Abstract
COVID-19 is spreading rapidly yet there is no clinically proven drug available now. Soil-derived Streptomyces sp. GMR22 has a large genome size (11.4 Mbp) and a huge BGCs (Biosynthetic Gene Clusters) encoding secondary metabolites. This bacterium is a potential source for producing a wide variety of compounds which are able to block SARS-CoV-2, the causative agent of COVID-19. This study aimed to predict the secondary metabolites of Streptomyces sp. GMR22 and to evaluate the ability as SARS-CoV-2 inhibitor. The AntiSMASH 5.0 was used for genome mining analysis and targeted liquid chromatography-high resolution mass spectrometry (LC-HRMS) was used for metabolite analysis. In silico molecular docking was performed on important target proteins of SARS-CoV-2 i.e., spike protein (PDB ID: 6LXT), Receptor Binding Domain (RBD)-ACE2 (Angiotensin-Converting Enzyme 2) (PDB ID: 6VW1), 3CLpro (3-chymotrypsin-like protease) (PDB ID: 6M2N), and RdRp (RNA-dependent RNA polymerase) (PDB ID: 6M71). Two compounds from GMR22 extract, echoside A and echoside B were confirmed by targeted LC-HRMS and potential as SARS-CoV-2 inhibitor. Echoside A and echoside B showed higher docking score than remdesivir as COVID-19 drug on four target proteins, i.e., spike protein (−7.9 kcal/mol and −7.8 kcal/mol), RBD-ACE2 (−7.5 kcal/mol and −8.2 kcal/mol), 3CLpro (−8.4 kcal/mol and −9.4 kcal/mol) and RdRp (−7.3 kcal/mol and −8.0 kcal/mol). A combination of genome mining and metabolomic approaches can be used as integrated strategy to elucidate the potential of GMR22 as a resource in the discovery of anti-COVID -19 compound.
Keywords: Streptomyces, Secondary metabolite, SARS-CoV-2, AntiSMASH, COVID-19
Streptomyces, secondary metabolite, SARS-CoV-2, AntiSMASH, COVID-19.
1. Introduction
Streptomyces is the most robust source for producing new bioactive compounds, antibiotics, and extracellular enzymes [1]. Previously identified Streptomyces from rhizosphere soil, Wanagama Forest, Gunungkidul, Yogyakarta, Indonesia, Streptomyces sp. GMR22 has 63 Biosynthetic Gene Clusters (BGCs), dominated by polyketide synthase (PKS) using AntiSMASH 3.0. Polyketide synthase involves in the production of polyketide compounds. Polyketide is a huge family of natural product class in which some clinically proven drugs, such as tetracycline, daunorubicin, erythromycin, rapamycin and lovastatin are included. Other than polyketide gene cluster, NRPS (Nonribosomal Peptide Synthetase), siderophore, RiPPs (Ribosomally Synthesized and Post-Translationally Modified Peptides) and terpene were also found [2]. NRPS is enzyme which synthesize non-ribosomal peptides [3]. Siderophore encodes iron-chelating compounds, such as Desferrioxamine B, which have been predicted 100% could be produced by GMR22. Desferrioxamine B possessed anti-cancer activity in the cells of colon cancer [4]. GMR22 also predicted to harbour RiPPs gene, enabled this strain to generate class of cyclic or linear peptidic natural products [5]. Triterpenes are class of natural products composed of three terpene units. Previous study revealed that triterpenes had antiplasmodial activity [6]. Moreover, among genus Streptomyces, strain GMR22 has the highest number of BGCs, indicating that it is a prominent source for producing a wide variety of compounds [7, 8].
Global public health is still facing crisis due to the new virus, SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus-2), which originally emerged in Wuhan, China in December, 2019. This virus was declared as pandemic as of March, 11 2020 by the WHO (World Health Organization) and currently named as COVID-19 (Coronavirus disease) [9]. This virus is spreading rapidly yet there is no clinically effective drug available now which specifically treats COVID-19. It has been suggested that SARS-CoV-2 relied on viral spike protein and ACE2 receptor in the human body which found in the heart, lung, vessels, gut, kidney, testis and the brain for cell entry [10]. As for RNA viruses, RdRp is important for RNA synthesize. While, 3CLPro mediates maturation of non-structural proteins that essential during virus replication [11]. Therefore, we used RdRp (PDB ID: 6M71), and 3CLpro (PDB ID: 6M2N) as targets to inhibit the viral replication, while spike protein (PDB ID: 6LXT) and RBD-ACE2 (PDB ID: 6VW1) as targets to block the entry of SARS-CoV-2 [12], [13]. The compound found in this study could answer the urgent demand to counter SARS-CoV-2 (Severe Acute Respiratory Syndrome Corona Virus 2).
Complementary approaches, namely metabologenomics, was employed in the present study to find the targeted compounds which were directly correlated with GMR22 [14]. The genome mining approach predicted the secondary metabolites from GMR22 using a website-based bioinformatics platform. This research was combined with the metabolomic approach using mass spectrometry, LC-HRMS (Liquid Chromatography-High Resolution Mass Spectrometry), for thoroughly profiling targeted secondary metabolites synthesized by GMR22. Aiming to obtain secondary metabolites as SARS-CoV-2 inhibitors from Streptomyces sp. GMR22, this study gained insight of the secondary metabolites profile of GMR22, then subjected to in silico molecular docking against COVID-19 target proteins.
2. Material and methods
2.1. Microorganism strain
The Streptomyces sp. GMR22 isolated from the Cajuput rhizospheric soil of Wanagama forest, Indonesia was used in this study [8]. Strain GMR22 was maintained in International Streptomyces Project-2 (ISP-2) agar medium (Difco, Sparks, USA).
