Summary
Activity of liver x receptor (LXR), the homeostatic regulator of cholesterol metabolism, is elevated in triple-negative breast cancer (BCa) relative to other BCa subtypes, driving drug resistance and metastatic gene signatures. The loci encoding LXRα and LXRβ produce multiple alternatively spliced proteins, but the true range of variants and their relevance to cancer remain poorly defined. Here, we report seven LXR splice variants, three of which have not previously been reported and five that were prognostic for disease-free survival. Expression of full-length LXRα splice variants was associated with poor prognosis, consistent with a role as an oncogenic driver of triple-negative tumor pathophysiology. Contrary to this was the observation that high expression of truncated LXRα splice variants or any LXRβ splice variant was associated with longer survival. These findings indicate that LXR isoform abundance is an important aspect of understanding the link between dysregulated cholesterol metabolism and cancer pathophysiology.
Subject areas: Molecular biology, Molecular mechanism of gene regulation, Cancer systems biology, Cancer
Graphical abstract
Highlights
-
•
LXR splicing is extensive in breast cancer
-
•
Three new LXR splice variants are confirmed at transcript and protein level
-
•
Full-length LXRα is associated with shorter disease-free survival in patients with TNBC
-
•
LXRβ or truncated LXRα variants associate with better prognosis
Molecular biology; Molecular mechanism of gene regulation; Cancer systems biology; Cancer
Introduction
Breast cancer (BCa) is the most commonly diagnosed cancer in women in the UK and is the cause of ∼600,000 cancer death worldwide (Bray et al., 2018). The triple-negative BCa (TNBC) subtype has higher rates of recurrence in the first three years after prognosis and increased mortality rates (Hudis and Gianni, 2011). TNBC is defined by low estrogen (ER), progesterone (PR), and Her2 receptor expression (Perou et al., 2000). Due to this lack of expression of therapeutic molecular targets, primary disease can only be systemically treated with cytotoxic chemotherapy, and TNBC remains a cancer of unmet clinical need; novel targeted therapies are urgently needed.
Nutritional and epidemiological studies indicate that cholesterol metabolism may play a role in the etiology and severity of TNBC (Cioccoloni et al., 2020; Jiang et al., 2019; Liu et al., 2017; WCRF/AICR, 2017). The liver x receptors (LXRα/NR1H3; LXRβ/NR1H2) are homeostatic regulators of cholesterol metabolism and as such have been suggested as potential therapeutic targets. LXRα activation by hydroxylated cholesterol (oxysterols) has been linked to poor survival by driving multi-drug resistance in patients with TNBC (Hutchinson et al., 2021) and increased metastasis in mouse models (Baek et al., 2017; Nelson et al., 2013). On the other hand, LXRβ activation by histamine conjugated oxysterols such as dendrogenin A can induce lethal autophagy in BCa (Poirot and Silvente-Poirot, 2018). Intriguingly, ER-negative and ER-positive BCa subtypes respond differently to LXR ligands (Hutchinson et al., 2019) even though oxysterol concentrations are similar between BCa subtypes (Solheim et al., 2019). Collectively, these data imply that the differential control over LXR’s transcriptional regulation between subtypes is not simply at the level of ligand type or concentration.
LXRα and LXRβ are each thought to be translated from their own single main transcript variants, producing 447 and 460 amino acid proteins, respectively (Shinar et al., 1994; Willy et al., 1995). These “full-length” isoforms harbor domains that provide distinct functions, including activation function 1 (AF1), hinge region (H), DNA-binding domain (DBD), and ligand-binding domain (LBD) (Willy et al., 1995), as is typical for many ligand-dependent nuclear receptors.
Aside from the canonical full-length LXRα1, four additional LXRα splice variants have been reported in human cell lines or tissue samples previously (Chen et al., 2005; Endo-Umeda et al., 2012), and a single report found one alternative splice variant to the major LXRβ1 isoform (Hashimoto et al., 2009). These studies found that alternative splicing of LXR disrupts the integrity of the DNA- and ligand-binding domains, rendering the proteins with reduced or even absent response to ligand (Chen et al., 2005; Endo-Umeda et al., 2012). In the case of LXRβ, a single known splice variant functions as an RNA co-activator (Hashimoto et al., 2009). Genomic and proteomic databases including National Center for Biotechnology Information (NCBI) (Pruitt et al., 2005), The Cancer Genome Atlas (TCGA) Splicing Variants database (TSVdb (Sun et al., 2018)), Ensembl (Aken et al., 2016), and Universal Protein Resource (UniProt) (Bairoch et al., 2005) indicate that the LXRα and LXRβ genes encode more splice variants than previously reported. For example, a study exploring some of these databases indicated up to 62 different LXRα transcript variants (Annalora et al., 2020), making it the most extensively spliced member of the nuclear receptor superfamily.
However, experimental evidence for the existence of such an array of isoforms is lacking. Given the strong links between cholesterol metabolism, for which the LXRs play vital regulatory roles, and TNBC etiology, the aim of this study was to describe the LXR splice repertoire and clinical significance in human triple-negative BCa.
Results
Database analysis of LXR transcript splice variance in breast cancer
To evaluate the variety of LXR splice variants, the NCBI and ENSEMBL databases were mined for mRNA transcript variants and UNIPROT database for protein variants. In total, this analysis indicated there were 64 LXRα transcript variants, of which 48 could code for 26 LXRα protein variants (see Figure S1). We found 11 LXRβ transcript variants that could code for 9 LXRβ proteins (see Figure S2). These included all nine variants (α1-α5, β1-β4) later observed in BCa cells and/or breast tumor tissue (summarized in Figure 1). Previous publications have used a consistent naming system for the LXRα isoforms but deviate from nomenclature of NCBI, TCGA Splicing (TSVdb), ENSEMBL, and UNIPROT databases. For example, NM_001130101 previously referred to as α3 (Chen et al., 2005; Endo-Umeda et al., 2012; Rondanino et al., 2014) is annotated as α2 in the NCBI database. NM_001130102 has previously been referred to as α2 (Chen et al., 2005; Rondanino et al., 2014) but is annotated as α3 in the NCBI database. Furthermore, due to the complexity of potential LXR transcript variants (Annalora et al., 2020), the naming system from previous publications (Chen et al., 2005; Endo-Umeda et al., 2012; Rondanino et al., 2014) was not appropriate, and instead, the NCBI annotation system was adopted. Details of all previous reported primer pairs used to detect LXR are available in Figure S3. Full details of how variants pertain to databases and previous studies are provided for LXRα in Figure S4 and Table S1 and for LXRβ in Figure S5 and Table S2. During this annotation and documentation process, some NCBI predicted LXR isoforms were found to be identical to curated isoforms. For example, the LXRαx5 amino acid sequence is derived from two NCBI predicted isoforms (XP_011518107.1 and XP_005252763.1) and is identical to the LXRα1 amino acid sequence (NP_005684.2). We confirmed this using Clustal Omega multiple sequence alignment software (https://www.ebi.ac.uk/Tools/msa/clustalo/) (EMBL-EBI, Dublin, Ireland) (Figure S6).
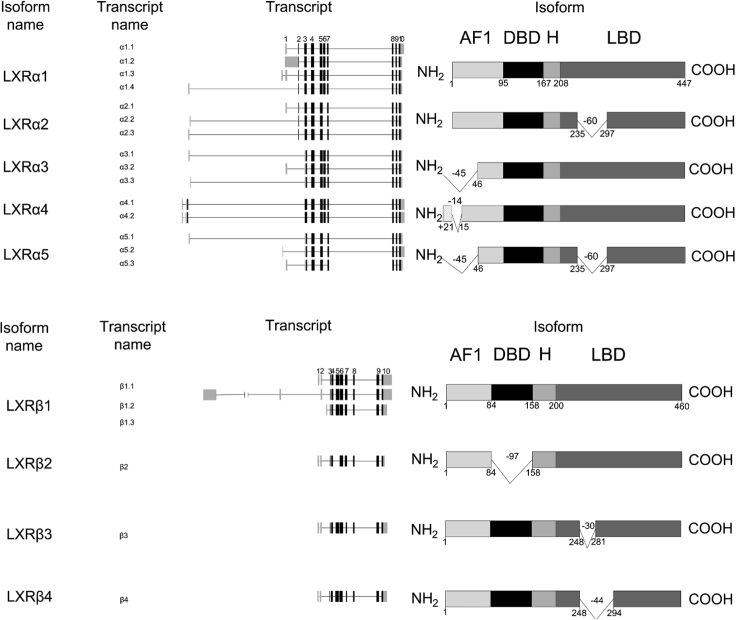
Figure 1.
Schematic diagram of LXR splice variants
(A and B) Schematic diagram of (A) LXRα and (B) LXRβ splice variants detected at RNA and protein level in this study. AF1 = activation function 1; DBD = DNA binding domain; H = hinge; LBD = ligand binding domain. The numbers right below the isoform domain boxes are represented the position of amino acids.
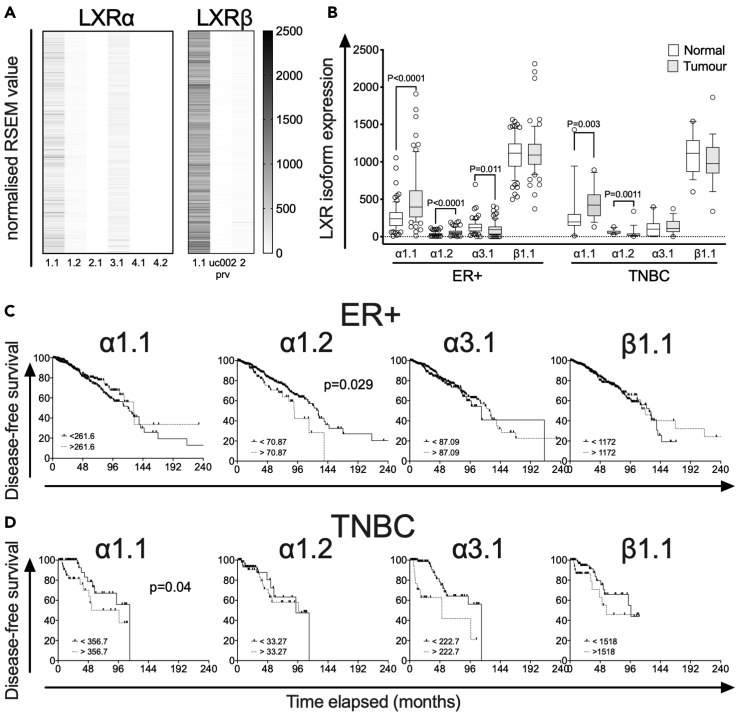
To investigate the expression of LXR splice variants in BCa, we first examined RNA-seq data from 1,103 BCa tumors in the TCGA Pan-Cancer Atlas utilizing the TSVdb web interface (Sun et al., 2018). These data showed that out of six LXRα and three LXRβ splice variants with RSEM reads (Li and Dewey, 2011), the α1.1 (median RSEM value = 231.52) and β1.1 (median RSEM value = 1158.53) variants were most highly expressed in primary BCa samples (Figure 2A). The α3.1 (median RSEM value = 115.42), α1.2 (median RSEM value = 31.88), and β2 (median RSEM value = 21.69) transcripts were expressed at low levels. No other transcript variants were detected in this database (Figure 2A).
