Abstract
Plant somatic cells can be reprogrammed into a pluripotent cell mass, called callus, which can be subsequently used for de novo shoot regeneration through a two-step in vitro tissue culture method. MET1-dependent CG methylation has been implicated in plant regeneration in Arabidopsis, because the met1-3 mutant exhibits increased shoot regeneration compared with the wild-type. To understand the role of MET1 in de novo shoot regeneration, we compared the genome-wide DNA methylomes and transcriptomes of wild-type and met1-3 callus and leaf. The CG methylation patterns were largely unchanged during leaf-to-callus transition, suggesting that the altered regeneration phenotype of met1-3 was caused by the constitutively hypomethylated genes, independent of the tissue type. In particular, MET1-dependent CG methylation was observed at the blue light receptor genes, CRYPTOCHROME 1 (CRY1) and CRY2, which reduced their expression. Coexpression network analysis revealed that the CRY1 gene was closely linked to cytokinin signaling genes. Consistently, functional enrichment analysis of differentially expressed genes in met1-3 showed that gene ontology terms related to light and hormone signaling were overrepresented. Overall, our findings indicate that MET1-dependent repression of light and cytokinin signaling influences plant regeneration capacity and shoot identity establishment.
Keywords: Arabidopsis, callus, cryptochrome 1, cytokinin, DNA methylation, MET1, shoot regeneration
INTRODUCTION
Plant somatic cells can be reprogrammed to form an unorganized pluripotent cell mass, called callus. Incubation on callus-inducing medium (CIM) activates cell proliferation, facilitating callus formation. Accumulating evidence shows that callus tissue resembles root primordium, regardless of the origin of tissue explants (Atta et al., 2009; Sugimoto et al., 2010). Consistently, callus formation is initiated from pericycle-like cells (Sugimoto et al., 2010). The founder cell undergoes asymmetric cell division and then enables the acquisition of root primordium identity (Dubrovsky et al., 2000; Sugimoto et al., 2010), with the activation of genes including WUSCHEL-RELATED HOMEOBOX 11 (WOX11) (Liu et al., 2014) and LATERAL ORGAN BOUNDARIES DOMAINs (LBDs) (Feng et al., 2012). Then, callus cells establish regeneration competence via the expression of root stem cell regulator genes, including PLETHORA 1 (PLT1), PLT2, SHORT-ROOT (SHR), SCARECROW (SCR), and WOX5 (Kareem et al., 2015; Sugimoto et al., 2010). After the acquisition of pluripotency, shoot regeneration can be triggered by incubating the callus on cytokinin-rich shoot-inducing medium (SIM). The cytokinin-inducible type-B ARABIDOPSIS RESPONSE REGULATOR (ARR)–WUSCHEL (WUS) module plays a crucial role in de novo shoot organogenesis from callus (Meng et al., 2017; Zhang et al., 2017). Shoot-specific physiological processes, including light signaling, also promote de novo shoot regeneration (Nameth et al., 2013).
Chemical modifications of DNA or core histone proteins alter chromatin structure, contributing to gene expression regulation, independent of the changes in DNA sequence. Methylation of the fifth carbon of cytosine residue is the most extensively studied epigenetic modification in both plants and mammals (Kim et al., 2021; Yoo et al., 2021). DNA methylation usually represses gene transcription (Chen et al., 2008; Jeddeloh et al., 1998; Zilberman et al., 2007), although increasing evidence shows that DNA methylation can also activate gene expression (Baubec et al., 2013; Brackertz et al., 2002; Fujita et al., 2003; Fukushige et al., 2006; Harris et al., 2018; Lang et al., 2017; Waterfield et al., 2014; Zemach and Grafi, 2003). The Arabidopsis thaliana genome is selectively methylated in CG, CHG, and CHH (H = A, T, or C) contexts. DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) catalyzes de novo methylation of cytosine residues in all sequence contexts (CG, CHG, and CHH) through the RNA-directed DNA methylation (RdDM) pathway (Cao and Jacobsen, 2002a). CG methylation is maintained by METHYLTRANSFERASE 1 (MET1) (Zubko et al., 2012), whereas the maintenance of CHG methylation requires CHROMOMETHYLASE 3 (CMT3) (Bartee et al., 2001; Lindroth et al., 2001). Maintenance of asymmetric CHH methylation is ensured by CMT2, DRM1, and DRM2 (Cao and Jacobsen, 2002b). In addition, cytosine methylation can be reversibly removed by the DNA glycosylase/lyase mechanism. In Arabidopsis, REPRESSOR OF SILENCING 1 (ROS1), DEMETER (DME), DME-LIKE 2 (DML2), and DML3 proteins facilitate DNA base excision repair (BER) as an active demethylation mechanism (Ortega-Galisteo et al., 2008; Penterman et al., 2007).
DNA methylation is closely associated with plant regeneration. For example, MET1-dependent DNA methylation negatively controls the expression of core shoot regeneration regulator genes, including WUS, and represses de novo shoot organogenesis on CIM (Li et al., 2011; Liu et al., 2018). The met1 mutant displays enhanced WUS expression and thereby higher rates of shoot regeneration without incubation on CIM. Moreover, spatiotemporal expression of MET1 is delicately regulated via the dual mode of action of cytokinin on SIM. At the early stage of shoot induction, MET1 expression in calli is induced by the cytokinin–CYCD3–E2FA module, repressing WUS expression. With increasing incubation time on SIM, MET1 expression is restricted to the outer cell layers of the callus, whereas WUS expression is activated by type-B ARRs in cell layers beneath the MET1-expressing regions (Liu et al., 2018).
In this study, we conducted whole-genome bisulfite sequencing (BS-seq) of the met1-3 mutant and wild-type, and compared the changes in CG methylation landscape between the two genotypes during callus formation to understand the role of MET1 in plant regeneration. Notably, CG methylation was largely unchanged during leaf-to-callus transition, regardless of genotypes. Thus, the enhanced shoot regeneration phenotype of met1-3 was caused by the constitutively hypomethylated genes. We particularly focused on MET1-dependent CG methylation at the blue light receptor loci, CRYPTOCHROME 1 (CRY1) and CRY2. MET1-dependent CG methylation repressed the expression of both CRY genes. CRY1 subsequently regulated cytokinin signaling, especially type-B ARR genes. Given that shoot regeneration requires cytokinin signaling, enhanced cytokinin signaling possibly led to enhanced de novo shoot regeneration in met1-3. Overall, our results suggest that MET1-dependent CG methylation negatively regulates light and cytokinin signaling and influences plant regeneration.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis met1-3 (Johnson et al., 2007), cry1-1 (Koornneef et al., 1980), and cry2/fha-1 (Koornneef et al., 1991) mutants have been described previously. Seeds were germinated on Murashige and Skoog (MS) medium at 22°C-23°C under a long-day photoperiod (16 h light/8 h dark) with fluorescent light (150 µmol photons/m2s). The third and fourth leaves of two-week-old seedlings were used as explants to induce callus on CIM (B5 medium supplemented with 0.5 µg/ml 2,4-dichlorophenoxyacetic acid [2,4-D] and 0.05 µg/ml kinetin). The plates were incubated at 22°C under continuous dark for 2 weeks (Fan et al., 2012). To induce de novo shoot regeneration, leaf explant-derived callus preincubated on CIM for 7 days was transferred to SIM (B5 medium supplemented with 0.9 µmol/L indole-3-acetic acid and 2.5 µmol/L 2-isopentenyladenine). The plates were incubated at 25°C under continuous light for up to 3 weeks to examine the shoot regeneration capacity of the callus.
