Abstract
Objectives
Maternal factors that are enriched in oocytes have attracted great interest as possible key factors in somatic cell reprogramming. We found that surfeit locus protein 4 (Surf4), a maternal factor, can facilitate the generation of induced pluripotent stem cells (iPSCs) previously, but the mechanism remains elusive.
Materials and Methods
In this study, we investigated the function and mechanism of Surf4 in somatic cell reprogramming using a secondary reprogramming system. Alkaline phosphatase (AP) staining, qPCR and immunofluorescence (IF) staining of expression of related markers were used to evaluate efficiency of iPSCs derived from mouse embryonic fibroblasts. Embryoid body and teratoma formation assays were performed to evaluate the differentiation ability of the iPSC lines. RNA‐seq, qPCR and western blot analysis were applied to validate the downstream targets of Surf4.
Results
Surf4 can significantly facilitate the generation of iPSCs in a proliferation‐independent manner. When co‐expressed with Oct4, Sox2, Klf4 and c‐Myc (OSKM), Surf4 can activate the response to endoplasmic reticulum (ER) stress at the early stage of reprogramming. We further demonstrated that Hspa5, a major ER chaperone, and the active spliced form of Xbp1 (sXbp1), a major mediator of ER stress, can mimic the effects of Surf4 on somatic cell reprogramming. Concordantly, blocking the unfolded protein response compromises the effect of Surf4 on reprogramming.
Conclusions
Surf4 promotes somatic cell reprogramming by activating the response to ER stress.
Maternal factor Surf4 can significantly facilitate the generation of induced pluripotent stem cells (iPSCs) in a proliferation‐independent manner. When co‐expressed with Oct4, Sox2, Klf4 and c‐Myc (OSKM), Surf4 can activate the response to endoplasmic reticulum (ER) stress at the early stage of reprogramming.
1. INTRODUCTION
Terminally differentiated somatic cells can be reprogrammed into a pluripotent state by Yamanaka factors (Oct3/4, Sox2, Klf4 and c‐Myc). 1 This transition is accompanied by global and dramatic changes at the transcriptional, epigenetic and metabolic levels. 2 , 3 , 4 Although many cellular mechanisms have been revealed to date, the process of reprogramming is still inefficient, time‐consuming and stochastic. 5 Somatic cell nuclear transfer provides a fast, relatively efficient and deterministic reprogramming model in which terminally differentiated somatic nuclei can be reprogrammed to a totipotent state by factors in the oocyte cytoplasm. 6 Therefore, the role of oocyte factors in somatic reprogramming has been widely studied, and an increasing number of studies have shown that oocyte factors can improve reprogramming efficiency. 7 , 8 We previously found that some oocyte‐enriched proteins identified through mass spectrometry 9 can enhance somatic cell reprogramming. 10 In this study, we further explored the function and mechanism of Surf4, which is one of these maternal factors.
SURF4, known as endoplasmic reticulum (ER)‐derived vesicles protein 29 (Erv29p) in Saccharomyces cerevisiae and surfeit locus protein 4 homolog (SFT‐4) in Caenorhabditis elegans, is an integral ER membrane protein 11 , 12 and is required for packaging soluble secretory proteins into ER‐derived transport vesicles. 11 , 13 Binding to the amino‐terminal hydrophobic tripeptide motifs of soluble cargo proteins with different affinities, SURF4 enables prioritization of their exit from the ER. 14 SURF4 circulates between the ER/ER‐Golgi intermediate compartment (ERGIC)/Golgi and mediates the anterograde or retrograde transport of cargo proteins. 13 , 15 , 16 , 17 , 18 , 19 Disruption of Surf4 trafficking results in a reduction in the number of ERGIC clusters 20 and accumulation of cargo proteins in the ER compartment. 21 Dysregulation of ER‐Golgi vesicle transport induces ER stress, 22 and in turn, when ER stress occurs, the expression of Erv29p significantly increases. 23 Deficiency of Surf4 in mice results in embryonic lethality after implantation. 21 A study of lipoprotein transport revealed that SURF4‐mediated ER export of lipoproteins controls lipid homeostasis in mice and humans. 24
During mouse preimplantation development, Surf4 was highly enriched in MII oocytes 9 , 25 and early embryos before the two‐cell stage. 25 , 26 In this paper, we demonstrate that Surf4 significantly promotes Yamanaka factor‐mediated iPSC generation via activation of the response to ER stress.
2. METHODS AND MATERIAL
2.1. Mice
R26rtTA; Col1a1‐4F2A mice (Jackson Laboratory stock number 011004) 27 were crossed with OG2 mice (Jackson Laboratory stock number 004654) to obtain R26rtTA; Col1a1‐4F2A; Oct4‐EGFP mice. The specific pathogen‐free mice were housed in the animal facility of Tongji University. All our study procedures were consistent with the Tongji University Guide for the care and use of laboratory animals.
2.2. Cell culture
Mouse embryonic fibroblasts (MEFs) were derived from 13.5‐dpc embryos. MEFs were maintained in Dulbecco's modified eagle medium (DMEM) (Sigma D5671) medium supplemented with 10% (vol/vol) foetal bovine serum (FBS) (Gibco 10270‐106) and 1 mM L‐glutamine (Merck Millipore TMS‐002‐C). Embryonic stem cells (ESCs) and iPSCs were cultured on mitomycin C (Sigma M4287) treated MEFs in Embryonic stem medium (ESM) containing DMEM (Sigma D5671) supplemented with 15% (v/v) FBS (Gibco 16000‐44), 1 mM L‐glutamine (Merck Millipore TMS‐002‐C), 0.1 mM mercaptoethanol (Merck Millipore ES‐007‐E), 1% nonessential amino acid (NEAA) stock (Merck Millipore TMS‐001‐C), and 1000 U/ml leukaemia inhibitory factor (LIF) (Merck Millipore ESGRO 1107).
