Summary
Nonalcoholic fatty liver disease (NAFLD) is a global health-care problem with limited therapeutic options. To obtain a cellular resolution of pathogenesis, 82,168 single-cell transcriptomes (scRNA-seq) across different NAFLD stages were profiled, identifying hepatocytes and 12 other non-parenchymal cell (NPC) types. scRNA-seq revealed insights into the cellular and molecular mechanisms of the disease. We discovered a dual role for hepatic stellate cells in gene expression regulation and in the potential to trans-differentiate into myofibroblasts. We uncovered distinct expression profiles of Kupffer cells versus monocyte-derived macrophages during NAFLD progression. Kupffer cells showed stronger immune responses, while monocyte-derived macrophages demonstrated a capability for differentiation. Three chimeric NPCs were identified including endothelial-chimeric stellate cells, hepatocyte-chimeric endothelial cells, and endothelial-chimeric Kupffer cells. Our work identified unanticipated aspects of mouse with NAFLD at the single-cell level and advanced the understanding of cellular heterogeneity in NAFLD livers.
Subject areas: Animal physiology, Cell biology, Transcriptomics
Graphical abstract

Highlights
-
•
Of all, 82,168 single-cell transcriptomes across different NAFLD stages were profiled
-
•
Hepatocytes and 12 non-parenchymal cell types were identified in mouse liver
-
•
Three chimeric NPCs were identified in mouse liver
Animal physiology; Cell biology; Transcriptomics
Introduction
The liver is the largest organ in the body that performs diverse functions essential for energy homeostasis, including synthesis of amino acids, lipids and glucose, and biotransformation of xenobiotics and endogenous byproducts. Hepatocytes account for about 60% of the total cell population in human liver, with non-parenchymal cells (NPCs) encompassing the other 40%. Among NPCs, liver sinusoidal endothelial cells (LSECs) contribute ∼40%, Kupffer cells (KCs) ∼30%, hepatic stellate cells (HSCs) 10%–25%, and other cells ∼5% (Braet et al., 2018; Wisse, 1977). Dysregulation of hepatic metabolism contributes to the pathogenesis of diabetes, dyslipidemia, and nonalcoholic fatty liver disease (NAFLD). Widespread modern unhealthy dietary habits have made NAFLD the most frequent chronic liver disease worldwide with ∼25% of the population, yet there are no approved pharmacological treatments (Friedman et al., 2018; Younossi et al., 2016).
NAFLD is an umbrella term that comprises a continuum of liver conditions varying from benign stage of steatosis alone as nonalcoholic fatty liver (NAFL), to a more severe process of steatosis plus inflammation and/or fibrosis as nonalcoholic steatohepatitis (NASH), or even worse stage of cirrhosis and hepatocellular carcinoma (HCC) (Friedman et al., 2018). NASH is currently the second most common liver disease among patients awaiting liver transplantation and is expected to become the most common cause by 2030 (Brunt et al., 2015; Estes et al., 2018; Shaker et al., 2014). Despite this substantial health and economic burden, therapeutic options for NAFLD remain limited, in part, owing to the lack of deep understanding of the cellular and molecular mechanisms that drive the transformation from benign to severe. The nutrition-overload liver injury model of NASH pathogenesis is the central and initial step. Responsive activation of local KCs, HSCs, LSECs, and infiltration of immune cells further accelerate the disease progression. The understanding of when and how the hepatocytes and different NPCs respond to this energy-overloaded lipotoxicity microenvironment is currently unclear. Single-cell transcriptomic technologies are transforming our understanding of cellular diversity and functions in health and disease, enabling interrogation of homeostatic and pathogenic cell populations in the liver (Ramachandran et al., 2020; Saviano et al., 2020). A single-cell expression atlas of healthy and NAFLD mouse livers is critical to understanding disease pathogenesis. Recently, several human liver single-cell RNA-sequencing (scRNA-seq) methods have been applied to investigate the landscape of normal liver, cirrhotic liver, and liver tumor (Aizarani et al., 2019; MacParland et al., 2018; Massalha et al., 2020; Ramachandran et al., 2019). More mouse liver single-cell gene analysis studies were reported which either provided insights into the intercellular crosstalk among NPCs or focused on subsets of liver cells, for example, hepatocytes, stellate cell, macrophages, or myeloid cells (Daemen et al., 2021; Xiong et al., 2019; Krenkel et al., 2019, 2020; Remmerie et al., 2020; Dobie et al., 2019; Park et al., 2021; Seidman et al., 2020). The cross comparison of different studies was limited due to different NAFLD diet induction paradigms and different treatment timelines used in these different reports. A comprehensive study to cover all liver cell types and different NAFLD disease stages is needed for the deeper understanding of disease pathogenesis.
In order to extend our understanding of hepatocytes and the subgroups of each cell type from the same model with a slow progressive NAFLD disease pathology crossing healthy, NAFL, and NASH, we performed scRNA-seq to dissect hepatocyte and NPCs transcriptomes in healthy and diet-induced NAFL and NASH mouse livers. Our analyses revealed the gene expression landscape of hepatocytes and NPCs in healthy and NAFLD mouse livers, as well as the gene regulation and transition that occurs with the onset of NASH. In addition, we identified several chimeric hepatic NPCs in healthy and NAFLD mouse livers based on results of in situ hybridization, flow cytometry, and scRNA-seq. This study sheds light on the complexity of liver cell heterogeneity and provides novel insights into the fundamental biology and pathology of NAFLD.
Results
Single-cell expression atlas of healthy and NAFLD mouse livers
Previous studies have shown that mice on high fat high fructose diet (HFHFD), also called AMLN diet, for more than 26 weeks recapitulate key features of human NASH (Clapper et al., 2013; Guo et al., 2017; Tolbol et al., 2018; Xiong et al., 2019). Whole liver bulk RNA-seq analysis uncovered the diet-induced upregulation of genes involved in lipid metabolism, inflammation, fibrosis, and cell death (Tolbol et al., 2018). In order to capture the pathological continuum of NAFLD, mice were fed a chow diet or a HFHFD diet. Consistent with previous studies, HFHFD induced liver injury by 15 weeks, referred to as NAFL mice, and fibrosis and inflammation by 30 weeks, referred to as NASH mice (Figure 1A). Consistent with previous studies, HFHFD-induced liver injury (as indicated by increased liver transaminase ALT and AST) showed dyslipidemia and steatosis at 15 and 30 weeks diet treatment (Figures S1A–S1G). More severe liver pathology of fibrosis and inflammation was induced by 30 weeks of HFHFD treatment (Figures S1A–S1G).
Figure 1.
Capturing, sequencing, and classification of liver cell types from healthy and NAFLD mouse liver
(A) The scheme of study design, cell isolation, capture, and RNA sequencing.
(B) Uniform Manifold Approximation and Projection (UMAP) plot of liver cell clusters based on 82,168 single-cell transcriptomes. Cell counts for hepatocytes (Hep), cholangiocyte/hepatic progenitor cells (Cho/HPCs), endothelial cells (Endo), hepatic stellate cells (HSCs), myofibroblast (Myo), monocyte or monocyte-derived macrophage (Mo/MoMF), dendritic cells (DCs), plasmacytoid DCs (pDCs), T/NK cells (T/NK), neutrophil (N), and B cells (B) are indicated in parentheses.
(C) Expression of representative enriched genes for each cell type. Gene expression violin plots were shown in log-scale UMI. Colors correspond to cell types in Figure 1B.
