Abstract
p270 is an integral member of human SWI-SNF complexes, first identified through its shared antigenic specificity with p300 and CREB binding protein. The deduced amino acid sequence of p270 reported here indicates that it is a member of an evolutionarily conserved family of proteins distinguished by the presence of a DNA binding motif termed ARID (AT-rich interactive domain). The ARID consensus and other structural features are common to both p270 and yeast SWI1, suggesting that p270 is a human counterpart of SWI1. The approximately 100-residue ARID sequence is present in a series of proteins strongly implicated in the regulation of cell growth, development, and tissue-specific gene expression. Although about a dozen ARID proteins can be identified from database searches, to date, only Bright (a regulator of B-cell-specific gene expression), dead ringer (a Drosophila melanogaster gene product required for normal development), and MRF-2 (which represses expression from the cytomegalovirus enhancer) have been analyzed directly in regard to their DNA binding properties. Each binds preferentially to AT-rich sites. In contrast, p270 shows no sequence preference in its DNA binding activity, thereby demonstrating that AT-rich binding is not an intrinsic property of ARID domains and that ARID family proteins may be involved in a wider range of DNA interactions.
SWI-SNF complexes were first identified in yeast cells, where they are involved in the regulation of an array of inducible genes including those required for the mating type switch and sucrose fermentation pathways (16, 28). More-recent studies suggest that these complexes have a more general role in the regulation of gene expression. The isolation and characterization of Drosophila melanogaster and mammalian homologs of many of the yeast complex members suggest that SWI-SNF complexes play fundamental roles in the regulation of gene expression during cell growth and development in all organisms (reviewed in reference 14; 17).
Although SWI-SNF complexes have demonstrated DNA binding capabilities (29), the source of this activity in the complexes remains unclear. The only DNA binding protein identified to date in mammalian SWI-SNF complexes is BAF-57, which has a DNA binding activity restricted to four-way junction DNA. SWI-SNF complexes lacking a functional BAF-57 retain DNA binding activity, indicating that other DNA binding components must be present (41).
p270 is an integral member of human SWI-SNF complexes, first identified through its shared antigenic specificity with p300 and CREB binding protein (CBP) (5, 6). The p300/CBP/p270 cross-reactive antibodies coprecipitate a series of proteins that includes the mammalian SWI-SNF complex components BRG1, BAF-170, BAF-155, and hSNF5/Ini1. Conversely, antibodies directed against the individual human SWI-SNF complex components BRG1 and BAF-155 immunoprecipitate p270, as demonstrated by reactivity with a p270-specific antibody (6). The sequence of p270 described here indicates that this protein contains a highly conserved DNA binding region termed the AT-rich interactive domain, or ARID, first recognized in the murine Bright and the Drosophila dead ringer (DRI) gene products (9, 11). The approximately 90-residue ARID sequence is present in a series of proteins strongly implicated in the regulation of cell growth, development, and tissue-specific gene expression (see Table 1). Bright is a regulator of B-cell-specific gene expression (11, 42), and DRI is a corepressor for dorsal, is implicated in the activation or repression of other transcriptional regulators, and is required for normal Drosophila development (9, 32, 37). The diverse array of regulatory proteins that contain an ARID consensus (reviewed in reference 19) includes the yeast SWI1 protein, a component of yeast SWI-SNF complexes that has been found to cross-link with DNA in vitro (29).
TABLE 1.
Proteins with ARID motifsa
| Protein(s) | Function (reference[s]) |
|---|---|
| Bright | B-cell-specific activator (11, 42) |
| DRI | Corepressor for Drosophila dorsal (9, 37); may function as an activator in other contexts (32) |
| MRF-1, MRF-2 | Modulator recognition factors; repressors of CMV enhancer activity (12, 35, 43) |
| RBP 1, RBP 2 | Retinoblastoma binding proteins (7); RBP 1 is a repressor of E2F (21, 22) |
| smcx, smcy | Sex-specific transcription factors (1, 15) |
| jumonji | Neural tube-specific transcription factor (33) |
| eyelid | Antagonist of Drosophila wingless (36, 38) |
| SWI1 | Component of yeast SWI-SNF complex (25, 28) |
| p270 | Component of human SWI-SNF complex (6) |
Partial list of known ARID proteins. A few others are recognizable on the basis of open reading frames in GenBank sequences but have no known function.
At present, more than a dozen ARID-bearing proteins can be identified from database sequences. However, only Bright, DRI, and MRF-2, a cytomegalovirus (CMV) enhancer binding protein, have been characterized directly in regard to their DNA binding properties. Bright binds to matrix attachment regions, which are highly AT-rich stretches of about 20 to 40 bases (11). DRI binds to a core ATTA motif similar to that present in homeodomain sites (9). MRF-2 binds preferentially to the sequence AATAC/T in binding site selection assays (43). While the properties of these proteins have suggested that ARIDs have intrinsic specificity for AT-rich sequences (albeit of somewhat different compositions), analysis of p270 indicates that sequence specificity is not intrinsic to the core ARID motif. p270 binds native duplex DNA in an ARID sequence-dependent manner but shows no discernible sequence preference in its DNA binding activity, indicating that DNA binding via ARID regions is not restricted to AT-rich sequences and consistent with suggestions that ARID family proteins are involved in a wider range of DNA interactions. The presence of a recognized DNA binding domain in p270 and the demonstrated ability of p270 to bind linear duplex DNA suggest that p270 contributes to the DNA binding activity in the SWI-SNF complex.
MATERIALS AND METHODS
cDNA isolation.
p270 was gel purified and subjected to peptide microsequence analysis as described in reference 6. The R29 peptide (pep29) (sequence shown in Fig. 1) was used to generate p270-specific rabbit antiserum (rabbit 67). A human HeLa λgt11 cDNA expression library (Clontech) randomly primed with oligo(dT) was screened with rabbit 67 antiserum using standard procedures. An insert from a positive clone identified in this initial screening process was then used to screen a human HeLa λZAPII oligo(dT)-primed cDNA library (Stratagene) by DNA hybridization, which yielded a clone containing approximately 2.5 kb of 3′ cDNA sequence. A 5′-rapid amplification of cDNA ends (5′-RACE) procedure was used to extend the 2.5-kb cDNA sequence. Total RNA obtained from WI-38 cells was subjected to sequential rounds of extension using a 5′-RACE kit in accordance with the manufacturer's instructions (Gibco BRL). After two rounds of nested PCR (Expand high-fidelity PCR system; Boehringer-Mannheim) using gene-specific primers and primers provided with the kit, the amplified products were digested with appropriate restriction enzymes and cloned into appropriate plasmid vectors. DNA sequence data were obtained from both strands of plasmid templates using both the fmol cycle sequencing system (Promega) and automated DNA sequencing procedures. Approximately 50% of the sequence could be confirmed by expressed sequence tag (EST) sequences already present in the data banks.
