Abstract
Cells can achieve error-free repair of DNA double-strand breaks (DSBs) by homologous recombination through gene conversion with or without crossover. In contrast, an alternative homology-dependent DSB repair pathway, single-strand annealing (SSA), results in deletions. In this study, we analyzed the effect of mRAD54, a gene involved in homologous recombination, on the repair of a site-specific I-SceI-induced DSB located in a repeated DNA sequence in the genome of mouse embryonic stem cells. We used six isogenic cell lines differing solely in the orientation of the repeats. The combination of the three recombination-test substrates used discriminated among SSA, intrachromatid gene conversion, and sister chromatid gene conversion. DSB repair was most efficient for the substrate that allowed recovery of SSA events. Gene conversion with crossover, indistinguishable from long tract gene conversion, preferentially involved the sister chromatid rather than the repeat on the same chromatid. Comparing DSB repair in mRAD54 wild-type and knockout cells revealed direct evidence for a role of mRAD54 in DSB repair. The substrate measuring SSA showed an increased efficiency of DSB repair in the absence of mRAD54. The substrate measuring sister chromatid gene conversion showed a decrease in gene conversion with and without crossover. Consistent with this observation, DNA damage-induced sister chromatid exchange was reduced in mRAD54-deficient cells. Our results suggest that mRAD54 promotes gene conversion with predominant use of the sister chromatid as the repair template at the expense of error-prone SSA.
DNA double-strand breaks (DSBs) form a major threat to the integrity of chromosomes and viability of cells. Unrepaired or incorrectly repaired DSBs may lead to translocations or loss of chromosomes, which could result in cell death or uncontrolled cell growth. Eukaryotes have developed several mechanisms to repair DSBs, including nonhomologous DNA end-joining (NHEJ) and homologous recombination (HR). In Saccharomyces cerevisiae, DSBs are efficiently repaired through HR by the RAD52 group genes, while a contribution of NHEJ to DSB repair is only observed in the absence of HR (32). In mammalian cells, NHEJ plays a major role in DSB repair (18). More recently, it has become clear that in addition to NHEJ, HR can play an important role in DSB repair in mammalian cells as well (22).
Several pathways of homology-dependent DSB repair have been described for S. cerevisiae (32). One of these pathways, single-strand annealing (SSA), specifically occurs when a DSB is made between directly repeated DNA sequences. The DSB is processed by removal of part of the 5′ strand on each side of the break, exposing long 3′ overhangs (25). The single-stranded DNA (ssDNA) overhangs anneal to a long complementary stretch of DNA, and nonhomologous ssDNA ends are removed. As a result, one of the repeats and the intervening sequence are deleted. In vertebrates, a similar pathway has been described (5).
An alternative homology-dependent DSB repair pathway, mediated by the RAD52 group genes, is gene conversion (GC) (32, 46). DSB repair through this pathway also requires the DNA around the DSB to be degraded to produce 3′ ssDNA overhangs. One or both of these ends invade a homologous DNA sequence, which can be found either on the homologous chromosome or, in the S and G2 phases of the cell cycle, on the sister chromatid. Several models for this invasion have been described, including DSB gap repair and synthesis-dependent strand annealing (11, 34). In a model for DSB gap repair, both ends invade the homologous duplex and the gap is filled by DNA synthesis. The resulting Holliday junctions are resolved either with or without crossover (CO). We will use the terms “CO” for events involving GC with CO and “GC” for GC without CO. In the simplest model for synthesis-dependent strand annealing, only one end invades the homologous sequence. After DNA synthesis primed from the invaded end, the newly synthesized strand reanneals with the other end of the DSB. Then, the second strand is synthesized, resulting in a strong bias towards non-CO (11). However, if a long tract of DNA is synthesized, the result will appear similar to CO. RAD52 is important for almost all GC and CO pathways (32). Other genes involved include RAD51, RAD54, and RDH54/TID1 (8, 26, 32). RAD51 and RAD54 are mainly required for GC. RDH54, a homologue of RAD54, is only required for GC using the homologous chromosome, while RAD54 is involved in GC with both the sister chromatid and the homologous chromosome (2, 26, 42). In mammalian cells, similar GC and CO pathways have been found, but very little is known about the genetic requirements of the different pathways. Most of the above-mentioned genes have a homologue in mammals (22). Nevertheless, the importance of each gene can differ in mammalian and S. cerevisiae cells. For example, the mouse RAD52 (mRAD52) gene can be mutated without a major effect on recombination, while it is the most important gene in S. cerevisiae (40).
One of the other RAD52 group genes, RAD54, is clearly important in mammalian cells. The Rad54 protein belongs to the SWI2/SNF2 protein family whose members modulate protein-DNA interactions in an ATP-dependent manner (23). The S. cerevisiae and human Rad54 proteins are double-stranded DNA-dependent ATPases that interact with Rad51, a key player in the search for homologous template DNA (6, 14, 20, 35, 45, 48). Compared to wild-type cells, RAD54-deficient mouse embryonic stem (ES) cells are two- to fourfold more sensitive to ionizing radiation, methyl methanesulfonate, and mitomycin C (MMC) (10). In addition, HR in mRAD54-deficient cells is 5- to 10-fold reduced, as measured by targeted integration of exogenous DNA (10). This reduction in HR can explain the sensitivity of cells lacking mRad54 to DSB-inducing DNA-damaging agents, although a direct involvement of mRad54 in DSB repair has not yet been demonstrated.
Much information concerning the mechanisms of DSB repair in S. cerevisiae has been obtained by using a site-specific DSB induced by rare-cutting endonucleases (15). Recently, it has been shown that the S. cerevisiae mitochondrial enzyme I-SceI, which recognizes and cuts a nonpalindromic 18-bp site, leaving 4-bp 3′ overhangs, works efficiently in mammalian cells, but is not toxic to these cells (17). Analysis of the repair products of the site-specific DSB allows quantitation of the relative contribution of NHEJ and different homology-dependent pathways of DSB repair in mammalian cells (7, 21, 27, 28, 47). In this study, we have investigated the relative contribution of different homology-dependent pathways to the repair of an I-SceI-induced chromosomal DSB in mouse ES cells that were either mRAD54-proficient or -deficient.
MATERIALS AND METHODS
Construction of mRAD54 targeting vectors.
Targeting vectors were constructed to integrate three different recombination-test substrates into the mRAD54 genomic locus. The substrates were cloned into the unique SfuI site of exon 4, thereby disrupting mRAD54. The first targeting vector was made by inserting the DRneo construct (28), linearized with XhoI, into the SfuI site of a 9-kb EcoRI fragment from mRAD54 encompassing exons 4, 5, and 6 (Fig. 1A) (10). The second and third targeting vectors were made by inserting the IRneo and SCneo recombination-test substrates in a similar manner (Fig. 1A) (21).
