Abstract
Purpose:
To develop a multi-modal model to automate glaucoma detection.
Design:
Development of a machine-learning glaucoma detection model.
Methods:
We selected a study cohort from the UK Biobank dataset with 1193 eyes of 863 healthy subjects and 1283 eyes of 771 subjects with glaucoma. We trained a multi-modal model that combines multiple deep neural nets, trained on macular optical coherence tomography volumes and color fundus photos, with demographic and clinical data. We performed an interpretability analysis to identify features the model relied on to detect glaucoma. We determined the importance of different features in detecting glaucoma using interpretable machine learning methods. We also evaluated the model on subjects who did not have a diagnosis of glaucoma on the day of imaging but were later diagnosed (progress-to-glaucoma, PTG).
Results:
Results show that a multi-modal model that combines imaging with demographic and clinical features is highly accurate (AUC 0.97). Interpretation of this model highlights biological features known to be related to the disease, such as age, intraocular pressure, and optic disc morphology. Our model also points to previously unknown or disputed features, such as pulmonary function and retinal outer layers. Accurate prediction in PTG highlights variables that change with progression to glaucoma – age and pulmonary function.
Conclusions:
The accuracy of our model suggests distinct sources of information in each imaging modality and in the different clinical and demographic variables. Interpretable machine learning methods elucidate subject-level prediction and help uncover the factors that lead to accurate predictions, pointing to potential disease mechanisms or variables related to the disease.
TOCStatement
Data from the UK Biobank was used to develop a machine learning model that accurately identifies glaucomatous eyes. It combines information from macular optical coherence tomography volumes and color fundus photos with demographic and clinical data. The model highlights biological features known to be related to the disease, such as age, intraocular pressure, and optic disc morphology. It also points to previously unknown or disputed features, such as pulmonary function and retinal outer layers.
Introduction
Glaucoma is the leading cause of irreversible blindness worldwide, affecting approximately 76 million people in 2020 and predicted to affect nearly 111.8 million by 20401. Glaucoma is typically asymptomatic in its early stages; several challenges exist that prevent timely and accurate diagnosis. First, considerable expertise is required to perform the appropriate clinical exam and to interpret several specialized tests, such as visual field testing and retina and optic nerve imaging. The demand for this expertise is outpacing the supply of experts available to interpret tests and make diagnoses2. Second, glaucoma is often asymptomatic until the advanced stages of the disease. In the United States, approximately 50% of the estimated 3 million people with glaucoma are undiagnosed and in other parts of the world, estimates are as high as 90%3,4,5,6,7. New diagnostic tools that improve the diagnostic efficiency of the existing clinical workforce are therefore vital for enabling earlier detection of the disease to facilitate early intervention8,9.
Although glaucoma is asymptomatic in its early stages, structural changes in the macula and RNFL precede the onset of clinically detectable vision loss10. Many studies have therefore attempted to automatically diagnose glaucoma using retinal imaging data. Most of these studies used either color fundus photos (CFPs) or features extracted from CFPs11,12,13,14,15,16,17,18,19,20. Other studies21,22 used features extracted from retinal B-scans obtained via Optical Coherence Tomography (OCT), a three-dimensional volumetric medical imaging technique used to image the retina. Macular OCT images are used to extract features such as thickness of the RNFL, ganglion cell-inner plexiform layer (GCIPL), or full macular thickness. Models evaluating changes in thickness of various retinal layers are promising since such changes, a direct result of tissue loss, are highly accurate disease predictors. However, thickness maps are derived automatically and, despite advances in OCT hardware and software, errors in segmenting retinal OCT images remain relatively common, with error estimates between 19.9% and 46.3%23,24,25. A study comparing a model built on raw macular OCT images with one built on thickness maps demonstrated that the former was significantly more accurate than the latter in detecting glaucoma26.
In this work, we built a new, multi-modal, feature-agnostic model that includes clinical data, CFPs and macular OCT B-scans. Data for our model came from the UK Biobank, a multi-year, large-scale effort to gather medical information and data, with the goal of characterizing the environmental and genetic factors that influence health and disease27. About 65,000 UK Biobank participants underwent ophthalmological imaging procedures, which provided both macular OCT and CFP data that we matched with clinical diagnoses and with many other demographic, systemic and ocular variables. Specifically, cardiovascular and pulmonary variables were chosen as markers of overall health. We used raw macular OCT and CFP data and did not rely on features extracted from these images. The use of machine learning, and particularly deep learning (DL), methods to analyze biomedical data has come under increased scrutiny because these methods can be difficult to interpret and interrogate28,29; therefore, we applied machine learning interpretability methods to demystify and explain specific data features that led to accurate model performance30. Finally, we validated our model by comparing it to expert clinicians’ interpretation of CFPs to provide an additional benchmark for the performance of our machine learning model relative to current clinical practice.
Methods
Data access:
We conducted an analysis of cross-sectional data from the UK Biobank. Data was obtained through the UK Biobank health research program. De-identified color fundus photos, OCT scans, and health data were downloaded from the UK Biobank repository and our study, which did not involve human subjects research, was exempt from IRB approval. The UK Biobank study was approved by an IRB under 11/NW/0382. The UK Biobank’s research ethics committee approval means that researchers wishing to use the resource do not need separate ethics approval, unless re-contact with participants is required (irrelevant in our case). Analysis of de-identified human data is not considered human subjects research by the University of Washington Institutional Review Board and does not require additional approval and an exemption was determined.
Data set and Cohort selection:
The UK Biobank is an ongoing prospective study of human health, for which data has been collected from over half a million individuals31. Participants throughout the UK were recruited between 2006 and 2010 and were aged 40–69 years at the time of recruitment. The data set contains information from questionnaires, multi-modal imaging measurements, and a wide range of genotypic and phenotypic assessments. Data collection is ongoing, allowing for longitudinal assessments. We analyzed a subset of the UK Biobank participants based on a snapshot of the repository that was created in the fall of 2017. This subset consisted of data from 96,020 subjects, 65,000 of which had retinal imaging data. This data set consisted of between one to three visits for each of the subjects. Color Fundus Photographs (CFP) data was available for only the first visit for these subjects. Retinal OCT data was available for first and second visits. The participants were given questionnaires to report various eye conditions, to which they could report healthy or chose one or more the following eye conditions: glaucoma, cataract, macular degeneration, diabetic retinopathy and injury for each eye. We used the answers provided to the questionnaire as the labels for each eye. We did not examine the images to determine or alter the labels associated with the retinal image and clinical data.
Cohort selection:
We selected a cohort from this data for the following three classes: A) subjects who in their first study visit report that they have been diagnosed with glaucoma and consistently report a glaucoma diagnosis in follow-up visits (glaucoma); B) subjects who in their first study visit report that they had no ocular conditions and consistently reported no ocular condition in follow-up visits (healthy); C) subjects who in their first visit report no ocular conditions, but in a subsequent visit report having received a diagnosis of glaucoma, labeled as the “progress to glaucoma” group (PTG). Ocular measurements were only available for the first two visits. The ocular data includes retinal imaging (both CFPs and macular OCTs) as well as IOP, Corneal hysteresis and Corneal resistance factor. However, a subset of the PTG group (n=21 eyes) received glaucoma diagnosis between the first and second visit and we used this subset to conduct statistical analysis of IOP. Systemic and pulmonary variables were available for the entire PTG group both pre- and post-diagnosis, and we were able to analyze the impact of diagnosis on these variables for the entire PTG group.
Exclusion criteria:
We excluded all subjects who preferred not to answer questions about their ocular conditions or did not know how to answer these questions. For glaucoma subjects, we excluded any subjects who listed any ocular conditions in addition to glaucoma, such as age-related macular degeneration, diabetic retinopathy, cataract, or injury. For the healthy subjects, we excluded any subjects whose visual acuity was recorded as worse than 20/30 vision in either eye. We also excluded any healthy subjects with any secondary health conditions (determined by primary diagnosis codes record in their hospital inpatient records). Finally, we excluded any retinal OCT scans from all three classes that could not be aligned using motion translation (x and/or y shift). Supplementary Figure 1 shows a flow chart of subject/image inclusion and distribution among subject groups. Supplementary Figure 2 shows a sample of excluded retinal OCT images. The final number of for the three groups was glaucoma (subjects=863, eyes=1193), healthy (subjects=771, eyes=1283), and PTG (subjects=55, eyes=98). Supplementary Figure 3 shows the age and gender distribution of subjects in each of these groups. CFP images were available for only 56 of the 98 eyes in the PTG group (retinal OCT images were available for all PTG subjects).
Test set:
The data was separated by subject such that both eyes and all visits of any subject belonged to either test, train or validation set. At the outset, we randomly selected 100 eyes, 50 healthy and 50 with glaucoma. These were set aside, as the test set on which we evaluated each of the models. An additional 170 eyes were assigned as a validation set for parameter tuning and model selection. The data was separated by subject such that both eyes of any subject belonged to either test, train or validation set. The test set was also rated by five glaucoma experts. Glaucoma experts used the CFPs for providing their scores. Glaucoma experts marked 13 CFPs from the test set as being of such poor quality as to preclude any assessment. All comparisons of clinician and model performance excludes these 13 eyes. Supplementary Figure 4 shows a sample of excluded CFPs.
Evaluating expert performance:
Five glaucoma-fellowship trained ophthalmologists were recruited for the study to evaluate CFP images from test set to provide an expert diagnosis. The glaucoma experts identified the eye in each CFP as either healthy or glaucoma and rated the confidence in the diagnosis from 1 to 5. A higher number indicated higher confidence in their diagnosis. This resulted in a 10-point scale for the diagnosis. We used this 10-point scale to create ROC curves for each expert.
