Abstract
Robust machine learning models based on radiomic features might allow for accurate diagnosis, prognosis, and medical decision-making. Unfortunately, the lack of standardized radiomic feature extraction has hampered their clinical use. Since the radiomic features tend to be affected by low voxel statistics in regions of interest, increasing the sample size would improve their robustness in clinical studies. Therefore, we propose a Generative Adversarial Network (GAN)-based lesion-focused framework for Computed Tomography (CT) image Super-Resolution (SR); for the lesion (i.e., cancer) patch-focused training, we incorporate Spatial Pyramid Pooling (SPP) into GAN-Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE). At SR, the proposed model achieved better perceptual quality with less blurring than the other considered state-of-the-art SR methods, while producing comparable results at SR. We also evaluated the robustness of our model’s radiomic feature in terms of quantization on a different lung cancer CT dataset using Principal Component Analysis (PCA). Intriguingly, the most important radiomic features in our PCA-based analysis were the most robust features extracted on the GAN-super-resolved images. These achievements pave the way for the application of GAN-based image Super-Resolution techniques for studies of radiomics for robust biomarker discovery.
Subject terms: Biomarkers, Cancer, Medical imaging, Biomarkers, Cancer imaging, Lung cancer, Biomedical engineering
Introduction
Recently, medical image analysis has been revolutionized by available large-scale datasets and technology advancements in statistics and artificial intelligence. In particular, combining radiomics1—an approach to extract quantitative features from medical images—and machine learning has obtained meaningful clinical insights. Robust machine learning models based on large-scale radiomic features might allow for accurate diagnosis, prognosis, and medical decision-making; of course, thoroughly considering the whole radiomic processes is essential to obtain these reliable models.
Despite the potential of radiomics, high quantitative feature variability across different software implementations has hampered its clinical use2,3. This phenomenon derives from the lack of standardized definitions and extraction of radiomic features with validated reference values. To tackle this limitation and facilitate clinical interpretation, the Image Biomarker Standardization Initiative2 produced and validated the reference values for commonly-used radiomic features. However, as the paper’s authors highlighted, image features still need to be robust against differences in acquisition, reconstruction, and segmentation to ensure reproducibility. For this reason, recent studies have investigated the robustness of radiomic features in several scenarios and applications using heterogeneous datasets. Several sources of variability have been assessed, such as image and region of interest (ROI) perturbations4,5, slice thickness variations6,7, and different resampling strategies8. Since the radiomic features might tend to be affected by low statistics in ROI voxels, we hypothesize that increasing such a sample size would increase the robustness of radiomic features in clinical studies. Therefore, we aim to apply image Super-Resolution (SR) to increase the number of voxels used in the computation of radiomic features.
Generative Adversarial Networks (GANs) have been commonly exploited for Data Augmentation (DA), along with image SR9, thanks to their ability to improve feature robustness. Sandfort et al.10 used CycleGAN11-based DA for Computed Tomography (CT) segmentation by translating contrast images into synthetic non-contrast ones. To maximize the DA effect with GAN combinations, Han et al.12 proposed a two-step GAN-based DA approach that generates and refines brain Magnetic Resonance (MR) images with/without tumors separately. Considering the GAN-based DA’s interpolation/extrapolation effect, GAN may remarkably help achieve reference values for radiomic features. The most prominent work on CT image SR is GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE)13, outperforming previous works14–17. GAN-CIRCLE can preserve anatomical information and suppress noise, leading to excellent diagnostic performance in terms of traditional image quality metrics13,18. For example, Guha et al.18 exploited GAN-CIRCLE to super-resolve trabecular bone microstructures and improved the structural similarity index. Meanwhile, GAN-based lesion-focused medical image SR can improve SR performance around lesions, especially for downstream radiomic analyses19. Along with GAN-based medical image SR, novel approaches based on progressive GANs20 and attention mechanisms21 have been recently applied to video SR.
For the first time, in this paper, we evaluate the robustness of radiomic features extracted from super-resolved images by GAN-SR and bicubic interpolation. The authors incorporated Spatial Pyramid Pooling (SPP)22 into the discriminator of GAN-CIRCLE13 to handle different input CT image sizes for patch-focused training in lesions; we cropped the input CT images to their lesion bounding boxes to reduce training costs and improve image quality (e.g., fewer artifacts)19. Along with perceptual quality evaluation, we also assessed the robustness of radiomics, in terms of quantization, for our model against a bicubic interpolation baseline on a separate lung cancer CT dataset. We found that the most important radiomic features in our Principal Component Analysis (PCA)-based examination were the most robust features extracted on the GAN-super-resolved images.
To summarize, this work provides the following contributions:
definition of the first GAN-based, lesion-focused, SR framework for CT images;
comparison with state-of-the-art SR techniques highlighting the suitability of the proposed framework;
at SR, the images are characterized by better perceptual quality, as suggested by the peak signal-to-noise ratio and structural similarity index measures, on a large-scale dataset;
at SR, the proposed GAN-based model achieves comparable results to the ones obtained by state-of-the-art SR techniques;
the proposed GAN-SR framework improves the robustness of the most important radiomic features in an independent lung CT dataset.
Materials and methods
Analyzed CT datasets
DeepLesion dataset
As a subset of the DeepLesion dataset23, which contains 10, 594 scans of 4, 427 patients, our study exploits 10, 000 CT slices with an image size of pixels and in-plane pixel spacing between 0.18 and 0.98 mm (median: 0.82 mm). The dataset contains diverse lesion images for various body parts with 2D lesion information on diameter measurements, bounding boxes, and semantic labels. We use the DeepLesion dataset to train a GAN-CIRCLE model for SR.