2.2. Genome mining analysis
A previous study had done genome sequencing for GMR22 [7]. This Whole Genome Shotgun project had been deposited at DDBJ/ENA/GenBank under accession JACGSQ000000000. The version described in this paper was version JACGSQ010000000. The genome of GMR2 was uploaded onto AntiSMASH 5.0 website (https://antismash.secondarymetabolite.org) to obtain gene cluster data [15].
2.3. Metabolomic profiling of extract
Strain GMR22 was grown on ISP-2 (International Streptomyces Project 2) agar for 7 days at 28 °C. The agar was cut into pieces (2 × 2 cm) and was put into 50 mL Tryptic Soy Broth (TSB) and incubated for 3 days in an incubator shaker at 28 °C. Five percent of inoculum was cultured in 1 L Erlenmeyer flask containing 20 g soluble starch; 0.5 g NaCl; 1 g KNO3; 0.5 g K2HPO4.3H2O; 0.5 g MgSO4.7H2O; 0.01 g FeSO4.7H2O for 8 days at 150 rpm on an incubator shaker at 28 °C. The cell and supernatant were separated for 10 min at 5000 rpm. The cell was extracted with methanol in low stirring for 30 min, then separated for 15 min at 4500 rpm. The supernatant was taken as methanol crude extract and was evaporated to yield a gum.
The methanol crude extract was run on The Ultimate 3000 RSLCnano UHPLC-Q Exactive Orbitrap HRMS with an injection volume of 5 μL (concentration 10 μg/mL), a flow rate of 6.5 μL/min, and a column temperature of 30 °C. The Thermo Scientific™ Hypersil GOLD™ aQ analytical column was used for separation (50 mm length, 1.0 mm internal diameter, 1.9 μm particle size). Mobile phase A was 0.1% formic acid in MS grade water and mobile phase B was 0.1% formic acid in MS grade acetonitrile. The gradient program started at 5–50% for 13 min and the total run time was 30 min. The ESI ion source was set as follows: spray voltage of 3.80 kV, capillary temperature of 300 °C, and S-lens RF level of 50%. The compounds were detected in positive mode, Full Scan MS resolution of 70,000, targeted mode. The targeted mode used compounds information from the genome mining data. The compound information was obtained as mol file from PubChem or ChemSpider. The compounds were identified by Compound Discoverer 3.1 software. The peak extraction was done automatically by the software. The confirmation of each targeted compounds with respect to their parent compounds was achieved by considering isotope pattern match. Isotope pattern match in LC-HRMS could identify various relative abundance of isotope in the targeted compounds. Additionally, composition change was taken into account. All compound IDs were matched within +/- 5 ppm mass tolerance.
2.4. In silico molecular docking
Molecular docking was used to predict the binding affinity between the GMR22 targeted new compound against the target proteins of COVID-19. The target proteins used in this study were downloaded from Protein Data Bank, i.e., spike protein (PDB ID: 6LXT), RBD-ACE2 (PDB ID: 6VW1), 3CLpro (PDB ID: 6M2N), and RdRp (PDB ID: 6M71). The target protein list is given in Table 1. The 2D conformers of the GMR22 targeted compound were obtained from ChemSpider, then converted into 3D conformer using MarvinSketch tools (https://chemaxon.com). All water, ions, and other small molecules were removed from the target proteins, and then the hydrogen atoms were added using auto dock tools. The molecular docking was carried out by using Autodock Vina [16]. The docking result was binding affinity (kcal/mol) with a root mean square deviation (RMSD) score of less than 2 Å. Meanwhile, DisCoVery studio visualizer (DS visualizer) tool was used for visualizing the binding interaction of the GMR22 targeted compound on the binding site of proteins [17]. The molecular docking was performed in the Windows 10 Operating system, AMD A8 7410 (Quad-core; 2,2 GHz), with 4 GB RAM.
Table 1.
SARS-CoV-2 target proteins.
| Target Protein | Protein ID | Activity |
|---|---|---|
| Spike glycoprotein | 6LXT | Viral protein |
| RBD-ACE2 | 6VW1 | Cell invasion |
| 3CL Protease | 6M2N | Viral Protein |
| RNA-dependent RNA polymerase | 6M71 | Viral Protein |
3. Results and discussion
3.1. Genome mining analysis
Genome mining tool, AntiSMASH 5.0. is a well-known online platform to predict secondary metabolites synthesized by bacteria and various gene clusters involved in the secondary metabolite biosynthesis, such as polyketide synthase, nonribosomal synthetase, terpene, and RiPPs (ribosomally synthesized and post-translationally modified peptides) [15]. AntiSMASH is linked automatically to MIBiG (Minimum Information about Biosynthetic Gene Cluster) 2.0, representing the percent of similarity to known products and its related references. Compared to the bioinformatics analysis from the previous study, this study had more actual data due to the periodic database update of AntiSMASH. The database update was based on the latest researches about secondary metabolites from bacteria. The genome mining result is shown in Table 2.
Table 2.