To elucidate liver cell complexity, heterogeneity, and their dynamic changes during NAFLD pathogenesis, we performed scRNA-seq on hepatocytes and liver NPCs isolated from mice in healthy group with chow diet (n = 6), NAFL with HFHFD for 15 weeks (n = 3), and NASH with HFHFD for 30 (n = 2 for hepatocytes and n = 4 for NPCs) or 34 weeks (n = 1 for NPCs), individually. Hepatocytes were enriched by 50g low-speed centrifuge from freshly perfused and isolated liver single cell mixture before the scRNA-seq assay. Gated DAPI fluorescent negative liver NPCs were fluorescence-activated cell sorting (FACS) sorted prior to the scRNA-seq experiment to remove dead cells and cell debris (Figure 1A). Each NPC cell had on average 147,476 reads and each hepatocyte had 291,359 reads. The exon reads took up 45% of the total reads in NPCs and 24% in hepatocytes. The hepatocyte portion from HFHFD for 34 weeks was excluded from the analysis due to the low exon reads. The 34-week hepatocytes showed in the results were collected from the NPCs submission. We suspect that lower exon reads in 34-week HFHFD hepatocytes were primarily due to the vulnerability of severe liver injury. Overall, 82,168 single-cell transcriptomes were collected and analyzed, including 25,578 from healthy controls, 26,833 from HFHFD for 15 weeks, 23,709 from HFHFD for 30 weeks, and 6,048 from HFHFD for 34 weeks (Figure 1B). Our atlas revealed 13 major liver cell types in both healthy and NAFLD livers (Figures 1B and 1C), namely, albumin-positive hepatocytes (Hep), Epithelial cell adhesion molecule (Epcam), positive cholangiocytes and hepatic progenitor cells (Cho/HPCs), regulator of G protein signaling (Rgs) 5 positive hepatic stellate cells (HSCs), Mesothelin (Msln) positive myofibroblasts (Myo), c-type lectin domain family 4 member (Clec4) g positive liver endothelial cells (Endo), Clec4f positive Kupffer cells (KCs), C-C chemokine receptor type (Ccr) 2 positive monocyte or monocyte-derived macrophages (Mo/MoMF), C-C Motif Chemokine Ligand (Ccl) 4 positive neutrophils (N), Fms-Like Tyrosine kinase (Flt) 3 positive dendritic cells (DCs), Ccr9 positive plasma dendritic cells (pDCs), Cd3g positive T cells (T), X-C motif chemokine ligand (Xcl) 1 positive natural killer cells (NK), and Cd79a-positive B cells (B). The top two differentially expressed genes for each identified cell type were listed in Figure 1C. The large number of single cells captured in this study enhanced the opportunity to identify low percentages of liver cells, especially HSCs. It served the goals to identify new cell subpopulations, disease differentially expressed genes (DEGs), enriched disease pathways, and to reveal underlying cell lineage relationships.
Liver cell composition and gene expression changes in major cell types during NASH progression
Whole liver bulk RNA-seq analyses have uncovered the NASH disease gene regulation in lipid metabolism, inflammation, fibrosis, and cell death (Tolbol et al., 2018); however, it was difficult to differentiate which cell subpopulation contributed to certain gene changes or whether there were cell subpopulation changes along NASH progression. Liver scRNA-seq is an appropriate approach to resolve the issues. The mouse liver cell expression atlas showed that infiltrated immune cells increased relatively compared with other cell types during NASH progression (Figure 2A). Except the resident KCs, all infiltrated immune cells increased significantly at NAFL and NASH stages. The increase of immune cell infiltration was confirmed by increased CD45 positive area percentage from 1% to 2.6% with 30 weeks HFHFD treatment (Figures S1D and S1F). The lack of hepatocytes at 34 weeks was due to the exclusion of low-quality hepatocytes collected at that time point. Relative gene expression changes for each major liver cell type including hepatocytes, endothelial cells, Kupffer cells, and stellate cells were shown in Figures 2B–2E. To further define the function regulation of NAFLD hepatocytes, the differently expressed genes (DEGs) (adjusted p value < 0.01) were selected for Reactome pathway enrichment analysis. Consistent with NAFLD liver bulk RNA-seq analysis, both fat metabolism and glucose/fructose and mannose metabolism were upregulated across NAFLD and NASH stages in hepatocytes (Figure 2B and Table S1). Similarly, the significant endothelial DEGs were enriched in immune response and cholesterol metabolism pathways (Figure 2C and Table S2). As expected, immune response, amino acid/carbon metabolism, and complement/coagulation cascades pathways were the main functions regulated in Kupffer cells (Figure 2D and Table S3). Vascular smooth muscle contraction, extracellular matrix, inflammation, and lysosome-associated pathways were regulated in stellate cells (Figure 2E and Table S4). The lack of stellate cells at 34 weeks was due to only 8 cells collected at that time point.
Figure 2.
Changes of liver cell clusters, gene regulation, and pathway enrichment along HFHFD treatment
(A) Changes of liver cell clusters along the timeline of HFHFD treatment. Colors correspond to cell types in Figure 1B.
(B–E) Differential expressed genes between 15, 30, and 34 weeks compared to chow in hepatocytes (B), endothelial cells (C), Kupffer cells (D), and stellate cells (E). Gene grouped based on their function. Genes with unknown function not shown. Hepatocytes from 34-week HFHFD were not included in the analysis. The number of stellate cells from 34 weeks were limited and not included in the analysis.
Distinct hepatic stellate subpopulations and myofibroblast in healthy and NAFLD livers
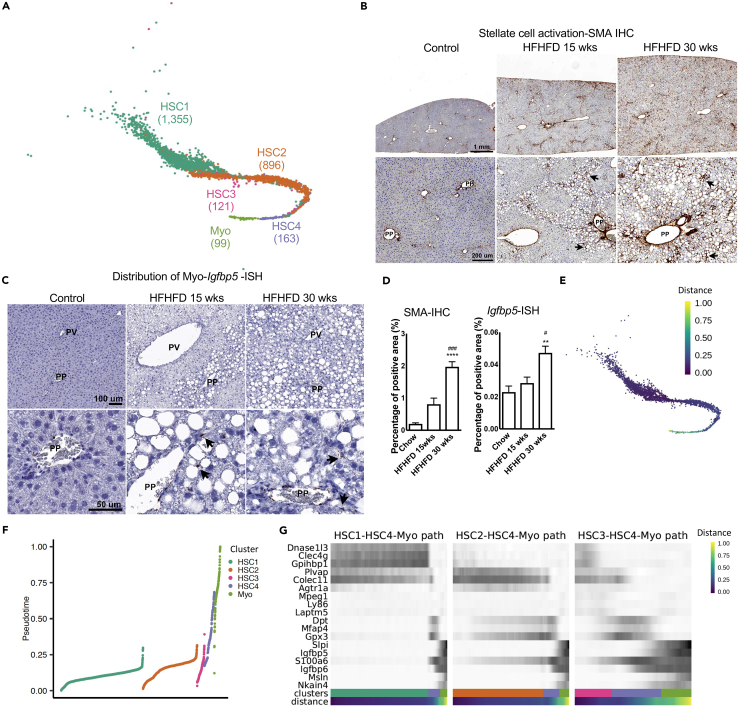
HSCs account for less than 10% healthy liver cells yet play important roles in fibrosis in chronic liver diseases (Friedman, 2008). scRNA-seq analysis provided further evidence to the complexity and heterogeneity of in vivo naive HSCs and Myo, identified HSCs subpopulations, and delineated underlying cell lineage relationships. Overall, 2,634 HSCs and Myo single-cell transcriptomes from healthy and NAFLD livers comprised four distinct HSCs populations and one Myo population (Figure 3A). The two main subpopulations: HSC1 and 2 composed 53.45% and 35.35% of HSCs, respectively. HSC3 and 4 only composed of 4.78 and 6.30%, respectively.
Figure 3.
Classification of cell subpopulations in stellate cells and myofibroblasts
(A) Clustering of stellate subpopulations and myofibroblasts.
(B) Representative SMA–IHC images to show the distribution of SMA in healthy and NASH livers. Arrows indicate positive staining. Images on top panel are same magnification, so do the ones on bottom panel. Scale bar on top panel indicates 1 mm. Scale bar on bottom panel indicates 200 um. PP: periportal; PV: perivenous.
(C) Representative Igfbp5–ISH images to show the distribution of Igfbp5 RNA in healthy and NASH livers. Arrows indicate positive staining. Images on top panel are same magnification, so do the ones on bottom panel. Scale bar on top panel indicates 100 um. Scale bar on bottom panel indicates 50 um.
(D) Quantification of SMA (n = 5–12 mice per group, 1 section per mouse) and Igfbp5 (n = 3 mice per group, 2 sections per mouse) staining. Data are represented as mean ± SEM. Statistical t tests were done by comparing to chow group, ∗∗: p < 0.01; ∗∗∗∗: p < 0.0001. Comparing to 15 weeks group, #: p < 0.05; ###: p < 0.001.
(E) Projection of pseudotime on UMAP generated by Seurat.
(F) Projection of pseudotime on each subpopulation.
(G) Expression of cluster enriched genes. Cells ordered by pseudotime within each stellate-to-myofibroblast transition path. Colors of cluster were matched with Figure 3A.