Figure 2.
LXR splicing events in BCa validated by TCGA Splicing Variants database (TSVdb)
(A) Heatmap visualization of normalized RSEM (RNA-Seq by Expectation Maximization) value expression of LXRα and LXRβ in 1,103 TCGA BCa samples. Note: Transcript uc002prv was only reported in TSVdb and is not recorded in NCBI, ENSEMBL, or UNIPROT databases.
(B) Comparison of LXR transcript variant expression, for α1.1, α1.2, α3.1, and β1.1, in matched tumor and normal tissues grouped by BCa subtype, ER+ (n = 78), and TNBC (n = 18). Statistical analysis by Mann-Whitney two-tailed U tests. P ≤ 0.05 was considered significant. The line shows the median value, the box shows 10th to 90th percentile, and whiskers show minimum-to-maximum value.
(C and D) Kaplan-Meier survival curves plotting disease-free survival of patients with TCGA BCa. The TCGA BCa samples (downloaded from TSvdb TCGA splicing database) were grouped by BCa subtype, (C) ER+ (n = 803), and (D) TNBC (n = 101). Survival curves of each subtype were separated into two groups: no event (ER+ = 675, TNBC = 79) and event (ER+ = 128; TNBC = 22) based on their overall survival data reported in cBioportal. p value is based on the log rank test, and data groups were considered significantly different if p < 0.05.
We further examined expression of the four variants (α1.1, α1.2, α3.1, β1.1) in matched tumor and normal samples from ER+ and TNBC subtypes. Both α1.1 and α1.2 were expressed at significantly higher levels in ER+ and TNBC tumor tissue compared to adjacent normal tissue (p < 0.01 for all; Figure 2B). α3.1 expression was lower in ER+ tumor tissue than adjacent normal (p = 0.011; Figure 2B), but there was no difference in expression of this isoform between tumor and normal tissue in TNBC disease. There was no difference in β1.1 expression in either ER+ or TNBC tumors relative to adjacent normal (p > 0.05; Figure 2B). There was no difference in expression of any individual LXR variants between ER+ and TNBC tumors (see Figure S7).
We then examined if transcript variant expression identified in TSVdb was associated with disease-free survival (DFS) in ER+ (Figure 2C) or patients with TNBC (Figure 2D). High α1.2 expression was associated with shorter DFS in ER+ patients (p = 0.029), while in patients with TNBC, high α1.1 expression was associated with shorter DFS (p = 0.04). Neither α3.1 nor LXRβ expression was associated with DFS. These data suggest that elevated LXRα1 (α1) may be linked to poor prognosis in patients with BCa.
LXR expression in breast cancer cell lines
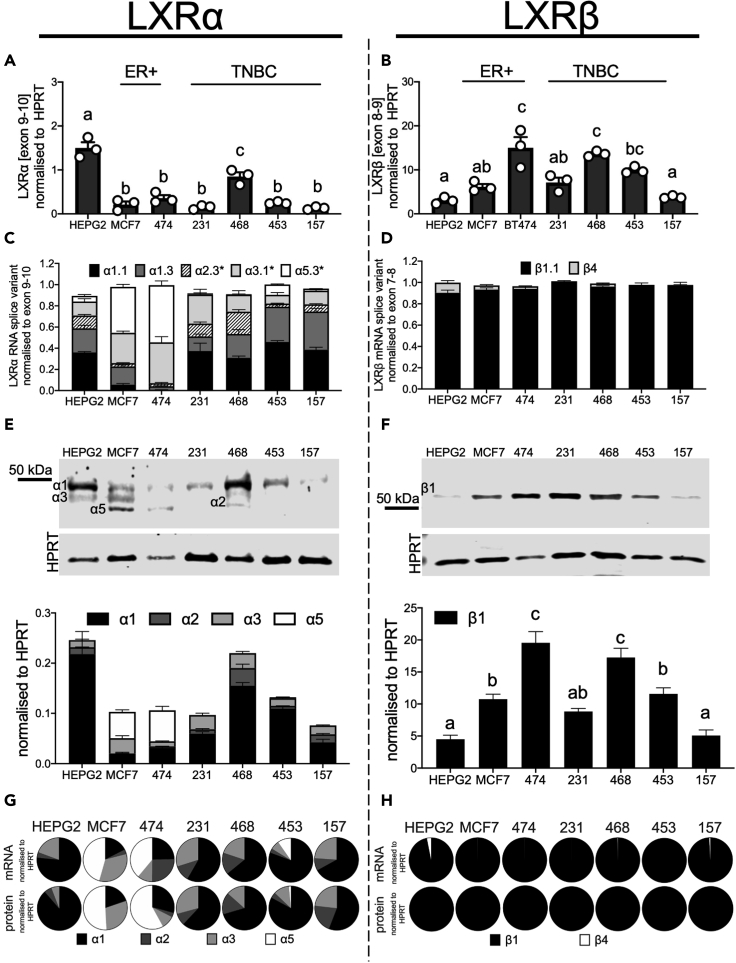
Having established evidence for LXR splice variant expression in the TSVdb dataset, we next examined transcript expression in a panel of BCa cell lines. HEPG2 cells express relatively high levels of both LXRα and LXRβ (Laffitte et al., 2001) so were included as a positive control. Total LXRα and LXRβ RNA expression was determined using SYBR green primers that target the exon 9–10 junction of LXRα and exon 8–9 junction for LXRβ. These exons are spliced together in all previously reported variants and in all variants described in this study (see Tables S1 and S2; Figures S1 and S2) and were used to estimate the total pool of LXR transcripts and for normalization of variant expression.
The claudin high TNBC cell line MDA.MB.468 had highest expression of LXRα of all BCa cell lines (p < 0.05). MDA.MB.468, MDA.MB.453, and BT474 had highest LXRβ (Figures 3A and 3B). In all cell lines analyzed, the expression of LXRα mRNA was significantly lower than that of LXRβ (all p < 0.05; Figure S8), recapitulating the observations from the TSVdb (Figure 2A). Across the cell lines, the transcripts measured accounted for between 91 and 100% of LXRα (exon 9–10) and 94–102% of LXRβ (exon 8–9) exon-exon boundaries (Figures 3C and 3D), indicating other variants were expressed at very low levels or did not contain the exon 9–10/7–8 boundaries. Across all cell lines, we detected five mRNA species (α1.1, α1.3, α2.3, α3.1, α5.3; Figure 3C) predicted to code for four LXRα protein variants (α1, α2, α3, α5) and two coding for LXRβ (β1, β4; Figure 3D). LXRα5 and LXRβ4 have not been reported in the literature previously.
Figure 3.
Diversity of LXR splice variants in breast cancer cell line panel
(A and B) RNA analyses of total LXRα (A) and LXRβ (B) expression.
(C and D) The stacked bar graphs show LXRα (C) and LXRβ (D) transcript variants normalized to ubiquitously expressed exon-exon junctions. Asterisks denote ambiguous amplicons.
(E and F) Representative blots and densitometry of LXRα (E) and LXRβ (F) protein variants observed in cell lines.
(G and H) Pie charts show contribution of each transcript variant and protein variant to the total amount of LXRα (G) and LXRβ (H). All data shown are mean of three independent replicates with SEM. Statistical significance was measured by two-tailed one-way analysis of variance (ANOVA), and significant differences are denoted with different letters if p ≤ 0.05.
α1 transcripts were dominant (50–80% of all LXRα) in HEPG2 and TNBC cells (Figure 3C: note black and dark gray sections corresponding to α1.1 and α1.3, respectively). In the ER+ cell lines, α5 (40–50%) and α3 (30–40%) were the majority species (Figure 3C: note light gray and white sections corresponding to α5.3 and α3.1, respectively). α2 comprised between 5 and 20% depending on the cell line (Figure 3C: hatched sections). LXRβ1.1 was detected in all BCa cell lines, with a very small amount of β4 transcript found in HEPG2 and the ER+ cell lines only (Figure 3D). We successfully detected over 90–100% of all the LXR variants (harboring the exon 9–10 and 7–8 junctions) present in the cell lines. From these data, we concluded that full-length α1 was the dominantly expressed variant in TNBC cells, truncated α5 was the dominant LXRα isoform in ER-positive cells, and β1 was the dominant LXRβ transcript across all cell types.
We next performed immunoblotting to establish the range of protein variants present. Representative blots and densitometry analysis show that bands corresponding to predicted sizes of LXRα1 (≈50kDa) and LXRβ1 (≈55kDa) were robustly and reproducibly detected in all cell lines (Figures 3E and 3F, respectively). However, optimization of immunoblotting experimental conditions, including antibody choice, indicated that multiple additional bands were present when probing for LXRα (Figure 3E). These bands closely corresponded to sizes of proteins that would be coded by the transcripts identified above, namely α1 (50 kDa), α2 (44 kDa), α3 (46 kDa), and α5 (39 kDa). The cell line-specific pattern of protein expression matched that of the transcript expression; α1 RNA and protein were highly expressed in HEPG2 and the TNBC cell lines, while α5 was dominant in the ER+ cells (Figure 3G). For LXRβ, only a single isoform β1 was present at both RNA and protein level (Figure 3H).
We concluded that in TNBC cell lines, full-length LXRα1 is the most abundant isoform comprising 50–80% of LXRα protein (depending on cell line), with two α1 transcript variants accounting for 51–75% of LXRα transcript. LXRα3, which lacks part of the AF-1 domain and has diminished response to ligand (Chen et al., 2005), was the next most abundant producing 15–30% of the protein and 8–27% of the transcript. LXRα2, which is non-responsive to ligand (Chen et al., 2005), made up 8–20% of the total LXRα protein coded by two transcripts producing 7–12% of the LXRα coding RNA. Only a small amount of LXRα5 was detected in TNBC (<10% in MDA.MB.453 cells and undetected in other TNBC lines). Interestingly, this “double-truncation” variant that lacks both the AF-1 region of α3 and the LBD region of α2 was the most abundant isoform in ER+ cells at protein (35–70%) and transcript level (43–54%).
