-mediated transcriptional activity in the presence of increasing concentrations of TSA was normalized to the no-ligand control for each TSA treatment used. Each data point represents the average of triplicate determinations (± standard error of the mean) from two separate experiments (n = 2).
To test whether histone deacetylases were also involved in hPRA repression of progesterone-responsive promoters, we transiently transfected HeLa cells with an expression vector for hPRA or hPRB together with the 2XPRE-TK-LUC reporter construct and induced them with 10−7 M R5020 alone or with increasing concentrations of TSA (Fig. 9B). hPRB-mediated transcriptional activity in the presence of ligand was not affected by increasing TSA concentrations. The basal activity of the 2XPRE-TK promoter was repressed (63%) by agonist-activated hPRA, as observed in Fig. 9A. Increasing concentrations of TSA reversed hPRA-mediated repression of basal activity in a dose-dependent manner. The increase in basal activity upon TSA treatment suggests that histone deacetylases are recruited to progesterone-responsive promoters by hPRA. Not surprisingly, even at the highest concentration of TSA used, hPRA was unable to activate transcription from the 2XPRE-TK promoter above the inherent basal level (Fig. 9B). Together, these findings suggest that inhibition of corepressor function is not sufficient to convert hPRA to a transcriptional activator. In addition, however, they demonstrate that agonist-activated hPRA can suppress basal transcription by possibly recruiting the SMRT repressor complex to the promoters of target genes.
hPRB, but not hPRA, interacts efficiently with the NR boxes of the coactivator proteins GRIP1 and SRC-1.
It follows, then, that one possible explanation for why agonist-bound hPRA fails to activate transcription is that unlike hPRB, hPRA fails to effectively recruit coactivators. Therefore, hPRB's ability to associate with coactivator proteins and displace corepressors results in an increase in PR transcriptional activity. Conversely, we propose that even when bound to agonist, hPRA fails to efficiently recruit coactivators and thus is unable to displace corepressors.
To analyze the ability of hPRA and hPRB to interact with coactivator proteins, we used the mammalian two-hybrid system. Specifically, we looked at the ability of full-length hPRA or hPRB fused to the heterologous VP16 acidic activation domain to interact with the NR box of either GRIP1 [GRIP1(NR)] or SRC-1 [SRC-1(NR)] fused to the Gal4DBD (Fig. 10A). Interaction between the two isoforms of PR and GRIP1(NR) and SRC-1(NR) respectively, was assayed by measuring the ability of VP16-hPRA, VP16-hPRB, and VP16-ΔhPRA fusions to activate transcription from a Gal4-responsive reporter plasmid (5XGal4-TATA-LUC) in the presence of R5020. VP16-ERα fusion protein was used as a positive control. As expected, ERα interacts with both GRIP1(NR) and SRC-1(NR) in the presence of estradiol. We also observed a slight interaction of ERα with the coactivators in the absence of hormone. This interaction disappeared with the addition of antagonist (data not shown). hPRB interacts with both SRC-1(NR) and GRIP1(NR) in the presence of R5020 (625- and 37-fold over control). Importantly, R5020-bound hPRA forms a weaker interaction with both SRC-1(NR) and GRIP1(NR) (50- and 8-fold over control) compared to hPRB. The deletion mutant of hPRA, lacking ID, which we previously showed to function as a transcriptional activator of progesterone-responsive promoters (18), formed a slightly stronger interaction with GRIP1(NR) and SRC-1(NR) compared to wild-type hPRA. The interaction of ΔhPRA with the coactivators was not as strong as that of hPRB with the coactivators, suggesting that the unique amino terminus of hPRB plays an important role in receptor-coactivator interaction. The mammalian two-hybrid data suggest that the ability of hPRB and hPRA to interact with coactivators correlates with the transcriptional activity of both receptors.
