Abstract
The translation of laboratory-scale bioprocess protocols and technologies to industrial scales and the application of human induced pluripotent stem cell (hiPSC) derivatives in clinical trials globally presents optimism for the future of stem-cell products to impact healthcare. However, while many promising therapeutic approaches are being tested in pre-clinical studies, hiPSC-derived products currently account for a small fraction of active clinical trials. The complexity and volatility of hiPSCs present several bioprocessing challenges, where the goal is to generate a sufficiently large, high-quality, homogeneous population for downstream differentiation—the derivatives of which must retain functional efficacy and meet regulatory safety criteria in application. It is argued herein that one of the major challenges currently faced in improving the robustness of routine stem-cell biomanufacturing is in utilizing continuous, meaningful assessments of molecular and cellular characteristics from process to application. This includes integrating process data with biological characteristic and functional assessment data to model the interplay between variables in the search for global optimization strategies. Coupling complete datasets with relevant computational methods will contribute significantly to model development and automation in achieving process robustness. This overarching approach is thus crucially important in realizing the potential of hiPSC biomanufacturing for transformation of regenerative medicine and the healthcare industry.
Subject terms: Induced pluripotent stem cells, Stem-cell biotechnology
Introduction
Modern bioprocessing technology and protocols have developed to the point of producing functional, clinically relevant numbers of pluripotent stem cells for use as cell and tissue source material1–3. However, optimization of manufacturing protocols is resource-intensive and costly, with characterization of phenotype being laborious and discontinuous. Coupled with in-process heterogeneity and the evolving regulatory framework surrounding assessment of stem-cell-derived therapeutics, it is difficult to develop scalable, robust processes that are strictly standardized and economically viable4,5. Clinical outcomes are dependent on biological function of the product, with quality hindered by obstacles such as a lack of reproducibility and robustness for scale-up and scale-out6. Utilization of human induced pluripotent stem cells (hiPSCs) and their derivatives for drug discovery, cell therapy, and disease modeling present a potentially invaluable resource, but existing limitations to their use include conceptual biases, technological limitations, process heterogeneity, rudimentary control over cell fate, and substantial cost7,8.
Addressing these limitations and obstacles in the development of stem-cell-based therapeutics includes the incorporation of technologies and methods capable of continuous monitoring and assessment of phenotype throughout the bioprocess, in conjunction with process control, standardization, and automation of protocols as they are developed. These advancements will require the integration of process systems, novel analytical technologies, and computational methods throughout development and optimization. This paper aims to explore the challenges and potential strategies to overcome obstacles faced in controlling pluripotent and differentiated phenotype in the context of functionality, efficacy, and safety throughout hiPSC biomanufacturing.
Highlighting challenges in the scalable and robust production of human iPSCs
Human pluripotent stem cells are promising candidates for therapeutic applications given their proliferative potential and capacity to differentiate into any cell type within the primary germ layers of an adult organism9. Naturally occurring populations of these cells are found within an early-stage embryonic blastocyst and can be isolated and expanded in vitro to generate a population of self-renewing human embryonic stem cells (ESCs)10. However, harvesting the cells requires destruction of the developing embryo. Utilization of hiPSCs as an alternative cell source effectively overcomes the legal, ethical, and moral barriers associated with the use of embryonic or epiblast stem cells11. However, conventional methods for the induction and maintenance of pluripotent state introduce a myriad of additional biological obstacles to effective application in stem-cell-derived therapeutics. These include an increased risk of mutation, retention of somatic epigenetic memory, potential for immunogenicity, and altered functional characteristics of differentiated phenotypes12–15.
Stem-cell research for drug development, disease modeling, and potential therapeutic applications has gained great momentum in recent years16–18. Progress has been made towards harnessing their curative potential and significant investment has been placed into assessment of therapeutic applications, with varying degrees of success19–21. The lack of success leading to suspension of many iPSC clinical trials specifically highlights a need to further elucidate the interplay between variables over the course of the bioprocess and their influence on stem-cell-derived product identity and function. Further, a lack of process robustness in conventional systems and protocols result in a high degree of in-process heterogeneity observed within batch populations and between-culture batches22. This heterogeneity reflects the high phenotypic plasticity of pluripotent stem cells, and the non-uniform spatiotemporal conditions observed across bioprocess systems23. These results are a strong reminder that process development strategies must be designed to strictly orchestrate both pluripotent and downstream phenotypes, with interventions made to minimize stochasticity and spatial gradient-induction within the process.
Biological complexities governing iPSC phenotype and downstream functionality
Transient behavior of iPSC populations cultured in vitro are dependent on genetic variants present in the derived source cell line and the culture environment in which they are sustained. Many cultured iPSC lines present copy-number variations, with considerable and non-uniform genetic load of single-nucleotide variants among clones within a particular population24,25. Several identified variants affect genes implicated in cancer, and clonal dominance observed in vitro suggest selection of clones with some phenotypic advantage that while desirable for their enhanced growth characteristics may present safety risks at the clinical stage26,27. Coupled with chromosomal instability often observed over extended in vitro culture, these considerations highlight the need for continuity of measurement to monitor and adapt both genetic and epigenetic characteristics of the cell population.
Evidence of immune response has been observed in autologous transplantations of iPSCs in mice and raises concerns for therapeutic potential in humans28–30. Many such studies have focused on elucidating the effect of reprogramming strategies on genetic and epigenetic defects in the induced pluripotent cell population, and their contributions to immunogenicity in therapeutic derivatives. However, given the potential for genetic and epigenetic abnormalities to arise in culture, there is only a paucity of studies that directly assess the contribution of in vitro conditions and bioprocess variables during iPSC expansion and subsequent differentiation on immunogenic potential31–33.
Epigenetic mechanisms play a pivotal role in the regulation of gene expression. These mechanisms are diverse and include behaviors such as histone protein modifications and DNA methylation34. Epigenetic memory is defined by inherited cellular behavior as a result of prior stimulus. These alterations result in reinforcement and suppression activity in gene regulatory networks, a “learned” identity in the cell and its progeny over successive generations35,36. This phenomenon is particularly well-noted in iPSC populations, given their propensity for epigenetic memory as a result of the somatic source cell identity, age, metabolic signature, and reprogramming strategy used for induction of pluripotency12,37. Indeed, somatic memory has been shown to result in preferential bias during differentiation towards certain lineages that affects phenotype and functionality in iPSC populations and their derivatives38. Further, these abnormal phenotypes raise questions of cellular stability, longevity, and tumorigenic risk39. More work is needed to understand the long-term epigenetic landscape of iPSC populations from reprogramming all the way through to their use as constituents within a functional therapeutic, and how these factors contribute to their safety and efficacy.