Biosynthetic gene clusters (BGCs) and secondary metabolites of Streptomyces sp. GMR22 based on the analysis of genome mining with AntiSMASH 5.0.
| Cluster | Type | Size (bp) | Most similar known cluster (%) | |
|---|---|---|---|---|
| Cluster 1 | T1PKS | 40,540 | Ansamitocin (19) | t1pks |
| Cluster 2 | NRPS | 53,306 | Actinomycin (7) | NRPS |
| Cluster 3 | T1PKS | 5,152 | Micromonolactam (100) | t1pks |
| Cluster 4 | T1PKS | 26,122 | Hygrocin (74) | t1pks |
| Cluster 5 | lanthipeptide | 23,293 | Reveromycin (9) | t1pks |
| Cluster 6 | T1PKS, hglE-KS, bacteriocin | 132,456 | Bafilomycin (50) | t1pks |
| Cluster 7 | T1PKS, NRPS-like | 47,644 | Ansatrienin (mycotrienin) (7) | nrps-t1pks |
| Cluster 8 | T2PKS | 72,503 | Hexaricin (33) | t2pks |
| Cluster 9 | terpene | 21,100 | Pristinol (100) | terpene |
| Cluster 10 | NRPS, lanthipeptide | 88,563 | Lysolipin (4) | t2pks |
| Cluster 11 | T1PKS, NRPS-like | 44,220 | Kedarcidin (1) | t1pks+t1pks |
| Cluster 12 | Lasso peptide | 22,385 | ||
| Cluster 13 | siderophore | 13,367 | Natamycin (9) | t1pks |
| Cluster 14 | butyrolactone | 10,989 | Salinipostin (22) | other |
| Cluster 15 | NRPS | 88,212 | Glycinocin A (53) | NRPS |
| Cluster 16 | T1PKS | 7,155 | Dihydrochalcomycin (19) | t1pks |
| Cluster 17 | T1PKS | 12,099 | Hygrocin (16) | t1pks |
| Cluster 18 | terpene | 18,679 | Hopene (53) | terpene |
| Cluster 19 | ectoine | 10,404 | Ectoine (100) | Other |
| Cluster 20 | T1PKS | 9,683 | Micromonolactam (100) | t1pks |
| Cluster 21 | T1PKS | 79,793 | Elaiophylin (87) | t1pks |
| Cluster 22 | hserlactone | 20,755 | Daptomycin (4) | NRPS |
| Cluster 23 | butyrolactone | 10,935 | Meilingmycin (2) | t1pks |
| Cluster 24 | NRPS, T3PKS, other | 100,063 | Feglymycin (78) | NRPS |
| Cluster 25 | NRPS | 39,173 | ||
| Cluster 26 | NRPS | 2,917 | ||
| Cluster 27 | T1PKS | 33,976 | Geldanamycin (69) | t1pks |
| Cluster 28 | T2PKS | 72,491 | Medermycin (50) | t2pks |
| Cluster 29 | T1PKS | 63,579 | Mediomycin A (46) | polyketide |
| Cluster 30 | Lasso peptide | 22,645 | Polyoxypeptin (5) | nrps-t1pks |
| Cluster 31 | terpene | 21,073 | BE-43547 A1-C2 (15) | nrps-t1pks |
| Cluster 32 | siderophore | 13,770 | ||
| Cluster 33 | terpene | 22,390 | Geosmin (100) | terpene |
| Cluster 34 | T1PKS | 114,376 | Mediomycin A (50) | polyketide |
| Cluster 35 | Aryl polyene, adderane | 42,383 | WS9326 (25) | NRPS |
| Cluster 36 | NRPS | 44,010 | Ochronotic pigment (75) | other |
| Cluster 37 | siderophore | 12,057 | ||
| Cluster 38 | bacteriocin | 11,340 | ||
| Cluster 39 | T1PKS | 62,547 | S56-p1 (11) | NRPS |
| Cluster 40 | siderophore | 11,787 | Desferrioxamine B (100) | other |
| Cluster 41 | fused, T3PKS, NRPS | 57,879 | Pheganomycin (38) | nrps-ripp |
| Cluster 42 | NRPS-like | 25,322 | Echosides (58) | NRPS |
| Cluster 43 | ladderane, aryl polyene, NRPS | 104,182 | Skyllamycin (46) | NRPS |
| Cluster 44 | NRPS | 34,118 | Meridamycin (18) | nrps-t1pks |
| Cluster 45 | indole | 21,142 | Terfestatin (61) | other |
| Cluster 46 | NRPS | 78,698 | Leinamycin (5) | nrps-t1pks+transatpks |
| Cluster 47 | terpene | 22,129 | Phenalinolactone (8) | terpene-saccharide |
| Cluster 48 | terpene | 20,908 | ||
| Cluster 49 | NRPS-like | 42,738 | Echosides (11) | NRPS |
| Cluster 50 | Beta-lactone | 28,726 | Sch47554/Sch47555 (7) | t2pks |
| Cluster 51 | lanthipeptide | 24,153 | ||
| Cluster 52 | CDPS | 20,650 | Bicyclomycin (100) | other |
| Cluster 53 | T1PKS | 44,651 | Mediomycin A (25) | polyketide |
| Cluster 54 | T1PKS | 59,316 | Elaiophylin (45) | t1pks |
| Cluster 55 | T1PKS | 77,674 | Nigericin (94) | t1pks |
| Cluster 56 | T1PKS | 126,000 | Azalomycin F (82) | t1pks |
| Cluster 57 | T3PKS, terpene, NRPS | 40,798 | Totopensamide (30) | nrps-t1pks |
| Cluster 58 | T2PKS | 72,515 | Spore pigment (83) | t2pks |
| Cluster 59 | terpene | 21,190 | 2-methylisoborneol (100) | terpene |
| Cluster 60 | PKS-like | 41,028 | Galbonolides (20) | t1pks |
| Cluster 61 | NRPS-like | 44,028 | Niphimycins C-E (6) | polyketide |
| Cluster 62 | T1PKS | 71,413 | Meridamycin (55) | nrps-t1pks |
| Cluster 63 | T1PKS, NRPS-like | 45,285 | Hygrocin (38) | t1pks |
| Cluster 64 | T1PKS | 61,962 | Concanamycin A (17) | t1pks |
| Cluster 65 | T1PKS | 46,362 | Salinomycin (8) | t1pks |
T1PKS = Type I Polyketide Synthase; T2PKS = Type II Polyketide Synthase; T3PKS = Type III Polyketide Synthase; NRPS = Nonribosomal Polyketide Synthase; CDPS = Cyclodipeptide Synthase.