Whole genome BS-seq
BS-seq libraries were constructed as described previously (Shim et al., 2021). Callus samples are heterogeneous and exhibit significant variation in gene expression; therefore, a large amount of sample (>1 g) was used to perform high-depth BS-seq (>70× coverage) for single biological replicate. The third and fourth leaves of in vitro-cultured 2-week-old seedlings were used for immediately harvesting leaf explants and for inducing callus on CIM. Using the cetyltrimethylammonium bromide (CTAB) method, genomic DNA was extracted from leaf explants and leaf explant-derived calli incubated on CIM for 2 weeks. Then, 5 µg of each genomic DNA sample was fragmented by Covaris shearing (Covaris, USA). Blunt-ended and phosphorylated fragments were adenylated at the 3'-ends, and ligated to a methylated adapter using the TruSeq DNA Library Preparation Kit (Illumina, USA). Then, ligation products were separated by agarose gel electrophoresis, and 275-350 bp products were purified. The purified fragments were bisulfite-treated using the EpiTect Bisulfite Conversion Kit (Qiagen, Germany), according to the manufacturer’s instructions. Bisulfite conversion was performed in a thermal cycler under the following conditions: (1) denaturation at 95°C for 5 min, (2) incubation at 60°C for 25 min, (3) denaturation at 95°C for 5 min, (4) incubation at 60°C for 85 min, (5) denaturation at 95°C for 5 min, (6) incubation at 60°C for 175 min, and (7) hold at 20°C. Bisulfite-converted DNA samples were purified twice using 20 µl of elution buffer included in the EpiTect kit. The bisulfite-treated fragments were amplified by polymerase chain reaction (PCR) using a primer cocktail included in the TruSeq kit to generate products with adaptors on both ends. The final products were used for constructing a BS-seq library, which was sequenced on the HiSeq2000 platform (Illumina).
BS-seq data analysis and differentially methylated region (DMR) identification
Raw BS-seq reads were analyzed as described previously (Smallwood et al., 2014), with slight modifications. Briefly, according to the Bismark Bisulfite Mapper guidelines (https://rawgit.com/FelixKrueger/Bismark/master/Docs/Bismark_User_Guide.html), the first 8 bp of raw BS-seq reads were trimmed using TrimGalore (parameters: --gzip --paired --clip_R1 8 --clip_R2 8) to prevent adaptor contamination (Krueger and Andrews, 2011). The trimmed reads were initially aligned to the TAIR10 version of the Arabidopsis reference genome sequence (https://www.arabidopsis.org/) using Bismark (Krueger and Andrews, 2011) and Bowtie2 (Langmead and Salzberg, 2012), with default parameters, according to the guideline of Bismark Bisulfite Mapper. Then, PCR duplicates in initial alignment files were purged using the deduplicate_bismark script. Bisulfite treatment was validated by calculating the fraction of unmethylated cytosine residues (C to T conversion rate > 99.0%) among the total number of mapped cytosines in the chloroplast genome. The average bisulfite conversion rate was greater than 98.4%, indicating successful bisulfite treatment. The mapped read statistics are summarized in Supplementary Table S1. Considering the cumulative number of cytosines in all examined samples, individual methyl-cytosines with more than five supporting reads were selected for further analysis (Supplementary Fig. S1).
To identify DMRs between met1-3 and wild-type genotypes, the Bismark output files were converted into bedGraph format using the bismark2bedGraph script, with –CX and –zero_based options. The output bedGraph files of met1-3 and wild-type leaf and callus samples were used as input for DMRfinder (Gaspar and Hart, 2017), and methylated regions for each methylation context were defined using the following criteria: maximum methylated region length, 500 bp; minimum number of methylcytosines, 3; maximum distance between methylcytosines, 100 bp; and minimum total read count > 20. Regions with significant difference in methylation levels (P < 0.05), i.e., more than 40%, 20%, and 10% absolute difference in the methylation of CG, CHG, and CHH contexts, respectively, between met1-3 and wild-type (for both leaf and callus tissues) were identified as DMRs, as reported previously (Bhatia et al., 2018; Chen et al., 2018; Liang et al., 2019; Liu et al., 2016; Stassen et al., 2018; Zhou et al., 2019).
To determine whether DMRs overlapped with genic regions (defined as the region encompassing the gene body and 1 kb sequence upstream of the transcription start site) and transposable element (TE) regions, the BS-seq data were investigated using BEDTools (Quinlan and Hall, 2010), based on the TAIR10 reference genome annotation. Genome-wide patterns of DNA methylation were visualized using Pandas, NumPy, SciPy, and pyplot libraries of Python. The cytosine conversion rate in genic regions was depicted using deepTools (Ramírez et al., 2014).
Functional enrichment and coexpression network analyses
To analyze the enriched biological functions of differentially methylated genes, the MapMan annotation was used for comparing the observed ratio of genes of interest (GOIs) in a selected gene group with the expected ratio of genes in the reference genome for a specific pathway through the hypergeometric test using a homemade Python script (Usadel et al., 2009). Moreover, to construct a coexpression network, the GOIs were inputted as queries into the NetworkDrawer implemented in ATTED-II (https://atted.jp) (Obayashi et al., 2018).
RNA-seq analysis
To analyze the impact of DNA methylation on gene expression in the wild-type and met1-3 mutant, the RNA-seq data of wild-type leaf explants generated previously (Lee et al., 2016) were used in this study. Additionally, high-depth RNA-seq analysis of met1-3 was performed in the current study using a large amount of leaf explants (>1 g) for single biological replicate.
Quantitative real-time reverse transcription PCR (RT-qPCR) analysis
Total RNA was extracted from met1-3 and wild-type leaf explants and callus tissues using the TRI reagent (TAKARA Bio, Japan), according to the manufacturer’s recommendations, and treated with an RNase-free DNase to eliminate genomic DNA contamination. The first-strand cDNA was synthesized from 2 μg of total RNA by reverse transcription using Moloney Murine Leukemia Virus reverse transcriptase (Dr. Protein, Korea) with oligo(dT18) primers. RT-qPCR experiments were performed on the Step-One Plus Real-Time PCR System (Applied Biosystems, USA). The comparative CT method was used to determine relative gene expression using the EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (eIF4A) gene (At3g13920) as an internal control. All RT-qPCR reactions were performed using three independent replicate samples. The specificity of RT-qPCR results was determined by melt curve analysis of the amplified products using the standard method.
Data availability
Whole genome BS-seq and RNA-seq data are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) data under the BioProject accession number PRJNA601842.
RESULTS
Enhanced shoot regeneration in met1-3
MET1 is involved in plant regeneration from root and pistil tissues (Li et al., 2011; Liu et al., 2018). To further analyze the functional impact of MET1 in plant regeneration, we performed the two-step in vitro plant regeneration process using leaf explants. The third and fourth leaves of 2-week-old seedlings were excised and placed on CIM. Upon callus emergence, callus tissues were transferred to SIM in order to induce shoot regeneration.