Machine learning models and training protocols:
We built separate DL models for each imaging modality (retinal OCT and CFP).
Retinal OCT model: the DL model built on the retinal OCT data took a single retinal OCT image as input and output a probability that the input image was from a subject with glaucoma. This model required individual B-scans. Each retinal OCT consisted of 128 B-scan images. This model was not provided any other additional information. This DL model was based on the Densenet architecture32, with four blocks with 6, 12, 48 and 32 layers each. We initialized model weights for this model with MSRA initialization33. Each retinal OCT B-scan is a gray scale 512 × 650 image. We flipped each right eye image left to right; we did this so that the optic nerve was on the same side for each scan. Additionally, we cropped each scan to an aspect ratio of 1:1 and down sampled to 224×224 pixels. Down-sampling is needed to enable use of limited GPU memory when fitting DL models and is common practice in applications of DL to OCT data34,35. We used a per pixel cross-entropy as the loss function with 0.1 label smoothing regularization36. We used Tensorflow37 with Adam optimizer38 and an initial learning rate of 1-e3 and epsilon of 0.1. We trained for 60 epochs (batch size 80) on one graphical processing unit (GPU). The hyper parameters for the training protocol were chosen by tuning on the validation data set. To improve the generalization ability of our model, we augmented the data by applying affine, elastic and intensity transformation over the input images.
CFP model: the DL model on the CFP took a single CFP image as input and outputs a probability that the input image was from a subject with glaucoma. This model was built with transfer learning39,40. We chose transfer learning as (a) we had 128X fewer CFP images, and (b) CFP are color images and transfer learning has been shown to be effective for detecting other pathology in fundus images41. We used the InceptionResnetV442 model, pre-trained on ImageNet data43. We used the Adam optimizer with an initial learning rate of 1-e5. We trained the model for 20 epochs, with a batch size of 400. During training, we kept the weights in 2/3 of the network (750 layers) frozen. We pre-processed each fundus image by flipping left CFP image so that optic nerve was on the same side of each image. We also subtracted local average color to reduce differences in lighting and cropped the images to contain the area around the optical nerve (Supplementary Figure 5). We augmented the CFP by applying affine, elastic and intensity transformations similar to the retinal OCT images.
Baseline models: modern gradient boosted decision trees often provide state-of-the-art performance on tabular style data sets where features are individually meaningful, as consistently demonstrated by open data science competitions44. We used gradient-boosted decision trees, implemented in XGBoost45, to build four baseline models (BM1, BM2, BM3 and BM4) based on demographic features: age, gender, ethnicity; systemic features: Body Mass Index (BMI), Forced Vital Capacity (FVC), Peak Expiratory Flow (PEF), heart rate, diastolic and systolic blood pressure, presence of diabetes, recent caffeine, and nicotine intake; and ocular features: Intraocular pressure (IOP), corneal hysteresis, and corneal resistance factor. We used IOPcc (corneal compensated IOP) in this study as it is thought to be less influenced by corneal measurements such as central corneal thickness, corneal hysteresis, and corneal resistance factor than other measures of IOP such as Goldmann applanation tonometry 46,47.We chose these factors a priori based on existing literature. The systemic features were chosen as markers of overall health. Our data set did not include data on direct smoking status. As there is evidence of smoking and pollution being linked with glaucoma48 we added pulmonary capacity variables: Forced Vital Capacity (FVC) and Peak Expiratory Flow (PEF) in addition to other systemic variables. We used the following hyper parameters for training: learning rate of 0.001, early stopping; regularization of 1.0, no regularization, no column sampling during training, and bagging sub-sampling of 70%. Hyper parameters were chosen by tuning on the validation data set.
Ensemble model: we combined clinical data with results from image-based models to build the final model. To combine data from image models we used the probability of glaucoma as estimated by the respective image model as the feature value for each image. We combined these (128 OCT slices and one fundus) to a 129-element vector as the results of the image-based models. This vector was then combined with all the features from BM3 for the final feature set. We used gradient-boosted decision trees to build this final model. The hyper parameters were chosen by tuning on the validation set and were as follows: learning rate 0.001, early stopping, bagging sub-sampling of 70%, L2 regularization of 1.0, no L1 regularization and no column sampling during training. As an additional control for potential over-fitting, we performed a shuffle-test49 where we repeated the training with randomly permuted labels (see Supplement Section: Shuffle Test Results).
Interpretability Methods: for pixel-level importance in the image-based DL models we used integrated gradients50 and SmoothGrad51 to determine salient pixels for the input images. For the tree-based models built using XGBoost, we used Tree explainer52 to calculate the SHAP values. The SHAP values were used to determine feature importance and feature interaction.
Statistical analysis:
We used bootstrapping53 to determine confidence intervals for AUC and accuracy displayed in Figures 2 and 5. We performed analysis of variance (ANOVA) test to analyze the differences in pulmonary function features (FVC and PEF) among the three groups: healthy, glaucoma and PTG. We used the Dunn Test54 with Bonferroni correction for pairwise comparison to determine differences between the three groups.
Figure 2:
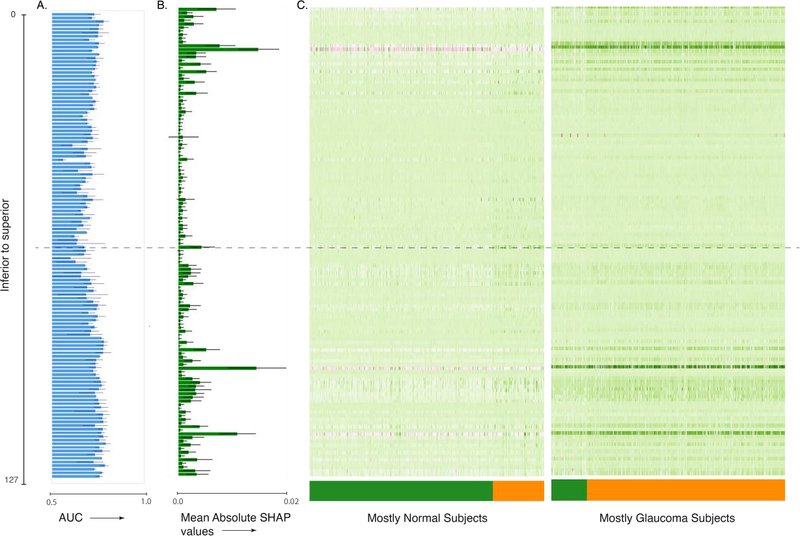
Interpretation of the ensemble model built on macular OCT images. (A) AUC for single image per retina models, (B) mean absolute SHAP values per retinal image for predicting glaucoma occurrence per retina, and (C) heat map of SHAP value per retinal image for predicting glaucoma occurrence per retina. The images are ordered from top to bottom and from superior to inferior retina. The dashed line indicates the central retinal image from the OCT volume.
Figure 5:
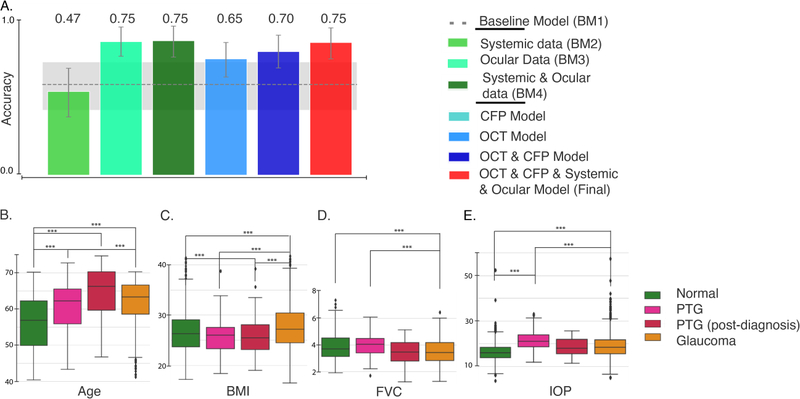
Evaluation of various models on the PTG cohort. (A) Accuracy of the models on the “progress-to-glaucoma” (PTG) cohort. The gray dashed line and shaded area denote the AUC and 95% CI for a base model built on demographics alone (age, gender and ethnicity; BM1). The bottom row shows the distribution of Age(B), BMI(C), FVC(D) and IOP(E) for healthy, PTG, PTG (after glaucoma diagnosis) and glaucoma. ***P < 0.0001.
Results
We built multiple models using clinical data to establish a baseline. Glaucoma is related to many biological features, the most important of which is age55. Thus, we built our first baseline model (BM1) on basic demographic characteristics of the patient and control populations. BM1 included age, gender, and ethnicity. Using these features, a boosted gradient tree-based model predicted an occurrence of glaucoma well above chance (area under the ROC: 0.81, 95% CI 0.71– 0.90).
