Abstract
Evi9 is a common site of retroviral integration in BXH2 murine myeloid leukemias. Here we show that Evi9 encodes a novel zinc finger protein with three tissue-specific isoforms: Evi9a (773 amino acids [aa]) contains two C2H2-type zinc finger motifs, a proline-rich region, and an acidic domain; Evi9b (486 aa) lacks the first zinc finger motif and part of the proline-rich region; Evi9c (239 aa) lacks all but the first zinc finger motif. Proviral integration sites are located in the first intron of the gene and lead to increased gene expression. Evi9a and Evi9c, but not Evi9b, show transforming activity for NIH 3T3 cells, suggesting that Evi9 is a dominantly acting proto-oncogene. Immunolocalization studies show that Evi9c is restricted to the cytoplasm whereas Evi9a and Evi9b are located in the nucleus, where they form a speckled localization pattern identical to that observed for BCL6, a human B-cell proto-oncogene product. Coimmunoprecipitation and glutathione S-transferase pull-down experiments show that Evi9a and Evi9b, but not Evi9c, physically interact with BCL6, while deletion mutagenesis localized the interaction domains in or near the second zinc finger and POZ domains of Evi9 and BCL6, respectively. These results suggest that Evi9 is a leukemia disease gene that functions, in part, through its interaction with BCL6.
Retroviral integration in murine hematopoietic cells can lead to the generation of leukemias by enhancing expression of cellular proto-oncogenes or by disrupting expression of tumor suppressor genes. Retroviral proviruses in murine leukemias thus provide powerful genetic tags for identifying leukemia disease genes (15).
One mouse strain that develops a high incidence of retrovirally induced leukemia is BXH2. More than 95% of BXH2 mice die of retrovirally induced myeloid leukemia by 1 year of age (2). A number of disease genes have been identified in BXH2 leukemias by proviral tagging. They include a tumor suppressor gene, Nf1 (neurofibromatosis type 1); a gene with homology to the lymphoid-restricted type II membrane protein Jaw1 Mrvi1 (Mrv integration site 1); a gene encoding a hematopoietic cell growth and differentiation factor, Myb (myeloblastosis oncogene); three homeobox genes, Hoxa7 (homeobox A7), Hoxa9 (homeobox A9), and Meis1 (myeloid ecotropic viral integration site 1); and a gene with homology to the Usp8 (ubiquitin-specific protease 8) oncogene and to genes encoding various cell cycle regulatory proteins, Evi5 (ecotropic viral integration site 5) (5, 21, 32). At least three of these genes are proven or suspected human disease genes: NF1 and HOXA9 are causally associated with myeloid leukemia, and EVI5 is associated with stage 4S neuroblastoma (5, 30), validating the usefulness of this approach for identifying human disease genes.
Although proviral tagging has successfully identified a number of leukemia disease genes, it appears that there are many more to be identified. For example, whereas seven disease genes have been identified in BXH2 leukemias to date, as many as 65% of BXH2 leukemias do not have a virally induced mutation in one of these genes. The expectation is that continued proviral tagging in BXH2 leukemias will identify additional disease genes and, based on previous studies, that a number of these genes will represent human disease genes.
In previous studies, we identified a new common site of proviral integration site in BXH2 leukemias that we called ecotropic viral integration site 9 (Evi9) (26). Proviral integrations at Evi9 are rare and were found in only 2 of 205 (1%) BXH2 leukemias tested. Chromosome and physical mapping studies showed that Evi9 is located near the c-rel proto-oncogene on chromosome 11 (but did not encode c-Rel) and suggested that Evi9 may represent a new leukemia disease gene. Here we show that this is likely to be the case by demonstrating that Evi9 encodes a novel zinc finger protein which transforms NIH 3T3 cells in vitro and binds to another zinc finger protein, BCL6, which itself is a known human B-cell proto-oncogene product.
MATERIALS AND METHODS
BXH2 mice and leukemic cell line.
