Abstract
Deletions that include the gene TAB2 and TAB2 loss-of-function variants have previously been associated with congenital heart defects and cardiomyopathy. However, other features, including short stature, facial dysmorphisms, connective tissue abnormalities and a variable degree of developmental delay, have only been mentioned occasionally in literature and thus far not linked to TAB2. In a large-scale, social media-based chromosome 6 study, we observed a shared phenotype in patients with a 6q25.1 deletion that includes TAB2. To confirm if this phenotype is caused by haploinsufficiency of TAB2 and to delineate a TAB2-related phenotype, we subsequently sequenced TAB2 in patients with matching phenotypes and recruited patients with pathogenic TAB2 variants detected by exome sequencing. This identified 11 patients with a deletion containing TAB2 (size 1.68–14.31 Mb) and 14 patients from six families with novel truncating TAB2 variants. Twenty (80%) patients had cardiac disease, often mitral valve defects and/or cardiomyopathy, 18 (72%) had short stature and 18 (72%) had hypermobility. Twenty patients (80%) had facial features suggestive for Noonan syndrome. No substantial phenotypic differences were noted between patients with deletions and those with intragenic variants. We then compared our patients to 45 patients from the literature. All literature patients had cardiac diseases, but syndromic features were reported infrequently. Our study shows that the phenotype in 6q25.1 deletions is caused by haploinsufficiency of TAB2 and that TAB2 is associated not just with cardiac disease, but also with a distinct phenotype, with features overlapping with Noonan syndrome. We propose the name “TAB2-related syndrome”.
Subject terms: Chromosome abnormality, Disease genetics, Mutation
Introduction
The gene TAB2 (TGFβ-activated kinase 1 binding protein 2, MIM*605101) is mapped to chromosome 6q25.1. Haploinsufficiency of TAB2 is associated with congenital heart defects (CHD) [1] and cardiomyopathy [2]. Other features have only been described occasionally, including short stature, facial dysmorphisms, connective tissue abnormalities and a variable degree of developmental delay [2, 3]. Cheng et al. reported 6q25.1 deletion patients who showed features overlapping with Noonan syndrome (NS) [2].
NS is a relatively common genetic disorder characterised by typical facial dysmorphisms, developmental delay, short stature and cardiac abnormalities (e.g., pulmonary valve stenosis, atrial septal defects (ASDs) and cardiomyopathy). More than 15 genes have been described in association with NS, all part of the Ras/MAPK (mitogen-activated protein kinase) signal transduction pathway. However, in approximately 20% of clinically diagnosed NS patients, no (likely) pathogenic variant in these genes is detected [4].
In a large social media-based project on chromosome 6 aberrations, we observed that individuals with a 6q25.1 deletion shared a distinct phenotype that could most likely be attributed to TAB2 haploinsufficiency. To explore this, we recruited a second cohort with matching phenotypes in whom we sequenced TAB2 and further included patients in whom a pathogenic TAB2 variant had been found by exome sequencing. This enabled us to further delineate the TAB2-related phenotype.
Methods
We describe patients with deletions containing TAB2 and patients with intragenic variants in TAB2. We compare these patients with literature case reports and define the TAB2-related phenotype.
Cohort 1: Patients with chromosome 6 aberrations recruited via social media
Patients were recruited via the Chromosome 6 Project, a parent-driven social media-based research project into chromosome 6 aberrations, as described previously [5]. Patients or their legal representatives were approached to participate via a Chromosome 6 Facebook group, Twitter (@C6study) and our website (https://www.chromosome6.org). Inclusion criteria were an isolated chromosome 6 aberration and availability of a microarray report. Microarray analyses were performed in diagnostic laboratories using different platforms, as specified in Table S1. Microarray results were converted to GRCh37/hg19 using the UCSC LiftOver Tool and visualised using the UCSC Genome Browser (http://genome.ucsc.edu).
Information on the phenotype was collected directly from patients or their legal representatives via a multilingual Chromosome 6 Questionnaire constructed using the MOLGENIS toolkit [6]. The questionnaire contains questions on congenital abnormalities, relevant dysmorphic features, development, behaviour and health-related problems (including cardiac disease) of the individual. Consent for publication was obtained from all participants. Consent for the use of photographs was optional. After consent, additional information was requested from the treating physicians. By focussing on patients with heart defects, we found a shared deletion region in 6q25.1.
Literature review chromosome 6q deletions
We used PubMed and Google Scholar to search for information on previously reported patients with a chromosome 6q deletion. Search terms included: chromosome 6q, 6q deletion, monosomy 6q and 6q*. Only publications reporting the current diagnostic standard procedures (including microarray results) were included.
By combining genotype data of patients with a heart defect from the Chromosome 6 Project with genotype data from patients described in the literature, we found a smallest region of deletion overlap within chromosome band 6q25.1. The only gene within this region is TAB2. For the present study, we only included patients and literature cases with a deletion that includes TAB2 (further referred to as: TAB2 deletion). Figure 1 visualises the microarray analysis results and the smallest region of overlap including TAB2. We then delineated a TAB2 deletion phenotype for this group of individuals.
Fig. 1. Overview of all deletions leading to our candidate gene TAB2.
Deletions including the gene TAB2 are shown for our social media cohort (horizontal black bar) and the literature (horizontal grey bar). The minimum and maximum extent of the deletions are shown, if these were known. The smallest region of deletion overlap only includes our candidate gene TAB2 (vertical black bar). Deletions visualised using the UCSC genome browser (https://genome.ucsc.edu). Literature cases were derived from 11 reports [1–3, 11–18]. See Tables S1 and S2 for details.
Data collected from TAB2 deletion patients in Cohort 1 was submitted to the DECIPHER database (decipher.sanger.ac.uk), IDs 425375 and 425379-425388.