FIG. 1.
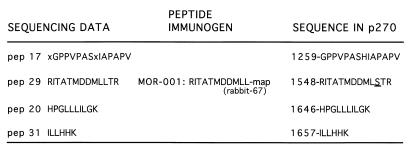
Sequences of peptides obtained from the microsequencing of p270. Left column, sequences of four peptides derived from the peptide sequencing of gel-purified p270 isolated from human cells; middle column, sequence of the multiple-antigen peptide (map) that was used to raise the p270-specific antibodies used in the cloning; right column, sequences of the corresponding regions deduced from the p270 cDNA clone, with the position number of the initial residue indicated for reference to Fig. 3. X represents uncertainty in reading the protein sequence. The underlining indicates a single discrepancy between the sequence obtained by direct sequencing and that obtained by sequencing the cDNA.
Northern blots.
Multiple-tissue Northern blots (Clontech) were screened with a 2.5-kb cDNA fragment spanning the 3′ terminus of the coding sequence for p270 (nucleotides 3678 to 6200). The probe was labeled by random priming with [α-32P]dATP. Hybridization and washing conditions were as described by the supplier.
Computer analysis.
DNA sequence compilation and open reading frame analysis were undertaken using programs of the Genetics Computer Group (GCG) sequence analysis software package (Madison, Wis.). The GCG Prettybox program was used to generate the alignment of the ARID motifs. Database amino acid sequence comparisons were undertaken using the BLASTp program with the low-complexity filtering option (2).
In vitro translation and DNA cellulose chromatography.
p270 cDNA fragments in appropriate plasmid vectors were used to generate [35S]methionine-labeled polypeptides using the TNT coupled-reticulocyte system (Promega). In vitro-translated p270 cDNA plasmid constructs used in this study include pPE14 (p270603–1927; encoding amino acids 603 to 1927), p110 (p2701237–1927), pNNE3 (p270543–1018), pMY6 (p270678–1018), and pNE9WY (p270543–1281 containing alanine substitutions at positions 715 and 738). In vitro-translated proteins were applied to native DNA cellulose columns (Pharmacia) equilibrated in loading buffer (10 mM potassium phosphate [pH 6.2], 0.5% NP40, 10% glycerol, 1 mM dithiothreitol [DTT], aprotinin [1 μg/ml], 50 mM NaCl); the bed volume was approximately 1 ml. The column was washed four times with 0.5 bed volumes of loading buffer and eluted stepwise with loading buffer adjusted to contain increasing concentrations of NaCl as indicated below. Aliquots of the flowthrough and each fraction were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis or in a nitrocellulose filter binding assay in a standard slot blot apparatus. Quantification was performed using a phosphorimager (Fuji) and associated software.
GST fusion proteins.
pNDX was generated by cloning a p270 cDNA fragment encoding residues 600 to 1018, containing the p270 ARID and flanking sequences, into pGEX4T3 (Pharmacia) to make a glutathione S-transferase (GST) fusion protein construct (GSTp270600–1018). The DRI plasmid p410 (GST-dri258–410) (9) (kind gift from R. Saint, University of Adelaide, South Australia, Australia) has been described previously. Fusion proteins were produced in Escherichia coli strain BL21 after induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and growth at 30 (GSTp270600–1018) or 37°C (GST-dri258–410). Fusion proteins were purified on glutathione-Sepharose beads using standard procedures.
Generation of amino acid substitution mutations.
Amino acid substitution mutants were generated using the QuikChange (Stratagene) system according to the manufacturer's instructions. The forward primer used to generate the Tyr→Ala substitution had the sequence CCTTGAAAAAGCAGGCTATCCAGTGTCTCTATGC. The forward primer used to generate the Trp→Ala substitution had the sequence GGTCAACAAGAACAAAAAAGCGCGGGAACTTGCAACC. The substituted bases in each primer are underlined. The sequence changes and the integrity of the surrounding sequence were verified by DNA sequencing.
PCR-assisted DNA binding selection from random oligonucleotides.
The GSTp270600–1018 fusion protein was used to determine the sequence specificity of the p270 ARID using PCR-assisted selection and amplification. A mixture of 50-base oligonucleotides, in which the middle 20 bases consisted of random nucleotides, was converted to double-stranded oligonucleotides by PCR, using 32P-end-labeled primers 1 (5′-GTACCCGGGGATCCT-3′) and 2 (5′-TTGCATGCCTGCAGG-3′) complementary to the flanking sequences in the 50-mers. Approximately 100 ng of double-stranded oligonucleotides was suspended in 250 μl of binding buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.5 mM EDTA, 10% glycerol, 1 mM DTT, 20 mg of bovine serum albumin (BSA)/ml, 2 mg of poly[dI-dC]/ml) and applied to approximately 100 ng of GSTp270600–1018 fusion protein attached to 10 ml of glutathione-Sepharose beads equilibrated in binding buffer. The mixture was rotated at 4°C for 1 h and then was centrifuged at 12,000 × g for 1 min. The supernatant was removed, and the pellet was washed twice with 1 ml of binding buffer with either 100 or 800 mM NaCl. This pellet was resuspended in 30 ml of water, boiled for 3 min, and centrifuged rapidly. This supernatant was digested with proteinase K for 3 h and extracted with phenol-chloroform, and the DNA was amplified by PCR with primers 1 and 2. The amplified DNA was gel purified and reapplied to the fusion protein–glutathione-Sepharose bead mixture. After four (for 100 mM NaCl washes) or six (for 800 mM NaCl washes) sequential cycles of elution, amplification, and reapplication, the double-stranded, gel-purified oligonucleotides were digested with BamHI and PstI, cloned into plasmid vectors, and sequenced.
EMSA.