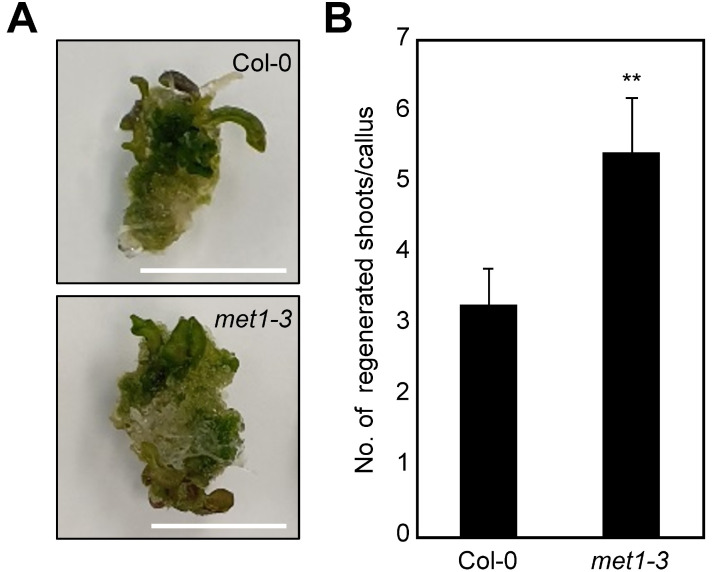
Plant regeneration capacity was determined by monitoring de novo shoot organogenesis from calli on SIM. The met1-3 mutant calli displayed greater shoot regeneration than wild-type calli (Fig. 1). These results suggest that MET1 possibly regulates de novo shoot organogenesis-related processes and affects plant regeneration.
Fig. 1. Increased de novo shoot regeneration efficiency in the met1-3 mutant.
(A) Shoot regeneration phenotypes. (B) Number of regenerated shoots. In (A) and (B), calli preincubated for 7 days on CIM were transferred to SIM. The number of regenerated shoots from calli was measured (n > 30) at 3 weeks after incubation on SIM. Error bars indicate the SEM. Statistically significant differences between wild-type and mutant calli are indicated by asterisks (Student’s t-test, **P < 0.01). Scale bars = 10 mm.
Global changes in DNA methylation in met1-3
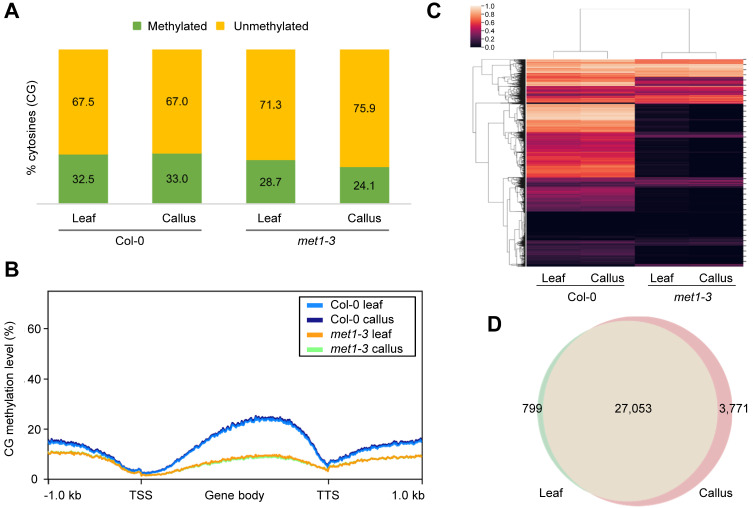
To understand the molecular basis of enhanced de novo shoot regeneration in met1-3, we conducted whole-genome BS-seq using met1-3 and wild-type leaf explants and leaf explant-derived calli (Fig. 2A, Supplementary Table S1, Supplementary Fig. S1). Because MET1 plays a major role in maintaining CG methylation in Arabidopsis (Zubko et al., 2012), we particularly focused on CG methylation in this study. The BS-seq results showed that CG methylation was extensively diminished in met1-3, regardless of the tissue type (Fig. 2B), whereas global CHG and CHH methylation levels were rather increased in met1-3 compared with the wild-type (Supplementary Fig. S2). Most DMRs in met1-3 were also concentrated in the CG context (Supplementary Fig. S2). Furthermore, during leaf-to-callus transition, CHG and CHH methylation was significantly changed particularly at TE regions in wild type (Supplementary Fig. S2B), consistent with a previous study showing that dynamic changes in CHG and CHH methylation levels in TE regions affect plant regeneration (Shim et al., 2021). However, the changes observed in CHG and CHH methylation levels during callus formation were maintained in met1-3 mutant (Supplementary Fig. S2), suggesting that the role of MET1 in plant regeneration is related primarily to CG methylation, independent of CHG and CHH methylation.
Fig. 2. Marginal changes in CG methylation during leaf-to-callus transition.
(A) Fractions of methylated and unmethylated cytosines in the CG context in wild-type and met1-3. (B) Average CG methylation levels over genic regions in wild-type and met1-3. TSS, transcription start site; TTS, transcription termination site. (C) Hierarchical clustering of genome-wide CG methylation patterns. The color bar indicates the methylation level. (D) Venn diagram of regions hypomethylated in each tissue of met1-3 relative to that of the wild-type.
Notably, the CG methylation level showed a negligible change during leaf-to-callus transition both in the wild-type and met1-3 mutant (Fig. 2B). Hierarchical clustering analysis also showed that CG methylation patterns for the same genotype were similar in both calli and leaves (Fig. 2C), consistent with the minor changes in CG methylation observed previously during plant growth and development (Ingouff et al., 2017; Ito et al., 2019; Saze et al., 2003). Because CG methylation levels were mostly reduced in met1-3 (Fig. 2C), we collected CG regions hypomethylated in met1-3 relative to the wild-type. The results showed that 27,852 and 30,824 genomic regions were hypomethylated in leaf (Supplementary Table S2) and callus (Supplementary Table S3) tissues, respectively, in met1-3. Notably, most of these regions (27,053) were commonly hypomethylated in both leaf (97.1%; 27,053 of 27,852) and callus (87.8%; 27,053 of 30,824) tissues (Fig. 2D), indicating that CG methylation is not dynamically reprogrammed during the course of callus formation.
Given that most of the hypomethylated CG-DMRs were found in genic regions (Supplementary Fig. S2), we collected genes containing hypomethylated CG regions in met1-3 within genic regions (Supplementary Tables S4 and S5). GO enrichment analysis revealed that MET1-dependent CG methylation regulates a variety of biological processes, and most of these processes were commonly enriched in both leaf and callus tissues (Supplementary Tables S6-S8). In particular, hypomethylated genes in met1-3 were enriched for GO terms related to light signaling (external stimuli response for red/far-red light [P value = 1.27 × 10–2 and 2.26 × 10–2 for leaf and callus tissues, respectively]) (Supplementary Table S6). Overall, these results suggest that the increased shoot regeneration efficiency of the met1-3 mutant is attributable to its constitutively hypomethylated regions, which are possibly associated with light signaling.
MET1-mediated CG methylation at the CRY1 and CRY2 loci
To further understand the biological processes regulated by MET1, we conducted RNA-seq analysis and identified genes differentially expressed between met1-3 and the wild-type. Because CG methylation levels did not change during callus formation in both genotypes, we used only leaf tissues to examine which genes were misregulated in met1-3 prior to callus formation. A total of 3,935 differentially expressed genes (DEGs) were identified. Among these, 1,769 genes were up-regulated and 2,166 genes were down-regulated in the met1-3 mutant compared with the wild-type (Supplementary Table S9).