FIG. 10.
hPRA interacts weakly with the NR boxes of the coactivator proteins GRIP1 and SRC-1. (A) HeLa cells were transiently transfected with 0.5 μg of 5XGal4-TATA-LUC, 50 ng of pBKC-βgal, 1 μg of either pM-GRIP1(NR) or pM-SRC-1(NR), 1 μg of either pVP16-T, pVP16-ERα, pVP16-hPRB, pVP16-hPRA, or pVP16-ΔhPRA, and 0.45 μg of pBSII-KS. Transcriptional activity of the luciferase gene was assayed on the 5XGal4-TATA-LUC reporter as in Fig. 2B. Transcriptional activity was measured following the addition of 10−7 M R5020 for PR or 10−7 M 17-β-estradiol for ERα (H); a control was done in the absence of ligands (NH). Transfections were normalized for efficiency as mentioned previously. The data are represented as fold induction over the control interaction between Gal4-GRIP1(NR) or Gal4–SRC-1(NR) and VP16-T for each ligand treatment group, which was normalized to 1.0. Each data point represents the average of triplicate determinations from three separate experiments. The average coefficient of variation at each point was <10% (n = 3). (B and C) GST pull-down assays. The fusion proteins GST-GRIP1(NR) (top) and GST-SRC-1(NR) (bottom) were immobilized on glutathione beads and incubated at 4°C for 24 h with in vitro-translated 35S-hPRA or 35S-hPRB in the presence of vehicle (NH), R5020, or RU486 (B) or baculovirus-purified hPRB bound to R5020 or RU486 (C). The bound baculovirus-purified receptors were analyzed by Western analysis using a polyclonal antibody against PR. An equimolar amount of GST alone was used as a negative control for each condition tested.
The ability of hPRA and hPRB to interact with SRC-1(NR) and GRIP1(NR) was also assessed in vitro using a GST pull-down assay (Fig. 10B). Specifically, we determined the ability of 35S-labeled hPRA or 35S-labeled hPRB to interact with either bacterially expressed GST alone, GST-GRIP1(NR), or GST-SRC-1(NR). These assays revealed a specific interaction between hPRB and both GRIP1(NR) and SRC-1(NR) in the presence of R5020 but not in the presence of RU486 or in the absence of ligands. Interestingly, under the same conditions, hPRA did not interact with GRIP1(NR) and SRC-1(NR), suggesting that additional factors, which are not present in the crude extracts, may be needed to account for the ability of hPRA to interact with the coactivators in vivo (Fig. 10A). To determine whether the association of hPRB with the coactivators was direct, we carried out a GST pull-down assay using GST–SRC-1(NR) incubated with baculovirus-purified receptors in the presence of R5020 or RU486 (Fig. 10C). This analysis revealed that hPRB interacts directly with SRC-1(NR) in the presence of the agonist R5020 but not the antagonist RU486 (Fig. 10C). hPR-A and SRC-1(NR) did not interact under the same conditions (data not shown). Together, these data indicate that hPRA and hPRB form different interactions with SRC-1(NR) and GRIP1(NR), implying that the failure of hPRA to activate transcription may be due to its inability to efficiently recruit coactivators as well as to its inherent higher affinity for corepressor proteins (Fig. 4 and 5).
hPRA is not targeted to the ERα coactivator complex.
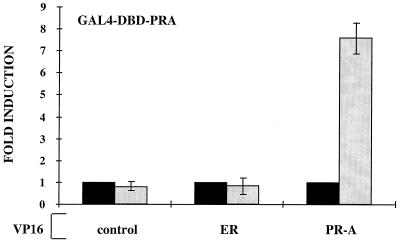
While the inability of hPRA to form productive interactions with coactivators may explain why hPRA is transcriptionally inactive, the inherent higher affinity of hPRA, but not hPRB, for SMRT may be important for hPRA-mediated inhibition of ERα transcriptional activity (Fig. 6). To further elucidate the mechanism of hPRA inhibition of ERα activity, we used both in vivo and in vitro binding assays to determine whether hPRA is targeted to the ERα transcription complex, thereby interfering with the ability of ERα to activate transcription. Specifically, we assessed the ability of VP16-ERα to bind to Gal4-hPRA in the presence of R5020 and estradiol in a mammalian two-hybrid system (Fig. 11). VP16-hPRA was used as a positive control in this assay. We tested for potential protein-protein interactions by assessing the ability of Gal4-PRA to recruit VP16-ERα or VP16-hPRA receptor fusions to DNA, using a mammalian two-hybrid assay. The results of this analysis indicated that hPRA-hPRA homodimers were capable of activating the Gal4-responsive promoter in the presence of R5020. No association between hPRA and ERα was observed under the same conditions (Fig. 11), suggesting that ERα is not targeted to hPRA in vivo. While it is possible that hPRA and ERα do exist in a complex within cells, it is possible that the mammalian two-hybrid assay is unable to detect such associations. Another possibility is that ERα needs to be bound to DNA in order to recruit hPRA. We are currently in the process of assessing these possibilities. In conclusion, we are currently unable to demonstrate a direct association of hPRA with ERα, suggesting that hPRA interferes with ERα signaling in another manner, possibly by preventing the association of ERα with a required factor.