In addition, we created three other models: The systemic data model (BM2) added cardiovascular and pulmonary variables – including Body Mass Index (BMI), Forced Vital Capacity (FVC), Peak Expiratory Flow (PEF), heart rate, diastolic and systolic blood pressure, and the presence of diabetes – to the demographic variables from BM1. These variables were chosen a priori based on small cohort studies that found relationships between glaucoma and BMI56,57,58 and age59, smoking and cardiovascular factors48 We also included transient factors, such as recent caffeine and nicotine intake, to account for any transient impact on blood pressure and heart rate. BM2 was more accurate then BM1 in detecting glaucoma (0.88 AUC, 95% CI: 0.79–0.96). In the third model (BM3), we added ocular data to BM1, including IOP, corneal hysteresis, and corneal resistance factor. We did not include visual acuity in BM3, as this factor was used in delineating our study groups: which individuals are patients and which are healthy controls, excluding as controls individuals with low visual acuity. BM3 performed similarly to BM2 (0.87 AUC, 95% CI:0.8–0.94). In the fourth model (BM4), we added systemic and ocular data to BM1. BM4 was more accurate than all three of the other baseline models, with a test set AUC of 0.92, 95% (CI: 0.87 – 0.96; Figure 1A, D).
Figure 1:
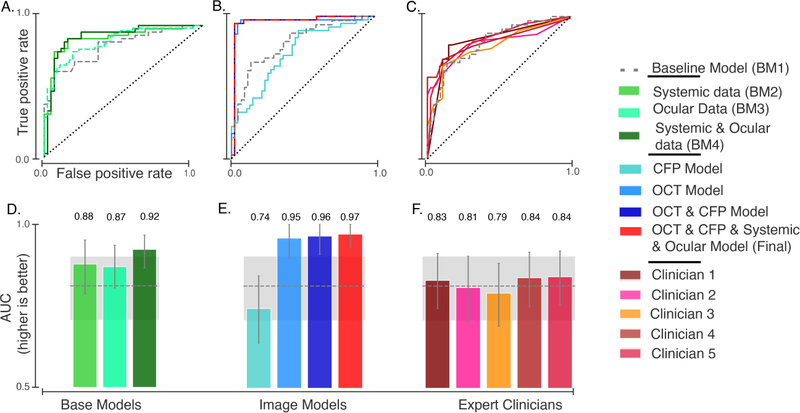
Results of glaucoma detection models. Receiver operating characteristic (ROC) curves are shown for (A) baseline models built with systemic and ocular data, (B) retinal imaging and final models, and (C) glaucoma expert ratings based on interpretation of CFPs. The corresponding area under the ROC curves (AUC) with (+/− 95% Confidence Interval) for models (D, E) and for clinician scores (F). The gray dashed line and shaded area denote the AUC and 95% CI for a base model (BM1) built on demographics (age, gender, and ethnicity).
We used SHapley Additive exPlanations (SHAP)60 to analyze the features that provide high predictive power in BM4. SHAP allocates optimal credit with local explanations using the classic Shapley values derived from game theory61 and provides a quantitative estimate of the contribution of different features to the predictive power of a model. A higher absolute SHAP value indicates greater feature impact on the model prediction and greater feature importance. The five features with the highest mean absolute SHAP values for BM4 were age, IOP, BMI, FVC and PEF. Supplementary Figures 6 and 7 show the most important features in BM4, as evaluated through SHAP, and the interaction effect among the top features.
We built a separate DL model on each retinal image modality. Glaucoma is characterized by structural changes in the optic disc and other parts of the retina. Visual examination of CFP and macular OCT images is therefore an important tool in current diagnostic practice62. Since our data set included both CFP and OCT images, we built separate DL models for each image modality (see Methods). The DL model built on CFP classified eyes diagnosed with glaucoma with modest accuracy (AUC: 0.74, 95% CI: 0.64–0.84; Figure 1B, E). The DL model built on macular OCT images was more accurate than all the baseline models and the model trained on CFP images (AUC: 0.95, 95% CI: 0.90– 1.0).
When we combined information from both the DL models trained on CFP and OCT via an ensemble, the resulting model was marginally more accurate than the DL model build on macular OCT images alone (AUC: 0.963, 95% CI:0.91–1.0). There are several studies which demonstrated the complementary nature of macular data and optic nerve-head data. Given the macular OCT does not contain the optic disc, it seems unlikely that CFP do not provide additional information.
We used several methods to interpret the DL models. DL models are notoriously inscrutable. However, several methods for interrogating these models have recently emerged63,64,65,66,50. To assess the features that lead to high performance of the image-based models, we first assessed which scan of the macular OCT provided the most information. We fit individual models to each scan of the macular OCT. Recall that macular OCTs are volumetric images; in the UK Biobank data set, each macular OCT consists of 128 scans. We found that models using scans from the inferior and superior macula were more accurate than those using the central portion of the macula (Figure 2A). Second, we built an ensemble model that used the results of the DL models of the individual macular OCT scans to predict glaucoma occurrence per retina. This model used each of the 128 macular OCT scans to make a prediction about the retina. Figure 2B shows the feature importance attributed to each scan via SHAP; it shows that scans from the inferior retina were deemed more important by this model. Large patient and control populations are heterogeneous, and we do not generally expect that information will consistently come from one particular part of the retina. Nevertheless, when considering the SHAP values of each macular OCT scan, we found that the data set broke into two major clusters based on the SHAP values from different retinal parts (Figure 2C). One cluster mostly contained retinas from healthy subjects and used scans from the inferior part of the retina as negative predictors of glaucoma. The second cluster mostly contained glaucomatous retinas, and SHAP values of these same scans from inferior and superior macula were used as positive predictors of glaucoma. This also explains why models fit only to scans from the inferior or the superior macula were more accurate.
In addition to the scan-by-scan analysis, image-based models can be evaluated pixel-by-pixel to determine the importance of specific image features to the DL models’ decision making. Using integrated gradients50, we generated saliency maps of the pixels responsible for DL model prediction. Figure 3 shows a macular OCT scan for an eye with glaucoma and a scan for a control eye along with the CFP images and CFP saliency maps for each eye.
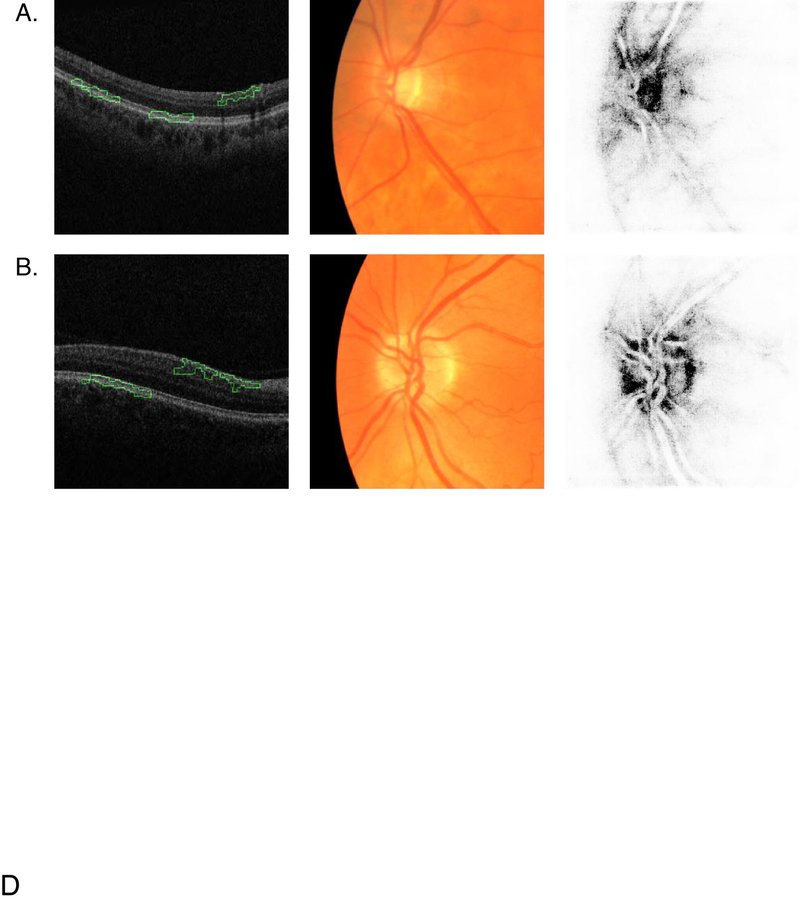
Figure 3:
Saliency maps for macular OCT and CFP. Columns left to right: macular OCT image overlaid with saliency map, cropped CFP input to the neural network, CFP saliency map. Each macular OCT image is laid out with its temporal side to the left. (A) Retina of a subject with glaucoma diagnosis. (B) Retina of healthy subject. The green outline on the OCT saliency map indicates the areas the model deems most important. The darker pixels on the CFP saliency map indicate the areas the model deems most important.
The CFP saliency maps typically highlight the optic nerve head in both normal and glaucomatous retinas. The saliency maps for OCT image typically highlight the nasal side of the RNFL and outer retinal layers.
We built the final model by combining both modalities of retinal imaging, demographic, systemic and ocular features. This model was an ensemble, which combined information from raw macular OCT B-scans and CFP images as well as all demographic, systemic and ocular data used in BM4. This final model had an AUC of 0.967, (95% CI: 0.93 – 1.0). Figure 4 shows the ten features with the highest mean absolute SHAP value over all observations in the data set. The most important features for this final model, as determined by their SHAP values, include age, IOP and FVC, in addition to the CFP and macular OCT scans from both inferior and superior macula. BMI is less significant than FVC in this final model. Further, IOP had a higher importance than age. This is a reversal in importance of features when compared to models built without information from retinal imaging. Unsurprisingly, this confirms that the CFP and OCT scans contain information that supersedes in importance the information provided by BMI and age.