Table 2 shows 65 biosynthetic gene clusters (BGCs) of Streptomyces sp. GMR22, involved in the secondary metabolism. The total number of BGCs is the highest among other Streptomyces [7] as the average number of BGCs in Streptomyces was 30 gene clusters [18]. More than a third of BGCs were dominated by polyketide synthase, consisting of pure 18 Type I polyketide synthase (T1PKS), 3 Type II polyketide synthase (T2PKS), 3 hybrid Type I polyketide synthase. and 3 hybrid Type III polyketide synthase. In addition to PKS gene cluster, pure NRPS, NRPS hybrid, siderophore, and terpene gene cluster were found as well. However, the number was much less than that of PKS gene cluster. Polyketide synthase (PKS) is well-recognized for producing diverse compounds with various bioactivity [4], for example, desferrioxamine B and elaiophylin. Both have been reported as anticancer [19, 20]. Six gene clusters had 100% similarity to known BGC products, namely pristinol (terpene), ectoine (ectoine), geosmin (terpene), desferrioxamine B (siderophore), bicyclomycin (cyclodipeptide synthase), 2-methylisoborneol (terpene), while others had mostly under 100% similarity to previously identified compounds. This genome mining result was able to predict the large variety of metabolites from GMR22.
3.2. Targeted metabolomic analysis
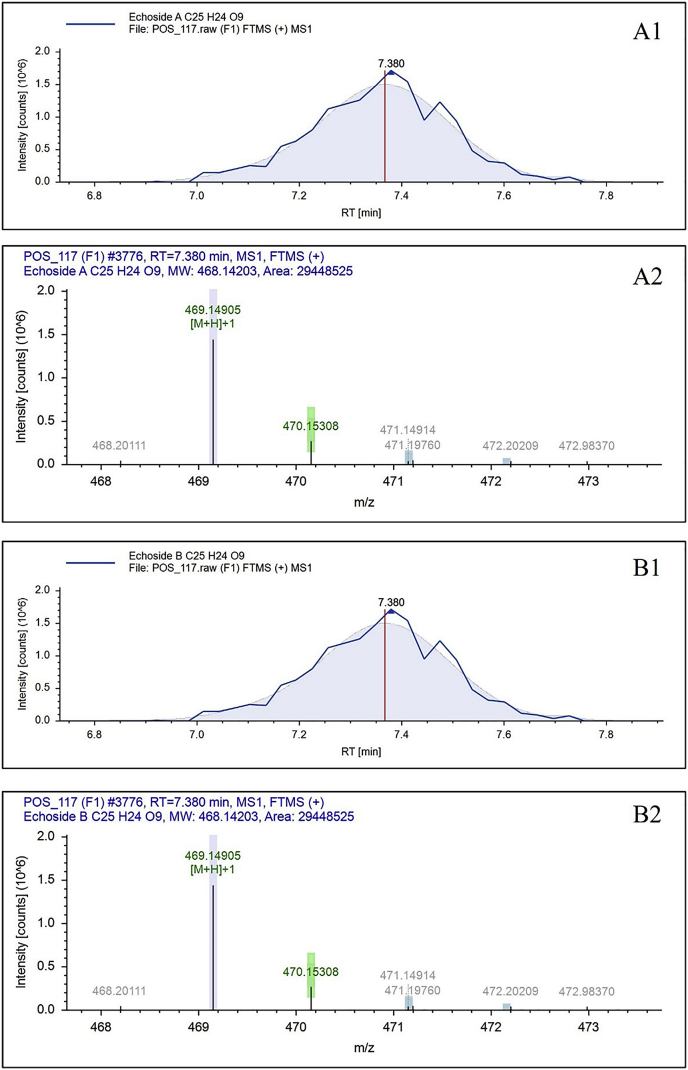
AntiSMASH 5.0 was used as a guide for the secondary metabolites profiling of GMR22 which was further confirmed by targeted metabolomic analysis with LC-HRMS. The compounds were detected by LC-HRMS showed in Table 3. The full spectra of targeted LC-HRMS showed in Supplementary Figure S1 and expected compounds data of targeted LC-HRMS showed in Supplementary Table S1. Most of the compounds detected by LC-HRMS underwent composition change of the target parent compound based on the genome mining analysis. The compounds predicted by using antiSMASH have similarity of 1–100% with BGCs from the database. This means that the compounds produced by GMR22 are not exactly the same as the compounds predicted by genome mining. Meanwhile, echoside A and echoside B were compounds which had been predicted in the genome mining analysis and had been detected in LC-HRMS. These compounds have C25H24O9 formula and 468.14203 of molecular weight (Da). The echoside A and echoside B spectrum and ionization showed in Figure 1. These compounds were detected at 7.3 min of RT and there was no composition change from parent compound as a target. These results confirmed that echoside A and echoside B were produced by Streptomyces sp. GMR22 based on both on genomic analysis using AntiSMASH and metabolomic analysis using LC-HRMS. Both appeared same, but the only different were their orientation. They are isomers.
Table 3.