LXR splice variant expression and clinical significance in triple-negative breast cancer
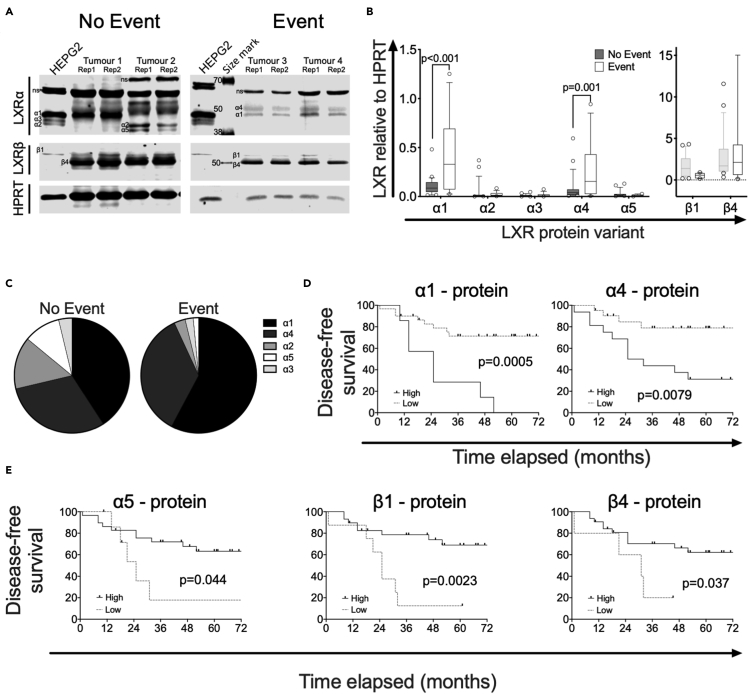
To evaluate LXR variant expression and significance in clinical samples, LXR protein expression was measured in a cohort of 38 fresh-frozen TNBC tumor samples (mean follow-up of three years), which has been reported previously (Hutchinson et al., 2019; Solheim et al., 2019) (Table S3). Patients were dichotomized into two groups: “No Event” patients defined as those who were alive and disease free at the time of last reporting or had died from unrelated reasons; “Event” patients were those who had died from their disease and/or had disease recurrence. Representative blots are shown in Figure 4A. Event patients had significantly higher expression than “No Event” patients of both full-length isoforms (α1: p < 0.0001; α4: p = 0.001; Figure 4B). Together, α1 and α4 comprised 70% of the total LXRα protein in the “No Event” group, but this rose to 93% in the “Event” group (Figure 4C). No significant difference was observed in the level of α2, α3, α5, β1, or β4 protein variants between groups (Figure 4B).
Figure 4.
LXRα1 and LXRα4 are highly expressed in patients with TNBC with poor survival
(A) Representative blots showing the LXR splicing events in tumors derived from 38 patients with TNBC, who were either free from disease (No Event; n = 23) or who had relapsed or died (Event; n = 15) after follow-up (A).
(B) Differential expressions of LXRα and LXRβ protein variants in No Event and Event groups were plotted in box and whiskers charts (B). The line shows the median value, the box shows 10th to 90th percentile, and whiskers show minimum-to-maximum values. Statistical analysis was established using multiple t-tests with Holm-Sidak for multiple correction; p ≤ 0.05 was considered significant.
(C) Pie charts show contribution of each transcript variant and protein variant to the total amount of LXRα in No Event and Event patients (C).
(D and E) Kaplan-Meier survival curves plotting disease-free survival of patients with TNBC dichotomized based on protein expression of full-length LXRα1 and LXRα4 transcripts (D) or truncated LXRα5 and LXRβ1 and LXRβ4 (E). All protein measured relative to HPRT. Data derived from the mean of two different slices of tumors. Significance determined by the Log rank (Mantel-cox) test where p ≤ 0.05 was considered significant.
When patients were dichotomized using Receiver Operating Characteristic (ROC) analysis, high protein expression of the full-length variants were, as expected, found associated with significantly shorter DFS (α1: p = 0.0005; α4: p = 0.0079; Figure 4D). Interestingly, in this more nuanced analysis, high expression of α5 (p = 0.044), LXRβ1 (p = 0.0023), and LXRβ4 (p = 0.037) was associated with significantly longer DFS (Figure 4E). Furthermore, dichotomizing based on RNA expression also indicated that high expression at the transcript level was associated with longer or shorter DFS in the same pattern as found for the protein (see Figure S9A). In addition, α3.1, which codes for intact LBD but harboring a deletion in the AF1 domain, was also associated with shorter DFS (p = 0.022; Figure S9A). The α2.3 transcript, which codes for a protein with the same LBD deletion as α5 was, like α5, associated with longer DFS (p = 0.031; Figure S9A). α2 and α3 protein were not associated with DFS (see Figure S9B). We concluded that full-length LXRα isoforms may exacerbate disease severity, whereas those lacking the full LBD or LXRβ isoforms were associated with reduced disease severity.
Validation of isoform identity
Bands representing LXR variants were reduced by targeted siRNA
To confirm if the protein bands were LXRα variants, we first performed siLXRα treatments in cell lines. Using previously validated (Hutchinson et al., 2021) siRNA duplexes (Origene trisilencers) against LXRα and LXRβ, we found that the protein bands predicted by size to be α1, α2, α3, α5, and β1 were all significantly reduced by targeted siRNA in all cell lines tested (all p < 0.05; Figure S10). Note that α4 and β4 were not expressed in cell lines and so could not be validated in this way. Furthermore, siLXRα did not reduce LXRβ expression and siLXRβ did not reduce expression of any LXRα isoform, and, as is characteristic of LXRs (Chen et al., 2005; Prüfer and Boudreaux, 2007), all protein variants were localized in the nucleus (see Figure S11).
Unique peptides representing α4 and β1 were identified by MS S-trap mass spectrophotometry
As was the case for cell line analyses, confirmation of the identity of the observed protein variants in tumor tissue was important, especially having identified the α4 variant which has not previously been reported in the literature. As siRNA application was not possible on the tumor samples, we performed S-trap (Hayoun et al., 2019) coupled mass spectrometry (St-MS) in cell line and tumor lysates samples selected to represent as much of the diversity in LXRα (Tables 1 and S4) and LXRβ (Tables 2 and S5). This included two tumors in duplicate: ligand-treated MDA.MB.468 cells (see Figure S12A) and in siCON, siLXRα, and siLXRβ (see Figure S12B). We found 15 peptides uniquely corresponding to α4 BLAST sequence alignment indicated they were high-quality hits (100% identity), including the two first exons coding for the alternative AF1 domain in this isoform (see Figure S12C – boxed amino acids) supporting our prior conclusion that α4 was expressed in some tumor samples. Unique peptides of β1 were also detectable in our tumor sample (see Figure S12D – boxed amino acids). However, unique peptides corresponding to α1, α2, α3, α5, or β4 were not produced by our enzymatic cleavage, and in silico analysis indicated these isoforms would not be distinguishable from each other (Table3). Our next approach was to perform immunoprecipitation. We found this successfully enriched each isoform (see Figure S12E), but limited amounts of tumor meant bands could not be visualized for excision on Coomassie gels. We concluded that α4 and β1 were present in tumor tissue.
Table 1.
LXRα peptides detected by S-trap column coupled with mass spectrometry (MS)
| Sample | Total identified peptides | −10IgP [LXRα] | Coverage LXRα |
Supporting peptides [LXRα] | Unique | Amino acids position |
|
|---|---|---|---|---|---|---|---|
| Peptides | Start | End | |||||
| Tumor 2 Rep 1 | 3500 | 52.32 | 13% | Q.AQ(+.98)GGSSC(+57.02)ILR.E | No | 32 | 41 |
| A.QGGSSC(+57.02)ILR.E | No | 33 | 41 | ||||
| R.MPHSAGGTAGV.G | No | 46 | 56 | ||||
| R.ASSPPQ(+.98)ILPQ.L | No | 196 | 205 | ||||
| R.ASSPPQILPQLSPEQ(+.98)LGMIEK.L | No | 196 | 216 | ||||
| R.AMN(+.98)ELQLN(+.98)DAEFALLI.A | No | 345 | 360 | ||||
| Tumor 2 Rep 2 |
3971 | 58.66 | 22% | K.C(+57.02)RQ(+.98)AGM(+15.99)REEC(+57.02)VLSEEQ(+.98)IRLK.K | No | 158 | 177 |
| R.QEEEQAHATSLPPR.A | No | 182 | 195 | ||||
| R.ASSPPQILPQLSPEQ(+.98)LGMIEK.L | No | 196 | 216 | ||||
| L.SPEQ(+.98)LGMIEK.L | No | 207 | 216 | ||||
| G.M(+15.99)IEKLVAAQQ(+.98)Q(+.98).C | No | 213 | 223 | ||||
| K.QLPGFLQLSR.E | No | 274 | 283 | ||||
| Q.VEFINPIFEFSR.A | No | 333 | 344 | ||||
| R.AMNELQLNDAEFALLI.A | No | 345 | 360 | ||||
| Tumor 5 Rep 1 |
3713 | 61.52 [α4 +LXRα homolog] | 19% | MQ(+.98)QTSWN(+.98)PLGGTC(+57.02)K.Q | Yes [α4] | −14 | −1 |
| 59.30 [LXRα homolog] | 16% | R.AEPPSEPTEIRPQ(+.98)K.R | No | 70 | 83 | ||
| KC (+57.02)RQAGMR.E | No | 196 | 208 | ||||
| R.ASSPPQILPQLSP.E | No | 196 | 208 | ||||
| R.ASSPPQILPQLSPEQ(+.98)LGMIEK.L | No | 196 | 216 | ||||
| G.MIEKLVAAQQQC(+57.02)NRR.S | No | 213 | 227 | ||||
| K.TSAIEVMLLETSRRYN(+.98)PG.S | No | 292 | 309 | ||||
| Tumor 5 Rep 2 |
3412 | 38.73 | 9% | C.VLSEEQIR.L | No | 168 | 175 |
| R.ASSPPQILPQLSPEQ(+.98)LGMIEK.L | No | 196 | 216 | ||||
| R.ASSPPQ(+.98)ILPQ.L | No | 196 | 205 | ||||
| R.EDQIALLK.T | No | 284 | 291 | ||||
Amino acids position numbers based on LXRα1 structure. The gray highlight indicated unique peptides of LXR variant detected by MS.
Table 2.