Figure 4:
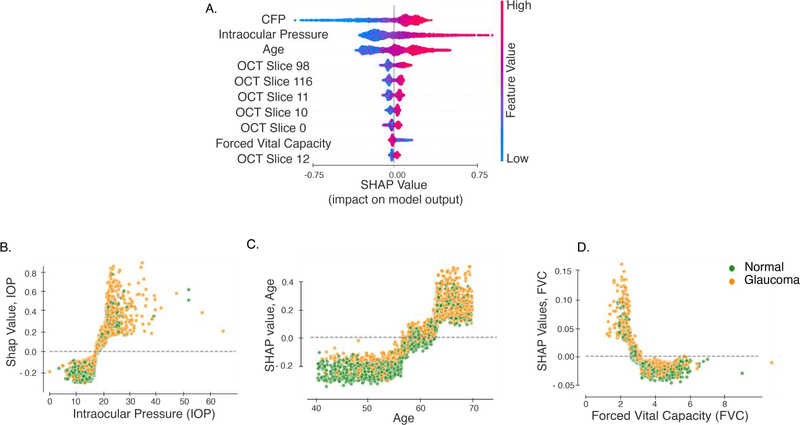
Interpretation of the final model built on image, demographic, systemic and ocular data. Interpretation for models built on medical and optometric features is based on SHAP values. (A) The 10 most important features from this model. SHAP values vs feature values for (B) Age, (C) IOP, (D) FVC and (D) BMI. Each point denotes a subject, and the color denotes whether the subject has been diagnosed with glaucoma.
We compared the performance of our model with ratings from glaucoma expert to provide a comparison to current clinical practice. To compare the performance of our final model to expert clinicians, five glaucoma experts evaluated CFPs of the test set. Initially, experts were also given access to OCT images for each subject. However, raw b-scans from macular OCTs are not an image modality that experts usually examine during regular clinical practice for glaucoma diagnosis. Since we did not have access to thickness maps, experts made the diagnoses using only the CFP data. (Figure 1C and D). The highest AUC for the expert rating was 0.84, and the lowest was 0.79. The average pairwise kappa for the five experts was 0.75, indicating a good level of agreement between experts about the diagnosis.
We validated our model by evaluating it on patients that progress to glaucoma. The UK Biobank data set contained several subjects who lacked a glaucoma diagnosis on their first study visit but received a diagnosis before a subsequent visit. These “progress-to-glaucoma” (PTG) subjects provide a unique opportunity to evaluate our model, which was built on data from glaucomatous and healthy subjects. Detection of glaucoma in the PTG cohort was tested using all our models (Figure 5A). Both BM1 (based on age, gender and ethnicity) and BM2 (added systemic variables) were indistinguishable from chance performance (BM1: 51 % correct: 95% CI [36% – 64%]; BM2: 47% correct: 95% CI [33%–60%]). BM4, which included ocular variables, achieved substantially higher accuracy at 75% correct (95% CI: [67%–83%]). The model trained on macular OCT images achieved slightly lower accuracy at 65% correct (95% CI [55% – 74.5%]), and the model trained on combined CFP and OCT achieved an accuracy of 69% (95% CI [60.2% –78.6%]). The final model trained on OCT, CFP and all other available features achieved an accuracy of 75% correct (95% CI [65% – 83%]).
This evaluation may provide additional insight into the biological features of the disease. For many of these features, including age and BMI, the PTG group lie between the normal and glaucoma groups (Figure 5B to E). We identified two interesting deviations from this pattern. First, for the pulmonary capacity variables (FVC and PEF), the PTG group was indistinguishable from the healthy subjects in our sample, and both healthy and PTG subjects significantly differed from patients with glaucoma. This difference is statistically significant even when controlling for age (see Supplementary Results). However, on a subsequent visit, after receiving a glaucoma diagnosis, the pulmonary capacity measurements of this group was indistinguishable from that of the glaucoma group. Second, the PTG group had a significantly higher IOP than the group diagnosed with glaucoma (Figure 5D; see Supplementary Results). The post-diagnosis IOP measurements of the PTG group shows similar trend with lower IOP values.
Finally, as the labels we used were based on self-report, we performed several analyses to ascertain the reliability of glaucoma labels (Supplementary Results).
Discussion
Automating glaucoma detection using imaging and clinical data may be an important and cost-effective strategy for providing population-level screening. In this study, we used machine learning to construct an interpretable machine learning model that combined clinical information with multi-modal retinal imaging to detect glaucoma. We created and compared several models based on clinical data to establish a baseline: BM1 used demographic data (age, gender, ethnicity), BM2 added systemic medical data (cardiovascular, pulmonary), and BM4 added ocular data (IOP, corneal hysteresis, corneal resistance factor). Our final model was an ensemble, which combined information from raw macular OCT B-scans and CFP images as well as all demographic, systemic and ocular data used in BM4. This final model had an AUC of 0.97.
In interpreting this final model, we found that CFP, age, IOP, macular OCT images from the inferior and superior macula, and FVC were the most important features (Figure. 4). The significance placed upon age and IOP by our final model reiterate previously known risk factors for glaucoma. The positive SHAP values for IOP in our model rapidly increased above an IOP of approximately 20. This is consistent with the fact that ocular hypertension, defined as IOP greater than 21, is a key risk factor for the disease and furthermore clinicians may be more likely to diagnose glaucoma in individuals who have an IOP greater than 2167,68,69,70,71. Age and IOP switched places in their relative importance in our final model, which includes retinal imaging, in addition to BM4 features. This suggests that retinal imaging includes information that supersedes or is redundant with information linked to age. This finding is consistent with previous research, which demonstrated the ability of CFP to predict cardiovascular risk factors including age72. Several population-based studies have already demonstrated an increase in the prevalence of glaucoma with age and have also identified differences in the prevalence of glaucoma among individuals of varying ethnicities73,74,75. In their study of polygenic risk scores for intraocular pressure using data from the UK Biobank data set, Gao et al76 calculated an AUC for the diagnosis of primary open angle glaucoma of 0.713 for a model including only age and sex. The addition of ethnicity in our model may explain why our AUC for BM1 was slightly higher at 0.81. We also observe two discontinuities in the age vs. SHAP values for age (Figure. 4B), at ages 57 and 65. At both ages the SHAP values for age increase at a higher rate than before. This could be both due to biological, as well as socio-economic factors (e.g., 65 is the age of retirement in the UK). However, it is difficult to make strong inferences about the relationship between age, diagnosis, and retinal imaging data, because these may be related in complicated manners. The fact that these individuals self-report that they have glaucoma is also mediated by clinical decision making of the clinicians who assessed these individuals and told them they had glaucoma, itself possibly affected by age. For more on issues with self-reported labels, see below.
The relationship between BMI and glaucoma is controversial, with studies citing evidence for no correlation57, positive correlation77, and negative correlation78 between the two. Consistent with the most comprehensive of these studies, the meta-analysis conducted by Liu et al.77, BM4 demonstrated a positive correlation between glaucoma and increased BMI (Supplementary Figure 6). The correlation between BMI and glaucoma might also be due to ascertainment bias, as subjects with high BMI are more likely to seek medical care (for non-glaucoma related health issues) leading to higher diagnosis of glaucoma in this population. An important novel finding of our study was the correlation of pulmonary measures, especially decreased FVC, with glaucoma. There are several possible explanations for this finding. First, a recent study by Chua et al. found a correlation between glaucoma and atmospheric particulate matter79. Chua et al.’s study did not include pulmonary function tests such as FVC and was correlational in nature, but other studies have linked exposure to particulate matter with decreased FVC80,81,82, suggesting common causes for reduced FVC and for glaucoma. Second, it may be that the treatment of glaucoma with topical beta blocker therapy has an impact on reducing FVC83. This idea receives further support from the findings in the PTG group, who do not have a diagnosis and have presumably not received any treatment. These individuals have FVC that is higher than the glaucoma group and is indistinguishable from the healthy group before a diagnosis is made. After a diagnosis is made, their FVC also decreases to levels indistinguishable from that of the glaucoma group. Thus, lower FVC values could indicate a result of glaucoma treatment.
Examination of the pixel-by-pixel importance of both retinal image modalities provided additional insight into what our model focused on when predicting glaucoma (Figure 3). For the CFPs, the model focused on the optic disc, a known source of information in the clinical diagnosis of glaucoma84. For the macular OCT B-scans, the model relied on previously validated retinal areas, including the inferior and superior macula85. In addition, the algorithm points to the nasal macular RNFL. The effect of glaucoma on RNFL integrity is well understood, and RNFL thickness maps are often used clinically to diagnose glaucoma. However, the automated algorithms that are used clinically have a high segmentation error rate, resulting in variable thickness estimates, which may in turn lead to errors in diagnosis23. By avoiding reliance on extracted features such as thickness maps, our approach enabled the discovery of other possible biological features of glaucoma. For example, consistent with recent results in the same data set86, the model also identified other (non-RNFL) parts of the inner retina as important (e.g., see Figure 3B).
