Abstract
Patients with rare central nervous system (CNS) tumors typically have a poor prognosis and limited therapeutic options. Historically, these cancers have been difficult to study due to small number of patients. Recent technological advances have identified molecular drivers of some of these rare cancers which we can now use to generate representative preclinical models of these diseases. In this review, we outline the advantages and disadvantages of different models, emphasizing the utility of various in vitro and ex vivo models for target discovery and mechanistic inquiry and multiple in vivo models for therapeutic validation. We also highlight recent literature on preclinical model generation and screening approaches for ependymomas, histone mutated high-grade gliomas, and atypical teratoid rhabdoid tumors, all of which are rare CNS cancers that have recently established genetic or epigenetic drivers. These preclinical models are critical to advancing targeted therapeutics for these rare CNS cancers that currently rely on conventional treatments.
Keywords: atypical teratoid rhabdoid tumors, diffuse midline glioma, ependymoma, preclinical cancer models, rare cancers
Introduction
Primary central nervous system (CNS) tumors form in the brain or spinal cord. An estimated 83,830 new cases, about 25,000 of those being malignant, were expected to be diagnosed in the US in 2020 based on the findings from the 2013–2017 Central Brain Tumor Registry of the United States (CBTRUS) report.1 Glioblastomas, meningiomas, nerve sheath tumors, and pituitary tumors account for about 80% of all primary CNS tumors diagnosed in the US, while the remaining primary CNS cancers are considered extremely rare and have not been extensively studied.1 Due to technological advancements, next-generation sequencing, and recent epigenetic and molecular studies, our understanding of the molecular drivers of these rare cancers has improved drastically. Despite these insights, the treatment standards have remained the same for most CNS cancers and survival has not significantly improved for patients with rare CNS tumors. The low incidence of these cancers compromises the ability to test new treatments in prospective clinical trials, underscoring the need for laboratory models to identify the most promising treatments before embarking on the few clinical trials that are feasible. In this review, we highlight three rare CNS cancers: ependymoma (EPN), diffuse midline glioma (DMG), and atypical teratoid rhabdoid tumors (ATRT), all of which have distinct molecular characteristics used to generate preclinical models that accurately recapitulate the biology of tumors. In vitro and ex vivo models are advantageous to understand cellular mechanisms of cancer growth and drug response and to identify therapeutic targets by screening approaches while in vivo models are critical to validate novel targets and therapeutics in biologically complex systems.
Ependymoma (EPN)
Ependymomas (EPN) arise from cells lining the spinal canal and the ventricles of the brain. While ependymomas occur in all age groups, they happen more frequently in children and adolescents compared to adults.1 Moreover, although the median 5-year survival rate is 83.4% across all age groups, the survival rate drops to 42–55% for infants diagnosed with ependymoma compared to 91% survival for adults age 20–44.2 Strikingly, up to 50% of children with intracranial ependymoma will have their cancer relapse, usually at the primary site of the tumor.3 Patients with relapsed ependymoma have a dismal 5-year survival rate of 25%. This suggests that current treatment regimens are not curative for the majority of pediatric patients and novel therapeutics are needed.
Ependymomas occur in three clinically and anatomically distinct locations: supratentorial (ST-EPN), infratentorial/posterior fossa (PF-EPN), and spinal ependymoma (SP-EPN). In adults, there are similar incidences of spinal and intracranial ependymoma; however, almost 80% of ependymomas in children are intracranial.1 When stratified by location, patients with intracranial ependymomas have an overall worse prognosis than those with spinal cord tumors and within intracranial tumors, ST-EPN has a worse progression-free survival compared with PF-EPN.4
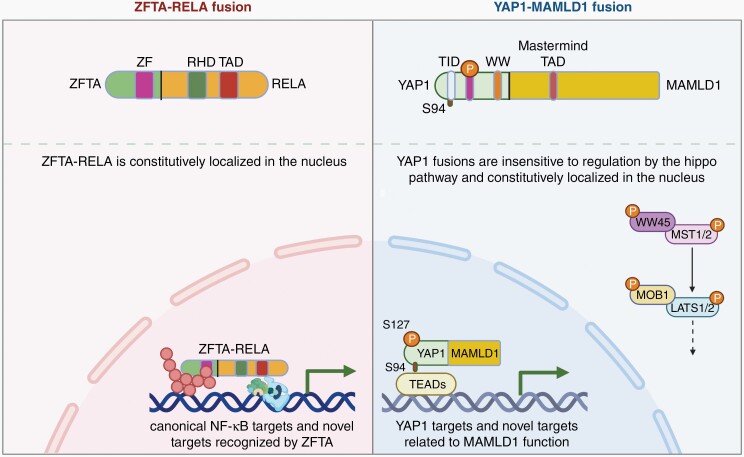
Recent genomic studies have subclassified ependymomas through their molecularly distinct signatures that are also location specific.5 ST-EPN are characterized as driven by RELA fusion or by YAP1 fusion products, most commonly occurring in children. About 70% of supratentorial ependymomas are marked by a chromothripsis event on chromosome 11q that fuses the ZFTA gene (previously known as C11orf95) to several different genes, most commonly RELA. The ZFTA-RELA fusion translocates to the nucleus and has been shown to activate both NF-κB and non-NFκB targets, in part by binding to a unique DNA motif recognized by the ZFTA (C11orf95) moiety of the fusion protein (Figure 1a).6 The remaining ST-EPN harbor various YAP1 gene fusions (YAP1-MAMLD1, YAP-FAM118B) with an otherwise relatively quiet genome, suggesting the fusion product is sufficient for oncogenesis and likely the initiation event. YAP1 is a downstream effector of the Hippo signaling pathway, commonly dysregulated in many cancers. Typically, a core kinase cascade regulates the phosphorylation, cellular localization, and co-factor transcriptional activity of YAP1; however, recent work has shown that YAP1 fusion proteins are constitutively active and expression of different YAP1 gene fusions are sufficient to generate EPN in mice, highlighting their role as oncogenic drivers (Figure 1b).7 PF-EPN are characterized by epigenetic changes, with few single nucleotide variations or DNA copy number alterations. Posterior fossa group A (PF-A) ependymoma have a greater extent of CpG island DNA methylation compared with group B (PF-B).8 PF-A is more common in young children and has a worse prognosis, compared with PF-B.9 Additionally, PF-A tumors are marked by loss of histone H3 lysine 27 trimethylation (H3K27me3) staining driven by aberrant DNA methylation.10
Fig. 1.