The BXH2 recombinant inbred strain was obtained from the Jackson Laboratory (Bar Harbor, Maine) and maintained at the NCI-Frederick Cancer Research and Development Center. BXH2 leukemic cell lines have been previously described (19).
Genomic and cDNA cloning.
Evi9 ecotropic proviral integration sites from BXH2 leukemia cell lines B162 and B139 locus were isolated according to previously described methods (26). A murine bacterial artificial chromosome (BAC) clone was purchased from Research Genetics after PCR-based screening for positive clones. Lambda phage clones derived from a 129/Sv mouse genomic library (Stratagene) were also isolated by hybridization. Exon trapping was performed using BAC clone B413N15 and the pSPL3 vector (Life Technology) according to the manufacturer's protocol. The trapped exons were used as a probe to screen an 11-day mouse cDNA library (Clontech). Positive clones were purified, subcloned into pBluescript, and sequenced.
RNA isolation, Northern blots, and reverse transcription-PCR (RT-PCR).
Poly(A)+ RNA was isolated from frozen leukemia cell suspensions using a FastTrack 2.0 kit (Invitrogen). Total RNA was extracted from mouse normal tissues or cultured cell suspensions by the RNAzol method (TelTest). Two micrograms of poly(A)+ RNA was fractionated by electrophoresis in 1.0% agarose gels containing formaldehyde and transferred to Hybond N+ membrane (Amersham). Mouse multiple-tissue Northern blots were purchased from Clontech. Membranes were hybridized and washed according to the method of Church and Gilbert (4). AA53, which is a 1.5-kb cDNA fragment which corresponds to nucleotides 838 to 2346 of Evi9, was used as a probe.
First-strand cDNA was synthesized from 1 μg of total RNA by using random hexamers and Superscript II reverse transcriptase (Life Technologies) in a total volume of 20 μl. The mixture was diluted to 50 μl, and 2 μl was subjected to PCR using a temperature cycling protocol of 94°C for 30 s, 60°C for 1 min, and 72°C for 4 min (35 cycles). PCR products were separated by electrophoresis through a 1.2% agarose gel, transferred to a Hybond N+ membrane, and hybridized with the AA53 probe. The PCR primers used were Evi9 RACE1 (5′-TGAACCGAGCCGTCGTCCGCACG-3′) and S2 (5′-TCCATCCGAAAACTGCCAC-3′).
Antibodies.
Rabbit polyclonal antisera were raised against the recombinant Evi9 proteins (Ab1 for amino acids [aa] 80 to 208 and Ab2 for aa 613 to 663). Anti-BCL6 antibodies were described previously (27). Anti-PML and anti-Sp100 antibodies were kind gifts from Thomas Sternsdorf. Anti-Myc epitope antibody 9E10 (Boehringer Mannheim) was also used.
Plasmid constructs.
Three cDNA isoforms of Evi9 were subcloned into the pCDNA3.1 expression plasmid (Invitrogen). Green fluorescent protein (GFP)-tagged constructs were generated by insertion of the isoforms in frame into the EGFPN1 vector (Clontech). Truncated proteins for glutathione S-transferase (GST) fusion constructs were generated by PCR of the full-length cDNA clone. The Myc-tagged BCL6 expression construct was a kind gift from Tohru Miki, and PML and Sp100 constructs were gifts of Thomas Sternsdorf. GST-BCL6 fusion constructs were described previously (25).
Transformation assays.
NIH 3T3 fibroblasts were seeded on six-well culture dishes at low cell density 18 h prior to transfection. pCDNA expression plasmids bearing each isoform of Evi9 were transfected into NIH 3T3 cells, using Lipofectamine and Opti-MEM (Life Technology). Transfectants were first selected with G418, and 2 × 104 cells of G418-resistant fibroblasts were suspended in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 0.3% agar in 3-cm-diameter dish. The cells were cultured at 37°C under 5% CO2, and colonies were counted after 3 weeks. The cells were plated in triplicate each for three independent experiments.
Transient transfection and immunofluorescence.
