Abstract
Small ruminants are suited to a wide variety of habitats and thus represent promising study models for identifying genes underlying adaptations. Here, we considered local Mediterranean breeds of goats (n = 17) and sheep (n = 25) from Italy, France and Spain. Based on historical archives, we selected the breeds potentially most linked to a territory and defined their original cradle (i.e., the geographical area in which the breed has emerged), including transhumant pastoral areas. We then used the programs PCAdapt and LFMM to identify signatures of artificial and environmental selection. Considering cradles instead of current GPS coordinates resulted in a greater number of signatures identified by the LFMM analysis. The results, combined with a systematic literature review, revealed a set of genes with potentially key adaptive roles in relation to the gradient of aridity and altitude. Some of these genes have been previously implicated in lipid metabolism (SUCLG2, BMP2), hypoxia stress/lung function (BMPR2), seasonal patterns (SOX2, DPH6) or neuronal function (TRPC4, TRPC6). Selection signatures involving the PCDH9 and KLH1 genes, as well as NBEA/NBEAL1, were identified in both species and thus could play an important adaptive role.
Subject terms: Population genetics, Animal breeding, Ecological genetics, Molecular biology
Introduction
Sheep and goats were among the first mammalian livestock species to be domesticated, in a process that started at ca. 10,000–9500 B.P. in Mesopotamia1. The subsequent expansion across the Mediterranean Basin via maritime transport included different waves of colonization. The remains testifying to these founding periods along the northern coasts of the western Mediterranean are dated from 8100 to 7700 B.P.2.
Waves of dispersal, which have dispatched small groups of individuals (founder effect) to very diverse habitats, are the starting point of the process that led to the emergence of breeds3. Traditional pastoralism, characterized by limited human intervention (i.e., “soft” artificial selection4), and in conjunction with a strong influence of natural selection, led over the millennia to populations genetically differentiated and adapted to various agro-climatic conditions5. Sheep and goats provide an interesting model for deepening our knowledge of adaptive mechanisms under extreme environmental conditions.
In industrialized countries, artificial breeding has intensified over the past 200 years4, focusing mainly on agronomic traits related to milk, meat production and fiber quality. Many breeds have experienced negative impacts on their ability to adapt, their level of hardiness and sometimes their fitness by crossbreeding and/or intensive artificial selection4, 6. In contrast, the so-called local breeds have remained at least partially, connected to their original environments. We suggest it is among these local breeds that selection signatures related to environmental adaptation will be most detectable. For example, studies on local sheep in Ethiopia7 and Tibet8, 9 have identified several genes involved in adaptation to thermal stress and hypoxia. Kim et al.10, considering both local goats and sheep of hot and arid environment, have highlighted genes implicated in the adaptation to thermal stress.
This study aimed at identifying the genomic bases of adaptation in local breeds of goats and sheep distributed along the Mediterranean arc, in Italy, the south of France and Spain. The area under consideration harbours contrasting environments, such as high altitude areas (Alps, Pyrenees, Corsica), coastal areas, wetlands and arid areas, with closely related sheep and goat breeds11–13. All these features provide an ideal situation for the identification of adaptation signatures.
We hypothesize that selection signatures relevant to environmental adaptation, will be more reliably identified by reference to the environmental conditions of the breed’s cradle of origin, rather than to those of the locations where the animals were actually sampled. Indeed, breeds of domestic animals are at the interface between anthropized and natural environments; their evolutionary trajectory is thus determined by both the environment and human societies. First, (i) sheep and goat breeds have originally emerged within socially structured human groups, sometimes referred to as clans or tribes, occupying a well-defined territory, and within which herds were transmitted from generation to generation14, 15. In industrialized countries, the traditional links between territory, social group and breeds has become distorted. Thus, herds of a given breed can be found outside of its original cradle. Hence, the search for a selection signature may be optimized by considering the environmental conditions of the breed’s cradle, where the genomes have been shaped over time, rather than the current location of herds. Moreover, (ii) we should consider that traditional pastoralism was largely based on transhumance. During these periods, the animals are kept outdoors, most often in mountain altitude summer pastures, such that the strength of natural selection is particularly pronounced. Even if transhumance practices are increasingly abandoned16, 17, they have fundamentally shaped the genetic adaptations of sheep and goats.
The objective of this study was to investigate the regions of the genome underlying environmental adaptations, i.e. selection signatures linked to environmental variables. Selection signatures are identified as genomic areas comprising a number of neighboring markers under selection. We used the PCAdapt18 and LFMM19 methods. PCAdapt uses principal component analysis to describe population structure and identifies candidate markers as outliers in terms of inferred population structure. The LFMM approach searches for significant associations with environmental factors while controlling for neutral population structure. These approaches were coupled with a thorough analysis of the literature to identify the signatures including genes most likely related to environment adaptations. Moreover, we used datasets that have been previously investigated, in order to assess if the use of historical and anthropological dimensions of breeds improves the detection of selection signatures. We reasoned that (i) the inclusion of authentic local breeds in the analysis will limit the background noise caused by breeds for which the link to the environment is weak or has eroded, for instance by crossbreeding or migration; and (ii) targeting the geographical area in which populations have evolved rather than the present distribution will optimize the identification of adaptive genomic regions. Next, the proposed approach was implemented to search for selection signatures underlying local adaptation. We attached particular importance to the selection signatures involving homologous genomic regions in sheep and goats.
Results
Breed analyses
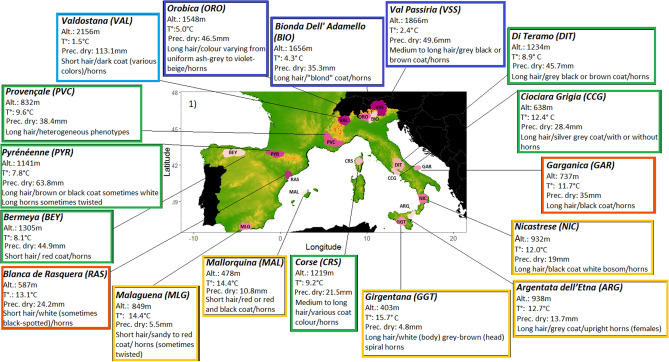
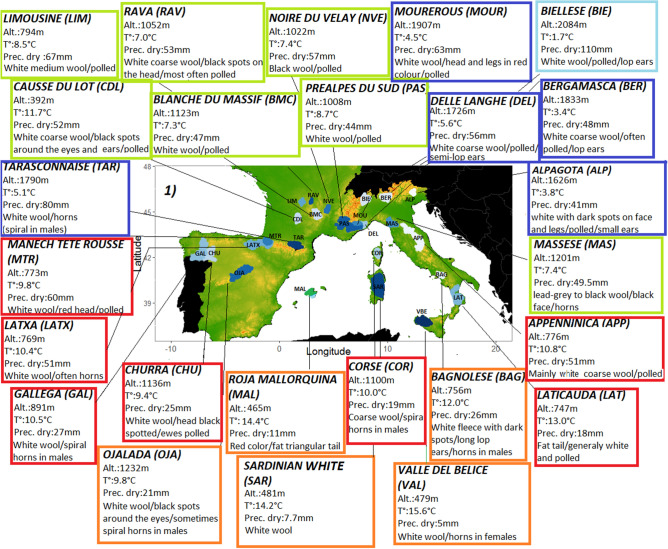
The dataset includes 17 goat breeds and 25 sheep breeds, selected on the basis of historical and anthropological documentation (Table 1, see Supplementary Table 1 for the initial list of 83 breeds and “Material and methods” section for an explanation of the choice of breeds for analysis). Figures 1 and 2 show the geographical cradles of the different breeds and provide details on the phenotypes of the animals and the environmental conditions at the cradle level.
Table 1.
Description of datasets.
| Breed name | Code | Country | Cradle location |
|---|---|---|---|
| Goat dataset | |||
| Bionda dell Adamello | BIO (n = 24) | Italy | The Alpine massif |
| Valdostana | VAL (n = 24) | Italy | The Alpine massif |
| Orobica | ORO (n = 24) | Italy | The Alpine massif |
| Val Passiria | VSS (n = 24) | Italy | The Alpine massif |
| Di Teramo | DIT (n = 24) | Italy | Abruzzo massif |
| Ciociara Grigia | CCG (n = 19) | Italy | Aurunci Mountains |
| Garganica | GAR (n = 20) | Italy | The Gargano promontory |
| Argentata Dell’Etna | ARG (n = 25) | Italy | Mount Etna |
| Nicastrese | NIC (n = 25) | Italy | The province of Catanzaro |
| Girgentana | GGT (n = 30) | Italy | The Agrigento coast in Sicily |
| Corse | CRS (n = 29) | France | Corsican massifs |
| Provençale | PVC (n = 18) | France | The "Provence of hills" |
| Pyrénéenne | PYR (n = 25) | France | Pyrenees |
| Bermeya | BEY (n = 24) | Spain | Asturias |
| Blanqua De Rasquera | RAS (n = 20) | Spain | Southern Catalonia |
| Malaguena | MLG (n = 41) | Spain | Malaga |
| Mallorquina | MAL (n = 20) | Spain | Island of Mallorca |
| Sheep dataset | |||
| Manech Tête Rousse | MTR (n = 25) | France | Basque hillsides |
| Tarasconnaise | TAR (n = 15) | France | Central Pyrenees |
| Causse Du Lot | CDL (n = 20) | France | Plateaus of the Lot |
| Limousine | LIM (n = 18) | France | Millevaches plateau |
| Rava | RAV (n = 20) | France | The Massif Central |
| Blanche Du Massif | BMC (n = 20) | France | The Massif Central |
| Noire Du Velay | NVE (n = 19) | France | The Massif Central |
| Préalpes Du Sud | PAS (n = 17) | France | The Southern Alps |
| Mourerous | MOUR (n = 16) | France | The Maritime Alps |
| Corse | COR (n = 16) | France | The Corsican massifs |
| Delle Langhe | DEL (n = 24) | Italy | The Alpine massif |
| Biellese | BIE (n = 21) | Italy | The Alpine massif |
| Bergamasca | BER (n = 24) | Italy | The Alpine massif |
| Alpagota | ALP (n = 24) | Italy | The Alpine massif |
| Massese | MAS (n = 24) | Italy | The Alpi Apuane mountains |
| Appenninica | APP (n = 24) | Italy | The Apennine mountains |
| Laticauda | LAT (n = 24) | Italy | Campania and Calabria |
| Bagnolese | BAG (n = 23) | Italy | Campania |
| Valle Del Bellice | VAL (n = 24) | Italy | Sicily |
| Sardinian White | SAR (n = 24) | Italy | Sardinia |
| Latxa | LATX (n = 24) | Spain | Pyrenees |
| Gallega | GAL (n = 27) | Spain | Gallicia |
| Churra | CHU (n = 30) | Spain | Castile and León |
| Ojalada | OJA (n = 24) | Spain | The province of Soria |
| Roja Mallorquina | (MAL, n = 28) | Spain | Island of Mallorca |
Breeds, number of individuals, cradle location.