NSCLC-radiomics dataset
The Non-Small Cell Lung Cancer-Radiomics (NSCLC-Radiomics) dataset24 is a well-established publicly available dataset that contains CT slices from 422 NSCLC patients. For careful and reliable radiomic analyses, our study uses a highly homogeneous subset composed of 142 CT scans, accounting for 17, 938 CT slices with an image size of pixels, in-plane pixel spacing of 0.98 mm, and slice thickness of 3.00 mm. The B19f convolution kernel was applied on all the scans for CT image reconstruction.
The dataset provides annotated 3D tumor segmentation masks and clinical outcome data. The images are used to assess our proposed lesion-focused CIRCLE-GAN framework in terms of radiomic feature robustness.
The proposed GAN-powered framework for radiomic feature robustness
Pre-processing
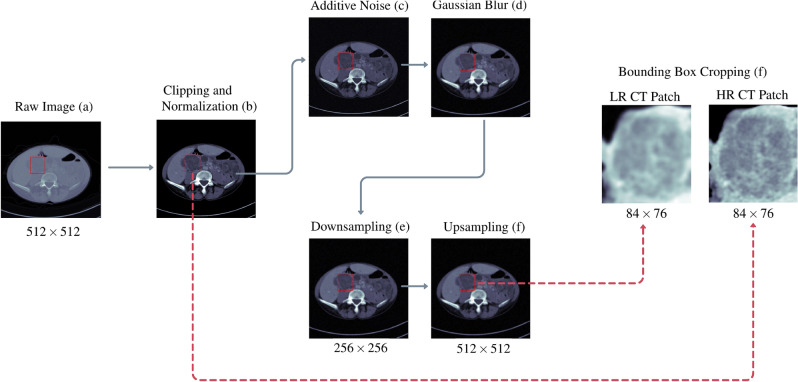
For all the implemented SR approaches, the range of intensity for raw CT volumes was clipped to Hounsfield Units (HU), and then normalized to [0, 1]. We generated the Low-Resolution CT (LRCT) counterparts from the High-Resolution CT (HRCT) images by degrading them through a Gaussian white noise process with a standard deviation of 0.25 and a Gaussian blur, with a kernel size of pixels and a bandwidth of 1.6. Afterwards, the images were downsampled with a scale of 2 and upsampled using the nearest neighbor interpolation, according to You et al.13. The upsampling step improves feature extraction by enforcing the same image size for LRCT and HRCT25. As in the original GAN-CIRCLE13, for convenience in the training of our proposed network, we upsampled the LR image via proximal interpolation to ensure that input and output have the same size. Image patches were then cropped based on the lesion bounding box annotations in the metadata—the cropping process leads to avoiding artifact generation out of the lesion area19. The preprocessing pipeline is displayed in Fig. 1.
Figure 1.
CT image preprocessing pipeline for GAN training. The HU values of input CT images (a) were clipped to the range HU and normalized to the unit range [0, 1] (b). To generate the low resolution CT image counterpart, the image was perturbed by noise addition (c) and Gaussian blurring (d), downsampled by a factor of (e) and then upsampled to the original dimension (f) using a nearest neighbor interpolation method. Finally, the HRCT patch and LRCT patch were extracted from the lesion bounding box crops (g).
By applying this procedure only on the Deeplesion dataset, we generated 10, 000 LRCT/HRCT patches with similar image sizes for training a CIRCLE-GAN-based SR model.
CIRCLE-GAN-based image super resolution
Network architecture
We used a modified version of CIRCLE-GAN13 to tackle the SR problem effectively. The CIRCLE-GAN is a cycle-consistent adversarial model consisting of two non-linear generative mappings and their respective discriminators that are trained jointly for optimal convergence.
The first generative mapping attempts to generate a realistic high-resolution image that a discriminator cannot distinguish from the real one, whereas a generative mapping is responsible for generating a realistic low-resolution image , not distinguishable by a discriminator . This minimax game is formulated as follows:
| 1 |
The generator networks G and F share the same architecture, which consists of networks for feature extraction and reconstruction. The feature extraction network consists of twelve layers (i.e., feature blocks) of convolution kernels, bias, Leaky Rectified Linear Unit (ReLU) activation, and dropout. Each block output is concatenated through skip connections before the reconstruction network to capture local/global image features. The number of output filters in each convolutional layer is set according to You et al.13. In the reconstruction network, two branches are stacked in a network-in-network fashion to increase non-linearity and potentially reduce the filter space dimension for faster computation. A transposed convolutional layer with is adopted for upsampling and the last convolutional layer combines all feature maps to produce the SR output.
The discriminators and also share the same network architecture, which is composed of four blocks of convolution kernel, bias, instance normalization, and Leaky ReLU activation followed by an SPP layer and then two dense layers. Inspired by He et al.26, the SPP layer was added to handle multi-sized LRCT/HRCT input patches, allowing for the training of a lesion patch-focused network. Figure 2 displays the discriminator and generator architectures used in our work.
Figure 2.
The discriminator and generator architectures devised for GAN-SR of medical images.
Similar to GAN-CIRCLE13, the loss function combines four different loss terms to regularize the training procedure by enforcing the desired mappings:
an adversarial loss term () to enforce the matching of empirical distributions in the source and target domains;
a -norm cycle-consistency loss term () to prevent degeneracy in the adversarial learning and promote forward and backward cycle consistency, defined as and ;
a -norm identity loss term () to regularize the training process and promote the relationships and ;
a joint sparsifying loss term () to promote image sparsity and reduced noise.
Thus, the overall loss function used for training is defined as:
| 2 |
where , and are weighting parameters to balance the different loss terms, respectively.
Implementation details
The proposed network was trained in an end-to-end fashion to optimize the loss function; the convolution layers’ weights were initialized with a zero-mean Gaussian distribution, with a standard deviation of 2/m, where , f is a filter size, and is the number of filters; this initialization can relieve diminishing gradients and improve the convergence of deep network architectures27.