2.3. Lentiviral vector construction and iPSCs derivation
Full‐length mouse cDNA of Surf4 (NM_011512), Hspa5 (NM_001163434.1), spliced form of Xbp1 (sXbp1) (NM_001271730.1) and a dominant negative form of Xbp1 (sXbp1‐ΔDBD) (deleting 553–606 bp in the sequence NM_001271730.1) 28 were cloned and inserted into the Fuw‐TETON vector and the shRNA sequences were constructed into pSicoR vector. The constructed plasmids (in Fuw‐TETON vector for overexpression and in pSicoR vector for knockdown) preparation and iPSCs induction procedure were performed according to a previously reported method. 29 Plasmids were extracted with Plasmid Mini Kit (Tiangen, China) and EndoFree Plasmid Maxi Kit (Cwbio). HEK293T cells were transfected with the plasmids along with the lentivirus packaging plasmids ps‐PAX‐2 and pMD2G using VigoFect transfection reagent. Fresh medium was changed 8–10 h after transfection, and the medium containing virus was collected at further 48 h. The reprogrammable MEFs were seeded in 12‐well plates at a density of 1.2 × 104 cells per well (unless otherwise indicated) and then were infected with virus‐containing medium for 8–12 h. Infected MEFs were cultured in ESM supplemented with 1 µg/ml doxycycline (Dox) for 2–3 weeks. The cells were observed and tested at indicated time points during reprogramming. After colonies formation, the cells were cultured in ESM without Dow for further 2–3 days, and then the colonies were mechanically picked for establishing the iPS cell lines.
2.4. Cell growth curve
The MEFs were plated onto 12‐well plates at a density of 1.2 × 104 cells per well and were harvested every 48 or 72 h and counted in a haemocytometer. Each group contained three replicates.
2.5. Alkaline phosphatase (AP) staining
Alkaline phosphatase staining kit (Beyotime, C3206, China) was used for AP staining according to the instructions of the manufacturer. In briefly, at the end of reprogramming, the cells were washed once by Dulbecco's Phosphate‐Buffered Saline (DPBS), and fixed by 10% formaldehyde solution for 5 min at room temperature. Then, the cells were washed once by deionized water and stained by the reagent provide by the kit.
2.6. Karyotype analysis
Cells were trypsinized and treated with potassium chloride (KCl) (0.4 M)/sodium citrate (0.4 M) (1:1) for 5 min at 37℃, and then prefixed with fixative composed of methanol/acetic acid (3:1) and resuspended in 1–5 ml of fixative. Cells were centrifuged 5 min at 1000 rpm before a final resuspension in 1–5 ml of fixative. Cells were then spread on slides and stained with Giemsa. A minimum of 15 metaphases were captured and analysed.
2.7. RNA isolate and real time PCR
Total RNA was extracted using TRNzol Universal Reagent (Tiangen) and reverse transcribed using the 5× All‐In‐One RT MasterMix (ABM). Quantitative reverse‐transcription PCR was performed with SYBR®FAST Universal qPCR Kit (KAPA) and the ABI7500 Fast Real‐time PCR system (Applied Biosystems) or QuantStudio5 (Applied Biosystems). The reactions were performed in triplicate and relative mRNA expression is normalized to β‐actin as an endogenous control using the ΔCT method. Primer sequences are available in the Table S1.
2.8. Immunofluorescence (IF) staining
Immunofluorescence staining was performed as previously described. 30 Cells growing on slides were fixed with 4% paraformaldehyde and were permeabilized by 0.5% Triton X‐100 (in DPBS) for 15 min at room temperature. The cells were blocked in 5% bovine serum albumin (BSA) in DPBS for 1 h at room temperature and incubated with the primary antibodies against OCT4 (1:500, Santa Cruz, SC‐5279), NANOG (1:500, Cosmo Bio, RCAB001P), SSEA1 (1:100, Millipore, MAB4301) in BSA/DPBS buffer overnight at 4℃. The samples were washed three time in DPBS and incubated with fluorochrome conjugated secondary antibodies Alexa Fluor 594 donkey anti‐mouse IgG (Thermo Fisher, A21203), or Alexa Fluor 594 donkey anti‐rabbit IgG (Thermo Fisher, A21207) in BSA/DPBS buffer for 2 h at room temperature. The cells were washed three times in DPBS and DNA was labelled with DAPI (1 µg/ml, Merck Millipore) in DPBS. The stained cells were observed using an LSM 880 microscope (Zeiss) with a Plan Neofluar 63×/1.4 Oil DIC objective.
2.9. Embryoid body (EB) differentiation
IPSCs were trypsinized and plated onto tissue culture plates for 15–30 min to deplete feeder cells. Floating cells were collected and were cultured total of 5 × 104 cells per drop in hanging drop for 2 days and transferred to ultra‐low cluster plates (Costar) in DMEM (Gibco) supplemented with 15% (v/v) FBS, 1 mM L‐glutamine (Merck Millipore), 0.1 mM mercaptoethanol (Merck Millipore), 1% NEAA stock (Merck Millipore), but without LIF. Five days later, EBs were collected and re‐plated onto gelatine‐coated tissue cultured dishes for 21 days. Total RNA of the cells was extracted and analysed for the markers for three embryonic germ layers by qPCR. The primer sequences are available in the Table S1.
2.10. Teratoma formation
The iPSCs were trypsinized and a total of 2–5 × 106 iPSCs were subcutaneously injected into the groin of SCID mice. Four to eight weeks post‐injection, teratomas formed and were very palpable. The tumour samples were dissected and processed for haematoxylin‐eosin staining.
2.11. Flow cytometry analysis
For Oct4 +‐GFP population test, the cells were dissociated into single‐cell suspension in FACS buffer (PBS+0.1% BSA), filtered and analysed by CytoFLEX S (Beckman Coulter).
For analysis of intermediates, the reprogramming cells on day 3 after induction with or without Surf4 overexpression (OE) were dissociated into single‐cell suspension in FACS buffer and incubated with 5 μl of PE/Cy7‐conjugated antibody against THY1.2 (BioLegend, 140310) and/or APC‐conjugated antibody against SSEA1 (BioLegend, 125608) in 100 μL FACS buffer per 106 cells. Cells were washed once in FACS buffer after 15–30 min staining on ice, suspended in FACS buffer and analysed by CytoFLEX S (Beckman Coulter).
2.12. RNA‐sequencing and data processing
Total RNA from independent biological replicates of MEFs, day 3 samples in reprogramming with or without Surf4, was isolated using a QIAGEN RNeasy Kit (14104, Germantown, US). The RNA sequencing libraries were generated using a KAPA Stranded RNA‐Seq Kit Illumina platform (KK8440, Wilmington, US). Paired‐end 150‐bp sequencing was further performed on a HiSeq 2500 (Illumina) at Berry Genomics Corporation.