The DNA binding specificities of p270 and DRI were examined by electrophoretic mobility shift assay (EMSA) using purified GSTp270600–1018 and GST-dri258–510 fusion proteins. Approximately 100 ng of fusion protein was incubated with 32P-labeled double-stranded PCR product (approximately 2 × 104 cpm) in 10 mM Tris (pH 7.5)–200 mM KCl–1 mM EDTA–0.2 mM DTT–10% glycerol–50 mg of BSA/ml at 30°C for 30 min. Poly(dI-dC) was added to individual reaction mixtures before incubation as indicated below. After electrophoresis on 5% acrylamide gels in Tris-glycine buffer, the gels were dried and autoradiographed. The sequences of the ∼20-bp inserts in the probes are as follows: dri.16, CATCAATAAATTAGAATTAA; p270.18, ACCGGGGGTGCCACACCGTTGA.
RESULTS
Peptide sequencing and isolation of p270 cDNA.
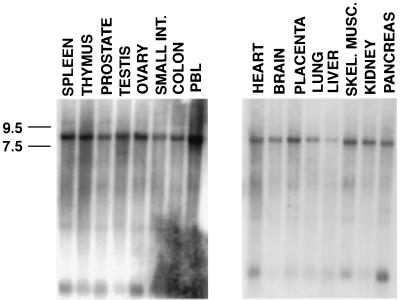
p270 was originally detected as a novel protein recognized by the p300-reactive monoclonal antibody NM1. Analysis of NM1 immune complexes indicated that p270 is an integral component of mammalian SWI-SNF complexes (5, 6). In order to characterize this protein further, p270 was isolated and purified to obtain the peptide sequence. Four p270 peptides obtained from this analysis are shown in Fig. 1. The sequence of peptide 29 was used to generate a synthetic peptide and raise rabbit antipeptide antibodies (rabbit 67). These antibodies recognize p270 specifically in NM1-, BRG1-, or BAF-155-specific immune precipitations (6). The rabbit 67 antibody was used to screen a HeLa cDNA expression library randomly primed with oligo(dT), which yielded a 650-bp fragment encoding a sequence that matched p270 peptide 29 in 12 of 13 residues. The fragment also contained a sequence encoding matches to two other peptides, pep20 and pep31, in the same open reading frame. This fragment was used to screen a second HeLa cDNA library. Several overlapping cDNAs were isolated, including a 2.5-kb clone containing additional cDNA sequences 5′ and 3′ to the original 650-bp fragment. The 2.5-kb fragment included a sequence encoding the pep17 peptide and contained a consensus polyadenylation signal (AATAAA) and a poly(A) tail. The cDNA hybridized to RNA species migrating at approximately 8.0 kb (Fig. 2), consistent with the expected size of the p270 mRNA. We concluded that the 2.5-kb fragment encoded an authentic p270 sequence and included the 3′ terminus of the p270 cDNA. In addition, as expected from the broad distribution patterns of other components of the SWI-SNF complexes, p270 expression was detected in all tissues examined (Fig. 2).
FIG. 2.
Hybridization of p270 cDNA sequence with an ∼8.0-kb RNA band in multitissue Northern blots. Blots were probed with a 2.5-kb cDNA corresponding to the 3′ terminus of the p270 mRNA. Molecular sizes (left) are according to the measurements provided by the supplier. INT, intestine; PBL, peripheral blood lymphocytes; SKEL. MUSC., skeletal muscle.
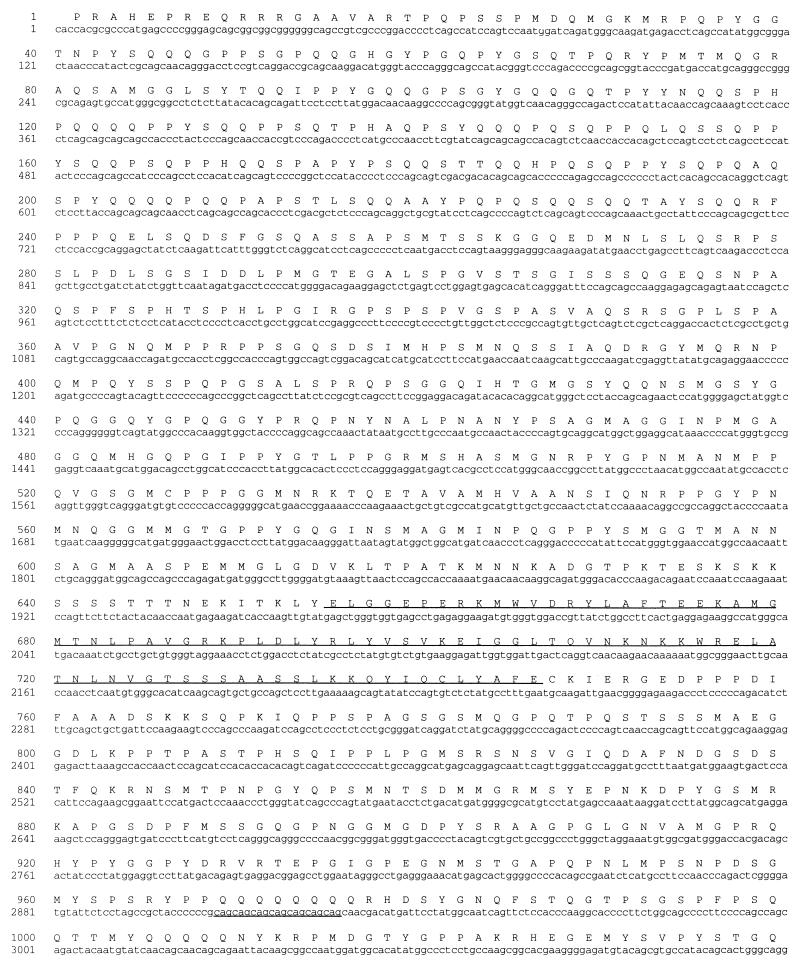
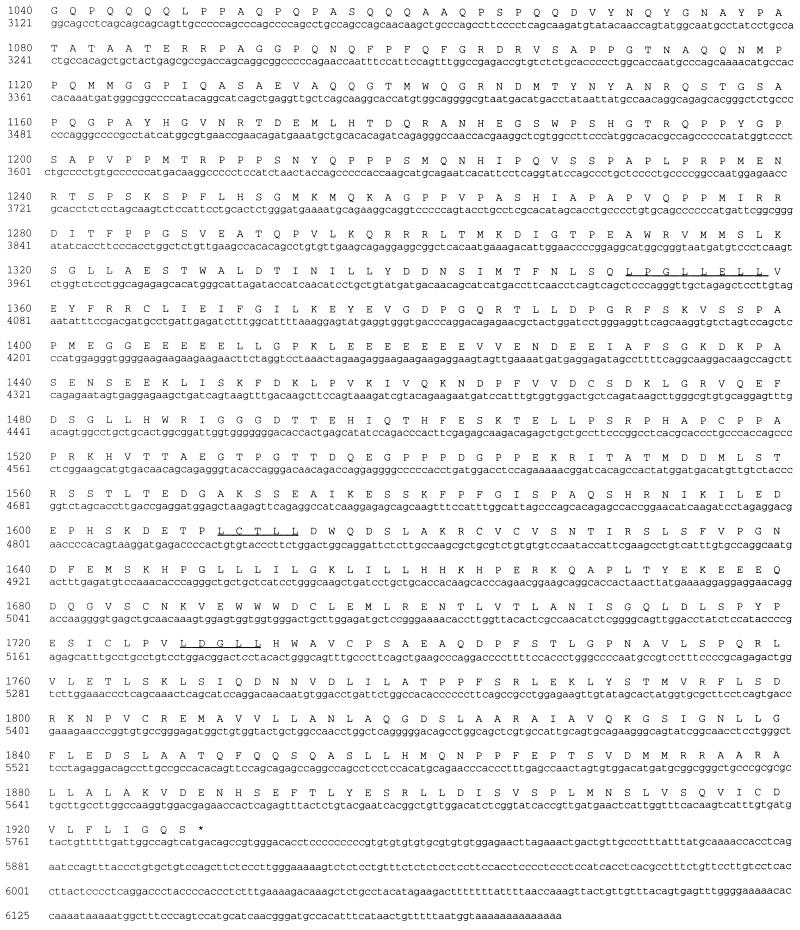
A 5′-RACE procedure on human WI-38 cell total RNA was used to extend the cDNA sequence. Successive rounds with this procedure yielded a total of 6.2 kb of sequence with an open reading frame encoding 1,927 amino acid residues. The 5′ terminus of this cDNA sequence is strikingly GC rich, which prevents analysis of the extreme N-terminal sequence. From its relative migration in protein gels, we estimate that p270 contains approximately 2,000 amino acid residues, which suggests that the clone we have is over 95% complete. This large clone reveals a number of structural features as discussed below. The sequence of the p270 clone and the deduced sequence of the corresponding open reading frame are shown in Fig. 3.
FIG. 3.