Schematic of known oncogenic mechanisms of ZFTA-RELA (a) and YAP1-MAMLD1 (b) fusions in supratentorial ependymomas. ZFTA-RELA fusion retains portion of ZFTA with zinc-finger (ZF) domain and portion of RELA with Rel homology domain (RHD) and transcriptional activation domain (TAD), resulting in constitutively active and nuclear localized fusion product and expression of canonical NF-kB and novel targets. YAP1-MAMLD1 fusion retains N-terminal YAP1 with S94 phosphorylation site within the YAP1-TEAD interaction domain (TID), S127 phosphorylation site, and WW domain (WW) along with C-terminal MAMLD1 containing nuclear localization signal (NLS) and transcriptional activation domain (TAD). YAP-MAMLD1 fusion is resistant to regulation by Hippo kinase cassette and constitutively nuclear, resulting in transcriptional activity of YAP1 and MAMLD1 targets. Created with BioRender.com
Histone-Mutated High-Grade Gliomas (HGG)
Diffuse Midline Glioma (DMG) is an aggressive type of brain tumor that most often forms in midline locations including the pons in the brainstem, thalamus, spinal cord, and cerebellum.1 DMG is characterized by the presence of K27M mutations in histone 3 genes, most frequently in H3F3A (encoding for H3.3) and at lower frequencies in HIST1H3B or HIST1H3C (encoding for H3.1) which lead to loss of the H3K27me3 mark. Frequently co-mutated genes include TP53, PPM1D, FGFR1, PIKC3A, NF1, PTEN, PDGFRA, and TCF12.11 Pediatric patients with H3 K27M-mutated DMGs have an extremely poor prognosis with a median survival <1 year after diagnosis; whereas, adults harboring DMG tumors with H3 K27M mutations have longer survival (median OS of 27.6 months).12 Additionally, adult tumors contain greater heterogeneity in origin site and molecular pathogenesis, compared to children.12
Another mutational variant of the H3F3A gene that results in HGG is a G34R mutation and a less frequent G34V mutation.13 In contrast to DMG, these tumors occur most often in cerebral hemispheres. G34R/V mutations of H3.3 result in impaired catalytic activity of SETD2 and inhibiting mismatch repair (MMR). Despite their heterogeneous histopathology, tumors with H3.3 G34 mutations display uniform epigenetic DNA methylation signatures and frequently have co-occurrent ATRX/DAXX and TP53 mutations.14 In this stratified group, the median overall survival is 22 months, slightly better than most other HGG molecular subtypes.14
Atypical Teratoid Rhabdoid Tumors (ATRT)
Atypical teratoid rhabdoid tumors (ATRT) are highly lethal and account for up to 50% of all CNS tumors for infants in the first year of life.1 Although the majority of cases are diagnosed in children under the age of three, cases of ATRT have been reported in adults and tend to occur in the cerebral hemisphere and sellar region.15 These tumors present as large, heterogeneous, highly dense, invasive, and vascular masses and may occur as multiple intracranial and/or with extracranial lesions.16 The presence of rhabdoid cells and multi-lineage differentiation helps to distinguish this tumor type from other CNS cancers.17
The majority of ATRT are characterized by loss of Chr22q11.2 with subsequent loss of SMARCB1, a core subunit of the SWI/SNF chromatin-remodeling complex, while a small proportion instead harbor mutations in SMARCA4, an ATPase subunit of the SWI/SNF complex.16 SMARCB1 loss leads to major epigenetic changes due to dysregulation of chromatin remodeling including loss of histone acetylation (H3K27Ac) at enhancer regions and dysfunction of PRC2 and histone methylation (H3K27Me) by EZH2.18,19 ATRT can also be further classified into three distinct molecular groups (ATRT-MYC, ATRT-SHH, and ATRT-TYR) that may facilitate the identification of targeted therapeutics.20
Treatment for Rare CNS Cancers Has Remained Static
Despite increased molecular insights in the past decade, rare CNS tumors are similarly treated with surgery and/or radiotherapy and sometimes with conventional chemotherapeutic agents used for other more common brain tumors. Proton beam therapy is used in children with promising results to reduce the dose of ionizing radiation administered with the hopes of preventing toxicity and damage.21 Chemotherapy has been used for these rare CNS cancers, especially in younger patients, to protect from long-term effects of radiation therapy. However, this treatment option has resulted in unclear results in terms of balancing efficacy in prolonged survival while maximizing quality of life.
Reliable Preclinical Models Are Needed for Developing Novel Therapeutics
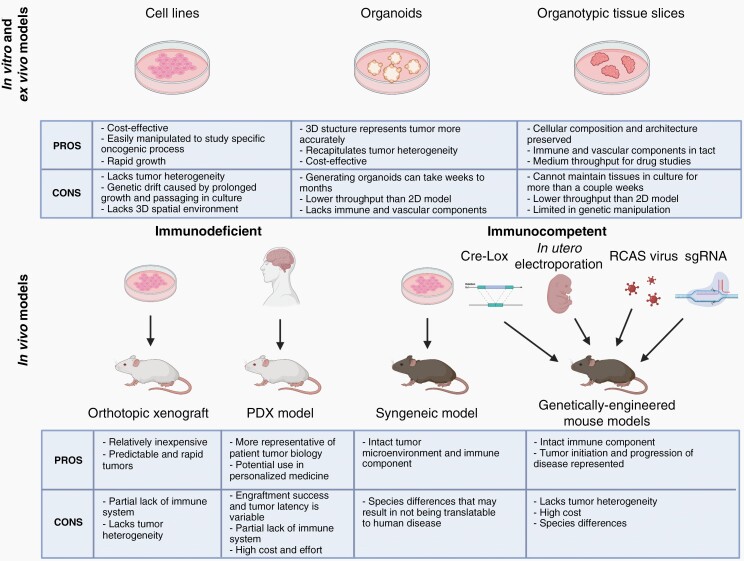
Treatment of rare CNS cancers has remained static, with few approved targeted treatment options. Due to the rarity of these diseases, it is extremely difficult to recruit sufficient numbers of patients for randomized, large clinical trials. Only recently have we begun to understand the genetic and molecular drivers of these diseases that would necessitate further stratification for targeted therapies. Thus, preclinical models that accurately replicate patient biology and predict response are critical for developing novel targeted therapeutics. In vitro and ex vivo models are advantageous for their low cost, high-throughput, reproducibility, and mechanistic insights. They have been critical for elucidating the underlying genes and pathways involved in CNS cancer progression and therapeutic resistance. However, they do not completely phenocopy the heterogeneity and complexity of human tumors. Thus, in vivo models are needed particularly in the late stages of therapeutic development before clinical use. No single model can currently recapitulate the progression, heterogeneity, and drug response of tumors; however, we can utilize different models together to gain greater insight before translational application (Figure 2).
Fig. 2.
Summary of advantages and disadvantages of preclinical models for rare cancers. In vitro and ex vivo models include culturing cell lines, organoids, and organotypic tissue slices. In vivo models consist of immunodeficient models including orthotopic xenograft and patient-derived xenograft (PDX), and immunocompetent models including syngeneic and genetically-engineered mouse models (by Cre-Lox system, in utero electroporation, RCAS/tv-a system, or CRISPR/Cas9 technology). Created with BioRender.com.
In Vitro Models
Use of in vitro models allows researchers to control and interrogate specific function of oncogenic drivers to understand the biology of tumor progression. Since patient samples for these CNS cancers are difficult to obtain, immortalized cell lines derived from tumors which maintain their genetic changes, have been critical for identifying the genetic and epigenetic mutations that contribute to tumor initiation. Cell lines derived from human or mouse tumors can be genetically and pharmacologically manipulated to elucidate the mechanisms of cancer cell growth, immune regulation, and therapeutic drug resistance. The rapid growth and ease of working with cell lines make them ideal for high-throughput screening and large-scale omics studies. Additionally, co-culture of tumor cells with stromal cells such as fibroblasts, endothelial, and immune cells can be more precisely controlled using cell lines.