Functional characteristics have been reported to vary among groups studying iPSC derivatives40,41. Combining the implications discussed so far in this paper, it is no surprise that the biological factors responsible for variability in the reprogrammed iPSC population extend to their progeny throughout expansion and subsequent differentiation. Furthermore, biological sex differences add additional considerations—X-inactivation in female cells results in differing regulatory dynamics from XY-active male cell lines. These differences are known to lead to diverse mechanisms in cell state de-regulation and cancer progression42–44. Given the disparity in available data on biological sex, it is critical that researchers recognize these differences and make every effort to ensure equality in process development. While relatively few studies have focused specifically on the downstream implications of the factors discussed, it is pertinent that scientists and engineers make a concerted effort to include this information in process development and optimization, using generated data to better understand and control these overlapping cause-and-effect interactions [Fig. 1].
Fig. 1. Considerations for clinical stem-cell manufacturing.
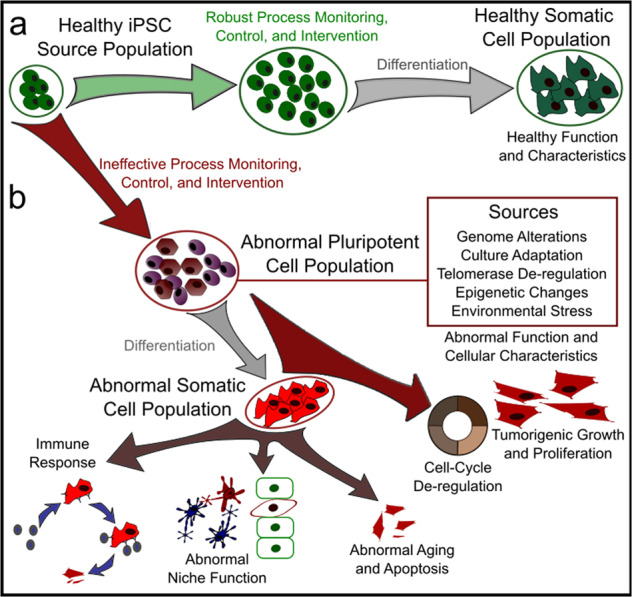
a Ideal stem-cell population dynamics in the presence of adequate process control and intervention strategies. Maintenance of high-quality phenotype is critical in ensuring safety and efficacy in derived products. b Erosion of phenotype in the presence of inadequate process control and intervention, the result of which may take the form of genetic defects, elicitation of immune response by the host, abnormal function, cell cycle de-regulation, and tumorigenesis.
Cellular interaction networks governing the pluripotent state
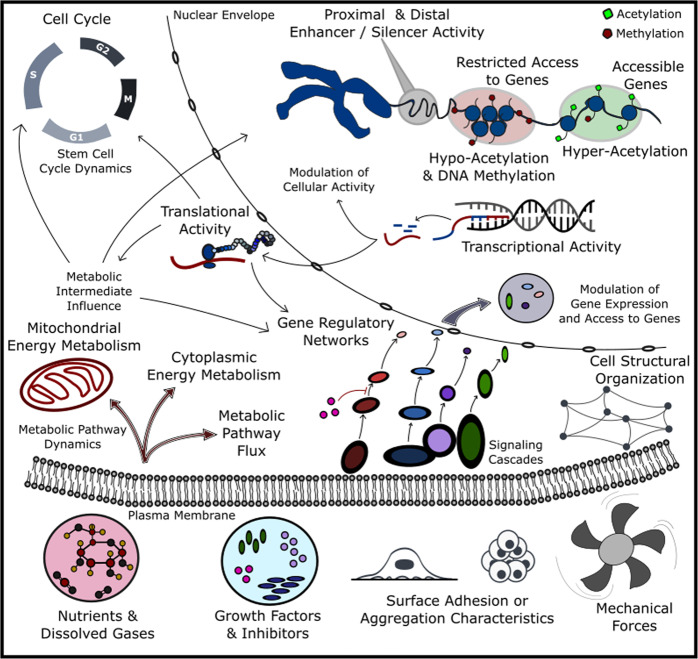
The challenges discussed so far reflect the complex cellular mechanisms and network interactions that govern phenotype. The origins of these mechanisms and their implications for stem-cell-derived products must be understood to facilitate effective design and optimization of bioprocesses. Cellular phenotype is a consequence of the inherited features in the population, combined with cellular regulation and environmental niche responses. This section will describe these interactions and the results of recent work to understand and control phenotype with regard to their respective mechanisms. Arguably, the interaction network arising from complex multi-system behavior within the cell results in a co-regulatory dynamic that is critical to the maintenance of pluripotent phenotype [Fig. 2].
Fig. 2. Process interaction networks at a glance.
Visualization of the interaction network at a high level of abstraction. Some of the critical cellular machinery dictating cell health and phenotype are shown. This machinery interacts with one another and influences from the bioprocess to continuously maintain and influence overall quality in the process population.
DNA methylation is known to play an important role in restricting genetic access by transcriptional machinery45. In naïve state pluripotency, hypomethylation of the global genome is observed46. As the cell transitions towards a primed state, hypermethylation of the genome occurs, an orchestrated restriction of access to genes by epigenetic machinery. IPSCs exhibit unique methylation patterns as a result of reprogramming, with outcomes dependent on source cell characteristics and the strategy used47. This somatic memory is a form of genomic imprinting and may result in aberrant patterns of activity not exhibited in naturally occurring pluripotent stem cells. Further, contributions to the genetic landscape from clonal inheritance through passaging have been shown to play an important role during reprogramming, expansion, and subsequent differentiation of the cell population28,48,49.
Pluripotent stem-cell populations exhibit unique chromatin activity that is fundamental to their plasticity50. In the earliest stages of pluripotency, nuclear chromatin in the naïve-state pluripotent cell maintains open euchromatin structures, supporting an enhanced ability of these cells to respond to a wide array of developmental signaling cues51. As these cells become primed to differentiate, interactions within gene regulatory networks result in a repressive heterochromatin state, diminishing cell potency in combination with other mechanisms as the cell transitions to a further developed lineage-committed phenotype. These distinct histone protein modifications and gene methylation patterns govern gene accessibility to the transcriptional machinery of the cell52. These phenomena are significant given the influence they exert over gene expression. However, they constitute a subset of mechanisms within the overall interaction network governing iPSC phenotype and are influenced by both genetic and metabolic activity within the network53.