Cohort 2: Patients with TAB2 variants
After recognising the TAB2 deletion phenotype, we identified families with normal microarray results but a similar phenotype in our own institution. After confirming that they had TAB2 variants, we recruited other patients with TAB2 variants identified by exome sequencing at two other Dutch hospitals. Consent was obtained for publication and for publication of photographs, if applicable.
TAB2 variants were submitted to the Global Variome shared LOVD and can be accessed here: databases.lovd.nl/shared/references/DOI:10.1038/s41431-021-00948-0.
Exome sequencing was performed in the genomic diagnostic laboratories of the three Dutch university hospitals, as described previously [7–9]. Sanger sequencing of TAB2 was performed in the Radboud University Medical Center, Nijmegen, the Netherlands. DNA was isolated from leucocytes by standard methods. The coding region and exon–intron boundaries of TAB2 (GenBank: NM_015093.5) were amplified from genomic DNA. Sanger sequencing was performed using the BigDye Terminator Sequencing Kit and ABI 3730 XL (Applied Biosystems). Primers are available on request.
Literature review TAB2 variants
PubMed and Google Scholar were used to search for previously reported patients with a pathogenic TAB2 variant. Search terms included: TAB2 and variant/mutation.
Clinical scoring system NS
The clinical scoring system for NS published by Van der Burgt et al. [10] was used to compare the clinical features of our patients with NS. The system scores six criteria, one of which is a typical or suggestive face for NS. This facial criterion includes six characteristics: broad forehead, hypertelorism, downslanting palpebral fissures, ptosis, low set and posteriorly angulated ears and broad and/or short neck. If all six facial characteristics were present in our patients, the face was defined as “typical”. If four of the six facial characteristics were present, the face was defined as “suggestive” for NS. In all patients, the craniofacial phenotype was evaluated by a clinical geneticist either in the outpatient clinic or using photographs.
Results
We describe 25 cases from 16 families with TAB2 deletions (n = 11) or TAB2 variants (n = 14) and compare these with data from 45 literature cases.
Identifying the candidate gene TAB2 in 6q25.1 deletion patients
Data from 36 deletion patients (Chromosome 6 Project, n = 11; literature case reports, n = 25) showed a cluster with a shared phenotype of CHD and short stature (<−2 SD) in the region 6q25.1. The size of the deletions ranged from 0.12 to 14.31 Mb. The smallest region of deletion overlap led to our candidate gene TAB2 (see Fig. 1).
Patient characteristics of cohort 1: TAB2 deletion cohort
The data on 11 patients (9 female, 2 male, median age 4 years (range 8 months to 40 years)) from 10 families collected via the Chromosome 6 Project are summarised in Table 1 (details in Table S1). Eight of these 11 patients (73%) had cardiac anomalies. Six had congenital valve defects involving one or more valves, with the mitral valve involved in five patients. An additional ASD and/or ventricular septal defect (VSD) was present in four patients. At age 1 week, patient Id134 had surgical closure of her VSD and a coarctation of the aorta was repaired. Four patients had cardiomyopathy, three in combination with a structural heart defect. Two children, Id134 and Id133, died due to dilated cardiomyopathy (DCM) at age 8 months and 4 years, respectively. All patients had similar facial characteristics including a broad forehead (100%), hypertelorism (91%), ptosis (91%) and low set ears (91%) (Fig. 2). Eight patients (73%) had a short stature, and in two this was disproportionate (short limbs). One patient was eligible for growth hormone therapy. Connective tissue abnormalities, including joint hypermobility, umbilical and inguinal hernias and skeletal and/or skin abnormalities, were reported in nine patients (82%) (data in Table S1). Nine patients had hypotonia (82%), five reported hearing loss (45%) and seven had mild-to-severe developmental delay (64%).
Table 1.
Genotype and clinical characteristics in TAB2 deletions and variants.
| TAB2 deletion cohort n = 11 | TAB2 deletion literature n = 25 | TAB2 variant cohort n = 14 | TAB2 variant literature n = 22 | |
|---|---|---|---|---|
| Deletion size | 8.67 Mb (1.68–14.31)a | 1.76 Mb (0.12–11.92)a | ||
| Age last follow up | 4 years (8 mos to 40 yrs)a | 9.5 years (15 days to 66 yrs)a | 11.5 years (2–46)a | 30 years (2–75)a |
| Gender (F/M) | 9/2 | 16/9 | 8/6 | 12/10 |
| Birth weight | 10th centile (3–90)a | 25th centile (2–75)a | 10th centile (150)a | |
| Height | −2.5 SD (−3.8 to −1)a | 3rd centile (1–27)a | −2.5 SD (−4.8 to −0.3)a | |
| Head circumference | +0.9 SD (−3 to +1.5)a | 50th centile (1–97)a | −0.8 SD (−1.5 to 0)a | |
| Facial characteristics | ||||
| Broad forehead | 11/11 | 15/25 | 13/14 | 9/22 |
| Hypertelorism | 10/11 | 6/25 | 4/14 | 2/22 |
| Up/downslant | 6/11 | 7/25 | 7/14 | 4/22 |
| Ptosis | 10/11 | 13/25 | 11/14 | 9/22 |
| Ears, abnormal position | 10/11 | 10/25 | 8/14 | 2/22 |
| Broad short neck | 1/11 | 2/25 | 3/14 | 1/22 |
| Heart | ||||
| CHD/TAA | 7/11 | 24/25 | 12/14 | 19/22 |
| Cardiomyopathy | 4/11 | 7/25 | 4/14 | 8/22 |
| Arrhythmia | 2/11 | 4/25 | 2/14 | 4/22 |
| Connective tissue/skeletal | ||||
| Hypermobility | 7/11 | 8/25 | 11/14 | 5/22 |
| Pedes planovalgi | 3/11 | 2/25 | 10/14 | 2/22 |
| Pectus excavatum | 1/11 | 1/25 | 6/14 | 0/22 |
| Hypotonia | 9/11 | 8/25 | 2/14 | 2/22 |
| Hearing loss | 5/11 | 1/25 | 5/14 | 5/22 |
| Developmental delay | 7/11 | 12/25 | 4/14 | 0/22 |
| Definite NSb | 6/11 | 7/14 | ||
For more detailed overview see Table S1: TAB2 deletion cohort, Table S2: TAB2 deletion literature, Table S3: TAB2 variant cohort and Table S4: TAB2 variant literature.