DNA sequence and deduced amino acid sequence of human p270. The nucleotide sequence of the human p270 cDNA is represented along with the deduced amino acid sequence of the product of the 5,781-bp open reading frame. The four LXXLL motifs and the ARID motif are underlined. Two Q-rich regions are present, spanning residues 45 to 253 and 969 to 1072. The sequence also includes a series of seven CAG repeats (34) indicated by underlining beginning at nucleotide 2907.
p270 is an ARID protein family member structurally related to yeast SWI1.
A filtered ungapped BLAST search program (2, 3) was used to search for proteins showing relationships to p270. This program identified several proteins with a statistically significant degree of relationship to p270, including p300. Other proteins with significant homology to p270 include (i) a cDNA product (B120) whose coding sequence was cloned as part of a search for genes containing multiple CAG repeats (34) (accession no. AB001895) and whose sequence appears to be a portion of the p270 sequence, but whose coding sequence differs from our sequence at the 5′ terminus and contains a frameshift that gives rise to a truncated-p270-encoding open reading frame; (ii) eyelid (eld), also referred to as osa, a ubiquitously expressed protein involved in embryonic growth and differentiation in Drosophila (36, 38), and (iii) a product of a Caenorhabditis elegans cDNA sequence (YK7C8.5; GenBank accession no. U80439) which appears from its overall degree of sequence relationship to be an authentic C. elegans homolog of p270 but which has not yet been ascribed any function (44). Most of the proteins with structural relationships to p270 have in common a recently described DNA binding motif termed ARID (9, 11). Proteins with ARID regions include human and murine Bright, Drosophila DRI and its human homolog DRIL1 (18), the CMV enhancer binding proteins MRF-1 and MRF-2, eyelid, retinoblastoma binding proteins (RBP) 1 and 2, and yeast SWI1 (p300 does not contain an ARID motif). A comparison of the ARID region from p270 with those of other ARID-containing proteins is shown in Fig. 4. The functions of the known ARID proteins are listed in Table 1. Outside of the ARID regions, proteins in this list are not generally homologous to one another.
FIG. 4.
A comparison of the ARID regions from p270 with those of other ARID-containing proteins. The boundaries of the ARID region were originally defined in Bright and DRI (9, 11). These sequences are aligned here together with others more recently added to the database. Black boxes, identical residues; gray boxes, conservative differences. The consensus sequence consists of 36 highly conserved residues with an approximately 95-residue stretch. All sequences were obtained from public protein databases. Accession numbers are as follows, eyelid, AF053091; SWI1, P09547; Bright, U60335; DRIL1, U88047; DRI, U62542; RBP 1, P29374; RBP 2, P29375; MRF-1, S27962; MRF-2, S27963; smcx, P41229; jumonji, U57592; Arabidopsis thaliana open reading frame (ORF) product f2202.20, AAC83071; C. elegans ORF products c01g8 and t23d8, U80438 and Z81128, respectively.
p270 does show an interesting degree of relationship with Drosophila eyelid, a protein involved in development, including embryonic segmentation and photoreceptor differentiation (36, 38). Like p270, eyelid is a large protein (2,715 residues) and contains an ARID consensus sequence. In addition to the ARID region, p270 and eyelid have a high degree of identity at their C termini. Residues 1277 to 1371 of the 1,927 residues encoded by the p270 gene open reading frame show 46% identity to eyelid residues 1768 to 1862. Another stretch, consisting of residues 1599 to 1809 in p270 and residues 2170 to 2384 in eyelid shows 48% identity. Two sets of human EST sequences present in the data banks were proposed by Treisman et al. (36) to be potential human homologs of eyelid; p270 is the origin of one of these EST sequences. However, the high degree of identity between p270 and eyelid does not extend to sequences N-terminal of the ARID region.