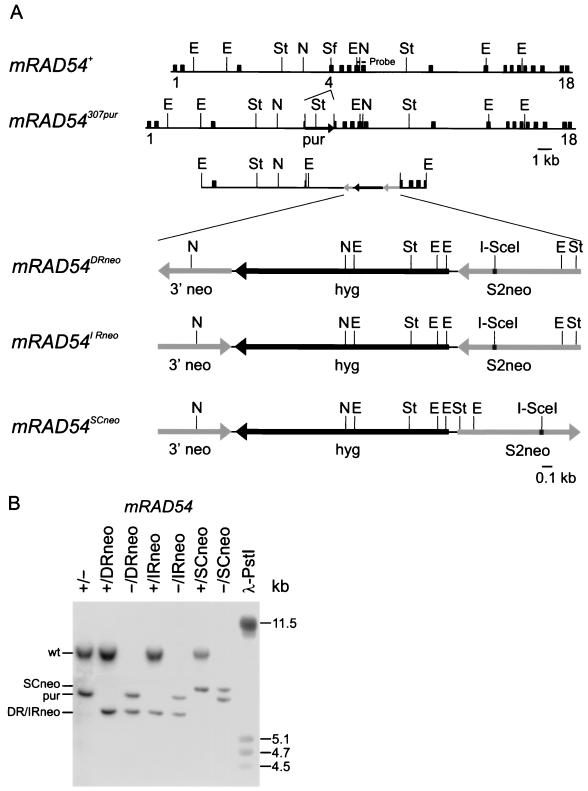
FIG. 1.
Generation of mRAD54+/− and mRAD54−/− ES cells containing recombination-test substrates. (A) Structure of the genomic mRAD54 locus and targeting vectors containing the substrates. The two upper lines represent the wild-type (mRAD54+) and the puromycin-targeted knockout (mRAD54307pur) alleles, respectively. The 18 exons that encode mRad54 are indicated by boxes. The dashed line above exons 7 and 8 indicates the position of the probe used to distinguish the different mRAD54 alleles after digestion of the genomic DNA with StuI. The arrow shows the position of the puromycin (pur) selectable marker gene. The locations of selected restriction sites are shown: E, EcoRI; N, NcoI; Sf, SfuI; St, StuI. The third line shows a generic representation of the targeting vectors. The three lower lines show the three different substrates inserted into the mRAD54 locus in more detail. The black arrow indicates the hygromycin (hyg)-selectable marker gene. The gray arrow on the left represents the 700-bp 3′ neomycin-selectable marker gene (3′ neo). The gray arrow on the right represents the full-length S2neo gene, which contains a 4-bp deletion at the 18-bp I-SceI site insertion (indicated in black). (B) DNA blot of ES cells containing wild-type (+) and knockout (−) mRAD54 alleles in addition to alleles with recombination-test substrates. Genomic DNA was digested with StuI. The DNA blot was hybridized with the probe indicated in panel A. Phage λ DNA digested with PstI was used as a size marker. The lengths of marker fragments are indicated in kilobases on the right and the positions of the different mRAD54 alleles are shown on the left.
ES cell culture and electroporation.
Heterozygous mRAD54 ES cells of the genotype mRAD54+/307pur were electroporated with the different targeting vectors and cultured on gelatinized dishes as described previously (10). The cells were split 24 h after electroporation, and hygromycin B (hygro) was added to a final concentration of 200 μg/ml. After 7 to 10 days, colonies were isolated and expanded. Genomic DNA from individual clones was digested with StuI and analyzed by DNA blotting using a flanking probe (Fig. 1B). The blot was rehybridized with a 700-bp 3′ neomycin (neo) fragment to confirm a single integration of the targeting vector.
I-SceI transfections.
ES cells containing the recombination-test substrates were cultured in medium containing hygro at a concentration of 200 μg/ml. Transfection of 3.2 × 106 cells was done by electroporation with 6 μg of either pPGK3xnlsI-SceI or pCBA3xnls-I-SceI, which transiently express I-SceI from the phosphoglycerate kinase I (PGK) or the chicken β-actin promoter, respectively (9, 39). To determine the transfection efficiency, 6 μg of pPGKCAS-eGFP, containing the green fluorescent protein (GFP) gene under the control of a PGK promoter, was cotransfected in a number of experiments. In parallel, cells were electroporated without DNA or with pBSIIKS or pPGKCAS-eGFP alone. After electroporation, 103 cells were plated without selection to determine the cloning efficiency. The remaining cells were grown for 1 day without selection before they were split and cultured in medium containing G418 (200 μg/ml) or G418 (200 μg/ml)–hygro (200 μg/ml). When pPGKCAS-eGFP had been cotransfected with the I-SceI-expressing plasmid, a portion of the cells was subjected to fluorescence-activated cell sorting analysis 1 day after transfection to determine the percentage of cells positive for GFP expression. After 8 to 11 days, cells were fixed, stained, and counted. The number of clones from the cells transfected with the I-SceI-expressing plasmid was corrected for the number of clones from the mock-transfected cells. To enable comparison between the number of clones from different cell lines and experiments, the absolute number of clones was divided by the cloning efficiency and transfection efficiency. The data on the number of G418- and G418-hygro-resistant clones is based on three to seven independent experiments, using two or three independent cell lines for each genotype. In several of the experiments, colonies were isolated and expanded. Genomic DNA from individual clones was analyzed for recombination events by digestion with either NcoI or EcoRI and DNA blotting using the 700-bp 3′ neo fragment as a probe. After analysis of DNA isolated from DRneo recombinants digested with NcoI, 20% of the clones showed, in addition to the banding patterns expected for SSA-CO or GC, the hybridization pattern of the original construct. These were scored as SSA-CO or GC, respectively. Colonies from all recombination substrates that had aberrations in the hybridization pattern which were difficult to interpret were not included in the analysis. Inclusion of these aberrant clones did not alter the conclusions.
SCEs.
Sister chromatid exchanges (SCEs) in ES cell lines of the genotypes mRAD54+/+, mRAD54+/−, and mRAD54−/− and a derivative of the mRAD54−/− line expressing the hRAD54 cDNA were analyzed (10, 45). The mRAD54 knockout allele in these lines was mRAD54307neo. The cell lines were coded to prevent bias in the analysis. SCE analysis was performed according to standard procedures, with the cells either mock treated or treated with 0.2 μg of MMC/ml. At least 40 metaphases per cell line were analyzed for both the number of chromosomes and SCEs.
RESULTS
The recombination-test substrates.