LXRβ peptides detected by S-trap column coupled with mass spectrometry (MS)
| Sample | Total identified peptides | −10IgP [LXRβ] | Coverage LXRβ |
Supporting peptides [LXRβ] | Unique | Amino acids position |
|
|---|---|---|---|---|---|---|---|
| Peptides | Start | End | |||||
| Tumor 2 Rep 1 |
3500 | 46.60 | 7% | L.MIQQLVAAQ.L | No | 226 | 234 |
| Q.Q(+.98)LVAAQLQC(+57.02)NK.R | No | 229 | 239 | ||||
| Q.VEFINPIFEFSR.A | No | 346 | 357 | ||||
| K.RPQDQ(+.98)LR.F | No | 410 | 416 | ||||
| Tumor 2 rep 2 | 3971 | 39.62 | 10% | R.RSVVRGGAR.R | No | 113 | 121 |
| K.EAGM(+15.99)REQ(+.98)C(+57.02)VLSEEQIRKK.K | No | 151 | 168 | ||||
| K.VTPWPLGADPQSR.D | Yes [β1] | 248 | 260 | ||||
| K.RPQDQ(+.98)LR.F | No | 410 | 416 | ||||
| Tumor 5 Rep 1 |
3713 | 56.33 | 14% | R.SVVRGGAR.R | No | 113 | 121 |
| E.LM(+15.99)IQQ(+.98)LVAAQ.L | No | 225 | 234 | ||||
| V.QLTAAQ(+.98)ELMIQ(+.98).Q | No | 218 | 228 | ||||
| Q.Q(+.98)LVAAQLQC(+57.02)NK.R | No | 229 | 239 | ||||
| K.RSFSDQ(+.98)PK.V | No | 240 | 247 | ||||
| K.VTPWPLGADPQ(+.98)SR.D | Yes [β1] | 248 | 260 | ||||
| A.LQQ(+.98)PYVEALLS.Y | No | 394 | 404 | ||||
| E.ALLSYTR.I | No | 401 | 407 | ||||
| Tumor 5 Rep 2 |
3412 | 59.82 | 20% | R.RSVVRGGAR.R | No | 113 | 121 |
| C.VLSEEQIR.K | No | 159 | 166 | ||||
| R.KQ(+.98)QQ(+.98)ESQSQSQSPVGPQG.S | No | 172 | 189 | ||||
| E.GVQLTAAQ(+.98)ELMIQ.Q | No | 216 | 228 | ||||
| E.LM(+15.99)IQQ(+.98)LVAAQ.L | No | 225 | 234 | ||||
| Q.QLVAAQ(+.98)LQ(+.98)C(+57.02)N(+.98)K.R | No | 229 | 239 | ||||
| K.RSFSDQ(+.98)PK.V | No | 240 | 247 | ||||
| R.EDQIALLK.A | No | 297 | 304 | ||||
| A.LQQ(+.98)PYVEALLS.Y | No | 394 | 404 | ||||
| K.RPQDQ(+.98)LR.F | No | 410 | 416 | ||||
Amino acids position numbers based on LXRβ1 structure. The gray highlight indicated unique peptides of LXR variant detected by MS.
Table 3.
Unique peptides of LXR variants
| LXR | Also, recognize | Peptide mass | Amino acid position (based on LXRa1 structure) |
Exon | Unique peptides (after trypsin digestion) | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| α1 | α2 | 1607.8137 | 1 | 15 | 2 | MSLWLGAPVPDIPPD |
| 1348.7140 | 293 | 300 | 6 | SAIEVMLLETSR | ||
| α2 | α5 | 1148.6343 | 235,236-297 | 300 | 5/7 | VT-VMLLETSR |
| α3 | α5 | 2307.1761 | 46 | 69 | 3 | MPHSAGGTAGVGLEAAEPTALLTR |
| α4 | – | 1550.7090 | −14 | −1 | -1d | MQQTSWNPLGGTCK |
| 554.3045 | −19 | −15 | -1d | QPPGR | ||
| XP_0243040XXX | – | 2942.3842 | Additional 64 amino acids due to the presence of a retained intron in between exon 6 and 7 | Exon -7a | GEAEWDYLWEGPPDIELGEP NLLGSR | |
| 1170.5538 | DEENRPPWK | |||||
| 1005.4669 | EVAGEGQGMK | |||||
| 741.3890 | TSPPSPR | |||||
| 590.3079 | RPCSK | |||||
| 510.2381 | FAACV | |||||
| β1 | 1423.7328 | 248 | 260 | Exon6/ | VTPWPLGADPQSR | |
| 1475.7892 | 268 | 279 | exon7abc | FAHFTELAIISVQ | ||
| β3 | 1021.5564 | 249–280 | 286 | Exon6/exon7b | V-TEIVDFAK | |
| β4 | 545.3405 | 249–293 | 299 | Exon6/Exon 7c | V-TLGREDQ | |
Amino acids position number based on α1 and/or β1 amino acid numbering position. Amino acid numbering position with “-” indicates the additional amino acid(s) coming before α1 and/or β1’s amino acid position number 1. The numerical exons represent the first discovered exons. The numerical exons followed with alphabets represent the later discovered exons.
Transcript-protein correlation analysis
It was not possible to detect unique peptides for all LXR splice variants using MS. As an alternative approach, we performed linear correlation analysis between RNA and protein expression of each variant in all cell lines (see Figure S13) and all primary breast tumor samples (see Figure S14). The primers used to detect different RNA species were usually specific to a single variant, but it was not possible to design unique primers for every variant. We hypothesized that where detection was ambiguous, RNA expression should correlate with expression of the protein for which it coded, thus identifying the correct RNA species and validating the protein isoform previously assigned only by mass/charge. Linear correlation analysis was performed in the seven cell lines comparing expression of each transcript variant against each protein band for which it could potentially code. There was a strong and significant positive correlation between transcript variants and the protein isoform for which they were predicted to code: α1 protein correlated with α1.1 (R2 = 0.97; p < 0.0001; Figure S13 top row) and α1.3 (R2 = 0.78; p = 0.0084; Figure S13 top row). β1 protein correlated with β1.1 transcript (R2 = 0.99; p < 0.0001; Figure S13 bottom row). During primer design it became clear that due to complexity arising from the large number of LXR coding transcripts, several transcripts were not distinguishable from each other due to sequence homology (denoted with asterisks in Figure 3C). Specifically, indistinguishable transcripts were α1.4 and α2.3; α3.1 and α5.1; and α3.3 and α5.3. We tested all potential protein isoforms for correlation with the ambiguous primer pairs: α1.4/α2.3 correlated with α2 protein (p = 0.0022; R2 = 0.97; Figure S13 row 2) but not α1 protein (p > 0.05); α3.1/α5.1 correlated with α3 protein (p = 0.034; R2 = 0.63; Figure S13 row 3) but not α5 protein (p > 0.05); and α3.3/α5.3 correlated with α5 protein (p = 0.029; R2 = 0.65; Figure S13 row 4) but not α3 protein (p > 0.05). We also designed primers against the exon 5 and 7 junction to simultaneously recognize the α2 and α5 variants that lack exon 6 and a portion of the LBD. These α2/α5 primers detected RNA that correlated with α5 protein (p = 0.0065; R2 = 0.8; Figure S13 fourth row) but not with α2 (p > 0.1; Figure S13 second row).
Next, although cytoplasmic extraction was not possible in frozen tumor samples as they had lost cellular substructure in the freezing process, given the number of replicates was larger (n = 38), we considered testing for correlations a reasonable approach (see Figure S14). As for cell lines, both α1.1 and α1.3 correlated with α1 protein (p < 0.05 for both); α2.3 correlated with α2 protein (p = 0.036); α3.1 correlated with α3 protein (p = 0.056); α4.1 correlated with α4 protein (p = 0.0037; note, this variant was not detected in cell lines); and α5.3 (p = 0.039) and α2/α5 (p = 0.054) correlated with α5 protein. For LXRβ, β1.1 transcript correlated with β1 protein (p = 0.022). β3 and β4 are almost identical in size (46 and 47 kDa, respectively) and so are indistinguishable by immunoblotting. β4 transcript (p = 0.031) but not β3 (p > 0.05) correlated with the band corresponding to β3/β4 protein (see Figure S14 bottom row) confirming this as LXRβ4. The correlative analysis we performed here, in combination with our other validation experiments, allows high confidence that the splice variants had been correctly identified. In combination with the siRNA in the cell line studies above and MS experiments, these correlative observations indicate that five LXRα variants and two LXRβ variants are expressed in TNBC.
LXR splice variants are differentially correlated with expression of target genes in Event and No Event patients
We previously reported that LXRα expression is positively correlated to target gene expression in ER-negative patients who had relapsed and/or died due to their disease (Event patients) but not those who survive disease free (No Event patients) (Hutchinson et al., 2021). Here, we tested the hypothesis that differential LXR splice variant expression between cancers may contribute to disease etiology via their ability to control expression of gene targets. Two canonical LXR target genes representing LXR’s roles in cholesterol metabolism (ABCA1) and chemoresistance (ABCB1) were tested for correlations with LXR splice variant expression (Hutchinson et al., 2021; Pan et al., 2019).
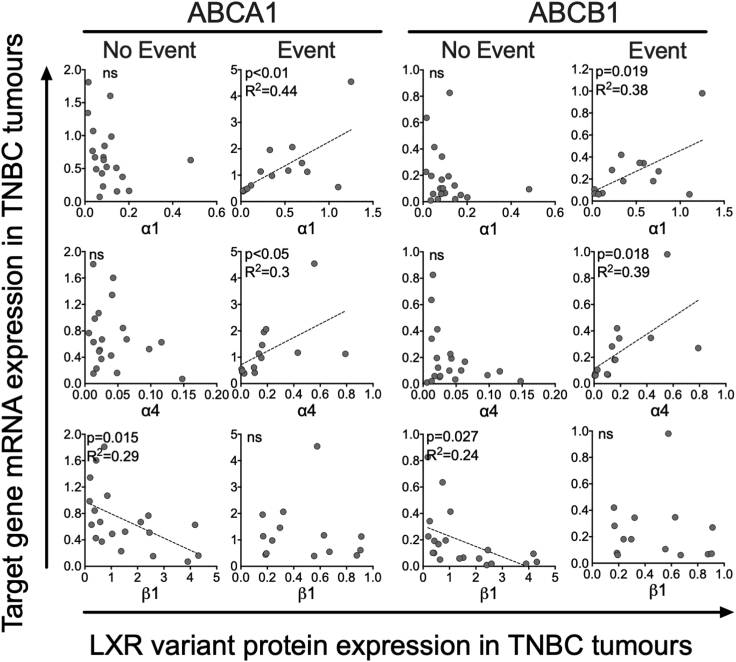
Expression of both full-length isoforms (α1 and α4) positively correlated with expression of ABCA1 and ABCB1 but interestingly only in Event patients (p < 0.05 for all; Figure 5). Strikingly, expression of β1 was inversely correlated with both ABCA1 and ABCB1 but only in No Event patients (p < 0.05; Figure 5). Statistically significant correlations between α2, α3, or α5 with target genes were not observed (see Figure S15). From these data, we concluded that while full-length LXRα is associated with activation of gene targets in patients who relapse or die; LXRβ is associated with inhibition of the same target genes in TNBC survivors.