GO enrichment analysis of the DEGs showed that GO terms related to light signaling and responses were significantly enriched for light signaling and responses (external stimuli response for UV-A/blue light [P value = 7.46 × 10–3] and UV-B light [P value = 2.41 × 10–2]) (Table 1), which is consistent with the fact that MET1-dependent CG-DMRs were enriched in genes involved in light signaling (Supplementary Table S6). In addition, it was notable that auxin (phytohormone action for auxin biosynthesis [P value = 1.41 × 10–2]) and cytokinin-related processes (phytohormone action for cytokinin perception and signal transduction [P value = 9.61 × 10–3] and cytokinin transport [P value = 2.09 × 10–2]) were also overrepresented (Table 1). These results suggest that DNA methylation-dependent regulation of light signaling ultimately affects hormone signaling, which may account for the altered plant regeneration phenotype of the met1-3 mutant (Fig. 1).
Table 1.
Functional enrichment analysis of DEGs in met1-3 relative to the wild-type
| Functional categorya | No. of query genes per category | Total No. of query genes | Observed value (%) | No. of reference genes per category | Total No. of reference genes | Expected value (%) | P value |
|---|---|---|---|---|---|---|---|
| RNA biosynthesis (transcriptional regulation) | 442 | 3,936 | 11.2 | 1,962 | 27,206 | 7.2 | 1.87E-23 |
| Enzyme classification (EC_2 transferases) | 100 | 3,936 | 2.5 | 382 | 27,206 | 1.4 | 1.22E-09 |
| Enzyme classification (EC_1 oxidoreductases) | 87 | 3,936 | 2.2 | 420 | 27,206 | 1.5 | 2.94E-04 |
| Solute transport (carrier-mediated transport) | 148 | 3,936 | 3.8 | 784 | 27,206 | 2.9 | 3.42E-04 |
| External stimuli response (pathogen) | 53 | 3,936 | 1.3 | 233 | 27,206 | 0.9 | 4.55E-04 |
| Vesicle trafficking (clathrin-independent machinery) | 3 | 3,936 | 0.1 | 3 | 27,206 | 0.0 | 3.03E-03 |
| Carbohydrate metabolism (fermentation) | 4 | 3,936 | 0.1 | 6 | 27,206 | 0.0 | 5.14E-03 |
| Carbohydrate metabolism (oligosaccharide metabolism) | 6 | 3,936 | 0.2 | 13 | 27,206 | 0.0 | 6.27E-03 |
| Phytohormone action (ethylene-biosynthesis) | 7 | 3,936 | 0.2 | 17 | 27,206 | 0.1 | 6.75E-03 |
| External stimuli response (light-UV-A/blue light) | 10 | 3,936 | 0.3 | 30 | 27,206 | 0.1 | 7.46E-03 |
| Protein modification (phosphorylation) | 195 | 3,936 | 5.0 | 1,148 | 27,206 | 4.2 | 8.34E-03 |
| Lipid metabolism (lipid bodies-associated activities) | 12 | 3,936 | 0.3 | 40 | 27,206 | 0.1 | 8.99E-03 |
| Cell wall organization (pectin) | 39 | 3,936 | 1.0 | 185 | 27,206 | 0.7 | 9.17E-03 |
| Phytohormone action (cytokinin-perception and signal transduction) | 10 | 3,936 | 0.3 | 31 | 27,206 | 0.1 | 9.61E-03 |
| Protein modification (S-glutathionylation) | 19 | 3,936 | 0.5 | 76 | 27,206 | 0.3 | 1.06E-02 |
| Phytohormone action (abscisic acid-conjugation and degradation) | 5 | 3,936 | 0.1 | 11 | 27,206 | 0.0 | 1.36E-02 |
| Phytohormone action (auxin-biosynthesis) | 6 | 3,936 | 0.2 | 15 | 27,206 | 0.1 | 1.41E-02 |
| External stimuli response (gravity) | 6 | 3,936 | 0.2 | 15 | 27,206 | 0.1 | 1.41E-02 |
| Multi-process regulation (programmed cell death [PCD] system) | 9 | 3,936 | 0.2 | 29 | 27,206 | 0.1 | 1.79E-02 |
| DNA damage response (photoreactivation) | 2 | 3,936 | 0.1 | 2 | 27,206 | 0.0 | 2.09E-02 |
| Phytohormone action (cytokinin-transport) | 2 | 3,936 | 0.1 | 2 | 27,206 | 0.0 | 2.09E-02 |
| Phytohormone action (signaling peptides) | 43 | 3,936 | 1.1 | 219 | 27,206 | 0.8 | 2.16E-02 |
| Phytohormone action (abscisic acid-transport) | 3 | 3,936 | 0.1 | 5 | 27,206 | 0.0 | 2.41E-02 |
| External stimuli response (light-UV-B light) | 3 | 3,936 | 0.1 | 5 | 27,206 | 0.0 | 2.41E-02 |
| Solute transport (channels) | 35 | 3,936 | 0.9 | 180 | 27,206 | 0.7 | 4.00E-02 |
| Phytohormone action (gibberellin-biosynthesis) | 5 | 3,936 | 0.1 | 14 | 27,206 | 0.1 | 4.07E-02 |
| Multi-process regulation (phosphatidylethanolamine-binding [PEB] protein-dependent signaling) | 3 | 3,936 | 0.1 | 6 | 27,206 | 0.0 | 4.30E-02 |
| Phytohormone action (salicylic acid-conjugation and degradation) | 3 | 3,936 | 0.1 | 6 | 27,206 | 0.0 | 4.30E-02 |
| Phytohormone action (gibberellin-perception and signal transduction) | 4 | 3,936 | 0.1 | 10 | 27,206 | 0.0 | 4.44E-02 |
| Multi-process regulation (SnRK1-kinase regulatory system) | 8 | 3,936 | 0.2 | 29 | 27,206 | 0.1 | 4.88E-02 |
Significantly overrepresented terms (P < 0.05) are presented. Statistical significance was determined by the hypergeometric test.
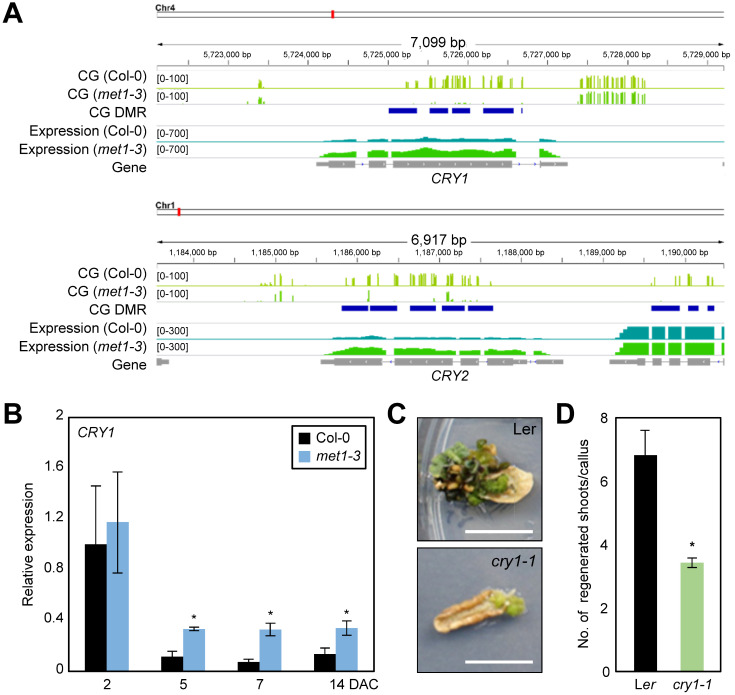
The biological impact of light signaling on plant regeneration remains unknown; therefore, we decided to investigate how light signaling regulates de novo shoot regeneration. We revisited the hypomethylated regions in met1-3, and found that key blue light receptor genes, including CRY1 and CRY2, were under the control of MET1-dependent CG methylation (Fig. 3A) in both leaf and callus tissues (Supplementary Tables S4 and S5). Expression levels of CRY1 and CRY2 were higher in met1-3 than in the wild-type (Fig. 3A, Supplementary Table S10), suggesting that CG methylation repressed the expression of these genes. To validate the transcriptional control over photoreceptor genes by MET1, we carried out RT-qPCR analysis using met1-3 and wild-type calli. The expression of CRY genes gradually decreased in the wild-type during callus formation, but the reduction was diminished in met1-3 (Fig. 3B, Supplementary Fig. S3), indicating that light signaling triggered by CRY1 and CRY2 is derepressed in met1-3. The contradicting results of CRY expression in leaf tissues between RNA-seq and RT-qPCR might be explained by the differential sensitivity of the two techniques.