The three substrates that were used to measure HR frequencies in mouse ES cells are schematically depicted in Fig. 1A. They contain a hygromycin selectable marker gene (hyg) flanked by two inactive neomycin selectable marker genes, S2neo and 3′ neo. One of the crippled neo genes, 3′ neo, consists of the 3′ 700 bp of the neo gene. The other, S2neo, is a full-length neo gene, which contains a 4-bp deletion and the 18-bp insertion of the I-SceI site at the position of the NcoI site at bp 576 of neo (28). Expression of the I-SceI enzyme can create a DSB in S2neo. By recombination between S2neo and 3′ neo, the original NcoI site of S2neo, which is present in 3′ neo, can be restored, creating an intact neo gene. The three recombination-test substrates differ solely in the relative orientation of the two crippled neo genes (Fig. 1A). DRneo contains both crippled neo genes as direct repeats. Transcription of S2neo occurs towards the 5′ end of 3′ neo. IRneo contains both genes as inverted repeats because 3′ neo has been inverted relative to its orientation on DRneo. SCneo contains the genes as direct repeats, but in contrast to DRneo, transcription of S2neo occurs away from the 3′ end of 3′ neo (21).
Homologous integration of the recombination-test substrates in the mRAD54 locus.
We targeted the recombination-test substrates to the mRAD54 gene of ES cells to obtain single integration of the substrates at a defined and transcriptionally active position in the genome. Consequently, the targeted cell lines are isogenic and differ only in the presence of an mRAD54+ or an mRAD54− allele and the orientation of the crippled neo genes of the substrates. To achieve this, the substrates were subcloned into exon 4 of the mRAD54 gene to create targeting vectors that would result in disruption of the gene (10). The resulting mRAD54 alleles are referred to as mRAD54DRneo, mRAD54IRneo, and mRAD54SCneo, respectively. The targeting vectors were transfected into mRAD54+/− ES cells of the genotype mRAD54+/307pur (10). After selection with hygro, targeted clones were identified by DNA blotting with a unique probe outside the targeting construct (Fig. 1).
The disruption of mRAD54 by the recombination-test substrates was confirmed by the hypersensitivity of mRAD54307pur/DRneo ES cells to γ-irradiation (data not shown). The survival curve of the mRAD54+/DRneo cell line after γ-irradiation was similar to that of wild-type cells, as expected, because heterozygote mRAD54 cells show no obvious phenotype (10). Immunoblot analysis using α-hRad54 showed that mRad54 protein was present in all mRAD54+ cell lines containing the substrates but could not be detected in any of the mRAD54 knockout cell lines (data not shown).
The DRneo substrate: DSB repair events.
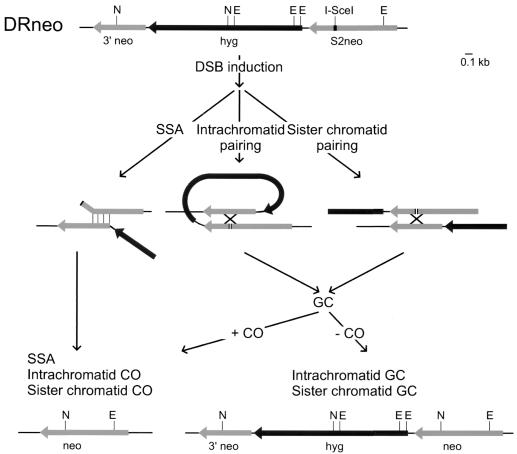
Transfection of an I-SceI-expressing plasmid in cells containing DRneo can result in a DSB in S2neo (Fig. 2). The DSB can be repaired by NHEJ with or without a deletion or insertion (27). NHEJ will not result in the restoration of an intact neo gene, and therefore NHEJ events will not be recovered. This is true for all substrates. An alternative repair pathway is SSA (Fig. 2). During SSA within DRneo, complementary strands of S2neo and 3′ neo will anneal, resulting in an intact neo gene and deletion of the intervening hyg gene. A third pathway to repair the DSB is HR by GC (Fig. 2). GC by recombination with S2neo on the sister chromatid will result in restoration of nonfunctional S2neo, and therefore, these events will not be recovered. To obtain an intact neo gene by GC or CO, the S2neo containing the DSB needs to pair with either 3′ neo on the same chromatid or, in the S and G2 phases of the cell cycle, with 3′ neo on the sister chromatid. These modes of homologous pairing are referred to as intrachromatid and sister chromatid pairing in Fig. 2. If the intermediate is resolved without a CO, the resulting clone will contain intact neo and hyg genes and a 3′ neo gene, and the cell will be resistant to G418 and hygro. On the other hand, if a single CO takes place or the GC tract continues beyond the neo genes, the resulting clone will contain an intact neo gene while the hyg gene and 3′ neo will be lost. The cell will only be resistant to G418. At the DNA level, the outcome of CO is therefore identical to the outcome of SSA (Table 1).
FIG. 2.
Model of possible mechanisms for homology-dependent DSB repair on DRneo. The DSB induced at the I-SceI site and indicated by the gap in S2neo can be repaired by different repair pathways that are depicted schematically. Only repair events yielding an intact neo gene are shown. A summary of all possible outcomes of DSB repair is given in Table 1, and the different pathways are described in detail in the text. The annealing of the complementary ssDNA during SSA is indicated by thin vertical lines. Pairing of S2neo and 3′ neo (indicated by the cross) can result in GC with or without CO. Symbols are the same as those in Fig. 1.
TABLE 1.
Possible outcomes of repair events for the different recombination-test substrates after induction of a DSB by I-SceI
| DSB repair event | Possible outcome for recombination-test substrates
|
|||||
|---|---|---|---|---|---|---|
| DRneo
|
IRneo
|
SCneo
|
||||
| Viabilitya | Resulting resistance geneb | Viabilitya | Resulting resistance geneb | Viabilitya | Resulting resistance geneb | |
| NHEJ | + | hyg | + | hyg | + | hyg |
| SSA | + | neo | − | + | None | |
| GC | + | neo, hyg | + | neo, hyg | + | neo, hyg |
| Intrachromatid CO | + | neo | + | neo, hyg | + | None |
| Sister chromatid CO | + | neo | − | + | neo, hyg | |
Plus and minus signs indicate whether repair through these pathways results in viable or inviable cells, respectively.
neo and hyg indicate the expected expression of the neo and hyg genes, respectively.
The DRneo substrate: relative efficiency of DSB repair events.