Metabolomic analysis of Streptomyces sp. GMR22 extract using targeted LC-HRMS based on predicted compound from genome mining analysis.
| Parent Compound | Formula | Molecular Weight | Composition Change | RT [min] | Area (Max.) |
|---|---|---|---|---|---|
| Natamycin | C6H11NO4 | 161.06881 | –(C27H36O9) | 4.606 | 321808049.5 |
| Elaiophylin | C6H12O4 | 148.07356 | –(C48H76O14) | 7.037 | 3873193.757 |
| Hygrocin A | C7H8O2 | 124.05243 | –(C21H23NO6) | 7.148 | 158292572.2 |
| Echoside A | C25H24O9 | 468.14203 | No | 7.368 | 29448524.78 |
| Echoside B | C25H24O9 | 468.14203 | No | 7.368 | 29448524.78 |
| Echoside C | C18H14O3 | 278.09429 | –(C6H8O6) | 8.975 | 55169603.62 |
| Desferrioxamine B | C11H20N2O5 | 260.13722 | –(C14H28N4O3) | 13.422 | 3710903011 |
| Kedarcidin | C8H15NO3 | 173.10519 | –(C45H45ClN2O13) | 14.324 | 122233428.1 |
| Geldamycin | C13H18O5 | 254.11542 | –(C17H24N2O3) | 19.514 | 25962945.37 |
| Bafilomycin B1 | C35H56O9 | 620.39243 | –(C9H9NO4) | 25.472 | 6677673.906 |
| Medermycin | C22H20O9 | 428.11073 | –(C2H7N) +(O) | 26.035 | 1251745.932 |
| Daptomycin | C10H21NO | 171.16231 | –(C62H80N16O25) | 26.352 | 15124300.16 |
| Skyllamycin A | C12H13NO | 187.09971 | –(C63H81N11O19) | 26.482 | 19901446.49 |
| Glycinocin A | C15H28O2 | 240.20893 | –(C42H62N12O17) | 26.523 | 135669155.6 |
RT: retention times (minutes).
Figure 1.
Echoside A (A) and Echoside B (B). Chromatogram (1) and mass spectra (2) of Streptomyces sp. GMR22 extract detected on LC-HRMS analysis.
3.3. In silico molecular docking
The number of target proteins that hold a pivotal role in viral entry and replication become attractive SARS-CoV-2 drug targets. Targeting RdRp, and 3CLpro was considered efficient to inhibit the enzyme function during SARS-CoV-2 intracellular replication, while targeting RBD-ACE2 and spike glycoprotein was efficient to prevent SARS-CoV-2 from entering and fusing to the host cell [12]. The target proteins of SARS-CoV-2 and the compound was prepared using Auto Dock Tools, then subjected to molecular docking using Autodock Vina and visualized with DS Visualizer. This study used a standard drug, remdesivir that had been reported to effectively inhibit SARS-CoV-2 in vitro [21]. The binding energy result is displayed in Table 4. Compared to the reference drug, echoside A and echoside B from GMR22 attributed better docking scores with all the selected target proteins. The lower of docking score means the stronger binding affinity between the ligand and protein target. The echoside A and echoside B compounds showed the strongest binding affinity with 3CLPro protein with docking score of −8.4, followed by spike protein, RBD-ACE2 and RdRp protein with docking score of −7.9, −7.5 and −7.3, respectively. Meanwhile, echoside B displayed strongest binding affinity with 3CLpro (−9.4 kcal/mol), followed by rbd (−8.2 kcal/mol), rdrp (−8.0 kcal/mol), spike protein (−7.8 kcal/mol). It shows stronger binding affinity compared with remdesivir as comparison drug with docking score of −6.5, −6.9, −6.7 and −6.4 in 3CLPro, spike protein, RBD-ACE2 and RdRp protein, respectively.
Table 4.
Binding energy of the GMR22 targeted compounds and reference compound against the target proteins of SARS-CoV-2.
| Compounds | Binding energy (kcal/mol) |
|||
|---|---|---|---|---|
| 6LXT (spike protein) |
6VW1 (RBD-ACE2) |
6M2N (3CLpro) |
6M71 (RdRp) |
|
| Remdesivir∗ | −6.9 | −6.7 | −6.5 | −6.4 |
| Natamycin | −9.3 | −9.6 | −9.9 | −8.8 |
| Elaiophylin | −10.2 | −10.9 | −10.9 | −9.9 |
| Hygrocin A | −9.2 | −9.4 | −9.7 | −8.9 |
| Echoside B | −7.8 | −8.2 | −9.4 | −8.0 |
| Echoside A | −7.9 | −7.5 | −8.4 | −7.3 |
| Echoside C | −8.1 | −7.7 | −8.6 | −8.1 |
| Desferrioxamine B | −4.9 | −5.6 | −6.2 | −4.8 |
| Kedarcidin | −10.0 | −10.7 | −9.4 | −10.0 |
| Geldanamycin | −6.6 | −7.4 | −7.0 | −7.1 |
| Bafilomycin B1 | −8.8 | −9.8 | −9.6 | −8.9 |
| Medermycin | −9.4 | −8.6 | −9.1 | −7.8 |
| Daptomycin | −8.9 | −9.1 | −9.3 | −8.9 |
| Skyllamycin A | −8.4 | −10.7 | −8.1 | −7.1 |
| Glycinocin A | −7.8 | −9.0 | −7.4 | −7.8 |
Reference drug.