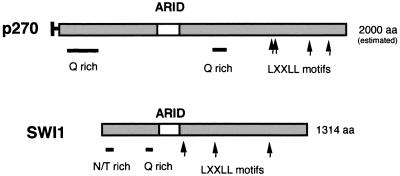
We were particularly interested by the appearance of yeast SWI1 (ADR6) (25, 27) in the list of proteins showing a relationship to p270. Our initial characterization of p270 shows that p270 is an integral member of human SWI-SNF complexes (6). Human homologs of yeast SWI2, SWI3, and SWP73 and SNF5 are known (23, 24, 39, 40), but a homolog of SWI1 has not previously been identified. Computer comparisons do not identify significant stretches of sequence homology between p270 and SWI1 outside of the ARID regions. However, a schematic comparison of p270 and SWI1 (Fig. 5) reveals additional shared, potentially functional motifs. Both contain Q-rich regions. Such regions have been implicated in transactivation functions (31). Both also contain multiple copies of amino acid motif LXXLL, which has been shown to be critical for the binding of a variety of nuclear proteins to liganded nuclear hormone receptors (10). The presence of p270 in human SWI-SNF complexes, combined with the ARID motif and other sequence similarities shared by p270 and SWI1, suggests that p270 is a human counterpart of the yeast SWI1 product.
FIG. 5.
Alignment of p270 and SWI1. p270 and SWI1 have an overall similarity of structure, in that they both contain ARID regions and they both have multiple LXXLL motifs (which are implicated in binding to nuclear hormone receptors), as well as Q-rich regions, which are often associated with transactivation regions. SWI1 has an unusual asparagine- and threonine-rich stretch near the N terminus; we do not yet know if this feature also occurs in p270. Other than these common features, p270 and SWI1 do not show direct sequence homology.
p270 contains an ARID-dependent DNA binding activity.
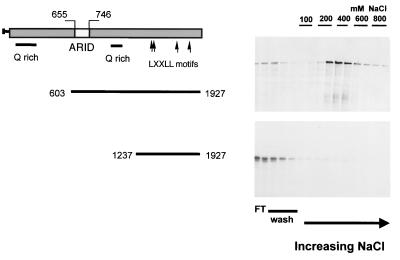
To assess the DNA binding ability of p270, in vitro-translated, 35S-labeled p270 peptides were applied to native DNA cellulose columns (Fig. 6). Initially, we tested p270 peptide p270603–1927 consisting of residues 603 to 1927 of the 1,927-residue open reading frame product. This peptide contains an intact ARID region and was retained on the column with an elution peak in the 400 mM fractions (Fig. 6, upper panel). Related peptide p2701237–1927 is N-terminally truncated relative to p270603–1927. It retains the LXXLL motifs but not the ARID region. This peptide showed no ability to bind native DNA; it passed through the column and was recovered in the wash fractions (Fig. 6 lower panel).
FIG. 6.
DNA binding activity in p270. Reticulocyte lysate-translated 35S-labeled p270 peptides were applied to native DNA cellulose columns. The columns were washed with loading buffer and then with increasing salt concentrations. Aliquots of the flowthrough (FT), wash, and eluted fractions were analyzed on SDS gels and visualized by autoradiography. A p270 peptide encompassing residues 603 to 1927 and containing an intact ARID region was retained on the column (upper panel). A related peptide, encompassing residues 1237 to 1927, with the ARID domain deleted, passed through the column and was recovered primarily in the flowthrough and wash fractions (lower panel). The sequences encompassed by the p270 peptides are indicated schematically to the left of the gels.
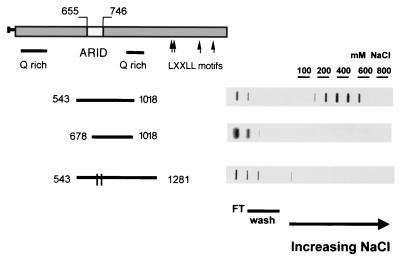
To test more specifically the relationship between the ARID region and p270 DNA binding activity, we constructed and tested additional peptides. Fractions from the DNA cellulose columns were assessed in a filter binding assay to facilitate quantification of the DNA binding activity. Peptide p270543–1018 contains the entire ARID region, which extends from approximately residue 655 to residue 746. This smaller peptide was retained on the DNA cellulose column with essentially the same affinity as the longer peptide p270603–1927 and showed an elution peak in the 400 mM fractions (Fig. 7, upper panel). The peptide p270678–1018 is N-terminally truncated relative to p270543–1018. As a result of this truncation, the ARID region is incomplete, missing the first 23 of the 91 residues that span the core consensus sequence. The resultant peptide is almost completely devoid of DNA binding activity. Like p2701237–1927, it is recovered almost entirely in the wash fractions (Fig. 7, middle panel). Quantification of the counts per minute recovered in respective fractions is indicated in the legend to Fig. 7.
FIG. 7.
ARID-dependent DNA binding activity in p270. Reticulocyte lysate-translated 35S-labeled p270 peptides were applied to native DNA cellulose columns and eluted as described for Fig. 6. Aliquots of the flowthrough (FT), wash, and eluted fractions were analyzed in a protein filter binding assay and visualized by autoradiography. The sequences encompassed by the p270 peptides are indicated schematically to the left of the gels. The parallel lines in the bar corresponding to the peptide comprising residues 523 to 1281 represent the two single-residue substitutions at positions 715 and 738. The elution profiles were quantified by phosphorimaging. With the p270543–1018 peptide (upper panel), 31.3% of the total counts per minute were recovered in the flowthrough and wash fractions, while 45.4% eluted in the 200 to 600 mM salt fractions. With peptide p270678–1018 (middle panel), 95.3% of the total counts per minute were recovered in the flowthrough and wash fractions while 2.28% eluted in the 200 to 600 mM salt fractions. With peptide p270523–1281WY (lower panel), 76.2% of the total counts per minute were recovered in the flowthrough and wash fractions while 16.7% eluted in the 200 to 600 mM salt fractions.