The relative contribution of the different homology-dependent DSB repair pathways was investigated by transfection of an I-SceI-expressing plasmid into mRAD54+/DRneo ES cells. As a control, a mock transfection was performed either with no DNA or with pBSIIKS or pPGKCAS-eGFP. Before transfection, the cells were grown by hygro selection to reduce the background due to spontaneous recombination events. After transfection, the cells were grown for 1 day without selection. Subsequently, they were divided over multiple dishes and cultured in either G418-containing medium or G418-hygro-containing medium. After 8 to 11 days, the cells were fixed and the number of colonies on each dish was counted. The frequency of spontaneously arising G418-resistant colonies varied between 10−5 and 10−6. No significant differences in the induction of G418-resistant colonies were found between transfection of a control plasmid or no DNA. The recombination frequency was increased 100- to 1,000-fold after transfection of an I-SceI-expressing plasmid.
G418-resistant colonies are obtained after all likely recombination events: SSA, GC, and CO. In contrast, G418-hygro-resistant colonies are only obtained after GC (Fig. 2). Therefore, the ratio of the number of G418-hygro-resistant colonies to G418-resistant colonies is an indication of the contribution of GC to all HR events. The advantage of this ratio is that it is an internal measure that can be compared directly between different cell lines and separate experiments. In addition, the ratio is not dependent on the transfection or the cloning efficiency of the cell line. In mRAD54+/DRneo ES cells that have no defect in HR (10), this ratio of G418-hygro- to G418-resistant colonies was 0.15 ± 0.01. Thus, around 15% of all recombination events consist of GC. The contribution of CO to the repair of a DSB is usually equal to or lower than the contribution of GC (4, 21, 32). Therefore, it is likely that SSA accounts for the majority of recombination events recovered from DRneo.
The DRneo substrate: the effect of mRAD54 on DSB repair.
Next, we determined the effect of mRAD54 on the repair of a DSB induced by I-SceI in DRneo by using mRAD54−/DRneo ES cell lines. The ratio of G418-hygro-resistant to G418-resistant colonies shifted from 0.15 ± 0.01, observed for mRAD54-proficient cells, to 0.077 ± 0.007 for mRAD54-deficient cells. Thus, the contribution of GC (G418-hygro-resistant clones) to the total number of recombination events (G418-resistant clones) was reduced in the absence of mRad54 protein. We conclude that the mRad54 protein is involved in repairing DSBs in vivo.
To confirm these results at the DNA level, we isolated DNA from both G418-resistant mRAD54+/DRneo and mRAD54−/DRneo ES cell colonies (Fig. 3). Most clones showed a hybridization pattern consistent with either GC or SSA and/or CO (Table 2). The ratio of GC to all recombination events was 0.175 for mRAD54+/DRneo and 0.095 for mRAD54−/DRneo ES cells (Table 2). Thus, an approximately twofold difference in the proportion of GC in the absence of mRAD54 was again observed. However, the ratio for each genotype was slightly, but not significantly (P > 0.2), higher when analyzed by DNA blotting, compared to the colony formation assay.
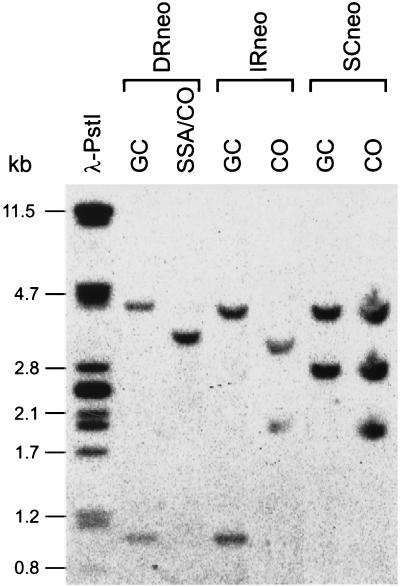
FIG. 3.
DNA blot analysis of I-SceI-induced DSB repair events in ES cells containing the different recombination-test substrates. mRAD54-proficient ES cells containing either DRneo, IRneo, or SCneo were transfected with an I-SceI-expressing plasmid. After selection with G418 or G418-hygro, genomic DNA from individual clones was digested with EcoRI. The outcome of repair of the I-SceI-induced DSB was analyzed by DNA blotting using a 700-bp 3′ neo probe. Only a selection of the clones listed in Table 2 is shown. As shown in Fig. 2 and 5, the sizes of the EcoRI fragments labeled with the neo probe indicate whether the DSB has been repaired by GC or CO. With DRneo, SSA results in the same molecular outcome as CO. Phage λ DNA digested with PstI was used as a size marker. The lengths of marker fragments are on the left.
TABLE 2.
Relative contribution of different homology-dependent repair events of I-SceI-induced DSBs in mRAD54-proficient and -deficient cells containing the recombinant-test substrates
| Genotype | Distribution of DSB repair eventsa
|
|||
|---|---|---|---|---|
| GC | CO (SSA) | Other | Total | |
| mRAD54+/DRneo | 17 | 80 | 1 | 98 |
| mRAD54−/DRneo | 6 | 57 | 3 | 66 |
| mRAD54+/IRneo | 124 | 2 | 4 | 130 |
| mRAD54−/IRneo | 112 | 0 | 8 | 120 |
| mRAD54+/SCneo | 50 | 54 | 24 | 128 |
| mRAD54−/SCneo | 55 | 50 | 37 | 142 |
Distribution of DSB repair events was determined by expanding colonies obtained after transfection of an I-SceI-expressing plasmid to ES cells of the indicated genotype and selection with either G418 (DRneo) or G418-hygro (IRneo, SCneo). DNA from the colonies was analyzed by DNA blotting after digestion with EcoRI or NcoI. The clones were classified according to their restriction pattern resulting from GC or CO. For DRneo, SSA was recovered in the same class as CO (CO-SSA). When the restriction pattern and intensity of the bands was different from the expected pattern of HR events, the clone was counted in the category “Other.”
We wished to determine whether the decrease in the ratio of GC to all recombination events observed in the absence of mRad54 was due to a decrease in the number of GCs, to an increase in SSA, or to both. Inclusion of additional controls and measuring the cloning and transfection efficiency allowed the comparison of the number of colonies obtained with different cell lines and separate experiments. The decrease in the proportion of GCs appeared to be due to both a significant increase (P < 0.05) in the recombination events yielding only G418 resistance, of which SSA is probably the most common, and a very slight, nonsignificant, decrease (P > 0.10) in the number of GCs (Fig. 4A and B). These results suggest that in the absence of mRad54 and the presence of direct repeats, ES cells shift their repair process from GC to SSA.
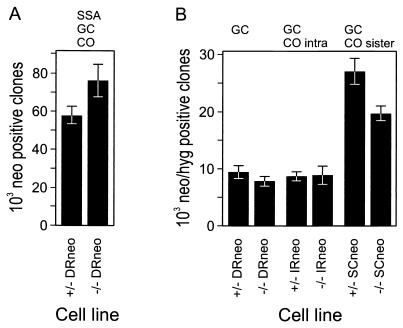
FIG. 4.