Figure 5.
Protein expression of LXR variants is differentially associated with LXR target gene expression in TNBC tumors
Patients with TNBC (n = 38) were divided into two groups: no event (n = 23) and event (n = 15) based on disease-free survival status. Linear regression was used to determine if the slope of the lines were significantly different from 0 (p ≤ 0.05). Line of best fit is shown if the relationship was significant. Circles represent mean protein expression of two tumor slices for individual patients with TNBC.
Discussion
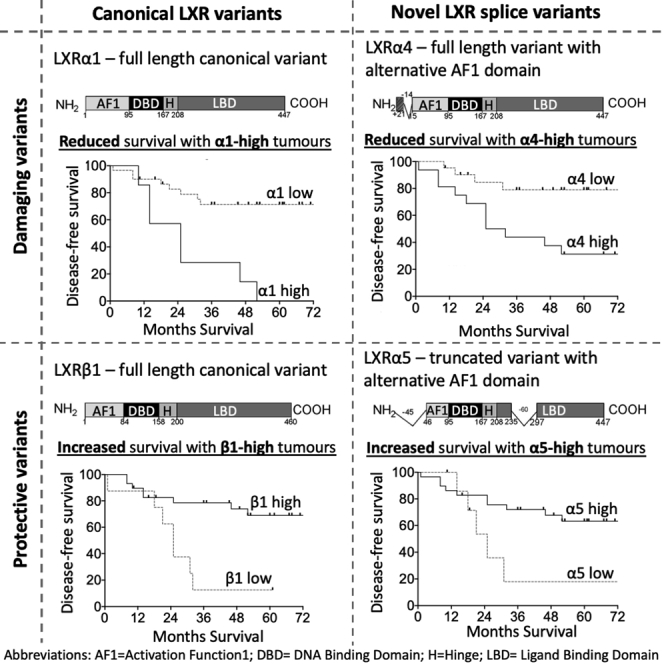
The objective of this study was to establish the repertoire, expression levels, and pathophysiological significance of LXR splice variants in BCa. In TNBC, we found evidence that five LXRα and two LXRβ splice variants are expressed at RNA and protein levels. Three of these isoforms, α4, α5, and β1, are new to the literature and do not have validated UNIPROT records. Our data demonstrate that LXR splice variants have clinical significance in TNBC; full-length LXRα (α1 and α4) is associated with shorter DFS, while LXRβ and LXRα variants with truncated ligand-binding domains were associated with longer DFS.
This current study’s findings may provide preliminary evidence for oncogenic or tumor suppressive functions of LXR splice variants. High expression of the full-length α1 was strongly associated with shorter DFS at both transcript and protein level. High expression of two other isoforms, α3 (transcript only) and α4 (transcript and protein), were also associated with shorter DFS. Although both have disruptions to their AF1 domains, they are otherwise homologous to full-length α1, including, importantly, an uninterrupted LBD. α5 skips the same exons as α2 and α3 and has the same deletions; 60 amino acids are missing from the LBD, and 45 amino acids of the AF1 domain are missing, respectively. These truncations have been shown separately to diminish (α3) or completely abolish (α2) transcriptional activity (Chen et al., 2005). Interestingly, high transcript expression of α2 was, like α5, linked to longer DFS. High α3 transcript on the other hand, like full-length α1 and α4, was linked to shorter DFS. These observations suggest that splice variants with disrupted LBD are advantageous, but the protein can tolerate disruption or even partial deletion of the AF1 domain and still be associated with worse prognosis. β1 (P55055-1) and β4 (M0R2F9) are generated by the mutually exclusive inclusion or exclusion of exon 7, again leading to a change in the LBD. With LXRβ however, high expression of either β1 or β4 was linked to longer DFS. The cellular role of this altered LBD remains to be determined.
Belorusova et al. reported peptides of LXRα comprising the H3 and partial part of H5 correlated to high ABCA1 expression (Belorusova et al., 2019). In addition, the differential relationship between target genes and LXRα isoforms in different patient groups also points to an oncogenic role for LXRα and a tumor suppressor role for LXRβ in TNBC tumors. Indeed, a beneficial role for LXRβ has been reported previously, albeit in non-TNBC subtypes. Both dendrogenin A and RGX-104 are tumor suppressors and selective LXRβ agonists (Poirot and Silvente-Poirot, 2018; Segala et al., 2017; Tavazoie et al., 2018). An understanding of the effect of these compounds on TNBC cells and tissue would help delineate if LXRβ or LXRα are predominant (given LXRβ’s higher expression levels) and if LXRβ is also a therapeutic target in TNBC disease. LXRβ may be the predominant isoform in TNBC and is normally able to supress LXRα signaling due to its expression levels. However, functional activity cannot be inferred by isoform expression only and other mechanisms of differential LXR isoform regulation are likely to be at play. For example, post-translational modifications, particularly at the S198 residue (Wu et al., 2015), can modify LXR transactivation potential. Our previous work found that co-factor expression levels modified LXR’s response to ligand (Hutchinson et al., 2019). It is also possible that promoter/chromatin accessibility could differentially alter the ability of the LXR isoforms to bind to target gene promoters. Endogenous ligand synthesis may also be important. Several non-cancer cell types found in the TNBC tumor microenvironment such as macrophages (Blanc et al., 2013), adipocytes (Li et al., 2014), and fibroblasts (Axelson and Larsson, 1995) are potent converters of cholesterol to oxysterols. Whether these cells drive activation of LXR isoforms in the adjacent tumor cells through paracrine or juxtacrine secretion of ligand has, to date, not been reported in the literature. Indeed, a key limitation of the current study is that it was not possible to establish if a single cell, or cell type, was expressing multiple or single splice variants. This would provide a mechanism where the cell could fine-tune its response to ligand.
To the best of our knowledge, α4 (NM_001934; NP_001238863; B4DXU5), α5 (NM_001363595.2; NP_001350524.1; B5MBY7), and β4 (M0R2F9) have not been reported in the literature before. α4 has an alternative 21 amino acids in the start of the AF1 domain producing a unique peptide mass signature that was detected with MS. These data support its inclusion in UNIPROT with a Q13133 prefix. As this variant has not been investigated before and we perform no functional studies herein, the significance of this partial AF1 substitution remains unknown. α5 was not detected by MS, but its presumed protein band (39 kDa) was lost after siLXRα treatment, its transcript was robustly expressed, and protein and transcript expression were strongly correlated. Although a unique peptide for β4 was predicted from in silico analyses, it was not detected with MS; however, its transcript was robustly expressed, and protein and transcript expression were strongly correlated.
Some of our results are at odds with prior observations. We were unable to detect expression of XP_02430405X (previously termed α4 (Endo-Umeda et al., 2012)), in any cell lines or tissues, at RNA or protein level, including in HEPG2 or MCF7 cell lines that were previously shown to express this isoform (Bunay et al., 2020; Endo-Umeda et al., 2012). Intriguingly, a variant previously reported to be α5 (Bunay et al., 2020; Endo-Umeda et al., 2012) did not correspond to any of the 64 LXRα transcripts from NCBI nor any LXRα UNIPROT entries. In this study (Endo-Umeda et al., 2012), the measurement of XP_02430405X and previously reported α5 relied on PCR primers that detected a retained intron between exon 6–7 and a retained intron between exon 7–8, respectively (see Figure S3). We went to extensive lengths to ensure primary RNA or gDNA did not contaminate our cDNA libraries, steps that surprisingly have not been reported in LXR splicing studies previously. Cytoplasmic RNA is preferred over total cell isolates when examining splice expression, and this has been reported previously to increase sensitivity of splice junction detection (Zaghlool et al., 2013). Previous detection of XP_02430405X may therefore be due to amplification of incompletely processed transcripts or contaminating gDNA. During our primer design stage, we found that the PCR primers previously used to measure α1 at the exon 2–3 junction (Chen et al., 2005; Endo-Umeda et al., 2012; Rondanino et al., 2014) also detect α2 (see Figure S3). In addition, primers previously used to measure α2 at the exon 5–7 junction (Chen et al., 2005; Endo-Umeda et al., 2012; Rondanino et al., 2014) also detect the previously unreported α5 variant (see Figure S3). Our correlation analyses in cell lines and primary tumor samples suggest the majority of exon 6 skipping transcripts actually code for α5 not α2. We also found α5 is significantly (20-fold) higher than α2 in MCF7 and equal to α2 in HEPG2, the same cell lines that in prior studies were used to show α2 was strongly expressed. It is likely that previous reports have over-estimated the contribution of α2 and under-estimated the contribution of α5 to the LXR pool.
In summary, this study provides critical insight into the pathophysiology of LXR splicing in BCa. Our data are consistent with the hypothesis that full-length LXRα is oncogenic but LXRβ and truncated LXRα variants may suppress tumor development. Functional studies should help resolve if the LXRs are drivers of tumor pathophysiology or simply passengers. We propose the existence of two new LXRα isoforms: α4 containing an alternative AF1 region and α5 which lacks large sections of the AF1 and LBD regulatory regions. TNBC is defined as a category of exclusion. The data presented here suggest that measuring LXRα1, LXRα5, and LXRβ at RNA or protein level may allow sub-stratification of patients with TNBC for increased accuracy of prognosis. Perhaps more importantly, given the array of pharmacological and dietary modulators of the LXR pathway, defining gene networks regulated by the opposing oncogenic and tumor suppressor LXR variants will aid in the development of lifestyle and pharmacological interventions to reduce incidence and improve survival in this challenging cancer of unmet clinical need.