CHD congenital heart disease, F female, M male, NS Noonan syndrome, SD standard deviation, TAA thoracic aorta aneurysm.
aMedian (range)
bSee Table S5 for details.
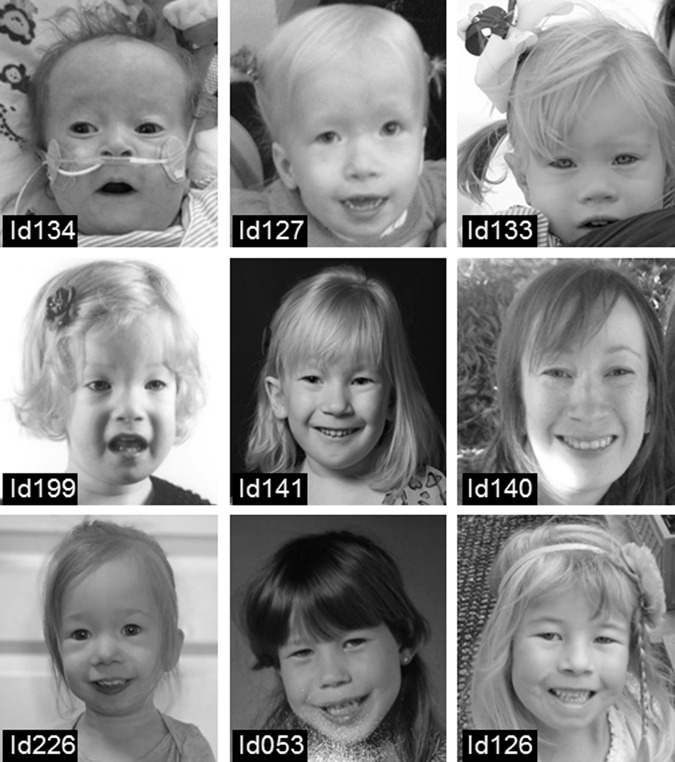
Fig. 2. Clinical photographs of patients with a TAB2 deletion.
Top row (left to right): patient Id134 at age 6 months, patient Id127 at age 4 years and patient Id133 at age 4 years. Middle row (left to right) patient Id199 at age 3 years, patient Id141 at age 6 years and patient Id140 at age 34 years. Bottom row (left to right): patient Id226 at age 2 years, patient Id053 at age 6 years and patient Id126 at age 8 years. Note the characteristic facial phenotype: a broad forehead, hypertelorism, ptosis, low set ears and an upslant in most patients. Written consent was given to the authors to publish the patient’s photographs.
Literature on TAB2 deletions
Twenty-five patients with a TAB2 deletion were described in 11 literature case reports [1–3, 11–18]. The literature data is summarised in Table 1 (details in Table S2). All patients had a cardiac anomaly (100%), and 24 had a structural malformation. Single or multiple valve defects were seen in 20 patients, and most had a mitral valve defect, often in combination with another valve defect. Thirteen patients had an (additional) ASD, VSD, or both. Hypoplastic left heart syndrome (HLHS) was reported in one patient (IV.3), who died shortly after birth [13]. Cardiac surgery was performed in five patients. Three young children had CHD repair including the closure of a VSD [2, 16, 17]. The pulmonic valve was also replaced for one of them. Her grandmother had a mitral valve replacement at age 22 years [17]. The fifth patient underwent aortic root repair and aortic valve replacement at age 51 years [2]. Seven patients developed a cardiomyopathy. Dysmorphic facial characteristics were reported in 22 patients (88%). Twenty-one patients had a short stature (84%). A patient reported by Stagi et al. received growth hormone therapy, with good results reported [18]. Connective tissue abnormalities were reported in 12 patients (48%) (Table S2), 8 patients (32%) had hypotonia and 12 patients (48%) had mild-to-moderate developmental delay.
Patient characteristics of cohort 2: TAB2 variants cohort
The data collected for 14 individuals (8 female, 6 male, median age 11.5 years (range 2‒46 years)) from six families with pathogenic TAB2 variants are summarised in Table 1 (details in Table S3).
After reviewing the facial phenotypes of the cluster of patients with CHD from the Chromosome 6 Project, author WK recognised the 6q25.1 deletion phenotype in two families (C and D) known at the department in Groningen. The patients in these families had normal microarray results.
The proband (D6) of family D was the first patient clinically suspected of a pathogenic TAB2 variant. Family D is a three-generation family with six individuals affected with mitral and pulmonary valve defects, disproportionate short stature, hypermobility, pes planus and hearing loss. NS had been considered, but no variants in known NS-related genes were detected. Exome sequencing with the request to first analyse TAB2 revealed a novel nonsense variant (c.899 C > A (p.(Ser300*))) in the proband, and Sanger sequencing confirmed the variant in another four affected family members. The deceased affected grandmother was an obligate carrier, and the variant was excluded in the grandfather and one non-affected child (Fig. S1D).
In family C, TAB2 was directly analysed by Sanger sequencing after clinical recognition. This is a two-generation family with three affected individuals with mitral valve defects, aortic dilatation, disproportionate short stature, hypermobility or hypotonia, pes planus and pectus excavatum. Sanger sequencing revealed a novel frameshift variant, predicted to result in a premature stop in TAB2 (c.885_886del (p.(Pro296fs))), in all three affected family members (Fig. S1C).