FIG. 11.
hPRA does not associate with the ERα transcription complex. HeLa cells were transiently transfected with 500 ng of 5XGal4-TATA-LUC, 50 ng of pBKC-βgal, 1,000 ng of pM-hPRA, and either 1,000 ng of pVP16-T (control), pVP16-hPRA, or pVP16-ERα (gray bars). Transcriptional activity was assayed on the 5XGal4-TATA-LUC reporter following the addition of 10−7 M 17-β-estradiol or 10−7 M R5020 and represents an indirect measure of the binding of the fusion proteins. Transfections were normalized for efficiency as mentioned above. The data are represented as fold induction over the control interaction between Gal4-hPRA and VP16-T in the absence of ligands, which was normalized to 1.0 (black bars). Each data point represents the average of triplicate determinations from three separate experiments.
DISCUSSION
Recent insights into the mechanism of steroid hormone action have advanced our understanding of PRA action significantly and suggested how the two forms of PR, hPRA and hPRB, manifest their unique regulatory activities in target cells. Specifically, it has been demonstrated that nuclear hormone receptors, upon binding their cognate ligands, undergo distinct conformational changes. This event permits the dissociation of the receptors from corepressor complexes, possessing histone deacetylase activity and facilitates their interaction with coactivator complexes, which display histone acetylase activity. As a consequence, the DNA-bound receptor is able to positively regulate target gene transcription (19, 23, 48, 50, 55, 60, 62). In support of this model, it has been shown that the ability of nuclear receptors to repress target gene transcription correlates with their ability to bind to the corepressors NCoR and SMRT (21, 60). Conversely, transcriptional activation by nuclear hormone receptors was observed to correlate with the recruitment of coactivators to the promoter region of target genes (19, 24, 29, 43, 49). To determine whether the opposing transcriptional activities of hPRA and hPRB were due to differential cofactor association, we examined the abilities of hPRA and hPRB to interact with different coactivators and corepressors and assessed the effects of these associations on the receptors' transcriptional activities. Using both in vivo and in vitro methodologies, we found that antagonist-bound hPRA has a higher affinity for the NR box of SMRT (C'SMRT) than antagonist-bound hPRB (Fig. 4 and 5). This interaction appears to be physiologically relevant since overexpression of C'SMRT (a dominant negative SMRT) effectively reverses hPRA-mediated transrepression of ERα transcriptional activity. In addition, overexpression of SMRT enhanced the ability of hPRA to inhibit hER-mediated transcriptional activity. Significantly, we also observed that unlike hPRB, hPRA did not associate efficiently with the coactivators SRC-1 and GRIP1. Thus, the robust interaction of hPRA with SMRT together with its inability to efficiently engage coactivators appears to explain why hPRA is a repressor of progesterone-responsive promoters.
Initially, it was proposed that the differences in the transcriptional activities of hPRA and hPRB were due to a third activation function, AF-3, present within the extreme amino terminus of hPRB, a region that is absent in hPRA (47). Thus, it was considered that functional synergy between the activation functions located in the amino terminus (AF-3 and AF-1) and the carboxyl terminus (AF-2) was required for maximal hPRB transcriptional activity. However, unlike AF-1 and AF-2, AF-3 does not demonstrate autonomous activity when fused to a heterologous DBD (40, 47), suggesting that instead of functioning as a classical AF, AF-3 might be required for proper AF-1 and AF-2 transcriptional activity. For instance AF-3 may contribute to hPRB transcriptional activity directly, by enhancing the activity of AF-1 or AF-2, or indirectly, by suppressing an inhibitory function contained within sequences common to both hPRA and hPRB (18, 30). Evidence in support of the latter hypothesis came from our studies, as well as those of others, which identified an ID within the amino terminus of hPRA which, when deleted, resulted in a receptor mutant functionally indistinguishable from hPRB (18, 25, 26). Specifically, it was demonstrated that the first 140 aa of hPRA are necessary for its ability to function as a transcriptional inhibitor as well as a transrepressor of heterologous steroid receptor transcriptional activity (18). Thus, one role of AF-3 is to override the function of the ID present within the amino terminus of the receptor, allowing hPRB to activate transcription (18, 25).