n: number of individuals.
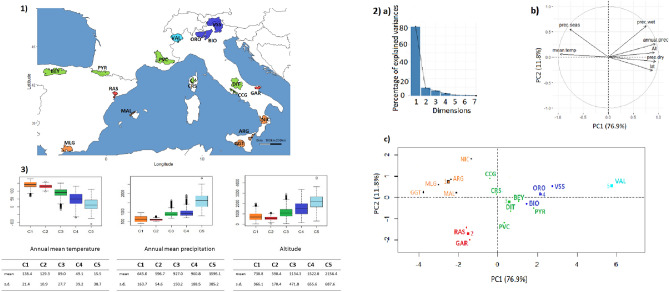
Figure 1.
Description of goat cradles as a function of environmental variables: synthetic descriptions of cradles and some characteristic breed phenotypic traits. The breed labels are color coded according to the clustering ranking defined by the PCA/HCPC analysis shown in Fig. 4.
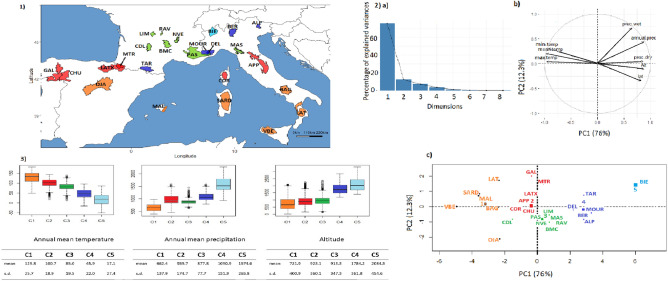
Figure 2.
Description of sheep cradles as a function of environmental variables: synthetic descriptions of cradles and some characteristic breed phenotypic traits. The breed labels are color coded according to the clustering ranking defined by the PCA/HCPC analysis shown in Fig. 6.
For 15 of the 42 selected sheep and goat breeds considered, we found documented evidence targeting ancestral populations at their origin, dating back several millennia (see Supplementary Table 2 for historical references). The remaining breeds have a history of at least several centuries, with the exception of 4 Italian goat breeds (DIT, CCG, GAR, ARG) and 3 Italian sheep breeds (DEL, BIE, VAL), which were recognized about a hundred years ago under their current names, but which have an ancient history characterized by mixtures of populations native to the territories considered. Seventeen breeds were classified as “critical” or “endangered” according to FAO20. Considering jointly the goat and sheep breeds, 30 breeds have been described as showing adaptations to stressful environments and 29 are, or were, transhumant. For most transhumant breeds the transhumance is, or was, vertical, so we included their summer mountain pasture in the geographical definition of the cradle (see Supplementary Table 2).
The Admixture analysis is detailed in Supplementary text 1. For both goats and sheep (Figure 3 in the Supplementary text 1), the cross-entropy curve decreases as K increases, which is characteristic of a distance isolation pattern21, moreover, the Mantel tests postulated distance isolation patterns (p value = 0.049 for goats and 0.001 for sheep). In this case, to correct for the limited genetic structure, we used K = 1 and K = 2, in the LFMM analyses, as recommended by Dalongeville et al.22, De Kort et al.23 and Capblancq et al.24.
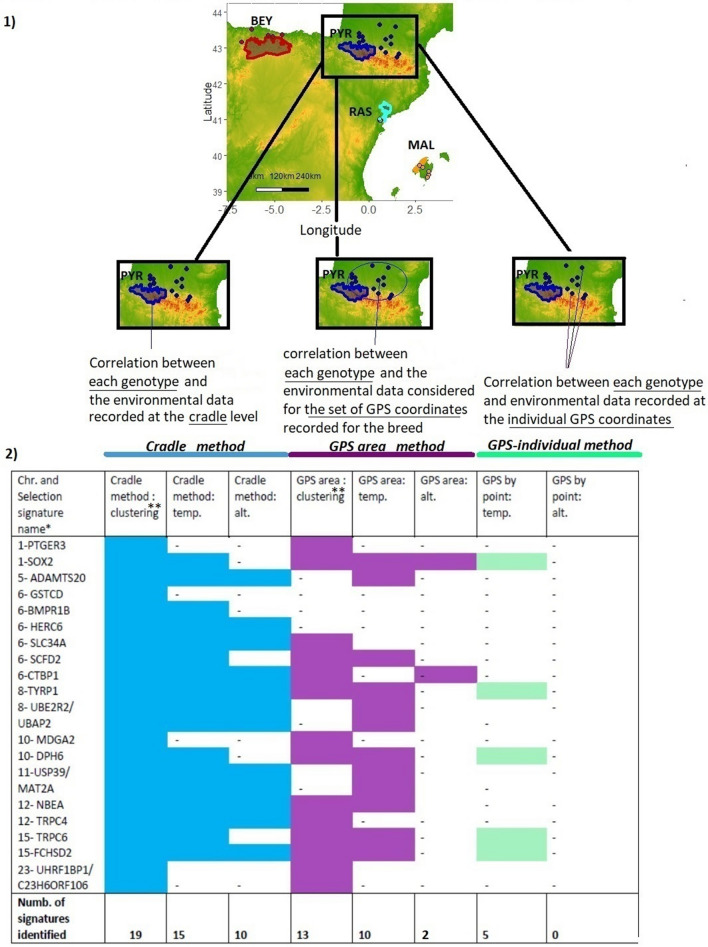
Comparison of approaches
We performed the LFMM analyses on the goat dataset using three approaches for characterising the environment of each breed (see “Identification and analysis of genomic selection signatures” and “Experimental test of the method” sections in “Material and methods” section and Fig. 3). The poor results obtained with the Annual Mean Precipitation are not shown (i.e., the different methods could only reveal two signatures each, with NBEA identified in all cases). Notably, for the Altitude, the individual GPS method did not detect any signatures; the GPS area method identified only two signatures, whereas 10 signatures were recorded with the cradle method. For the Mean Annual Temperature, five signatures were observed with the individual GPS method, 10 with the GPS area method, and 15 by the cradle method. The process of clustering the environments allowed the detection of 13 signatures via the GPS areas and 19 (maximum value) via the cradles (see details in Supplementary Figures 3 and 4).
Figure 3.
Experimental test of the method. The signature search was done via LFMM. The environment was characterized either at the level of the GPS coordinates (individual GPS method) or considering all the GPS points recorded for a given breed (GPS area method), or at the level of the cradle (cradle method). The environmental variables Altitude and Mean Annual Temperature were considered individually. Chr.: chromosome number. *Selection signature are named according to the gene(s) most likely targeted by the selection process, taking into account the distance of the SNPs from the genes, the associated p values and the number of SNPs near or in the gene(s); it is possible to know in detail the genes associated with each selection signature (see Supplementary Table 3). **Clustering: the environment was characterized considering all the variables at the same time and according to the PCA/HCPC process.
Goat dataset: track for selection signatures
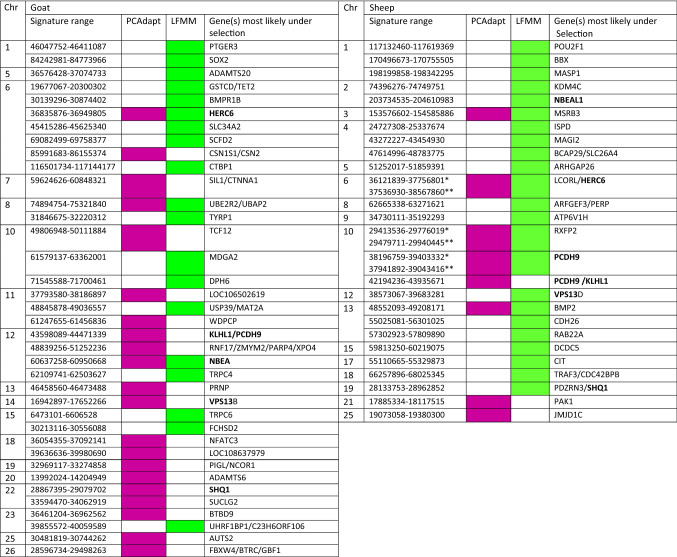
From the 37 signatures, 21 were identified by PCAdap, 19 by LFMM; 3 signatures were supported by both methods (Table 2). Seven signatures were observed on chromosome 6, and four on chromosome 12. Genes associated with these signatures have been documented in the literature (Supplementary Table 3). Of the 21 signatures highlighted by PCAdapt, 15 were found to be involved in adaptive processes and of the 19 signatures highlighted by LFMM, 15 were linked to adaptive processes according to the literature.