HSCs activation and/or their trans-differentiation to myofibroblasts (Myo) due to hepatocyte injury and chronic hepatic inflammation is one of the hallmarks of liver fibrosis (Alegre et al., 2017; Elpek, 2014; Hernandez-Gea and Friedman, 2011; Higashi et al., 2017). Alpha-smooth muscle actin (αSMA) (James et al., 2009) was one of the most commonly used markers to identify activated stellate cells in vitro or in vivo (Hernandez-Gea and Friedman, 2011; Puche et al., 2013a, 2013b). Consistent with previous reports, gradually increased SMA immunohistochemistry (IHC) and Myo marker insulin-like growth factor-binding protein (Igfbp) 5 in situ hybridization (Zheng et al., 2017) positive areas were observed in livers from healthy to NASH (Figures 3B–3D). Expression of activated HSCs marker Acta2 (SMA gene symbol) and Myo marker Igfbp5 were shown in Figure S2C for each individual stellate cell subpopulation and time points of diet treatment. Acta2 and Igfbp5 expression and regulation were distinctively different along NASH progression. Acta2 was upregulated in several subgroups of stellate cell but downregulated in Myo along NASH progression. Igfbp5 was barely expressed in stellate cell subgroups except 34 weeks HSC4 but highly expressed in Myo. The different expression pattern indicates that stellate cell activation was not simply gene expression regulation or cell trans-differentiate to Myo. Instead, it is the combination of two processes in response to NASH progression. SMA IHC was conducted to check the distribution of SMA-positive cells. Healthy livers had very low SMA-positive signal about 0.2% area and the positive staining was mostly localized around periportal (PP) area where there is presence of bile ducts and blood vessels. After 15 or 30 weeks of diet treatment, SMA IHC signals were significantly increased to 0.8% and 2% area, respectively, which was not only expanded around PP area but also detected around hepatocytes especially those injured hepatocytes with macro-vesicles (Figures 3B and 3D). The distribution of Igfbp5-positive Myo in healthy liver was around PP area and much less, only 0.02% positive area. After 15 or 30 weeks of diet treatment, Igfbp5-positive Myo were not only increased around PP area but also extended out to hepatocyte area especially around injured hepatocytes with macro-vesicles, 0.03% and 0.05% positive area, respectively (Figures 3C and 3D).
Pseudotime analysis is usually used to characterize cell developmental trajectory based on transcriptional similarities (Campbell and Yau, 2018). Here, we employed it to further study the lineage relationship of HSCs (Figure 3E). The results suggested a plausible cell trans-differentiation trajectory from stellate cells to Myo. State 1 cells were from HSC1, HSC2, and HSC3, state 2 cells were from HSC4, while state 3 cells were from Myo (Figures 3E–3G). The proximal pseudotime ordering between HSC4 and Myo suggested that HSC4 was an intermediate stage for the transition from stellate cells to Myo. Three apparent HSC-to-Myo transition paths were defined based on the pseudotime trajectory (Figure 3G). Along the paths, HSCs gradually lost common stellate cell markers such as Reelin (Reln) and extracellular matrix protein (Ecm) 1 and gained Myo cell markers such as Igfbp5, Igfbp6, S100a6, and keratin 19. Igfbp family was reported to mark HSCs to Myo trans-differentiation, especially the increased expression of Igfbp5 and Igfbp6 in Myo (Boers et al., 2006). Similarly, S100a6 was identified as a key marker of activated Myo (Krenkel et al., 2019). Gene transition pattern of HSC3 and HSC2 to Myo was much smoother comparing with HSC1 to Myo (Figure 3G). The expression of S100a6 increased gradually from HSC2 and HSC3 to HSC4 then Myo. However, HSC1 did not have similar basal expression of the gene, instead showed somewhat sudden transition to the expression in Myo. These data indicated that HSC3 and HSC2 might be easier to trans-differentiate to Myo than HSC1 (Figure 3G).
HSC1 and HSC2 share some common stellate markers Rgs5, Reln, Lum, and Ecm1 (Figure S2A). However, HSC1 showed interesting endothelial and stellate cell chimeric marker expression, which not only expressed those common stellate cell markers listed above but also many endothelial cell markers like protein tyrosine phosphatase receptor type B (Ptprb) and Clec4g (Figure 4A). The DEGs of HSC1 and HSC2 were involved in endocrine-regulated calcium reabsorption, renin secretion et al. pathways (Figure 4A). The expression pattern of endothelial marker Ptprb and stellate cell marker Lum showed the differences between these two subpopulations (Figure 4B). Similar subpopulations of HSCs were not reported in the manuscript but were integrated by us in Xiong's dataset with much fewer cells in each subpopulation (Xiong et al., 2019; GEO: GSE129516) (Figure S3). In order to check whether HSC1-endothelial-chimeric stellate cells and HSC2 distributed differently across the healthy and NASH liver, Ptprb (green florescence), and Lum (red fluorescence) double in situ hybridization was performed on healthy and NASH livers. In healthy liver, both Ptprb and Lum-positive cells were evenly distributed across PP and perivenous (PV) areas. Double-labeled HSC1 shared the similar distribution pattern as Lum single-labeled HSC2. However, their distributions in NASH liver were different. Ptprb-positive endothelial cells still maintained the even distribution pattern in NASH liver, but Lum-positive stellate cells tended to cluster around focal lesion areas which is consistent with the observation that HSCs were usually activated by inflammation. There were still double-positive HSC1 cells around both PP and PV areas in NASH liver, although not as extensive as healthy liver. They tended to have clustered distribution pattern like activated HSCs (Figures 4C–4D).
Figure 4.
The identification of HSC1 endothelial-chimeric stellate cell subpopulation
(A) Volcano plot of differentially expressed genes in endothelial-chimeric stellate cells HSC1 compared to HSC2. DEGs were colored based on enriched pathways.
(B) Expression profile of stellate cell marker Lum and endothelial cell marker Ptprb in endothelial, stellate, and endothelial-chimeric stellate cells.
(C) Representative Ptprb and Lum double RNA–ISH images to show the distribution of HSC1 and HSC2 in healthy liver. Arrows indicate double stained cells. Images are same magnification. Scale bar indicates 20 um. PP: periportal; PV: perivenous.
(D) Representative Ptprb and Lum double RNA–ISH images to show the distribution of HSC1 and HSC2 in NASH liver. Bottom panel showed the clustered HSC1 double-positive cells. Arrows indicate double stained cells. Images are same magnification. Scale bar indicates 20 um.
Taken together, our data demonstrated the heterogeneity of HSCs and Myo in vivo and highlighted the differences between activated HSCs and Myo. Pseudotime ordering demonstrated the HSC-to-Myo transition trajectories.
Classification of immune cells in healthy and NAFLD mouse livers
Overall, 30,182 single-cell transcriptomes from immune cells were derived from healthy and NAFLD livers (Figure 5A). Liver immune cells are heterogeneous and consist of multiple cell types with various immune functions. ScRNA-seq data identified eight distinct immune cell populations, including 2,852 DCs, 660 pDCs, 2380 T cells, 518 B cells, 574 NK cells, 110 neutrophils (N), 9,210 monocyte and monocyte derived macrophages (Mo/MoMF), and 13,878 KCs (Figure 5A). Except liver resident KCs, all infiltrated immune cells increased significantly at NAFL and NASH stages (Figure 2A). The increase of immune cell infiltration was consistent with the diet-induced inflammation (Figures S1D and S1F). Selected cell linage marker genes were shown in Figure 1C.
Figure 5.
Classification of immune cells in mouse liver
(A) Clustering of immune cells.
(B) Gene expression of M1 (Mrc1 and Cd163) and M2 (Fcgr1 and Fcgr2b) markers in immune cells. Color bars indicate the expression level in log-scale UMI.
(C) Gene expression of Kupffer cell marker Vsig4 and Mo/MoMF marker Ccr2. Color bars indicate the expression level in log-scale UMI.
(D) Quantification of Vsig4-ISH of Kupffer cells and Ccr2-ISH of Mo/MoMF (n = 3 mice per group, 2 sections per mouse). Data are represented as mean ± SEM. Statistical t tests were performed comparing to chow group, ∗: p < 0.05; ∗∗∗∗: p < 0.0001. Comparing to 15 wks group, ###: p < 0.001.
(E) Representative Vsig4-ISH images to show the distribution of Kupffer cells in healthy and NASH livers. Arrows indicate positive stained cells. Top and bottom panel Images are same magnification. Scale bar on the top panel indicates 100 um. Scale bar on the bottom panel indicates 100 um. PP: periportal.
(F) Representative Ccr2-ISH images to show the distribution of Mo/MoMF in healthy and NASH livers. Arrows indicate positive stained cells. Top and bottom panel Images are same magnification. Scale bar on the top panel indicates 100 um. Scale bar on the bottom panel indicates 20 um.