Consecutively, another four novel truncating TAB2 variants were detected by exome sequencing in families A, B, E and F (Table S3). Their pedigrees are shown in Fig. S1, and the positions of the variants are shown in Fig. S2.
In summary, in our six families with a TAB2 variant, 12 out of 14 patients had heart disease (86%). In one patient, a cardiac workup was not performed. Nine patients had mitral valve defects, and four also had a cardiomyopathy. Thoracic aortic aneurysms were reported in three patients from two families. Patient B1 had a VSD, which was surgically closed at age 7 months. Patient D1 had an aortic valve replacement at age 39 years and died at age 46 years, cause unknown. All 14 patients share specific facial characteristics: broad forehead (93%), up/downslant (50%), ptosis (79%) and low set ears (57%) (Fig. 3). Ten patients (71%) had short stature, mostly disproportionate (relatively short limbs). Connective tissue abnormalities were seen in 13 patients (93%) (Table S3): 11 had joint hypermobility, 10 had pes planus and six had pectus excavatum. Hearing loss was reported in five patients (36%) and mild developmental delay in four (29%).
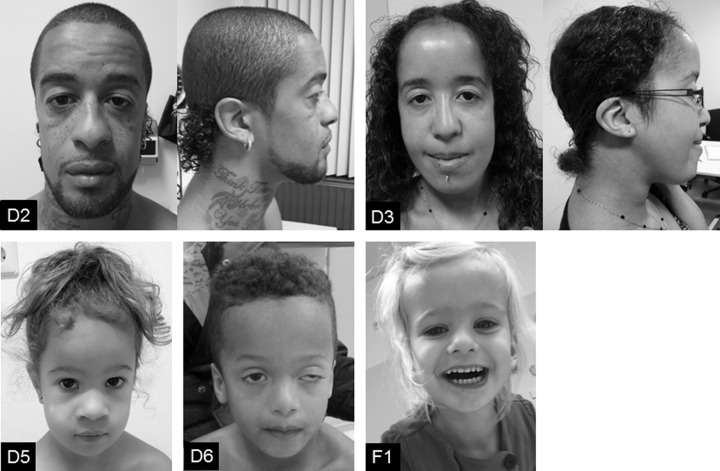
Fig. 3. Clinical photographs of patients with a TAB2 variant.
Top row (left to right): Patient D2 at age 32 years and patient D3 at age 24 years. Bottom row (left to right): patient D5 at age 4 years, patient D6 at age 5 years and patient F1 at age 3 years. Note the characteristic facial phenotype: a broad forehead, an up/downslant, ptosis and low set ears. Written consent was given to the authors to publish the patient’s photographs.
Literature on TAB2 variants
Four familial and four sporadic TAB2 variants in 20 patients were reported in the literature (Fig. S2) [1, 3, 19–23]. The data extracted from the literature reports are summarised in Table 1 (details Table S4). Six variants were truncating, two were missense variants. All 20 literature patients had cardiac diseases (100%). Fourteen patients had mitral valve defects, eight had cardiomyopathy and five had both. One patient had a mitral valve replacement at age 75 years, and his son had a mitral and aortic valve replacement at age 51 years [19]. A 39-year-old man had DCM and a pulmonary artery aneurysm. This aneurysm was resected at age 35 years and a pulmonary homograft valve was implanted. Due to heart failure, he is now on the heart transplant list [21]. One girl was diagnosed with DCM at age 7 months. She received a heart transplant at age 9 months and died at age 2.5 years [22]. A 60-year-old man and a 61-year-old woman died due to sudden cardiac arrythmia and heart failure, respectively [1, 3]. Ten of the 20 patients (50%) were reported to have dysmorphic facial characteristics, 10 had a short stature (50%) and seven had connective tissue abnormalities (35%) (Table S4). Hearing loss was reported in four patients (20%). Developmental delay has not been reported.
In addition to the 20 patients discussed above, Wade et al. described two unrelated individuals with missense variants with a completely different phenotype of frontometaphyseal dysplasia type 3 that was suggested to be caused by gain-of-function variants in TAB2. These patients did not display cardiac anomalies or features overlapping with NS [24, 25].
Clinical overlap with NS in patients with a TAB2 variant or deletion
In four of our families with a TAB2 variant, NS had been considered as a diagnosis, but no variants in NS-associated genes were detected. In one of our TAB2 deletion patients, NS was considered as a diagnosis, but testing was not performed after the abnormal microarray result. In the literature, NS-associated genes were tested in two TAB2 variant cases and two TAB2 deletion cases, but no pathogenic variants were identified [3, 13, 23] (Tables S1–S4).
We retrospectively scored all patients with a TAB2 variant or deletion in our cohorts for the diagnostic criteria of NS. Twenty patients (80%) had facial features suggestive for NS. Six TAB2 deletion patients (55%) and seven TAB2 variant patients (50%) fulfilled the criteria for definite NS (Table S5).
Combining the data from our two cohorts and the literature reports (n = 70), we observed cardiac disease in 93% of patients, mainly mitral valve anomalies (66%) and/or cardiomyopathy (34%); short stature in 70%; connective tissue features including hypermobility in 59% and dysmorphic facial features in 81%. Surgical interventions for cardiac anomalies are reported in 13 patients (19%). Four patients died from the (late) effects of structural heart disease, and three patients with DCM died in infancy.
Discussion
According to Online Mendelian Inheritance in Man (OMIM), TAB2 is associated with non-syndromic CHD (MIM#614980) [26]. However, our study clearly demonstrates that the phenotype is not restricted to the heart but is a recognisable syndrome that shows considerable overlap with NS. Our study further demonstrates how social media recruitment can be a very powerful tool to delineate rare diseases.