Several networks have been identified as critical in sustaining the pluripotent state. It is important to highlight that these networks differ depending on the developmental state of the cell53. While the key pathways regulating Oct4, Sox2, and Nanog are important across pluripotent phenotypes, the naïve state is associated with gene expression activity distinct from the primed state, with differential regulation of TFCP2L1, OTX2, DUSP6, ZIC, DPPA, and Klf genes recently reported as indicators of pluripotent state51. Interactions between the cell and environmental niche modulate their expression, while also exerting influence over the expression of genes governing cell cycle and metabolic pathway activity54,55. This genetic modulation is enabled via key pathways including FGF, LIF, TGF-β, WNT, IGF, and BMP56. These pathways influence their target genes directly, but also cohesively and antagonistically interact with one another, resulting in complex activation and inhibition activity that guides cell phenotype in the context of pluripotency.
Metabolic intermediates also play important roles in cell state homeostasis and fate commitment57. Alpha-ketoglutarate is a prime example, as it inhibits histone and DNA methylation through its role in the upregulation of jumonji-c (JmjC) domain histone demethylase and ten-eleven translocation methylcytosine dioxygenase (TET1) acetyl mark deposition58. Similarly, acetyl Coenzyme A has been shown to contribute to the activation of histone acetyltransferase (HAT). Further, nicotine adenine dinucleotide (NAD+) activity is associated with SIRT1 upregulation59. These types of behaviors are intuitive in the discussion of cell phenotype, given the shifts in mitochondrial activity and dominant metabolic pathway activity observed as cell state shifts from naïve towards a primed pluripotent state, and onwards into lineage commitment60. This also raises questions about the extent to which the metabolic abnormalities present in iPSCs influence phenotype—a contributing factor that has yet to be fully explored48.
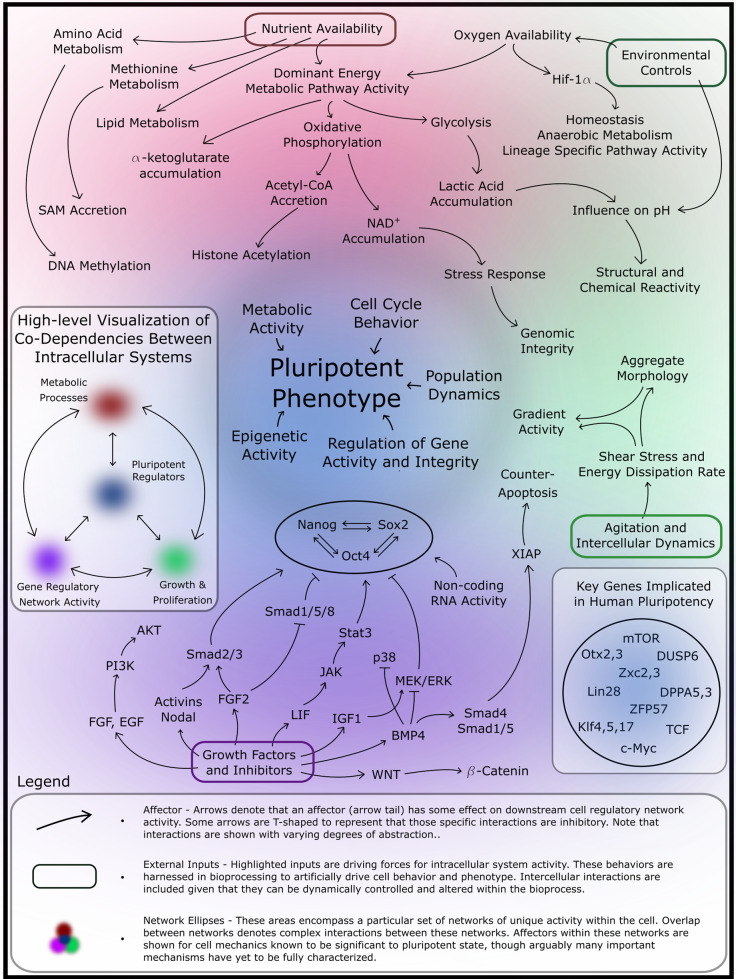
This evidence indicates that maintenance and transition of the cell state is highly dependent on metabolic network activity. In parallel, gene regulatory networks (GRNs) contribute to regulation of epigenetics and gene expression. Coupling metabolism with GRNs and regulation of the epigenetic structure of DNA within the cell results in a complex interaction network. These variables must be considered in the development and optimization of systems and protocols to produce safe, effective stem-cell-derived therapeutics. A number of these variables, their influences, and the implication for phenotype and cell health are visualized below [Fig. 3].
Fig. 3. Complex interplay between internal regulatory processes and the influence of local environmental cues.
Orchestration of phenotype is complex, depicted above in a conceptual interaction network. These interactions are illustrated to visualize the interplay between fundamental cellular processes. Ideal process control must be optimized to maintain strict homeostasis of the embryonic niche in pluripotent cell expansion bioprocessing by guiding cell network activity.
Interplay between cell state and the bioprocess environment
Given the preceding discussion regarding the various factors influencing adoption or maintenance of cell phenotype, the obvious question becomes how to effectively control these events within the context of clinical manufacturing. Lessons in developmental biology have elucidated to some extent the mechanisms driving manifestation of the pluripotent state and its subsequent loss as a combination of cellular programs and influence by the dynamic niche environment61. Methodologies to reprogram, maintain, expand, and differentiate pluripotent cell populations have borrowed heavily from the mechanisms governing embryonic development. Reprogramming and differentiation strategies will not be discussed herein, although the reader is encouraged to explore several relevant reviews62,63.
Maintenance of the pluripotent state and successful large-scale expansion of a pluripotent population requires careful design and implementation of the process environment, with considerations made for the entirety of the cell product pipeline. The most effective systems for large-scale expansion maximize the available space for cell cultivation. Three-dimensional stirred-tank environments are a strong choice for expansion given their spatial optimization, maintenance of a well-mixed environment, and feasibility for inclusion of sensing modalities5. This is particularly important in maintaining dissolved gas, signaling factor, and nutrient concentration throughout the vessel.
The addition of shear stress and vorticity presents both complication and opportunity given evidence of a supporting role for mechanical forces in stimulating pathway activity, affecting growth rates, and influencing aggregate formation64. Influence over aggregate formation is paramount given the gradients observed throughout individual aggregates. While microcarriers present an alternative approach to minimize gradients, they reduce cultivation capacity and present additional downstream purification challenges5,64.