The requirement for the ARID region in the DNA binding function was also tested by analysis of amino acid substitution mutations. Two of the most highly conserved amino acids in the core ARID motif are the tryptophan at position 715 in the p270 sequence (Fig. 3 and 4) and the tyrosine at position 738. Site-directed mutagenesis was used to show that each of these residues was changed to an alanine in a background construct expressing residues 543 to 1281. The wild-type p270543–1281 peptide showed the same DNA binding affinity as the other ARID-containing peptides tested, with a peak elution in the 400 mM fractions (not shown). Individually, each of the substitutions resulted in a partial impairment of DNA binding activity (not shown). In combination, these two substitutions almost completely abrogated the DNA binding activity of the p270 peptide (Fig. 7, lower panel; quantification is reported in the legend).
The marked effect of the limited deletion or of the two single-amino-acid changes in the p270 ARID domain on the DNA binding activity of p270 supports the conclusion that the DNA binding activity observed in p270 derives from the presence of the ARID consensus. Analysis with p270-specific antibodies of fractions from whole-cell lysates passed over native DNA cellulose columns showed p270 eluting with a profile similar to that seen with the in vitro-translated ARID-containing peptides (not shown).
The p270 ARID region does not show a preference for AT-rich sequences.
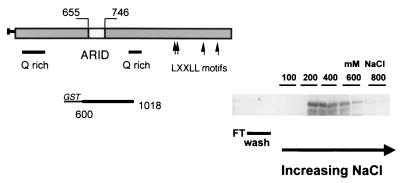
To determine whether the ARID region of p270 has sequence-specific DNA binding activity, a GST fusion protein containing p270 residues 600 to 1018, GSTp270600–1018, was subjected to a PCR-based selection and amplification procedure directly analogous to that used to assess the DNA binding properties of DRI. Before beginning the PCR selection, we first verified that the GST modification does not abrogate the DNA binding activity of the p270 peptide. The p270 fusion protein was eluted from the glutathione-Sepharose beads, passed over a DNA cellulose column, and collected in a series of increasing salt fractions in the manner described above. Aliquots of each fraction were separated on SDS-polyacrylamide gels. The electrophoresed proteins were transferred to nitrocellulose blots and probed with GST-specific antibodies. The DNA binding profile of the fusion protein (Fig. 8) was essentially the same as that seen with the reticulocyte lysate-translated peptides shown above. GST alone passed through the columns and was recovered entirely in the flowthrough and wash fractions (not shown).
FIG. 8.
DNA binding activity of a GST-p270 fusion peptide. GST fusion protein GSTp270600–1018 was passed over a DNA cellulose column and collected in a series of increasing salt fractions as described for Fig. 6. Aliquots of each fraction were separated on SDS-polyacrylamide gels. The electrophoresed proteins were transferred to nitrocellulose blots, probed with GST-specific antibodies, and visualized by chemiluminescence. The fusion protein eluted primarily in the 400 mM salt fractions. The sequences encompassed by the fusion peptide are indicated schematically to the left of the gel.
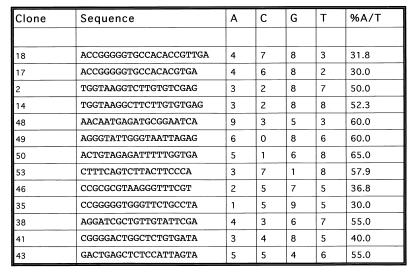
Having verified that the fusion protein binds to DNA, we used this construct as the basis for the PCR-based selection of preferred DNA binding sequences. The fusion protein, bound to glutathione-Sepharose beads, was incubated with a pool of oligomers containing stretches of 20 random bases. Unbound oligomers were washed off and the remaining pool was amplified by PCR. This process was repeated four times, and the remaining oligomers were cloned and sequenced. In this original analysis, unbound oligomers were washed off in 100 mM NaCl in a standard cyclic amplification and selection procedure. When this protocol failed to reveal any sequence preference in the final cloning, the procedure was repeated at higher stringency. In the second protocol unbound oligomers were washed off in 800 mM NaCl and the process was repeated through six cycles. Despite this high-stringency selection, the resulting clones revealed no preference for specific sequences. The resulting sequences are shown in Fig. 9. Twenty-five independent clones were sequenced and yielded 13 unique sequences. The overall percentage of A+T residues in the 13 unique sequences is 47.9%. This is only marginally higher than the AT content of the original oligonucleotide pool, which we estimate to be 44.0% based on sequences obtained from 16 oligonucleotides cloned without selection from the original batch (not shown). The selected sequences were analyzed by the Wisconsin GCG PileUp program to search for similarity, but no common motifs were discernible.
FIG. 9.
Sequences of oligonucleotides bound by p270. Shown are sequences bound by the p270 fusion protein after six cycles of amplification and selection. The overall percentage of AT residues is 47.9%. The sequences were analyzed by the Wisconsin GCG PileUp program, but no common motifs were detected.
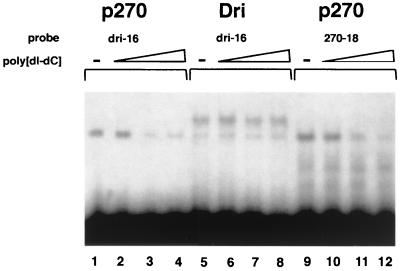
These results are in sharp contrast to the strong sequence specificity of DRI. We therefore used EMSAs to compare p270 and DRI directly. The GST-p270600–1018 fusion protein was compared with the GST-dri258–410 fusion protein, the construct used to determine DRI specificity. Lane 1 in Fig. 10 shows GST-p270600–1018 in the absence of poly(dI-dC) shifting a probe containing the AT-rich, DRI-selected consensus sequence (AATTAA). Lanes 2 through 4 show aliquots of the same reaction mixture incubated with increasing amounts of cold poly(dI-dC). The poly(dI-dC) competes effectively with the DRI consensus sequence for binding to the p270 peptide. Lanes 5 through 8 show the same series of reactions, except that GST-dri258–410 was used in place of the p270 peptide. When the DRI-selected sequence is bound to DRI, poly(dI-dC) does not compete effectively. Lane 9 shows the GST-p270600–1019 fusion protein binding to a probe corresponding to clone 18 from the p270 selection. This probe is competed by poly(dI-dC) as effectively as the probe based on the DRI-selected sequence (compare lanes 10 to 12 with lanes 2 to 4). Thus, in direct comparison to DRI, p270 does not show a specific preference for this AT-rich sequence. Moreover, p270 does not show specific preference for a sequence obtained from the p270 selection protocol. The DNA binding properties of p270 indicate that AT preference is not an intrinsic property of ARID regions. The consensus ARID sequence appears to specify general DNA binding properties, while any preference for specific sequences is likely to derive from other residues in the ARID region.