Only two earlier reports discussed the possibility of a broader phenotype [2, 3]. Other reports occasionally mentioned dysmorphic features, but a TAB2-related syndrome was not considered. This may be due to a lack of detailed clinical data in these other reports, or to variability in gene expression. A milder phenotype, restricted to the heart, in the two missense variants reported by Thienpont et al. cannot be excluded [1]. As the features we describe are recurrently seen in both TAB2 deletion patients and patients with pathogenic TAB2 variants, they may be caused by loss-of-function (LOF) of one TAB2 allele. However, the genetic heterogeneity in TAB2-related disease is intriguing. The completely different phenotypes reported by Wade et al. associated with TAB2 missense variants are assumed to be caused by gain of function of TAB2 [24, 25].
Data from 70 patients (25 described by us and 45 from literature) demonstrates that the recurrent TAB2-related phenotype should be included in the differential diagnosis of NS.
Heart disease is a key feature of the TAB2-related phenotype. All patients with TAB2 variants or deletions described in the literature had cardiac anomalies. Our data confirm that cardiac disease is a frequent feature, however, five of our patients did not show cardiac disease (three patients with a TAB2 deletion and two with a TAB2 variant). An investigative and/or publication bias towards patients with heart disease in literature and the limited information on genotypes and phenotypes of relatives in most reports may explain the difference. However, this difference may also partly be due to the slightly younger median age of our patients compared to the literature (4 vs 9.5 years in deletions; 11.5 vs 30 years in variants), which may have resulted in a lower number of patients with (late onset) cardiomyopathy. In our cohorts and in the literature, a variety of cardiac anomalies are observed, with mitral valve defects reported most frequently, often in combination with defects of one or more of the other valves. ASDs and/or VSDs are reported more often in TAB2 deletions, both in our cohort and in the literature, in comparison to patients with a TAB2 variant, suggesting involvement of other genes. Interestingly, CITED2 (CBP/p300-interacting transactivator, with glu/asp-rich c-terminal domain 2, MIM*602937, 6q24.1) is known to be associated with ASDs and VSDs, but this gene was only deleted in one patient (Id134). In the other 16 TAB2 deletion patients with ASDs and/or VSDs, the deletion did not include genes known to cause cardiac defects other than TAB2. Furthermore, cardiomyopathy, often DCM, is present in 34% of the patients overall, and the prevalence could actually be higher, as cardiomyopathy can occur later in life and most of the reported patients are children.
The most severe structural cardiac disease was reported by Cheng et al. [2, 13]. Two children in one family were born with HLHS and died shortly after birth. One of these children had the familial TAB2 deletion; the other was not tested. Considering that this is the only report of HLHS associated with a TAB2 variant, HLHS may either be the most severe presentation in the spectrum of cardiac valve defects, or another genetic variant might be involved in this family. Unfortunately, exome sequencing was not performed [2, 13].
The cardiac anomalies observed in our cohorts and in previously reported patients required surgery in some cases and caused early death in a few patients. TAB2-associated cardiac anomalies appear more severe than those observed in NS. In addition, the mitral valve is more frequently involved in TAB2-associated cardiac anomalies as opposed to the pulmonary valve in NS.
Short stature (disproportionate in most reports) is the second most frequent characteristic of the TAB2-related phenotype. Apart from TAB2, two genes have been suggested as candidate genes for short stature in deletion cases: LATS1 (large tumour suppressor kinase 1, MIM*603473, 6q25.1) [15] and PLAGL1 (pleomorphic adenoma gene-like 1, MIM*603044, 6q24.2) [18]. Because the prevalence of (disproportionate) short stature was equally high in our two cohorts, we conclude that the short stature in patients with 6q25.1 deletions is most likely caused by TAB2 haploinsufficiency.
Typical facial dysmorphisms are also part of the TAB2-related phenotype and were similar in our two cohorts. Twenty of the 25 patients (80%) had a face suggestive of NS. Facial characteristics overlapping with those in NS were reported previously, most often in TAB2 deletion patients. Connective tissue anomalies, most often joint hypermobility, were reported in 82% of our TAB2 deletion patients and 93% of our TAB2 variant patients. In the literature, this was 48% and 35%, respectively. The gene UST (uronyl 2-sulfotransferase, MIM*610752) has been suggested to cause the connective tissue anomalies seen in TAB2 deletion patients [15]. However, we do not find clear differences between our two cohorts, which suggests that LOF of TAB2 is the most probable explanation for these features.
Developmental delay in individuals with a deletion including TAB2 is variable. Moderate-to-severe developmental delay was only reported in individuals with a deletion larger than 6.47 Mb. This might very well be caused by the LOF effect of other genes or regulatory sequences involved in the deletion. However, four patients in our TAB2 variant cohort also had mild developmental delay. This may suggest TAB2 plays a role in psychomotor development, but the numbers are too low to be conclusive given that this has not been reported in 20 patients with TAB2 variants in previous publications.
Finally, hearing loss and decreased vision are present in several of our patients and the patients reported in the literature. It remains unclear whether these problems are part of the TAB2-related phenotype, or coincidental, as the numbers are low and detailed clinical data are not available [21].
In our two cohorts, all patients had a syndromic phenotype. Non-syndromic TAB2-associated CHD has been reported in the literature, but these studies unfortunately do not include clinical photographs [1, 17, 20]. The primary focus on heart disease in these studies might have hampered the recognition of other mild features, including facial characteristics, connective tissue abnormalities or developmental delay, although we cannot rule out substantial variability in expression.