All of the RNA‐Seq sequencing reads were processed using BBDuk (version 38.34) to remove adapters and low‐quality reads. 31 The filtered reads were mapped to the mouse reference genome using STAR (version 0.6.0) with the default parameters except for the ‘quantMode GeneCounts’ parameter. 32 Gene expression for each sample was quantified by FPKM using StringTie (version 1.3.3b). 33
A clustered heat map of Pearson correlation and principal component analysis (PCA) was implemented using the R function procomp. Differentially expressed genes (DEGs) were selected on the basis of a fold change >1.5 and false discovery rate (FDR) <0.05 using limma. The DEGs were clustered based on their expression levels in the samples. Gene Ontology (GO) enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) web‐accessible tool. Gene ontology terms for each function cluster were summarized to a representative term, and p‐values were plotted to show the significance.
2.13. Western blot analysis
Cells were washed once with PBS and lysed by cell lysis buffer (KeyGEN, KGP701‐100) containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X‐100 and protease inhibitors for 30 min ice, and then ultrasonicated. The samples were boiled to 100℃ for 10–15 min in loading buffer (EpiZyme, LT101S) with 2% β‐mercaptoethanol (Amersham, CT). Anti α‐TUBULIN (1:10000, Proteintech, 66031‐1‐Ig) was used as endogenous control and anti‐SURF4 (1:1000, Bioswamp, PAB44330), anti‐XBP1 (1:1000, ABclonal, A1731) and anti‐DDIT3 (1:1000, Novus, NB600‐1335) was used. ECL peroxidase‐labelled sheep anti‐mouse antibody (GE Healthcare, NA931V) or HRP‐labelled goat anti‐rabbit antibody (Beyotime, A0208) were used as secondary antibodies.
2.14. Treatment with endoplasmic reticulum stress inducers and inhibitors
MEFs were treated with ER stress inducers Brefeldin A (Sigma, B5936), Tunicamycin (MedChemExpress, HY‐A0098), Thapsigargin (MedChemExpress, HY‐13433) or inhibitors TUDCA (MedChemExpress, HY‐19696A), Salubrinal (MedChemExpress, HY‐15486), Azoramide (MedChemExpress, HY‐18705), when they were subjected to reprogramming after with or without transduction of lentivirus.
2.15. Statistical analysis
The statistical data are presented as the mean ± SEM of at least three independent experiments. Significance was calculated using Student's t tests.
3. RESULTS
3.1. Surf4 can facilitate iPSCs induction
In our previous study, by mining proteomic data of preimplantation embryos, we found that several maternal factor candidates can facilitate somatic cell reprogramming. 10 In the present study, we aimed to explore the function and mechanism of one of the maternal factors, Surf4, in somatic cell reprogramming. We employed a secondary reprogramming system based on the drug‐inducible expression of the four Yamanaka factors (Figure S1A). Mouse embryonic fibroblasts were derived from R26rtTA; Col1a1‐4F2A; Oct4‐EGFP mice, 27 which harbour the doxycycline‐inducible polycistronic 4F2A cassette (Oct4, Sox2, Klf4 and c‐Myc), constitutively expressed reverse tetracycline transactivator (rtTA) 27 and expressed green fluorescent protein (GFP) under the control of the Oct4 promoter and distal enhancer. The induced expression of O, S, K, M under the addition of Doxcycline (Dox) could reprogram the MEFs into Oct4‐GFP+ iPSCs.
As indicated by the Oct4‐GFP signal, many more iPSC colonies appeared in the Surf4 OE group after day 9 (Figure 1A). At the end of reprogramming, Surf4 caused an approximate 4‐ to 8‐fold increase in iPSC colony number and up to a 20% increase in the percentage of Oct4‐GFP+ cells (Figure 1B). Together with improving reprogramming efficiency, we also observed that OE of Surf4 reduced cell proliferation during the process (Figure S1B). However, this phenomenon did not recur in MEFs (Figure S1C), which suggested that the proliferation attenuation by Surf4 was dependent on reprogramming. Therefore, we monitored the reprogramming kinetics with or without the OEof Surf4 and found that OE of Surf4 significantly reduced the THY1+ cell population (Figure S1D) and increased the SSEA1+ cell percentage (Figure S1E) during reprogramming. The primary iPS colonies in the Surf4 OE group exhibited normal morphology with a multiplied colony number compared with the control group (Figure 1D), as presented by AP staining (Figure 1E). In contrast, knockdown of Surf4 (Surf4 KD) led to a decrease in the number of AP+ or Oct4‐GFP+ colonies and the percentage of Oct4‐GFP+ cells (Figures 1C and S1F) without influencing the morphology of the iPSCs (Figure S1G).
FIGURE 1.
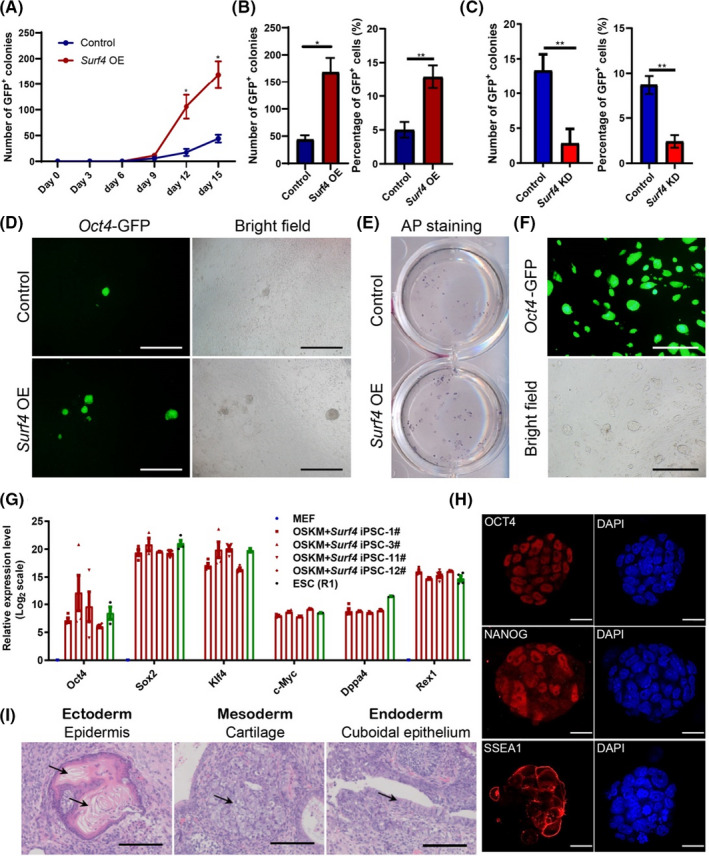
Surf4 Promotes iPSCs Generation. (A) Kinetics of Oct4‐GFP+ colony formation with or without exogenous Surf4 during the reprogramming process (n = 3). (B) The number of Oct4‐GFP+ colonies and the percentage of Oct4‐GFP+ cells 15 days after induction induced with or without exogenous Surf4 (n = 3, *p < 0.05, by Student's t test for comparison). (C) Cell proliferation curve with or without exogenous Surf4 during reprogramming. (D) Morphology of primary iPS colonies. Scale bars, 1000 μm. Magnification: ×40. (E) Alkaline phosphatase (AP) staining of the primary iPS colonies. (F) Morphology of an established OSKM + Surf4‐iPSC cell line. Scale bars, 1000 μm. Magnification: ×40. (G) Quantitative PCR (qPCR) analysis of pluripotent genes in OSKM + Surf4‐iPSCs. The data are presented as the means ± SEM (n = 3). (H) Immunostaining of pluripotent gene products OCT4, NANOG and SSEA1 in OSKM + Surf4‐iPSCs. The nuclei were stained with DAPI. Scale bars, 50 μm. (I) Haematoxylin and eosin (H&E) staining of teratomas generated from OSKM + Surf4‐iPSCs. Scale bars, 100 μm. See also Figure S1 and Table S1
Established iPS cell lines derived upon the OE of Surf4 (OSKM + Surf4‐iPSCs) displayed typical dome‐shaped morphology resembling embryonic stem cells (ESCs) (Figure 1F). Most of the iPS cell lines possessed a normal karyotype (Figure S1H). The pluripotent genes were activated at the RNA and protein levels (Figure 1G,H). We also evaluated the differentiation ability of these iPS cell lines in vitro and in vivo. The cells can differentiate into cells from all three germ layers through embryoid body (EB) formation (Figure S1I). They could also form teratomas consisting of cells from three germ layers after subcutaneous injection into SCID mice (Figure 1I). Thus, Surf4 can facilitate iPSC generation without influencing pluripotency.