Cultured cells have the ability to form 3D spheroids through cell-cell contacts in ultra-low attachment conditions either with or without the addition of extracellular matrix components. Spheroids recapitulate cell-cell junctions and cell polarity and more accurately mimic the structures within a tumor while retaining the advantages of cell culture studies. Neurosphere cultures from ependymomas maintain cancer stem cell characteristics, contain 3D interactions and display tumor gradients of oxygen and nutrients.22 However, prolonged growth and passaging in culture, particularly on plastic dishes, can cause genetic drift in cancer cells thus decreasing replicative traits of the tumors in which they were derived.23
An approach to model cancer initiation and growth for tumors with known molecular drivers is to introduce genetic changes in human embryonic stem cells or induced pluripotent stem cells (iPSCs). Human iPSCs were used by Haag et al. to investigate the cell of origin for H3K27M-driven tumor development by engineering an inducible H3.3-K27M allele in the endogenous locus of different disease-relevant neural cell types; they found that only neural stem cells gave rise to tumors upon induction of H3.3-K27M and TP53 inactivation.24
A major drawback of these models is that they lack both intertumoral and intratumoral heterogeneity seen in human patients, which can result in vast differences in response between in vitro studies and clinical trials. To overcome the lack of intertumoral heterogeneity, multiple cell lines derived from different sources should be used to confirm experimental findings. Interestingly, Kinker et al. performed single-cell RNA-seq studies in 198 cancer cell lines representing 22 cancer types and found considerable intratumoral heterogeneity particularly in biological processes related to cell cycle, senescence, and epithelial-mesenchymal transition.25
Ex Vivo Models
Tumor organoids are grown in 3D culture with surrounding extracellular matrix that better recapitulates the tumor composition with varying progenitor and differentiated cells.26 Organoids are established directly from patient tumors or by genetic engineering of healthy tissue organoids or stem cells; those generated from patient tumor samples retain the genomic profile as well as cellular and molecular phenotype of the tumor it was derived from. They can easily be expanded and cultured long term as well as be cryopreserved for storage. Stromal cells can also be incorporated in tumor organoids to better recapitulate the tumor microenvironment. Organoid culture is cost-effective compared with in vivo models and can allow for drug screening while resembling tumor heterogeneity and composition. To study SMARCB1 loss throughout the differentiation process in ATRT, a recent study introduced an inducible SMARCB1 loss-of-function system into human iPSCs differentiated into cerebral spheroids.27 This study identified substantial differences in downstream effects of SMARCB1 loss depending on the differentiation state, providing a novel platform for studying the heterogeneity of ATRT tumorigenesis.27 The organoid model system is promising for better modeling rare CNS cancers; however, the process of generating organoids can take weeks to months and have lower throughput capabilities for screening compared with 2D cell culture. Additionally, organoids are established using a relatively small number of cells from a tumor, which may have implications on clonality and heterogeneity, with propagation of only cells that can survive in culture.
Organotypic tissues slices of the brain were first described in the 1960s, where the mouse brain was thinly sliced (200–250 μm) and maintained in culture ex vivo.28 This technique has been adapted for studying tumor biology and has proven effective at preserving the histological and 3D structure of a tumor, including maintaining inter- and extra-cellular interactions, cell matrix components, and presence of stromal and immune cells.29 Many slices can be generated from a tumor and the cellular composition and architecture are preserved for days to weeks. Tumor tissue slices can then be used to test the effects of cancer therapeutics in a sample that maintains normal architecture and an intact native tumor microenvironment.30,31 Moreover, effects on not only tumor cells but also on the stromal cells can be assessed in this system using microscopy and biochemical techniques.32 Recently, the Gujral lab adapted the tissue slice protocol by preparing 400 μm slices with an automated vibratome followed by using a tissue chopper to create 400 × 400 × 400 μm3 cuboids.33 Use of tumor cuboids is ideal for utilizing limited samples and increasing screening throughput while maintaining the complexity of the whole tissue.
In Vivo Models
Transplant Models
Syngeneic or xenograft transplant models where murine or human cell lines, primary tumor cells or tumor tissue are implanted into mice are relatively inexpensive compared to other in vivo models and generate predictable and rapid tumors. Cells or tissue can be engrafted subcutaneously to easily monitor tumor growth or orthotopically in the native tumor site, which more accurately models the tumor microenvironment. Orthotopic graft models have a translational advantage but are more labor-intensive and technically challenging than subcutaneous engraftment and often require advanced imaging methods to monitor tumor growth. Syngeneic models engrafting mouse cancer cell lines into mice have a more realistic microenvironment and immune system since the tumor sample is genetically and immunologically compatible with the host. Human cell line xenografts represent the human biology more closely but require immunocompromised mice, which lack functional immune components. To overcome this drawback, immunocompromised mice can have their immune system “humanized” by engraftment of human immune cells, either with cord blood-derived hematopoietic stem cells or with peripheral blood mononuclear cells (PBMCs).34
Multiple modeling approaches have used human iPSC-derived neural stem cells to study the impact of ectopic H3.3-K27M or H3.3-G34R expression – combined with p53 loss and/or PDGFRA activation on the transcriptome and epigenome. Importantly, these studies have demonstrated the role of H3K27M to cause glioma formation in vivo using transplant models.24,35–37 A syngeneic model of ST-EPN-RELA tumors was generated by the intracranial injection of in vitro-transduced ZFTA-RELA-expressing murine Blbp-GFP/Cdkn2a-/- neural stem cells into CD-1 nude mice.38 A high percentage of injected mice developed tumors recapitulating “vascular-variant” human supratentorial ependymomas that stained positive for phospho-Ser276-RELA (a marker for transcriptionally active RELA), as well as CCND1 and L1CAM.
Patient-derived xenografts (PDX) involve implanting small pieces of tumors directly from patients or organoids derived from patient tumors. These models can be used to assess the efficacy of novel treatments and to inform preclinical trials. Recently, Smith et al. successfully developed 37 orthotopic xenograft models derived from CNS tumors from pediatric patients, including EPN and ATRT. These models exhibited high fidelity and molecular heterogeneity with their respective parental tumors based on histopathology, whole-genome and whole-exome sequencing, RNA sequencing, and DNA methylation assays.39 PDX models sustain the heterogeneity of patient samples; however, the success rate of tumor cell engraftment and the tumor latency is variable. A disadvantage of patient-derived orthotopic xenografts is that this model lacks the functional immune component, critical for preclinical and immunotherapy testing. The use of patient-matched humanized mice along with patient-derived xenografts have huge implications for personalized medicine; however, the high costs and limited immune function are still prohibitive for wide-scaled use. Additionally, human stroma from the patient tumor becomes replaced with murine stroma over time, therefore potentially losing critical interactions for tumor growth and progression.
Genetically Engineered Mouse Models (GEMMs) of Rare CNS Tumors
Transplant models are relatively fast, but they rely on using samples of cancer cells or tumors that are fully transformed in terms of malignancy. Thus, it is difficult to study the process of tumor initiation and progression, including the process of metastasis, in these models. Genetically engineered mouse models (GEMMs) can be created by introducing specific changes in the mouse genome by the Cre-Lox system, in utero electroporation, RCAS/tv-a system, or with Clustered Regularly Interspaced Short Palindormic Repeats (CRISPR)/Cas9 technology. Here, we describe current GEMMs of rare CNS cancers.