Table 2.
Selection signatures identified in goats and sheep by LFMM and PCAdapt using the cradle method.
Each signature was named according to the gene(s) most likely targeted by the selection process, taking into account the distance of the SNPs from the genes, the associated p values and the number of SNPs near or in the gene(s). All annotated genes associated with each selection signature are highlighted in Supplementary Tables 3 and 4.
*PCAdapt signature; **LFMM signature; Chr.: chromosome number, in bold genes identified in both datasets.
The LFMM analysis was based on the PCA (Principal Component Analysis)/HCPC (Hierarchical Clustering on Principal Components) ranking in which the cradles were clustered according to the similarity of their environments (Fig. 4). The PCA analysis was largely driven by the first component (76.9% of variance explained by PC1). The cradles appeared to be distributed along a South–North axis, following a decreasing temperature and aridity gradient towards the negative values of PC1, and an increasing altitudinal gradient towards the positive ones. HCPC clustering, indicating K = 5 as the optimal number of clusters, was as follows: the first group (1) clustered breeds from southern Italy, Sicily, southern Spain and the island of Majorca, i.e. regions with high annual mean temperatures and very low rainfall during the dry season. Group (2) clustered the Spanish (Blanca de Rasquera, RAS) and Italian (Garganica, GAR) breeds, raised under low rainfall also during the wet season. The third group (3) clustered Spanish, French and Italian goat breeds, in an intermediate position with regard to the aridity gradient during the dry season. Groups (4) and (5) corresponded to Italian breeds in regions of high altitude, low average temperatures and high precipitation (Fig. 4). PCA/HCPC processed with environmental data recorded at the GPS area level (see Supplementary Figure 4) also showed five clusters that followed a temperature and precipitation gradient, but without an altitude gradient (Supplementary Figure 4.3), resulting in a different ranking clustering than the one obtained by the cradle method (Fig. 4) and a lower number of signatures highlighted by LFMM (Fig. 3).
Figure 4.
Goat cradles analyses as a function of environmental variables. (1) Mapping display of the cradles coloured according to the result of the clustering. (2) HCPC analysis. (a) Scree plot. (b) PCA correlation circle. (c) PCA score plot of goat cradles. Colours represent the main clusters obtained by HCPC (K = 5). PC 1 and PC 2 represent respectively principal components 1 and 2; numbers in brackets show the variance explained by each PC. (3) Boxplot representation for the variables Mean Annual Temperature, Mean Annual Precipitation and Altitude for the different HCPC groups and associated numerical values. Alt.: Altitude in meters, T°: Temperature in °C × 10, prec.: precipitation in millimeters, min.: minimal, max.: maximal, s.d.: standard deviation.
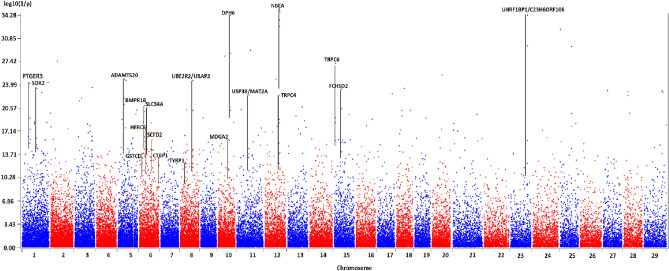
The LFMM analysis, based on the cluster ranking obtained by the cradle method (Fig. 5), showed several strong signals near genes that have been implicated in adaptation: SOX2, ADAMTS20, UBE2R2/UBAP2, DPH6, NBEA, TRPC4, TRPC6, and UHRF1BP1/C23H6ORF106. Strong signals detected by PCAdapt analysis and also found in the literature as implicated in adaptive processes corresponded to the genes SIL1/CTNNA1, UBE2R2/UBAP2, SHQ1, SUCLG2 as well as a large area on chromosome 12 containing RNF17, ZMYM2, PARP4 and XPO4 and the vast intergenic area between KLH1 and PCDH9. A particularly strong signal was found on chromosome 18 at locus LOC108637979. The detailed results can be viewed in Supplementary Tables 3, 5 and 6.
Figure 5.
Genome scan for selection signatures in local goat breeds obtained by the LFMM approach and via the cradle method. Each signature was named according to the gene(s) most likely targeted by the selection process, taking into account the distance of the SNPs from the genes, the associated p values and the number of SNPs near or in the gene(s).
Sheep dataset: track for selection signatures
From the 26 selection signatures, 8 were identified by PCAdapt, 23 by LFMM; 5 signatures were detected by both methods (Table 2). Genes associated with these signatures have been documented in the literature (Supplementary Table 4), all PCAdapt signatures and 18 out of 23 LFMM signatures could be linked to environmental adaptation.
Figure 6 shows the PCA/HCPC results used for the LFMM analysis. As in Fig. 4 for goats, PC1 accounted for a large part of the variation (76.0%) and showed similar South-North gradients of temperature and aridity (Fig. 6). The optimal K = 5 generated the following clusters: group (1) concerned regions with highest aridity and temperature levels, including the Sicilian, Southern Italian, the island of Majorca and Central Spanish breeds. Groups (2) and (3) were in an intermediate position, in term of dryness and wetness. Group (2) corresponded to regions showing slightly higher average temperatures than group (3) and high levels of precipitation during the wet season (especially for Gallega GAL, Manech Tête Rousse MTR and Latxa LATX), while group (3) included regions of higher altitudes (except for Causse du Lot CDL). Groups (4) and (5), including the Pyrenean and Alpine breeds of the three countries, corresponded to areas of high altitudes and significant precipitation levels (Fig. 6). LFMM on the basis of this cluster ranking obtained by the cradle method (Fig. 7) showed several strong signals near genes that previously have been implicated in environmental adaptation: NBEAL1, MSRB3, RXFP2, BMP2. Several other regions identified by LFMM included genes not previously been implicated in environmental adaptation (Supplementary Table 4).
Figure 6.
Sheep cradles analyses as a function of environmental variables. (1) Mapping display of the cradles coloured according to the result of the clustering. (2) HCPC analysis. (a) Scree plot. (b) PCA correlation circle. (c) PCA score plot of goat cradles. Colours represent the main clusters obtained by HCPC (K = 5). PC 1 and PC 2 represent respectively principal components 1 and 2; numbers in brackets show the variance explained by each PC. (3) Boxplot representation for the variables Mean Annual Temperature, Mean Annual Precipitation and Altitude for the different HCPC groups and associated numerical values. Alt.: Altitude in meters, T°: Temperature in °C × 10, prec.: precipitation in millimeters, min.: minimal, max.: maximal, s.d.: standard deviation.
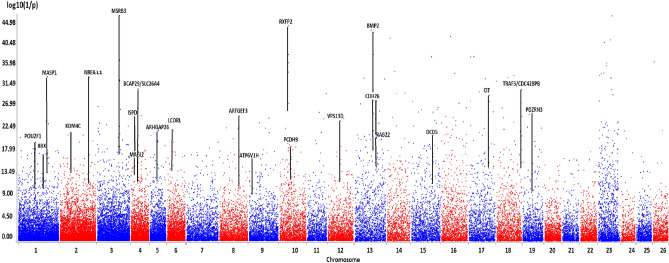
Figure 7.
Genome scan for selection signature in local sheep breeds obtained by the LFMM approach and via the cradle method. Each signature was named according to the gene(s) most likely targeted by the selection process, taking into account the distance of the SNPs from the genes, the associated p values and the number of SNPs near or in the gene(s).
The strongest PCAdapt signals corresponded to: PAK1 and the KLHL1/PCDH9 intergenic region, both described previously as being involved in environmental adaptation. A broad signal was on chromosome 6, near the genes HERC6, SPP1, LAP3, also previously involved in adaptation. The detailed results can be viewed in Supplementary Tables 4, 5 and 7.
Comparison of sheep and goat signatures
The genes NBEA (NBEA in goat and NBEAL1 in sheep), SHQ1 and VPS13 genes (goat VPS13B and sheep VPS13D), the regions containing HERC6 and ABCG2 as well as the KLHL1/PCDH9 intergenic region appeared to be under selection in both goat and sheep (Table 2). Most of the selection signatures (74%), or genes included in the signatures, were found in the literature on mammals and birds as being related to environmental adaptation (Supplementary Tables 3 and 4). Of these, the most noteworthy include the following: 16 goat and 14 sheep selection signatures previously identified in Chinese sheep adapted to extreme environments9, four signatures in goats and three in sheep found in Ethiopian sheep kept at high-altitude7 and three goat and six sheep signatures detected in Egyptian sheep and goats from arid environments10.
Discussion
This study proposed a new genomic landscape approach for the identification of selection signatures in livestock breeds. The key aspects of this approach are (i) filtering the breeds to be included in the study and (ii) matching the genotypes to the environmental conditions recorded not at the current coordinates of the individuals, but at the level of the areas identified as their cradle of origin.