In addition to hPR, several other transcription factors have been shown to contain both activation and repression functions (2, 3, 8, 15, 17, 21, 34). Of particular relevance to our studies of hPRA, it has been shown in vitro that the ability of RORα to repress transcription correlates with the ability of the inhibitory domain within RORα to recruit the corepressors NCoR and SMRT (21). In addition, RORα was shown to preferentially associate with NCoR and not SMRT in vivo. When we tested the ability of hPRA and hPRB to interact with NCoR and SMRT in the presence of antagonist, we found that while both receptors associate with NCoR, hPRA has a higher affinity for SMRT than hPRB (Fig. 5C). Furthermore, a deletion mutant lacking the inhibitory domain, ΔhPRA, does not interact efficiently with SMRT. This implies that like the case for RORα, a specific domain within hPRA is required for corepressor interactions.
The ability of agonist-activated nuclear receptors to activate transcription correlates with their ability to displace corepressors and engage coactivators (reviewed in reference 55). Not surprisingly, therefore, we were able to show in this study that agonist-bound hPRB, but not hPRA, can form a productive interaction with coactivators, thus allowing hPRB to activate transcription from progesterone-responsive promoters. This suggested that hPRA may be unable to completely dissociate from corepressors and thus may not be able to recruit coactivators. However, the fact that R5020-bound hPRA was unable to activate transcription in cells expressing a dominant negative variant of SMRT, or in the presence of the histone deacetylase inhibitor TSA, suggests that dissociation from corepressors is not sufficient for hPRA to activate transcription (Fig. 8). This observation, together with the mammalian two-hybrid data, implies that agonist-bound hPRA, unlike hPRB, does not efficiently recruit coactivators. It appears, therefore, that the unique sequences present at the amino terminus of hPRB are required for proper transcriptional activation.
In most cell and promoter contexts, the transcriptional activity of steroid hormone receptors appears to require the functional synergy between the amino and carboxyl termini of each individual receptor (6, 39, 40, 46, 53, 57). This synergy occurs as a consequence of an agonist-dependent association between the amino and carboxyl AFs of ERα (31), the androgen receptor (5, 14, 27), and hPRA and hPRB, respectively (54). Interestingly, in the case of hPRA and hPRB, the amino terminus of hPRB containing AF-3 was shown to interact more efficiently with the carboxyl terminus of the receptor than the amino terminus of hPRA lacking AF-3 (54). This agonist-dependent interaction was enhanced by the addition of SRC-1 and CBP, while dominant negative variants of SRC-1 and CBP completely abolished this interaction, suggesting that these coactivators may be required for transcriptional synergy between the amino-terminal and carboxyl-terminal AFs of the receptor (54). Interestingly, a role for coactivators as bridging factors between the amino and carboxyl AFs of receptors is supported by the observation that SRC-1 can interact with both the amino and carboxyl termini of PR (44).
Previously, we have shown that the agonist-dependent interaction of the PR carboxyl terminus with the amino terminus of hPRB is more robust than that with the amino terminus of hPRA, an activity which mirrors their activity as transcriptional activators (18, 25, 26, 38, 56, 58, 60). Thus, the ability of hPRB to function as an activator of transcription could be due to the fact that hPRB, but not hPRA, undergoes a conformational change which is conducive to coactivator binding. The ability of hPRA and hPRB to adopt different conformations within the cell is also supported by our peptide analysis (Fig. 2 and 3). The peptide competition data presented in Fig. 3 also suggest that the two receptors are bound to different cellular factors which may, in turn, explain their distinct functions within the cell. For example, hPRB, unlike hPRA, is likely to be associated with AF-2-type coactivators. It is not surprising, then, that the LX-H10 peptide, which contains an LXXLL motif common to these coactivators, was an efficient inhibitor of hPRB activity but had no effect on hPRA activity when overexpressed in cells along with the receptors (Fig. 3). This hypothesis is further supported by additional findings which show that agonist-bound hPRB, but not agonist-bound hPRA, directly interacts with the NR boxes of the coactivators GRIP1 and SRC-1 in vitro (Fig. 10B). Although our studies focused on the ability of hPRA to interact specifically with the previously defined NR boxes of SRC-1 and GRIP1, it has been reported previously that both hPRA and hPRB interact with full-length SRC-1A in the presence of agonist (44). However, in these latter studies it was not determined whether hPRA and hPRB can bind directly to the sequences of SRC-1 used in this study. In addition, the SRC-1(NR) protein used in our studies did not contain the fourth LXXLL motif found in SRC-1A. Together, these observations suggest that the two PR isoforms do not interact in the same manner with SRC-1.