The discriminators’ learning rate was set to equally for and , while the learning rate for the generators G and F was set to , following the Two Times Update Rule (TTUR)28, to improve GAN convergence under mild assumptions. Dropout regularization layers, applied in the generators, were initialized with the rate . Leaky ReLU layers were initialized with the negative slope coefficient . The loss weights , , and were set to 1, 0.5 and 0.00001, respectively.
The training used the Adam optimizer with exponential decay rates of and during 100 epochs with batches of 16 images. On average, the training took 9-11 hours per iteration, using TensorFlow (version 2.3.0) on a shared HPC workspace with an Nvidia Tesla P100 Graphics Processing Unit (GPU). The implemented code is available under the GNU license on https://github.com/erickcfarias/SR-CIRCLE-GAN.
Model evaluation and comparisons
To evaluate the trained model, conventional quantitative metrics—namely, Peak Signal-to-Noise Ratio (PSNR) and Structural Similarity Index Measure (SSIM)—were calculated on 1, 000 CT images held out for performance evaluation. As a baseline for comparison, we also resampled the images using a Bicubic interpolation method.
To test the effectiveness of our framework, we compared it with other state-of-the-art methods, namely: Image Super-Resolution Network with an Expectation-Maximization Attention Mechanism (EMASRN21), Enhanced Deep Super-Resolution (EDSR29), Cascading Residual Network (CARN30) and Super-Resolution based on Dictionary Learning and Sparse Representation (DLSR31). For the EMASRN model, we relied on the implementation available at https://github.com/xyzhu1/EMASRN, optimizing the network for -norm loss during 1000 epochs with , a batch size of 16, and a learning rate of halved every 200 epochs. For the EDSR model, we trained the network with the Adam optimizer with , , optimizing for -norm loss during 500 epochs, a batch size of 16, and a learning rate of halved every 100 epochs. For the CARN model, we trained the network with the Adam optimizer with , , optimizing for -norm loss during 500 epochs, a batch size of 16, and a learning rate of halved every 100 epochs. For the DLSR model, we trained the dictionaries with a size of 2048 atoms, using 100, 000 randomly sampled patches, a sparsity regularization parameter and -pixel patches with an overlap of 4 pixels between adjacent patches. We varied the upscale rate to generate the and versions for all the tested models.
To further assess the performance of the proposed GAN-CIRCLE-based SR method, at SR, we compared the native GAN-CIRCLE SR against the sequential application of two GAN-CIRCLE instances at SR, denoted as GAN-CIRCLEx.
Radiomic feature extraction
The radiomic features considered in this study were computed using PyRadiomics (version 2.2.0)32, an open-source Python package widely used for this purpose. Since this software requires image input to be in the Neuroimaging Informatics Technology Initiative (NIfTI) format33, a preliminary step was performed to convert the original Digital Imaging and Communications in Medicine (DICOM) scan and segmentation files to this format using custom software written in MATLAB (The Mathworks Inc., Natick, MA, USA) version R2019b.
Excluding the shape-based features and first-order features (since they are independent of the rebinning), 75 3D radiomic texture features were calculated without any image filters applied from the following categories: Gray-Level Co-occurrence Matrix features (GLCM)34–36 (24), Gray-Level Dependence Matrix (GLDM)37 (14), Gray-Level Run Length Matrix (GLRLM)38 (16), Gray-Level Size Zone Matrix (GLSZM)39 (16) and Neighboring Gray-Tone Difference Matrix Features (NGTDM)40 (5).
The radiomic features were extracted from the NSCLC radiomics CT dataset by using different quantization configurations: the number of bins varied in . By relying upon the slice thickness, which is the same for all CT scans included in this homogeneous subset of the whole NSCLC dataset, 3D feature computation without any resampling was used to avoid interpolation artifacts.
Radiomic feature robustness analysis
The intraclass correlation coefficient (ICC) was computed to identify which features are correlated with the number of bins used during the quantization step. Given k multiple measurements to be compared (i.e., 6 different rebinnings), 41 for a two-way random-effects (or mixed effects) model was used:
| 3 |
where and are the mean square for rows and mean square for error, respectively.
According to the ICC values42, we divided the features into:
Poor robustness: ICC 0.5;
Moderate robustness: 0.5 < ICC 0.75;
Good robustness: 0.75 < ICC 0.9;
Excellent robustness: ICC > 0.9.
We investigated how the robustness of the textural features (in terms of ICC) varies according to the different groups of images. For each group, with the aim of identifying the most robust features, the ICC was calculated by varying the number of bins considered . By doing so, we determined the number of robust features by varying the number of bins in the quantization step. After determining the features showing excellent robustness, we aimed to identify the most relevant features for the analysis at hand; for this purpose, we used in an agnostic way the most best known technique of dimensionality reduction: the PCA43. For this purpose, we had to select a specific quantization setting binning; therefore, the different number of bins were perturbed, via mathematical morphology operations, to select the most robust setting. With more details, the original ROIs were perturbed using morphological operators (opening and closing with a 3D spherical structuring element of 1-pixel radius). Accordingly, we produced three versions for each ROI (i.e., original, opening, and closing). This procedure simulates ROI variations through consideration of intra-/inter-reader dependence during manual contouring44. The optimal number of bins was selected after the ROI perturbation process, by considering the rebinning with the highest number of robust features. It is worth noting that the optimal binning was selected on the Original images and not on the super-resolved ones, thus adopting the most conservative choice for fair comparisons.
With the goal of carefully analyzing these variations in terms of ICC, and after the selection of the optimal rebinning setting, we assessed the importance of these features by means of a ranking procedure: we performed a PCA and we calculated a weighted average of the features extracted from the Original images, according to the first three Principal Components (PCs), to assess their relative importance. In particular, we calculated the correlation matrix (as well as the eigenvectors and eigenvalues of the correlation matrix) to identify the PCs. PCs represent the directions of the data that explain a maximum amount of variance, i.e., the directions that capture most of the relevant and non-redundant information in the data. Then, to determine the relative importance of the features for the PCs considered, we used a quadrature sum for the individual features related to the different PCs. In this way, we determined a ranking of the features by the study of their relative weights in the main components considered.