Limitations of the study
There were three important limitations of this study. It was not possible to separate all novel (or canonical) LXR isoforms using S-trap mass spectrometry alone. Secondly, limiting tissue amounts meant some experiments were not possible, for example, testing for correlations between the LXR isoforms with additional target genes such as SREBP and APOE. The limited tissue also meant LXR purification with immunoprecipitation and/or Coomassie approaches prior to mass spectrometry was not feasible. This would have allowed us to assign LXR-specific peptides as belonging to different LXR proteins that had been identified by mass-charge analysis with western blotting, for example, β4. Finally, as mentioned above, isoforms α4 and β4 were identified in tumor tissue but not BCa cell lines. It is possible that the source of these isoforms is non-tumor cells of the tumor microenvironment, such as fibroblasts, adipocytes, or immune infiltrate, and not the tumor cells per se. Cell sorting with cell surface markers of freshly collected tumor samples is required to delineate the source of these primary material isoforms specifically.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LXRα | R&D System | Cat. PP-PPZ0412-00; RRID:AB_2154888 |
| LXRβ | Active Motif | Cat. 61177; RRID:AB_2614980 |
| HPRT | Santa Cruz | Cat. Sc-376938; RRID: n/a |
| IRDye 800CV goat anti-mouse | LI-COR Biosciences | Cat. 926-68170; RRID:AB_10956589 |
| IRDye 680RD goat anti-rabbit | LI-COR Biosciences | Cat. 926-68071; RRID:AB_10956166 |
| Isotype IgG2a | Cell Signalling | Cat. 61656S; RRID:AB_2799613 |
| Biological samples | ||
| Frozen tumor samples (n=38), see Table S3 | Leeds Breast Research Tissue Bank | Ethic approval ID: 15/HY/0025 and LBTB_TAC_1/17 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle Medium (DMEM) | Thermo Fisher | Cat. 31966047 |
| Fetal Bovine Serum (FBS) | Thermo Fisher | Cat. 11560636 |
| Lipofectamine RNAiMAX | Thermo Fisher | Cat. 13778030 |
| OptiMeM | Thermo Fisher | Cat. 31985062 |
| bis(sulfosuccinimidyl)suberate | Thermo Fisher | Cat. A39266 |
| GW3965 | Tocris | Cat. 2474 |
| Dynabeads Protein A | Thermo Fisher | Cat. 100002D |
| PMSF protease and phosphatase inhibitor | Thermo Fisher | Cat. 78440 |
| NUPAGE LDS sample loading buffer | Thermo Fisher | Cat. NP0007 |
| DTT reducing agent | Thermo Fisher | Cat. NP0004 |
| TBS blocking buffer | Li-COR | Cat. 927-60001 |
| Critical commercial assays | ||
| ReliaPrepTM RNA Cell Miniprep System | Promega | Cat. Z6012 |
| Cytoplasmic & Nuclear RNA Purification Kit | Norgen | Cat. 21000 |
| GoScriptTM Reverse Transcription kit | Promega | Cat. A5003 |
| BCA kit | Thermo Fisher | Cat. 23227 |
| Experimental models: Cell lines | ||
| HEPG2 | ATCC | HB-8065 |
| MCF7 | ATCC | HTB-22 |
| BT474 | ATCC | HTB-20 |
| MDA.MB.231 | ATCC | CRM-HTB-26 |
| MDA.MB.468 | ATCC | HTB-132 |
| MDA.MB.453 | ATCC | HTB-131 |
| MDA.MB.157 | ATCC | HTB-24 |
| Oligonucleotides | ||
| CONTROL siRNA | Origene | Cat. SR30004 |
| siLXRα | Origene | Cat. SR322981 |
| siLXRβ | Origene | Cat. SR305039 |
| Primers for qPCR, see Table S4 | This paper | N/A |
| Software and algorithms | ||
| Clustal Omega-Multiple sequence alignment | EMBL-EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| PeptideMass tool | Swiss Institute of Bioinformatics | http://www.expasy.org/tools/peptide-mass.html |
| Diagram Venn | Bioinformatics & Evolutionary Genomics | http://bioinformatics.psb.ugent.be/webtools/Venn/ |
| GraphPad Prism v8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Image Studio™ Lite | LI-COR Biosciences | https://www.licor.com/bio/image-studio/ |
| ApE-A plasmid Editor | M.Wayne Davis | https://jorgensen.biology.utah.edu/wayned/ape/ |
| AutoCAD | Autodesk | https://www.autodesk.co.uk |
| Other | ||
| NCBI | National Center for Biotechnology Information | https://www.ncbi.nlm.nih.gov |
| ENSEMBL | European Bioinformatics Institute (EMBL-EBI) | https://www.ensembl.org/index.html |
| UNIPROT | EMBL-EBI, Swiss Institute of Bioinformatics (SIB), and Protein Information Resources (PIR) | https://www.uniprot.org |
| cBioportal | Memorial Sloan Kettering Cancer Center | https://www.cbioportal.org |
| TCGA Splicing Database (TSVdb) | Key Laboratory of Disease Proteomics of Zhejiang Province | http://www.tsvdb.com |
Resource availability
Lead contact
Further information and requests for resources, reagents, data, and/or experimental protocols reported in this paper should be directed to and will be fulfilled by the lead contact, Dr James L Thorne (J.L.Thorne@leeds.ac.uk).
Materials availability statement
There are restrictions to the availability of Tumor tissues due to MTA restrictions and limiting sample amounts. This study did not generate other unique reagents.
Experimental models and subject details
Cell lines
The HEPG2, MCF7, BT474, MDA.MB.231, MDA.MB.468, MDA.MB.453, and MDA.MB.157 cell lines were originally obtained from ATCC (Manassas, USA). Cell lines were routinely cultured in Dulbecco's Modified Eagle Medium (DMEM; Thermo Fisher, UK, Cat. 31966047), supplemented with 10% Fetal Bovine Serum (FBS; Thermo Fisher, UK, Cat. 11560636) and incubated in 37°C, 5% CO2. Cells were periodically checked and confirmed to be mycoplasma free.
Human subjects
Thirty-eight frozen tumor samples were obtained with ethical approval from the Leeds Breast Research Tissue Bank (15/HY/0025 and LBTB_TAC_1/17). The clinicopathological features and selection criteria of these cohorts have been described previously (Broad et al., 2021; Hutchinson et al., 2019, 2021; Solheim et al., 2019) (details in Table S3). All subjects were female. The Leeds TNBC tumor cohorts were grouped based on their disease-free survival (DFS) status. TNBC patients who did not have disease relapse during follow-up (median follow-up time was 96 months) were categorized to ‘No Event' group. Meanwhile, TNBC patients who had relapsed and/or died due to their disease were grouped to an 'Event' group (median follow-up time was 20 months).
Method details
Systematic transcript variant analysis in public databases
The NR1H3/LXRα and NR1H2/LXRβ spliced variant sequences were assessed in NCBI (Pruitt et al., 2005) and ENSEMBL (Aken et al., 2016) databases. Sequence annotation records of LXRα and LXRβ spliced variant read from databases were stored in a feature library created with ApE plasmid editor (Davis, 2012). This feature library can then be used to scan any nucleotide and/or amino acid sequence that are complementary to all alternative forms of the LXRα and/or LXRβ sequences recorded in this library. The number of nucleotides in each exon and intron were counted in ApE plasmid editor to determine the proportions for drawing squares and lines represented exons and introns. The schematic diagrams of LXR variant structures were drawn using AutoCAD (Autodesk, US) software are shown in Figures 1, S1 (LXRα), and S2 (LXRβ). Variants, their database links and names, and previously reported names, are listed in S1 (LXRα) and S2 (LXRβ).
The cancer genome atlas (TCGA) splicing database analyses
TCGA splicing variants database (TSVdb) was searched and LXR splicing expression data in BCa tumor tissue, reported as normalized RNA-Seq by Expectation Maximization (RSEM) values, were downloaded using the TSVdb webtool (http://www.tsvdb.com) (Sun et al., 2018). Unreported and/or missing clinical status of the deposited TCGA BCa tumor samples were obtained from patient tumor sample information in cBioportal (http://cBioportal.org) (Cerami et al., 2012).
siRNA transfection
siRNA transfection was performed as previously reported (Thorne et al., 2018). Briefly, 1x105 cells were plated in 6-well plates and incubated overnight. Lipofectamine RNAiMAX (Thermo Fisher, UK, Cat. 13778030), siLXRα (Origene, USA, Cat. SR322981) and siLXRβ (Origene, USA, Cat. SR305039) or the scrambled siRNA (Origene, USA, Cat. SR30004) were diluted in OptiMeM (Thermo Fisher, UK, Cat. 31985062), and added to the cells at a final concentration of 30 nM. The cells were incubated and after 20 h the media was removed and fresh DMEM added. Knockdown was confirmed at the protein level 48 h post-transfection.
mRNA extraction, cDNA synthesis, qPCR and primer design
Total RNA was isolated from approximately 5 × 105 cells using the ReliaPrepTM RNA Cell Miniprep System (Promega, UK, Cat. Z6012) following product guidelines, including on column digestion of gDNA. Cytoplasmic RNA was isolated from approximately 2 × 106 cells using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA, Cat. 21000). The concentration of isolated RNA was determined spectrophotometrically, and RNA purity was evaluated by 260/230 nm and 260/280 nm ratios using a CLARIOstar plate reader (BMG LABTECH, Germany). cDNA was synthesized from 2 μg RNA using the GoScriptTM Reverse Transcription kit (Promega, UK, Cat. A5003). Primers were designed using NCBI BLAST primer design or, where transcript variants were not available in the NCBI database, primer 3 software was used (Untergasser et al., 2012). Cytoplasmic RNA was extracted to ensure only fully processed LXR transcripts were measured. Amplicon size was restricted between 80 to 150 bp and amplicons were required to span an exon-exon boundary. Primer efficiency was measured from qPCR standard curves, and primers were redesigned if amplification efficiency did not fall within 90-110% and/or a single peak was not observed in melting temperature analysis.
During primer design, it became clear that due to complexity arising from the large number of LXR coding transcripts, several transcripts were not distinguishable from each other due to the sequence homology and coding redundancy. Thus, some primer pairs detected ambiguous amplicons that could belong to two or sometimes three different splice variants. This ambiguity has not been addressed in the literature before as for example, the PCR primers previously used to measure α1 (Chen et al., 2005; Endo-Umeda et al., 2012; Rondanino et al., 2014) also amplify α2. Primers previously used to measure α2 via the exon 6 skipping (5-7 junction) (Chen et al., 2005; Endo-Umeda et al., 2012; Rondanino et al., 2014) also detect the previously unreported α5 variant. By documenting all possible LXR transcript variants (Figures S1 and S2; Tables S1 and S2), any potentially ambiguous amplicons were identified and unique primer sets against exon-exon junction could be designed for most transcript variants. Primers were designed using NCBI BLAST primer design or, where transcript variants were not available in the NCBI database, primer 3 software was used (Untergasser et al., 2012). According to the gene sequences from NCBI and ENSEMBL, the amplicon size was restricted between 80 to 150 bp. The standard curve from 1-5 ug each concentration cDNA ratio was carried to determine the efficiency of the primers. Primer pairs were chosen based on specificity (single peak in melting temperature analysis) and amplification efficiency. For the purpose of comparison, we summarized primer locations used in previous studies (see Figure S3). All primer sequences used in this study are shown in Table S6 and their locations within the LXR loci are shown graphically for LXRα in Figure S4 and for LXRβ in Figure S5.