Fig. 3. MET1-dependent CG methylation at the CRY loci.
(A) CG methylation patterns at the chromatin of CRY1 and CRY2 genes. DNA methylation states and mRNA levels are depicted. (B) Transcript level of CRY1 in met1-3 determined by RT-qPCR. Three independent biological replicates were averaged. Asterisks indicate significant differences (Student’s t-test; *P < 0.05). DAC, days after incubation on CIM. (C and D) Shoot regeneration capacity of the cry1-1 mutant. Calli preincubated for 7 days on CIM were transferred to SIM. The number of regenerated shoots from calli was measured (n > 30) at 3 weeks after incubation on SIM. Statistically significant differences between wild-type and mutant calli are indicated by asterisks (Student’s t-test, *P < 0.05). Scale bars = 10 mm.
Next, we investigated whether shoot regeneration efficiency of met1-3 was indeed associated with increased expression of CRYs. To test this possibility, we employed cry1-1 and cry2/fha-1 mutants, and examined their de novo shoot regeneration capacity. The cry1-1 mutant particularly displayed reduced shoot regeneration (Figs. 3C and 3D), while cry2/fha-1 showed a statistically insignificant increase in shoot regeneration efficiency (Supplementary Fig. S3), compared with wild type. Given that CRY1 is responsible for sensing both low and high intensities of blue light, whereas CRY2 mainly perceives a low intensity of blue light (Lin et al., 1998; Yu et al., 2010), it is reasonable to speculate that CRY1 has a broad and greater impact during plant regeneration. Together, these data suggest that MET1-dependent CG methylation represses CRY1 expression and influences the plant regeneration process.
Coexpression of CRY1 and cytokinin signaling genes
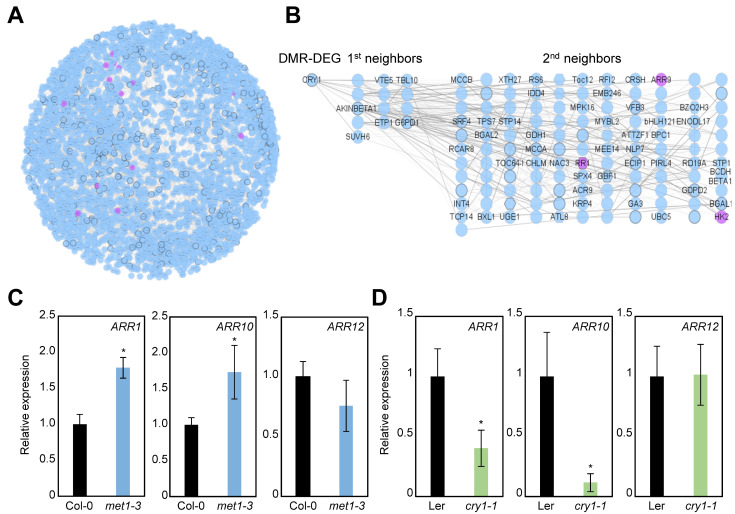
To estimate the extent of the biological impact of MET1 during plant regeneration, we identified genes both hypomethylated and up-regulated in met1-3 (Supplementary Table S10); these genes included CRY1 and CRY2. A total of 386 genes were identified, which were then subjected to coexpression network analysis. Of the 386 genes, 314 were included in a scaled-subnetwork consisting of 3,390 genes (Fig. 4A, Supplementary Table S11). To characterize the network, we performed functional enrichment analysis of all 3,390 genes. The results showed that cytokinin signaling was particularly overrepresented (phytohormone action for cytokinin-perception and signal transduction [P value = 3.95 × 10–5]) (Table 2). Consistently, core cytokinin signaling genes, such as ARR1 and ARR9, were found in the coexpression network (Fig. 4A).
Fig. 4. Potential connection of CRY1 and cytokinin signaling components.
(A) Coexpression network of genes hypomethylated and up-regulated in met1-3. Cytokinin signaling genes are indicated by purple nodes. (B) Close neighbor genes of CRY1 in the coexpression subnetwork. Cytokinin signaling genes are indicated by purple nodes. (C and D) Transcript levels of type-B ARR genes in met1-3 (C) and cry1-1 (D) calli determined by RT-qPCR. Three independent biological replicates were averaged. Asterisks indicate significant differences (Student’s t-test; *P < 0.05).
Table 2.