Results
Image super-resolution results
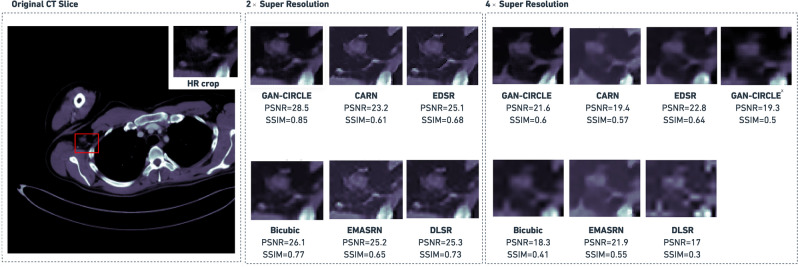
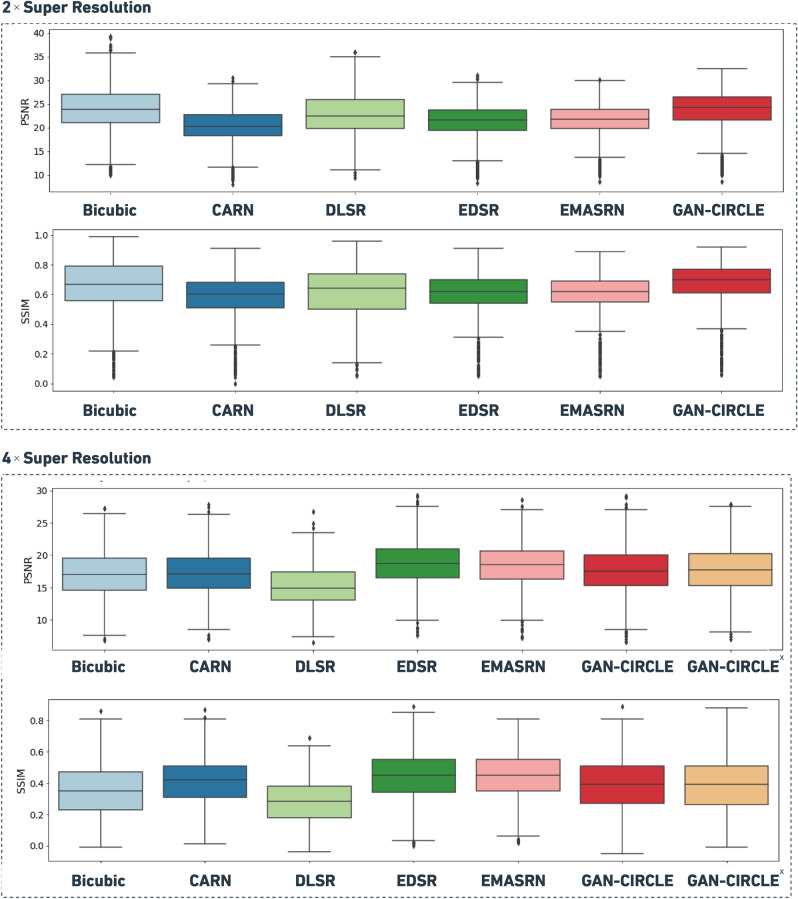
Figure 3 shows an example of both and super-resolved images obtained by the considered methods. This example provides a qualitative visual assessment of the super-resolved images. Figure 4 reports the boxplots of the PSNR/SSIM metrics for 1, 000 CT images. From the analysis of Fig. 4, one can see that, at SR, the proposed GAN-CIRCLE-based method achieved higher median values than the other competitors for both the considered metrics (i.e., PSNR and SSIM). On the other hand, at SR, the best SSIM and PSNR values were obtained with the EDSR and EMASRN SR methods. To assess the statistical significance of these results, we performed a Mann–Whitney test for pairwise comparisons (using ). The p-values were adjusted via the Benjamini–Hochberg method for multiple comparisons. Based on the p-values yielded by the statistical test, at SR, GAN-CIRCLE achieved significantly higher PSNR and SSIM values than the other competitors. The only exception is represented by the Bicubic interpolation for which the differences of the median SSIM and PSNR values were not statistically significant. At SR, GAN-CIRCLE showed statistically significant differences, in terms of SSIM and PSNR, when compared against the Bicubic interpolation method and DLSR. The differences were not statistically significant when we compared GAN-CIRCLE against EDSR, EMASRN, and CARN. Finally, at SR, GAN-CIRCLEx produced results comparable to the ones achieved with GAN-CIRCLE.
Figure 3.
Perceptual quality comparison, on the DeepLesion test images and super-resolved held out for performance evaluation, obtained by the investigated SR methods. The PSNR and SSIM values are shown at the bottom of each super-resolved image. In the case of SR, GAN-CIRCLEx denotes the sequential application of two GAN-CIRCLE instances at SR.
Figure 4.
Boxplots comparing PSNR and SSIM metrics for 1000 CT images held out for performance evaluation, super-resolved at and by using the investigated SR methods. In the case of SR, GAN-CIRCLEx denotes the sequential application of two GAN-CIRCLE instances at SR.
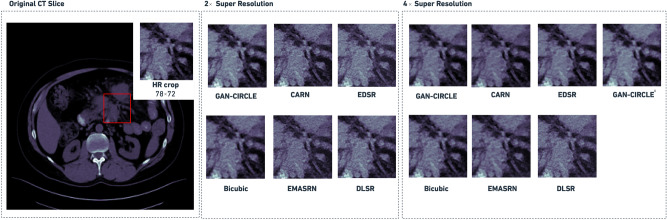
Figure 5 shows a randomly selected example from the Deeplesion dataset to endorse the quality of the produced images and assess the generalization ability of the investigated SR methods. Although PSNR/SSIM are widely adopted evaluation metrics, some studies19,45 have demonstrated their limitations on medical image SR tasks since images with low perceptual quality could exhibit high PSNR/SSIM values. Overall, at both and SR, the GAN-generated images were less blurry, with better texture, sharper edges, and visually more similar to the ground truth, as shown in Figs. 3 and 5.