3.2. Global profile of the effects of Surf4 on reprogramming
To investigate the effect of Surf4 on reprogramming, we analysed the transcriptomes of reprogramming cells with or without Surf4 on day 3 and MEFs. Based on the correlation matrix (Figure 2A) and PCA (Figure S2A), the two reprogramming cell samples were distinct from MEFs. When compared to MEFs individually, the reprogramming cells with or without OE of Surf4 had 3880 and 4077 differentially expressed genes (DEGs), with more than one‐half of these DEGs shared between both cell samples (Figure 2B). These shared genes were related to reprogramming: The upregulated genes were mainly enriched in keratinization, suggesting that the mesenchymal‐to‐epithelial transition (MET) occurred, and the downregulated genes were related to focal adhesion and development (Figure S2B).
FIGURE 2.
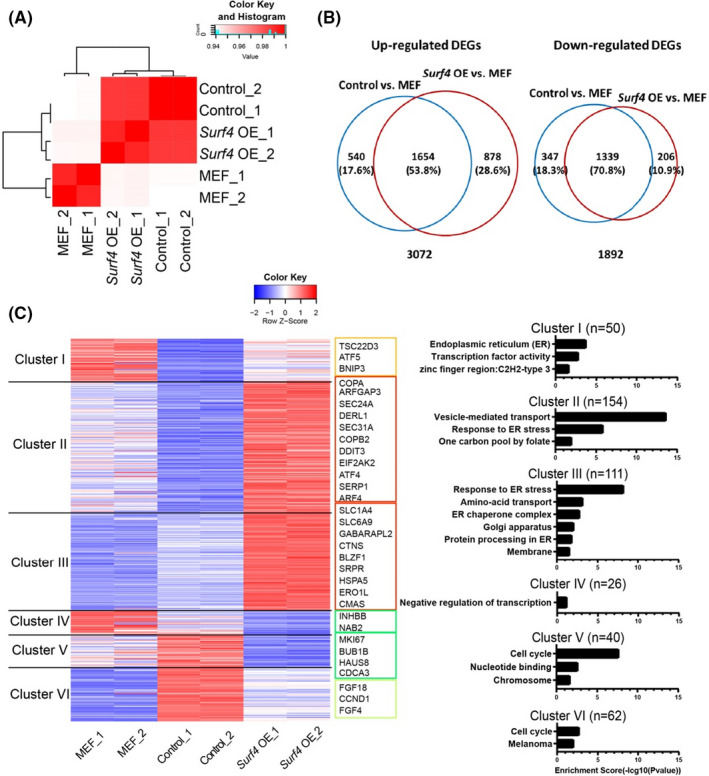
Transcriptional Changes Induced by Surf4 in Reprogramming. (A) Heat map of Pearson's correlation coefficients between MEFs and the reprogramming cells induced by OSKM with or without exogenous Surf4 at day 3. (B) Venn diagram showing overlap of upregulated genes and downregulated genes in reprogramming with or without Surf4 overexpression compared with the MEF group. (C) Heat map of clustering of differentially expressed genes among samples [MEFs and reprogramming cells (Control: OSKM + Vector or Surf4 OE: OSKM + Surf4)] on reprogramming day 3 (left). Gene ontology analysis of the corresponding clusters (right). See also Figure S2, Tables S1 and S2
By comparing the three cell groups, we obtained 443 DEGs (fold change >1.5, FDR <0.05), which were clustered into six groups (Figure 2C and Table S2). A large number of genes downregulated or mildly upregulated in the early stage of reprogramming (as the control group showed) were markedly upregulated by Surf4 OE (Cluster I, 50 genes; Cluster II, ~150 genes and Cluster III, ~110 genes). These genes were mainly enriched in vesicle‐mediated transport and response to ER stress, which were closely related to the function and localization of SURF4. In addition, the expression of dozens of cell cycle genes that was upregulated at the early phase of reprogramming but was decreased in the Surf4 OE group (Cluster V and Cluster VI), which was consistent with the proliferation suppression function of Surf4 (Figure 1C). The expression levels of the DEGs were also confirmed by qPCR (Figures S2C,D).