Functional enrichment analysis of all genes contained in the coexpression subnetwork shown in Fig.4A
| Functional categorya | No. of query genes per category | Total No. of query genes | Observed value (%) | No. of reference genes per category | Total No. of reference genes | Expected value (%) | P value |
|---|---|---|---|---|---|---|---|
| External stimuli response (pathogen) | 66 | 3,391 | 1.9 | 233 | 27,206 | 0.9 | 6.60E-11 |
| Nutrient uptake (iron uptake) | 26 | 3,391 | 0.8 | 61 | 27,206 | 0.2 | 4.10E-09 |
| Protein modification (phosphorylation) | 207 | 3,391 | 6.1 | 1,148 | 27,206 | 4.2 | 1.85E-08 |
| Enzyme classification (EC_2 transferases) | 80 | 3,391 | 2.4 | 382 | 27,206 | 1.4 | 1.88E-06 |
| Carbohydrate metabolism (starch metabolism) | 19 | 3,391 | 0.6 | 52 | 27,206 | 0.2 | 7.85E-06 |
| Phytohormone action (cytokinin-perception and signal transduction) | 13 | 3,391 | 0.4 | 31 | 27,206 | 0.1 | 3.95E-05 |
| Amino acid metabolism (degradation) | 19 | 3,391 | 0.6 | 58 | 27,206 | 0.2 | 4.64E-05 |
| Protein homeostasis (autophagy) | 17 | 3,391 | 0.5 | 50 | 27,206 | 0.2 | 6.78E-05 |
| Protein modification (S-glutathionylation) | 20 | 3,391 | 0.6 | 76 | 27,206 | 0.3 | 8.02E-04 |
| Cytoskeleton organization (microtubular network) | 29 | 3,391 | 0.9 | 132 | 27,206 | 0.5 | 1.56E-03 |
| Nutrient uptake (copper uptake) | 9 | 3,391 | 0.3 | 24 | 27,206 | 0.1 | 1.61E-03 |
| Phytohormone action (jasmonic acid-biosynthesis) | 8 | 3,391 | 0.2 | 21 | 27,206 | 0.1 | 2.61E-03 |
| Cell cycle organization (cytokinesis) | 18 | 3,391 | 0.5 | 74 | 27,206 | 0.3 | 3.66E-03 |
| Phytohormone action (salicylic acid-perception and signal transduction) | 3 | 3,391 | 0.1 | 4 | 27,206 | 0.0 | 7.02E-03 |
| Solute transport (channels) | 34 | 3,391 | 1.0 | 180 | 27,206 | 0.7 | 8.53E-03 |
| Solute transport (carrier-mediated transport) | 120 | 3,391 | 3.5 | 784 | 27,206 | 2.9 | 9.78E-03 |
| Multi-process regulation (target of rapamycin [TOR] signaling) | 5 | 3,391 | 0.1 | 12 | 27,206 | 0.0 | 1.11E-02 |
| Phytohormone action (salicylic acid-biosynthesis) | 2 | 3,391 | 0.1 | 2 | 27,206 | 0.0 | 1.55E-02 |
| Nutrient uptake (sulfur assimilation) | 5 | 3,391 | 0.1 | 13 | 27,206 | 0.0 | 1.63E-02 |
| Enzyme classification (EC_1 oxidoreductases) | 67 | 3,391 | 2.0 | 420 | 27,206 | 1.5 | 2.01E-02 |
| Cell cycle organization (DNA replication) | 16 | 3,391 | 0.5 | 76 | 27,206 | 0.3 | 2.36E-02 |
| Vesicle trafficking (target membrane tethering) | 17 | 3,391 | 0.5 | 84 | 27,206 | 0.3 | 2.87E-02 |
| Phytohormone action (salicylic acid-conjugation and degradation) | 3 | 3,391 | 0.1 | 6 | 27,206 | 0.0 | 2.89E-02 |
| Enzyme classification (EC_3 hydrolases) | 41 | 3,391 | 1.2 | 251 | 27,206 | 0.9 | 4.22E-02 |
| Vesicle trafficking (clathrin-independent machinery) | 2 | 3,391 | 0.1 | 3 | 27,206 | 0.0 | 4.27E-02 |
| Chromatin organization (chromatin remodeling complexes) | 13 | 3,391 | 0.4 | 63 | 27,206 | 0.2 | 4.48E-02 |
Significantly overrepresented terms (P < 0.05) were shown. Statistical significance was determined by the hypergeometric test.
In agreement with the finding that blue light receptor genes were included in the coexpression network of genes regulated by MET1-dependent CG methylation (Fig. 4A), the CRY1 gene was also closely connected to cytokinin signaling. Gene regulatory network analysis showed that type-A and type-B ARR genes were particularly coexpressed with CRY1 in a relation of second neighbor (Fig. 4B). Given that photoreceptors are involved in shoot identity establishment (Ikeuchi et al., 2016; Nameth et al., 2013), which requires cytokinin biosynthesis and signaling (Meng et al., 2017; Skoog and Miller, 1957; Zhang et al., 2017), it is plausible that CRY1 is functionally associated with cytokinin signaling.
Considering that enhanced cytokinin signaling promotes de novo shoot organogenesis (Atta et al., 2009; Skoog and Miller, 1957; Sugimoto et al., 2010), the functions of MET1 and CRY1 in shoot regeneration were likely owing to altered cytokinin signaling. To test this possibility, we examined whether the transcript levels of cytokinin signaling genes were affected in met1-3 and cry1-1 mutants. Type-B ARR1 and ARR10 genes were up-regulated in met1-3 (Fig. 4C) but were significantly repressed in cry1-1 (Fig. 4D), as expected. These results indicate that MET1-dependent CG methylation represses CRY1 and consequently cytokinin signaling, thus affecting plant regeneration.
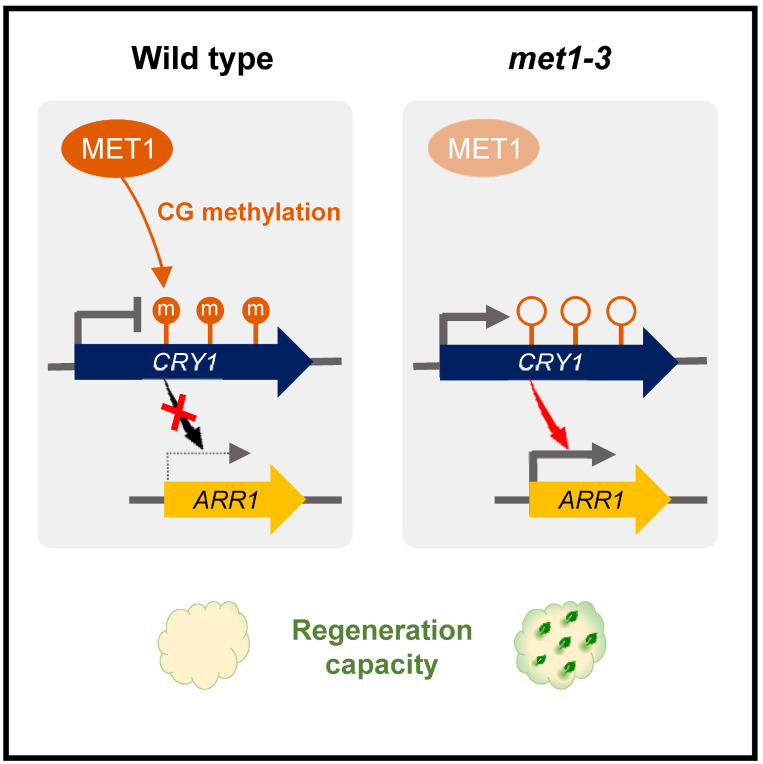
Taken together, our findings demonstrate that MET1-dependent CG methylation represses CRY1 expression and consequently reduces cytokinin signaling. In the met1-3 mutant, methylation-free contexts at the CRY1 locus activate gene expression and enhance cytokinin signaling (Fig. 5). The signaling axis encompassing MET1 and CRY1 influences the plant regeneration process, which is intrinsically based on the balance between auxin and cytokinin signaling.
Fig. 5. The role of MET1 in de novo shoot regeneration.
The CRY1 gene is silenced by MET1-dependent DNA methylation to ensure homeostasis of light signaling in wild-type leaves. In met1-3, the CG methylation-free CRY1 locus is transcriptionally activated, which stimulates the expression of cytokinin signaling genes, such as type-B ARRs. Increased cytokinin signaling promotes de novo shoot regeneration.
DISCUSSION
In vitro plant regeneration involves two steps: callus formation and de novo shoot regeneration. Callus tissue is analogous to the lateral root primordium (Sugimoto et al., 2010); therefore, callus formation is initiated by genes involved in lateral root formation, including LBDs. Callus cells then establish the root stem cell niche through the expression of pluripotency regulators, PLTs and WOXs (Kareem et al., 2015; Sugimoto et al., 2010). PLT- and WOX-induced root stem cell identity is considered as the cellular nature of pluripotency (Gaillochet and Lohmann, 2015; Lee and Seo, 2018), facilitating consequent tissue regeneration. After pluripotency acquisition, de novo shoot organogenesis can be initiated on SIM through the activation of type-B ARRs (Meng et al., 2017) and subsequently WUS, which defines the shoot developmental program (Zhang et al., 2017).