Homologous recombination frequencies for the recombination-test substrates. As described in Materials and Methods, 1.6 × 106 mRAD54-proficient and -deficient ES cells containing the indicated substrates in the identical genomic location were transfected with pCBA3xnls-I-SceI and processed. Shown is the normalized number of G418- or G418-hygro-resistant colonies ± standard error of the mean for three independent experiments with two cell lines from all six genotypes. (A) HR frequency of mRAD54+/DRneo (+/−) and mRAD54−/DRneo (−/−) ES cells. Colonies containing an intact neo gene were obtained after repair of the I-SceI-induced DSB by SSA, GC, and CO. (B) Frequencies of GC and CO for ES cells containing the substrates. For all three substrates, neo- and hyg-containing colonies were obtained after repair of the I-SceI-induced DSB by intrachromatid GC and sister chromatid GC. The IRneo- and SCneo-containing cell lines each have one additional possibility to yield G418-hygro-resistant colonies. In the IRneo-containing lines, these clones can be formed by intrachromatid CO. For the SCneo-containing lines, they can be formed by CO after pairing with the sister chromatid.
The IRneo and SCneo substrates: DSB repair events.
The experiments with DRneo-containing cell lines yielded useful information on the frequency of GC. However, because SSA and CO result in clones that are identical at the DNA level, the relative frequencies of these repair events could not be determined. To obtain information on the usage of CO either within the same chromatid or with the sister chromatid, we constructed mRAD54+/− and mRAD54−/− cell lines containing IRneo and SCneo.
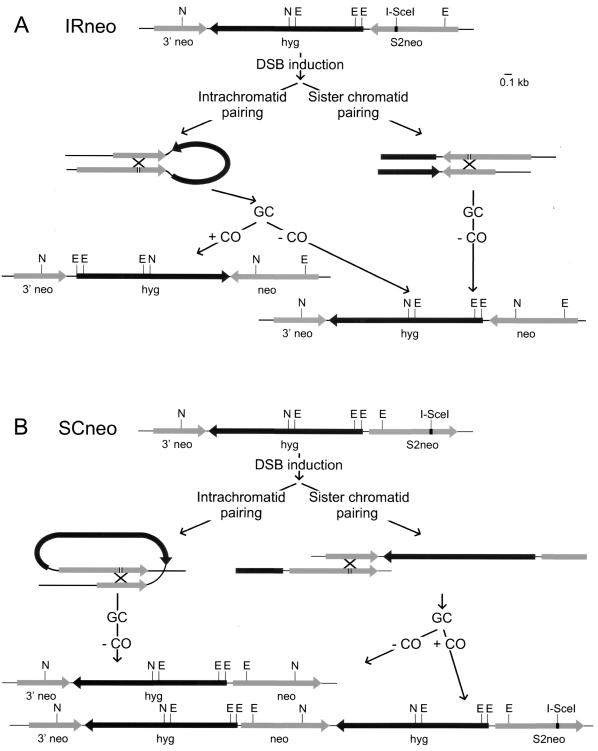
IRneo contains S2neo and 3′ neo as inverted repeats (Fig. 5A). In contrast to DRneo, the I-SceI-induced DSB in IRneo cannot be repaired through SSA. Due to the inverse orientation of the crippled neo genes, nucleolytic processing of the DSB will expose identical rather than complementary ssDNA tails. However, repair of the DSB by recombination is possible through several different routes (Fig. 5A and Table 1). Both GC and CO can occur by using 3′ neo on either the same chromatid or the sister chromatid as a template. Both GC events will result in G418-hygro-resistant cells. In contrast, CO involving the sister chromatid results in a dicentric chromosome and an acentric chromosome, which is incompatible with cell survival. DSB repair through CO after pairing with 3′ neo on the same chromatid results in G418-hygro-resistant cells. The orientation of the hyg gene will be inverted by the CO. Thus, CO can be distinguished from GC at the DNA level (Fig. 3).
FIG. 5.
Schematic representation of possible homology-dependent DSB repair pathways for IRneo and SCneo. Only repair events yielding an intact neo gene are depicted. A summary of all possible outcomes of DSB repair is given in Table 1, and the different pathways are described in detail in the text. Symbols are the same as in Fig. 1. The I-SceI-induced DSB is indicated by the gap in S2neo. Recombination between S2neo and 3′ neo, indicated by the cross, can lead to restoration of the original NcoI site resulting in an intact neo gene by GC with or without CO. Concerning the COs, only the product that results in an intact neo gene is shown. Shown are the outcomes of DSB repair events on IRneo (A) and SCneo (B).
SCneo contains S2neo and 3′ neo as direct repeats (Fig. 5B). In contrast to DRneo, transcription of S2neo occurs away from the 3′ end of 3′ neo, which has implications for the outcome of DSB repair. Expression of I-SceI in a cell containing SCneo can result in a DSB in S2neo. SSA is a possible repair event and will pair the 5′ end of 3′ neo to the 3′ end of S2neo. As a result, the DSB is repaired and 3′ neo is recovered, which will not be detected because the cell will remain sensitive to G418. Similar to the other substrates, GC can occur by pairing with 3′ neo on the same chromatid or the sister chromatid. In this way, an intact neo gene will be obtained and the hyg gene will be retained. CO after pairing with 3′ neo on the same chromatid will yield the same outcome at the DNA level as SSA, namely a single 3′ neo gene which will not be recovered. On the other hand, CO after unequal pairing with 3′ neo on the sister chromatid will result in an intact neo gene with a partial duplication of the rest of the construct resulting in two intact hyg genes (Fig. 5B and Table 1). This event can be distinguished from GC by DNA blotting (Fig. 3).
The IRneo and SCneo substrates: relative efficiency of DSB repair events.
To investigate the relative efficiency of the different HR repair pathways, mRAD54+/IRneo and mRAD54+/SCneo ES cells were transfected with an I-SceI-expressing plasmid, as described above for the DRneo-containing cell lines. The spontaneous recombination frequency was 10−5 to 10−6 (data not shown). The number of colonies on the pCBA3xnls-I-SceI-transfected dishes was normalized for the number of colonies on the mock-transfected dishes, the cloning efficiency, and the transfection efficiency. The resulting recombination frequency was about 10−2. In the SCneo-containing cell lines, not all recombination events are recovered, as SSA, GC using S2neo on the sister chromatid, and CO after pairing with 3′ neo on the same chromatid do not result in G418 resistance (Table 1). Nevertheless, transfection of the I-SceI-expressing plasmid into mRAD54+/SCneo cells resulted in three times more colonies than transfection into mRAD54+/IRneo cells aid (Fig. 4B). The number of G418-hygro-resistant colonies obtained after transfection of mRAD54+/IRneo cells with pCBA3xnls-I-SceI was comparable to the number obtained after transfection of mRAD54+/DRneo cells (Fig. 4B).