Ependymoma (EPN)
Ozawa et al. facilitated the RCAS/tv-a model as well as a lentiviral system for somatic cell gene transfer to express different variants of ZFTA-RELAFUS in mice.40 Expression of ZFTA-RELAFUS1 lead to the de novo formation of ST-Ependymoma-like tumors in both Cdkn2a null and wild-type mice and tumors displayed several histological and histopathological features characteristic of ependymoma. RNA sequencing analysis furthermore showed that the transcriptomes of these ZFTA-RELAFUS1-driven mouse tumors up-regulated both NF-κB and non-NF-κB targets and were significantly enriched for gene expression signatures of human ST-EPN-RELA tumors specifically, but not for the other molecular EPN subtypes. Interestingly, ZFTA-RELAFUS3 and ZFTA-RELAFUS4, both retaining two ZFTA Zink finger domains, induced tumor formation at lower rates and a longer latency compared to ZFTA-RELAFUS1/2 expression.41ZFTA-RELAFUS8, lacking the majority of the REL homology domain of RELA, was unable to induce tumor formation.41 Finally, by introducing a series of point mutations into the sequence of ZFTA-RELAFUS1, Ozawa et al. could show that the Rel homology domain of RELA is essential for the oncogenic functions of ZFTA-RELAFUS1 as mutagenesis of serine to glutamic acid of ZFTA-RELAFUS1 Ser486 (corresponding to RELA Ser276) abolished its oncogenic potential.
A smaller subset of ST-EPN patient tumors harbor YAP1 gene fusions, most prominently YAP1-MAMLD1 and to a lesser degree YAP1-FAM118B.5 Intracranial expression of either YAP1-MAMLD1 or YAP1-FAM118B in Nestin-expressing cells was sufficient to induce tumor formation in neonatal mice, whereas in embryonic brains, only YAP1-MAMLD1, but not YAP1-FAM118B was able to induce tumor formation.7,41,42 Tumors formed by the intracranial expression of either YAP1-MAMLD1 or YAP1-FAM118B had a spindle-cell histomorphology and tended to grow diffusely into the brain parenchyma which is in contrast with the typical circumscribed growth pattern of RELAFUS-driven tumors.7,41 RNA-Seq and ChIP-Seq experiments of YAP1-MAMLD1-driven mouse tumors showed significant overlaps in the gene expression patterns and genome occupancy of human ST-EPN-YAP1 tumors.7,42
The activity of wild-type YAP1 is controlled by the Hippo signaling pathway under normal physiological conditions and activated Hippo signaling results in the phosphorylation of YAP1 at several serine residues and the subsequent nuclear exclusion and proteasomal degradation.43 These YAP1 gene fusions represent activating mutations of YAP1 that are resistant to inhibitory Hippo signaling. Consequently, the intracranial expression of constitutively active YAP1 (S127/397A-YAP1) in Nestin-expressing cells also resulted in tumor formation. Similarly, expression of 5SA-YAP1 (S61/109/127/164/397A-YAP1) or co-deletion of Lats1/2 in Neurod6-expressing cells in the developing embryo resulted in the formation of ependymoma-like tumors shortly after birth.44
Diffuse Midline Glioma (DMG)
The Becher group utilized the RCAS/tv-a system to deliver DMG-specific genetic alterations to generate a GEMM by expressing RCAS-PDGFB, RCAS-Cre, RCAS-H3.3K27M in the pons of Nestin-tv-a/p53fl/fl mice.45 They have since then used this model to investigate the influence of additional genetic alterations (such as ACVR1 R206H) or test the efficacy of different treatment strategies in inhibiting tumor growth.45–48
Several other studies have generated transgenic mice that express H3.3-K27M in different embryonic or neonatal cell populations, in conjunction with p53/Atrx loss and/or PDGFRα activation, leading to the development of DMG-like tumors.49–51 Pathania et al. found that expression of H3.3-K27M under the H3f3a locus in embryonic stem cells is lethal and zygotes did not grow past the four-cell stage.51 By contrast, expression of H3.3-K27M with combined loss of p53 in neural progenitor cells around E12.5-E13.5 led to the formation of DMG-like tumors after 6–8 months. Tumor growth could be further accelerated by co-expression of wild type Pdgfra and/or loss of Atrx. Similarly, Pajovic et al. found that germline expression of H3.3-K27M under the Fabp7 promoter combined with p53 loss resulted in the formation DMG-like tumors as well as other cancers in a subset of mice.49 The same study also found that H3.3-K27M expression leads to the activation of the RAS/MYC axis in both human and mouse DMGs that can be pharmacologically exploited by either directly targeting MYC or by inhibiting the MAPK or PI3K/AKT arms of the RAS cascade.
Atypical Teratoid Rhabdoid Tumors (ATRTs)
Complete loss of Smarcb1 expression in either all somatic cells or only in Nestin-expressing cells is embryonically lethal in mice.52,53 Similarly, ubiquitous loss of Smarcb1 expression in neonates is also lethal and does not lead to tumor formation.53 By contrast, reduced deletion of Smarcb1 – induced by the injection of Smarcb1flox/flox-Rosa26-CreERT2 mice with a low dose of Tamoxifen – between E6-E18 resulted in the formation of ATRT-like tumors at an average latency of 90 days.53 Similarly, combined loss of Smarcb1 and p53 in GFAP-expressing cells also resulted in the formation of ATRT-like tumors starting at around 1 month of age.52 These tumors stained negative for BAF47 and NG2, markers that are typically also absent in human ATRT.
Before our current understanding of the genetic and epigenetic alterations that drive these rare CNS cancers, cross-species genomic comparative analysis of murine models to genomic subgroups of human tumors was used to model disease. Particularly in tumors with high mutational burden and unknown cellular origin, cross-species genomics is a tool that can provide insight into the mechanisms of oncogenesis. To rigorously compare transcriptome information from two different biological conditions (including different species), the agreement of differential expression (AGDEX) was developed. AGDEX can be used to: (1) identify individual genes and gene-sets that are differentially expressed in each experiment; (2) meta-analytically integrate results across experiments to identify differentially expressed genes and gene-sets; and (3) characterize and determine the statistical significance of similarities of differential expression profiles across the two experiments for the entire transcriptome and for specific gene-sets.54 Accordingly, Johnson and colleagues matched the transcriptomes of human ependymoma tumors with mouse neural stem cells.55 They revealed the amplification of EPHB2 and deletion of INK4A/ARF in human tumors matched only that of embryonic cerebral Ink4a/Arf-/- neural stem cells and consequently generated the first high-fidelity mouse model of ST-EPN driven by Ephb2 signaling.55 AGDEX can also be used to confirm that a cancer mouse model recapitulates the same gene signatures as human tumors, as described in the mouse model of ATRT generated by Han et al.53 This approach is notably applicable and promising for other rare CNS cancers where the oncogenic drivers remain unknown.
Summary on Model Systems
Each preclinical model has its advantages and disadvantages as outlined, to contribute to our understanding of the basic biology of tumors and predict clinical outcomes of drug response, identify biomarkers and study therapeutic resistance. In vitro and ex vivo models are ideal for high-throughput screening to identify therapeutics that can then be validated in vivo. Combining multiple preclinical models can give us the greatest confidence to efficiently translate therapeutics from the lab to the clinic.