In addition to the RNFL and inner retina, the model relied on the outer layers of the retina for glaucoma diagnosis. The involvement of the retinal outer layers in glaucoma is controversial. In a typical analysis of OCT images that focuses on the thickness of different parts of the retinal layers, glaucoma effects are usually not found in outer layers87,88,89, but an association between age, IOP and retinal pigment epithelium thickness is sometimes detected90. Some anatomical studies do not find any differences in the outer retinal layer between healthy and glaucomatous eyes91. Other studies, using psychophysical methods in human subjects with glaucoma92,93, using histological methods in human eyes94,95 have shown the involvement of the retinal outer layer in glaucoma. In addition, Choi et al.96 used high-resolution imaging techniques (ultrahigh-resolution Fourier-domain optical coherence tomography, UHR-FD-OCT, and adaptive optics, AO) to image glaucomatous retinas. They found a loss of retinal cone receptor cells that correspond to visual field loss. This loss of cones could cause subtle changes in the appearance of this part of the retina, that are not reflected in changes in thickness but are still captured by the DL model (e.g., changes in texture). Ha et. al found that the retinal photoreceptor ellipsoid zone intensity in SD-OCT was decreased in glaucomatous eyes, and this decrease correlated with the stage of glaucoma. They were able to create an automated model that could quantify the changes in ellipsoid zone intensity. This was not an agnostic model, but it suggests that there is a quantifiable change in the outer retina in glaucoma that a deep learning model may be able to identify97,98. On the other hand, the changes to outer retina that are used by the DL model could also be an artifact related to thinning of the inner retina without a specific biological basis in outer retina.
The ability of DL models to use visual cues that are not apparent to the human eye has been previously demonstrated in another study in which retinal angiograms were generated from OCT images99. This finding is also consistent with a recent study that used unsegmented OCT scans and reported the involvement of outer retinal layers in a DL model that detects glaucoma26,100. Nevertheless, this result could also be a consequence of more prosaic effects of DL sensitivity: One possibility is that the DL model is relying on the outer retina OCT signal, because changes to more superficial layers such as the RNFL lead to signal hyperintensity or increased speckle scatter in the deeper layers. Another possibility is that the outer retinal layers are being used as an anatomic reference point for the model to use overall retinal thickness.
Our final model detected the occurrence of glaucoma with an accuracy of 75% on a cohort that had not yet been clinically diagnosed at the time of their testing (“progress-to-glaucoma”, PTG). This does not constitute early detection: even though the individuals were not clinically diagnosed, they may already have significant disease progression, since many patients are undiagnosed even in relatively late stages of the disease10. The median IOP value was higher for the PTG cohort than for the subjects diagnosed with glaucoma, possibly because treatments for glaucoma are designed to decrease IOP. The PTG group also tended to be younger than those diagnosed with glaucoma. Interestingly, FVC in the PTG group was higher than in the glaucoma group and was indistinguishable from healthy subjects. This finding helps explain why BM2, which relied heavily on PVC and PEF, performed relatively poorly on the PTG cohort, achieving an AUC of 47% (Figure 5A). It also provides possible evidence against a causal relationship between FVC and glaucoma, as mentioned above. Furthermore, in a post-diagnosis visit, pulmonary factors (FVC and PEF) in these individuals were lower and indistinguishable from that of the patients with glaucoma, further supporting a possible treatment effect. This area warrants further investigation. The results obtained with this cohort are somewhat limited. This is because the size of this cohort is rather small – only 55 participants. Due to this limited sample size, this group was only used as a further validation and not used to fit any of the models.
Before our model can be considered for use in a real-world setting, several limitations should be considered and addressed. A major limitation of our study was the veracity of ground truth labels used to train the model. Labels used were based on self-report, which may be problematic101. In particular, there is a concern that labels may include significant proportions of false negatives (i.e., people who have glaucoma, but do not report so; glaucoma is generally under-reported7) and false positives (i.e., people who do not have glaucoma, but report that they do. For example, because they confuse different eye disorders). Self-reported labels were particularly pernicious for assessment of PTG individuals, as this designation relied on several self-reported labels for each individual. To further explore these concerns, we examine several factors (Supplemental Results). A previous study that used self-report as labels for models of glaucoma with this dataset79 confirmed that the distributional characteristics of the UK Biobank participants who self-reported glaucoma matched the demographic distribution of those from other population studies such as the Blue Mountain Eye Study102, the Rotterdam Eye Study103, and the Baltimore Eye Study104. In general, it would have been better to use the gold-standard ICD 9/10 available for UK Biobank participants who underwent inpatient procedures. However, this poses significant challenges too: the proportion of participants who met this criterion is too low for machine learning approaches that take advantage of retinal imaging data. Additionally, this population would be biased since they warranted inpatient clinical care and therefore would potentially represent a subset of glaucoma that has increased severity of the disease, compared to other individuals with glaucoma. Nevertheless, the presence of these labels provides an additional opportunity to evaluate the veracity of self-report. When a gold-standard ICD-10 diagnostic code of glaucoma is available, it is always consistent with self-report, suggesting a low prevalence of false negatives in this group. In addition, concerns about self-report labels are mitigated by the high test-retest reliability of self-report: only 0.3% of individuals with repeat visits provide inconsistent self-report. Furthermore, all but one individual who reported that they were prescribed medication that is used for treatment of glaucoma, self-reported that they have glaucoma, which suggests a low prevalence of false negatives in this group. Still, while we eliminated any subject who had inconsistent answers or declined to answer, the generally high rate of undiagnosed glaucoma and the potential for recollection error means that some participants may have been incorrectly labeled. Considering the effects of such mislabeling, we note that a high prevalence of false positives (i.e., a substantial portion of glaucoma suspects or ocular hypertensive participants mistakenly reporting having glaucoma) would weaken the associations and effects that we are reporting, since they would systematically bias the effects towards the null hypothesis. While we agree that this may affect our classification model, we believe that the systemic risk factors that we have found as a positive effect may be an under-estimate of the true effects. Nevertheless, the issue of self-reported labels cannot be fully overcome with these data, and these results would have to be confirmed in a dataset in which ground-truth labels are available.
Another limitation of the present study is that we included only subjects without other ocular disorders. In the general population, glaucoma may coexist with other ocular comorbidities, and it is unclear what effect this may have on the model’s ability to detect glaucoma accurately. This also limits the translation of this model to real-world use. Nevertheless, selecting subjects with only a glaucoma diagnosis and no other ocular morbidities instills confidence that the model we built is glaucoma-specific: delineates the boundaries between these groups, and identifies the features specific to glaucoma. Furthermore, there is some evidence that the UK Biobank has a slight healthy volunteer bias105, potentially biasing inferences to the general population from a model based only on this data. In our results, this bias would probably also induce a bias to the null. Nevertheless, taken together, these factors suggest that a model such as the one proposed here would need to be optimized with more data and data that is more representative of the general population and patient populations before it could be utilized in practice.
Finally, features of the optic disc are clinically important in diagnosing glaucoma. The limited quantity and poor quality of the CFPs in the UK Biobank data set likely contributed to the low AUC of both the CFP DL model and the expert clinician grading. In addition, clinicians did not have access to any additional information about the individuals, and it is very likely that clinicians informally use demographic information, such as patient age, and self-reported daily activities as additional information when making diagnostic determination. This means that in other data, or in clinical settings where CFP have very high quality, and additional information is synthesized into clinical decision making, these data may improve the input of models that use CFP. Furthermore, as demonstrated in our combined model, synthesizing multiple sources of information helps us draw clinical insights into the pathogenesis of the disease.
Our study combined information from multiple sources – including two different retinal imaging modalities (CFP and OCT), demographic data, and systemic and ocular measurement – to build a model that detects glaucoma. This approach yielded not only very accurate detection, but it also enabled us to isolate and interpret critical variables that helped us draw clinical insights into the pathogenesis of the disease.
Supplementary Material
Acknowledgments and disclosures
AR was supported through a grant from the Betty & Gordon Moore Foundation and the Alfred P. Sloan Foundation to the University of Washington eScience Institute Data Science Environment. AYL was support by NIH/NEI K23EY029246 and an unrestricted grant from Research to Prevent Blindness and the Lowy Medical Research Institute. PM and MB were supported by NSF grants AITF 1535565, NSF grants IIS 1524535 and a gift from Intel. SL was supported by NIH R35 GM 128638.