3.3. Activation of the response to ER stress facilitates reprogramming
To determine the effect of protein transport and ER stress on reprogramming, we employed brefeldin (BFA), a specific inhibitor of protein trafficking, and found that it could enhance reprogramming in a dose‐dependent manner (Figure 3A). BFA is an ER‐Golgi transport inhibitor that has been shown to cause protein accumulation in the ER and lead to ER stress. 34 Then, we tested two other ER stress inducers: tunicamycin (Tm), which inhibits N‐linked glycosylation and disrupts protein maturation in the ER, 35 and thapsigargin (Tg), which inhibits sarcoplasmic and ER Ca2+‐ATPase (SERCA), which subsequently depletes Ca2+ stores in the ER. 36 , 37 Tm and Tg both promoted reprogramming at proper concentrations but suppressed reprogramming at high concentrations owing to impaired cell survival (Figure S3A,B). However, when we tried an ER stress inhibitor, tauroursodeoxycholic acid (TUDCA), which functions as a chemical chaperone, reduces stress‐induced aggregation of proteins and inhibits the PERK pathway to prevent unfolded protein response (UPR) dysfunction, 38 , 39 we found that it negligibly affected reprogramming efficiency (Figure S3B). Thus, we speculated that the response to ER stress can promote reprogramming, and it may not through PERK pathway.
FIGURE 3.
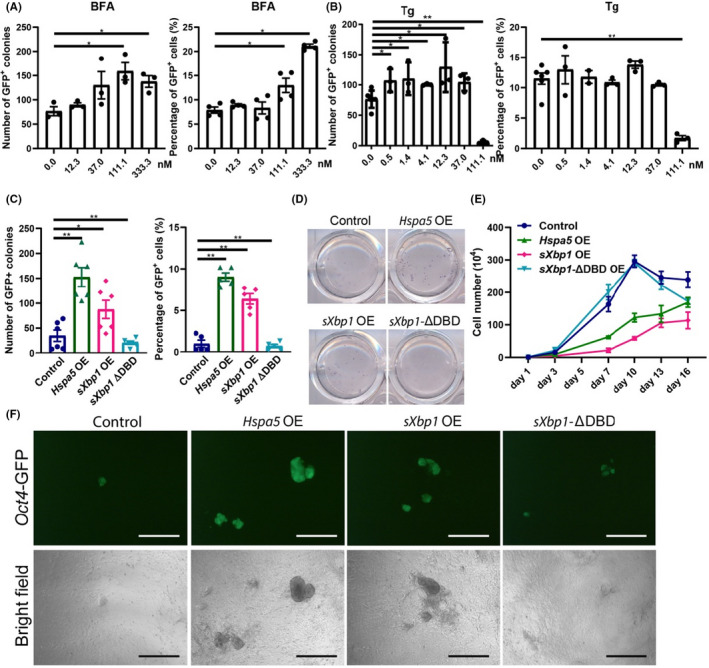
Activation of the Response to ER Stress Facilitates Reprogramming. (A) The number of Oct4‐GFP+ colonies and the percentage of Oct4‐GFP+ cells induced by OSKM in the presence of the UPR inducer brefeldin A (BFA). The cells were seeded in 12‐well plates at a density of 1.6×104 cells per well. (B) The number of Oct4‐GFP+ colonies and the percentage of Oct4‐GFP+ cells induced by OSKM in the presence of the UPR inducer thapsigargin (Tg). The cells were seeded in 12‐well plates at a density of 1.6 × 104 cells per well. (C) The number of Oct4‐GFP+ colonies and the percentage of Oct4‐GFP+ cells induced by OSKM plus the effectors of UPR, Hspa5, the spliced form of Xbp1 (sXpb1) and a dominant negative form of Xbp1 (sXbp1‐ΔDBD), which lacks the DNA‐binding domain of sXbp1. (D) AP staining of primary iPS colonies induced by OSKM plus effectors of the UPR. (E) Cell proliferation curve with or without exogenous Hspa5, sXbp1 and its dominant negative mutant during reprogramming. (F) Oct4‐GFP+ represents the morphology of the primary colonies. Scale bars, 200 μm. Magnification: ×40. See also Figure S3 and Table S1
Upon ER stress, the UPR is triggered and mediated by the IRE1‐XBP1, PERK‐eIF2α or ATF6 pathways to activate downstream transcription factors to reduce global protein synthesis and enhance the cellular protein‐folding capacity. Eventually, these factors relieve stress and re‐establish ER homoeostasis or lead to apoptosis if they fail to recover. Then, we examined the effects of the main mediators of the response to ER stress on re‐programming. OE of Hspa5 or the active spliced form of Xbp1 (sXbp1) dramatically increased the number of Oct4‐GFP+ or AP+ iPSC colonies and the percentage of Oct4‐GFP+ iPS cells (Figure 3C,D). Similar to Surf4, OE of Hspa5 and sXbp1 reduced cell proliferation during the process (Figure 3E). In contrast, sXbp1‐ΔDBD, the dominant negative mutant version lacking the DNA‐binding domain, markedly reduced the efficiency of iPSC generation (Figure 3C,D) without affecting the morphology of the iPSC colonies (Figure 3F). These data strongly suggest that the response to ER stress, especially the IRE1‐XBP1 pathway, is required for reprogramming.
3.4. Response to ER stress mediates the reprogramming facilitation by Surf4
To further investigate the relationship between Surf4 and the UPR in the ER (UPRER), we examined the expression level of ER stress‐induced effectors at day 3 during reprogramming with or without Surf4. These effectors, such as Ddit3, Hsp5a and Atf4, were boosted by exogenous Surf4 at the RNA and protein levels (Figure 4A,B). In the whole reprogramming process, these ER stress‐related genes exhibited transient increases at the early phase and the surge appeared at day 6, while Surf4 caused earlier increases (Figure S3E), as the expression level of Surf4 was upregulated during reprogramming (Figure S3C,D). These results suggested that Surf4 might facilitate reprogramming by activating UPRER at an early phase.
FIGURE 4.