Utilizing Preclinical Models to Identify Molecular Drivers and Therapeutic Options
In Silico Modeling
An inexpensive and rapid initial approach to predict drug response is through in silico tumor modeling. Computational modeling of drug sensitivity using available data sets can be used alongside experimental approaches to optimize the effort and costs associated with high-throughput screens. In silico modeling relies on inhibitor response data collected from in vitro and in vivo studies. Thus, controlled and well-executed preclinical studies are integral to generating accurate in silico models. This model is limited by predicting only cell-intrinsic effects; future work integrating toxicities on the tumor microenvironment will improve the accuracy of these models. Donson et al. combined in silico prediction of drug sensitivity alongside in vitro high-throughput screening of FDA-approved oncology drugs for rapid clinical application. This study used Posterior Fossa ependymoma cell lines established from patients with metastatic intracranial recurrence characterized by a high-risk chromosomal 1q gain and identified 3 drug classes (fluorinated pyrimidines, retinoids, and a subset of RTK inhibitors) effective at inhibiting PF-EPN cell proliferation.56
Direct Targeting of Oncogenic Drivers
Using preclinical models, oncogenic drivers have recently been identified and validated in some rare CNS cancers. For example, genetic profiling identified YAP1 fusion-driven ependymomas and recent studies have shown that overexpression of YAP1-MAMLD1 or YAP1-FAM118B is sufficient to generate ependymoma in mice.5,7,42 These YAP1 gene fusions retain the TEAD-binding domain and the WW domains, but are truncated before Ser397.5,7,42,43 Serine to alanine mutation of Ser94 of YAP1-MAMLD1 results in decreased YAP activity and a significantly decreased oncogenic potential when expressed in mice.7,42 Additionally, pharmacological inhibition of the YAP1-TEAD interaction by Verteporfin was sufficient to inhibit the growth of YAP1-FAM118B-driven murine tumor cells in vitro,7 indicating that the interaction with TEAD transcription factors is essential for the oncogenic functions of YAP1 fusion proteins. Thus, targeting these fusion proteins could result in inhibiting YAP1 fusion-driven ependymoma. In ZFTA-RELA fusion-positive supratentorial ependymoma, the fusion product contains the p65 subunit of NFκB, which is normally inhibited by IκB and degraded by the proteasome. Broad-spectrum proteasome inhibitors can thus be utilized to block NFκB signaling caused by the RELA fusion protein. Marizomib, a second-generation irreversible proteasome inhibitor that penetrates the blood–brain-barrier, has undergone preliminary testing in clinical trial to treat ependymoma in adults (NCT03727841).57 Another approach to directly targeting a known oncogenic protein is through PRoteolysis TArgeting Chimeras (PROTACs), which binds a target protein and triggers ubiquitin-mediated protein degradation. Many pharmaceutical and biotechnology companies are employing this technology to degrade “undruggable” protein targets.58
Screening Approaches
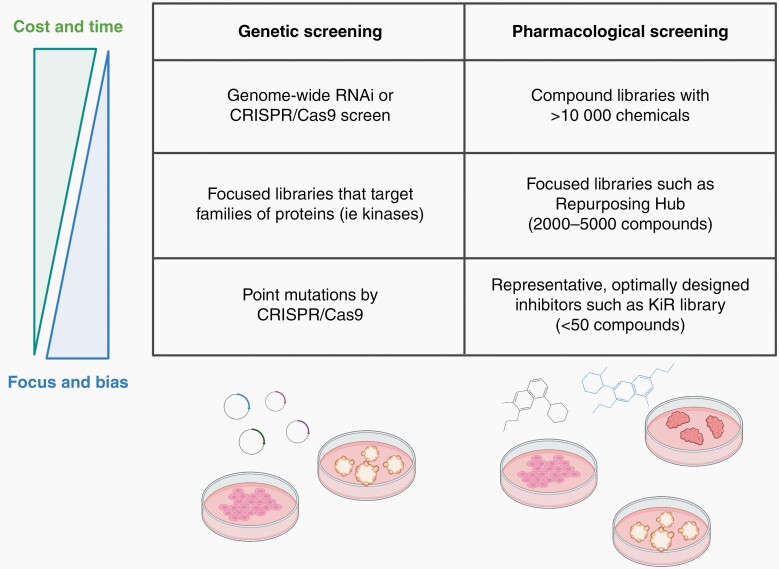
Oftentimes, directly inhibiting the oncogenic driver of tumors has proven difficult or impossible, particularly in cases that result in high heterogeneity such as mutations in epigenetic regulation in DMG. Additionally, loss of tumor suppressor genes such as SMARCB1 loss in ATRT is critical for cancer progression and cannot be directly targeted. As previously mentioned, cell lines derived from human CNS tumors are easy and cost-effective to use for high-throughput screening. We can use unbiased genetic and/or pharmacological screening methods to identify proliferation, survival, and resistance pathways that tumor cells rely on and follow up with these studies in other preclinical models (Figure 3).
Fig. 3.
High-throughput genetic and pharmacological screening approaches to identify therapeutic targets. Broad genome-wide RNA interference (RNAi) and Clustered Regularly Interspaced Short Palindormic Repeats (CRISPR)/Cas9 screens as well as large compound libraries provide an unbiased approach but have high costs and efforts. Use of smaller libraries (both genetic and pharmacological) are more focused and less cost- and labor-intensive. Created with BioRender.com.
Genetic Screening
RNA interference (RNAi) high-throughput screening approaches use genome-wide siRNA or shRNA libraries to systematically target individual genes, combined with phenotypic reporting, for the fundamental discovery of key molecular targets. These screens have been integral to our understanding of the underpinning biology of cancer cell growth. The high reproducibility and large-scale ability of this technology have led to unbiased identification, validation, and characterization of target genes and development of new lead compounds. Schramm et al. recently performed a large-scale genetic screen on patient-derived DMG cell lines with the DECIPHER pooled shRNA library Module 1, which targets 5,042 signaling pathway genes with multiple shRNAs per gene.59 Fibroblast growth factor receptor (FGFR) signaling and serine/threonine protein phosphatase 2A (PP2A) were identified as top hits involved in DMG proliferation in the screen and validated by genetic approaches and pharmacological inhibition in vitro.59 Other groups have taken focused RNAi screening approaches to identify novel targets for DMG growth, including using a kinome-wide shRNA screen in DMG stem cell cultures,60 an epigenomic/chromatin-associated shRNA screen in patient-derived DMG cultured cells,61 and a RAS signaling pathway-targeted siRNA screen in both DMG stem cell and patient-derived cultures.62 The latter two studies validated their findings in vivo using orthotopic xenograft models.61,62 This technology, however, has been limited to assessing the molecular targets intrinsic to tumor cells and does not account for the diverse and complex extrinsic interactions of cancer cells with the tumor microenvironment. Additionally, there are technical challenges to RNAi screens, including the transfection process, resulting in incomplete penetrance and potentially only partial knockdown. The transfection process can also cause stress and toxicity, resulting in changes to cell phenotype.
The advent of CRISPR/Cas9 technology has allowed for more precise and reliable genome editing capabilities. Using well-curated genome-scale sgRNA CRISPR libraries, many studies have identified genes involved in tumor growth, drug sensitivity or resistance, and immune response.63 Specific sgRNA libraries targeting families of proteins such as kinases or proteins involved in epigenetic regulation have also been utilized. Additionally, CRISPR screens are more versatile than RNAi approaches in that this system can be used to introduce point mutations in genes and activate gene transcription in addition to a gene interference approach.63 Merck and colleagues performed a genome-wide CRISPR/Cas9 knock-out screen in 6 human ATRT cell lines and identified 671 context-specific essential genes.64 They used this data to rationally design a library of small molecule inhibitors and assessed cell growth in these cells, identifying CDK4/6 as a target to inhibit ATRT tumor growth.64 Although much improved from RNAi strategies, the sgRNAs used in CRISPR screens may still result in off-target effects. Additionally, the results of these genomic screens may identify candidates that lack specific inhibitor compounds or that are difficult to target therapeutically.