Protein lysate extraction
Five mg fresh-frozen tumor biopsy samples were homogenized with a 1 mL Dounce tissue grinder (SLS, UK, Cat. HOM3580). Cell pellets and/or tumor samples were lysed in RIPA buffer (10 mM Tris-HCl pH 8, 140 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA, 0.5 mM EGTA), with 1 mM PMSF (Thermo Fisher, UK, Cat. 78440) added fresh prior to use. The lysed pellets were incubated on ice for 5 min and centrifuged at 11,500xg at 4°C for 10 min. The protein lysates concentrations were determined using a BCA kit (Thermo Fisher, UK, Cat. 23227) and lysates were kept in −80°C until further analyses. Each tumor sample was split in two and duplicate extractions were made. For each protein quantified both duplicates were run and the average of each was used for analysis. Concordance of duplicates was high (see Figure S16).
Cytoplasmic and nuclear protein extraction
For the cytoplasmic and nuclear protein extractions, the REAP (Rapid Efficient And Practical) protocol (Suzuki et al., 2010) was followed with a slight modification. In brief, 1 × 107 cells were suspended in 1 mL ice cold PBS with 1 mM PMSF. 200 μL of cell lysate was taken, put into a new chilled Eppendorf tube and centrifuged at 1,500 rpm for 3 min at 4°C. Supernatant was removed. The cell pellet (from 200 μL cell lysate) was resuspended in RIPA buffer with 1 mM PMSF (Thermo Fisher, UK, Cat. 78440) added fresh prior to use and labeled as a “whole cell fraction”. The remaining 800 μL cell lysate was centrifuged at 11,500×g at 4°C for 10 s. The supernatant was removed. The cell pellet (from 800 μL cell lysate) was resuspended in 200 μL ice-cold PBS+0.1 NP40 with 1 mM PMSF and centrifuged at 11,500xg at 4°C for 10 s. The supernatant was taken into a new chilled Eppendorf tube and labeled as a “cytoplasmic fraction”. The cell pellet was then resuspended in 200 μL ice-cold PBS+0.1 NP40 with 1 mM PMSF and centrifuged at 11,500×g at 4°C for 10 s. The supernatant was removed and cell pellet resuspended in 50 uL RIPA buffer with 1 mM PMSF. Cell lysate was sonicated with a water-bath sonicator (Diagenode Bioruptor Pico, Belgium, Germany) with cycles of 10 s on and 10 s off for five cycles, followed by centrifugation at 11,500 g at 4°C for 10 min. The supernatant was taken, put into a new chilled Eppendorf tube, and labeled as a “nuclear fraction”. The protein lysates concentrations were determined using a BCA kit and lysates were kept in −80°C until further analyses.
Immunoblotting
Forty-five micrograms of protein lysate combined with NUPAGE LDS sample loading buffer (Thermo Fisher, UK, Cat. NP0007) and DTT reducing agent (Thermo Fisher, UK, Cat. NP0004) was heated at 70°C for 10 min. For LXR variant expression, the protein lysate was loaded onto a 10% SDS polyacrylamide gel, electrophoresed at constant 80 V for 150 min, and transferred onto a PVDF membrane (Merck, UK, Cat. IPFL00010). The membrane was then blocked with TBS Odyssey Blocking Buffer (LI-COR Biosciences, UK, Cat. 92750000) for 1 h. Proteins were probed with anti-LXRα (R&D Systems, USA, Cat. PP-PPZ0412-00, dilution 1/1000), anti-LXRβ (Active Motif, Germany, Cat. 61177, dilution 1/1000), and anti-HPRT (Santa Cruz Biotechnology, Cat. sc-376938, dilution 1/100) overnight at 4°C. The membrane was then blocked and probed with LICOR secondary antibodies (IRDye 800CW goat anti-mouse Cat. 926-68170, IRDye 680RD goat anti-rabbit Cat. 926-68071; dilution 1/15,000, LI-COR Biosciences, UK) for 1 h and signal was visualized using the Odyssey system (LI-COR Biosciences, UK).
Immunoprecipitation
One mg protein sample was precleared with Dynabeads Protein A (Thermo Fisher, UK, Cat.100002D) and 1 μg isotype IgG2a (Cell Signalling, USA, Cat. 61656S). Immunoprecipitation was performed by incubating 40 μL of Dynabeads Protein A coupled to 2 μg of anti-IgG2a or 2μg of anti-LXRα using bis(sulfosuccinimidyl)suberate (Thermo Fisher, UK; Cat. A39266) with 1 mg protein sample overnight at 4°C. Dynabeads were washed 3 times with 10 mM Tris-HCl, 50 mM KCl (pH 7.5) and sample eluted in 20 μL of NUPAGE LDS sample loading buffer containing 100 mM DTT, then heated at 70°C for 10 min 10 at 70°C. The supernatant was transferred to a new Eppendorf tube after being separated from Dynabeads using a magnetic separator (Promega, UK, Cat. CD4002).
In-silico peptide mass prediction
The amino acid sequences of LXR variants downloaded from NCBI, ENSEMBL, and/or UNIPROT databases were subjected to the PeptideMass tool (http://www.expasy.org/tools/peptide-mass.html; Swiss Institute of Bioinformatics, Switzerland) (Gasteiger et al., 2003) to perform theoretical peptides from trypsin digestion in silico. Trypsin is the most preferred protease choice for peptide generation due to its high proteolytic activity and cleavage specificity (Saveliev et al., 2013). A set of peptides from each LXR variant were then subjected to Diagram Venn (http://bioinformatics.psb.ugent.be/webtools/Venn; Bioinformatics & Evolutionary Genomics, Belgium) in order to find the unique peptides of each LXR variant.
S-trap column coupled mass spectrometry (MS)
Protein samples were processed using the S-TRAP Micro column (PROTIFI, NY, USA) following the manufacturer's instructions. Proteins were fully solubilized by adding 20 μL of 10% SDS solution to 20 μL sample in RIPA buffer. Reduction and alkylation were then performed. DTT was added to a final concentration of 20 mM before heating to 56°C for 15 min with shaking. The sample was removed from heat and allowed to cool for 5 min. Iodoacetamide was then added to a final concentration of 40 mM. The sample was maintained at 20°C for 15 min with shaking in the dark. Phosphoric acid was then added to a final concentration of 1.2%, to ensure inactivation of all enzymatic activity and maximize sensitivity to proteolysis. Samples were then diluted with S-Trap binding buffer (100 mM TEAB pH 7.1 in methanol), and 1ug of trypsin, reconstituted in 50mM triethylamonium bicarbonate (TEAB), was added before the sample was quickly loaded onto the S-trap column. Proteins were captured within the submicron pores of the three-dimensional trap. Proteins captured within the trap present exceptionally high surface area allowing them to be washed free of contaminants. The S-trap was washed by adding 150 μL binding buffer before being spun at 4000xg for 30 s. 30 μL of 0.02 μg/μL Trypsin (Promega, WI, USA) was then added to the top of the S-trap. S-traps were loosely capped and placed in a 1.5mL Eppendorf and heated to 46°C for 15 min with no shaking. Digested peptides were eluted by first spinning the S-trap at 4,000 g for 1 min. Further elution was performed in 40 μL 50mM TEAB, 40 μL 0.2% formic acid, and 30 μL 50% acetonitrile with 0.2% formic acid prior to centrifugation. Eluent was combined then dried down prior to resuspension in 0.2% formic acid.
Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism v8. The expression of LXR splice variants in TCGA tumors compared with the adjacent normal tissues was assessed using a Mann-Whitney two-tailed U test. Differential LXR splicing expression levels in both TCGA BCa and Leeds TNBC tumor samples were established using multiple t-tests with Holm-Sidak for multiple correction. The knockdown siRNA experiment was analyzed using a two-tailed one-way ANOVA. The relationship between protein isoform variants and their mRNA transcripts were assessed using Spearman’s correlation and linear regression. Receiver operating characteristic (ROC) curves were used to establish the expression cut offs for high and low expression levels of each LXR variant. The true positive rate (the proportion of patients with the disease correctly diagnosed) was plotted as a function of the false positive rate (proportion of patients without the disease who are incorrectly diagnosed as having the disease) and the cut-off between ‘high’ and ‘low’ expression was determined by maximizing the clinical sensitivity (fraction of true positives to all patients with the disease correctly diagnosed) and specificity (fraction of true negatives to all patients without the disease correctly diagnosed). Tumor expression of each LXR variant was then assessed alongside patient survival to assess whether expression is predictive of survival in Kaplan Meier graphs. Patient survival was analyzed using log-rank test. Densitometry was performed using Image Studio™ Lite (LI-COR Biosciences, UK) software.
Acknowledgments
The mass spectrometry was performed and analysis was supported by Dr. James Ault and Dr. Rachel George at the Biomolecular Mass Spectrometry Facility, Faculty of Biological Sciences, University of Leeds. P.L. was jointly funded by a Leeds International Doctoral Scholarship and the University of Leeds School of Food Science and Nutrition.
Author contributions
Conceptualization, J.L.T.; data curation, P.L. and J.L.T.; formal analysis, P.L. and J.L.T.; funding acquisition, J.L.T.; methodology, P.L. and J.L.T.; project administration, J.L.T.; resources, T.A.H. and J.L.T.; supervision, J.B.M., T.A.H., and J.L.T.; experiments P.L. and S.A.H.; visualization, P.L. and J.L.T.; writing–original draft, P.L. and J.L.T.; writing–review & editing, P.L., J.B.M., T.A.H., and J.L.T.
Conflict of interest
The authors declare no competing interests.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103212.
Supplemental information
Data and code availability
All NGS data used in this study were freely available in public repositories at the date of publication. Links to repositories can be found via inline citations.