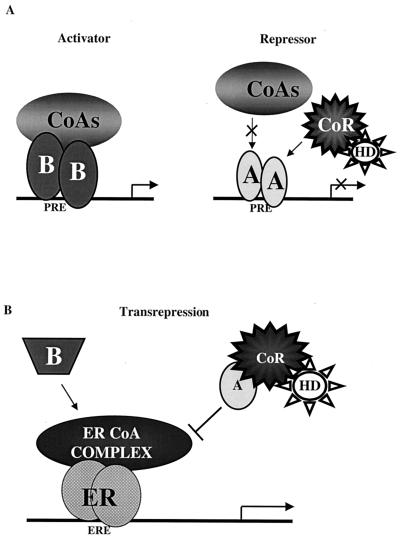
Our working model to explain the opposing transcriptional activities of hPRA and hPRB is depicted in Fig. 12A. We propose that hPRB is a transcriptional activator of progesterone-responsive promoters, since upon binding hormone, hPRB undergoes a conformational change which allows it to dissociate from corepressor proteins and recruit coactivators. This productive interaction with the coactivators allows the receptor to activate transcription from the promoters of target genes. Conversely, under the same conditions, hPRA is transcriptionally inactive because, unlike hPRB, it does not effectively recruit coactivators to the promoters of target genes. Thus, the inability of hPRA to activate target gene transcription does not appear to be related to its ability to associate with corepressors such as SMRT. However, our data reveal a central role for SMRT in hPRA-mediated repression of ERα transcriptional activity. Thus, we propose that the hPRA-SMRT complex blocks estrogen action by interfering with the assembly or function of the ERα-coactivator complex.
FIG. 12.
Two distinct models are required to describe the molecular mechanism of action of hPRA. (A) Transcriptional activation. Based on the in vivo and the in vitro binding studies, we propose that hPRA interacts more efficiently with corepressors and less efficiently with coactivators than hPRB. In the presence of hormone, hPRB, but not hPRA, undergoes a favorable conformational change which allows it to displace corepressors (CoR) and recruit coactivator proteins (CoA), thus allowing hPRB to activate transcription from progesterone-responsive promoters. HD, histone deacetylase; A, hPRA; B, hPRB. (B) Transrepression. Based on our in vivo transrepression data, we propose that hPRA transrepresses ERα-mediated transcription by a transcriptional interference mechanism. In this model, ERα activates transcription by recruiting a complex of coactivator proteins (ERα CoA complex) to the regulatory region of target genes. hPRA (A), but not hPRB (B), targets and sequesters a member of the ERα CoA complex, thus preventing ERα from activating transcription. hPRA transrepression of ERα transcriptional activity is further enhanced by the recruitment by hPRA of the corepressor SMRT (CoR).
Whereas the results of these studies explain why hPRB acts as a strong transcriptional activator of progesterone-responsive promoters and why hPRA is transcriptionally inactive in these contexts, it remains to be determined how the hPRA-SMRT complex can transrepress ERα transcriptional activity. We believe that agonist-bound hPRB can interfere with ERα transcriptional activity by squelching a required coactivator protein (i.e., p160 family of coactivators). It does not appear, however, that hPRAs transrepression function involves a direct competition between hPRA and ERα for coactivators. It may well be that hPRA inhibits the activity of a cofactor required for ERα action by binding directly at a site distinct from the ERα-interacting site or indirectly by binding to other proteins within the ERα-coactivator complex. Distinguishing between these possibilities is the subject of our current investigations.