It has been the mainstream practice to define macrophages based on their activation states as classical (M1) or alternative (M2) activation (Murray et al., 2014). According to this terminology, M1 cells are microbicidal and tumoricidal, and favor Th1 proinflammatory responses. The common markers for M1 macrophages were mannose receptor C type 1 (Mrc1) and Cd163. On the other hand, M2 macrophages favor Th2 responses, have anti-inflammatory effects and promote tissue repair. The common markers for M2 macrophages were immunoglobulin gamma Fc region receptor (Fcgr) 1 and Fcgr2b (Montoya et al., 2019). In order to examine these features in liver macrophages, the expression level of M1 markers and M2 markers were shown in Figure 5B. Unfortunately, no distinct clusters of M1 or M2 population were observed in liver scRNA-seq. It seems that M1 markers were mainly expressed by KCs and a small proportion of Mo/MoMF. M2 markers were mainly expressed by Mo/MoMF and some KCs (Figure 5B). It raised the question whether the classical M1 versus M2 dichotomic view in liver was precise, and whether there are better ways to understand macrophage populations in the liver (Guillot and Tacke, 2019; Koyama and Brenner, 2017).
It is worthwhile to look deep into the two main subpopulations of macrophages identified in our data set, KCs and Mo/MoMF. The expression of several commonly used macrophage markers, V-set and immunoglobulin domain containing (Vsig) 4, Clec4f, Lyz1, and Ccr2 were shown in violin plots (Figure S4C). It is clear that Lyz1 is a common marker for both KCs and Mo/MoMF. Ccr2 is enriched in Mo/MoMF while Clec4f and Vsig4 are specifically expressed by KCs. Vsig4 or Ccr2-positive cells were very well aligned with KCs or Mo/MoMF subpopulation (Figure 5C). The quantification of Vsig4-ISH positive areas maintained around 0.4% in different stages of NAFLD liver (Figure 5D). It indicated that Kupffer cells maintained similar density along the NAFLD progression. On the other hand, the quantification of Ccr2-ISH positive area increased significantly from 0.05% area in healthy liver to 0.2% area in NAFL liver, then further increased to 0.4% area in NASH liver (Figure 5D). It indicated that monocytes were continuously recruited to liver while the disease was getting more severe. Although the density of KCs was not regulated by disease stages, the distribution was altered. KCs distributed evenly across the healthy liver. In the diseased liver, the homogenous distribution vanished, instead many KCs accumulated around the injured hepatocytes to form unique crown-like structures (Figure 5E). On the other hand, unlike KCs, Mo/MoMF was very rare in healthy liver, and they were sparse around PP area. After diet induction, more Mo/MoMF infiltrated into liver tissues and accumulated around the injured hepatocytes (Figure 5F).
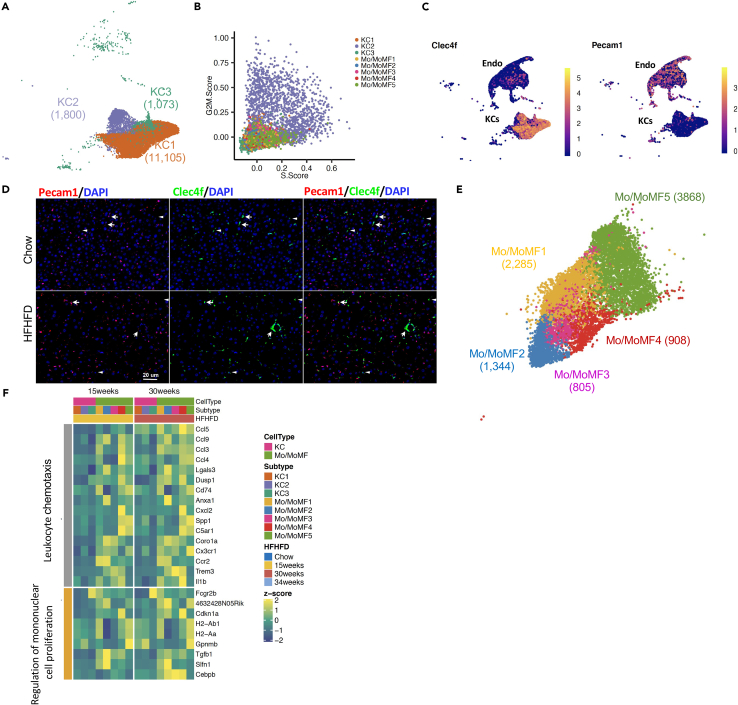
Classification of residential Kupffer cells and infiltrated monocyte-derived macrophages in healthy and NAFLD mouse livers
We identified three Kupffer cell subpopulations based on the clustering of 13,878 KCs (Figure 6A). KC1 is the main population with 11,005, 79.30% of KCs, whereas KC2 and 3 only have composed of 12.97% and 7.73% of KCs. Cell subpopulation markers and expression percentages were shown in Figure S4A. There was no disease-specific KC subpopulation, but its composition was regulated by NAFLD. KC1 maintained ∼80% KCs through all disease stages. KC2 population was increased from 5.25% in healthy liver to 13.3% in NASH liver. KC3 was decreased from 11.78% in healthy liver to 3.39% in NASH liver. Cell division genes were expressed higher by KC2 comparing with the other two subpopulations. Higher cell division genes led to higher S and G2M scores (Figure 6B), indicating KC2 had higher proliferating capacity. The increased percentage of KC2 at disease stages reflected the increased KC proliferation in response to the damage. The average density of KCs based on Vsig4-ISH positive area was not changed, but the overall number was increased due to the enlarged liver size induced by diet. KC3 was a unique subpopulation with chimeric expression of both endothelial cells and Kupffer cells markers as shown by the Pecam1 and Clec4f ISH double staining (Figures 6C and 6D). It is consistent with recent published data on endothelial cell heterogeneity in liver tumors (Zhao et al., 2020) and analysis using Xiong's data (Figure S5). In healthy liver, Clec4f and Pecam1 ISH double-stained KC3-endothelial-chimeric Kupffer cells distributed evenly throughout the liver. In NASH liver, KC3 was still distributed across the liver but formed the crown-like structure around injured hepatocytes (Figure 6D). The function of KC3 needs to be further studied.
Figure 6.
Classification of Kupffer cells and Mo/MoMF in mouse liver
(A) Clustering of Kupffer cell subpopulations.
(B) Proliferating scores of KCs and Mo/MoMF.
(C) Gene expression of Kupffer cell marker Clec4f and endothelial cell marker Pecam1 in endothelial, Kupffer, and endothelial-chimeric Kupffer cells. Color bars indicate the expression level in log-scale UMI.
(D) Representative Clecf4 and Pecam1 double RNA-ISH images for the distribution of endothelial-chimeric Kupffer cells in healthy and NASH livers. Arrows indicate double stained cells. Arrowheads indicate Clec4f single stained cells. Images are same magnification. Scale bar indicates 20 um.
(E) UMAP plot of Mo/MoMF subpopulations.
(F) Top enriched pathways of differentially expression genes between KCs and Mo/MoMF at HFHFD 15 and 30 weeks.
9,210 Mo/MoMF cells were isolated from healthy and NAFLD livers and 252 of total Mo/MoMF cells were from healthy liver. The set of highly regulated genes between Mo/MoMF and KCs generated by Sakai et al., including Ccr2, Cleg4f, et al., were used to map the Mo/MoMF subpopulations (Figures 6E and S4B) (Sakai et al., 2019). The naming of the Mo/MoMF subgroups was based on the distance from monocyte to macrophage reported by two recent papers (Sakai et al., 2019; Bonnardel et al., 2019). None of Mo/MoMF subpopulations showed increased S and/or G2M score as KC2 did (Figure 6B). Differentially expressed genes between Mo/MoMF and KCs indicated increased leukocyte chemotaxis and cell proliferation (Figure 6F). In short, gene expression changes signify the monocyte differentiation process.
In order to further understand the Kupffer and Mo/MoMF subpopulations, we did an integrative analysis with a recent data set published by Remmerie et al. (Figure S6) (Remmerie et al., 2020; GEO: GSE156052). It showed that most Kupffer cells and Mo/MoMF subpopulations were identified in the Remmerie et al. dataset as well. Mo/MoMF 1–3 subtypes were aligned very well with patrolling monocytes, Ly6Chi monocytes, and transitioning monocytes, respectively.