Since both GC and CO result in G418-hygro resistance of IRneo and SCneo, we investigated the distribution of these events by DNA blotting. GC and CO can be discriminated because they result in a different restriction pattern after digestion with EcoRI (Fig. 3 and 5). IRneo almost exclusively showed GC, which implies that CO within the same chromatid between inverted repeats is a rare event in ES cells (Table 2). In SCneo, GC and CO contributed equally to the recovered HR events (Table 2). Thus, CO after pairing with the sister chromatid, which usually does not lead to deleterious chromosome rearrangements, is a common event. A relatively high number of SCneo-derived clones showed restriction patterns that could not be explained by GC or CO (Table 2). These clones were excluded from the analysis, but their inclusion did not alter the conclusions.
The IRneo and SCneo substrates: the effect of mRAD54 on DSB repair.
With DRneo-containing cell lines, we observed, in the absence of mRad54 protein, a significant increase in SSA with a concomitant very slight reduction of GC. Therefore, we investigated the effect of mRAD54 on HR in the other substrates. We transfected pCBA3xnls-I-SceI into mRAD54−/IRneo and mRAD54−/SCneo cells and analyzed the colonies obtained as described above. DNA blot analysis revealed that there was no difference in the relative distribution of HR events between mRAD54-proficient and -deficient ES cell lines containing IRneo or SCneo (Table 2). mRAD54−/IRneo cells showed only GC, and mRAD54−/SCneo cells showed an equal number of GCs and COs.
The number of colonies obtained from mRAD54−/IRneo cells did not differ from the number of colonies from mRAD54+/IRneo cells (Fig. 4B). Thus, no indication was obtained for an involvement of mRAD54 in the repair of a DSB between inverted repeats by GC. However, mRAD54−/SCneo ES cells gave rise to fewer colonies than mRAD54+/SCneo ES cells after transfection of an I-SceI-expressing plasmid (Fig. 4B). There was a consistent, statistically significant (P < 0.05) decrease to approximately 70% of the number of colonies obtained with mRAD54-proficient cell lines containing SCneo. This indicates a role for mRAD54 in GC and CO with the sister chromatid in DSB repair in this substrate.
Influence of mRAD54 on the induction of SCEs.
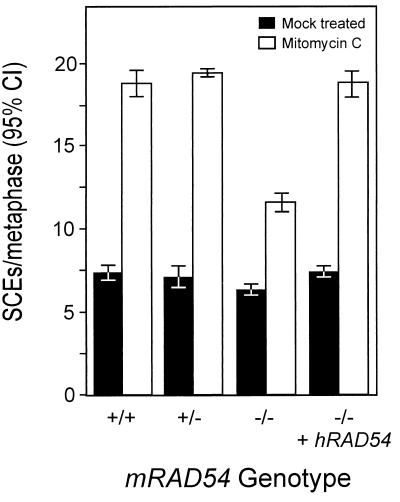
To obtain independent evidence for a role of mRAD54 in sister chromatid recombination, we measured the spontaneous and DNA damage-induced levels of SCEs in mRAD54-proficient and -deficient ES cells. ES cells of the genotypes mRAD54+/+, mRAD54+/−, and mRAD54−/− were analyzed. The spontaneous level of SCEs found in the mRAD54−/− cell line was slightly reduced compared to that observed in the mRAD54-proficient control cell lines (Fig. 6). In all cell lines, no numerical or gross structural chromosomal abnormalities were observed. DNA damage inflicted by the DNA interstrand cross-linking agent MMC increased the number of SCEs. Treatment of the cells with 0.2 μg of MMC/ml for 1 h increased the number of SCEs 2.6-fold in the mRAD54+/+ and mRAD54+/− ES cell lines. In the mRAD54−/− cell line, the increase in SCEs was only 1.8-fold. The difference in the average number of SCEs among mRAD54+/+, mRAD54+/−, and mRAD54−/− cells was significant (Fig. 6; P < 0.05). In addition, we included a derivative of the mRAD54−/− cell line that expressed the hRAD54 cDNA in the SCE analysis as a control. Expression of this cDNA rescues the DNA damage sensitivities of mRAD54−/− cells (45). The expression of hRAD54 returned the number of SCEs in the mRAD54−/− ES cell line to wild-type levels, both spontaneously and after treatment with MMC. In all cell lines treated with MMC, no apparent chromosomal changes were observed.
FIG. 6.
Induction of SCEs by MMC in mRAD54-proficient and -deficient ES cells. ES cells of the indicated genotypes were either mock treated or treated with 0.2 μg of MMC/ml for 1 to 2 h, and metaphase spreads were prepared. Forty to 95 metaphases per sample were scored for the number of SCEs per cell. The frequency of spontaneous SCEs is shown in black, while the frequency of SCEs after treatment with MMC is shown in white. The error bars indicate the 95% confidence intervals.
DISCUSSION
The major homology-dependent DSB repair pathway for DRneo is SSA.
In this study, we have analyzed HR in mouse ES cells. Using DRneo, a distinction can be made between DSB repair through SSA and CO on one hand and GC on the other hand (Fig. 2). Our results show that approximately 15% of all homology-dependent DSB repair events within DRneo in mouse ES cells occur through recombination between the direct repeats via GC. These results appear not to be specific for mouse ES cells, because 25% of the DSB repair events in CHO cells containing integrated DRneo occur through GC (27). The separate contributions of SSA and CO to DSB repair cannot be determined with DRneo. However, a comparison to the results with SCneo, which also contains direct repeats, suggests that GC and CO occur at similar frequencies (Table 2). This implies that SSA accounts for 70% of all homology-dependent DSB repair events within DRneo in ES cells.
Consistent with our results, recombination between direct repeats through GC, when compared to SSA, accounts for the minority of detected events in a number of other assay systems, including DSB-induced events on plasmids and in chromosomes in S. cerevisiae and vertebrate cells (12, 19, 27, 31, 41). However, there are a number of exceptions, both in S. cerevisiae and mammalian cells, in which GC accounts for the majority of spontaneous and DSB-induced events (4, 13, 30, 38, 47). Variables that might contribute to observed differences among assay systems include the length and sequence context of the repeats, the distance between the repeats, the position of the DSB, and heterology in the repeats or at the ends (12, 13, 31, 33, 47). Both GC and SSA use 3′ ssDNA tails as intermediates. GC requires search for homology followed by joint molecule formation actively mediated by Rad51-coated ssDNA. On the other hand, SSA involves the more passive process of annealing of complementary single-strands, although it does also, at least partially, depend on Rad52. Depending on the presence of nonhomologous ends and the length and sequence context of the repeat, these two processes might be affected differentially. Finally, the distribution of repair events may also be dependent on the stage of the cell cycle, as GC using the sister chromatid is only possible in S and G2.