COS7 and HeLa cells were seeded on poly-l-lysine-coated culture slides (Falcon) 18 h prior to transfection. Transfection of expression plasmids was carried out using Lipofectamine and Opti-MEM. After overnight culture, cells were fixed in methanol for 30 min at −20°C, washed three times with phosphate-buffered saline, blocked with 10% normal goat serum, and incubated with primary antibodies. Positive signals were visualized with fluorescein isothiocyanate or rhodamine. Nuclear DNA was counterstained with Hoechst 33342. The cells were examined by confocal laser-scanning microscopy (Olympus) or Carl Zeiss Axiophoto II fluorescence microscopy.
Coimmunoprecipitation and in vitro binding experiments.
A total of 107 B162, Raji, or Jurkat cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, 1 mg of leupeptin per ml, 1 mg of aprotinin per ml). The cell suspensions were sonicated and clarified by centrifugation. Immunoprecipitation was carried out by using anti-Evi9 antibody Ab1 or preimmune serum and then by adding protein A-Sepharose (Pharmacia). Immunoprecipitates were Western blotted with the anti-BCL6 antibody by the enhanced chemiluminescence method (Amersham).
For in vitro binding experiments, bacterially expressed GST fusion proteins were purified according to methods previously described (33). In vitro-translated Evi9 and BCL6 proteins were prepared by using pSP64 plasmids bearing cDNAs, the TnT coupled reticulocyte lysate system (Promega), and [35S]methionine. Similar quantities of GST or GST fusion proteins immobilized on glutathione-Sepharose beads were incubated with in vitro-translated proteins in buffer A (50 mM Tris [pH 7.5], 0.5 mM EDTA, 0.4 M NaCl, 5 mM MgCl2, 5% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 0.1 mM dithiothreitol) for 1 h at 4°C with gentle agitation. Bound proteins were then washed four times in 1 ml of buffer A, eluted in SDS sample buffer, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE).
Nucleotide sequence accession numbers.
The sequence data reported have been submitted to the GenBank database under accession no. AF051525, AF169036, and AF169037.
RESULTS
Evi9 transcript identification.
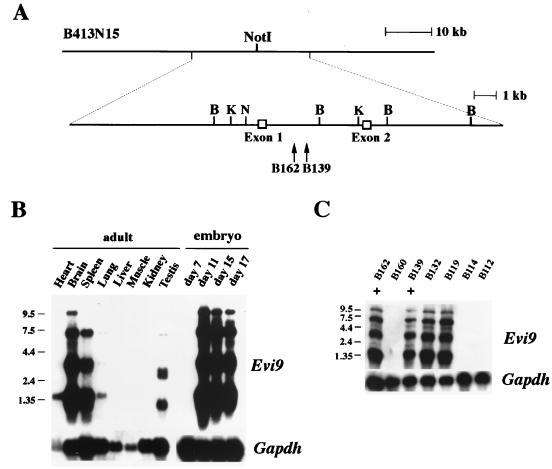
Genomic restriction analysis of DNA from two BXH2 myeloid leukemias with Evi9 proviral integrations (B139 and B162) showed that the proviral integrations are located about 1 kb apart from each other (Fig. 1A). A 90-kb BAC clone (B413N15) which covers the integration site was then isolated and exon trapped. Two exons of 171 bp (exon 1) and 198 bp (exon 2) were identified (Fig. 1A). Sequence analysis showed that the proviral integrations are located within the first intron of the gene in the reverse transcriptional orientation (Fig. 1A), a location and orientation frequently found for retroviral integrations that activate gene expression through an enhancer mechanism (15).
FIG. 1.
Identification of Evi9 transcripts. (A) Partial genomic structure of the 5′ region of Evi9. Exons 1 and 2 were trapped from the 90-kb BAC clone B413N15. B; BamHI, K; KpnI, N; NotI. Locations of the proviral integration sites in BXH2 leukemias B139 and B162 are shown by arrows. (B and C) Evi9 expression in normal adult and embryonic mouse tissue (B) and BXH2 leukemias (+ indicates leukemias with proviral integration at Evi9) (C). The blots were probed with Gapdh to normalize for RNA loading. Sizes of molecular weight markers in kilobases are shown on the left of each panel.