Hepatocyte and endothelial subpopulations in healthy and NAFLD mouse livers
Hepatocytes account for 60% of liver cells and play essential roles in detoxification, lipogenesis, protein metabolism, and gluconeogenesis (McEnerney et al., 2017). We obtained single-cell transcriptomes from 25,848 hepatocytes derived from healthy and NAFLD liver and identified seven distinct hepatocyte subpopulations, namely, Hep1 to 7 (Figure 7A). Top enriched genes in each cluster were shown on Figure S7B. Hep6 and 7 had very limited cell numbers which are 306, 1.18% and 258, 1.00% individually. Hep2 to 4 accounted 58.50% of hepatocytes and shared very similar enriched gene profile but with distinct expression profiles (Figure S7B). Hepatocyte-zoned gene expression pattern was consistent with the previous study (Halpern et al., 2017), for example, perivenous gene Cyp2e1 were highly expressed by Hep1, 10.7% of hepatocytes, while PP gene Gls2 were highly expressed by Hep2 to 5, 87.10% of hepatocytes (Figure S7A).
Figure 7.
Classification of hepatocytes and endothelial cell subpopulations in mouse liver
(A) Clustering of hepatocytes.
(B) Clustering of endothelial cells.
(C) Gene expression of hepatocyte marker Ass1 and endothelial marker Dnase1l3 in hepatocytes, endothelial, and hepatocyte-chimeric endothelial cells. Color bars indicate the expression level in log-scale UMI.
(D) Representative Dnase 1l3 and Ass1 double RNA–ISH images for the distribution of hepatocyte-chimeric endothelia cells in healthy and NASH livers. Arrows indicate double stained cells. Images are same magnification. Scale bar indicates 20 um.
(E) Double-positive chimeric cells were identified with imaging flow cytometer.
(F) The scheme of liver cell landscape in healthy and NAFLD livers. Hep: hepatocytes. Endo: endothelial cells. KC: Kupffer cells. Cho: cholangiocytes. Myo: myofibroblasts. Mo/MoMF: monocyte or monocyte-derived macrophage.
Hepatic endothelial cells composed of more than 20% of liver cells and are known to regulate fibrogenesis. We obtained single-cell transcriptomes from 23,037 endothelial cells (Endo) derived from healthy and NAFLD liver (Figure 7B). Clustering of liver endothelial cells identified five subpopulations (Figure 7B). Classical endothelial cell markers, Clec4g and deoxyribonuclease 1-like 3 (Dnase) 1l3, did not differentiate among the five clusters. To fully annotate endothelial subpopulations, differentially expressed markers were identified (Figure S7C). Endothelial-zoned gene expression pattern was consistent with the previous study. For example, WNT signal gene, R-spondin-3 (Rspo3), was highly expressed in Endo3 (7.31% of the Endo cells), and PP gene lymphocyte antigen (Ly6a) was highly expressed in Endo1 (9.30%) (Figures S6 and 7C). Endo2 was formed by sinusoidal endothelial cells (79.70%) (Figure 7B). A small subpopulation of Endo4 (1.85%) showed chimeric gene signatures for both Endo and Hep characterized by sinusoidal endothelial marker Dnas1l3 and hepatic marker arginosuccinate synthase 1 (Figures 7C and 7D).
The chimeric Endo-hepatocyte-chimeric endothelial cell cluster was also identified in Xiong's data (Figure S7D). In order to assess the spatial distribution of Endo4 cells, endothelial marker Dnase1l3 (red fluorescent) and hepatocyte marker Ass1 (green fluorescent) double fluorescent ISH staining were conducted. In both healthy and NASH liver, Dnase1l3-positive Endo cells were distributed around Ass1-positive hepatocytes. Several Dnase1l3-positive Endo cells had weak Ass1-positive staining, as arrowheads pointed out in the Figure 7D. The double-positive cells were distributed randomly among Endo cells and did not show any distinct distribution pattern.
We conducted imaging flow cytometry to further validate the chimeric cells are indeed distinct cell types and not the doublets and/or phagocytosis. Several surface marker antibodies were used to label different liver cell types. Asialoglycoprotein receptor (ASGR) 1 was used as hepatocyte surface marker, CD31 as endothelial cell surface marker, CD140b as stellate cell surface marker, and F4/80 as Kupffer cell surface marker. The single cell images indicated that these double-positive cell surface marker cells were single cells as shown in the bright field instead of doublets and/or phagocytosis (Figure 7E).
Discussion
Our study revealed the transcriptomic landscape of hepatocytes and NPCs in healthy and NAFLD mouse livers, their gene regulation, and transition with NASH progression. Thirteen major cell lineages and 3–7 subpopulations in each lineage were identified in the liver. Chronic liver disease was never a hepatocyte stand-alone pathology process, although they represent 60% of liver cells and serve energy metabolism and detoxication functions. Cell interactions between hepatocytes and among NPCs form and maintain the microenvironment to keep homeostasis or break the balance under pathologic environment (Xiong et al., 2019). Particularly for NAFLD, the pathology was initiated from toxic lipid accumulation in hepatocytes. Injured hepatocytes release a signal to trigger the early proinflammatory step. KCs recognize the injury and contribute to the recruitment of blood circulating immune cells. Sustained liver inflammation activates stellate cells. Activated stellate cells produce more deposition of collagen and other extracellular matrix proteins, which lead to severe fibrosis (Figure 7F) (Friedman et al., 2018). A better understanding of these mechanisms, and the roles of different cell types in this process, is critically important for the prevention and management of NAFLD liver diseases. Here the large-scale dataset and deep analysis of scRNA-seq opened the door to truly appreciate the heterogeneity and complexity of the NAFLD disease progression and built a tool to study signal crosstalk between and within the cell types. This will elucidate the understanding of NAFLD disease mechanism and identifying new potential targets.
We would like to point out a caveat that scRNA-seq procedure may have a bias in collecting cell types that can survive through the manipulations. Particularly in this study, in order to collect high quality cells for scRNA-seq, we harvested hepatocytes and NPCs from separate but identically treated mice because of the harsher conditions of the NPCs isolation process made it not ideal for hepatocyte isolation. In short, the collagenase conditions for the NPC digestion are harsher, since the NPC cells need to be released from perisinusoidal space as opposed to hepatocyte digestion, which does not need as harsh conditions due to the superficial location of the hepatocytes compared with NPCs. Furthermore, stellate cells were harder to be isolated from fibrotic liver. Any cell composition regulation suggested by scRNA-seq needs to be verified by orthogonal methods, such as cell marker IHC or ISH on intact tissue sections.
The limitation of small number of in vivo HSCs raised the question about unrecognized, functionally, and genetically diverse subgroups of HSCs. Benefited from this relatively naive and less manipulated HSCs scRNA-seq study, the novel endothelial-chimeric stellate cell subpopulation was identified; and the same chimeric subpopulation was mapped in Xiong's data although with fewer cells. The chances for the chimeric cells being doublets are low based on following observations. (1) The average detected gene numbers per cell are comparable between HSC1 and HSC2 (Figure S8), on the other hand, the detected gene numbers from doublets captured by HASH, were almost the sum of the detected gene numbers from two cell types. (2) The identification of double ISH-stained cells of endothelial marker Ptprb and stellate cell marker Lum further confirmed the existence of chimeric cells. HSCs are considered to be liver-specific pericytes (Hellerbrand, 2013; Pinzani, 1995a, 1995b) and, like pericytes in other tissues, are physically connected with endothelial cells and directly interact with them in a reciprocal manner (Poisson et al., 2017). Further experiments are needed to test whether this subpopulation is more pericyte-like and whether they serve certain complimentary endothelial functions. In addition, several other chimeric hepatic NPCs in healthy and NAFLD mouse livers were identified including hepatocyte-chimeric endothelial cell subpopulation and endothelial-chimeric Kupffer cell subpopulation (Figures 7E, S9A–S9C). Similar comparative pathway enrichment analyses were done for all the chimeric cell subpopulations. Hepatocyte-chimeric endothelial cell subpopulation maintains endothelial cell-related functions—fluid shear stress and atherosclerosis—and share similar PPAR signaling processes like their surrounding hepatocytes. Endothelial-chimeric Kupffer cell subpopulation maintains Kupffer cell-associated lysosome and leishmaniasis functions and added some of endothelial cell-related functions, like fluid shear stress and atherosclerosis. The similar cell subpopulation was reported in orthotopic liver tumors scRNA-seq study (Zhao et al., 2020). It was described as Kupffer-ECs (expressing both Kupffer and Endo markers) or macrophage-ECs (expressing macrophage and Endo markers). Since macrophage-ECs were significantly enriched in tumor-associated samples and were phenotypically closer to tumor-associated macrophages, it seems that this subpopulation might be regulated in severe liver disease environment.