We confirmed that the TAB2-related phenotype has overlap with NS, as suggested previously by Cheng et al. in patients with TAB2 deletions [2]. More than half of our patients could be clinically diagnosed with definite NS based on the scoring system of Van der Burgt et al. [10]. The face was suggestive of NS in 80%. However, in our opinion the typical NS “Gestalt” is different from the facial features in the TAB2-related phenotype, especially the eyes appear different, with quite narrow palpebral fissures as most clearly shown in patients Id140, Id141 and F1. Our patients also present with hypermobility and increased skin folds, which is not typical for NS. Since the TAB2-related phenotype is equally present in both TAB2 deletion patients and patients with pathogenic TAB2 variants, we expect the TAB2-related phenotype to be a result of LOF of TAB2.
Several functional and expression studies support that a TAB2 variant may be related to NS/RASopathies. An in-silico analysis showed that TAB2 is in strong co-expression with SOS1 (Fig. S3) [27], one of the many genes of the MAPK pathway known to be associated with NS [28]. TAB2 is also an activator of MAP3K7 (mitogen-activated protein kinase kinase kinase 7, MIM*602614, coding for the TAK1 protein), another member of the MAPK pathway. These data suggest that the TAB2-related phenotype may be associated with the RASopathies, although further research is needed to confirm this hypothesis. The role of Tab2 in cardiac development has also been shown in mice and zebrafish [1, 29]. In humans, TAB2 is expressed in endothelial cells of the ventricular trabeculae and the endocardial cushions of human embryos [1]. The endocardial cushions are involved in cardiac valve formation, and cardiac valve abnormalities are the most common cardiac problem in our TAB2-related phenotype. Morlino et al. demonstrated that a TAB2 variant (c.1398dup (p.(Thr467fs))) encodes a truncated protein that is unable to bind to TAK1 [27]. They showed reduced proliferation in patient’s fibroblasts, which could explain some clinical features we observed, such as short stature, skeletal underdevelopment and facial characteristics [30].
Zhang et al. have shown that TAB2 expression is essential for prolonged activation of the protein TAK1 [31]. Haploinsufficiency of TAB2 might therefore result in a decreased activation of TAK1. An intronic variant in MAP3K7 causing a two amino-acid insertion in the kinase domain of TAK1 has been reported in a patient with cardiospondylocarpofacial syndrome (CSCFS, MIM#157800), which has overlap with the TAB2-related phenotype, specifically polyvalvular cardiac anomalies, short stature and connective tissue abnormalities [30]. Thus far it is claimed that MAP3K7 deletion patients do not have any overlap with CSCFS [30]. However, we previously reviewed patients with deletions including MAP3K7 (n = 14) and found characteristics similar to those seen in the TAB2-related phenotype: short stature, hypermobility and cardiac anomalies (mainly ASDs) but no cardiac valve defects [5]. All the MAP3K7 deletions we reviewed were >6.43 Mb and contained other genes that might explain the phenotype. Since no valve defects are seen in patients with a MAP3K7 deletion, the interaction of TAB2 and MAP3K7 must be more complex than a simple lack of activation of MAP3K7.
Our report of 25 patients with TAB2 variants or deletions has some limitations. The data of deletion patients collected via online recruitment is self-reported information provided by parents, which may not always be accurate. However, these data have been compared with information collected from medical professionals and appear to be highly consistent (unpublished data). Another limitation may be that our TAB2 variant patients were collected retrospectively. Therefore, data on the overall prevalence of TAB2 variants is not available. In addition, the reported phenotype might be biased towards the more severe end of the spectrum, as these patients are more likely to undergo exome sequencing or microarray testing.
We further demonstrated that recruiting patients via social media can result in efficient case finding. The data on 6q25.1 deletion patients collected via social media was sufficiently detailed and complete to allow us to recognise this phenotype in families with normal microarray results. We were then able to prove that TAB2 is responsible for the phenotype in 6q25.1 deletion patients by identifying TAB2 variants in six families. Combining data from our cohorts and from literature allowed us to delineate the TAB2-related phenotype.
Based on this phenotype, we recommend that TAB2 be added to the gene panels used in genetic work-up of patients with congenital heart disease, cardiomyopathy, short stature and connective tissue disease, and those suspected for NS, as well as to neonatology rapid sequencing panels. We also recommend CNV analysis for deletions including TAB2 in these patients. Based on the clinical and molecular relationship with MAP3K7, analysis of this gene should also be considered in these patients.
For individuals with a newly detected TAB2 variant or deletion, we recommend cardiac evaluation followed by regular follow-up, as valvular heart disease, thoracic aneurysm and cardiomyopathy may manifest later in life. We also recommend evaluation of overall development, hearing, vision and connective tissue abnormalities. Genetic counselling should be offered to all individuals with a known TAB2 variant or deletion. Further studies are required to delineate the role of TAB2 in causing short stature and to examine the feasibility of growth hormone therapy.
In conclusion, we successfully used social media to delineate the TAB2-related phenotype as a syndrome with cardiac disease, short stature, specific facial appearance, connective tissue abnormalities (mainly hypermobility) and occasionally mild intellectual disability. This phenotype may be caused by heterogeneous genetic conditions, either a deletion including the gene TAB2, or a truncating, or missense variant in TAB2. We propose naming this phenotype “TAB2-related syndrome”. We recommend adding TAB2 to appropriate gene panels and testing for deletions including TAB2 in patients who have congenital heart disease, cardiomyopathy, short stature and/or connective tissue disease and/or are suspected for NS. We also recommend thorough evaluation and follow-up in patients diagnosed with a TAB2-related syndrome, including regular screening for cardiomyopathy and thoracic aneurysm. More data are needed to develop detailed follow-up guidelines for the TAB2-related syndrome.