Performance of cradle versus GPS method
We targeted French, Spanish and Italian breeds with strong historical links to their territory of origin, in order to build datasets for analysis. For most of the breeds, this link was present for several centuries. The majority of the 63 signatures identified in the current study had not been detected in the initial studies of these datasets24–30. A high proportion (74%) of the highlighted selection signatures were previously identified by studies on environmental genetic adaptation in birds and mammals from different parts of the world, providing support for our approach. Further corroboration came from the many signatures identified in our study that have been previously reported in sheep or goats from extreme environments in different parts of the world (7, 9, 10, 31, 32, see Supplementary Tables 3 and 4 for a complete list), and also from the several signatures found in both sheep and goats. We applied particularly stringent screens and reinforced our results with a thorough analysis of the literature in order to limit false positives as much as possible. Although our results suggest that the conducted approach can complement classical approaches by providing new results, formal mathematical modeling as well as the identification of candidate mutations and the appropriate functional analyses are required to validate the results.
Results from our approach highlighted a few points. First, the vast majority of small ruminant breeds have been shaped by traditional transhumant breeding practices. However, the individual GPS method will be able to consider summer grazing areas (i) only for breeds and herds always managed according to these practices while transhumance practices are declining and (ii) only if the sampling is carried out during the summer period. Consistent with this, we observed that GPS methods revealed very few signatures with the Altitude variable, whereas it was possible to detect 53% of the total signatures identified with the cradle approach by the consideration of this single variable. Secondly, considering the Mean Annual Temperature variable, (i) the difference between the number of signatures detected by the GPS methods and the cradle approach was clear but less pronounced than for the Altitude variable and (ii) the correlation of genotypes with environmental variables recorded at the level of the area defined by the set of the breed’s GPS coordinates (GPS area method) detected more signatures than considering the environmental variable at individual GPS coordinates (individual GPS method). This suggested that there are benefits of smoothing environmental data at the area level rather than considering it at individual points. Finally, the greatest number of selection signatures was identified by considering all the environmental variables at the same time and using the cradle method including transhumant pasture areas.
Thus, there are several strengths of our novel cradle method. First, taking into account the history of the breeds avoids including breeds that could interfere with the signal (i.e. breeds which do have a strong link with a given environment or whose link has been distorted). Secondly, in industrialized countries, agricultural intensification in recent decades has led to major shifts in practices, particularly the abandonment of transhumance, and also significantly changed the distribution areas of herds. Thus, the environmental characteristics recorded at the current GPS points are different from those of the original cradle (see Supplementary Text 2 for details). Considering the cradle of the breed rather than the current distribution thus links with more accuracy the genetic determinants of adaptation with the environment that has mainly shaped the breed's genome. Thirdly, we combined the PCAdapt algorithm, which is particularly suited to the detection of selection signatures in heterogeneous populations18, with LFMM, which identifies the links between genomic and environmental variation. On the basis of the scientific literature we eliminated PCAdapt signatures that appeared to be related to agronomic selection and researched the links established in the literature between other signatures and environmental adaptation. It can be hypothesized that PCAdapt signals not detected by LFMM reflect local adaptation to patchy environmental conditions unrelated to the environmental gradient. The two methods thus provide complementary information. It can be noted that the cradles were organized in both datasets according to a gradient of temperature and altitude, as shown by the PCA analyses; LFMM thus highlighted areas of the genome whose variation was linked to this gradient. The correlation between temperature/humidity variables and altitude is a classic issue in ecology that can be addressed by including microclimatic variables33 and/or considering datasets designed in such a way as to allow the evaluation of temperature variations while altitude remains unchanged, or vice versa. These considerations provide an opening for further research that would improve our ability to link signatures more specifically to a specific factor.
It should be noted that the breeds, considered today, come from ancestral populations whose histories are largely unknown. Indeed, the various traces and historical documents indicating the existence of breeds on territories for several centuries, or even millennia, refer to the ancestral populations from which they originated. Hence, selection signatures identified may predate the formation of the breed or alternatively, reflect rapid recent changes. In addition, environmental features of a cradle may not be unique and may also have changed over time. The environmental characterization was inferred from climatic data recorded during the twentieth century. This constitutes a bias, which in the context of current climate change, must be carefully considered. We expect that our work will contribute to the synthesis of landscape genomics studies (e.g.34) by focusing on the particularities of livestock at the interface of the natural and anthropized environment.
Selection signatures identified in this study
The highlighted selection signatures can be categorized according to physiology and phenotypes. In Supplementary Text 3, we discussed the genes that seem to be involved (i) in lipid storage, (ii) in seasonal patterns and circadian behaviors, (iii) in coat colour and horns, (iv) in hypoxia and/or heat stress response, (v) in immunity, (vi) in lung function, and (vii) in neuronal function. We also discussed selection signatures that seemed difficult to classify because of their length and the diversity of the genes within them. Here we only focus on the most robust signals, considering both the p values and the number of SNPs involved in the selection signatures, and for which other studies have previously shown a link between the genes involved in the signatures and environmental adaptation.
Goat selection signatures
We identified SUCLG2 in goat by the PCAdapt approach. It was found under selection in sheep in arid Egyptian environments10 and in the Chinese desert9, but also in yaks adapted to extreme altitudes35. The gene is involved in the propanoate metabolic pathway and was significantly associated with growth in pigs36. Moreover, the latter study suggested that SUCLG2 plays a key role in the regulation of POU1F1, which is well known to be involved in growth function, and belongs to the same family as POU2F1, detected in sheep by our study. The study by Schmidt et al.37 in the blind subterranean mole rat, known to show remarkable tolerance to hypoxia and cancer resistance, also highlighted SUCLG2 and POU1F1 in the liver transcriptome. It was hypothesized that the energy-saving responses triggered in hepatic metabolic pathways may be crucial adaptations to low oxygen levels. Finally, the study of Tian et al.38, assessing energy metabolism-related genes in hypoxia-tolerant mammals, identified SUCLG2 as candidate gene of interest in the liver of cetaceans.
SOX2 and DPH6 were found correlated with the environmental gradient in goats by LFMM. In mice, SOX2 is expressed in adult SCN neurons and positively regulates transcription of the core clock gene, Period2, implicated in behavioral rhythms linked to environmental light cycles39. A link between SOX2 and cold adaptation has been found in marmots40. Interestingly, BBX, another Sox protein that belongs to the HMG box superfamily of DNA-binding proteins, was also identified in sheep in our study. DPH6 belongs to the circadian rhythm-related GO categories. Moreover it was found under selection in northern European cattle41 and yaks42, and associated with plateau conditions in Chinese sheep9.
It is interesting to note that the TRPC4 gene, on chromosome12, and the TRPC6 gene, on chromosome 15, are among the strongest signatures highlighted by the LFMM approach. Transient receptor potential channel (TRPC) proteins have been characterized as molecular substrates mediating receptor-activated cation influx. TRPC4 and TRPC6, in particular, have been shown to strongly contribute to synaptic information transfer in neuronal dendrites via the Ca2+-dependent release of neurotransmitter43. The increase in dendritic γ-aminobutyric acid (GABA) release from thalamic interneurons appears critically dependent on these TRP proteins44. Hence, these genes may mediate hot and cold sensation and affect endothelial-dependent regulation of vascular tone45. Moreover, there is strong evidence for an important function of TRPC6 in pulmonary vasculature46 and a functional role in hypoxia induction, potentially via the metabolism regulation of HIF-1α31, 32, 47, 48. In addition, TRPC6 was associated with high altitude adaptation in Tibetan highlanders49 and TRPC4 was implicated in body temperature regulation in cattle50.
Sheep selection signatures
Both PCAdapt and LFMM approaches identified the gene MSRB3. This gene has been subject to high selection pressure in sheep51 and has been suggested as a functional candidate for associations with ear morphology in numerous studies. It has been reported in large-eared sheep52. Its link with ears was also highlighted in pigs53, 54 and dogs55. Our study showed a correlation between SNPs near this gene and the environmental gradient. Interestingly, a recent study56 associated genetic variation in this gene and fat deposition in sheep. Furthermore, Webster et al.55 revealed that this gene was in genetic linkage with variants in HMGA2, a neighbouring gene, which influences body mass in dogs. Moreover, HMGA2 is known to be involved in adipose tissue, development and obesity in mouse57. This selection signature, for which the role of MSRB3 and HGMA2 is not yet elucidated, appears to be of central importance in terms of adaptation, because it was identified under selection in highland Ethiopian sheep7, in Tibetan sheep8, in Tibetan Yaks35 and in Tibetan dogs58.
Like MSRB3, both methods (PCAdapt and LFMM) implicated BMP2, which initiates osteoblast and adipocyte differentiation. It has previously been found in association with dry conditions in Chinese9 and Egyptian sheep10. It was also suggested to be under selection at high altitude, in Tibetan sheep8 and in yaks35. Adaptive mechanisms governed by BMP2 could be linked to lipid storage capacities, particularly at the level of the tail56, 59, 60.
Finally, RXFP2, well known to be under strong selection due to its role in horn development26, 51, 61, 62, was identified by both PCAdapt and LFMM approaches (see details in Supplementary Text 3).