Pathway stimulation, stochastic behavior, and gradient induction are important concepts in stem-cell bioprocessing. In vivo, the niche environment undergoes dynamic spatiotemporal changes during development that result in distinct oxygen, nutrient concentrations, and growth factor composition as the cells transition out of pluripotency61. This is important to highlight given that their plasticity extends to in vitro culture and is reflected in the distinct results observed in varied process implementations. Media formulations have been developed to regulate primed or naïve pathways in pluripotent stem cells, with varied results and prominent population heterogeneity65. While great effort has been placed in derivation of optimized media composition and replacement protocols, studies on dissolved oxygen availability, energy dissipation, and nutrient balance have only recently gained momentum in the field. Environmental oxygen is particularly interesting given the evidence of interactions of HIF family proteins with the epigenetic framework of the cell, and their direct transcriptional regulation of target genes involved in metabolism and cell cycle66. Modulation of available oxygen plays a role in growth and dominant metabolic activity of the population, an important consideration given the overall interaction network governing the state and health of the process population67.
Monitoring and characterization of pluripotent cell populations
What are the defining characteristics of a high-quality, safe, and effective pluripotent stem-cell product? Given the evidence presented, this definition will continue to evolve as researchers further elucidate mechanisms involved in the interaction network. Assessment of phenotype and function from a manufacturing perspective must balance between quantity and quality information, cost, process invasiveness, time to results, and sample representation of the population. This balancing act must satisfy safety and efficacy requirements in a clinical context and provide sufficient information to guide process development and optimization. Further, this information is necessary to assess cause-and-effect in subsequent differentiation and application.
The reader is referred to several techniques commonly used to assess the genome with varying levels of resolution68. Choice of technology and time points for repeated measurement should consider process dynamics, as well as the evidence for culture adaptation, mutability, and clonal dominance over the course of process progression26,27. While FACS analyses of surface and intracellular markers of pluripotency are commonly used, the exact targets of these analyses with respect to naive versus primed phenotypes continue to change as scientists uncover a more complete understanding of pluripotent state51. Further, the qualitative nature of high-throughput systems such as FACS makes it difficult to assess relative expression levels of key indicators of cell state transition within pluripotent populations.
Outside of targeted studies, dominant metabolic pathway activity has garnered significantly less attention in the bioprocessing sector. With evidence to suggest synergy between metabolism, epigenetic activity, and gene expression in stem-cell populations, a more complete focus on cell state should include integrated analysis of continuous in-process measurements and repeated offline characterization of the environment and cell population. Advancement in mass spectrometry systems and the momentum gained by online systems to assess dissolved gas and metabolic analytes has provided new opportunities to integrate these data alongside conventional characterization strategies.
It is still unclear what role metabolism and epigenetics play in the context of downstream function and phenotype, though the effect of donor variance on process heterogeneity suggests that methylation and acetylation data could go a long way in elucidating potential focal points for the research community, when combined with other characteristic and functional data. Functional assessments of human pluripotent stem cells exist, with varying degrees of rigor, dependent on the downstream application. While this paper will not present these methods in detail, it is important that interpretation of the results considers their limitations69.
Bioprocess design and the therapeutic cell product
Given the infancy of stem-cell-derived product usage in therapeutic application, there remain many unknowns not only from a fundamental science perspective but also in terms of safety and efficacy for clinical use70,71. Maintaining a balance within the environmental niche in which the population is induced, expanded, differentiated, and maintained prior to cell product use is paramount72. Given the large number of cells required for a single patient (106–1012) this balance becomes even more precarious when coupled with the necessity to expand, maintain, and influence large quantities of cells in the most resource-constrained (efficient) way possible5. Media formulations and shear stress application focused on maximum cell replication and expression of a small number of phenotypic markers are not sufficient to guarantee quality in the resultant cell population73.
These design considerations raise important questions about process specifications and the quality control metrics used throughout the entirety of the process. The choice of whether to use a surface-adherent or suspended aggregate approach inevitably impacts the cell population. The culture surface (or lack thereof) influences structural and morphological characteristics of the cell and alters gene regulatory pathways that are implicated alongside other pathways in order to balance phenotype74–76. This in conjunction with intracellular organization results in the gradient dynamics discussed earlier in this paper. Further, proliferation capacity and ability to sense and control the environmental niche are constrained by the process system used, affecting fluid dynamic setpoints, feeding regimen, timelines, and characterization strategies.
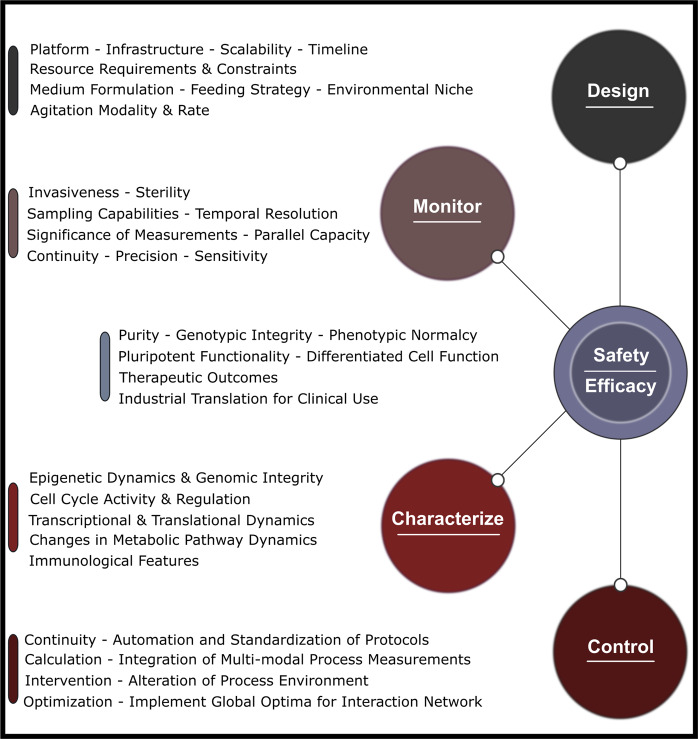
Given the spatial constraints of static systems, the strategy used to generate clinically relevant cell product numbers involves scaling out cell production by culturing replicates of a particular vessel in parallel77. Alternatively, dynamic suspension culture systems enable scale-up, increasing the capacity of the system within a singular vessel78. Both fundamental modalities include their own challenges, requiring assessment of resource constraints, bioprocess efficiency, robustness, reproducibility, capacity for monitoring, control, intervention, and tolerance of the process to donor variability and biological heterogeneity74–76,78. Considerations must also be made for sustainability and cost, as growing challenges for the future. A summary of these considerations is presented in Fig. 4.
Fig. 4. Overview of several important components and characteristics in bioprocess design, development, and optimization.