Plant regeneration involves substantial changes in the epigenetic landscape. Histone H3 modifications, including trimethylation of lysine 4 and lysine 36 residues (H3K4me3 and H3K36me3, respectively) and acetylation, exhibit dynamic changes during leaf-to-callus transition (Kim et al., 2018; Lee et al., 2017; Li et al., 2011). Various chromatin modifiers and remodelers have been identified as key regulators of cell fate transition (He et al., 2012; Ishihara et al., 2019). DNA methylation is also altered during the process of plant regeneration. In particular, genome-wide CHG and CHH methylation levels change during callus formation (Shim et al., 2021). By contrast, in this study, CG methylation was largely insensitive to cell fate transition, and CG methylation landscapes were mostly maintained during leaf-to-callus transition. It is plausible that CG methylation evolved to ensure genome integrity rather than to regulate gene expression.
Given the stable nature of CG methylation during leaf-to-callus transition, the altered de novo shoot regeneration capacity of met1-3 was attributable to its constitutively hypomethylated genomic regions, independent of the tissue type. It has been demonstrated that the WUS gene, a key regulator of shoot stem cell formation, is constitutively activated in met1, which accounts for their enhanced de novo shoot regeneration capability (Li et al., 2011). In this study, we found additional targets of MET1-dependent CG methylation. The CG-methylated and MET1-down-regulated genes were enriched for light and cytokinin signaling. In particular, CRY1, which is CG-methylated by MET1, was linked to cytokinin signaling genes, as suggested previously (Gangappa and Botto, 2016; Vandenbussche et al., 2007), although detailed molecular connections need to be elucidated.
Each step of the plant regeneration process requires a balance between auxin and cytokinin signaling. A high auxin-to-cytokinin ratio promotes callus formation (Atta et al., 2009; Skoog and Miller, 1957; Sugimoto et al., 2010), whereas a low auxin-to-cytokinin ratio promotes de novo shoot regeneration (Skoog and Miller, 1957). MET1-dependent CG methylation suppresses the blue light receptor gene CRY1, and subsequently inhibits cytokinin signaling and shoot identity establishment in calli. The met1-3 mutant displayed higher expression of CRY1 and consequently that of cytokinin signaling genes. Consistently, shoot regeneration was reduced in the cry1-1 mutant, possibly because of attenuated cytokinin signaling, but increased in the met1-3 mutant. Overall, our data suggest that MET1 catalyzes CG methylation and silences light signaling, in part, via the repression of CRY1. Light signaling exhibits extensive crosstalk with hormone signaling pathways, especially cytokinin signaling. In met1-3, enhanced light and cytokinin signaling promotes shoot identity establishment and de novo shoot organogenesis.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research (NRF-2019R1I1A1A01061376 to S.S.; NRF-2019R1A2C2006915 to P.J.S.) and Basic Research Laboratory (NRF-2020R1A4A2002901 to P.J.S.) programs provided by the National Research Foundation of Korea, by the National Research Foundation of Korea, and by the Creative-Pioneering Researchers Program through Seoul National University (0409-20200281 to P.J.S.).
Footnotes
AUTHOR CONTRIBUTIONS
P.J.S. conceived the project. S.S. conducted bioinformatics analyses. H.G.L. performed experiments. S.S. wrote the first draft of manuscript. P.J.S. revised the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Atta R., Laurens L., Boucheron-Dubuisson E., Guivarc’h A., Carnero E., Giraudat-Pautot V., Rech P., Chriqui D. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro . Plant J. 2009;57:626–644. doi: 10.1111/j.1365-313X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- Bartee L., Malagnac F., Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T., Ivánek R., Lienert F., Schübeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Bhatia H., Khemka N., Jain M., Garg R. Genome-wide bisulphite-sequencing reveals organ-specific methylation patterns in chickpea. Sci. Rep. 2018;8:9704. doi: 10.1038/s41598-018-27979-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackertz M., Boeke J., Zhang R., Renkawitz R. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J. Biol. Chem. 2002;277:40958–40966. doi: 10.1074/jbc.M207467200. [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002a;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. U. S. A. 2002b;99(Suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Ha M., Lackey E., Wang J., Chen Z.J. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics. 2008;178:1845–1858. doi: 10.1534/genetics.107.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Schönberger B., Menz J., Ludewig U. Plasticity of DNA methylation and gene expression under zinc deficiency in Arabidopsis roots. Plant Cell Physiol. 2018;59:1790–1802. doi: 10.1093/pcp/pcy100. [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Doerner P.W., Colón-Carmona A., Rost T.L. Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 2000;124:1648–1657. doi: 10.1104/pp.124.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M., Xu C., Xu K., Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012;22:1169–1180. doi: 10.1038/cr.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhu J., Du X., Cui X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana . Planta. 2012;236:1227–1237. doi: 10.1007/s00425-012-1673-3. [DOI] [PubMed] [Google Scholar]
- Fujita N., Watanabe S., Ichimura T., Ohkuma Y., Chiba T., Saya H., Nakao M. MCAF mediates MBD1-dependent transcriptional repression. Mol. Cell. Biol. 2003;23:2834–2843. doi: 10.1128/MCB.23.8.2834-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige S., Kondo E., Gu Z., Suzuki H., Horii A. RET finger protein enhances MBD2- and MBD4-dependent transcriptional repression. Biochem. Biophys. Res. Commun. 2006;351:85–92. doi: 10.1016/j.bbrc.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Gaillochet C., Lohmann J.U. The never-ending story: from pluripotency to plant developmental plasticity. Development. 2015;142:2237–2249. doi: 10.1242/dev.117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Botto J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant. 2016;9:1353–1365. doi: 10.1016/j.molp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Gaspar J.M., Hart R.P. DMRfinder: efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinformatics. 2017;18:528. doi: 10.1186/s12859-017-1909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C.J., Scheibe M., Wongpalee S.P., Liu W., Cornett E.M., Vaughan R.M., Li X., Chen W., Xue Y., Zhong Z., et al. A DNA methylation reader complex that enhances gene transcription. Science. 2018;362:1182–1186. doi: 10.1126/science.aar7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Chen X., Huang H., Xu L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 2012;8:e1002911. doi: 10.1371/journal.pgen.1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. Plant regeneration: cellular origins and molecular mechanisms. Development. 2016;143:1442–1451. doi: 10.1242/dev.134668. [DOI] [PubMed] [Google Scholar]