DSB repair associated with DNA COs occurs mostly from the sister chromatid.
DSB repair products of both IRneo and SCneo differ depending on whether they have been generated through GC or CO (Fig. 5). While IRneo detects intrachromatid COs, SCneo detects unequal COs between sister chromatids. It should be noted, however, that if a long tract of DNA is synthesized during GC, the result appears similar to a CO. With IRneo, DSB repair through CO occurs only in 1.5% of the analyzed repair events, while GC accounts for over 95% of the events (Table 2). This also indicates that GC tracts are generally shorter than 2.7 kb, because otherwise the outcome would have been scored as a CO. In contrast to the lack of COs with IRneo, we find that GC and CO contribute equally to DSB repair using SCneo. Thus, it appears that COs preferably arise when the sister chromatid, instead of a homologous sequence on the same chromatid, is used as the repair template. A large contribution of COs using SCneo has also been found in CHO cells, after induction of a DSB (21). A similar preference for CO using the sister chromatid has been observed in mouse L cells during spontaneous recombination between repeated sequences (4). In contrast, in S. cerevisiae, a preference for intrachromatid interactions has been found, as indicated by a low percentage of COs in an SCneo-like substrate (24). A high percentage of COs after intrachromatid interactions has also been found in the repair of an induced or spontaneous DSB using inverted repeats in S. cerevisiae (1, 37, 41, 44).
The preference for sister chromatid interactions during HR in mammalian cells could have arisen because sister chromatid recombination is, in general, less prone to the generation of chromosomal rearrangements than intrachromatid recombination. A significant fraction of mammalian genomes consists of repetitive DNA sequences. CO between these sequences will result in deleterious chromosomal rearrangements, except when the same sequence on the sister chromatid is used. Evidence thus far suggests that genome rearrangements are indeed suppressed during recombination between sequence repeats on nonhomologous chromosomes (39). Furthermore, the presence of mismatches between the repeats also prevents recombination, due to the mismatch repair system (49). S. cerevisiae contains hardly any repetitive sequences and will undergo less selection against allowing intrachromatid recombination. The preference for the sister chromatid in mammalian cells might occasionally result in unequal CO between sister chromatids, but if those events occur in a limited region, their potential deleterious effects could be minimized.
Comparison of the frequency of DSB repair events on the different substrates.
We find that the frequency of GC is comparable among the different substrates (around 6 × 10−3; Table 2 and Fig. 4). It seems reasonable to assume that with all three substrates, a similar fraction of the cells receives a DSB and that a similar fraction of these DSBs is channeled into a homology-dependent repair pathway. Repair by SSA or CO using the sister chromatid will result in correct DSB repair in cells containing DRneo or SCneo. However, these events are apparently aborted in cells containing IRneo. They may cause cell death instead of resulting in GC. Otherwise, more GC events should have been recovered with IRneo. This finding of less efficient recombinational repair between inverted repeats is not unique to our assay. Both after DSB induction in S. cerevisiae and spontaneously in mouse cells, the frequency of recombination between direct repeats is higher than between inverted repeats (3, 41).
mRad54 influences the repair of DSBs in DRneo.
A role for the mRad54 protein in the repair of DSBs has been postulated based on the ionizing radiation sensitivity and HR deficiency of mRAD54−/− ES cells (10). The results of our study provide direct evidence that mRad54 is involved in DSB repair in vivo. The difference in DSB repair between mRAD54-proficient and -deficient cells is most clearly seen when the DSB is induced between direct repeats, as is the case with DRneo and SCneo (Fig. 4A and B). The absence of mRAD54 causes a very slight reduction in GC during DSB repair of DRneo. This reduction is accompanied by a statistically significant increase in the number of COs and SSA, the latter of which is the most frequent. In S. cerevisiae, a similar increase in HR is seen in rad54 mutants, both with direct repeats on plasmids and in chromosomes (16, 29, 42). The frequency of SSA (or CO) is 1.9- to 27-fold higher in rad54 cells than in wild-type cells, while cell survival and the frequency of GC are decreased (16, 29, 42). These results suggest that there might be competition between SSA and GC (see below).
mRad54 influences recombination between sister chromatids.
In cells containing SCneo, the effect of mRAD54 on GC is more pronounced than in cells containing DRneo. Repair of the DSB through SSA is possible in SCneo, although those events are not detected. A statistically significant 27% decrease in the frequency of GC and CO is observed in the absence of mRAD54 (Fig. 4B). Since all COs take place after pairing with the sister chromatid, mRAD54 is clearly involved in sister chromatid recombination. This is also the case for S. cerevisiae RAD54 (2). The contribution of GC and CO remains about equal in mRAD54−/− cells, which indicates that mRAD54 is involved in both GC and CO (Table 2). In contrast to these results, S. cerevisiae RAD54 appears to be mainly involved in GC, although this has been investigated only with inverted repeats (37).
COs resulting in restoration of the neo gene in SCneo are the consequence of interactions with the sister chromatid and result in SCEs at the chromosomal level. Therefore, our results with SCneo predict a reduction in the level of SCEs in the absence of mRAD54. Indeed, we find a slightly lower level of spontaneous SCEs in mRAD54−/− ES cells compared to that in mRAD54+/+ ES cells (Fig. 6). Because SCEs are induced by DNA-damaging agents, we have also tested whether mRAD54−/− ES cells respond differently to MMC treatment in the SCE assay than mRAD54+/+ or mRAD54+/− cells. Treatment of mRAD54−/− cells with MMC yields a 1.5-fold lower induction of SCEs, compared to mRAD54+/+ cells (Fig. 6). This effect of RAD54 on spontaneous and DNA damage-induced SCEs corresponds to results obtained with chicken-derived cells, in which a reduction in the frequency of SCEs in RAD54- and RAD51-deficient chicken B lymphocytes is observed (43). From these results, we conclude that genes required for HR are also involved in promoting SCEs. The decrease in SCEs induced by DNA damage corresponds to the similar decrease in the number of COs during DSB repair on SCneo.
The observation that mRAD54 influences DNA damage-induced SCEs adds significantly to our results with the recombination-test substrates. The results of the SCE experiments show that mRAD54 is involved in homology-dependent DNA repair of DNA damages that are present in naturally occurring genomic sequences. The SCE experiments overcome two restrictions of the experiments with recombination-test substrates. First, the restriction enzyme-induced DSB that initiates repair in the experiments involving the recombination-test substrates might be recognized differently from other types of DNA damages, including DSBs introduced by ionizing radiation or DNA interstrand cross-linking agents. Second, in the experiments involving the recombination-test substrates, the introduction of repeated DNA sequences is necessary in order to select for successful DSB repair events. However, the presence of these repeated sequences will influence the distribution of observed repair events. SSA relies especially on the presence of repeated sequences and will be used less frequently in a more physiological situation.