Considerations must be made for the infrastructure and technology used to monitor, control, and intervene in the process. These considerations must be integrated with those made for characterization of the cell population to build a complete model for artificial manipulation of phenotype to ensure safety and efficacy downstream of expansion.
Conventional bioprocess trajectories are operator and protocol dependent, with relatively basic interventions occurring within the process itself5. Modern process development infrastructures generate large quantities of meaningful environmental data relevant to cell population dynamics, and these data are commonly used in a closed-loop, automated configuration to maintain pH, oxygen, and perfuse the culture medium. This environmental data are coupled with rigorous characterization through multi-omics approaches to characterize population phenotype throughout the developmental stages of a process68,73. However, there is generally a disconnect between how offline omics and in-process data are utilized, with datasets independently scrutinized to assess a particular characteristic of the process or cell population. The two have yet to be meaningfully integrated.
Advances in machine learning have provided the means to assess large quantities of multi-dimensional datasets with the aim of developing predictive models and intervention strategies for biological systems79–81. There exist unique opportunities to apply concepts in machine learning and computational modeling to advance stem-cell bioprocess development and clinical manufacturing. Several works within basic research and clinical biomanufacturing spheres have been published with this mindset, though the field of iPSC biomanufacturing has yet to rigorously explore these opportunities82–84. Robust iPSC expansion and differentiation processes are closed-loop systems, and the incorporation of predictive models and real-time intervention strategies has the potential to greatly enhance predictability and online process control over cell health and phenotype, resulting in safer and more effective iPSC-derived cell therapeutics.
Clinical trials and the current state of iPSC-derived therapeutics
Safety and efficacy remain key challenges to overcome in the utilization of pluripotent cell-derived therapeutics for routine use in clinical applications. This is represented in the lack of interventional studies using ESCs or iPSCs as the derivational source relative to other cell types85. As of August 2021, only 1.2% of documented stem-cell-derived therapeutic clinical trials in the United States alone utilized iPSCs. This number is even lower for ESCs, at 0.8%. Further, the vast majority of trials utilizing pluripotent stem cells involve in vitro disease modeling. While these studies exemplify the importance of PSCs in medicine and underline critical steps in the derivation of therapeutics for clinical application, the lack of interventional studies utilizing autologous or allogeneic PSC-derived cell therapies highlight the work that is still needed to bring them to the forefront of modern medicine. Recent work to spotlight interventional studies involving PSC-derived therapeutics show some promising progress and results in the sphere of PSC utilization86. A number of these trials have been further reviewed to bring clinical context to the challenges and recommendations outlined in this paper [Table 1].
Table 1.
Bioprocess characteristics and intervention outcomes for clinical trials involving pluripotent stem-cell-derived therapeutics.
| ID number | Derived cell type | Bioprocess characteristics | Manufacturing timeline | Population characterization strategies | Clinical outcomes/trial status |
|---|---|---|---|---|---|
| NCT01625559 | hESC-derived retinal pigment epithelium (Allogeneic) | hESC on mouse-feeder in static 2D culture, differentiation strategy not disclosed, in-process monitoring not disclosed | Not disclosed | Morphology, cell counts, pigmentation, karyotyping (g-banding), FACS (hPSC/hRPE markers), cell function assays, short tandem repeat analysis, sterility testing, mycoplasma testing, endotoxin analysis, GMP compliance | Phase I complete, results deemed favorable87 |
| NCT02923375 | iPSC-mesanchymoangioblast-derived mesenchymal stem cells (allogeneic) | Cymerus platform (2D system) in serum-free, feeder-free conditions, in-process monitoring not disclosed93 | Not disclosed | mRNA qPCR, antibody staining flow cytometry, comparative genomic hybridization, single-nucleotide polymorphism analysis, global transcriptome analysis, GMP-compliant manufacturing | Phase I complete. Short-term safety verified. Confounding factors obscured potential effectiveness89 |
| NCT02057900 | hESC-derived cardiovascular progenitors (allogeneic) | hESC on mouse-feeder in static 2D culture, treatment of cultures with BMP2 + SU5402 medium, downstream filtration of SSEA-1+ cells, in-process monitoring not disclosed | Not disclosed | Morphology, cell counts, marker staining, microscopic imaging, FACS (hESC/cardiac progenitor markers), HLA phenotyping, GMP compliance | Phase I complete, short-term safety verified, slightly improved symptomatic results94 |
| JPRNUMIN000011929 | iPSC-derived retinal pigment epithelium sheets (autologous) | 2D hiPSC culture, differentiation to hiPSC-RPE, seeding onto collagen type I gel, formation of monolayer, in-process monitoring not disclosed | 6 weeks to RPE sheet formation from hiPSC-RPE | Morphology, pigmentation, RT-PCR for RPE markers, assays for growth factors (ELISA), microarray genomics, MHC histocompatibility testing, DNA methylation testing, copy-number variation analysis, GMP compliance95 | Trial halted after 2 patients. Deemed ineffective, with safety concerns88 |
| NCT03482050 | ESC-derived astrocytes (allogeneic) | Not disclosed | Not disclosed | Not disclosed | Trial complete, no results posted |
| NCT03119636 | ESC-derived neural precursor cells (NPCs) (allogeneic) | EB neural differentiation strategy, not effectively disclosed for pre-clinical human ESC-derived NPCs in primates96 | 3 weeks to early neural rosette formation | Not disclosed | Unknown status |
Available data from interventional clinical trials utilizing PSC-derived therapeutics present mixed results from safety and efficacy standpoints. While a recent trial by Sung et al.87 reported short-term safety and improved outcomes as a result of treatment with ESC-derived retinal pigment epithelium (RPEs), a similar strategy utilizing iPSC-derived RPEs resulted in trial suspension over safety concerns88. The most prominent difference between these two trials is the use of allogeneic ESC-derived RPEs in the Sung trial, versus autologous iPSC-derived RPEs in the Mandai trial. While confounding variables exist in the clinical application stages, a critically important detail is the presence of copy-number variations in iPSC-derived RPEs revealed during genomic analysis, resulting in trial suspension citing safety concerns in adherence with Japanese iPSC laws. Considering the similarity in their downstream use of two-dimensional static culture systems, well-established protocols, and current good manufacturing practice (CGMP), observation of genomic aberrations highlights the complexity of the iPSC pipeline from harvest, reprogramming, and expansion, through to target cell derivation. While allogeneic iPSC-derived therapeutics have recently been shown to exhibit short-term safety in the derivation of mesenchymal stem cells for host-versus-graft disease, they pose innate immune rejection risks of their own, resulting in suppression of the host immune-system following transplantation89. Whether allogeneic iPSCs present a more favorable cell source material remains to be seen in a clinical context.