Figure 5.
SR example ( and factor) using the investigated SR methods from a sample slice randomly selected from the Deeplesion dataset (held-out set). In the case of SR, GAN-CIRCLEx denotes the sequential application of two GAN-CIRCLE instances at SR.
In the downstream radiomic analyses, we focused our attention on the Original images, the super-resolved images via the proposed GAN-SR framework (based on SPP and GAN-CIRCLE), and the Bicubic interpolation method. The Bicubic interpolation method obtained, at SR, the best performance (i.e., in terms of PSNR and SSIM) among the considered SR techniques. Moreover, it is commonly available and used in medical image processing.
Results of the robustness analysis
In this section, we describe and discuss the results of the robustness analysis related to the textural features (in terms of ICC) according to different image groups (i.e., Original, Bicubic, and GAN-SR). Table 1 reports the features with excellent robustness for the considered methods. According to these values, one can observe that all the techniques taken into account produced ten features with excellent robustness. Interestingly, our GAN-SR method shows superior performance in terms of ICC for four features. Moreover, the GAN-SR technique, as well as the Bicubic interpolation, achieved moderate to good robustness for GLRLM LongRunLowGrayLevelEmphasis and GLDM DependenceEntropy, while the features extracted from the Original images resulted in excellent robustness.
Table 1.
Features that obtained an excellent robustness for at least of the Original, Cubic and GAN-SR image groups.
| Feature ID | Feature name | Original | Bicubic | GAN-SR |
|---|---|---|---|---|
| #1 | GLCM Correlation | 0.980 | 0.979 | 0.984 |
| #2 | GLCM DifferenceEntropy | 0.846 | 0.911 | 0.910 |
| #3 | GLCM IDMN | 0.996 | 0.996 | 0.997 |
| #4 | GLCM ID | 0.997 | 0.995 | 0.998 |
| #5 | GLCM MCC | 0.633 | 0.938 | 0.923 |
| #6 | GLCM SumEntropy | 0.822 | 0.897 | 0.905 |
| #7 | GLRLM LongRunLowGrayLevelEmphasis | 0.926 | 0.560 | 0.631 |
| #8 | GLRLM LowGrayLevelRunEmphasis | 0.967 | 0.952 | 0.944 |
| #9 | GLRLM ShortRunLowGrayLevelEmphasis | 0.97 | 0.973 | 0.925 |
| #10 | GLDM DependenceEntropy | 0.910 | 0.870 | 0.895 |
| #11 | GLDM LargeDependenceLowGrayLevelEmphasis | 0.985 | 0.976 | 0.890 |
| #12 | GLDM LowGrayLevelEmphasis | 0.986 | 0.986 | 0.950 |
| #13 | GLDM SmallDependenceLowGrayLevelEmphasis | 0.902 | 0.955 | 0.946 |
Table 2 reports the most important features according to the implemented PCA-based procedure. These four features are related to the GLCM matrix (the GLCM characterizes the texture of an image by calculating the occurrences of voxel pairs with specific values in a defined spatial relationship36) and, in particular, are the following: Correlation, IDMN, IDN, SumEntropy (Feature IDs: #1, #3, #4, #6). Of particular interest is the SumEntropy feature, defined as the sum of neighborhood intensity value differences, which showed excellent robustness with the GAN-SR method, while it showed good robustness in Original and Bicubic.
Table 2.
Relative difference (in terms of ICC) of the GAN-SR against the Original and Bicubic versions on the most important radiomic features according to PCA analysis.
| Feature name | Original | Bicubic | GAN-SR | GAN-SR vs. Original (%) | GAN-SR vs. Bicubic (%) |
|---|---|---|---|---|---|
| GLCM Correlation | 0.980 | 0.979 | 0.984 | 0.41 | 0.51 |
| GLCM IDMN | 0.996 | 0.996 | 0.997 | 0.1 | 0.1 |
| GLCM IDN | 0.997 | 0.995 | 0.998 | 0.1 | 0.3 |
| GLCM SumEntropy | 0.822 | 0.897 | 0.905 | 10.1 | 0.89 |
Table 2 shows the relative difference (in terms of ICC) on the most important radiomic features between GAN-SR and the Original/Bicubic versions. With reference to the most important features, the GLCM Correlation denotes the linear dependency of gray-level values to their respective voxels in the GLCM; the Inverse Difference Moment Normalized (IDMN) is a measure of the local homogeneity of an image that normalizes the square of the difference between neighboring intensity values by dividing over the square of the total number of discrete intensity values; the Inverse Difference Normalized (IDN) is another measure of the local homogeneity of an image that normalizes the difference between the neighboring intensity values by dividing over the total number of discrete intensity values.
According to the procedure designed for robustness in the radiomic feature, the optimal binning was found with 64 bins after the perturbation process.