Selection signatures highlighted in both species
An interesting result concerns the area between the PCDH9 and KLHL1 genes that was identified as a selection signature in both sheep and goats. This region appears to be a genetic desert that extends over a syntenic segment conserved on bovine chromosome 12, goat chromosome 12 and sheep chromosome 10. Kim et al.10 also found this genomic region to show evidence of selection in both sheep and goats that are adapted to arid environments, and suggested a major role for this region. PCDH9 has been found to be implicated in autism disorders63. Protocadherins are thought to be implicated in various aspects of neuronal development and functions. PCDH9 in particular is involved in synaptic cell adhesion64, 65. KLHL1 was found related to neuron motion and neuromuscular process66 and it may play a role in organizing the actin cytoskeleton of the brain cells.
Another selection signature common to both species and most interesting is the neurobeachin signal (NBEA in goat, and NBEAL1 in sheep). In the present study, LFMM revealed a correlation of NBEA and the environmental gradient, in both goats and sheep; in goat it was also identified by PCAdapt. NBEA was previously reported to be associated with high altitude in Ethiopian sheep7, Chinese sheep9, cattle67, and yaks35. Furthermore, it was previously associated with body temperature regulation in cattle50 and was suggested to be under selection in Ugandan and Moroccan goats68, 69. NBEA is a brain specific A-kinase anchor protein (AKAP), which is required for synaptic surface expression of glutamate and GABA receptors. Neurons lacking the BEACH (beige-Chediak/Higashi) domain protein Neurobeachin (NBEA) show strongly reduced synaptic responses caused by a reduction in surface levels of glutamate and GABAA receptors. Hence, NBEA plays an essential role in thermal adaptation through the regulation of synaptic transmission70, 71.
Interestingly, Alberto et al.72 identified NBEA, and also, HERC6 and SLC34A2, all detected in the current study, as involved in selective sweeps that differentiate domestic from wild sheep and goat populations, possibly indicating predomestic selection on these genomic areas.
Finally, the signature near NBEAL1 is bordered in sheep by BMPR2, for which mutations have been associated with high Altitude Pulmonary Hypertension (APH) in Kyrgyz Highlanders73 and in cattle74. Moreover, this gene was found to be associated with desert and plateau conditions in Chinese sheep9. BMP2, which belongs to the TGF-β superfamily and is able to activate BMPR2, was observed as a major signal in sheep in this study (see above). Hypoxia may act on BMPR2 activity75 and on BMP2 signaling in the pulmonary vasculature76.
Conclusion
Our study provides an approach for integrating ecological, historical and cultural approaches in the search for selection signatures in small ruminants, which as domestic animals are at the very interface of natural and anthropized environments. One of the key step is the characterization of the environment at the cradle level, thus targeting the geographic area, including transhumant summer pastures, where the breed has evolved by traditional practices. Local breeds are derived from populations selected for centuries and sometimes millennia and have thus developed long-standing links between genome and environment. In current times, local breeds are increasingly neglected and have become endangered, or in some cases have even disappeared. For those that remain, traditional practices, including the transhumance that has been one of the main driving forces in their development, are being abandoned16, 17. This study emphasizes that local breeds are invaluable resources of identifying environmental adaptation in the context of climate changes.
Material and methods
Methodology
A summary diagram of the main steps of the proposed approach is shown in Supplementary Figure 1.
Selection of breeds
The aim was to keep breeds with the highest probability of having retained strong adaptations to the local environment, i.e. breeds least affected by the disruptions inherent to the intensification of agricultural practices. For the study, we have targeted the Mediterranean arc in France, Italy and Spain. This region has the advantage of harboring closely related sheep and goat breeds despite large environmental contrasts11–13. We initially, considered 32 breeds of goats and 51 breeds of sheep (Supplementary Table 1). Subsequently, we selected 17 goat breeds and 25 sheep breeds (Figs. 1 and 2, and Table 1) on the basis of historical and anthropological documentation and according to the following criteria: (i) the breeds are local (i.e. cosmopolitan breeds were excluded); (ii) the breeds have featured in the local tradition for at least one century; (iii) any recorded introgression of exotic breeds was limited and occurred more than 100 years ago. If available, recent evaluations of admixture were also used to select breeds to include (see25, 26–29, 77). Outline descriptions of the selected breeds are in Supplementary Table 2.
Geographic definition of cradle of origin
We determined cradles of origin on the basis of historical and anthropological documentation. Particular attention has been paid to transhumance practices (see Figs. 1 and 2 and Supplementary Table 2 for details). We used ArcGIS78 to map the cradles in such a way as to include areas of summer pastures when so-called vertical transhumance was recorded. The horizontal transhumance, involving movements covering long distances without significant altitude differences79, cannot be directly taken into account because a large horizontal range would complicate the definition of the cradles.
Environmental characterization
Bioclimatic data were obtained from the WorldClim database (v.1.480) covering the period from 1960 to 1990, with a spatial resolution of 30 arc-seconds in the WGS84 datum. We considered the bioclimatic variables most relevant to highlight the major contrasts in temperature and humidity between the cradles, i.e.: BIO1 = Annual Mean Temperature, BIO5 = Maximum Temperature of Warmest Month, BIO6 = Minimum Temperature of Coldest Month, BIO12 = Annual Precipitation, BIO13 = Precipitation of Wettest Month, BIO14 = Precipitation of Driest Month, BIO15 = Precipitation Seasonality (Coefficient of Variation). Altitude information was collected from the SRTM 90 m Digital Elevation Database (v.4.1)81. Finally, latitude was taken as a proxy for luminosity and seasonality. This procedure was performed in R 3.5.282 using the R package RSAGA83.
Because our objective was not to estimate allele-environment correlations between each SNP and each environmental variable at a time, but to capture the relation between the genome and the environment considered as a whole, the cradles were clustered according to their climatic profiles, following this procedure:
For each cradle, the environmental variables of 10,000 random location points were extracted, using R packages sp84, 85 and rgdal86.
In order to identify groups of breeds with relatively similar environmental conditions in their cradle area, principal component analysis (PCA) was performed on the environmental variables. Prior to the analysis, Spearman correlation coefficient identified highly correlated variables (i.e. correlation coefficient r ≥ 0.9), in which case the biologically less relevant variable was removed to facilitate graphical presentation. The PCA analysis was followed by hierarchical clustering on principal components (HCPC), using Euclidian distances and Ward’s method. These multivariate analyses were performed with R software, using the FactoMineR package87.
The LFMM19 analyses were conducted following the cluster ranking of the cradles obtained through the PCA/HCPC procedure. Indeed, this procedure, allows classifying the cradles by considering their climatic resemblance defined by the set of all the recorded climatic variables. This ranking is then used to establish the link between genome and environment by LFMM, in order to optimize the statistical power of the analysis.
Genotyping
For goats, we used the AdaptMap dataset, including breeds genotyped with the Caprine SNP50 BeadChip (available via Dryad: https://doi.org/10.5061/dryad.v8g21pt), which contains genotypes for 53,547 SNPs. For sheep, we merged a dataset of French breeds obtained with the Illumina Ovine HD SNP Beadchip (Zenodo repository https://doi.org/10.5281/zenodo.237116) with a dataset of Italian breeds (77, provided by the authors) and a dataset of Spanish breeds (27, provided by the authors), obtained with the Illumina Ovine SNP50 BeadChip. SNP data from French breeds were extracted from the 600 K variation using the Ovine SNP50 BeadChip coordinates of SNPs on the OAR v3.1 reference genome assembly using Vcftools88. Merging with the SNP50 data resulted in 40,455 genotypes. SNPs and animals were pruned with PLINK v1.0789 using the following filtering thresholds: (i) SNP call rate ≤ 97%; (ii) SNP minor allele frequency (MAF) ≤ 1%; (iii) animals displaying ≥ 10% of missing genotypes. After filtration of the merged datasets we retained 50,329 genotypes for 416 goats and 32,168 genotypes for 555 sheep (initial datasets included 420 and 576 individuals, respectively).
Identification and analysis of genomic selection signatures
We identified loci that may contribute to local adaptation by two methods: (i) the individual-based multivariate approach PCAdapt18, via the pcadapt R package, and (ii) the latent factor mixed models LFMM, as implemented in the R package LEA90, for testing genotype-environment association.
PCAdapt scans the genome to detect outliers with respect to the population structure. Unlike population-based approaches, the package can incorporate admixed individuals and does not assume prior knowledge of population structure. As recommended by Luu et al.18, the optimal number of principal components (i.e., K, the optimal number of genetic groups) was determined using the graphical PCAdapt function, by varying K from 1 to 35, and following “Cattell’s rule”, i.e., keeping PCs that correspond to eigenvalues to the left of the lower straight line in the screeplot91. This led us to choose K = 15 for goats and K = 20 for sheep (see Supplementary Figure 6). Candidate SNPs were identified by calculating the False Discovery Rate (FDR, α = 0.05) of the p values associated with Mahalanobis distance estimated by PCAdapt, using the R package q value92.
LFMM software assesses associations between genetic variation (response variable) and environmental factors (explanatory variables) using a linear mixed model and controlling for neutral genetic structure, such as population history and isolation-by-distance, with (random) latent factors via an MCMC algorithm. The number of latent factors was chosen in accordance with the Admixture93 analyses. We ran LFMM using 1000 sweeps for burn-in and 9000 additional sweeps. Since LFMM uses a stochastic algorithm, ten runs with different seeds were performed. We combined the Z-scores from the ten repetitions and computed the median. We then recalibrated Z-scores by dividing by the inflation factor before computing the p values. We then chose significant associations based on FDR (α = 0.05) using the R package qvalue. LFMM was used to test for the correlation between the genetic variations and the environment taken as a whole (i.e. the variables were not considered one by one, except to test the method), but the analysis was performed once, analyzing the correlation between genomic variations and the ranking obtained by the PCA/HPCP clustering of the cradles environments.