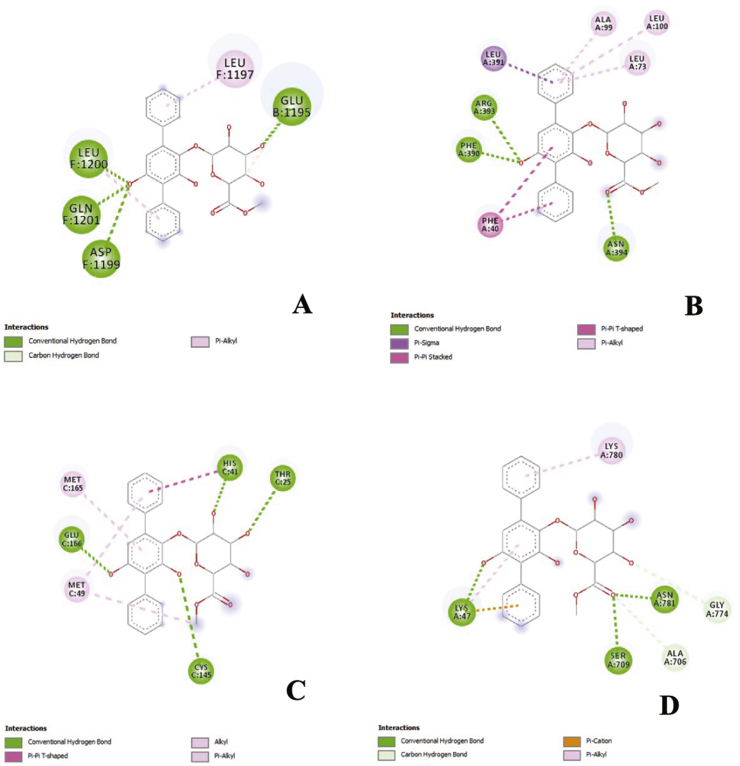
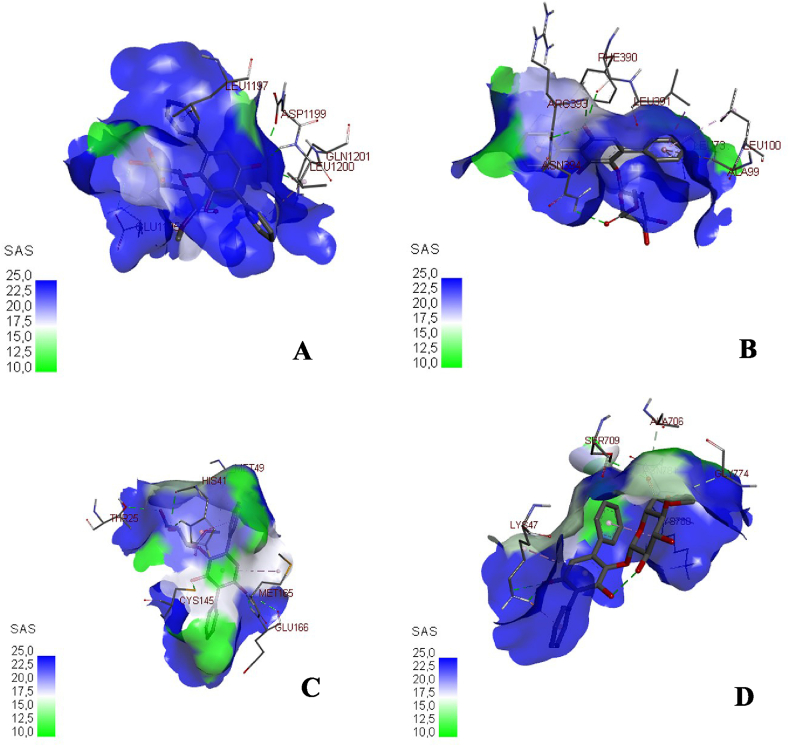
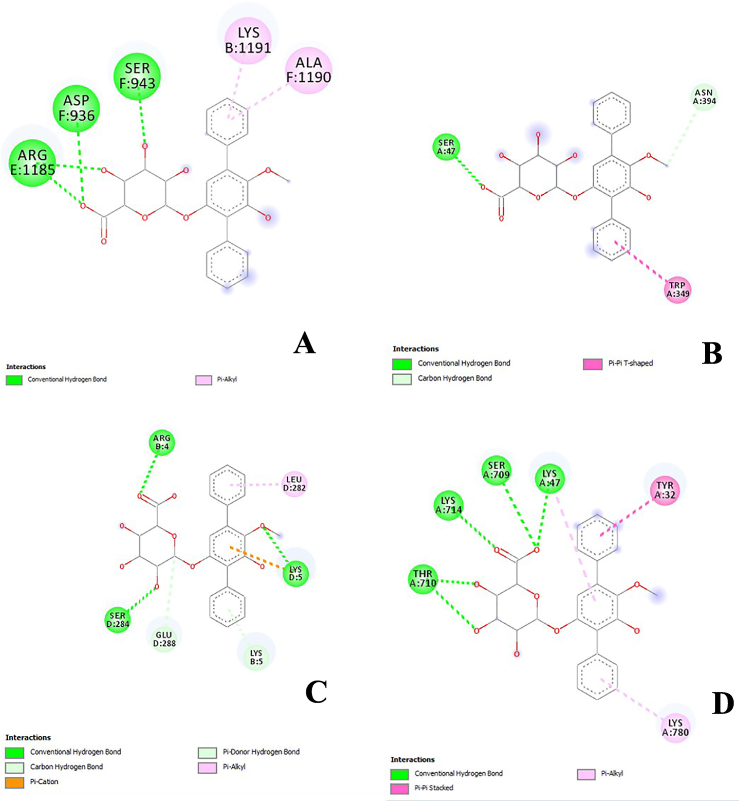
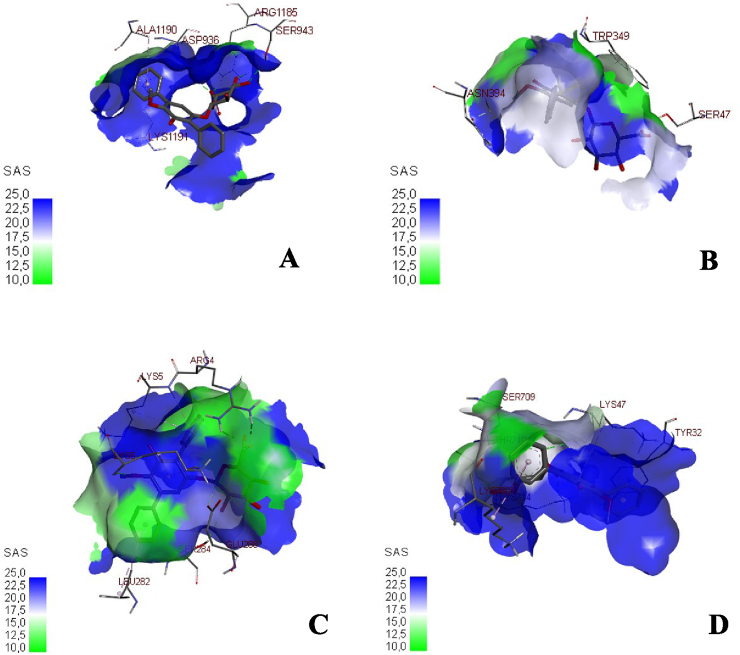
In the previous study, echosides, isolated from Streptomyces sp. LZ35, represent a class of para-terphenyl natural products that displayed DNA topoisomerase I and IIα inhibitory activities. This compound was produced by a tri-domain non-ribosomal peptide synthetase (NRPS)-like enzyme which encoded by the echA-gene [22]. Anthracyclines, including doxorubicin and daunorubicin, are well-documented chemotherapeutic drugs that have the same topoisomerase II inhibitory activity as echosides. These compounds were shown to stimulate IFN (Interferons) response that eventually suppressed the viral replication of Ebola VP35 in an ATM-kinase dependent manner and a cGAS (cyclic cGMP-AMP) - STING (Stimulator of Interferon Genes) mechanism [23]. Hence, in the molecular docking analysis, and echoside B could be considered as a potential anti-SARS-CoV-2 from GMR22. As this compound has higher affinity than remdesivir as COVID-19 drug. The visualization of binding between echoside A and target proteins is displayed in Figure 2 and Figure 3, while echoside B in Figure 4 and Figure 5. Echoside A formed interaction with the following residues of spike protein, LEU (1197) showed interaction with echoside A in pi-alkyl bond, ASP (1199), GLN (1201), and LEU (1200) interact with hydrogen bond (Figure 2A). In RBD-ACE2 protein, Echoside A formed with PHE (390), ARG (393), and ASN (394) in hydrogen bond, LEU (391) in pi-sigma bond, LEU (73), LEU (100), ALA (99) in pi-sigma bond and PHE (40) in pi-pi stacked bond (Figure 2B). Echoside A formed with THR (25), HIS (41), CYS (145) and GLU (166) in hydrogen bond, while MET (165) and MET (25) in pi-alkyl bond with the target site (3CLPro) (Figure 2C). In RdRp protein, Echoside A formed with LYS (47), SER (709), and