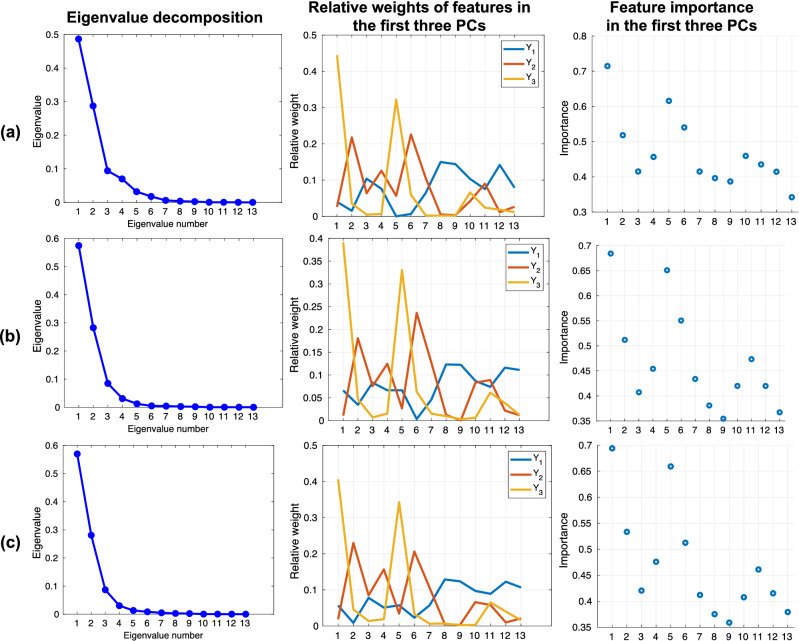
In Fig. 6, the plots in the left column justify the use of the first three PCs, as the first three eigenvalues cover at least of the trace of the covariance matrix in each group. The plots in the second column show the weights of the original features on the first three PCs, while the third column shows the relative importance of the features in the first three PCs. The most important features (in descending order), for the three groups of images, were as follows:
Original: #1, #5, #6, #2, #10;
Bicubic: #1, #5, #6, #2, #11;
GAN-SR: #1, #5, #2, #6, #4.
Figure 6.
PCA-based analysis of the importance of radiomic features for all image types: (a) Original; (b) Cubic; (c) GAN-SR. The first column shows the line plots of the values of the eigenvalues as a function of the number of eigenvalues. This is useful for the evaluation of the PCs required. The second column shows the relative weights of the original features on the first of three PCs, while the third column depicts the relative importance of the features (according to the IDs defined in Table 1) in the first three PCs.
Intriguingly, the features with a lower ICC in the GAN-SR method were those of less importance in terms of the PCA. Our GAN-SR method, therefore, increased the robustness of the most important features, compared to the Original and Cubic groups. These highly robust features are expected to generalize well on other and unseen imaging datasets.
Discussion
This paper presented the first application of GAN-based image SR to radiomic studies. As a proof-of-concept, CT images were considered. In particular, the DeepLesion23 dataset was used for training and testing the GAN-SR performance in terms of PSNR and SSIM. The performance of the proposed method was compared against recent state-of-the-art methods for image SR. To quantitatively assess the performance of the proposed framework and compared it against the considered state-of-the-art SR techniques, we relied on two commonly used metrics: PSNR and SSIM. Moreover, to carefully assess the performance of the proposed GAN-CIRCLE-based SR method at SR, we compared the native GAN-CIRCLE SR against the sequential application of two GAN-CIRCLE instances at SR (i.e., GAN-CIRCLEx). Experimental results showed that, at SR, the proposed GAN-CIRCLE-based method achieved better performance (with statistical significance, except for the Bicubic interpolation) than the other competitors for both the considered metrics. On the other hand, at SR, the best SSIM and PSNR values were obtained with the EDSR and EMASRN SR methods. Still, the performance of the proposed framework was comparable (i.e., no statistically significant difference) to the two best performers. According to the results achieved, we can state that the proposed SR framework can obtain competitive performance with respect to the considered competitors across the tested SR factors. Additionally, the visual assessment of the super-resolved images showed that, in general, the GAN-CIRCLE-based method produced images with better texture and sharper edges, and they looked visually more similar to the ground truth HRCT.
The experimental evidence allowed us to choose the proposed GAN-CIRCLE framework, integrating the SPP, as the most suitable approach for evaluating the impact of advanced image SR methods in oncological imaging. Therefore, the resulting GAN-SR model was leveraged to assess the robustness of the radiomic features extracted from the images of the TCIA NSCLC CT dataset46. This assessment required the computation of the ICC to identify the most robust features against the variations of the number of bins used in the quantization step. The ICC values, calculated for three different image groups (i.e., Original, Bicubic, and GAN-SR), showed that all the techniques obtained ten texture features with excellent robustness. Still, the proposed GAN-SR method presented superior ICC values in four of the ten features with excellent robustness. Finally, a PCA was performed to identify the relative importance of the radiomic features in the proposed GAN-SR technique. The results obtained from this analysis are particularly interesting as the features with the lowest ICC values are the ones deemed less relevant in terms of the PCA analysis. On the contrary, GAN-SR increased the robustness of the most important features compared to the Original and Bicubic groups. The result is relevant because the highly robust features identified by GAN-SR might generalize well on other CT datasets. The results of this study could pave the way for the application of GAN-based image SR techniques for studies of radiomics for robust biomarker discovery47,48.
Along with the novelties in lesion-focused GAN-based SR, this work belongs to the research strand dedicated to the analysis of robustness in radiomic features, with particular interest in oncological imaging. As a matter of fact, the investigation techniques used in our study were consistent with the state-of-the-art: the ICC was adopted in radiomic feature robustness analyses that assessed the impact of different imaging acquisition and reconstruction parameters6,7,49, as well as image perturbations4,5,8. Moreover, we identified the most important features in an agnostic manner, which is independent on a particular classification/prediction task at hand, by using a PCA-based investigation43.
The main limitation of the proposed SR method is inherent to its lesion-focused approach, which relies on a lesion detection step for ROI identification that limits the application of this method to datasets with a pre-existing mapping of ROIs. Regarding this matter, our methodological approach could be extended to include a lesion detection task as in19, to allow for CT images without lesion annotations in the training process. Considering that our GAN-SR method currently performs only in-plane 2D image SR, to avoid the effect of slice thickness variability6,7, GAN-based SR along the z-axis (i.e., yielding thinner slices) might relieve the problem related to highly anisotropic voxels50,51. Moreover, since our GAN-SR model does not remarkably improve PSNR/SSIM values, we could conduct feature recalibration, such as via self-attention mechanisms, to obtain features more similar to the ones of the original images21,52–54. Concerning future radiomics applications, since we showed the results on a homogeneous subset of the NSCLC-Radiomics dataset, we plan to test the generalization ability of GAN-extracted radiomic features on the whole dataset, considering variations on CT image acquisition and reconstruction parameters. In particular, a classification/prediction modeling task for NSCLC staging and type would be beneficial24.