Footnotes
Members of the UK Biobank Eye and Vision Consortium include: Tariq Aslam, Manchester University, Sarah A. Barman, Kingston University, Jenny H. Barrett, University of Leeds, Paul Bishop, Manchester University, Peter Blows, NIHR Biomedical Research Center (BRC), Catey Bunce, King’s College London, Roxana O. Carare, University of Southampton, Usha Chakravarthy, Queens University Belfast, Michelle Chan, NIHR BRC, Sharon Y.L. Chua, NIHR BRC, David P. Crabb, UCL, Philippa M. Cumberland, MSc, UCL Great Ormond Street Institute of Child Health, Alexander Day, NIHR BRC, Parul Desai, NIHR BRC, Bal Dhillon, University of Edinburgh, Andrew D. Dick, University of Bristol, Cathy Egan, NIHR BRC, Sarah Ennis, University of Southampton, Paul Foster, NIHR BRC, Marcus Fruttiger, NIHR BRC, John E. J. Gallacher, University of Oxford, David F. Garway-Heath, NIHR BRC, Jane Gibson, University of Southampton, Dan Gore, NIHR BRC, Jeremy A. Guggenheim, Cardiff University, Chris J. Hammond, King’s College London, Alison Hardcastle, NIHR BRC, Simon P. Harding, University of Liverpool, Ruth E. Hogg, Queens University Belfast, Pirro Hysi, King’s College London, Pearse A. Keane, NIHR BRC, Sir Peng T. Khaw, NIHR BRC, Anthony P. Khawaja, NIHR BRC, Gerassimos Lascaratos, NIHR BRC, Andrew J. Lotery, University of Southampton, Tom Macgillivray, University of Edinburgh, Sarah Mackie, University of Leeds, Keith Martin, University of Cambridge, Michelle McGaughey, Queen’s University Belfast, Bernadette McGuinness, Queen’s University Belfast, Gareth J. McKay, Queen’s University Belfast, Martin McKibbin, Leeds Teaching Hospitals NHS Trust, Danny Mitry, NIHR BRC, Tony Moore, NIHR BRC, James E. Morgan, Cardiff University, Zaynah A. Muthy, NIHR BRC, Eoin O’Sullivan, King’s College Hospital NHS Foundation Trust, Chris G. Owen, University of London, Praveen Patel, NIHR BRC, Euan Paterson, Queens University Belfast, Tunde Peto, Queen’s University Belfast, Axel Petzold, UCL, Jugnoo S. Rahi, UCL Great Ormond Street Institute of Child Health, Alicja R. Rudnikca, University of London, Jay Self, University of Southampton, Sobha Sivaprasad, NIHR BRC, David Steel, Newcastle University, Irene Stratton, MSc, Gloucestershire Hospitals, NHS Foundation Trust, Nicholas Strouthidis, NIHR BRC, Cathie Sudlow, University of Edinburgh, Dhanes Thomas, NIHR BRC, Emanuele Trucco, University of Dundee, Adnan Tufail, NIHR BRC, Veronique Vitart, University of Edinburgh, Stephen A. Vernon, DM, Nottingham University Hospitals, NHS Trust, Ananth C. Viswanathan, NIHR BRC, Cathy Williams, University of Bristol, Katie Williams, King’s College London, Jayne V. Woodside, Queen’s University Belfast, Max M. Yates, University of East Anglia, Jennifer Yip, University of Cambridge, and Yalin Zheng, University of Liverpool.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–2090. doi: 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 2.Foot B, MacEwen C. Surveillance of sight loss due to delay in ophthalmic treatment or review: frequency, cause and outcome. Eye (Lond). 2017. May;31(5):771–775. doi: 10.1038/eye.2017.1. Epub 2017 Jan 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaucoma Research Foundation: Glaucoma Facts and Stats. https://www.glaucoma.org/glaucoma/glaucoma-facts-and-stats.php. (Accessed December 2019).
- 4.Shen SY, Wong TY, Foster PJ, et al. The prevalence and types of glaucoma in malay people: the Singapore Malay eye study. Invest Ophthalmol Vis Sci. 2008;49(9):3846–3851. doi: 10.1167/iovs.08-1759 [DOI] [PubMed] [Google Scholar]
- 5.Foster PJ, Oen FT, Machin D, et al. The prevalence of glaucoma in Chinese residents of Singapore: a cross-sectional population survey of the Tanjong Pagar district. Arch Ophthalmol. 2000;118(8):1105–1111. doi: 10.1001/archopht.118.8.1105 [DOI] [PubMed] [Google Scholar]
- 6.Dirani M, Crowston JG, Taylor PS, et al. Economic impact of primary open-angle glaucoma in Australia. Clin Exp Ophthalmol. 2011;39(7):623–632. doi: 10.1111/j.1442-9071.2011.02530.x [DOI] [PubMed] [Google Scholar]
- 7.Susanna R Jr, De Moraes CG, Cioffi GA, Ritch R. Why Do People (Still) Go Blind from Glaucoma?. Transl Vis Sci Technol. 2015;4(2):1. Published 2015 Mar 9. doi: 10.1167/tvst.4.2.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heijl A, Leske MC, Bengtsson B, et al. Reduction of intraocular pressure and glaucoma progression: results from the Early Manifest Glaucoma Trial. Arch Ophthalmol. 2002;120(10):1268–1279. doi: 10.1001/archopht.120.10.1268 [DOI] [PubMed] [Google Scholar]
- 9.Boland MV, Ervin AM, Friedman D, et al. Treatment for Glaucoma: Comparative Effectiveness. Rockville (MD): Agency for Healthcare Research and Quality (US); April 2012. [PubMed] [Google Scholar]
- 10.Hood DC, Kardon RH. A framework for comparing structural and functional measures of glaucomatous damage. Prog Retin Eye Res. 2007;26(6):688–710. doi: 10.1016/j.preteyeres.2007.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bock R, Meier J, Nyúl LG, Hornegger J, Michelson G. Glaucoma risk index: automated glaucoma detection from color fundus images. Med Image Anal. 2010;14(3):471–481. doi: 10.1016/j.media.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 12.Carrillo J, Bautista L, Villamizar J, Rueda J, Sanchez M, and rueda D Glaucoma Detection Using Fundus. 2019. Images of The Eye. In: 2019 XXII Symposium on Image, Signal Processing and Artificial Vision (STSIVA) doi: 10.1109/STSIVA.2019.8730250. [DOI] [Google Scholar]
- 13.Septiarini A, Khairina DM, Kridalaksana AH, Hamdani H. Automatic Glaucoma Detection Method Applying a Statistical Approach to Fundus Images. Healthc Inform Res. 2018;24(1):53–60. doi: 10.4258/hir.2018.24.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nayak J, Acharya UR, Bhat PS, Shetty N, Lim TC. Automated diagnosis of glaucoma using digital fundus images. J Med Syst. 2009;33(5):337–346. doi: 10.1007/s10916-008-9195-z [DOI] [PubMed] [Google Scholar]
- 15.Chen Xiangyu, Xu Yanwu, Kee Wong Damon Wing, Wong Tien Yin, Liu Jiang. Glaucoma detection based on deep convolutional neural network. Annu Int Conf IEEE Eng Med Biol Soc. 2015;2015:715–718. doi: 10.1109/EMBC.2015.7318462 [DOI] [PubMed] [Google Scholar]
- 16.Orlando José Ignacio, Prokofyeva Elena, del Mariana Fresno, Blaschko Matthew B.. Convolutional neural network transfer for automated glaucoma identification. Proc. SPIE 10160, 12th International Symposium on Medical Information Processing and Analysis, 101600U (26 January 2017); 10.1117/12.2255740 [DOI] [Google Scholar]
- 17.Phene S, Dunn RC, Hammel N, et al. Deep Learning and Glaucoma Specialists: The Relative Importance of Optic Disc Features to Predict Glaucoma Referral in Fundus Photographs. Ophthalmology. 2019;126(12):1627–1639. doi: 10.1016/j.ophtha.2019.07.024 [DOI] [PubMed] [Google Scholar]
- 18.Ting DSW, Cheung CY, Lim G, et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA. 2017;318(22):2211–2223. doi: 10.1001/jama.2017.18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Li L, Wormstone IM, et al. Development and Validation of a Deep Learning System to Detect Glaucomatous Optic Neuropathy Using Fundus Photographs [published online ahead of print, 2019 Sep 12] [published correction appears in JAMA Ophthalmol. 2019 Dec 1;137(12):1468]. JAMA Ophthalmol. 2019;137(12):1353–1360. doi: 10.1001/jamaophthalmol.2019.3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology. 2018;125(8):1199–1206. doi: 10.1016/j.ophtha.2018.01.023 [DOI] [PubMed] [Google Scholar]
- 21.Mwanza JC, Oakley JD, Budenz DL, Chang RT, Knight OJ, Feuer WJ. Macular ganglion cell-inner plexiform layer: automated detection and thickness reproducibility with spectral domain-optical coherence tomography in glaucoma. Invest Ophthalmol Vis Sci. 2011;52(11):8323–8329. Published 2011 Oct 21. doi: 10.1167/iovs.11-7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muhammad H, Fuchs TJ, De Cuir N, et al. Hybrid Deep Learning on Single Wide-field Optical Coherence tomography Scans Accurately Classifies Glaucoma Suspects. J Glaucoma. 2017;26(12):1086–1094. doi: 10.1097/IJG.0000000000000765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansberger SL, Menda SA, Fortune BA, Gardiner SK, Demirel S. Automated Segmentation Errors When Using Optical Coherence Tomography to Measure Retinal Nerve Fiber Layer Thickness in Glaucoma. Am J Ophthalmol. 2017;174:1–8. doi: 10.1016/j.ajo.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miki A, Kumoi M, Usui S, et al. Prevalence and Associated Factors of Segmentation Errors in the Peripapillary Retinal Nerve Fiber Layer and Macular Ganglion Cell Complex in Spectral-domain Optical Coherence Tomography Images. J Glaucoma. 2017;26(11):995–1000. doi: 10.1097/IJG.0000000000000771 [DOI] [PubMed] [Google Scholar]
- 25.Asrani S, Essaid L, Alder BD, Santiago-Turla C. Artifacts in spectral-domain optical coherence tomography measurements in glaucoma. JAMA Ophthalmol. 2014;132(4):396–402. doi: 10.1001/jamaophthalmol.2013.7974 [DOI] [PubMed] [Google Scholar]
- 26.Maetschke S, Antony B, Ishikawa H, Wollstein G, Schuman J, Garnavi R. A feature agnostic approach for glaucoma detection in OCT volumes. PLoS One. 2019;14(7):e0219126. Published 2019 Jul 1. doi: 10.1371/journal.pone.0219126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UK Biobank. https://www.ukbiobank.ac.uk/. Online; accessed September,2019.