ASN (781) in hydrogen bond, ALA (706) and GLY (774) in carbon hydrogen bond, while LYS (780) showed pi-alkyl bond with protein target site (Figure 2D). The 3D conformation of echoside A and target proteins showed the amino acid interaction in 3D protein conformation with solvent accessible surface (SAS) for this binding site is computed. SAS is often used when calculating the transfer free energy required to move a biomolecule from aqueous solvent to a non-polar solvent such as a lipid environment. The SAS value of the echoside A with the target proteins scored between 10.0 – 22.5 (Figure 3). Echoside B formed interaction with the following residues of spike protein, LYS (1191) and ALA (1190) showed interaction with echoside B in pi-alkyl bond, ASP (936), SER (943), and ARG (1185) interact with hydrogen bond (Figure 4A). In RBD-ACE2 protein, Echoside B formed with SER (47) in hydrogen bond, ASN (394) in carbon hydrogen bond, TRP (349) in pi-pi T-shaped bond (Figure 4B). Echoside B formed with ARG (4), SER (284), LYS (5) in hydrogen bond, LYS (5) and GLU (288) in carbon hydrogen bond, and LEU (282) in pi-alkyl bond with the target site (3CLPro) (Figure 4C). In RdRp protein, Echoside B formed with LYS (47), SER (709), LYS (714), and THR (710) in hydrogen bond, TYR (32) in pi-pi stacked bond, while LYS (780) showed pi-alkyl bond with protein target site (Figure 4D). The amino acid interaction of echoside B with the targets protein in 3D protein conformation is displayed in Figure 5. The SAS value of the echoside B with the targets protein scored between 10.0 – 22.5.
Figure 2.
Binding visualization of echoside A with spike protein (A), RBD-ACE2 (B), 3CLpro (C), and RdRp (D), respective interacting amino acid residues.
Figure 3.
3D visualization of binding between echoside A with spike protein (A), RBD-ACE2 (B), 3CLpro (C), and RdRp (D), respective interacting amino acid residues.
Figure 4.