Pharmacological Screening
Pharmacological phenotypic-based screening utilizes well-characterized and already available small molecule compounds and can be applied for novel drug discovery and repurposing as well as for drug combination studies. The EphB2-driven ST-EPN mouse model developed by the Gilbertson group through cross-species genomics was used to screen 5,303 unique compounds, including 275 FDA-approved compounds.65 This study also utilized a kinome-wide binding assay to elucidate further signaling pathways involved in ependymoma growth and validate their findings in a syngeneic orthotopic model.65 IGF signaling and centrosome cycle pathways were identified as regulators of ependymoma, and fluorinated pyrimidines, including 5-fluorouracil (5-FU), found to be an ependymoma-selective compound; this finding resulted in a clinical trial for bolus administration of 5-FU in children and young adults with recurrent EPN (NCT01498783).66
The process of drug discovery, optimization, preclinical development, and clinical trials is both lengthy and costly, and these drugs often fail in clinical trials. The Broad Institute’s Cancer Program, Center for the Development of Therapeutics, and Connectivity Map project have created the Drug Repurposing Hub, an open-access repository of annotated FDA-approved and clinical trial drugs and pre-clinical tool compounds. This resource is available to researchers to accelerate drug development for diseases and is one example of an optimized library of small molecules that can quickly be repurposed. To discover therapeutics that could inhibit the growth of ATRT, which lacks targetable genomic alterations, Singh et al. employed a panel of 129 small molecule inhibitors from multiple pharmaceutical pipeline libraries against three ATRT human cell lines.67 Through target modulation, antibody array analysis, drug combination and in vivo studies, lapatinib was validated as an effective agent in treating ATRT.67 Use of FDA-approved therapeutics bypasses the need to evaluate the safety profile of these therapeutics in most cases, allowing for accelerated and immediate prioritization in clinical trials and faster translational application.
The Gujral lab has developed an approach to combine the inexpensive and predictive potential of computational methods with the validation and integration of in vitro data. Protein kinases are dysregulated in many types of cancer and as such, 46 of the 52 currently FDA-approved kinase inhibitors are used as cancer therapeutics.68 Small-molecule kinase inhibitors often target multiple targets at variable efficacies due to the conserved structural similarities of this family of proteins, a property called polypharmacology. Many groups have extensively profiled the activity of these kinase inhibitors, building our understanding of the mechanistic targets of these inhibitors.69–73 We can take advantage of the polypharmacology conveyed from these publicly available datasets combined with linear regression-based or machine learning approaches to build models for predicting the efficacy of many inhibitors on a given phenotype.74–76 Gujral et al. first utilized this systems polypharmacology approach using elastic net regularization combined with kinase mRNA expression profiling to identify kinases critical to cell migration, called Kinome Regularization (KiR).75 In this approach, six different cancer cell lines were treated with 8 doses each of 32 computationally-chosen kinase inhibitors, which have broad specificity, and cell migration was assessed by scratch wound assay. A response to this representative subset of the drugs is combined with the previously determined quantitative in vitro profiling data to generate regularized regression models that can predict the underlying kinases and response to untested kinase inhibitors. This model relies on linear combinations of the contributions of kinases on cellular behavior.75 However, kinase biology and cell signaling is highly complex and dynamic; therefore, the Kinase Inhibitor prediction using Deep Neural Network (KiDNN) approach was developed to reflect this complexity. KiDNN takes advantage of the DNN framework to produce non-linear, multilayer feed-forward networks that closely mimic the complex and dynamic nature of the cancer signaling pathways. Vijay and Gujral applied KiDNN to predict the effect of ~200 kinase inhibitors on migration of breast and liver cancer cells.76 They experimentally tested a subset of the inhibitors and showed that KiDNN predictions outperform predictions from the linear KiR model. Results from these focused-screening approaches (KiR or KiDNN) can then be validated in vitro and in vivo,77–79 representing a method that allows for the rational design of cheaper and more effective screening compared with unbiased testing of large compound libraries.
Combined Genetic and Pharmacological Screening
To exploit the advantages of both genome editing and pharmacological screening, Oberlick and colleagues performed a high-throughput small molecule inhibitor screen complemented by a genome-scale CRISPR/Cas9 knock-out screen in pediatric ATRT cell lines and they revealed several receptor tyrosine kinases as potential therapeutic targets.80 To interrogate the epigenetic regulators of DMG, a recent study used small molecules that target epigenetic regulators alongside a chromatin-focused CRISPR/Cas9 screen of 1,354 candidate genes and validated the use of Corin, an inhibitor of HDACs and LSD1, in blocking DMG cell growth in vitro and in in vivo xenografts.81 This rapid approach to identifying novel therapeutics can provide a rationale for testing these compounds’ efficacy in other preclinical models, such as GEMM and PDX in vivo models.
Discussion
Recent advances in sequencing and molecular biology technology have allowed us to identify and validate the tumorigenic events that occur in rare CNS cancers. Tumors driven by some specific and sufficient mutational or chromosomal events provide potential therapeutic targets. For example, in Chronic Myeloid Leukemia (CML), discovery of the BCR-ABL oncoprotein with high tyrosine kinase activity led to the development of Imatinib, a tyrosine kinase inhibitor (TKI), as an extremely successful therapeutic followed by use of second- and third-generation TKIs to treat resistant CML.82 However, rare tumors caused by these genetic events are difficult to study due to limited tumor tissue samples; furthermore, the oncogenic driver may not be directly targetable. An additional obstacle in developing novel therapeutics for rare cancers are that large, randomized clinical trials are impossible due to the low number of patients.
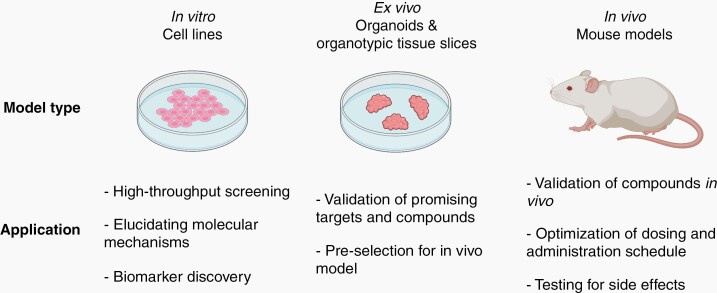
To overcome these difficulties, accurate preclinical models are needed to understand the biology behind these rare CNS cancers. Although conventional in vitro studies have helped in validating oncogenic events, more complex and heterogeneous models such as organoid and organotypic tissue slices are better representations of patient tumor biology. In vivo modeling systems that recreate tumor formation from the same sufficient mutations or fusions in genetically relatively quiet backgrounds provide both unlimited tumor tissue and large numbers of tumor bearing experimental animals for pre- and co-clinical trials. Cell lines containing representative molecular signatures can be applied in the initial stages of target discovery by screening approaches as well as investigating the molecular mechanisms of disease progression and murine models can be used for validation while ex vivo models provide a bridge between cell culture and mouse studies (Figure 4). Such systems can help us understand the biology of therapeutic response as well as mechanisms of resistance and recurrence. Using a combination of in vitro, ex vivo and in vivo models can minimize cost and effort while recapitulating tumor biology for targeted therapy development.
Fig. 4.
Summary of application of preclinical models (in vitro, ex vivo, and in vivo) at different stages of drug development. Highlights the use of in vitro and ex vivo models in the early drug discovery and development phase while ex vivo models are critical for validation of therapeutics and informing clinical trial design. Created with BioRender.com.