FIG. 10.
p270 shows no sequence preference in an EMSA. The DNA binding activity of p270 (lanes 1 to 4 and 9 to 12) and DRI (lanes 5 to 8) was assayed by gel shift using 32P-labeled oligonucleotide probes p270.18 (lanes 1 to 4) and dri.16 (lanes 5 to 12). Competition assays included unlabeled poly(dI-dC) at a molar excess of approximately 20:1 (lanes 2, 6, and 10), 140:1 (lanes 3, 7, and 11), and 600:1 (lanes 4, 8, and 12).
DISCUSSION
p270 was first recognized through its shared antigenic specificity with p300 and CBP. Cloning and sequencing p270, however, reveals that this protein is most closely related to an emerging family of proteins distinguished by the presence of a novel DNA binding region, termed ARID. The functions of ARID proteins as they are understood to date (summarized in Table 1) suggest that this family consists of transcriptional activators, coactivators, and corepressors. Bright and DRI are the founding members of the ARID family, and both bind to AT-rich sequences. In agreement with the expectation that the ARID consensus specifies DNA binding activity, p270 has an intrinsic ability to bind linear duplex DNA. This activity is dependent on the integrity of the consensus sequence; replacement of two consensus residues with alanine severely disrupts the DNA binding activity. However, unlike Bright and DRI, p270 does not bind preferentially to AT-rich sites; indeed p270 shows no sequence preference in its DNA binding activity. This property of p270 reveals that the ARID consensus sequence does not in itself specify a preference for AT-rich DNA. More likely, a binding site preference, or the lack thereof, is determined by the nature of the less conserved residues within ARID regions. It is unlikely that sequence specificity is determined by residues very distal to the ARID region, because the sequence specificity of DRI is contained within a 152-residue peptide (dri258–410) containing only the ARID region and residues closely flanking it (9). Likewise, the sequence specificity of MRF-2 is contained within a 108-residue peptide encompassing only the ARID region (35).
This demonstration that the ARID consensus does not in itself specify a preference for AT-rich DNA is consistent with the first structural studies on ARID proteins. Solution structures have been published for both MRF-2 (46) and DRI (13). Although the proposed structures differ in some respects, both are consistent with a wider range of DNA binding activity than a simple preference for AT-rich sites. The solution structure for the ARID region of MRF-2 suggests that ARID regions have features resembling the helices in homeodomains, but it also indicates that the structure shares a more significant degree of homology with DNA binding proteins such as DNA replication and repair nucleases and polymerases, which do not exhibit sequence specificity. The presumptive DNA contact residues in the ARID structure of DRI (the MKY sequence immediately following the invariant Y corresponding to position 738 in p270) are conserved between Bright and DRI but are not conserved in ARID proteins generally or in p270. It is clear from Fig. 4 that the region of high homology between Bright and DRI extends both N terminally and C terminally to the core consensus sequence. These “extended” ARID sequences appear to define a subgroup within the ARID family and may contribute to sequence-specific binding (9, 13, 19). The behavior of p270 yields new insight into the properties of ARID family members outside the Bright and DRI subgroup.
A general DNA binding function for p270 is more consistent with the presumptive role of the mammalian SWI-SNF complexes, which have not shown sequence specificity in their DNA binding. These complexes contain an ATP-dependent nucleosome remodelling activity that is associated with general transcriptional activation. The source of the required ATPase activity in the complex is believed to be BRG1 or other human members of the yeast SWI2 family. p270 is stably associated with human SWI-SNF complexes in vivo, as indicated by immune complex isolation with antibodies directed against at least three different components of the complexes (6). Wang et al. (39, 40) have also noted a 250-kDa protein band associated with human SWI-SNF immune complexes. This BRG1-associated factor (BAF-250) corresponds to p270 based on our evaluation of BRG1-associated complexes using the same antibodies (6). Biochemical purification also yields a complex containing a 250- to 270-kDa protein (hSWI-SNF complex A) (20). An alternative purification procedure yields a complex without a 250- to 270-kDa band (HSWI-SNF complex B), which retains nucleosome remodelling activity in vitro (20, 30). Thus p270 is not required for this aspect of hSWI-SNF complex function.
The presence of similar proteins, p270 and SWI1, in both human and yeast SWI-SNF complexes argues that the function of these proteins is intrinsic to the respective complexes. The identification of p270 as a member of the emerging family of ARID-containing proteins suggests that p270 functions as a coactivator or corepressor, perhaps in combination with nuclear hormone receptors, as implied by the presence of the LXXLL motifs. SWI-SNF complexes are linked with regulation in response to multiple steroid hormone receptors (see, e.g., references 4, 8, 24, 26, and 45). However, the manner in which the function of the hSWI-SNF complexes is integrated with specific transactivation complexes is not at all clear. In a recent review, Kingston and Narlikar (17) suggested that hSWI-SNF complexes have a general nucleosome-remodelling activity that can be upregulated in response to various signals. This model proposes that the actual activation of any promoter depends on the presence in the cell of specific transcription factor complexes, which must gain access to the DNA during the dynamic process of nucleosome remodelling. The properties of p270 suggest that a protein with potential coactivator or corepressor activity is available within the hSWI-SNF complex to bind to the newly accessible DNA. p270 may play an intermediary role, possibly regulated by interaction with specific activators, such as nuclear hormone receptors, before true activation complexes, including such coactivators as p300 and CBP, are formed at specific promoter sites.
ACKNOWLEDGMENTS
We thank Robert Saint and Phil Tucker for reagents and helpful discussions and Susan Rhodes, Simon Gregory, and Peter S. White for help with sequence analysis. We also thank Danny Orozco, Kimberly Day, Valery Audige, Patty Baxter, and Annette Heagy for excellent technical assistance and Scott Shore, Xavier Graña, Atul Kumar, George Beck, and Xiaomei Wang for additional advice and discussions.
This work was supported by PHS grant CA53592 (E.M.) from the NIH.