- 28.Winkler JK, Fink C, Toberer F, et al. Association Between Surgical Skin Markings in Dermoscopic Images and Diagnostic Performance of a Deep Learning Convolutional Neural Network for Melanoma Recognition [published online ahead of print, 2019 Aug 14]. JAMA Dermatol. 2019;155(10):1135–1141. doi: 10.1001/jamadermatol.2019.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badgeley MA, Zech JR, Oakden-Rayner L, et al. Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ Digit Med. 2019;2:31. Published 2019 Apr 30. doi: 10.1038/s41746-019-0105-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murdoch WJ, Singh C, Kumbier K, Abbasi-Asl R, Yu B. Definitions, methods, and applications in interpretable machine learning. Proc Natl Acad Sci U S A. 2019;116(44):22071–22080. doi: 10.1073/pnas.1900654116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. Published 2015 Mar 31. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang G, Liu Z, Pleiss G, Van Der Maaten L, Weinberger K. Convolutional Networks with Dense Connectivity. IEEE Trans Pattern Anal Mach Intell. 2019; doi: 10.1109/TPAMI.2019.2918284 [DOI] [PubMed] [Google Scholar]
- 33.He K, Zhang X, Ren S, and Sun J Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification. In: Proceedings of the IEEE international conference on computer vision. 2015, pages 1026–1034. [Google Scholar]
- 34.Yanagihara RT, Lee CS, Ting DSW, Lee AY. Methodological Challenges of Deep Learning in Optical Coherence Tomography for Retinal Diseases: A Review. Transl Vis Sci Technol. 2020;9(2):11. Published 2020 Feb 18. doi: 10.1167/tvst.9.2.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–1350. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 36.Müller R, Kornblith S, and Hinton GE. When does label smoothing help? In: Advances in Neural Information Processing Systems. 2019, pages 4696–4705. [Google Scholar]
- 37.Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, 1603.04467. [Google Scholar]
- 38.Kingma DP and Ba J Adam: A method for stochastic optimization. arXiv 2014. 1412.6980. [Google Scholar]
- 39.Pan SJ, & Yang Q A survey on transfer learning. IEEE Transactions on knowledge and data engineering, 2009. 22(10), 1345–1359. doi: 10.1109/TKDE.2009.191 [DOI] [Google Scholar]
- 40.Ahmed A, Yu K, Xu W, Gong Y, Xing E Training Hierarchical Feed-Forward Visual Recognition Models Using Transfer Learning from Pseudo-Tasks. In: Forsyth D, Torr P, Zisserman A (eds) Computer Vision – ECCV 2008. ECCV 2008. Lecture Notes in Computer Science, vol 5304. Springer, Berlin, Heidelberg. 10.1007/978-3-540-88690-7_6 [DOI] [Google Scholar]
- 41.Li X, Pang T, Xiong B, Liu W, Liang P and Wang T Convolutional neural networks based transfer learning for diabetic retinopathy fundus image classification. 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 2017, pp. 1–11, doi: 10.1109/CISP-BMEI.2017.8301998. [DOI] [Google Scholar]
- 42.Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. AAAI [Internet]. 2017. https://ojs.aaai.org/index.php/AAAI/article/view/11231 [Google Scholar]
- 43.http://www.image-net.org/.
- 44.Friedman JH. Greedy function approximation: a gradient boosting machine. Annals of statistics 2001; 29: 1189–1232. [Google Scholar]
- 45.Chen T and Guestrin C 2016. XGBoost: A Scalable Tree Boosting System. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ‘16). Association for Computing Machinery, New York, NY, USA, 785–794. doi: 10.1145/2939672.2939785 [DOI] [Google Scholar]
- 46.Medeiros FA, Weinreb RN. Evaluation of the influence of corneal biomechanical properties on intraocular pressure measurements using the ocular response analyzer. J Glaucoma. 2006;15(5):364–370. doi: 10.1097/01.ijg.0000212268.42606.97 [DOI] [PubMed] [Google Scholar]
- 47.Ehrlich JR, Radcliffe NM, Shimmyo M. Goldmann applanation tonometry compared with corneal-compensated intraocular pressure in the evaluation of primary open-angle Glaucoma. BMC Ophthalmol. 2012;12:52. Published 2012 Sep 25. doi: 10.1186/1471-2415-12-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonovas S, Filioussi K, Tsantes A, Peponis V. Epidemiological association between cigarette smoking and primary open-angle glaucoma: a meta-analysis. Public Health. 2004;118(4):256–261. doi: 10.1016/j.puhe.2003.09.009 [DOI] [PubMed] [Google Scholar]
- 49.Wood M How sure are we? Two approaches to statistical inference. arXiv 2018. 1803.06214. [Google Scholar]
- 50.Sundararajan M, Taly A, Yan Q. Axiomatic Attribution for Deep Networks. arXiv 2017. 1703.01365. [Google Scholar]
- 51.Smilkov D, Thorat N, Kim B, Viégas F, and Wattenberg M Smoothgrad: removing noise by adding noise. arXiv 2017. 1706.03825. [Google Scholar]
- 52.Lundberg SM, Erion G, Chen H, et al. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat Mach Intell. 2020;2(1):56–67. doi: 10.1038/s42256-019-0138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efron B and Tibshirani R Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. In: Statistical science (1986), pages 54–75. [Google Scholar]
- 54.Dunn OJ. Multiple comparisons using rank sums. Technometrics 1964. 6.3: 241–252. [Google Scholar]
- 55.Guedes G, Tsai JC, Loewen NA. Glaucoma and aging. Curr Aging Sci. 2011;4(2):110–117. doi: 10.2174/1874609811104020110 [DOI] [PubMed] [Google Scholar]
- 56.Jung Y, Han K, Park HYL, Lee SH, Park CK. Metabolic Health, Obesity, and the Risk of Developing Open-Angle Glaucoma: Metabolically Healthy Obese Patients versus Metabolically Unhealthy but Normal Weight Patients. Diabetes Metab J. 2020;44(3):414–425. doi: 10.4093/dmj.2019.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gasser P, Stümpfig D, Schötzau A, Ackermann-Liebrich U, Flammer J. Body mass index in glaucoma. J Glaucoma. 1999;8(1):8–11. [PubMed] [Google Scholar]
- 58.Lee JY, Kim TW, Kim HT, et al. Relationship between anthropometric parameters and open angle glaucoma: The Korea National Health and Nutrition Examination Survey. PLoS One. 2017;12(5):e0176894. Published 2017 May 8. doi: 10.1371/journal.pone.0176894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klein R, Lee KE, Gangnon RE, Klein BE. Relation of smoking, drinking, and physical activity to changes in vision over a 20-year period: the Beaver Dam Eye Study. Ophthalmology. 2014;121(6):1220–1228. doi: 10.1016/j.ophtha.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lundberg SM and Lee SI. A unified approach to interpreting model predictions. In: Advances in Neural Information Processing Systems. 2017, pages 4765–4774. [Google Scholar]
- 61.Roth AE. The Shapley value: essays in honor of Lloyd S. Shapley. Cambridge University Press, 1988. [Google Scholar]
- 62.Newman-Casey PA, Verkade AJ, Oren G, Robin AL. Gaps in Glaucoma care: A systematic review of monoscopic disc photos to screen for glaucoma. Expert Rev Ophthalmol. 2014;9(6):467–474. doi: 10.1586/17469899.2014.967218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, and Rabinovich A Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015, pages 1–9. [Google Scholar]
- 64.Olah C, Satyanarayan A, Johnson I, Carter S, Schubert L, Ye K, Mordvintsev A The building blocks of interpretability. Distill, 2018. 3(3), e10. [Google Scholar]
- 65.Olah C, Mordvintsev A and Schubert L, Feature visualization. Distill, 2017, 2(11), p.e7. [Google Scholar]
- 66.Shrikumar A, Greenside P, Kundaje A Learning Important Features Through Propagating Activation Differences. Proceedings of the 34th International Conference on Machine Learning, in Proceedings of Machine Learning Research 2017. 70:3145–3153 [Google Scholar]
- 67.Kass MA, Heuer DK, Higginbotham EJ, et al. The Ocular Hypertension Treatment Study: a randomized trial determines that topical ocular hypotensive medication delays or prevents the onset of primary open-angle glaucoma. Arch Ophthalmol. 2002;120(6):701–830. doi: 10.1001/archopht.120.6.701 [DOI] [PubMed] [Google Scholar]
- 68.Perkins ES. The Bedford glaucoma survey. I. Long-term follow-up of borderline cases. Br J Ophthalmol. 1973;57(3):179–185. doi: 10.1136/bjo.57.3.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hart WM Jr, Yablonski M, Kass MA, Becker B. Multivariate analysis of the risk of glaucomatous visual field loss. Arch Ophthalmol. 1979;97(8):1455–1458. doi: 10.1001/archopht.1979.01020020117005 [DOI] [PubMed] [Google Scholar]
- 70.Armaly MF, Krueger DE, Maunder L, et al. Biostatistical analysis of the collaborative glaucoma study. I. Summary report of the risk factors for glaucomatous visual-field defects. Arch Ophthalmol. 1980;98(12):2163–2171. doi: 10.1001/archopht.1980.01020041015002 [DOI] [PubMed] [Google Scholar]
- 71.Quigley HA, Enger C, Katz J, Sommer A, Scott R, Gilbert D. Risk factors for the development of glaucomatous visual field loss in ocular hypertension. Arch Ophthalmol. 1994;112(5):644–649. doi: 10.1001/archopht.1994.01090170088028 [DOI] [PubMed] [Google Scholar]