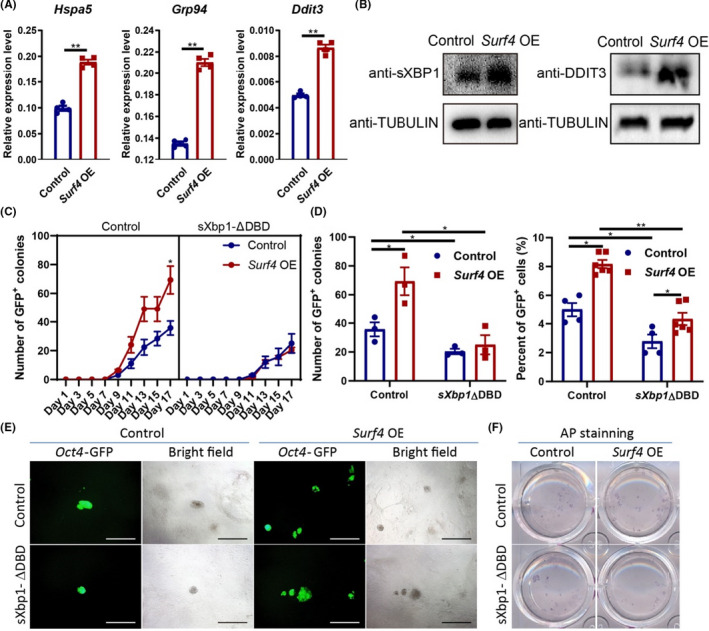
Response to ER Stress Mediates the Reprogramming Facilitation by Surf4. (A) The RNA level of ER stress‐related genes on day 3 of reprogramming with or without exogenous Surf4. Relative expression of these genes relative to β‐actin (n = 3, average ±SEM). (B) The protein level of ER stress‐related genes on day 3 of reprogramming with or without exogenous Surf4. (C) Kinetics of Oct4‐GFP+colony formation with or without exogenous Surf4 and sXbp1‐ΔDBD during reprogramming. (D) The number of Oct4‐GFP+ colonies and the percentage of Oct4‐GFP+ cells induced by OSKM plus Surf4 and sXbp1‐ΔDBD. (E) Morphology of the primary colonies induced by OSKM plus Surf4 and sXbp1‐ΔDBD. Scale bars, 1000 μm. Magnification: ×40. (F) AP staining of the primary iPS colonies. See also Figure S4 and Table S1
We speculated that Surf4 may improve the efficiency of reprogramming by temporarily activating the response to ER stress. To test our hypothesis, we introduced Surf4 and sXbp1‐ΔDBD at the same time in our reprogramming system to examine their function in iPSC generation. OE of sXbp1‐ΔDBD blocked the activation effect of Surf4 on reprogramming (Figure 4C–F). This result suggested that the ability of Surf4 to enhance reprogramming relies on the IRE‐XBP1 pathway.
It was recently reported that two small‐molecule modulators of the UPR, salubrinal (Sal) and azoramide (Azo), enhanced reprogramming. 40 Sal selectively inhibits eIF2alpha dephosphorylation 41 and activates the PERK/eIF2α branch of the UPR pathway, 42 while Azo improves ER protein‐folding ability and stimulates the expression of ER chaperones. 40 Therefore, we evaluated whether they can reverse the reduction in reprogramming efficiency caused by Surf4 KD. As shown in Figure S4A–C, the impaired reprogramming elicited by Surf4 KD was not reversed by these two activators, suggesting that activation of the PERK‐eIF2α pathway cannot rescue the reprogramming efficiency that was decreased upon Surf4 KD. Thus, these results suggested that downstream of ER stress, the IRE1‐XBP1 pathway mediated the effect of Surf4 in facilitating reprogramming.
4. DISCUSSION
Surf4 is enriched in mouse MII oocytes and zygotes 9 , 25 and significantly decreases from the 2‐cell stage during mouse preimplantation embryonic development. 25 , 26 We had previously found this maternal factor can promote somatic cell reprogramming, but its mechanism had not been elucidated. In this study, we demonstrate that Surf4 promotes reprogramming by activating the response to ER stress. This activation may cause a transient increase in the expression of UPR‐related genes, and the blockade of XBP1 impaired the effect of Surf4 on reprogramming.
It has been reported that Erv29p (homologous gene of Surf4 in S. cerevisiae) is involved in the degradation of soluble ER quality control substrates and is upregulated transcriptionally in response to ER stress. 11 It is possible excessive Surf4 may disturb the balance of protein transport flow and protein folding and processing in the ER, and subsequently, triggered the UPRER at the early stage of reprogramming. Such UPRER activation adapted to the stress, eventually restored of ER homeostasis or programmed cell death to protect the remaining cells.
The role of UPR‐related genes in reprogramming was consistent with a recent study, that reported transient activation of the UPRER is required for the acquisition of pluripotency. 43 In our study, we also observed that exogenous expression of an appropriate amount of Hspa5 or sXbp1 contributed to reprogramming. These UPRER effectors, as the downstream of Surf4, were transiently activated at the early stage of reprogramming. However, over‐high concentrations of ER stress inducers did not promote reprogramming owing to excessively reduced cell mount during the process. ‘Hyperactivated ER stress’ led to a decrease in reprogramming efficiency as a strong inducer of cell death. 40
During reprogramming, transient activation of the UPRER (we prefer to term it UPRER surge) at early phase is necessary and sufficient to promote reprogramming to somatic cells to a pluripotent state. 43 In our reprogramming system, UPRER surge occurred at day 6 in control group, and Surf4 brought such surge at day 3, which is probably why Surf4 facilitates reprogramming. Although the expression levels of sXpb1 and Ddit3 were significantly lower than those of the control at day 6 (Figure S3E), the increase in their expression levels on day 3 was sufficient to promote reprogramming.
Although the expression level of most ER stress‐relative genes were transiently upregulated by exogenous Surf4, not each of these genes overexpression can promote reprogramming efficiency. We have tried to overexpress Ddit3, but it did not facilitate somatic reprogramming (data not shown). In contrast, sXbp1 and Hspa5 can significantly increase reprogramming efficiency. Furthermore, the activation effect of Surf4 on reprogramming can be blocked by sXbp1‐ΔDBD, a dominant negative form of Xbp1. This means that the IRE‐XBP1 signal plays an important role in reprogramming.
The mechanism through which the activation of IRE‐XBP1 pathway, increases reprogramming efficiency remains to be elucidated. The UPR mainly alleviate ER stress by increasing the amount of molecular chaperones (such as HSPA5), ER luminal space and other folding catalysts to restore homeostasis, or to initiates apoptosis. 44 IRE1 mediated adaptive events, such as activation of XBP1s to upregulated expression levels of target genes, ER‐associated degradation (ERAD) of unfolded proteins, and IRE1‐dependent decay (RIDD) of cytosolic mRNAs. 45 These adaptive remodelling to ameliorate imbalances in ER proteostasis may benefit to the somatic signature turn off and allow pluripotent network to be set.
Previous studies have implicated Erv29p in ER quality control and are transcriptionally upregulated upon ER stress. 11 In this study, we found that overexpression of Surf4 in turn activates UPR at early phase in reprogramming. Although the exact mechanism of UPRER activation by Surf4 still needs to be investigated, activation of UPRER can promote reprogramming of human somatic cells to a pluripotent state. 43 We supported that overexpression of SURF4 may improve the reprogramming efficiency of human cells.