References
- Aken B.L., Ayling S., Barrell D., Clarke L., Curwen V., Fairley S., Fernandez Banet J., Billis K., García Girón C., Hourlier T. The ensembl gene annotation system. Database. 2016;2016:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annalora A.J., Marcus C.B., Iversen P.L. Alternative splicing in the nuclear receptor superfamily expands gene function to refine endo-xenobiotic metabolism. Drug Metab. Dispos. 2020;48:272–287. doi: 10.1124/dmd.119.089102. [DOI] [PubMed] [Google Scholar]
- Axelson M., Larsson O. Low density lipoprotein (LDL) cholesterol is converted to 27-hydroxycholesterol in human fibroblasts. Evidence that 27-hydroxycholesterol can be an important intracellular mediator between LDL and the suppression of cholesterol production. J. Biol. Chem. 1995;270:15102–15110. doi: 10.1074/jbc.270.25.15102. [DOI] [PubMed] [Google Scholar]
- Baek A.E., Yen-Rei A.Y., He S., Wardell S.E., Chang C.-Y., Kwon S., Pillai R.V., McDowell H.B., Thompson J.W., Dubois L.G. The cholesterol metabolite 27 hydroxycholesterol facilitates breast cancer metastasis through its actions on immune cells. Nat. Commun. 2017;8:1–11. doi: 10.1038/s41467-017-00910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belorusova A.Y., Evertsson E., Hovdal D., Sandmark J., Bratt E., Maxvall I., Schulman I.G., Åkerblad P., Lindstedt E.-L. Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands. Commun. Biol. 2019;2:1–13. doi: 10.1038/s42003-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M., Hsieh W.Y., Robertson K.A., Kropp K.A., Forster T., Shui G., Lacaze P., Watterson S., Griffiths S.J., Spann N.J. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity. 2013;38:106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clinic. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Broad R.V., Jones S.J., Teske M.C., Wastall L.M., Hanby A.M., Thorne J.L., Hughes T.A. Inhibition of interferon-signalling halts cancer-associated fibroblast-dependent protection of breast cancer cells from chemotherapy. Br. J. Cancer. 2021;124:1110–1120. doi: 10.1038/s41416-020-01226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunay J., Fouache A., Trousson A., de Joussineau C., Bouchareb E., Zhekun Z., Kocer A., Morel L., Baron S., Lobaccaro J.-M.A. Screening for Liver X Receptor modulators: where are we and for what use? Br. J. Pharmacol. 2020;178:3277–3293. doi: 10.1111/bph.15286. [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. AACR; 2012. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Beaven S., Tontonoz P. Identification and characterization of two alternatively spliced transcript variants of human liver X receptor alpha. J. Lipid Res. 2005;46:2570–2579. doi: 10.1194/jlr.M500157-JLR200. [DOI] [PubMed] [Google Scholar]
- Cioccoloni G., Soteriou C., Websdale A., Wallis L., Zulyniak M.A., Thorne J.L. Phytosterols and phytostanols and the hallmarks of cancer in model organisms: a systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 2020;1:21. doi: 10.1080/10408398.2020.1835820. [DOI] [PubMed] [Google Scholar]
- Davis, M.W. (2012). ApE—a Plasmid editor. [DOI] [PMC free article] [PubMed]
- Endo-Umeda K., Uno S., Fujimori K., Naito Y., Saito K., Yamagishi K., Jeong Y., Miyachi H., Tokiwa H., Yamada S., Makishima M. Differential expression and function of alternative splicing variants of human liver X receptor alpha. Mol. Pharmacol. 2012;81:800–810. doi: 10.1124/mol.111.077206. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M., Appel R., Bairoch A. Humana Press; 2003. The Proteomics Protocols Handbook: Protein Identification and Analysis Tools on the ExPASy Server. [Google Scholar]
- Hashimoto K., Ishida E., Matsumoto S., Shibusawa N., Okada S., Monden T., Satoh T., Yamada M., Mori M. A liver X receptor (LXR)-β alternative splicing variant (LXRBSV) acts as an RNA co-activator of LXR-β. Biochem. Biophys. Res. Commun. 2009;390:1260–1265. doi: 10.1016/j.bbrc.2009.10.132. [DOI] [PubMed] [Google Scholar]
- Hayoun K., Gouveia D., Grenga L., Pible O., Armengaud J. Evaluation of sample preparation methods for fast proteotyping of microorganisms by tandem mass spectrometry. Front. Microbiol. 2019;10:1985. doi: 10.3389/fmicb.2019.01985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudis C.A., Gianni L. Triple-negative breast cancer: an unmet medical need. Oncologist. 2011;16:1–11. doi: 10.1634/theoncologist.2011-S1-01. [DOI] [PubMed] [Google Scholar]
- Hutchinson S.A., Lianto P., Roberg-Larsen H., Battaglia S., Hughes T.A., Thorne J.L. ER-negative breast cancer is highly responsive to cholesterol metabolite signalling. Nutrients. 2019;11:2618. doi: 10.3390/nu11112618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson S.A., Websdale A., Cioccoloni G., Roberg-Larsen H., Lianto P., Kim B., Rose A., Soteriou C., Pramanik A., Wastall L.M. Liver x receptor alpha drives chemoresistance in response to side-chain hydroxycholesterols in triple negative breast cancer. Oncogene. 2021;40:2872–2883. doi: 10.1038/s41388-021-01720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhao X., Xu J., Li C., Yu Y., Wang W., Zhu L. The protective effect of dietary phytosterols on cancer risk: a systematic meta-analysis. J. Oncol. 2019;2019:11. doi: 10.1155/2019/7479518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte B.A., Joseph S.B., Walczak R., Pei L., Wilpitz D.C., Collins J.L., Tontonoz P. Autoregulation of the human liver X receptor α promoter. Mol. Cell. Biol. 2001;21:7558–7568. doi: 10.1128/MCB.21.22.7558-7568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:1–16. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Daly E., Campioli E., Wabitsch M., Papadopoulos V. De novo synthesis of steroids and oxysterols in adipocytes. J. Biol. Chem. 2014;289:747–764. doi: 10.1074/jbc.M113.534172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Yi Z., Guan X., Zeng Y.X., Ma F. The relationship between statins and breast cancer prognosis varies by statin type and exposure time: a meta-analysis. Breast Cancer Res. Treat. 2017;164:1–11. doi: 10.1007/s10549-017-4246-0. [DOI] [PubMed] [Google Scholar]
- Nelson E.R., Wardell S.E., Jasper J.S., Park S., Suchindran S., Howe M.K., Carver N.J., Pillai R.V., Sullivan P.M., Sondhi V. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science. 2013;342:1094–1098. doi: 10.1126/science.1241908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Zheng Y., Pan Q., Chen H., Chen F., Wu J., Di D. Expression of LXR-β, ABCA1 and ABCG1 in human triple-negative breast cancer tissues. Oncol. Rep. 2019;42:1869–1877. doi: 10.3892/or.2019.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C.M., Sørlie T., Eisen M.B., Van De Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Poirot M., Silvente-Poirot S. The tumor-suppressor cholesterol metabolite, dendrogenin A, is a new class of LXR modulator activating lethal autophagy in cancers. Biochem. Pharmacol. 2018;153:75–81. doi: 10.1016/j.bcp.2018.01.046. [DOI] [PubMed] [Google Scholar]
- Prüfer K., Boudreaux J. Nuclear localization of liver X receptor α and β is differentially regulated. J. Cell. Biochem. 2007;100:69–85. doi: 10.1002/jcb.21006. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondanino C., Ouchchane L., Chauffour C., Marceau G., Dechelotte P., Sion B., Pons-Rejraji H., Janny L., Volle D.H., Lobaccaro J.M., Brugnon F. Levels of liver X receptors in testicular biopsies of patients with azoospermia. Fertil. Steril. 2014;102:361–371.e365. doi: 10.1016/j.fertnstert.2014.04.033. [DOI] [PubMed] [Google Scholar]
- Saveliev S., Bratz M., Zubarev R., Szapacs M., Budamgunta H., Urh M. Trypsin/Lys-C protease mix for enhanced protein mass spectrometry analysis. Nat. Methods. 2013;10 i–ii. [Google Scholar]
- Segala G., David M., de Medina P., Poirot M.C., Serhan N., Vergez F., Mougel A., Saland E., Carayon K., Leignadier J. Dendrogenin A drives LXR to trigger lethal autophagy in cancers. Nat. Commun. 2017;8:1–17. doi: 10.1038/s41467-017-01948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinar D.M., Endo N., Rutledge S.J., Vogel R., Rodan G.A., Schmidt A. NER, a new member of the gene family encoding the human steroid hormone nuclear receptor. Gene. 1994;147:273–276. doi: 10.1016/0378-1119(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Solheim S., Hutchinson S.A., Lundanes E., Wilson S.R., Thorne J.L., Roberg-Larsen H. Fast liquid chromatography-mass spectrometry reveals side chain oxysterol heterogeneity in breast cancer tumour samples. J. Steroid Biochem. Mol. Biol. 2019;192:105309. doi: 10.1016/j.jsbmb.2019.02.004. [DOI] [PubMed] [Google Scholar]
- Sun W., Duan T., Ye P., Chen K., Zhang G., Lai M., Zhang H. TSVdb: a web-tool for TCGA splicing variants analysis. BMC Genomics. 2018;19:405. doi: 10.1186/s12864-018-4775-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Bose P., Leong-Quong R.Y., Fujita D.J., Riabowol K. REAP: a two minute cell fractionation method. BMC Res. Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M.F., Pollack I., Tanqueco R., Ostendorf B.N., Reis B.S., Gonsalves F.C., Kurth I., Andreu-Agullo C., Derbyshire M.L., Posada J. LXR/ApoE activation restricts innate immune suppression in cancer. Cell. 2018;172:825–840.e818. doi: 10.1016/j.cell.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne J.L., Battaglia S., Baxter D.E., Hayes J.L., Hutchinson S.A., Jana S., Millican-Slater R.A., Smith L., Teske M.C., Wastall L.M., Hughes T.A. MiR-19b non-canonical binding is directed by HuR and confers chemosensitivity through regulation of P-glycoprotein in breast cancer. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 2018 doi: 10.1016/j.bbagrm.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WCRF/AICR . World Cancer Research Fund International/American Institute for Cancer Research; 2017. Continuous Update Project Report: Diet, Nutrition, Physical Actvity and Cancer.wcrf.org/breast-cancer-2017 [Google Scholar]
- Willy P.J., Umesono K., Ong E.S., Evans R.M., Heyman R.A., Mangelsdorf D.J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Wu C., Hussein M.A., Shrestha E., Leone S., Aiyegbo M.S., Lambert W.M., Pourcet B., Cardozo T., Gustafson J.A., Fisher E.A. Modulation of macrophage gene expression via liver X receptor alpha serine 198 phosphorylation. Mol. Cell Biol. 2015;35:2024–2034. doi: 10.1128/MCB.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghlool A., Ameur A., Nyberg L., Halvardson J., Grabherr M., Cavelier L., Feuk L. Efficient cellular fractionation improves RNA sequencing analysis of mature and nascent transcripts from human tissues. BMC Biotechnol. 2013;13:99. doi: 10.1186/1472-6750-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All NGS data used in this study were freely available in public repositories at the date of publication. Links to repositories can be found via inline citations.