A lack of public disclosure in the majority of clinical trials is evident, with very few trials citing publications with reference to pre-clinical work, bioprocess strategies, characterization specifics, manufacturing details, or other pipeline information. In cases where public disclosure is provided, details are scarce. Aside from trial-specific information, publication of data to publicly accessible databases is at the discretion of the trial leads and in many cases is not available85. This unfortunately makes an objective comparison of manufacturing design, monitoring, control, and characterization across trials near impossible to perform. A set of minimal quality criteria does exist for constitution of a clinical-grade iPSC-derived therapeutic throughout the pipeline, and while a framework exists for therapeutic generation through CGMP there are no standardized requirements for systems implemented and protocols used, leading to differing process effectiveness, robustness, and outcomes90,91. While most documented trials involve conventional two-dimensional static systems for both PSC and differentiation strategies, the adoption of three-dimensional, computer-controlled systems has resulted in a paradigm shift both for PSC maintenance and expansion as well as differentiation where cell types and current differentiation strategies permit. Coupled with increased capacity for process monitoring in process environments, pending trials currently in the recruitment and active stages are expected to provide far greater detail and insight into process influence on therapeutic safety and efficacy in a clinical context across systems92. While a lack of standardization across the field drives innovation in biomanufacturing, methods to compute the dynamics of interactions between process and target cell population will inevitably be required to assess their interplay, and disclosure of data necessary to objectively identify best practices as the field evolves.
Conclusion and recommendations for the future
The global stem-cell research community has made leaps and bounds in the pursuit of knowledge pertaining to cellular and niche control over stem-cell phenotype. In parallel, pressure for novel medical approaches to cure disease and industrial push to implement clinical-scale engineering solutions have helped drive stem-cell science and technology over the last several decades. The number of active stem-cell-derived products in clinical trials has increased exponentially, and biopharmaceuticals companies have taken advantage of work on organoid development and disease modeling to amplify drug discovery capabilities by improving experimental capacity and scalability.
Given the interdisciplinary nature of the field and the pressure for clinical success, advancement is continuing across sectors, ranging from technological developments to increased-scale bioprocessing and clinical infrastructures. Despite these advancements, the challenges discussed in this paper have yet to be overcome in pursuit of safe and effective cell and tissue therapies to cure disease and regenerate tissue after injury. The mechanisms governing pluripotent phenotype are complex, and indicators of cell population quality extend far beyond marker presence or capacity for self-renewal and differentiation. Further, the mechanisms contributing to variance and uniqueness in pluripotent populations extend to their progeny and differentiated cell types.
Consolidating the nexus of gene expression, epigenetic activity, metabolism, cell cycle, and cell structural organization is crucial to properly influence phenotype to minimize variance and maximize cell quality. Integrating the full range of considerations and compiling more rigorous characterization and process analytics datasets across the process pipeline are necessary to enable advanced systems design. Expanding on these insights from a clinical perspective by integrating advanced medium formulations, process control strategies, online monitoring, real-time interventions, and automation alongside technological innovations will push the threshold of our capabilities to adapt iPSCs for widespread medical use in the coming decades. Coupling technology with rigorous clinical outcome measures in patients will ultimately allow the scientific community to ascertain and guide fate, function, safety, and efficacy.
Acknowledgements
This research was funded by a CIHR project grant. J.C. was funded by the NSERC PGS-D national scholarship program and the AGES program through the Alberta provincial government.
Author contributions
J.C. conceived of the presented ideas and perspectives. J.C. carried out a thorough review of literature and prepared the manuscript with support from K.M., M.S.K. and J.B. All authors reviewed, edited, and made significant contributions to the final manuscript.
Data availability
Any and all data are available upon request to the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kirouac DC, Zandstra PW. The systematic production of cells for cell therapies. Cell Stem Cell. 2008;3:369–381. doi: 10.1016/j.stem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins MJ, Farid SS. Human pluripotent stem cell-derived products: advances towards robust, scalable and cost-effective manufacturing strategies. Biotechnol. J. 2014;10:83–95. doi: 10.1002/biot.201400348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adil MM, Schaffer DV. Expansion of human pluripotent stem cells. Curr. Opin. Chem. Eng. 2017;15:24–35. doi: 10.1016/j.coche.2016.11.002. [DOI] [Google Scholar]
- 4.Kempf H, Andree B, Zweigerdt R. Large-scale production of human pluripotent stem cell derived cardiomyocytes. Adv. Drug Deliv. Rev. 2016;96:18–30. doi: 10.1016/j.addr.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Kropp C, Massai D, Zweigerdt R. Progress and challenges in large-scale expansion of human pluripotent stem cells. Process Biochem. 2017;59:244–254. doi: 10.1016/j.procbio.2016.09.032. [DOI] [Google Scholar]
- 6.Pellegrini G, et al. From discovery to approval of an advanced therapy medicinal product-containing stem cells, in the EU. Regen. Med. 2016;11:407–420. doi: 10.2217/rme-2015-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin X, et al. Stem cell organoid engineering. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen KG, et al. Pluripotent stem cell platforms for drug discovery. Trends Mol. Med. 2018;24:805–820. doi: 10.1016/j.molmed.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romito A, Cobellis G. Pluripotent stem cells: current understanding and future directions. Stem Cells Int. 2016;2016:1–20. doi: 10.1155/2016/9451492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 11.Caulfield T, et al. International stem cell environments: a world of difference. Nat. Rep. Stem Cells. 2009;1:1–5. [Google Scholar]
- 12.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:1–9. doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volarevic V, et al. Ethical and safety issues of stem cell-based therapy. Int. J. Med. Sci. 2018;15:36–45. doi: 10.7150/ijms.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Bai L, et al. Hepatocyte growth factor mediates mesenchymal stem cell-induced recovery in multiple sclerosis models. Nat. Neurosci. 2012;15:862–870. doi: 10.1038/nn.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trounson A, McDonald C. Stem cell therapies in clinical trials: progress and challenges. Cell Stem Cell. 2015;17:11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Garbern JC, Lee RT. Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell. 2013;12:689–698. doi: 10.1016/j.stem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trounson A, DeWitt ND. Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 2016;17:194–200. doi: 10.1038/nrm.2016.10. [DOI] [PubMed] [Google Scholar]