Screening approaches highlighted in this review can then be used in optimized in vitro and ex vivo preclinical models followed by validation in representative in vivo models. In so doing, targets for synthetic lethality can be predicted, identified, and experimentally addressed. Utilizing FDA-approved inhibitors and clinical-grade molecules can accelerate drug development and repurposing these safe existing drugs allows for immediate clinical application, an important aspect for treating patients with these rare CNS cancers that have a very poor prognosis. Moreover, these modeling and targeted therapy discovery approaches can help identify individual and combined therapeutic strategies for other types of rare cancers, not only rare CNS cancers.
Acknowledgments
This publication reflects the views of the individual authors and should not be construed to represent official views or policies of the US Food and Drug Administration or the National Cancer Institute (NCI). The views expressed in this article are the personal views of the author(s) and may not be understood or quoted as being made on behalf of or reflecting the position of the European Medicines Agency or one of its committees or working parties. Medical writing support was provided by Kendall Morgan, and funded by Neuro-Oncology Branch, Center for Cancer Research, NCI, National Institutes of Health (US Government contract).
Funding
The NCI Comprehensive Oncology Network for Evaluating Rare CNS Tumors (NCI-CONNECT) is a program within the Rare Tumor Patient Engagement Network (RTPEN), an initiative supported by Cancer Moonshot℠ funds and managed at the National Institutes of Health, National Cancer Institute, Center for Cancer Research, Neuro-Oncology Branch. This research was funded in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748.
Supplement sponsorship.
This supplement was sponsored by the National Institutes of Health.
Conflict of interest statement.
The authors declare no conflict of interests.
References
- 1. Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro Oncol. 2020; 22(12 Suppl 2):iv1–iv96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gatta G, Botta L, Rossi S, et al. Childhood cancer survival in Europe 1999–2007: results of EUROCARE-5--a population-based study. Lancet Oncol. 2014;15(1):35–47. [DOI] [PubMed] [Google Scholar]
- 3. Marinoff AE, Ma C, Guo D, et al. Rethinking childhood ependymoma: a retrospective, multi-center analysis reveals poor long-term overall survival. J Neurooncol. 2017;135(1):201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vera-Bolanos E, Aldape K, Yuan Y, et al. ; CERN Foundation . Clinical course and progression-free survival of adult intracranial and spinal ependymoma patients. Neuro Oncol. 2015;17(3):440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ozawa T, Kaneko S, Szulzewsky F, et al. C11orf95-RELA fusion drives aberrant gene expression through the unique epigenetic regulation for ependymoma formation. Acta Neuropathol Commun. 2021;9(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szulzewsky F, Arora S, Hoellerbauer P, et al. Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes Dev. 2020;34(15-16):1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mack SC, Witt H, Piro RM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20(2):143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Panwalkar P, Clark J, Ramaswamy V, et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017;134(5):705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen LH, Pan C, Diplas BH, et al. The integrated genomic and epigenomic landscape of brainstem glioma. Nat Commun. 2020;11(1):3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schulte JD, Buerki RA, Lapointe S, et al. Clinical, radiologic, and genetic characteristics of histone H3 K27M-mutant diffuse midline gliomas in adults. Neurooncol Adv. 2020;2(1):vdaa142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lowe BR, Maxham LA, Hamey JJ, Wilkins MR, Partridge JF. Histone H3 mutations: an updated view of their role in chromatin deregulation and cancer. Cancers (Basel). 2019;11(5):1– 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Korshunov A, Jakobiec FA, Eberhart CG, et al. Comparative integrated molecular analysis of intraocular medulloepitheliomas and central nervous system embryonal tumors with multilayered rosettes confirms that they are distinct nosologic entities. Neuropathology. 2015;35(6):538–544. [DOI] [PubMed] [Google Scholar]
- 15. Chan V, Marro A, Findlay JM, Schmitt LM, Das S. A systematic review of atypical teratoid rhabdoid tumor in adults. Front Oncol. 2018;8:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nesvick CL, Lafay-Cousin L, Raghunathan A, Bouffet E, Huang AA, Daniels DJ. Atypical teratoid rhabdoid tumor: molecular insights and translation to novel therapeutics. J Neurooncol. 2020;150(1):47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 18. Alver BH, Kim KH, Lu P, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erkek S, Johann PD, Finetti MA, et al. Comprehensive analysis of chromatin states in atypical teratoid/rhabdoid tumor identifies diverging roles for SWI/SNF and polycomb in gene regulation. Cancer Cell. 2019;35(1):95–110 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ho B, Johann PD, Grabovska Y, et al. Molecular subgrouping of atypical teratoid/rhabdoid tumors-a reinvestigation and current consensus. Neuro Oncol. 2020;22(5):613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jaramillo S, Grosshans DR, Philip N, et al. Radiation for ETMR: literature review and case series of patients treated with proton therapy. Clin Transl Radiat Oncol. 2019;15:31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Witt Hamer PC, Van Tilborg AA, Eijk PP, et al. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene. 2008;27(14):2091–2096. [DOI] [PubMed] [Google Scholar]
- 24. Haag D, Mack N, Benites Goncalves da Silva P, et al. H3.3-K27M drives neural stem cell-specific gliomagenesis in a human iPSC-derived model. Cancer Cell. 2021;39(3):407–422 e413. [DOI] [PubMed] [Google Scholar]
- 25. Kinker GS, Greenwald AC, Tal R, et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat Genet. 2020;52(11):1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18(7):407–418. [DOI] [PubMed] [Google Scholar]
- 27. Parisian AD, Koga T, Miki S, et al. SMARCB1 loss interacts with neuronal differentiation state to block maturation and impact cell stability. Genes Dev. 2020;34(19-20):1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Humpel C. Organotypic brain slice cultures: a review. Neuroscience. 2015;305:86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kenerson HL, Sullivan KM, Seo YD, et al. Tumor slice culture as a biologic surrogate of human cancer. Ann Transl Med. 2020;8(4):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nishida-Aoki N, Gujral TS. Emerging approaches to study cell-cell interactions in tumor microenvironment. Oncotarget. 2019;10(7):785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nishida-Aoki N, Bondesson AJ, Gujral TS. Measuring real-time drug response in organotypic tumor tissue slices. J Vis Exp. 2020;( 159): e61036. [DOI] [PubMed] [Google Scholar]
- 32. Sivakumar R, Chan M, Shin JS, et al. Organotypic tumor slice cultures provide a versatile platform for immuno-oncology and drug discovery. Oncoimmunology. 2019;8(12):e1670019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horowitz LF, Rodriguez AD, Au-Yeung A, et al. Microdissected “cuboids” for microfluidic drug testing of intact tissues. Lab Chip. 2021;21(1):122–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shultz LD, Keck J, Burzenski L, et al. Humanized mouse models of immunological diseases and precision medicine. Mamm Genome. 2019;30(5-6):123–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346(6216):1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Funato K, Smith RC, Saito Y, Tabar V. Dissecting the impact of regional identity and the oncogenic role of human-specific NOTCH2NL in an hESC model of H3.3G34R-mutant glioma. Cell Stem Cell. 2021;28(5):894–905 e897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen CCL, Deshmukh S, Jessa S, et al. Histone H3.3G34-mutant interneuron progenitors Co-opt PDGFRA for gliomagenesis. Cell. 2020;183(6):1617–1633 e1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506(7489):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith KS, Xu K, Mercer KS, et al. Patient-derived orthotopic xenografts of pediatric brain tumors: a St. Jude resource. Acta Neuropathol. 