The liver is the first defensive immune surveillance for detecting exogenous pathogens and antigens from gastrointestinal tract dietary and microbial products but also is tasked with facilitating immune tolerance which protects the host from antigenic overload of dietary components and bacterial products originating in the gut (Trivedi and Adams, 2016). Liver scRNA-seq dissects the unique milieu and function of hepatic immune cells and was essential for understanding liver-specific immunological properties and immune-mediated events implicated in NAFLD (Gill et al., 2018, 2019; Heymann and Tacke, 2016). Hepatic macrophages, a key player in maintaining homeostasis, the control of inflammation and repair of the liver, are a heterogeneous population consisting of cells derived from different origins and with different phenotypes and functions (Ju and Tacke, 2016). The possibility to reprogram macrophages into different phenotypes and functions (Montoya et al., 2019) holds great potential for targeting these cells for the treatment of liver diseases (Ju and Tacke, 2016). Classic M1 and M2 phenotypic macrophage may serve well for adipose macrophages but did not depict liver macrophages very well as indicated by our data. KCs were the main macrophages in healthy liver, while Mo/MoMF were recruited and formed a second macrophage population in the NAFLD liver. KCs self-renew from liver-resident cells originated from the fetal yolk sack (Perdiguero and Geissmann, 2016; Perdiguero et al., 2015). They are non-migratory cells, occupying a fixed position in the hepatic sinusoidal endothelium (Gordon and Pluddemann, 2017; Jenne and Kubes, 2013). On the other hand, Mo/MoMF are recruited from circulation monocytes and differentiated to macrophages after they are resident in the liver (Figure 7E) (Sakai et al., 2019; Bonnardel et al., 2019). Mo/MoMF helps KCs to maintain the liver homeostasis under liver disease condition. We recommend to group macrophages into KCs and Mo/MoMF first, then further subgroup KCs and Mo/MoMF based on their differential gene profiles. Early 2021, Daemetn et al. (Daemen et al., 2021) demonstrated VSIG4pos, TIM4neg monocyte-derived KCs (Mo-KCs) form hepatic crown-like structures and influence tissue fibrosis in mouse NASH liver. The structures are quite similar as the Vsig4-ISH positive crown-like structures identified in Figure 5E. We suspect that they might be the same subgroup of the macrophages since both are Vsig4-positive.
In summary, this study sheds light on liver cellular heterogeneity and provides novel insights into fundamental biology and pathology of NAFLD. With more understanding of the NAFLD gene regulation in different cell types, our next goal is to understand the landscape of the intrahepatic ligand-receptor signaling network.
Limitations of the study
We would like to point out a caveat that scRNA-seq procedure may have a bias in collecting cell types that can survive through the manipulations. Particularly in this study, in order to collect high quality cells for scRNA-seq, we harvested hepatocytes and NPCs from separate but identically treated mice because of the harsher conditions of the NPCs isolation process made it not ideal for hepatocyte isolation. In short, the collagenase conditions for the NPC digestion are harsher, since the NPC cells need to be released from perisinusoidal space as opposed to hepatocyte digestion, which does not need as harsh conditions due to the superficial location of the hepatocytes compared to NPCs. Furthermore, stellate cells were harder to be isolated from fibrotic liver. Any cell composition regulation suggested by scRNA-seq needs to be verified by orthogonal methods, such as cell marker IHC or ISH on intact tissue sections.
Benefited from this relatively big scale and naive scRNA-seq study, 3 chimeric NPCs were identified. However, the understanding of the chimeric NPCs is very limited. More specialized experiments need to be conducted to shed lights on their functions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-α-SMA | Abcam | Cat# ab124964; RRID: AB_11129103 |
| Rabbit polyclonal anti-CD45 | Abcam | Cat# ab10558; RRID: AB_442810 |
| Rat monoclonal anti-mouse CD31-BB700 | BD Biosciences | cat# 566490; RRID: AB_2869771 |
| Rat monoclonal anti-mouse CD45-APC-Cy7 | BD Biosciences | cat# 557659; RRID: AB_396774 |
| Rat monoclonal anti-mouse F4/80-Alexa Fluor 647 | BD Biosciences | cat# 565853; RRID: AB_2744474 |
| Rat monoclonal anti-mouse CD140b-PE | Biolegend | cat#136006; RRID: AB_1953271 |
| Mouse monoclonal ASGR1 antibody | This paper | N/A |
| RNAscope probes | ||
| Vsig4 | ACD Bio | Cat#520309 |
| Clec4f | ACD Bio | Cat#480421 |
| Ccr2 | ACD Bio | Cat#433271 |
| Ptprb | ACD Bio | Cat#481391 |
| Dnase 1l3 | ACD Bio | Cat#819171 |
| Lum1 | ACD Bio | Cat#480361 |
| Ass1 | ACD Bio | Cat#447991 |
| Igfbp5 | ACD Bio | Cat#425731 |
| Pecam1 | ACD Bio | Cat#316721 |
| Deposited data | ||
| Raw and BAM files | This paper | GSE166504 |
| Software and algorithms | ||
| 10x Genomics Cell Ranger | Zheng, Grace X.Y., et al., Nature Communications, 2017 | https://www.10xgenomics.com/ |
| Seurat V3 | Stuart, Butler, et al., Cell 2019 | https://satijalab.org/seurat/ |
Resource availability
Lead contact
-
•
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiping Cheng (xiping.cheng@regeneron.com).
Materials availability
-
•
This study did not generate new unique reagents.
Experimental model and subject details
Animals and diet information
Adult male C57BL/6 mice were housed under 12 hours of light per day in a temperature-controlled environment (22 ± 1°C, 60–70% humidity). Animals had free access to chow diet (5R53, Purina Lab, St. Louis, MO) or HFHFD (D09100301, Research Diets, New Brunswick, NJ). All procedures were conducted in compliance with protocols approved by the Regeneron Institutional Animal Care and Use Committee.
Method details
Single cell isolation and sequencing
Mice were anesthetized using isoflurane and perfused with two-step liver perfusion protocol (Kreamer et al., 1986; Seglen, 1976). After perfusion, hepatocytes were spun and collected at 50g, then processed for 10x Genomics single cell processing. The suspension went through 50% and 20% Percoll gradient centrifuge and NPCs were collected. FACS sorted unstained/DAPI- cells were loaded on a Chromium Single Cell Instrument (10x Genomics). RNA-seq libraries were prepared with the Chromium Single Cell 39 Library protocol (10x Genomics). Sequencing was performed on a NextSeq 500 (Illumina).
Chimeric cells identification with cell-surface marker staining and imaging flow cytometry
Non-parenchymal cells were isolated from wild type male mice on chow and subsequently stained with different cell surface markers: rat anti-mouse CD31-BB700 for endothelial cells (1:150, BD Biosciences, cat# 566490), rat anti-mouse CD45-APC-Cy7 for immune cells (1:500, BD Biosciences, cat# 557659), rat anti-mouse F4/80-Alexa Fluor 647 for Kupffer cells (1:200, BD Biosciences, cat# 565853), rat anti-mouse CD140b-PE for stellate cells (1:20, Biolegend, cat#136006) and mouse monoclonal ASGR1 antibody (6.7 μg/mL, in house) for hepatocytes. Then cells were stained with rat anti-mouse IgG-BB515 secondary antibody (1:500, BD Biosciences, cat#565104). Fluorescent minus one (FMO) staining was performed in parallel for each antibody. Cells were acquired on the Amnis® ImageStream®X Mk II imaging flow cytometer (Luminex Corporation, Austin, TX) using INSPIRE version 200.1.620 software at 60× magnification and the lowest speed setting. DAPI positive cells were excluded from gating. Focused, single-cell images were sub-gated and double-positive chimeric cells were identified and evaluated for co-expression.
Blood chemistry and liver lipid measurements
Blood glucose was determined using AlphaTRAK 2 (Zoetis, Parsippany, NJ). Plasma ALT, AST, triglycerides, cholesterol, HDL, LDL, and NEFA analytes were assayed in a Siemens ADVIA Chemistry XPTB Clinical System. Triglyceride, cholesterol, and NEFA measurements in a liver tissue were measured by Infinity Triglyceride, Infinity Cholesterol reagent (Thermo Fisher Scientific, Waltham, MA) and NEFA reagent (Wako Diagnostics, Mountain View, CA) respectively.