- 21.Braganca J, Lopes JA, Mendes-Silva L, Santos JMA. Induced pluripotent stem cells, a giant leap for mankind therapeutic applications. World J. Stem Cells. 2019;11:421–430. doi: 10.4252/wjsc.v11.i7.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M, Kino-oka M. Bioengineering considerations for a nurturing way to enhance scalable expansion of human pluripotent stem cells. Biotechnol. J. 2020;15:1–16. doi: 10.1002/biot.201900314. [DOI] [PubMed] [Google Scholar]
- 23.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat. Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund RJ, Narva E, Lahesmaa R. Understanding the genetics behind complex human disease with large-scale iPSC collections. Nat. Rev. Genet. 2012;13:732–744. doi: 10.1038/nrg3271. [DOI] [PubMed] [Google Scholar]
- 25.Popp B, et al. Need for high-resolution Genetic Analysis in iPSC: results and lessons from the ForIPS consortium. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-35506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkle FT, et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature. 2017;545:229–233. doi: 10.1038/nature22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao J, et al. Cells derived from iPSC can be immunogenic—yes or no? Protein Cell. 2014;5:1–3. doi: 10.1007/s13238-013-0003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Li W, Fu X, Xu Y. The immunogenicity and immune tolerance of pluripotent stem cell derivatives. Front. Immunol. 2017;8:1–6. doi: 10.3389/fimmu.2017.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chhabra A. Inherent immunogenicity or lack thereof of pluripotent stem cells: implications for cell replacement therapy. Front. Immunol. 2017;8:1–11. doi: 10.3389/fimmu.2017.00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pera MF. Stem cells: the dark side of induced pluripotency. Nature. 2011;471:46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- 32.Nishino K, et al. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:1–14. doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youssef AA, et al. The promise and challenge of induced pluripotent stem cells for cardiovascular applications. JACC Basic Transl. Sci. 2016;1:510–523. doi: 10.1016/j.jacbts.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Urso A, Brickner JH. Mechanisms of epigenetic memory. Trends Genet. 2014;30:230–236. doi: 10.1016/j.tig.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khoo TS, et al. Retention of somatic memory associated with cell identity, age and metabolism in induced pluripotent stem (iPS) cells reprogramming. Stem Cell Rev. Rep. 2020;16:251–261. doi: 10.1007/s12015-020-09956-x. [DOI] [PubMed] [Google Scholar]
- 36.Lee JE, Lee DR. Human embryonic stem cells: derivation, maintenance and cryopreservation. Int. J. Stem Cells. 2011;4:1–8. doi: 10.15283/ijsc.2011.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Kyttala A, et al. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Rep. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim M, Costello J. DNA methylation: an epigenetic mark of cellular memory. Exp. Mol. Med. 2017;49:1–8. doi: 10.1038/emm.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishizawa M, et al. Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell. 2016;19:341–354. doi: 10.1016/j.stem.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 41.Schwartzentruber J, et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat. Genet. 2018;50:54–61. doi: 10.1038/s41588-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anguera MC, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronen D, Benvenisty N. Sex-dependent gene expression in human pluripotent stem cells. Cell Stem Cell. 2014;8:923–932. doi: 10.1016/j.celrep.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Rubin JB, et al. Sex differences in cancer mechanisms. Biol. Sex. Differences. 2020;11:1–29. doi: 10.1186/s13293-020-00291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/S0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 46.Perrera V, Martello G. How does reprogramming to pluripotency affect genomic imprinting? Front. Cell Dev. Biol. 2019;7:1–16. doi: 10.3389/fcell.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pick M, et al. Clone‐ and gene‐specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 48.Panoupoulos AD, et al. Aberrant DNA methylation in human iPSCs associates with MYC-binding motifs in a clone-specific manner independent of genetics. Cell Stem Cell. 2017;20:505–517. doi: 10.1016/j.stem.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Assou S, et al. Recurrent genetic abnormalities in human pluripotent stem cells: definition and routine detection in culture supernatant by targeted droplet digital PCR. Stem Cell Rep. 2020;14:1–8. doi: 10.1016/j.stemcr.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collier AJ, Rugg-Gunn PJ. Identifying human naïve pluripotent stem cells−evaluating state‐specific reporter lines and cell‐surface markers. Bioessays. 2018;40:1–12. doi: 10.1002/bies.201700239. [DOI] [PubMed] [Google Scholar]
- 52.Efroni S, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harvey A, et al. Interplay between metabolites and the epigenome in regulating embryonic and adult stem cell potency and maintenance. Stem Cell Rep. 2019;13:573–589. doi: 10.1016/j.stemcr.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kapinas K, et al. The abbreviated pluripotent cell cycle. J. Cell Physiol. 2013;1:9–20. doi: 10.1002/jcp.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathieu J, Ruohola-Baker H. Metabolic remodeling during the loss and acquisition of pluripotency. Development. 2017;144:541–555. doi: 10.1242/dev.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hassani S, et al. Inhibition of TGFβ signaling promotes ground state pluripotency. Stem Cell Rev. Rep. 2014;10:16–30. doi: 10.1007/s12015-013-9473-0. [DOI] [PubMed] [Google Scholar]
- 57.Burgess RJ, Agathocleous M, Morrison SJ. Metabolic regulation of stem cell function. J. Intern. Med. 2014;276:12–24. doi: 10.1111/joim.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carey BW, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;6:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Baker D, et al. Detecting genetic mosaicism in cultures of human pluripotent stem cells. Stem Cell Rep. 2016;7:998–1012. doi: 10.1016/j.stemcr.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J, Yamauchi T, Belmonte JCI. An overview of mammalian pluripotency. Development. 2016;143:1644–1648. doi: 10.1242/dev.132928. [DOI] [PubMed] [Google Scholar]
- 61.Pennings S, Liu KJ, Qian H. The stem cell niche: interactions between stem cells and their environment. Stem Cells Int. 2018;2018:1–3. doi: 10.1155/2018/4879379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Efthymiou AG, Chen G, Rao M, Chen G, Boehm M. Self-renewal and cell lineage differentiation strategies in human embryonic stem cells and induced pluripotent stem cells. Expert Opin. Biol. Ther. 2014;14:1333–1344. doi: 10.1517/14712598.2014.922533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X, et al. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature. 2020;586:101–107. doi: 10.1038/s41586-020-2734-6. [DOI] [PubMed] [Google Scholar]