At the molecular level, SURF4 can interact with STIM1 in the ER to modulate store‐operated Ca2+ entry (SOCE). 46 In the present study, during reprogramming, SOCE was found to be reduced gradually and was further reduced by Surf4 at the early stage (data not shown). Whether SOCE is another barrier to reprogramming needs further investigation.
In the mature oocyte, many nucleic acids (mainly RNA) and proteins accumulate, which constitute the maternal material for early embryonic development. These factors not only drive sperm or somatic nuclei into totipotent embryos but also augment the efficiency of iPSCs. In recent years, an increasing number of oocyte factors have been found to promote somatic cell reprogramming through various mechanisms, including metabolic switching, 47 , 48 chromatin remodelling 49 , 50 and global epigenetic transformation. 51 , 52 , 53 , 54 Wider and deeper exploration of the action of maternal factors will pave the way to understanding somatic cell reprogramming.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
L.W. and S.H. designed the study, analysed the data and wrote the manuscript; J.S., K.Z., Y.Z. performed bioinformatics analysis; W.Y., R.Z., G.C., J.W, S.C., K.C., C.X., X.K., Y.Z., R.L. and H.W. performed some experiments and contributed to the discussion; S.G. and L.K. supervised the study and contributed to writing.
Supporting information
Supplementary Material
Table S1
Table S2
Table S3
ACKNOWLEDGEMENTS
We appreciate Professor Jia Li and Dr. Yubo Zhou from Shanghai Institute of Materia Medica, Chinese Academy of Sciences, for their advice and for providing anti‐sXBP1, anti‐ATF4 and anti‐DDIT3 antibodies. We thank Jiqing Yin and Chunxia Chen for their help with flow cytometry and Gang Chen for his help with experiments. We are also grateful to our laboratory colleagues for their assistance with experiments and advice. This work was supported by grants from the National Key Research and Development Program of China (2018YFC1004001 and 2018YFA0800101) and the National Natural Science Foundation of China (31721003, 31871489, and 31801243).
Wu L, He S, Ye W, et al. Surf4 facilitates reprogramming by activating the cellular response to endoplasmic reticulum stress. Cell Prolif. 2021;54:e13133. 10.1111/cpr.13133
Li Wu and Shengxiang He contributed equally.
Contributor Information
Lan Kang, Email: kanglan@tongji.edu.cn.
Shaorong Gao, Email: gaoshaorong@tongji.edu.cn.
DATA AVAILABILITY STATEMENT
The sequencing data sets have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through the GEO accession number GSE176177.
REFERENCES
- 1. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663‐676. [DOI] [PubMed] [Google Scholar]
- 2. Ryall JG, Cliff T, Dalton S, Sartorelli V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell. 2015;17(6):651‐662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu J, Ocampo A, Belmonte JCI. Cellular metabolism and induced pluripotency. Cell. 2016;166(6):1371‐1385. [DOI] [PubMed] [Google Scholar]
- 4. Kim KP, Choi J, Yoon J, et al. Permissive epigenomes endow reprogramming competence to transcriptional regulators. Nat Chem Biol. 2021;17(1):47‐56. [DOI] [PubMed] [Google Scholar]
- 5. Hanna J, Saha K, Pando B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462(7273):595‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jullien J, Pasque V, Halley‐Stott RP, Miyamoto K, Gurdon JB. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol. 2011;12(7):453‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maekawa M, Yamaguchi K, Nakamura T, et al. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474(7350):225‐229. [DOI] [PubMed] [Google Scholar]
- 8. Shinagawa T, Takagi T, Tsukamoto D, et al. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14(2):217‐227. [DOI] [PubMed] [Google Scholar]
- 9. Wang S, Kou Z, Jing Z, et al. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA. 2010;107(41):17639‐17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu L, Wu Y, Peng B, et al. Oocyte‐specific homeobox 1, Obox1, facilitates reprogramming by promoting mesenchymal‐to‐epithelial transition and mitigating cell hyperproliferation. Stem Cell Reports. 2017;9(5):1692‐1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem. 2001;276(26):23296‐23303. [DOI] [PubMed] [Google Scholar]
- 12. Foley DA, Sharpe HJ, Otte S. Membrane topology of the endoplasmic reticulum to Golgi transport factor Erv29p. Mol Membr Biol. 2007;24(4):259‐268. [DOI] [PubMed] [Google Scholar]
- 13. Belden WJ, Barlowe C. Role of Erv29p in collecting soluble secretory proteins into ER‐derived transport vesicles. Science. 2001;294(5546):1528‐1531. [DOI] [PubMed] [Google Scholar]
- 14. Yin Y, Garcia MR, Novak AJ, et al. Surf4 (Erv29p) binds amino‐terminal tripeptide motifs of soluble cargo proteins with different affinities, enabling prioritization of their exit from the endoplasmic reticulum. PLoS Biol. 2018;16(8):e2005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saegusa K, Sato M, Morooka N, Hara T, Sato K. SFT‐4/Surf4 control ER export of soluble cargo proteins and participate in ER exit site organization. J Cell Biol. 2018;217(6):2073‐2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Wang H, Xu B, et al. Receptor‐mediated ER export of lipoproteins controls lipid homeostasis in mice and humans. Cell Metab. 2020;33(2):350‐366. [DOI] [PubMed] [Google Scholar]
- 17. Lin Z, King R, Tang V, et al. The endoplasmic reticulum cargo receptor SURF4 facilitates efficient erythropoietin secretion. Mol Cell Biol. 2020;40(23):e00180‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emmer BT, Hesketh GG, Kotnik E, et al. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. eLife. 2018;7:e38839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mukai K, Ogawa E, Uematsu R, et al. Homeostatic regulation of STING by retrograde membrane traffic to the ER. Nat Commun. 2021;12(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitrovic S, Ben‐Tekaya H, Koegler E, Gruenberg J, Hauri HP. The cargo receptors Surf4, endoplasmic reticulum‐Golgi intermediate compartment (ERGIC)‐53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol Biol Cell. 2008;19(5):1976‐1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emmer BT, Lascuna PJ, Tang VT, et al. Murine Surf4 is essential for early embryonic development. PLoS One. 2020;15(1):e0227450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao X, Guo X, Tang X, et al. Misregulation of ER‐Golgi vesicle transport Induces ER stress and affects seed vigor and stress response. Front Plant Sci. 2018;9:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Low YS, Bircham PW, Maass DR, Atkinson PH. Kinetochore genes are required to fully activate secretory pathway expansion in S. cerevisiae under induced ER stress. Mol BioSyst. 2014;10(7):1790‐1802. [DOI] [PubMed] [Google Scholar]