For both PCAdapt and LFMM, we applied a stringent screen to identify selection signatures: candidate-selected regions were required to have at least 3 SNPs ≤ 500 kb apart, previously identified based on FDR and showing p value ≤ 10−9. The window was chosen on the basis of previous evidence that LD in sheep and goats does not persist beyond 500 kb (see Supplementary Figure 2). It should be noted that the size of the window considered is widely used and that the selection criteria retained are among the most stringent (see53, 54, 94–98). For each analysis, genes within a region spanning 100 kb upstream and downstream of the candidate selection regions, were annotated in such a way that genes including one or more SNPs identified as selected were specifically studied, but genes bordered by SNPs identified as selected were also considered. The chromosomal regions under selection pressure were inspected using the NCBI Genome Data Viewer (https://www.ncbi.nlm.nih.gov/genome/gdv/), Oar_v3.1 for sheep and ARS1 for goats. Biological functions of the genes identified were inferred from the literature. Studies focusing on environmental adaptation signatures involving the same genes as those identified here were compiled. As top candidate genes for adaptation, we considered genes identified by our study and by at least one other study focusing on environmental adaptation.
Experimental test of the method
We compared the cradle-based method with the use of the individual GPS coordinates. This comparison was carried out only for the goat dataset, for which the AdaptMap Consortium provided us with the geographical locations for all breeds considered.
For each GPS coordinate, the environmental variables were extracted for the circular region of radius of 5 km centered on the GPS point. We considered the most discriminating variables, Annual Mean Temperature, Annual Mean Precipitation and Altitude, in order to compare the average information obtained by breed from the GPS coordinates with that obtained from the cradles.
The LFMM analysis proceeded as follows (see Fig. 3):
The individual GPS method: we assessed the LFMM correlation between individual genotypes and the variables Annual Mean Temperature, Annual Mean Precipitation and Altitude, considered separately and extracted for each individual GPS coordinate.
The GPS area method: we considered environmental data for the set of all individual GPS coordinates recorded for the breed. (i) The LFMM correlation was calculated between each genotype and the average value obtained for the different variables considered separately (Annual Mean Temperature, Annual Mean Precipitation and Altitude). (ii) As an alternative approach, in order to group the different GPS areas according to their similarity, we used all the environmental variables recorded at the set of the breed GPS coordinates and followed the PCA/HCPC process as explained in “Environmental characterization” section. Then we tested LFMM correlation between the genotypes and the resulting cluster ranking.
The novel cradle method: For each breed, we considered environmental data at the cradle level. The LFMM correlation was tested in two steps, as for the GPS area method.
Complementary analyses
LD decay
To provide an insight into the overall levels of LD in the different breeds, genome-wide pairwise r2 values of SNPs separated by a maximum distance of 10 Mb, were calculated with PLINK software.
Admixture analyses
To assess population genetic structure we used ADMIXTURE software99. Prior to these analyses, SNP pruning was used to select a subset of SNPs with minimal linkage disequilibrium via the—indep option of PLINK with the following parameters: 50 SNPs per window, a shift of five SNPs between windows, and a Variation Inflation Factor threshold of two (corresponding to r2 > 0.5). SNPs identified by the study as potentially selected were removed. For K = 2 to K = 25, ten independent runs were performed. The entropy criterion was calculated via the sNMF function implemented in the R package LEA to assess the number of ancestral populations that best explains the genotypic data100, 101. We used the program CLUMPAK (102, http://clumpak.tau.ac.il) to analyze the multiple independent runs at a single K and visualize the results.
Mantel test
Finally, we assessed the correlation between genetic distances and geographic distances using the Mantel test via the mantel.randtest function, implemented in the R adegenet package103.
Supplementary Information
Acknowledgements
The authors would like to thank the AdaptMap consortium; Vincenzo Landi, Philippe Teinturier, Claire Jouannaux and the “Collectif CORAM” for their valuable insights into the Spanish and French local breeds. The calculations presented in this article were performed on (i) the CALI calculator of the University of Limoges, funded by the Limousin region, the European Union, the XLIM institutes, IPAM, GEIST, and the University of Limoges, and on (ii) the genotoul bioinformatics platform Toulouse Occitanie (Bioinfo Genotoul, https://doi.org/10.15454/1.5572369328961167E12). This work was supported by the French region Nouvelle-Aquitaine (PAPAGENO project, 2016–2018), and the AAP CNRS 2020: “Adaptation du Vivant à son Environnement” (EMPREINTES project, 2020–2021).
Author contributions
B.S. and M.C.: contributed to data analyses; P.V.: performed anthropological analyses; D.T.-M.: performed historical analyses; E.C., B.S., C.M.-R. and G.T.-K.: acquisition of data; M.B. and E.R.: contributed to the conception of the study; J.A.L.: acquisition of data/review and editing; S.J.G.H.: review and editing; F.P.: contributed to the conception of the study/review and editing; B.B.: contributed to data analyses/review and editing; A.D.S.: designed research/analyzed data/wrote the paper; all authors contributed to the final manuscript.
Data availability
For goats, we used the AdaptMap dataset, shared on Dryad: https://doi.org/10.5061/dryad.v8g21pt. For sheep, we used the French dataset online at the Zenodo repository https://doi.org/10.5281/zenodo.237116, and the Italian dataset partially available in the Figshare repository via https://doi.org/10.23644/uu.8947346 (for the few missing breeds please contact Elena Ciani: elena.ciani@uniba.it and Johannes A. Lenstra: j.a.lenstra@uu.nl).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bruno Serranito and Marco Cavalazzi.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-00682-z.
References
- 1.Vigne J-D. Early domestication and farming: What should we know or do for a better understanding? Anthropozoologica. 2015;50(2):123–150. doi: 10.5252/az2015n2a5. [DOI] [Google Scholar]
- 2.Zeder MA. Animal domestication in the Zagros: An update and directions for future research. MOM Édit. 2008;49(1):243–277. [Google Scholar]
- 3.Sponenberg DP, Bixby DE. Managing Breeds for a Secure Future: Strategies for Breeders and Breed Associations. ALBC; 2007. [Google Scholar]
- 4.Taberlet P, Valentini A, Rezaei HR, Naderi S, Pompanon F, Negrini R, Ajmone-Marsan P. Are cattle, sheep, and goats endangered species? Mol. Ecol. 2008;17(1):275–284. doi: 10.1111/j.1365-294X.2007.03475.x. [DOI] [PubMed] [Google Scholar]
- 5.Berihulay H, Abied A, He X, Jiang L, Ma Y. Adaptation mechanisms of small ruminants to environmental heat stress. Anim. Open Access J. MDPI. 2019;9(3):75. doi: 10.3390/ani9030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leroy G, Baumung R, Boettcher P, Scherf B, Hoffmann I. Review: Sustainability of crossbreeding in developing countries; definitely not like crossing a meadow…. Animal. 2016;10(2):262–273. doi: 10.1017/S175173111500213X. [DOI] [PubMed] [Google Scholar]
- 7.Edea Z, Dadi H, Dessie T, Kim K-S. Genomic signatures of high-altitude adaptation in Ethiopian sheep populations. Genes Genomics. 2019;41(8):973–981. doi: 10.1007/s13258-019-00820-y. [DOI] [PubMed] [Google Scholar]
- 8.Wei C, Wang H, Liu G, Zhao F, Kijas JW, Ma Y. Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 2016;6(1):26770. doi: 10.1038/srep26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Li W-R, Lv F-H, He S-G, Tian S-L, Peng W-F. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol. Biol. Evol. 2016;33(10):2576–2592. doi: 10.1093/molbev/msw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim ES, Elbeltagy AR, Aboul-Naga AM, Rischkowsky B, Sayre B, Mwacharo JM, Rothschild MF. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity. 2016;116(3):255–264. doi: 10.1038/hdy.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciani E, Mastrangelo S, Da Silva A, Marroni F, Ferenčaković M, Ajmone-Marsan P. On the origin of European sheep as revealed by the diversity of the Balkan breeds and by optimizing population-genetic analysis tools. Genet. Sel. Evol. GSE. 2020;52:1–14. doi: 10.1186/s12711-020-00545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colli L, Milanesi M, Talenti A, Bertolini F, Chen M, Crisà A. Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes. Genet. Sel. Evol. GSE. 2018;50:1–20. doi: 10.1186/s12711-018-0422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kijas JW, Townley D, Dalrymple BP, Heaton MP, Maddox JF, McGrath A. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE. 2009;4(3):e4668. doi: 10.1371/journal.pone.0004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brisebarre, A. Races ovines, systèmes d’élevage et représentations des éleveurs. in Développement rural, environnement et enjeux territoriaux. Regards croisés Oriental marocain et Sud-Est tunisien (dir. Bonte, P., Elloumi, M., Guillaume, H. & Mahdi, M.) 63–78 (Cérès Ed., 2009).