The absence of mRad54 has no influence on recombination within IRneo.
We find no change in the frequency of GC after induction of a DSB in IRneo in mRAD54-deficient cells compared to that in mRAD54-proficient cells (Fig. 4B). COs using the 3′ neo on the same chromatid are rare in mRAD54+/IRneo cells and have not been detected in mRAD54−/IRneo cells (Table 2). Similar to our results with chromosomal substrates in ES cells, disruption of RAD54 in S. cerevisiae cells has no effect on the repair of an induced DSB in inverted repeats located on a plasmid (16). In contrast, the rate of spontaneous GC between chromosomal inverted repeats is decreased 25-fold in a rad54 S. cerevisiae strain (37). Because S. cerevisiae cells display a different distribution of events, with a predominance of COs, a direct comparison between S. cerevisiae rad54 and mRAD54−/− ES cells is difficult (37). The lack of an effect of mRAD54 on DSB repair between inverted repeats in ES cells also contrasts with the effects of mRAD54 on DSB repair between direct repeats (Fig. 4). As we will discuss below, this could be due to the possibility to repair a DSB in direct repeats by SSA, which is not possible in inverted repeats.
Does mRad54 promote GC at the expense of SSA?
ssDNA tails are formed as a common intermediate in SSA and GC with or without CO. Because of this common intermediate, it is likely that a certain degree of competition exists between these two pathways (12). mRad54 could have a role in promoting GC, either directly or indirectly by blocking DSBs from being processed through the SSA pathway. This would explain the increase in the number of G418-resistant colonies in mRAD54-deficient cells with DRneo, which results from an increase in SSA. It would also explain the decrease in G418-hygro-resistant colonies with SCneo, because an increase in SSA, which is not recovered, would cause a decrease in the recovered GC events. With IRneo, SSA is not possible, and therefore, lack of mRad54 would not have any effect on DSB repair in this substrate. Mammalian chromosomes contain a significant amount of repetitive sequences that could be used to repair a DSB by SSA, thereby resulting in deletions. Inhibition of SSA by mRad54 is therefore even more relevant in mammalian cells than in S. cerevisiae, where similar effects of Rad54 on SSA have been found. Direct stimulation of GC pathways by mRad54, possibly by its interaction with mRad51, would decrease the contribution of SSA to DSB repair. Alternatively, mRad54 could suppress SSA directly. It has been shown recently that the purified human and S. cerevisiae Rad54 proteins have ATP-dependent DNA unwinding activity (36, 48). This activity would be ideally suited for the destabilization of intermediates in SSA or the stimulation of mRad51-mediated homologous DNA pairing and strand exchange (35).
ACKNOWLEDGMENTS
We thank M. de Bruijn for technical support and J. Essers for mRAD54+/− cells.
This work was supported by grants from The Netherlands Organization for Scientific Research, the Dutch Cancer Society, and the Human Frontier Science Program Organization. R.K. is a fellow of the Royal Netherlands Academy of Arts and Sciences.
REFERENCES
- 1.Aguilera A, Klein H L. Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics. 1989;123:683–694. doi: 10.1093/genetics/123.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollag R J, Liskay R M. Conservative intrachromosomal recombination between inverted repeats in mouse cells: association between reciprocal exchange and gene conversion. Genetics. 1988;119:161–169. doi: 10.1093/genetics/119.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag R J, Liskay R M. Direct-repeat analysis of chromatid interactions during intrachromosomal recombination in mouse cells. Mol Cell Biol. 1991;11:4839–4845. doi: 10.1128/mcb.11.10.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll D. Homologous genetic recombination in Xenopus: mechanism and implications for gene manipulation. Prog Nucleic Acid Res Mol Biol. 1996;54:101–125. doi: 10.1016/s0079-6603(08)60361-x. [DOI] [PubMed] [Google Scholar]
- 6.Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer W-D. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dresser M E, Ewing D J, Conrad M N, Dominguez A M, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fishman-Lobell J, Rudin N, Haber J E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godwin A R, Liskay R M. The effects of insertions on mammalian intrachromosomal recombination. Genetics. 1994;136:607–617. doi: 10.1093/genetics/136.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golub E I, Kovalenko O V, Gupta R C, Ward D C, Radding C M. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haber J E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov E L, Sugawara N, Fishman-Lobell J, Haber J E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 18.Jeggo P A, Taccioli G E, Jackson S P. Menage à trois: double strand break repair, V(D)J recombination and DNA-PK. Bioessays. 1995;17:949–957. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- 19.Jeong-Yu S J, Carroll D. Test of the double-strand-break repair model of recombination in Xenopus laevis oocytes. Mol Cell Biol. 1992;12:112–119. doi: 10.1128/mcb.12.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen U H, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 21.Johnson R D, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 22.Kanaar R, Hoeijmakers J H J, van Gent D C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 23.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 24.Klein H L. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988;120:367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein H L. Genetic control of intrachromosomal recombination. Bioessays. 1995;17:147–159. doi: 10.1002/bies.950170210. [DOI] [PubMed] [Google Scholar]
- 26.Klein H L. RAD54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang F, Han M, Romanienko P J, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liefshitz B, Parket A, Maya R, Kupiec M. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics. 1995;140:1199–1211. doi: 10.1093/genetics/140.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickoloff J A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol Cell Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickoloff J A, Singer J D, Hoekstra M F, Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989;207:527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- 32.Paques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paques F, Leung W Y, Haber J E. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 36.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 37.Rattray A J, Symington L S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ray A, Siddiqi I, Kolodkin A L, Stahl F W. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J Mol Biol. 1988;201:247–260. doi: 10.1016/0022-2836(88)90136-2. [DOI] [PubMed] [Google Scholar]
- 39.Richardson C, Moynahan M E, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rijkers T, Van Den Ouweland J, Morolli B, Rolink A G, Baarends W M, Van Sloun P P H, Lohman P H M, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudin N, Sugarman E, Haber J E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinohara M, Shita-Yamaguchi E, Buerstedde J-M, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonoda E, Sasaki M S, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugawara N, Paques F, Colaiacovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swagemakers S M A, Essers J, de Wit J, Hoeijmakers J H J, Kanaar R. The human Rad54 recombinational DNA repair protein is a double-stranded DNA-dependent. ATPase. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 46.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 47.Taghian D G, Nickoloff J A. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan T L, Essers J, Citterio E, Swagemakers S M, de Wit J, Benson F E, Hoeijmakers J H, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 49.te Riele H, Maandag E R, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]