ACKNOWLEDGMENTS
We thank N. Weigel, J. D. Chen, D. D. Moore, B. O'Malley, M. Stallcup, and X.-F. Wang for providing plasmids and reagents. We also acknowledge A. Vandongen, I. Tcherepanova-Freedman, C.-Y. Chang, X. Li, J. D. Norris (Department of Pharmacology and Cancer Biology, DUMC, Durham, N.C.), and James McNamara II (Department of Neurobiology, DUMC). Lori Sherman (Department of Pathology, UCHSC) provided technical assistance in the purification of the recombinant progesterone receptors.
This research was supported in part by National Institutes of Health grants DK50495 (D.P.M.) and DK49030 (D.P.E.), NCI Cancer Center core grant P30 CA46934 (D.P.E.), and predoctoral fellowships from USAMRMC (P.H.G. and E.A.K.).
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for NCoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Baichwal V R, Park A, Tjian R. The cell-type specific activator region of c-Jun juxtaposes constitutive and negatively regulated domains. Genes Dev. 1992;6:1493–1502. doi: 10.1101/gad.6.8.1493. [DOI] [PubMed] [Google Scholar]
- 3.Baichwal V R, Park A, Tjian R. Control of c-Jun activity by interaction of a cell-specific inhibitor with regulatory domain delta: differences between v- and c-Jun. Cell. 1990;63:815–825. doi: 10.1016/0092-8674(90)90147-7. [DOI] [PubMed] [Google Scholar]
- 4.Beato M, Arnemann G, Chalepakis E S, Willmann T. Gene regulation by steroid hormones. J Steroid Biochem. 1987;27:9–14. doi: 10.1016/0022-4731(87)90288-3. [DOI] [PubMed] [Google Scholar]
- 5.Berrevoets C A, Doesburg P, Steketee K, Trapman J, Brinkmann A O. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor2) Mol Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 6.Bocquel M T, Kumar V, Stricker C, Chambon P, Gronemeyer H. The contribution of the N- and C-terminal regions of steroid receptors to activation of transcription is both receptor and cell specific. Nucleic Acids Res. 1989;17:2581–2595. doi: 10.1093/nar/17.7.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi M E, Taraseviciene L, Nordeen S K, Allegretto E A, Edwards D P. High-mobility-group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown H J, Sutherland A, Cook A, Bannister A J, Kouzarides T. An inhibitor domain in c-Fos regulates activation domains containing the HOB1 motif. EMBO J. 1995;14:124–131. doi: 10.1002/j.1460-2075.1995.tb06982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalbous D, Galtier F. Differential effect of forms A and B of human progesterone receptor estradiol-dependent transcription. J Biol Chem. 1994;269:23007–23012. [PubMed] [Google Scholar]
- 10.Chang C-Y, Norris J D, Gron H, Paige L A, Hamilton P T, Kenan D J, Fowlkes D, McDonnell D P. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors α and β. Mol Cell Biol. 1999;19:8226–8239. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen K, Estes P A, Onate S A, Beck C A, DeMarzo A, Altmann M, Lieberman B A, St. John J, Nordeen S K, Edwards D P. Characterization and functional properties of the A and B forms of human progesterone receptors synthesized in a baculovirus system. Mol Endocrinol. 1991;5:1755–1770. doi: 10.1210/mend-5-11-1755. [DOI] [PubMed] [Google Scholar]
- 12.Clemm D L, Sherman L, Boonyaratanakornkit V, Schrader W T, Weigel N L, Edwards D P. Differential hormone-dependent phosphorylation of progesterone receptor A and B forms revealed by a phosphoserine site-specific monoclonal antibody. Mol Endocrinol. 2000;14:52–65. doi: 10.1210/mend.14.1.0413. [DOI] [PubMed] [Google Scholar]
- 13.Dana S L, Hoener P A, Wheeler D A, Lawrence C B, McDonnell D P. Novel estrogen response elements identified by genetic selection in yeast are differentially responsive to estrogens and antiestrogens in mammalian cells. Mol Endocrinol. 1994;8:1193–1207. doi: 10.1210/mend.8.9.7838152. [DOI] [PubMed] [Google Scholar]
- 14.Doesburg P, Kuil C W, Berrevoets C A, Steketee K, Faber P W, Mulder E, Brinkmann A O, Trapman J. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36:1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 15.Dubendorff J W, Whittaker L J, Eltman J T, Lipsick J S. Carboxyl-terminal elements of cMyb negatively regulate transcriptional activation in cis and in trans. Genes Dev. 1992;6:2524–2535. doi: 10.1101/gad.6.12b.2524. [DOI] [PubMed] [Google Scholar]