2020;140(2):209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ozawa T, Arora S, Szulzewsky F, et al. A De Novo Mouse Model of C11orf95-RELA fusion-driven ependymoma identifies driver functions in addition to NF-κB. Cell Rep. 2018;23(13):3787–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takadera M, Satomi K, Szulzewsky F, et al. Phenotypic characterization with somatic genome editing and gene transfer reveals the diverse oncogenicity of ependymoma fusion genes. Acta Neuropathol Commun. 2020;8(1):203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pajtler KW, Wei Y, Okonechnikov K, et al. YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat Commun. 2019;10(1):3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szulzewsky F, Holland EC, Vasioukhin V. YAP1 and its fusion proteins in cancer initiation, progression and therapeutic resistance. Dev Biol. 2021;475: 205– 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eder N, Roncaroli F, Domart MC, et al. YAP1/TAZ drives ependymoma-like tumour formation in mice. Nat Commun. 2020;11(1):2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lewis PW, Müller MM, Koletsky MS, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoeman CM, Cordero FJ, Hu G, et al. ACVR1 R206H cooperates with H3.1K27M in promoting diffuse intrinsic pontine glioma pathogenesis. Nat Commun. 2019;10(1):1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martínez-Vélez N, Garcia-Moure M, Marigil M, et al. The oncolytic virus Delta-24-RGD elicits an antitumor effect in pediatric glioma and DIPG mouse models. Nat Commun. 2019;10(1):2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mittapalli RK, Chung AH, Parrish KE, et al. ABCG2 and ABCB1 limit the efficacy of dasatinib in a PDGF-B-driven brainstem glioma model. Mol Cancer Ther. 2016;15(5):819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pajovic S, Siddaway R, Bridge T, et al. Epigenetic activation of a RAS/MYC axis in H3.3K27M-driven cancer. Nat Commun. 2020;11(1):6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Larson JD, Kasper LH, Paugh BS, et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019;35(1):140–155 e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pathania M, De Jay N, Maestro N, et al. H3.3(K27M) Cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas. Cancer Cell. 2017;32(5):684–700 e689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ng JM, Martinez D, Marsh ED, et al. Generation of a mouse model of atypical teratoid/rhabdoid tumor of the central nervous system through combined deletion of Snf5 and p53. Cancer Res. 2015;75(21):4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Han ZY, Richer W, Fréneaux P, et al. The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nat Commun. 2016;7:10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pounds S, Gao CL, Johnson RA, et al. A procedure to statistically evaluate agreement of differential expression for cross-species genomics. Bioinformatics. 2011;27(15):2098–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466(7306):632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Donson AM, Amani V, Warner EA, et al. Identification of FDA-approved oncology drugs with selective potency in high-risk childhood ependymoma. Mol Cancer Ther. 2018;17(9):1984–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Di K, Lloyd GK, Abraham V, et al. Marizomib activity as a single agent in malignant gliomas: ability to cross the blood-brain barrier. Neuro Oncol. 2016;18(6):840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gao H, Sun X, Rao Y. PROTAC technology: opportunities and challenges. ACS Med Chem Lett. 2020;11(3):237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schramm K, Iskar M, Statz B, et al. DECIPHER pooled shRNA library screen identifies PP2A and FGFR signaling as potential therapeutic targets for diffuse intrinsic pontine gliomas. Neuro Oncol. 2019;21(7):867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Silva-Evangelista C, Barret E, Ménez V, et al. A kinome-wide shRNA screen uncovers vaccinia-related kinase 3 (VRK3) as an essential gene for diffuse intrinsic pontine glioma survival. Oncogene. 2019;38(38):6479–6490. [DOI] [PubMed] [Google Scholar]
- 61. Dahl NA, Danis E, Balakrishnan I, et al. Super elongation complex as a targetable dependency in diffuse midline glioma. Cell Rep. 2020;31(1):107485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koncar RF, Dey BR, Stanton AJ, et al. Identification of novel RAS signaling therapeutic vulnerabilities in diffuse intrinsic pontine gliomas. Cancer Res. 2019;79(16):4026–4041. [DOI] [PubMed] [Google Scholar]
- 63. Yin H, Xue W, Anderson DG. CRISPR-Cas: a tool for cancer research and therapeutics. Nat Rev Clin Oncol. 2019;16(5):281–295. [DOI] [PubMed] [Google Scholar]
- 64. Merk DJ, Hirsch S, Tsiami F, et al. Genome-wide CRISPR and small-molecule screens uncover targetable dependencies in ATRT. bioRxiv. 2020. doi: 10.1101/2020.12.09.417378. [DOI] [Google Scholar]
- 65. Atkinson JM, Shelat AA, Carcaboso AM, et al. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell. 2011;20(3):384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wright KD, Daryani VM, Turner DC, et al. Phase I study of 5-fluorouracil in children and young adults with recurrent ependymoma. Neuro Oncol. 2015;17(12):1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Singh A, Lun X, Jayanthan A, et al. Profiling pathway-specific novel therapeutics in preclinical assessment for central nervous system atypical teratoid rhabdoid tumors (CNS ATRT): favorable activity of targeting EGFR- ErbB2 signaling with lapatinib. Mol Oncol. 2013;7(3):497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roskoski R Jr. Properties of FDA-approved small molecule protein kinase inhibitors: a 2020 update. Pharmacol Res. 2020;152:104609. [DOI] [PubMed] [Google Scholar]
- 69. Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Davis MI, Hunt JP, Herrgard S, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1046–1051. [DOI] [PubMed] [Google Scholar]
- 71. Duong-Ly KC, Devarajan K, Liang S, et al. Kinase Inhibitor Profiling Reveals Unexpected Opportunities to Inhibit Disease-Associated Mutant Kinases. Cell Rep. 2016;14(4):772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klaeger S, Heinzlmeir S, Wilhelm M, et al. The target landscape of clinical kinase drugs. Science. 2017; 358(6367): eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rata S, Gruver JS, Trikoz N, et al. An optimal set of inhibitors for reverse engineering via kinase regularization. bioRxiv. 2020. doi: 10.1101/2020.09.26.312348. [DOI] [Google Scholar]
- 74. Bello T, Gujral TS. KInhibition: a kinase inhibitor selection portal. iScience. 2018;8:49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gujral TS, Peshkin L, Kirschner MW. Exploiting polypharmacology for drug target deconvolution. Proc Natl Acad Sci U S A. 2014;111(13):5048–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vijay S, Gujral TS. Non-linear deep neural network for rapid and accurate prediction of phenotypic responses to kinase inhibitors. iScience. 2020;23(5):101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, MacBeath G. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell. 2014;159(4):844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Arang N, Kain HS, Glennon EK, et al. Identifying host regulators and inhibitors of liver stage malaria infection using kinase activity profiles. Nat Commun. 2017;8(1):1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Golkowski M, Lau HT, Chan M, et al. Pharmacoproteomics identifies kinase pathways that drive the epithelial-mesenchymal transition and drug resistance in hepatocellular carcinoma. Cell Syst. 2020;11(2):196–207 e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Oberlick EM, Rees MG, Seashore-Ludlow B, et al. Small-molecule and CRISPR screening converge to reveal receptor tyrosine kinase dependencies in pediatric rhabdoid tumors. Cell Rep. 2019;28(9):2331–2344 e2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Anastas JN, Zee BM, Kalin JH, et al. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG. Cancer Cell. 2019;36(5):528–544 e510. [DOI] [PubMed] [Google Scholar]
- 82. Sampaio MM, Santos MLC, Marques HS, et al. Chronic myeloid leukemia-from the Philadelphia chromosome to specific target drugs: a literature review. World J Clin Oncol. 2021;12(2):69–94. [DOI] [PMC free article] [PubMed] [Google Scholar]