Acknowledgements
This work was partially supported by The Mark Foundation for Cancer Research and Cancer Research UK Cambridge Centre [C9685/A25177], the Wellcome Trust Innovator Award, UK [215733/Z/19/Z] and the CRUK National Cancer Imaging Translational Accelerator (NCITA) [C42780/A27066]. Additional support was also provided by the National Institute of Health Research (NIHR) Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care. This work was partially supported by national funds through the FCT (Fundação para a Ciência e a Tecnologia) by the Projects GADgET (DSAIPA/DS/0022/2018) and the financial support from the Slovenian Research Agency (research core funding no. P5-0410).
Author contributions
All authors: Conceptualization, Writing—Review and Editing; E.C.F., C.d.N., C.H., L.R. and M.C. Methodology, Investigation, Software; E.C.F., C.d.N., C.H., M.C. and L.R. Formal analysis; E.C.F., M.C. and L.R. Writing—Original Draft; E.C.F., M.C. and L.R. Validation, Resources (computing resources and analysis tools); E.S., M.C. and L.R. Resources; E.C.F., C.d.N. and L.R. Data Curation; E.S., M.C. and L.R. Supervision and Project administration.
All authors reviewed the work, contributed to its overall scientific content and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mauro Castelli and Leonardo Rundo.
Contributor Information
Mauro Castelli, Email: mcastelli@novaims.unl.pt.
Leonardo Rundo, Email: lr495@cam.ac.uk.
References
- 1.Gillies R, Kinahan P, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2015;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwanenburg A, et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fornacon-Wood I, et al. Reliability and prognostic value of radiomic features are highly dependent on choice of feature extraction platform. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06957-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zwanenburg A, et al. Assessing robustness of radiomic features by image perturbation. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-018-36938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mottola M, et al. Reproducibility of CT-based radiomic features against image resampling and perturbations for tumour and healthy kidney in renal cancer patients. Sci. Rep. 2021;11:11542. doi: 10.1038/s41598-021-90985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shafiq-Ul-Hassan M, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017;44:1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escudero Sanchez L, et al. Robustness of radiomic features in CT images with different slice thickness, comparing liver tumour and muscle. Sci. Rep. 2021;11:8262. doi: 10.1038/s41598-021-87598-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le EP, et al. Assessing robustness of carotid artery ct angiography radiomics in the identification of culprit lesions in cerebrovascular events. Sci. Rep. 2021;11:3499. doi: 10.1038/s41598-021-82760-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahapatra D, Bozorgtabar B, Garnavi R. Image super-resolution using progressive generative adversarial networks for medical image analysis. Comput. Med. Imaging Graph. 2019;71:30–39. doi: 10.1016/j.compmedimag.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Sandfort V, Yan K, Pickhardt PJ, Summers RM. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019;9:16884. doi: 10.1038/s41598-019-52737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proc. IEEE International Conference on Computer Vision 2223–2232 (IEEE, 2017). 10.1109/ICCV.2017.244.
- 12.Han C, et al. Combining noise-to-image and image-to-image GANs: Brain MR image augmentation for tumor detection. IEEE Access. 2019;7:156966–156977. doi: 10.1109/ACCESS.2019.2947606. [DOI] [Google Scholar]
- 13.You C, et al. CT super-resolution GAN constrained by the identical, residual, and cycle learning ensemble (GAN-CIRCLE) IEEE Trans. Med. Imaging. 2020;39:188–203. doi: 10.1109/TMI.2019.2922960. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y. et al. Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network. In Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 91–99 (Springer, 2018). 10.1007/978-3-030-00928-1_11.
- 15.Yu, H. et al. Computed tomography super-resolution using convolutional neural networks. In IEEE International Conference on Image Processing (ICIP) 3944–3948 (IEEE, 2017). 10.1109/ICIP.2017.8297022.
- 16.Park J, et al. Computed tomography super-resolution using deep convolutional neural network. Phys. Med. Biol. 2018;63:145011. doi: 10.1088/1361-6560/aacdd4. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhari AS, et al. Super-resolution musculoskeletal MRI using deep learning. Magn. Reson. Med. 2018;80:2139–2154. doi: 10.1002/mrm.27178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guha, I. et al. Deep learning based high-resolution reconstruction of trabecular bone microstructures from low-resolution CT scans using GAN-CIRCLE. In Medical Imaging 2020: Biomedical Applications in Molecular, Structural, and Functional Imaging, Vol. 11317, 113170U (International Society for Optics and Photonics, 2020). 10.1117/12.2549318. [DOI] [PMC free article] [PubMed]
- 19.Zhu, J., Yang, G. & Lio, P. How can we make GAN perform better in single medical image super-resolution? A lesion focused multi-scale approach. In Proc. IEEE 16th International Symposium on Biomedical Imaging (ISBI) 1669–1673 (IEEE, 2019). 10.1109/ISBI.2019.8759517.
- 20.Yi P, et al. A progressive fusion generative adversarial network for realistic and consistent video super-resolution. IEEE Trans. Pattern Anal. Mach. Intell. 2020 doi: 10.1109/TPAMI.2020.3042298. [DOI] [PubMed] [Google Scholar]
- 21.Zhu X, et al. Lightweight image super-resolution with expectation-maximization attention mechanism. IEEE Trans. Circuits Syst. Video Technol. 2021 doi: 10.1109/TCSVT.2021.3078436. [DOI] [Google Scholar]
- 22.Ouyang, X., Cheng, Y., Jiang, Y., Li, C.-L. & Zhou, P. Pedestrian-synthesis-GAN: Generating pedestrian data in real scene and beyond. arXiv preprintarXiv:1804.02047 (2018).