- 16.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedl E M, Matthias P. Mapping of the transcriptional repression domain of the lymphoid specific transcription factor oct-2A. Eur J Biochem. 1995;234:308–316. doi: 10.1111/j.1432-1033.1995.308_c.x. [DOI] [PubMed] [Google Scholar]
- 18.Giangrande P H, Pollio G, McDonnell D P. Mapping and characterization of the functional domains responsible for the differential activity of the A and B isoforms of the human progesterone receptor. J Biol Chem. 1997;272:32889–32900. doi: 10.1074/jbc.272.52.32889. [DOI] [PubMed] [Google Scholar]
- 19.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 20.Gronemeyer H. Transcription activation by estrogen and progesterone receptors. Annu Rev Genet. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- 21.Harding H P, Atkins G B, Jaffe A B, Seo W J, Lazar M A. Transcriptional activation and repression by RORα and orphan nuclear receptor required for cerebellar development. Mol Endocrinol. 1997;11:1737–1746. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- 22.Heinzel T, Lavinski R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie G R, Seto E, Eisenman R N, W. R D, Glass C K, Rosenfeld M G. A complex containing NCoR, mSin3, and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 23.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 25.Hovland A R, Powel R L, Takimoto G S, Tung L, Horwitz K B. An N-terminal inhibitory function, IF, suppresses transcription by the A-isoform, but not the B-isoform, of human progesterone receptors. J Biol Chem. 1998;273:5455–5460. doi: 10.1074/jbc.273.10.5455. [DOI] [PubMed] [Google Scholar]
- 26.Huse B, Verca S B, Matthey P, Rusconi S. Definition of a negative modulation domain in the human progesterone receptor. Mol Endocrinol. 1998;12:1334–1342. doi: 10.1210/mend.12.9.0164. [DOI] [PubMed] [Google Scholar]
- 27.Ikonen T, Palvimo J J, Janne O A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 28.Ing N, Beekman J, Tsai S, Tsai M-J, O'Malley B. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 29.Jenster G, Spencer T E, Burcin M M, Tsai S Y, Tsai M-J, O'Malley B W. Steroid receptor induction of gene transcription: a two step model. Proc Natl Acad Sci USA. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraus W L, Weis K E, Katzenellenbogen B S. Inhibitory cross-talk between steroid hormone receptors: differential targeting of estrogen receptor in the repression of its transcriptional activity by agonist- and antagonist-occupied progestin receptors. Mol Cell Biol. 1995;15:1847–1857. doi: 10.1128/mcb.15.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonhardt S A, Altmann M, Edwards D P. Agonist and antagonists induce homodimerization and mixed ligand heterodimerization of human progesterone receptors in vivo by a mammalian two-hybrid assay. Mol Endocrinol. 1998;12:1914–1930. doi: 10.1210/mend.12.12.0210. [DOI] [PubMed] [Google Scholar]
- 33.Lessey B A, Alexander P S, Horwitz K B. The subunit characterization of human breast cancer progesterone receptors: characterization by chromatography and photoaffinity labelling. Endocrinology. 1983;112:1267–1274. doi: 10.1210/endo-112-4-1267. [DOI] [PubMed] [Google Scholar]
- 34.Li X Y, Green M R. Intracmolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes Dev. 1996;10:517–527. doi: 10.1101/gad.10.5.517. [DOI] [PubMed] [Google Scholar]
- 35.MacGregor G R, Caskey C T. Construction of plasmids that express E. coli β-galactosidase in mammalian cells. Nucleic Acids Res. 1989;17:2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonnell D P, Clemm D L, Hermann T, Goldman M E, Pike J W. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- 37.McDonnell D P, Clemm D L, Imhof M O. Definition of the cellular mechanisms which distinguish between hormone and antihormone activated steroid receptors. Semin Cancer Biol. 1994;5:327–336. [PubMed] [Google Scholar]
- 38.McDonnell D P, Goldman M E. RU486 exerts antiestrogenic activities through a novel progesterone receptor A form-mediated mechanism. J Biol Chem. 1994;269:11945–11949. [PubMed] [Google Scholar]