- 15.Hall SJG. Livestock biodiversity as interface between people, landscapes and nature. People Nat. 2019;1(3):284–290. doi: 10.1002/pan3.23. [DOI] [Google Scholar]
- 16.Caballero R, Fernández-González F, Badia RP, Molle G, Roggero PP, Bagella S, D’Ottavio P, Papanastasis VP. Grazing Systems and Biodiversity in Mediterranean Areas: Spain, Italy and Greece. Pastos; 2011. [Google Scholar]
- 17.Collantes F. The demise of European Mountain Pastoralism: Spain 1500–2000. Nomadic People. 2009;13(2):124–145. doi: 10.3167/np.2009.130208. [DOI] [Google Scholar]
- 18.Luu K, Bazin E, Blum MGB. pcadapt: An R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 2017;17(1):67–77. doi: 10.1111/1755-0998.12592. [DOI] [PubMed] [Google Scholar]
- 19.Frichot E, Schoville SD, Bouchard G, François O. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol. Biol. Evol. 2013;30(7):1687–1699. doi: 10.1093/molbev/mst063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.FAO. The State of the World’s Animal Genetic Resources for Food and Agriculture, edited by B. Rischkowsky & D. Pilling. Rome. (2007).
- 21.François O. Running Structure-Like Population Genetic Analyses with R. R Tutorials in Population Genetics. U. Grenoble-Alpes; 2016. pp. 1–9. [Google Scholar]
- 22.Dalongeville A, Benestan L, Mouillot D, Lobreaux S, Manel S. Combining six genome scan methods to detect candidate genes to salinity in the Mediterranean striped red mullet (Mullus surmuletus) BMC Genomics. 2018;19:1–13. doi: 10.1186/s12864-018-4579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Kort H, Vandepitte K, Mergeay J, Mijnsbrugge KV, Honnay O. The population genomic signature of environmental selection in the widespread insect-pollinated tree species Frangula alnus at different geographical scales. Heredity. 2015;115(5):415–425. doi: 10.1038/hdy.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capblancq T, Luu K, Blum MGB, Bazin E. Evaluation of redundancy analysis to identify signatures of local adaptation. Mol. Ecol. Resour. 2018;18(6):1223–1233. doi: 10.1111/1755-0998.12906. [DOI] [PubMed] [Google Scholar]
- 25.Bertolini F, Servin B, Talenti A, Rochat E, Kim ES, Oget C. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018;50(1):57. doi: 10.1186/s12711-018-0421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fariello M-I, Servin B, Tosser-Klopp G, Rupp R, Moreno C, Consortium, I. S. G. Selection signatures in worldwide sheep populations. PLoS ONE. 2014;9(8):e103813. doi: 10.1371/journal.pone.0103813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manunza A, Cardoso TF, Noce A, Martínez A, Pons A, Bermejo LA. Population structure of eleven Spanish ovine breeds and detection of selective sweeps with BayeScan and hapFLK. Sci. Rep. 2016;6(1):1–10. doi: 10.1038/srep27296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oget C, Servin B, Palhière I. Genetic diversity analysis of French goat populations reveals selective sweeps involved in their differentiation. Anim. Genet. 2019;50(1):54–63. doi: 10.1111/age.12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rochus CM, Tortereau F, Plisson-Petit F, Restoux G, Moreno-Romieux C, Tosser-Klopp G, Servin B. Revealing the selection history of adaptive loci using genome-wide scans for selection: An example from domestic sheep. BMC Genomics. 2018;19(1):71. doi: 10.1186/s12864-018-4447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz-Larrañaga O, Langa J, Rendo F, Manzano C, Iriondo M, Estonba A. Genomic selection signatures in sheep from the Western Pyrenees. Genet. Sel. Evol. GSE. 2018;50:1–12. doi: 10.1186/s12711-018-0378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Wang D, Yan G, Sun L, Tang C. TRPC6 is required for hypoxia-induced basal intracellular calcium concentration elevation, and for the proliferation and migration of rat distal pulmonary venous smooth muscle cells. Mol. Med. Rep. 2016;13(2):1577–1585. doi: 10.3892/mmr.2015.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Liu J, Zhou G, Guo J, Yan H, Niu Y. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Sci. Rep. 2016;6:38932. doi: 10.1038/srep38932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graae B, Frenne P, Kolb A, Brunet J, Chabrerie O, Verheyen K. On the use of weather data in ecological studies along altitudinal and latitudinal gradients. Oikos. 2011;121:3–19. doi: 10.1111/j.1600-0706.2011.19694.x. [DOI] [Google Scholar]
- 34.Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 2015;24(17):4348–4370. doi: 10.1111/mec.13322. [DOI] [PubMed] [Google Scholar]
- 35.Qi X, Zhang Q, He Y, Yang L, Zhang X, Shi P. The transcriptomic landscape of yaks reveals molecular pathways for high altitude adaptation. Genome Biol. Evol. 2019;11(1):72–85. doi: 10.1093/gbe/evy264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang F, Wang Q, Wang M, He K, Pan Y. Associations between gene polymorphisms in two crucial metabolic pathways and growth traits in pigs. Chin. Sci. Bull. 2012;57(21):2733–2740. doi: 10.1007/s11434-012-5328-3. [DOI] [Google Scholar]
- 37.Schmidt H, Malik A, Bicker A, Poetzsch G, Avivi A, Shams I, Hankeln T. Hypoxia tolerance, longevity and cancer-resistance in the mole rat Spalax—A liver transcriptomics approach. Sci. Rep. 2017 doi: 10.1038/s41598-017-13905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian R, Yin D, Liu Y, Seim I, Xu S, Yang G. Adaptive evolution of energy metabolism-related genes in hypoxia-tolerant mammals. Front. Genet. 2017;8:205. doi: 10.3389/fgene.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng AH, Bouchard-Cannon P, Hegazi S, Lowden C, Fung SW, Chiang C-K. SOX2-dependent transcription in clock neurons promotes the robustness of the central circadian pacemaker. Cell Rep. 2019;26(12):3191–3202.e8. doi: 10.1016/j.celrep.2019.02.068. [DOI] [PubMed] [Google Scholar]
- 40.Bai L, Liu B, Ji C, Zhao S, Liu S, Wang R. Hypoxic and cold adaptation insights from the Himalayan Marmot Genome. IScience. 2019;11:519–530. doi: 10.1016/j.isci.2018.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stronen AV, Pertoldi C, Iacolina L, Kadarmideen HN, Kristensen TN. Genomic analyses suggest adaptive differentiation of northern European native cattle breeds. Evol. Appl. 2019 doi: 10.1111/eva.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan D, Xiong X, Mipam T-D, Fu C, Li Q, Ai Y. Genetic diversity, molecular phylogeny, and selection evidence of Jinchuan Yak revealed by whole-genome resequencing. G3 (Bethesda, Md.) 2018;8(3):945–952. doi: 10.1534/g3.118.300572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Li Z, Hatcher JT, Chen Q-H, Chen L, Wurster RD. Deletion of TRPC6 attenuates NMDA receptor-mediated Ca2+ entry and Ca2+-induced neurotoxicity following cerebral ischemia and oxygen-glucose deprivation. Front. Neurosci. 2017;11:138. doi: 10.3389/fnins.2017.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munsch T, Freichel M, Flockerzi V, Pape H-C. Contribution of transient receptor potential channels to the control of GABA release from dendrites. Proc. Natl. Acad. Sci. U. S. A. 2003;100(26):16065–16070. doi: 10.1073/pnas.2535311100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan J, Li J, Zeng B, Chen G-L, Peng X, Zhang Y. Structure of the mouse TRPC4 ion channel. Nat. Commun. 2018;9:1–10. doi: 10.1101/282715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malczyk M, Erb A, Veith C, Ghofrani HA, Schermuly RT, Gudermann T. The role of transient receptor potential channel 6 channels in the pulmonary vasculature. Front. Immunol. 2017;8:707. doi: 10.3389/fimmu.2017.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Wang J, Wei Y, Liu Y, Ding X, Dong B. Crucial role of TRPC6 in maintaining the stability of HIF-1α in glioma cells under hypoxia. J. Cell Sci. 2015;128(17):3317–3329. doi: 10.1242/jcs.173161. [DOI] [PubMed] [Google Scholar]
- 48.Xu L, Chen Y, Yang K, Wang Y, Tian L, Zhang J. Chronic hypoxia increases TRPC6 expression and basal intracellular Ca2+ concentration in rat distal pulmonary venous smooth muscle. PLoS ONE. 2014;9(11):e112007. doi: 10.1371/journal.pone.0112007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng L, Zhang C, Yuan K, Gao Y, Pan Y, Ge X. Prioritizing natural-selection signals from the deep-sequencing genomic data suggests multi-variant adaptation in Tibetan highlanders. Natl. Sci. Rev. 2019;6(6):1201–1222. doi: 10.1093/nsr/nwz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howard JT, Kachman SD, Snelling WM, Pollak EJ, Ciobanu DC, Kuehn LA, Spangler ML. Beef cattle body temperature during climatic stress: A genome-wide association study. Int. J. Biometeorol. 2013;58:1665–1672. doi: 10.1007/s00484-013-0773-5. [DOI] [PubMed] [Google Scholar]
- 51.Kijas JW, Lenstra JA, Hayes B, Boitard S, Neto LRP, Cristobal MS. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012;10(2):e1001258. doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei C, Wang H, Liu G, Wu M, Cao J, Liu Z. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genomics. 2015;16(1):1–12. doi: 10.1186/s12864-015-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen M, Wang J, Wang Y, Wu Y, Fu J, Liu J. Genome-wide detection of selection signatures in Chinese indigenous Laiwu pigs revealed candidate genes regulating fat deposition in muscle. BMC Genet. 2018;19:1–9. doi: 10.1186/s12863-018-0622-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C, Liu C, Xiong X, Fang S, Yang H, Zhang Z. Copy number variation in the MSRB3 gene enlarges porcine ear size through a mechanism involving miR-584-5p. Genet. Sel. Evol. GSE. 2018;50:1–18. doi: 10.1186/s12711-018-0442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webster MT, Kamgari N, Perloski M, Hoeppner MP, Axelsson E, Hedhammar Å. Linked genetic variants on chromosome 10 control ear morphology and body mass among dog breeds. BMC Genomics. 2015;16:474. doi: 10.1186/s12864-015-1702-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mastrangelo S, Bahbahani H, Moioli B, Ahbara A, Abri MA, Almathen F. Novel and known signals of selection for fat deposition in domestic sheep breeds from Africa and Eurasia. PLoS ONE. 2019;14(6):e0209632. doi: 10.1371/journal.pone.0209632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xi Y, Shen W, Ma L, Zhao M, Zheng J, Bu S. HMGA2 promotes adipogenesis by activating C/EBPβ-mediated expression of PPARγ. Biochem. Biophys. Res. Commun. 2016;472(4):617–623. doi: 10.1016/j.bbrc.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 58.Gou X, Wang Z, Li N, Qiu F, Xu Z, Yan D. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 2014;24(8):1308–1315. doi: 10.1101/gr.171876.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan Z, Liu E, Liu Z, Kijas JW, Zhu C, Hu S. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2017;48(1):55–66. doi: 10.1111/age.12477. [DOI] [PubMed] [Google Scholar]
- 60.Zhu, C. et al.GWAS and Post-GWAS to Identification of Genes Associated with Sheep Tail Fat Deposition. Retrieved from https://www.preprints.org/manuscript/201906.0093/v1 (2019).