Supplementary information
Supplementary Table S5 and Figures S1-S3
Table S1. Genotype and phenotype details of TAB2 deletions
Table S2. Genotype and phenotype details of TAB2 deletions literature
Table S3. Genotype and phenotype details of TAB2 variants
Table S4. Genotype and phenotype details of TAB2 variants literature
Acknowledgements
We would like to express our gratitude to all the children and families for their participation. We also thank Kate McIntyre for editing the manuscript and the members of the Groningen Genomic Coordination Centre. Our special thanks go to Pauline Bouman, our contact for the Chromosome 6 Facebook group.
Author contributions
Conceptualization: AE, CMAR, WSK; Data curation: AE; Funding acquisition: AE, BF, CMAR, WSK; Investigation: AE, CMAR, WSK; Methodology: AE, CMAR, WSK; Resources: AE, EKSML, BF, PAT, KL, BBAV, TD, YJV, TR, MPB, MTRR, WSK; Supervision: WSK; Visualization: AE, PD; Writing – original draft: AE; Writing – review & editing: AE, EKSML, BF, PAT, KL, BBAV, TD, YJV, TR, MPB, MTRR, CMAR, WSK.
Funding
This work was supported by a grant from ZonMw (113312101) and by crowd-funding organised by Chromosome 6 parents. AE is recipient of a Junior Scientific Masterclass MD/PhD scholarship of the University Medical Center Groningen.
Competing interests
The authors declare no competing interests.
Ethical approval
The accredited Medical Ethics Review Committee of the University Medical Center Groningen waived full ethical evaluation because, according to Dutch guidelines, no ethical approval is necessary if medical information that was already available is used anonymously and no extra tests have to be performed.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00948-0.
References
- 1.Thienpont B, Zhang L, Postma AV, Breckpot J, Tranchevent L, Van Loo P, et al. Haploinsufficiency of TAB2 causes congenital heart defects in humans. Am J Hum Genet. 2010;86:839–49. doi: 10.1016/j.ajhg.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng A, Dinulos MBP, Neufeld‐Kaiser W, Rosenfeld J, Kyriss M, Madan‐Khetarpal S, et al. 6q25.1 (TAB2) microdeletion syndrome: congenital heart defects and cardiomyopathy. Am J Med Genet Part A. 2017;173:1848–57. doi: 10.1002/ajmg.a.38254. [DOI] [PubMed] [Google Scholar]
- 3.Ritelli M, Morlino S, Giacopuzzi E, Bernardini L, Torres B, Santoro G, et al. A recognizable systemic connective tissue disorder with polyvalvular heart dystrophy and dysmorphism associated with TAB2 mutations. Clin Genet. 2018;93:126–33. doi: 10.1111/cge.13032. [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Yin J, Yu H, Yuan T, Fernandez M, Yung CK, et al. Next-generation sequencing identifies rare variants associated with Noonan syndrome. Proc Natl Acad Sci USA. 2014;111:11473–8. doi: 10.1073/pnas.1324128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engwerda A, Frentz B, den Ouden AL, Flapper BCT, Swertz MA, Gerkes EH, et al. The phenotypic spectrum of proximal 6q deletions based on a large cohort derived from social media and literature reports. Eur J Hum Genet. 2018;26:1478–89. [DOI] [PMC free article] [PubMed]
- 6.Swertz MA, Dijkstra M, Adamusiak T, van der Velde JK, Kanterakis A, Roos ET, et al. The MOLGENIS toolkit: rapid prototyping of biosoftware at the push of a button. BMC Bioinform. 2010;11:S12–S12. doi: 10.1186/1471-2105-11-S12-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corsten-Janssen N, Bouman K, Diphoorn JCD, Scheper AJ, Kinds R, El Mecky J, et al. A prospective study on rapid exome sequencing as a diagnostic test for multiple congenital anomalies on fetal ultrasound. Prenat Diagn. 2020;40:1300–9. [DOI] [PMC free article] [PubMed]
- 8.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 9.Terhal PA, Vlaar JM, Middelkamp S, Nievelstein RAJ, Nikkels PGJ, Ross J, et al. Biallelic variants in POLR3GL cause endosteal hyperostosis and oligodontia. Eur J Hum Genet. 2020;28:31–39. doi: 10.1038/s41431-019-0427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Der Burgt I, Berends E, Lommen E, Van Beersum S, Hamel B, Mariman E. Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am J Med Genet. 1994;53:187–91. doi: 10.1002/ajmg.1320530213. [DOI] [PubMed] [Google Scholar]
- 11.Nowaczyk MJM, Carter MT, Xu J, Huggins M, Raca G, Das S, et al. Paternal deletion 6q24.3: A new congenital anomaly syndrome associated with intrauterine growth failure, early developmental delay and characteristic facial appearance. Am J Med Genet Part A. 2008;146A:354–60. doi: 10.1002/ajmg.a.32144. [DOI] [PubMed] [Google Scholar]
- 12.Meloni VA, Guilherme RS, Oliveira MM, Migliavacca M, Takeno SS, Sobreira NLM, et al. Cytogenomic delineation and clinical follow‐up of two siblings with an 8.5 Mb 6q24.2‐q25.2 deletion inherited from a paternal insertion. Am J Med Genet Part A. 2014;164:2378–84. doi: 10.1002/ajmg.a.36631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng A, Neufeld-Kaiser W, Byers PH, Liu YJ. 6q25.1 (TAB2) microdeletion is a risk factor for hypoplastic left heart: a case report that expands the phenotype. BMC Cardiovas Disord. 2020;20:137. doi: 10.1186/s12872-020-01404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caselli R, Mencarelli MA, Papa FT, Uliana V, Schiavone S, Strambi M, et al. A 2.6 Mb deletion of 6q24.3–25.1 in a patient with growth failure, cardiac septal defect, thin upperlip and asymmetric dysmorphic ears. Eur J Med Genet. 2007;50:315–21. doi: 10.1016/j.ejmg.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Salpietro V, Ruggieri M, Mankad K, Di Rosa G, Granata F, Loddo I, et al. A de novo 0.63 Mb 6q25.1 deletion associated with growth failure, congenital heart defect, underdeveloped cerebellar vermis, abnormal cutaneous elasticity and joint laxity. Am J Med Genet Part A. 2015;167:2042–51. doi: 10.1002/ajmg.a.37118. [DOI] [PubMed] [Google Scholar]