- 23.Yan K, Wang X, Lu L, Summers RM. DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J. Med. Imaging. 2018;5:1. doi: 10.1117/1.JMI.5.3.036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aerts H, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyu Q, Shan H, Wang G. MRI super-resolution with ensemble learning and complementary priors. IEEE Trans. Comput. Imaging. 2020;6:615–624. doi: 10.1109/TCI.2020.2964201. [DOI] [Google Scholar]
- 26.He K, Zhang X, Ren S, Sun J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015;37:1904–1916. doi: 10.1109/TPAMI.2015.2389824. [DOI] [PubMed] [Google Scholar]
- 27.He, K., Zhang, X., Ren, S. & Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proc. IEEE International Conference on Computer Vision (ICCV) 1026–1034 (IEEE, 2015). 10.1109/ICCV.2015.123.
- 28.Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B. & Hochreiter, S. GANs trained by a two time-scale update rule converge to a local Nash equilibrium. In Proc. 31st International Conference on Neural Information Processing Systems (NIPS) (2017).
- 29.Lim, B., Son, S., Kim, H., Nah, S. & Mu Lee, K. Enhanced deep residual networks for single image super-resolution. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops 136 –144 (2017). 10.1109/TCYB.2019.2952710.
- 30.Ahn, N., Kang, B. & Sohn, K.-A. Fast, accurate, and lightweight super-resolution with cascading residual network. In Proc. European Conference on Computer Vision (ECCV) 252–268 (2018). 10.1007/978-3-030-01249-6_16.
- 31.Jiang C, Zhang Q, Fan R, Hu Z. Super-resolution CT image reconstruction based on dictionary learning and sparse representation. Sci. Rep. 2018;8:8799. doi: 10.1038/s41598-018-27261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Griethuysen J, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox R, et al. A (sort of) new image data format standard: NIfTI-1. Neuroimage. 2004;22:1–30. doi: 10.1016/j.neuroimage.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC. 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 35.Haralick RM. Statistical and structural approaches to texture. Proc. IEEE. 1979;67:786–804. doi: 10.1109/PROC.1979.11328. [DOI] [Google Scholar]
- 36.Rundo, L. et al. HaraliCU: GPU-powered Haralick feature extraction on medical images exploiting the full dynamics of gray-scale levels. In Proc. International Conference on Parallel Computing Technologies (PaCT), Vol. 11657 of LNCS 304–318, 978-3-030-25636-4\_24 (ed. Malyshkin, V.) (Springer International Publishing, 2019).
- 37.Sun C, Wee WG. Neighboring gray level dependence matrix for texture classification. Comput. Vis. Graph. Image Process. 1983;23:341–352. doi: 10.1016/0734-189X(83)90032-4. [DOI] [Google Scholar]
- 38.Galloway MM. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975;4:172–179. doi: 10.1016/S0146-664X(75)80008-6. [DOI] [Google Scholar]
- 39.Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: Application to cell classification. IEEE Trans. Biomed. Eng. 2013;61:630–637. doi: 10.1109/TBME.2013.2284600. [DOI] [PubMed] [Google Scholar]
- 40.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern. 1989;19:1264–1274. doi: 10.1109/21.44046. [DOI] [Google Scholar]
- 41.Shrout P, Fleiss J. Intraclass correlations: Uses in assessing rater reliability. Psychol. Bull. 1979;86:420–428. doi: 10.1037/0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 42.Scalco E, et al. T2w-MRI signal normalization affects radiomics features reproducibility. Med. Phys. 2020;47:1680–1691. doi: 10.1002/mp.14038. [DOI] [PubMed] [Google Scholar]
- 43.Jolliffe I. Principal component analysis. Encycl. Stat. Behav. Sci. 2005 doi: 10.1002/0470013192.bsa501. [DOI] [Google Scholar]
- 44.Sushentsev N, et al. MRI-derived radiomics model for baseline prediction of prostate cancer progression on active surveillance. Sci. Rep. 2021;11:12917. doi: 10.1038/s41598-021-92341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ledig, C. et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 105–114 (IEEE, 2017). 10.1109/CVPR.2017.19.
- 46.Clark K, et al. The cancer imaging archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papanikolaou N, Matos C, Koh DM. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imaging. 2020;20:33. doi: 10.1186/s40644-020-00311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castiglioni I, et al. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021;83:9–24. doi: 10.1016/j.ejmp.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Shafiq-ul-Hassan M, et al. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-28895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kudo, A., Kitamura, Y., Li, Y., Iizuka, S. & Simo-Serra, E. Virtual thin slice: 3D conditional gan-based super-resolution for CT slice interval. In International Workshop on Machine Learning for Medical Image Reconstruction 91–100 (Springer, 2019). 10.1007/978-3-030-33843-5_9.
- 51.Zhang, K. et al. SOUP-GAN: Super-resolution MRI using generative adversarial networks. arXiv preprintarXiv:2106.02599 (2021). [DOI] [PMC free article] [PubMed]
- 52.Li, Y. et al. Super-resolution and self-attention with generative adversarial network for improving malignancy characterization of hepatocellular carcinoma. In Proc. IEEE International Symposium on Biomedical Imaging (ISBI) 1556–1560 (IEEE, 2020). 10.1109/ISBI45749.2020.9098705.
- 53.Li M, Hsu W, Xie X, Cong J, Gao W. SACNN: Self-attention convolutional neural network for low-dose CT denoising with self-supervised perceptual loss network. IEEE Trans. Med. Imaging. 2020;39:2289–2301. doi: 10.1109/TMI.2020.2968472. [DOI] [PubMed] [Google Scholar]
- 54.Han C, et al. MADGAN: Unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinform. 2021;22:31. doi: 10.1186/s12859-020-03936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]