- 39.McDonnell D P, Vegeto E, O'Malley B W. Identification of a negative regulatory function for steroid receptors. Proc Natl Acad Sci USA. 1992;89:10563–10567. doi: 10.1073/pnas.89.22.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer M-E, Quirin-Stricker C, Lerouge T, Bocquel M-T, Gronemeyer H. A limiting factor mediates the differential activation of promoters by the human progesterone receptor isoforms. J Biol Chem. 1992;267:10882–10887. [PubMed] [Google Scholar]
- 41.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 42.Norris J, Fan D, Aleman C, Marks J R, Futreal P A, Wiseman R W, Iglehart J D, Deininger P L, McDonnell D P. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- 43.Norris J D, Paige L A, Christensen D J, Chang C-Y, Huacani M R, Fan D, Hamilton P T, Fowlkes D M, McDonnell D P. Peptide antagonists of the human estrogen receptor. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- 44.Onate S A, Boonyaratanakonrnkit V, Spencer T E, Tsai S Y, Tsai M J, Edwards D P, O'Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of the steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 45.Onate S A, Tsai S, Tsai M-J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 46.Pham T A, Hwung Y P, Santiso-Mere D, McDonnell D P, O'Malley B W. Ligand-dependent and -independent function of the transactivation regions of the human estrogen receptor in yeast. Mol Endocrinol. 1992;6:1043–1050. doi: 10.1210/mend.6.7.1508220. [DOI] [PubMed] [Google Scholar]
- 47.Sartorius C A, Melville M Y, Hovland A R, Tung L, Takimoto G S, Horwitz K B. A third transactivation function (AF3) of human progesterone receptors located in the unique N-terminal segment of the B-isoform. Mol Endocrinol. 1994;8:1347–1360. doi: 10.1210/mend.8.10.7854352. [DOI] [PubMed] [Google Scholar]
- 48.Seol W, Mahon M J, Lee Y-K, Moore D D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/NCoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 49.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O'Malley B W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 50.Smith C L, Nawaz Z, O'Malley B W. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 51.Smith C L, Onate S A, Tsai M J, O'Malley B W. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith D F, Faber L E, Toft D O. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J Biol Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- 53.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 54.Tetel M J, Giangrande P H, Leonhardt S A, McDonnell D P, Edwards D P. Hormone dependent interaction between the amino- and carboxyl-terminal domains of the progesterone receptor in vitro and in vivo. Mol Endocrinol. 1999;13:910–924. doi: 10.1210/mend.13.6.0300. [DOI] [PubMed] [Google Scholar]
- 55.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 56.Tung L, Mohamed M K, Hoeffler J P, Takimoto G S, Horwitz K B. Antagonist-occupied human progesterone B-receptors activate transcription without binding to progesterone response elements and are dominantly inhibited by A-receptors. Mol Endocrinol. 1993;7:1256–1265. doi: 10.1210/mend.7.10.8123133. [DOI] [PubMed] [Google Scholar]
- 57.Tzukerman M T, Esty A, Santiso-Mere D, Danielian P, Parker M G, Stein R B, Pike J W, McDonnell D P. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- 58.Vegeto E, Shahbaz M M, Wen D X, Goldman M E, O'Malley B W, McDonnell D P. Human progesterone receptor A form is a cell- and promoter-specific repressor of human progesterone receptor B function. Mol Endocrinol. 1993;7:1244–1255. doi: 10.1210/mend.7.10.8264658. [DOI] [PubMed] [Google Scholar]
- 59.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner B L, Norris J D, Knotts T A, Weigel N L, McDonnell D P. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol Cell Biol. 1998;18:1369–1378. doi: 10.1128/mcb.18.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wen D X, Xu Y F, Mais D E, Goldman M E, McDonnell D P. The A and B isoforms of the human progesterone receptor operate through distinct signaling pathways within target cells. Mol Cell Biol. 1994;14:8356–8364. doi: 10.1128/mcb.14.12.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M G, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]