- 16.Bisgaard A, Kirchhoff M, Tümer Z, Jepsen B, Brøndum‐Nielsen K, Cohen M, et al. Additional chromosomal abnormalities in patients with a previously detected abnormal karyotype, mental retardation, and dysmorphic features. Am J Med Genet Part A. 2006;140A:2180–7. doi: 10.1002/ajmg.a.31425. [DOI] [PubMed] [Google Scholar]
- 17.Weiss K, Applegate C, Wang T, Batista DAS. Familial TAB2 microdeletion and congenital heart defects including unusual valve dysplasia and tetralogy of fallot. Am J Med Genet Part A. 2015;167:2702–6. doi: 10.1002/ajmg.a.37210. [DOI] [PubMed] [Google Scholar]
- 18.Stagi S, Lapi E, Pantaleo M, Carella M, Petracca A, De Crescenzo A, et al. A new case of de novo 6q24.2-q25.2 deletion on paternal chromosome 6 with growth hormone deficiency: a twelve-year follow-up and literature review. BMC Med Genet. 2015;16:69–z. doi: 10.1186/s12881-015-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Permanyer E, Laurie S, Blasco-Lucas A, Maldonado G, Amador-Catalan A, Ferrer-Curriu G, et al. A single nucleotide deletion resulting in a frameshift in exon 4 of TAB2 is associated with a polyvalular syndrome. Eur J Med Genet. 2020;63:103854. [DOI] [PubMed]
- 20.Chen J, Yuan H, Xie K, Wang X, Tan L, Zou Y, et al. A novel TAB2 nonsense mutation (p.S149X) causing autosomal dominant congenital heart defects: a case report of a Chinese family. BMC Cardiovasc Disord. 2020;20:27. doi: 10.1186/s12872-019-01322-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caulfield TR, Richter JE, Brown EE, Mohammad AN, Judge DP, Atwal PS. Protein molecular modeling techniques investigating novel TAB2 variant R347X causing cardiomyopathy and congenital heart defects in multigenerational family. Mol Genet Genom Med. 2018;6:666–72. doi: 10.1002/mgg3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasilescu C, Ojala TH, Brilhante V, Ojanen S, Hinterding HM, Palin E, et al. Genetic basis of severe childhood-onset cardiomyopathies. J Am Coll Cardiol. 2018;72:2324–38. doi: 10.1016/j.jacc.2018.08.2171. [DOI] [PubMed] [Google Scholar]
- 23.Ackerman JP, Smestad JA, Tester DJ, Qureshi MY, Crabb BA, Mendelsohn NJ, et al. Whole exome sequencing, familial genomic triangulation, and systems biology converge to identify a novel nonsense mutation in TAB2‐encoded TGF‐beta activated kinase 1 in a child with Polyvalvular Syndrome. Congenit Heart Dis. 2016;11:452–61. doi: 10.1111/chd.12400. [DOI] [PubMed] [Google Scholar]
- 24.Wade E, Daniel P, Jenkins Z, McInerney-Leo A, Leo P, Morgan T, et al. Mutations in MAP3K7 that Alter the activity of the TAK1 signaling complex cause frontometaphyseal dysplasia. Am J Hum Genet. 2016;99:392–406. doi: 10.1016/j.ajhg.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade EM, Jenkins ZA, Daniel PB, Morgan T, Addor MC, Adés LC, et al. Autosomal dominant frontometaphyseal dysplasia: Delineation of the clinical phenotype. Am J Med Genet Part A. 2017;173:1739–46. doi: 10.1002/ajmg.a.38267. [DOI] [PubMed] [Google Scholar]
- 26.McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Online Mendelian Inheritance in Man, OMIM®. https://omim.org/. Accessed 1 July 2020.
- 27.Deelen P, van Dam S, Herkert JC, Karjalainen JM, Brugge H, Abbott KM, et al. Improving the diagnostic yield of exome- sequencing by predicting gene-phenotype associations using large-scale gene expression analysis. Nat Commun. 2019;10:2837–4. doi: 10.1038/s41467-019-10649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepri F, De Luca A, Stella L, Rossi C, Baldassarre G, Pantaleoni F, et al. SOS1 mutations in Noonan syndrome: molecular spectrum, structural insights on pathogenic effects, and genotype–phenotype correlations. Hum Mutat. 2011;32:760–72. doi: 10.1002/humu.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orelio C, Dzierzak E. Identification of 2 novel genes developmentally regulated in the mouse aorta-gonad-mesonephros region. Blood. 2003;101:2246–9. doi: 10.1182/blood-2002-07-2260. [DOI] [PubMed] [Google Scholar]
- 30.Morlino S, Castori M, Dordoni C, Cinquina V, Santoro G, Grammatico P, et al. A novel MAP3K7 splice mutation causes cardiospondylocarpofacial syndrome with features of hereditary connective tissue disorder. Eur J Hum Genet: EJHG. 2018;26:582–6. doi: 10.1038/s41431-017-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Macartney T, Peggie M, Cohen P. Interleukin-1 and TRAF6-dependent activation of TAK1 in the absence of TAB2 and TAB3. Biochem J. 2017;474:2235–48. doi: 10.1042/BCJ20170288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S5 and Figures S1-S3
Table S1. Genotype and phenotype details of TAB2 deletions
Table S2. Genotype and phenotype details of TAB2 deletions literature
Table S3. Genotype and phenotype details of TAB2 variants
Table S4. Genotype and phenotype details of TAB2 variants literature