- 64.Liu M, Liu N, Zang R, Li Y, Yang S. Engineering stem cell niches in bioreactors. World J. Stem Cells. 2013;5:124–135. doi: 10.4252/wjsc.v5.i4.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kilens S, et al. Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-017-02107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saito S, et al. Emerging roles of hypoxia-inducible factors and reactive oxygen species in cancer and pluripotent stem cells. Kaohsiung J. Med. Sci. 2015;31:279–286. doi: 10.1016/j.kjms.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borys BS, et al. Optimized serial expansion of human induced pluripotent stem cells using low‐density inoculation to generate clinically relevant quantities in vertical‐wheel bioreactors. Stem Cells Transl. Med. 2020;9:1036–1052. doi: 10.1002/sctm.19-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Assou S, Bouckenheimer J, De Vos J. Concise Review: assessing the genome integrity of human induced pluripotent stem cells: what quality control metrics? Stem Cells Transl. Med. 2018;36:814–821. doi: 10.1002/stem.2797. [DOI] [PubMed] [Google Scholar]
- 69.Buta C, et al. Reconsidering pluripotency tests: do we still need teratoma assays? Stem Cell Res. 2013;11:552–562. doi: 10.1016/j.scr.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mao AS, Mooney DJ. Regenerative medicine: current therapies and future directions. Proc. Natl Acad. Sci. USA. 2015;112:14452–14459. doi: 10.1073/pnas.1508520112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li K, Zhang M, Xie F, Liu P, Xu S. Differentiation of pluripotent stem cells for regenerative medicine. Biochem. Biophys. Res. Commun. 2016;471:1–4. doi: 10.1016/j.bbrc.2016.01.182. [DOI] [PubMed] [Google Scholar]
- 72.Joddar B, Ito Y. Artificial niche substrates for embryonic and induced pluripotent stem cell cultures. J. Biotechnol. 2013;168:218–228. doi: 10.1016/j.jbiotec.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 73.Baghbaderani BA, et al. Detailed characterization of human induced pluripotent stem cells manufactured for therapeutic applications. Stem Cell Rev. Rep. 2016;12:394–420. doi: 10.1007/s12015-016-9662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sart S, Bejoy J, Li Y. Characterization of 3D pluripotent stem cell aggregates and the impact of their properties on bioprocessing. Process Biochem. 2017;59:276–288. doi: 10.1016/j.procbio.2016.05.024. [DOI] [Google Scholar]
- 75.Galvanauskas V, et al. Current state and perspectives in modeling and control of human pluripotent stem cell expansion processes in stirred-tank bioreactors. Biotechnol. Prog. 2017;33:355–364. doi: 10.1002/btpr.2431. [DOI] [PubMed] [Google Scholar]
- 76.Polanco A, Kuang B, Yoon S. Bioprocess technologies that preserve the quality of iPSCs. Trends Biotechnol. 2020;38:1128–1140. doi: 10.1016/j.tibtech.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 77.Tohyama S, et al. Efficient large-scale 2D culture system for human induced pluripotent stem cells and differentiated cardiomyocytes. Stem Cell Rep. 2017;9:1406–1414. doi: 10.1016/j.stemcr.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lavon, N., Zimerman, M. & Itskovitz-Eldor, J. In Advances in Biochemical Engineering/Biotechnology Book Series, ABE 163, 22–37 (2017). Edited by Scheper, T., Zhong, J. Published by Springer-Verlag Berlin Heidelberg NewYork.
- 79.Oyetunde T, Bao FS, Chen JW, Martin HC, Tang YJ. Leveraging knowledge engineering and machine learning for microbial manufacturing. Biotechnol. Adv. 2018;36:1308–1315. doi: 10.1016/j.biotechadv.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 80.Zitnik M, et al. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf. Fusion. 2019;50:71–91. doi: 10.1016/j.inffus.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doulgkeroglou M, et al. Automation, monitoring, and standardization of cell product manufacturing. Front. Bioeng. Biotechnol. 2020;8:1–12. doi: 10.3389/fbioe.2020.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malta TM, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sugimoto K. Machine learning-driven label-free cell sorting for CAR-T cell manufacturing. Cytotherapy. 2019;21:S39. doi: 10.1016/j.jcyt.2019.03.376. [DOI] [Google Scholar]
- 84.Williams B, et al. Prediction of human induced pluripotent stem cell cardiac differentiation outcome by multifactorial process modeling. Front. Bioeng. Biotechnol. 2020;8:1–15. doi: 10.3389/fbioe.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobold S, et al. A manually curated database on clinical studies involving cell products derived from human pluripotent stem cells. Stem Cell Rep. 2020;15:546–555. doi: 10.1016/j.stemcr.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deinsberger J, Reisinger D, Weber B. Global trends in clinical trials involving pluripotent stem cells: a systematic multi-database analysis. npj Regen. Med. 2020;5:1–13. doi: 10.1038/s41536-020-00100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sung Y, et al. Long-term safety and tolerability of subretinal transplantation of embryonic stem cell-derived retinal pigment epithelium in Asian Stargardt disease patients. Br. J. Opthalmol. 2021;105:829–837. doi: 10.1136/bjophthalmol-2020-316225. [DOI] [PubMed] [Google Scholar]
- 88.Mandai M, et al. Autologous induced stem cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- 89.Bloor AJC, et al. Production, safety and efficacy of iPSC-derived mesenchymal stromal cells in acute steroid-resistant graft versus host disease: a phase I, multicenter, open-label, dose-escalation study. Nat. Med. 2020;26:1720–1725. doi: 10.1038/s41591-020-1050-x. [DOI] [PubMed] [Google Scholar]
- 90.Doss MX, Sachinidis A. Current challenges of iPSC-based modeling and therapeutic implications. Cells. 2019;8:1–16. doi: 10.3390/cells8050403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dashtban M, Panchalingam KM, Shafa M, Baghbaderani BA. Addressing manufacturing challenges for commercialization of iPSC-based therapies. Methods Mol. Biol. 2021;2286:179–198. doi: 10.1007/7651_2020_288. [DOI] [PubMed] [Google Scholar]
- 92.Yamanaka S. Pluripotent stem cell-based cell therapy – promise and challenges. Cell Stem Cell. 2020;27:523–531. doi: 10.1016/j.stem.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 93.Ozay EI, et al. Cymerus iPSC-MSCs significantly prolong survival in a pre-clinical, humanized mouse model of Graft-vs-host disease. Stem Cell Res. 2019;35:1–13. doi: 10.1016/j.scr.2019.101401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Menasche P, et al. Transplantation of human embryonic stem cell-derived cardiovascular progenitors for severe ischemic left ventricular dysfunction. J. Am. Coll. Cardiol. 2018;71:429–438. doi: 10.1016/j.jacc.2017.11.047. [DOI] [PubMed] [Google Scholar]
- 95.Kamao H, et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, et al. Human clinical-grade parthenogenetic ESC-derived dopaminergic neurons recover locomotive defects of nonhuman primate models of Parkinson’s disease. Stem Cell Rep. 2018;11:171–182. doi: 10.1016/j.stemcr.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Any and all data are available upon request to the corresponding author.