Tissue morphology
Liver samples were collected, fixed in 10% Formalin for 24 hours and then changed to 70% Ethanol prior to paraffin embedding. 5 um paraffin liver sections were prepared for histology. IHC and RNA-ISH were done on separate sections. For RNA-ISH, sections were permeabilized and hybridized with mRNA probes to Vsig4, Clec4f, Ccr2, Ptprb, Pecam1, Dnase 1l3, Lum1, Ass1 and Igfbp5 (ACD Bio, Newark, CA). Following probe hybridization and amplification, mRNA was detected using RNAscope 2.5 HD Assay brown kit or RNAscope Multiplex Fluorescent V2 assay (n = 3–6 mice from each group). For IHC, sections from each animal were stained with anti-SMA and anti-CD45. Sirius Red staining slides were in Picro-Sirius Red solution 0.25% Direct Red 80 (Sigma-Aldrich, St. Louis, MO) in saturated aqueous solution of picric acid for 1 h then followed by 3 washes of 0.5% acetic acid. Slides were scanned using Aperio AT2 Brightfield Slide Scanner or Zeiss Axio Scan Z1 slide scanner and replicates of each animal were analyzed using Halo software (Indica Labs, Albuquerque, NM).
Alignment, barcode assignment, and UMI counting
The Single Cell Software Cell Ranger Suite, version 2, was used to perform sample de-multiplexing, barcode processing, and single-cell gene UMI (unique molecular index) quantification (http://software.10xgenomics.com/single-cell/overview/welcome).
Cell clustering, differential expression, and pathway analysis
To eliminate batch effect among different batches, we used Seurat 3 (https://github.com/satijalab/seurat/) integration workflow. Cells were removed based on the following criteria: cells with fewer than 300 detected genes, more than 60,000 detected UMI, and a very high (>0.8) mitochondrial genome transcript ratio were filtered. The top 2,000 variable genes (HVGs) were identified using the function FindVariableFeatures method. Canonical Correlation Analysis (Berry et al., 2017) was used to identify common anchors between cells among batches. The first 30 dimensions of the CCA were chosen to integrate chow, 15 weeks, 30 weeks, and 34 weeks hepatocytes and NPCs. After integration, gene expression was scaled and centered for each HVG, followed by principle component analysis to reduce the number of dimensions representing each cell. The number of components used was determined based on the elbow plot. The top 30 PCs were selected for dimension reduction by Uniform Manifold Approximation and Projection (UMAP). Clusters were identified using function FindCluster in Seurat. FindAllMarkers function in Seurat was performed to call cell type-specific genes and differentially expressed genes. Top-ranked genes were ordered by fold change under a threshold of expressed in at least 25% of cells, larger than 1.5-fold change and adjusted p value <0.01 (Bonferroni correction).
Differential gene expression analysis between HFHFD mice and chow mice were performed using the function FindMarker in Seuart, using a Wilcoxon rank sum test. Genes with an adjusted p value less than 0.01, expressed in at least 25% of cells, larger than 1.5-fold change were considered to be differentially expressed. All differentially expressed genes were subjected to Reactome Pathway database (REACTOME) enrichment analysis. The significance of enriched REACTOME gene set was assessed by clusterProfiler (Yu et al., 2012).
Pseudotime analysis
To estimate the pseudotime we used Scanpy's partitioned-based graph abstraction function (PAGA) to order stellate cells and myofibroblast, mapped resulting pseudotime value onto original UMAP generated by Seurat described above. (Wolf et al., 2019)(https://github.com/theislab/paga).
Quantification and statistical analysis
Statistical analysis
All quantification data are mean ± SEM. Statistical analyses were performed utilizing GraphPad software Prism 6.0. All parameters were analyzed by student's t-test, one-way ANOVA or two-way ANOVA; a threshold of p < 0.05 was considered statistically significant.
Acknowledgments
Author contributions
Conceptualization, Y.X. and X.C.; methodology and investigation, S.Y.K., F.A., C.A., M.N., Y.W., and Y.Z.; data curation, Q.S. and Y.X.; writing – original draft, X.C.; writing – review & editing, M.E.B., M.A., M.W.S., A.J.M., Y.X., and X.C.
Declaration of interests
All authors are and were employees and shareholders of Regeneron Pharmaceuticals.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103233.
Contributor Information
Yurong Xin, Email: yrxinres@gmail.com.
Xiping Cheng, Email: xiping.cheng@regeneron.com.
Supplementalinformation
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
The accession number for the scRNA-seq data reported in this paper is GSE166504.
References
- Aizarani N., Saviano A., Mailly L., Durand S., Herman J.S., Pessaux P., Baumert T.F., Grun D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre F., Pelegrin P., Feldstein A.E. Inflammasomes in liver fibrosis. Semin. Liver Dis. 2017;37:119–127. doi: 10.1055/s-0037-1601350. [DOI] [PubMed] [Google Scholar]
- Berry S.A., Longo N., Diaz G.A., McCandless S.E., Smith W.E., Harding C.O., Zori R., Ficicioglu C., Lichter-Konecki U., Robinson B., Vockley J. Safety and efficacy of glycerol phenylbutyrate for management of urea cycle disorders in patients aged 2months to 2years. Mol. Genet. Metab. 2017;122:46–53. doi: 10.1016/j.ymgme.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Boers W., Aarrass S., Linthorst C., Pinzani M., Elferink R.O., Bosma P. Transcriptional profiling reveals novel markers of liver fibrogenesis: gremlin and insulin-like growth factor-binding proteins. J. Biol.Chem. 2006;281:16289–16295. doi: 10.1074/jbc.M600711200. [DOI] [PubMed] [Google Scholar]
- Bonnardel J., T'Jonck W., Gaublomme D., Browaeys R., Scott C.L., Martens L., Vanneste B., De Prijck S., Nedospasov S.A., Kremer A. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity. 2019;51:638–654.e639. doi: 10.1016/j.immuni.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braet F., Taatjes D.J., Wisse E. Probing the unseen structure and function of liver cells through atomic force microscopy. Semin.Cell Dev Biol. 2018;73:13–30. doi: 10.1016/j.semcdb.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Brunt E.M., Wong V.W., Nobili V., Day C.P., Sookoian S., Maher J.J., Bugianesi E., Sirlin C.B., Neuschwander-Tetri B.A., Rinella M.E. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers. 2015;1:15080. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- Campbell K.R., Yau C. Uncovering pseudotemporal trajectories with covariates from single cell and bulk expression data. Nat. Commun. 2018;9:2442. doi: 10.1038/s41467-018-04696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapper J.R., Hendricks M.D., Gu G., Wittmer C., Dolman C.S., Herich J., Athanacio J., Villescaz C., Ghosh S.S., Heilig J.S. Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G483–G495. doi: 10.1152/ajpgi.00079.2013. [DOI] [PubMed] [Google Scholar]
- Daemen S., Gainullina A., Kalugotla G., He L., Chan M.M., Beals J.W., Liss K.H., Klein S., Feldstein A.E., Finck B.N. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep. 2021;34:108626. doi: 10.1016/j.celrep.2020.108626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie R., Wilson-Kanamori J.R., Henderson B.E.P., Smith J.R., Matchett K.P., Portman J.R., Wallenborg K., Picelli S., Zagorska A., Pendem S.V. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 2019;29:1832–1847. doi: 10.1016/j.celrep.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elpek G.O. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: an update. World J. Gastroenterol. 2014;20:7260–7276. doi: 10.3748/wjg.v20.i23.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes C., Razavi H., Loomba R., Younossi Z., Sanyal A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67:123–133. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat.Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill U.S., Pallett L.J., Kennedy P.T.F., Maini M.K. Liver sampling: a vital window into HBV pathogenesis on the path to functional cure. Gut. 2018;67:767–775. doi: 10.1136/gutjnl-2017-314873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill U.S., Pallett L.J., Thomas N., Burton A.R., Patel A.A., Yona S., Kennedy P.T.F., Maini M.K. Fine needle aspirates comprehensively sample intrahepatic immunity. Gut. 2019;68:1493–1503. doi: 10.1136/gutjnl-2018-317071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S., Pluddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. 2017;15:53. doi: 10.1186/s12915-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot A., Tacke F. Liver macrophages: old dogmas and new insights. Hepatol. Commun. 2019;3:730–743. doi: 10.1002/hep4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X.F., Gao J.L., Li J.M., Li D. fat-1 mice prevent high-fat plus high-sugar diet-induced non-alcoholic fatty liver disease. Food Funct. 2017;8:4053–4061. doi: 10.1039/c7fo01050h. [DOI] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Toth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerbrand C. Hepatic stellate cells--the pericytes in the liver. Pflugers Arch. 2013;465:775–778. doi: 10.1007/s00424-012-1209-5. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gea V., Friedman S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- Heymann F., Tacke F. Immunology in the liver--from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 2016;13:88–110. doi: 10.1038/nrgastro.2015.200. [DOI] [PubMed] [Google Scholar]