REFERENCES
- 1.Agulnik A I, Bishop C E, Lerner J L, Agulnik S I, Solovyev V V. Analysis of mutation rates in the SMCY/SMCX genes shows that mammalian evolution is male driven. Mamm Genome. 1997;8:134–138. doi: 10.1007/s003359900372. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Boguski M S, Gish W, Wootton J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lippman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallas P B, Yaciuk P, Moran E. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J Virol. 1997;71:1726–1731. doi: 10.1128/jvi.71.2.1726-1731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallas P B, Cheney I W, Liao D, Bowrin V, Byam W, Pacchione S, Kobayashi R, Yaciuk P, Moran E. p300/CREB binding protein-related protein p270 is a component of mammalian SWI-SNF complexes. Mol Cell Biol. 1998;18:3596–3603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fattaey A R, Helin K, Dembski M S, Dyson N, Harlow E, Vuocolo G A, Hanobik M G, Haskell K M, Oliff A, Defeo-Jones D, Jones R E. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- 8.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 9.Gregory S L, Kortschak R D, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA binding proteins. Mol Cell Biol. 1996;16:792–799. doi: 10.1128/mcb.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heery D, Kalkhoven M E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 11.Herrscher R F, Kaplan M H, Lelsz D L, Das C, Scheuermann R, Tucker P W. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 12.Huang T H, Oka T, Asai T, Okada T, Merrills B W, Gertson P N, Whitson R H, Itakura K. Repression via differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1695–1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwahara J, Clubb R T. Solution structure of the DNA binding domain from Dead ringer, a sequence-specific AT-rich interaction domain (ARID) EMBO J. 1999;18:6084–6094. doi: 10.1093/emboj/18.21.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 15.Kent-First M G, Maffitt M, Muallen A, Brisco P, Schultz J, Ekenberg S, Agulnik A I, Agulnik I, Schramm D, Bavister B, Abdul-Mawgood A, Vandeberg J. Gene sequence and evolutionary conservation of human SMCY. Nat Genet. 1996;14:128–129. doi: 10.1038/ng1096-128. [DOI] [PubMed] [Google Scholar]
- 16.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 17.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 18.Kortschak R D, Reimann H, Zimmer M, Eyre H J, Saint R, Jenne D E. The human dead ringer/bright homolog, DRIL1: cDNA cloning, gene structure, and mapping to D19S886, a marker on 19p13.3 that is strictly linked to the Peutz-Jeghers syndrome. Genomics. 1998;51:288–292. doi: 10.1006/geno.1998.5259. [DOI] [PubMed] [Google Scholar]
- 19.Kortschak, R. D., P. W. Tucker, and R. Saint. ARID proteins come in from the desert. •••, in press. [DOI] [PubMed]
- 20.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 21.Lai A, Marcellus R C, Corbell H B, Branton P E. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene. 1999;18:2091–2100. doi: 10.1038/sj.onc.1202520. [DOI] [PubMed] [Google Scholar]
- 22.Lai A, Lee J M, Yang W-M, DeCaprio J A, Kaelin W G, Jr, Seto E, Branton P E. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol Cell Biol. 1999;19:6632–6641. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Hara P J, Horowitz H, Eichinger G, Young E T. The yeast ADR6 gene encodes homopolymeric amino acid sequences and a potential metal-binding domain. Nucleic Acids Res. 1988;16:10153–10169. doi: 10.1093/nar/16.21.10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostlund-Farrants A-K, Blomquist P, Kwon H, Wrange O. Glucocorticoid receptor-mediated response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 28.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodelling machine. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 29.Quinn J, Fryberg A M, Ganster R W, Schmidt M C, Petersen C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 30.Schnitzler G, Sif S, Kingston R. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 31.Seipel K, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shandala T, Kortshak R D, Gregory S, Saint R. The Drosophila dead ringer gene is required for early embryonic patterning through regulation of argos and buttonhead expression. Development. 1999;126:4341–4349. doi: 10.1242/dev.126.19.4341. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi T, Yamazaki Y, Katoh-fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi T, Bing-Kun C, Qiu Y, Sonobe H, Ohtsuki Y. Molecular cloning and expression of a novel human cDNA containing CAG repeats. Gene. 1997;204:71–77. doi: 10.1016/s0378-1119(97)00525-8. [DOI] [PubMed] [Google Scholar]
- 35.Thrower A R, Bullock G C, Bissell J E, Stinski M F. Regulation of a human cytomegalovirus immediate-early gene (US3) by a silencer-enhancer combination. J Virol. 1996;70:91–100. doi: 10.1128/jvi.70.1.91-100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treisman J E, Luk A, Rubin G M, Heberlein U. eyelid antagonizes wingless signaling during Drosophila development and has homology to the bright family of DNA-binding proteins. Genes Dev. 1997;11:1949–1962. doi: 10.1101/gad.11.15.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentine S A, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey A J. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez M, Moore L, Kennison J A. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126:733–742. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Calpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Goldstein A, Zong R T, Lin D, Neufeld E J, Scheuermann R H, Tucker P W. Cux/CDP homeoprotein is a component of NF-muNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription activator. Mol Cell Biol. 1999;19:284–295. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitson R H, Huang T, Itakura K. The novel Mrf-2 DNA-binding domain recognizes a five-base core sequence through major and minor-groove contacts. Biochem Biophys Res Commun. 1999;258:326–331. doi: 10.1006/bbrc.1999.0643. [DOI] [PubMed] [Google Scholar]
- 44.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfeld J, Burton J, Connell M, Copsey T, Cooper J, Coulson A, Craxton M, Dear S, Du Z, Durbin R, Favello A, Fraser A, Fulton L, Gardner A, Green P, Hawkins T, Hillier L, Jier M, Johnston L, Jones M, Kershaw J, Kirsten J, Laisster N, Latreille P, Lightning J, Lloyd C, Mortimore B, O'Callaghan M, Parsons J, Percy C, Rifken L, Roopra A, Saunders D, Shownkeen R, Sims M, Smaldon N, Smith A, Smith M, Sonnhammer E, Staden R, Sulston J, Thierry-Mieg J, Thomas K, Vaudin M, Vaughan K, Waterston R, Watson A, Weinstock L, Wilkinson-Sproat J, Wohldman P. 2.3 mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 45.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 46.Yuan Y-C, Whitson R H, Liu Q, Itakura K, Chen Y. A novel DNA-binding motif shares structural homology to DNA replication and repair nucleases and polymerases. Nat Struct Biol. 1998;5:959–964. doi: 10.1038/2934. [DOI] [PubMed] [Google Scholar]