- 24. Wang X, Wang H, Xu B, et al. Receptor‐mediated ER export of lipoproteins controls lipid homeostasis in mice and humans. Cell Metab. 2021;33(2):350‐66.e7. [DOI] [PubMed] [Google Scholar]
- 25. Wang B, Pfeiffer MJ, Drexler HC, Fuellen G, Boiani M. Proteomic analysis of mouse oocytes identifies PRMT7 as a reprogramming factor that replaces SOX2 in the induction of pluripotent stem cells. J Proteome Res. 2016;15(8):2407‐2421. [DOI] [PubMed] [Google Scholar]
- 26. Gao Y, Liu X, Tang B, et al. Protein expression landscape of mouse embryos during pre‐implantation development. Cell Rep. 2017;21(13):3957‐3969. [DOI] [PubMed] [Google Scholar]
- 27. Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single‐gene transgenic mouse strains for reprogramming adult somatic cells. Nat Methods. 2010;7(1):56‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y, Lee J, Reno CM, et al. Regulation of glucose homeostasis through a XBP‐1‐FoxO1 interaction. Nat Med. 2011;17(3):356‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full‐term development of tetraploid blastocyst‐complemented embryos. Cell Stem Cell. 2009;5(2):135‐138. [DOI] [PubMed] [Google Scholar]
- 30. Gao Y, Chen J, Li K, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12(4):453‐469. [DOI] [PubMed] [Google Scholar]
- 31. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics. 2009;25(9):1105‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. 2013;29(1):15‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nat Biotechnol. 2015;33(3):290‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chardin P, McCormick F. Brefeldin A: the advantage of being uncompetitive. Cell. 1999;97(2):153‐155. [DOI] [PubMed] [Google Scholar]
- 35. Mizrahi A, O'Malley JA, Carter WA, Takatsuki A, Tamura G, Sulkowski E. Glycosylation of interferons. Effects of tunicamycin on human immune interferon. J Biol Chem. 1978;253(21):7612‐7615. [PubMed] [Google Scholar]
- 36. Jackson TR, Patterson SI, Thastrup O, Hanley MR. A novel tumour promoter, thapsigargin, transiently increases cytoplasmic free Ca2+ without generation of inositol phosphates in NG115‐401L neuronal cells. Biochem J. 1988;253(1):81‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, Dawson AP. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)‐ATPase. Proc Natl Acad Sci USA. 1990;87(7):2466‐2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xie Q, Khaoustov VI, Chung CC, et al. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress‐induced caspase‐12 activation. Hepatology (Baltimore, MD). 2002;36(3):592‐601. [DOI] [PubMed] [Google Scholar]
- 39. Liu F, Cui Y, Ge P, Luan J, Zhou X, Han J. Tauroursodeoxycholic acid attenuates inorganic phosphate‐induced osteoblastic differentiation and mineralization in NIH3T3 fibroblasts by inhibiting the ER stress response PERK‐eIF2α‐ATF4 pathway. Drug Discov Ther. 2015;9(1):38‐44. [DOI] [PubMed] [Google Scholar]
- 40. Guallar D, Fuentes‐Iglesias A, Souto Y, et al. ADAR1‐Dependent RNA Editing Promotes MET and iPSC Reprogramming by Alleviating ER Stress. Cell Stem Cell. 2020;27(2):300‐14.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boyce M, Bryant KF, Jousse C, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307(5711):935‐939. [DOI] [PubMed] [Google Scholar]
- 42. Lee DY, Lee KS, Lee HJ, et al. Activation of PERK signaling attenuates Abeta‐mediated ER stress. PLoS One. 2010;5(5):e10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simic MS, Moehle EA, Schinzel RT, et al. Transient activation of the UPR(ER) is an essential step in the acquisition of pluripotency during reprogramming. Sci Adv. 2019;5(4):eaaw0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bashir S, Banday M, Qadri O, et al. The molecular mechanism and functional diversity of UPR signaling sensor IRE1. Life Sci. 2021;265:118740. [DOI] [PubMed] [Google Scholar]
- 45. Chang TK, Lawrence DA, Lu M, et al. Coordination between two branches of the unfolded protein response determines apoptotic cell fate. Mol Cell. 2018;71(4):629‐36.e5. [DOI] [PubMed] [Google Scholar]
- 46. Fujii Y, Shiota M, Ohkawa Y, et al. Surf4 modulates STIM1‐dependent calcium entry. Biochem Biophys Res Comm. 2012;422(4):615‐620. [DOI] [PubMed] [Google Scholar]
- 47. Khaw SL, Min‐Wen C, Koh CG, Lim B, Shyh‐Chang N. Oocyte factors suppress mitochondrial polynucleotide phosphorylase to remodel the metabolome and enhance reprogramming. Cell Rep. 2015;12(7):1080‐1088. [DOI] [PubMed] [Google Scholar]
- 48. Mathieu J, Zhou W, Xing Y, et al. Hypoxia‐inducible factors have distinct and stage‐specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014;14(5):592‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gonzalez‐Muñoz E, Arboleda‐Estudillo Y, Otu HH, Cibelli JB. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science. 2014;345(6198):822‐825. [DOI] [PubMed] [Google Scholar]
- 50. Kunitomi A, Yuasa S, Sugiyama F, et al. H1foo has a pivotal role in qualifying induced pluripotent stem cells. Stem Cell Reports. 2016;6(6):825‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jiang Q, Huang X, Hu X, et al. Histone demethylase KDM6A promotes somatic cell reprogramming by epigenetically regulating the PTEN and IL‐6 signal pathways. Stem Cells (Dayton, Ohio). 2020;38(8):960‐972. [DOI] [PubMed] [Google Scholar]
- 52. Ang YS, Tsai SY, Lee DF, et al. Wdr5 mediates self‐renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hernandez C, Wang Z, Ramazanov B, et al. Dppa2/4 facilitate epigenetic remodeling during reprogramming to pluripotency. Cell Stem Cell. 2018;23(3):396‐411.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang Y, Zhang H, Wang L, et al. JMJD3 acts in tandem with KLF4 to facilitate reprogramming to pluripotency. Nat Commun. 2020;11(1):5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1
Table S2
Table S3
Data Availability Statement
The sequencing data sets have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through the GEO accession number GSE176177.