2D visualization of binding between echoside B with spike protein (A), RBD-ACE2 (B), 3CLpro (C), and RdRp (D), respective interacting amino acid residues.
Figure 5.
3D visualization of binding between echoside B with spike protein (A), RBD-ACE2 (B), 3CLpro (C), and RdRp (D), respective interacting amino acid residues.
4. Conclusions
The present study was carried to explore novel inhibitors of SARS-CoV-2 directly correlated with soil-derived bacterium, Streptomyces sp. GMR22, by applying both genome mining and metabolomic approaches. Echoside A and Echoside B showed the best binding interaction with three target proteins, i.e., spike protein, RBD-ACE2, and 3CLpro. Furthermore, echoside A and echoside B displayed better docking score than remdesivir, showed high potential as antiviral, and have been tested as antiviral in other previous study, thus echoside A and echoside B could be utilized for further exploration as an antiviral agent for SARS-CoV-2.
Declarations
Author contribution statement
Yohana Nadia Melinda: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Jaka Widada: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Tutik Dwi Wahyuningsih: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Rifki Febriansah: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Ema Damayanti: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mustofa Mustofa: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by Indonesian Ministry of Research, Technology and Higher Education (3402/UNI.DITLIT/DIT-LIT/PT/2020).
Data availability statement
Data associated with this study has been deposited at GenBank under the accession number JACGSQ010000000.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors thank Tri Purwanti and Hendy Dwi Warmiko for the technical support.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Olanrewaju O.S., Babalola O.O. Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019;103(3):1179–1188. doi: 10.1007/s00253-018-09577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herdini C., et al. Secondary bioactive metabolite gene clusters identification of anticandida-producing streptomyces Sp. GMR22 isolated from Wanagama forest as revealed by Genome mining approach. Indones. J. Pharm. 2017;28(1):26–33. [Google Scholar]
- 3.Wang H., Fewer D.P., Holm L., Rouhiainen L., Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. U. S. A. 2014;111(25):9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salis O., Bedir A., Kilinc V., Alacam H., Gulten S., Okuyucu A. The anticancer effects of desferrioxamine on human breast adenocarcinoma and hepatocellular carcinoma cells. Cancer Biomarkers. 2014;14(6):419–426. doi: 10.3233/CBM-140422. [DOI] [PubMed] [Google Scholar]
- 5.Zhong Z., He B., Li J., Li Y.X. Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs) Synth. Syst. Biotechnol. 2020;5(3):155–172. doi: 10.1016/j.synbio.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaka M., Yangchum A., Rachtawee P., Komwijit S., Lutthisungneon A. Hopane-type triterpenes and binaphthopyrones from the scale insect pathogenic fungus aschersonia paraphysata BCC 11964. J. Nat. Prod. 2010;73(4):688–692. doi: 10.1021/np1000363. [DOI] [PubMed] [Google Scholar]
- 7.Herdini C., et al. Diversity of nonribosomal peptide synthetase genes in the Anticancer Producing actinomycetes isolated from marine sediment in Indonesia. Indones. J. Biotechnol. 2016;20(1):34. [Google Scholar]
- 8.Nurjasmi R., Widada J., Ngadiman N. Diversity of actinomycetes at several forest types in Wanagama I Yogyakarta and their potency as a producer of antifungal compound. Indones. J. Biotechnol. 2009;14(2):1196–1205. [Google Scholar]
- 9.Bhat E.A., et al. SARS-CoV-2: insight in genome structure, pathogenesis and viral receptor binding analysis – an updated review. Int. Immunopharm. 2021;95:107493. doi: 10.1016/j.intimp.2021.107493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X., et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joshi S., Joshi M., Degani M.S. Tackling SARS-CoV-2: proposed targets and repurposed drugs. Future Med. Chem. 2020;12(7):1579–1601. doi: 10.4155/fmc-2020-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall D.C., Ji H.F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Trav. Med. Infect. Dis. 2020;35:101646. doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosse J.T., Ghosh S., Sproule A., Overy D., Cheeptham N., Boddy C.N. Whole genome sequencing and metabolomic study of cave Streptomyces isolates ICC1 and ICC4. Front. Microbiol. 2019;10:1–12. doi: 10.3389/fmicb.2019.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blin K., et al. AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trott A.J., Olson O., “Autodock vina Improving the speed and accuracy of docking. J. Comput. Chem. 2019;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studio D. Accelrys Softw. Inc.; 2015. Dassault Systemes BIOVIA, Discovery Studio Modelling Environment, Release 4.5. [Google Scholar]
- 18.Ishaque N.M., et al. Isolation, genomic and metabolomic characterization of Streptomyces tendae VITAKN with quorum sensing inhibitory activity from Southern India. Microorganisms. 2020;8(1) doi: 10.3390/microorganisms8010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X., et al. Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy. 2015;11(10):1849–1863. doi: 10.1080/15548627.2015.1017185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Heel A.J., De Jong A., Song C., Viel J.H., Kok J., Kuipers O.P. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J., et al. Identification and catalytic characterization of a nonribosomal peptide synthetase-like (NRPS-like) enzyme involved in the biosynthesis of echosides from Streptomyces sp. LZ35. Gene. 2014;54(2):352–358. doi: 10.1016/j.gene.2014.05.053. [DOI] [PubMed] [Google Scholar]
- 23.Luthra P., et al. Topoisomerase II inhibitors induce DNA damage-dependent interferon responses circumventing ebola virus immune evasion. mBio. 2017;8(2) doi: 10.1128/mBio.00368-17. e00368-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at GenBank under the accession number JACGSQ010000000.