- Ingouff M., Selles B., Michaud C., Vu T.M., Berger F., Schorn A.J., Autran D., Van Durme M., Nowack M.K., Martienssen R.A., et al. Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev. 2017;31:72–83. doi: 10.1101/gad.289397.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Sugimoto K., Tarr P.T., Temman H., Kadokura S., Inui Y., Sakamoto T., Sasaki T., Aida M., Suzuki T., et al. Primed histone demethylation regulates shoot regenerative competency. Nat. Commun. 2019;10:1786. doi: 10.1038/s41467-019-09386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Nishio H., Tarutani Y., Emura N., Honjo M.N., Toyoda A., Fujiyama A., Kakutani T., Kudoh H. Seasonal stability and dynamics of DNA methylation in plants in a natural environment. Genes. 2019;10:544. doi: 10.3390/genes10070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J.A., Bender J., Richards E.J. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis . Genes Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Bostick M., Zhang X., Kraft E., Henderson I., Callis J., Jacobsen S.E. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareem A., Durgaprasad K., Sugimoto K., Du Y., Pulianmackal A.J., Trivedi Z.B., Abhayadev P.V., Pinon V., Meyerowitz E.M., Scheres B., et al. PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 2015;25:1017–1030. doi: 10.1016/j.cub.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yang W., Forner J., Lohmann J.U., Noh B., Noh Y. Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis . EMBO J. 2018;37:e98726. doi: 10.15252/embj.201798726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Lee H.J., Choi M.Y., Kang S.S., Kim Y.S., Shin J.K., Choi W.S. UHRF1 induces methylation of the TXNIP promoter and down-regulates gene expression in cervical cancer. Mol. Cells. 2021;44:146–159. doi: 10.14348/molcells.2021.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana . Mol. Gen. Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Rolff E., Spruit C.J.P. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L. ) Heynh. Z. Pflanzenphysiol. 1980;100:147–160. doi: 10.1016/S0044-328X(80)80208-X. [DOI] [Google Scholar]
- Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z., Wang Y., Tang K., Tang D., Datsenka T., Cheng J., Zhang Y., Handa A.K., Zhu J.K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E4511–E4519. doi: 10.1073/pnas.1705233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Park O.S., Seo P.J. RNA-seq analysis of the Arabidopsis transcriptome in pluripotent calli. Mol. Cells. 2016;39:484–494. doi: 10.14348/molcells.2016.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Park O.S., Seo P.J. Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci. Signal. 2017;10:eaan0316. doi: 10.1126/scisignal.aan0316. [DOI] [PubMed] [Google Scholar]
- Lee K., Seo P.J. Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 2018;23:235–247. doi: 10.1016/j.tplants.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Li W., Liu H., Cheng Z.J., Su Y.H., Han H.N., Zhang Y., Zhang X.S. DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 2011;7:e1002243. doi: 10.1371/journal.pgen.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Chang Y., Lu J., Wu X., Liu Q., Zhang W., Su X., Zhang B. Global methylomic and transcriptomic analyses reveal the broad participation of DNA methylation in daily gene expression regulation of Populus trichocarpa . Front. Plant Sci. 2019;10:243. doi: 10.3389/fpls.2019.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Yang H., Guo H., Mockler T., Chen J., Cashmore A.R. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Liu H., Zhang H., Dong Y.X., Hao Y.J., Zhang X.S. DNA METHYLTRANSFERASE1-mediated shoot regeneration is regulated by cytokinin-induced cell cycle in Arabidopsis . New Phytol. 2018;217:219–232. doi: 10.1111/nph.14814. [DOI] [PubMed] [Google Scholar]
- Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., Xu L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis . Plant Cell. 2014;26:1081–1093. doi: 10.1105/tpc.114.122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.W., Zhou J.X., Huang H.W., Li Y.Q., Shao C.R., Li L., Cai T., Chen S., He X.J. Two components of the RNA-directed DNA methylation pathway associate with MORC6 and silence loci targeted by MORC6 in Arabidopsis . PLoS Genet. 2016;12:e1006026. doi: 10.1371/journal.pgen.1006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W.J., Cheng Z.J., Sang Y.L., Zhang M.M., Rong X.F., Wang Z.W., Tang Y.Y., Zhang X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL . Plant Cell. 2017;29:1357–1372. doi: 10.1105/tpc.16.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameth B., Dinka S.J., Chatfield S.P., Morris A., English J., Lewis D., Oro R., Raizada M.N. The shoot regeneration capacity of excised Arabidopsis cotyledons is established during the initial hours after injury and is modulated by a complex genetic network of light signalling. Plant Cell Environ. 2013;36:68–86. doi: 10.1111/j.1365-3040.2012.02554.x. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Aoki Y., Tadaka S., Kagaya Y., Kinoshita K. ATTED-II in 2018: a plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol. 2018;59:e3. doi: 10.1093/pcp/pcx191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo A.P., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- Penterman J., Zilberman D., Huh J.H., Ballinger T., Henikoff S., Fischer R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42((Web Server issue)):W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Scheid O., Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Shim S., Lee H.G., Park O.S., Shin H., Lee K., Lee H., Huh J.H., Seo P.J. Dynamic changes in DNA methylation occur in TE regions and affect cell proliferation during leaf-to-callus transition in Arabidopsis. Epigenetics. 2021 Jan 15; doi: 10.1080/15592294.2021.1872927. [Epub]. https://doi.org/10.1080/15592294.2021.1872927 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 1957;11:118–130. [PubMed] [Google Scholar]
- Smallwood S.A., Lee H.J., Angermueller C., Krueger F., Saadeh H., Peat J., Andrews S.R., Stegle O., Reik W., Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen J.H.M., López A., Jain R., Pascual-Pardo D., Luna E., Smith L.M., Ton J. The relationship between transgenerational acquired resistance and global DNA methylation in Arabidopsis. Sci. Rep. 2018;8:14761. doi: 10.1038/s41598-018-32448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Jiao Y., Meyerowitz E.M. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell. 2010;18:463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Usadel B., Poree F., Nagel A., Lohse M., Czedik-Eysenberg A., Stitt M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32:1211–1129. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Habricot Y., Condiff A.S., Maldiney R., Straeten D.V.D., Ahmad M. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana . Plant J. 2007;49:428–441. doi: 10.1111/j.1365-313X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- Waterfield M., Khan I.S., Cortez J.T., Fan U., Metzger T., Greer A., Fasano K., Martinez-Llordella M., Pollack J.L., Erle D.J., et al. The transcriptional regulator Aire co-opts the repressive ATF7ip-MBD1 complex for induction of immune tolerance. Nat. Immunol. 2014;15:258–265. doi: 10.1038/ni.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H., Park K., Lee J., Lee S., Choi Y. An optimized method for the construction of a DNA methylome from small quantities of tissue or purified DNA from Arabidopsis embryo. Mol. Cells. 2021;44:602–612. doi: 10.14348/molcells.2021.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Liu H., Klejnot J., Lin C. The cryptochrome blue light receptors. Arabidopsis Book. 2010;8:e0135. doi: 10.1199/tab.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Grafi G. Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 2003;34:565–572. doi: 10.1046/j.1365-313x.2003.01756.x. [DOI] [PubMed] [Google Scholar]
- Zhang T.Q., Lian H., Zhou C.M., Xu L., Jiao Y., Wang J.W. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell. 2017;29:1073–1087. doi: 10.1105/tpc.16.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Sng N.J., LeFrois C.E., Paul A.L., Ferl R.J. Epigenomics in an extraterrestrial environment: organ-specific alteration of DNA methylation and gene expression elicited by spaceflight in Arabidopsis thaliana . BMC Genomics. 2019;20:205. doi: 10.1186/s12864-019-5554-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- Zubko E., Gentry M., Kunova A., Meyer P. De novo DNA methylation activity of METHYLTRANSFERASE 1 (MET1) partially restores body methylation in Arabidopsis thaliana . Plant J. 2012;71:1029–1037. doi: 10.1111/j.1365-313X.2012.05051.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome BS-seq and RNA-seq data are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) data under the BioProject accession number PRJNA601842.