- 61.Allais-Bonnet A, Grohs C, Medugorac I, Krebs S, Djari A, Graf A. Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae. PLoS ONE. 2013;8(5):e63512. doi: 10.1371/journal.pone.0063512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnston SE, McEwan JC, Pickering NK, Kijas JW, Beraldi D, Pilkington JG. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol. Ecol. 2011;20(12):2555–2566. doi: 10.1111/j.1365-294X.2011.05076.x. [DOI] [PubMed] [Google Scholar]
- 63.Oksenberg N, Stevison L, Wall JD, Ahituv N. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 2013;9(1):e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayashi S, Takeichi M. Emerging roles of protocadherins: From self-avoidance to enhancement of motility. J. Cell Sci. 2015;128(8):1455–1464. doi: 10.1242/jcs.166306. [DOI] [PubMed] [Google Scholar]
- 65.Seong E, Yuan L, Arikkath J. Cadherins and catenins in dendrite and synapse morphogenesis. Cell Adhes. Migr. 2015;9(3):202–213. doi: 10.4161/19336918.2014.994919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shin D-H, Lee H-J, Cho S, Kim HJ, Hwang JY, Lee C-K. Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level. BMC Genomics. 2014;15(1):240. doi: 10.1186/1471-2164-15-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng, X. Angus Cattle at High Altitude: Pulmonary Arterial Pressure, Estimated Breeding Value and Genome-Wide Association Study (PhD thesis). (Colorado State University, 2017).
- 68.Benjelloun, B. Diversité des génomes et adaptation locale des petits ruminants d’un pays méditerranéen : le Maroc (PhD thesis) (Université Grenoble Alpes, France, 2015).
- 69.Onzima RB, Upadhyay MR, Doekes HP, Brito LF, Bosse M, Kanis E. Genome-wide characterization of selection signatures and runs of homozygosity in Ugandan Goat Breeds. Front. Genet. 2018;9:318. doi: 10.3389/fgene.2018.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farzana F, Zalm R, Chen N, Li KW, Grant SGN, Smit AB. Neurobeachin regulates glutamate- and GABA-receptor targeting to synapses via distinct pathways. Mol. Neurobiol. 2016;53(4):2112–2123. doi: 10.1007/s12035-015-9164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nair R, Lauks J, Jung S, Cooke NE, de Wit H, Brose N. Neurobeachin regulates neurotransmitter receptor trafficking to synapses. J. Cell Biol. 2013;200(1):61–80. doi: 10.1083/jcb.201207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alberto FJ, Boyer F, Orozco-terWengel P, Streeter I, Servin B, de Villemereuil P. Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 2018;9:1–9. doi: 10.1038/s41467-018-03206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iranmehr A, Stobdan T, Zhou D, Poulsen O, Strohl KP, Aldashev A. Novel insight into the genetic basis of high-altitude pulmonary hypertension in Kyrgyz highlanders. Eur. J. Hum. Genet. EJHG. 2019;27(1):150–159. doi: 10.1038/s41431-018-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Newman JH, Holt TN, Hedges LK, Womack B, Memon SS, Willers ED. High-altitude pulmonary hypertension in cattle (Brisket disease): Candidate genes and gene expression profiling of peripheral blood mononuclear cells. Pulmon. Circ. 2011;1(4):462–469. doi: 10.4103/2045-8932.93545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, X., Kong, Q., Zhao, C., Cai, Z., & Wang, M. New pathogenic variant of BMPR2 in pulmonary arterial hypertension. Cardiology in the Young, 29(4), 462–466. 10.1017/S1047951119000015 (2019). [DOI] [PubMed]
- 76.Anderson L, Lowery JW, Frank DB, Novitskaya T, Jones M, Mortlock DP. Bmp2 and Bmp4 exert opposing effects in hypoxic pulmonary hypertension. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;298(3):R833–R842. doi: 10.1152/ajpregu.00534.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ciani E, Crepaldi P, Nicoloso L, Lasagna E, Sarti FM, Moioli B. Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim. Genet. 2014;45(2):256–266. doi: 10.1111/age.12106. [DOI] [PubMed] [Google Scholar]
- 78.ESRI . ArcGIS Desktop: Release 10. Environmental Systems Research Institute; 2011. [Google Scholar]
- 79.Ruiz M, Ruiz JP. Ecological history of transhumance in Spain. Biol. Conserv. 1986;37:73–86. doi: 10.1016/0006-3207(86)90035-2. [DOI] [Google Scholar]
- 80.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 81.Jarvis, A., Reuter, H. I., Nelson, A. & Guevara, E. Hole-filled seamless SRTM dataV4, International Centre for Tropical Agriculture (CIAT). Available from https://srtm.csi.cgiar.org (2008).
- 82.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018). https://www.R-project.org.
- 83.Brenning A. Statistical geocomputing combining R and SAGA: The example of landslide susceptibility analysis with generalized additive models. In: Böhner J, Blaschke T, Montanarella L, editors. Hamburger Beitraege zur Physischen Geographie und Landschaftsoekologie. SAGA; 2008. pp. 23–32. [Google Scholar]
- 84.Bivand, R. S., Pebesma, E. & Gomez-Rubio, V. Applied Spatial Data Analysis with R 2nd edn (Springer, 2013). http://www.asdar-book.org/.
- 85.Pebesma, E. J. & Bivand, R. S. Classes and methods for spatial data in R. R News5(2), 9–13. https://CRAN.R-project.org/doc/Rnews/ (2005).
- 86.Keitt, T. H., Bivand, R., Pebesma, E. & Rowlingson, B. rgdal: Bindings for the geospatial data abstraction library. Copy at http://www.tinyurl.com/h8w8n29 (2010).
- 87.Le S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008;25(1):1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 88.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frichot E, François O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015;6(8):925–929. doi: 10.1111/2041-210X.12382. [DOI] [Google Scholar]
- 91.Cattell RB. The Scree plot test for the number of factors. Multivar. Behav. Res. 1966;1:140–161. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- 92.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401(6755):788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 94.Ablondi M, Viklund Å, Lindgren G, Eriksson S, Mikko S. Signatures of selection in the genome of Swedish warmblood horses selected for sport performance. BMC Genomics. 2019;20(1):717. doi: 10.1186/s12864-019-6079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Avila F, Mickelson JR, Schaefer RJ, McCue ME. Genome-wide signatures of selection reveal genes associated with performance in American Quarter Horse subpopulations. Front. Genet. 2018;9:249. doi: 10.3389/fgene.2018.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen M, Pan D, Ren H, Fu J, Li J, Su G. Identification of selective sweeps reveals divergent selection between Chinese Holstein and Simmental cattle populations. Genet. Sel. Evol. 2016;48(1):76. doi: 10.1186/s12711-016-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheruiyot EK, Bett RC, Amimo JO, Zhang Y, Mrode R, Mujibi FDN. Signatures of selection in admixed dairy cattle in Tanzania. Front. Genet. 2018;9:607. doi: 10.3389/fgene.2018.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.López ME, Linderoth T, Norris A, Lhorente JP, Neira R, Yáñez JM. Multiple selection signatures in farmed Atlantic Salmon adapted to different environments across hemispheres. Front. Genet. 2019;10:901. doi: 10.3389/fgene.2019.00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011;12:246. doi: 10.1186/1471-2105-12-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frichot E, Mathieu F, Trouillon T, Bouchard G, Francois O. Fast and efficient estimation of individual ancestry coefficients. Genetics. 2014;196:973–983. doi: 10.1534/genetics.113.160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015;15(5):1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jombart T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For goats, we used the AdaptMap dataset, shared on Dryad: https://doi.org/10.5061/dryad.v8g21pt. For sheep, we used the French dataset online at the Zenodo repository https://doi.org/10.5281/zenodo.237116, and the Italian dataset partially available in the Figshare repository via https://doi.org/10.23644/uu.8947346 (for the few missing breeds please contact Elena Ciani: elena.ciani@uniba.it and Johannes A. Lenstra: j.a.lenstra@uu.nl).