- Higashi T., Friedman S.L., Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv. Drug Deliv. Rev. 2017;121:27–42. doi: 10.1016/j.addr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C., Kapoor R.R., Ismail D., Hussain K. The genetic basis of congenital hyperinsulinism. J. Med. Genet. 2009;46:289–299. doi: 10.1136/jmg.2008.064337. [DOI] [PubMed] [Google Scholar]
- Jenne C.N., Kubes P. Immune surveillance by the liver. Nat. Immunol. 2013;14:996–1006. doi: 10.1038/ni.2691. [DOI] [PubMed] [Google Scholar]
- Ju C., Tacke F. Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell Mol Immunol. 2016;13:316–327. doi: 10.1038/cmi.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama Y., Brenner D.A. Liver inflammation and fibrosis. J. Clin.Invest. 2017;127:55–64. doi: 10.1172/JCI88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreamer B.L., Staecker J.L., Sawada N., Sattler G.L., Hsia M.T., Pitot H.C. Use of a low-speed, iso-density percoll centrifugation method to increase the viability of isolated rat hepatocyte preparations. In Vitro Cell Dev Biol. 1986;22:201–211. doi: 10.1007/BF02623304. [DOI] [PubMed] [Google Scholar]
- Krenkel O., Hundertmark J., Abdallah A.T., Kohlhepp M., Puengel T., Roth T., Branco D.P.P., Mossanen J.C., Luedde T., Trautwein C. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut. 2020;69:551–563. doi: 10.1136/gutjnl-2019-318382. [DOI] [PubMed] [Google Scholar]
- Krenkel O., Hundertmark J., Ritz T.P., Weiskirchen R., Tacke F. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells. 2019;8:503. doi: 10.3390/cells8050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland S.A., Liu J.C., Ma X.Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massalha H., Bahar Halpern K., Abu-Gazala S., Jana T., Massasa E.E., Moor A.E., Buchauer L., Rozenberg M., Pikarsky E., Amit I. A single cell atlas of the human liver tumor microenvironment. Mol. Syst.Biol. 2020;16:e9682. doi: 10.15252/msb.20209682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEnerney L., Duncan K., Bang B.R., Elmasry S., Li M., Miki T., Ramakrishnan S.K., Shah Y.M., Saito T. Dual modulation of human hepatic zonation via canonical and non-canonical Wnt pathways. Exp. Mol.Med. 2017;49:e413. doi: 10.1038/emm.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya D., Mehta M., Ferguson B.G., Teles R.M.B., Krutzik S.R., Cruz D., Pellegrini M., Modlin R.L. Plasticity of antimicrobial and phagocytic programs in human macrophages. Immunology. 2019;156:164–173. doi: 10.1111/imm.13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.R., Cho C.S., Xi J., Kang H.M., Lee J.H. Holistic characterization of single-hepatocyte transcriptome responses to high-fat diet. Am J Physiol Endocrinol Metab. 2021;320:E244–E258. doi: 10.1152/ajpendo.00391.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E.G., Geissmann F. The development and maintenance of resident macrophages. Nat. Immunol. 2016;17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E.G., Klapproth K., Schulz C., Busch K., de Bruijn M., Rodewald H.R., Geissmann F. The origin of tissue-resident macrophages: when an erythro-myeloid progenitor is an erythro-myeloid progenitor. Immunity. 2015;43:1023–1024. doi: 10.1016/j.immuni.2015.11.022. [DOI] [PubMed] [Google Scholar]
- Pinzani M. Hepatic stellate (ITO) cells: expanding roles for a liver-specific pericyte. J. Hepatol. 1995;22:700–706. doi: 10.1016/0168-8278(95)80227-4. [DOI] [PubMed] [Google Scholar]
- Pinzani M. Novel insights into the biology and physiology of the Ito cell. Pharmacol. Ther. 1995;66:387–412. doi: 10.1016/0163-7258(94)00072-b. [DOI] [PubMed] [Google Scholar]
- Poisson J., Lemoinne S., Boulanger C., Durand F., Moreau R., Valla D., Rautou P.E. Liver sinusoidal endothelial cells: physiology and role in liver diseases. J. Hepatol. 2017;66:212–227. doi: 10.1016/j.jhep.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Puche J.E., Lee Y.A., Jiao J., Aloman C., Fiel M.I., Munoz U., Kraus T., Lee T., Yee H.F., Jr., Friedman S.L. A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology. 2013;57:339–350. doi: 10.1002/hep.26053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puche J.E., Saiman Y., Friedman S.L. Hepatic stellate cells and liver fibrosis. Compr.Physiol. 2013;3:1473–1492. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- Ramachandran P., Dobie R., Wilson-Kanamori J.R., Dora E.F., Henderson B.E.P., Luu N.T., Portman J.R., Matchett K.P., Brice M., Marwick J.A. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P., Matchett K.P., Dobie R., Wilson-Kanamori J.R., Henderson N.C. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat. Rev. Gastroenterol. Hepatol. 2020;17:457–472. doi: 10.1038/s41575-020-0304-x. [DOI] [PubMed] [Google Scholar]
- Remmerie A., Martens L., Thone T., Castoldi A., Seurinck R., Pavie B., Roels J., Vanneste B., De Prijck S., Vanhockerhout M. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity. 2020;53:641–657.e614. doi: 10.1016/j.immuni.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M., Troutman T.D., Seidman J.S., Ouyang Z., Spann N.J., Abe Y., Ego K.M., Bruni C.M., Deng Z., Schlachetzki J.C.M. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain Kupffer cell identity. Immunity. 2019;51:655–670.e658. doi: 10.1016/j.immuni.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviano A., Henderson N.C., Baumert T.F. Single-cell genomics and spatial transcriptomics: discovery of novel cell states and cellular interactions in liver physiology and disease biology. J. Hepatol. 2020;73:1219–1230. doi: 10.1016/j.jhep.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P.O. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- Seidman J.S., Troutman T.D., Sakai M., Gola A., Spann N.J., Bennett H., Bruni C.M., Ouyang Z., Li R.Z., Sun X. Niche-specific peprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity. 2020;52:1057–1074. doi: 10.1016/j.immuni.2020.04.001. e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaker M., Tabbaa A., Albeldawi M., Alkhouri N. Liver transplantation for nonalcoholic fatty liver disease: new challenges and new opportunities. World J. Gastroenterol. 2014;20:5320–5330. doi: 10.3748/wjg.v20.i18.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbol K.S., Kristiansen M.N., Hansen H.H., Veidal S.S., Rigbolt K.T., Gillum M.P., Jelsing J., Vrang N., Feigh M. Metabolic and hepatic effects of liraglutide, obeticholic acid and elafibranor in diet-induced obese mouse models of biopsy-confirmed nonalcoholic steatohepatitis. World J. Gastroenterol. 2018;24:179–194. doi: 10.3748/wjg.v24.i2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P.J., Adams D.H. Gut-liver immunity. J. Hepatol. 2016;64:1187–1189. doi: 10.1016/j.jhep.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Wisse E. Ultrastructure and function of Kupffer cells and other sinusoidal cells in the liver. Med. Chir Dig. 1977;6:409–418. [PubMed] [Google Scholar]
- Wolf F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Gottgens B., Rajewsky N., Simon L., Theis F.J. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Kuang H., Ansari S., Liu T., Gong J., Wang S., Zhao X.Y., Ji Y., Li C., Guo L. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol.Cell. 2019;75:644–660 e645. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi Z.M., Blissett D., Blissett R., Henry L., Stepanova M., Younossi Y., Racila A., Hunt S., Beckerman R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology. 2016;64:1577–1586. doi: 10.1002/hep.28785. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Molina-Portela M.D.P., Parveen A., Adler A., Adler C., WangH.E. W., Ni M., Wei Y., Atwal G. Heterogeneity and chimerism of endothelial cells revealed by single-cell transcriptome in orthotopic liver tumors. Angiogenesis. 2020;23:581–597. doi: 10.1007/s10456-020-09727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
The accession number for the scRNA-seq data reported in this paper is GSE166504.