- 72.Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2(3):158–164. doi: 10.1038/s41551-018-0195-0 [DOI] [PubMed] [Google Scholar]
- 73.Ryskulova A, Turczyn K, Makuc DM, Cotch MF, Klein RJ, Janiszewski R. Self-reported age-related eye diseases and visual impairment in the United States: results of the 2002 national health interview survey. Am J Public Health. 2008;98(3):454–461. doi: 10.2105/AJPH.2006.098202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varma R, Ying-Lai M, Francis BA, et al. Prevalence of open-angle glaucoma and ocular hypertension in Latinos: the Los Angeles Latino Eye Study. Ophthalmology. 2004;111(8):1439–1448. doi: 10.1016/j.ophtha.2004.01.025 [DOI] [PubMed] [Google Scholar]
- 75.Friedman DS, Wolfs RC, O’Colmain BJ, et al. Prevalence of open-angle glaucoma among adults in the United States [published correction appears in Arch Ophthalmol. 2011 Sep;129(9):1224]. Arch Ophthalmol. 2004;122(4):532–538. doi: 10.1001/archopht.122.4.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao XR, Huang H, Kim H. Polygenic Risk Score Is Associated With Intraocular Pressure and Improves Glaucoma Prediction in the UK Biobank Cohort. Transl Vis Sci Technol. 2019;8(2):10. Published 2019 Apr 4. doi: 10.1167/tvst.8.2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu W, Ling J, Chen Y, Wu Y, Lu P. The Association between Adiposity and the Risk of Glaucoma: A Meta-Analysis. J Ophthalmol. 2017;2017:9787450. doi: 10.1155/2017/9787450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin SC, Pasquale LR, Singh K, Lin SC. The Association Between Body Mass Index and Open-angle Glaucoma in a South Korean Population-based Sample. J Glaucoma. 2018;27(3):239–245. doi: 10.1097/IJG.0000000000000867 [DOI] [PubMed] [Google Scholar]
- 79.Chua SYL, Khawaja AP, Morgan J, et al. The Relationship Between Ambient Atmospheric Fine Particulate Matter (PM2.5) and Glaucoma in a Large Community Cohort. Invest Ophthalmol Vis Sci. 2019;60(14):4915–4923. doi: 10.1167/iovs.19-28346 [DOI] [PubMed] [Google Scholar]
- 80.Chestnut LG, Schwartz J, Savitz DA, Burchfiel CM. Pulmonary function and ambient particulate matter: epidemiological evidence from NHANES I. Arch Environ Health. 1991;46(3):135–144. doi: 10.1080/00039896.1991.9937440 [DOI] [PubMed] [Google Scholar]
- 81.Wang HJ, Li Q, Guo Y, Song JY, Wang Z, Ma J. Geographic variation in Chinese children’ forced vital capacity and its association with long-term exposure to local PM10: a national cross-sectional study. Environ Sci Pollut Res Int. 2017;24(28):22442–22449. doi: 10.1007/s11356-017-9812-9 [DOI] [PubMed] [Google Scholar]
- 82.Havet A, Hulo S, Cuny D, et al. Residential exposure to outdoor air pollution and adult lung function, with focus on small airway obstruction. Environ Res. 2020;183:109161. doi: 10.1016/j.envres.2020.109161 [DOI] [PubMed] [Google Scholar]
- 83.Pavia D, Bateman JR, Lennard-Jones AM, Agnew JE, Clarke SW. Effect of selective and non-selective beta blockade on pulmonary function and tracheobronchial mucociliary clearance in healthy subjects. Thorax. 1986;41(4):301–305. doi: 10.1136/thx.41.4.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Armaly MF. Optic cup in normal and glaucomatous eyes. Invest Ophthalmol. 1970;9(6):425–429. [PubMed] [Google Scholar]
- 85.Patel SB, Reddy N, Lin X, Whitson JT. Optical coherence tomography retinal nerve fiber layer analysis in eyes with long axial lengths. Clin Ophthalmol. 2018;12:827–832. Published 2018 May 3. doi: 10.2147/OPTH.S162023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khawaja AP, Chua S, Hysi PG, et al. Comparison of Associations with Different Macular Inner Retinal Thickness Parameters in a Large Cohort: The UK Biobank. Ophthalmology. 2020;127(1):62–71. doi: 10.1016/j.ophtha.2019.08.015 [DOI] [PubMed] [Google Scholar]
- 87.Unterlauft JD, Rehak M, Böhm MRR, Rauscher FG. Analyzing the impact of glaucoma on the macular architecture using spectral-domain optical coherence tomography. PLoS One. 2018;13(12):e0209610. Published 2018 Dec 31. doi: 10.1371/journal.pone.0209610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cifuentes-Canorea P, Ruiz-Medrano J, Gutierrez-Bonet R, et al. Analysis of inner and outer retinal layers using spectral domain optical coherence tomography automated segmentation software in ocular hypertensive and glaucoma patients. PLoS One. 2018;13(4):e0196112. Published 2018 Apr 19. doi: 10.1371/journal.pone.0196112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kita Y, Anraku A, Kita R, Goldberg I. The clinical utility of measuring the macular outer retinal thickness in patients with glaucoma. Eur J Ophthalmol. 2016;26(2):118–123. doi: 10.5301/ejo.5000678 [DOI] [PubMed] [Google Scholar]
- 90.Ko F, Foster PJ, Strouthidis NG, et al. Associations with Retinal Pigment Epithelium Thickness Measures in a Large Cohort: Results from the UK Biobank. Ophthalmology. 2017;124(1):105–117. doi: 10.1016/j.ophtha.2016.07.033 [DOI] [PubMed] [Google Scholar]
- 91.Kendell KR, Quigley HA, Kerrigan LA, Pease ME, Quigley EN. Primary open-angle glaucoma is not associated with photoreceptor loss. Invest Ophthalmol Vis Sci. 1995;36(1):200–205. [PubMed] [Google Scholar]
- 92.Holopigian K, Seiple W, Mayron C, Koty R, Lorenzo M. Electrophysiological and psychophysical flicker sensitivity in patients with primary open-angle glaucoma and ocular hypertension. Invest Ophthalmol Vis Sci. 1990;31(9):1863–1868. [PubMed] [Google Scholar]
- 93.Odom JV, Feghali JG, Jin JC, Weinstein GW. Visual function deficits in glaucoma. Electroretinogram pattern and luminance nonlinearities. Arch Ophthalmol. 1990;108(2):222–227. doi: 10.1001/archopht.1990.01070040074034 [DOI] [PubMed] [Google Scholar]
- 94.Panda S, Jonas JB. Decreased photoreceptor count in human eyes with secondary angle-closure glaucoma. Invest Ophthalmol Vis Sci. 1992;33(8):2532–2536. [PubMed] [Google Scholar]
- 95.Nork TM, Ver Hoeve JN, Poulsen GL, et al. Swelling and loss of photoreceptors in chronic human and experimental glaucomas. Arch Ophthalmol. 2000;118(2):235–245. doi: 10.1001/archopht.118.2.235 [DOI] [PubMed] [Google Scholar]
- 96.Choi SS, Zawadzki RJ, Lim MC, et al. Evidence of outer retinal changes in glaucoma patients as revealed by ultrahigh-resolution in vivo retinal imaging. Br J Ophthalmol. 2011;95(1):131–141. doi: 10.1136/bjo.2010.183756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ha A, Kim YK, Jeoung JW, Park KH. Ellipsoid Zone Change According to Glaucoma Stage Advancement. Am J Ophthalmol. 2018;192:1–9. doi: 10.1016/j.ajo.2018.04.025 [DOI] [PubMed] [Google Scholar]
- 98.Ha A, Sun S, Kim YK, Jeoung JW, Kim HC, Park KH. Automated Quantification of Macular Ellipsoid Zone Intensity in Glaucoma Patients: the Method and its Comparison with Manual Quantification. Sci Rep. 2019;9(1):19771. Published 2019 Dec 24. doi: 10.1038/s41598-019-56337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee CS, Tyring AJ, Wu Y, et al. Generating retinal flow maps from structural optical coherence tomography with artificial intelligence. Sci Rep. 2019;9(1):5694. Published 2019 Apr 5. doi: 10.1038/s41598-019-42042-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thompson AC, Jammal AA, Berchuck SI, Mariottoni EB, Medeiros FA. Assessment of a Segmentation-Free Deep Learning Algorithm for Diagnosing Glaucoma From Optical Coherence Tomography Scans. JAMA Ophthalmol. 2020;138(4):333–339. doi: 10.1001/jamaophthalmol.2019.5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Foreman J, Xie J, Keel S, van Wijngaarden P, Taylor HR, Dirani M. The validity of self-report of eye diseases in participants with vision loss in the National Eye Health Survey. Sci Rep. 2017;7(1):8757. Published 2017 Aug 18. doi: 10.1038/s41598-017-09421-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mitchell P, Smith W, Attebo K, Healey PR. Prevalence of open-angle glaucoma in Australia. The Blue Mountains Eye Study. Ophthalmology. 1996;103(10):1661–1669. doi: 10.1016/s0161-6420(96)30449-1 [DOI] [PubMed] [Google Scholar]
- 103.Dielemans I, Vingerling JR, Wolfs RC, Hofman A, Grobbee DE, de Jong PT. The prevalence of primary open-angle glaucoma in a population-based study in The Netherlands. The Rotterdam Study. Ophthalmology. 1994;101(11):1851–1855. doi: 10.1016/s0161-6420(94)31090-6 [DOI] [PubMed] [Google Scholar]
- 104.Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma. The Baltimore Eye Survey. JAMA. 1991;266(3):369–374. [PubMed] [Google Scholar]
- 105.Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am J Epidemiol. 2017;186(9):1026–1034. doi: 10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.