Hybridization of adult and embryonic tissue Northern blots using the two exons as a probe identified four major Evi9 transcripts 9.5, 7.4, 3.2, and 1.5 kb in size. In the adult, the highest Evi9 expression was found in the brain and spleen with lower levels in the testis and lung (Fig. 1B). In the embryo, little expression of Evi9 was detected at day 7. However, by day 11 high levels of Evi9 expression were observed, which continued until day 17, the last embryonic stage examined.
Northern blots containing RNA from a number of BXH2 myeloid leukemia cell lines were also examined for Evi9 expression. In the two cell lines containing viral integrations at Evi9, high expression of Evi9 was observed (Fig. 1C). In contrast, cell lines like B160, B114, and B112, which lack proviral integrations at Evi9, do not express Evi9 at detectable levels. These results are consistent with the hypothesis that proviral integrations at Evi9 upregulate the expression of Evi9. In a few cell lines like B132 and B119, which do not have proviral integrations at Evi9, high expression of Evi9 was also observed. While the significance of this result is unclear, it is possible that these cells lines contain proviral integrations at Evi9 that were undetected because they lie outside the region screened for proviral integrations or, alternatively, there are other mechanisms for upregulating Evi9 expression in leukemic cells besides proviral integration.
Evi9 encodes a novel Krüppel-like zinc finger protein.
Evi9 cDNAs were identified by screening an 11-day mouse embryonic cDNA library, using exons 1 and 2 as the probe. Sequence analysis of the positive clones yielded a 3,002-bp continuous sequence containing a 2,319-bp open reading frame, 273 bp of 5′ untranslated region (UTR) sequence, and 410 bp of 3′ UTR sequence (Fig. 2). The deduced amino acid sequence contains both single and double Cys2-His2-type zinc finger motifs, indicating that Evi9 encodes a member of the Krüppel-like family of zinc finger proteins. Outside of these zinc finger domains, Evi9 has no homology to anything in current databases, indicating that it encodes a novel protein. Known structures such as a KRAB box, POZ domain, or FAX domain, sometimes seen in Krüppel-type zinc finger proteins (1, 10, 17, 29, 41), are missing. The only other notable features of the Evi9, beside the zinc fingers, are a 186-aa proline-rich region located between the two zinc finger motifs and an acidic domain located downstream of the second zinc finger motif. The presence of an acidic domain suggests that Evi9 may act as a transcriptional activator in some cell contexts (24). As shown in Fig. 1B and C, Evi9 has large, alternatively spliced transcripts. Sequence analysis of multiple clones revealed that there are additional 3′ untranslated sequences which contribute larger transcripts (data not shown).
FIG. 2.
Nucleotide and deduced amino acid sequence of Evi9 (GenBank accession no. AF051525). Zinc finger motifs are in bold. The first and second exons shown in Fig. 1A are boxed. A short acidic-rich region is doubly underlined. Two potential poly(A) signal sequences in the 3′ UTR are singly underlined.
Multiple Evi9 isoforms.
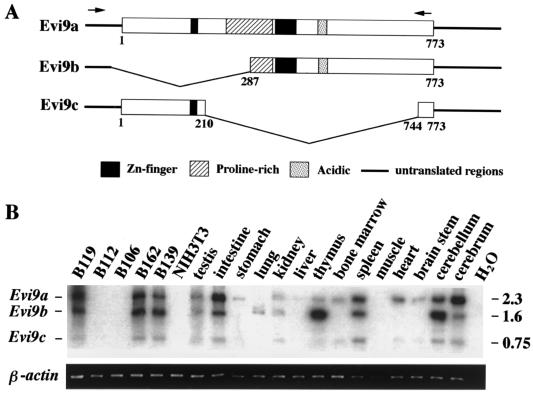
Evi9 cDNA sequencing suggested that Evi9 may be alternatively spliced. RT-PCR experiments confirmed this prediction and identified two alternatively spliced Evi9 isoforms designated Evi9b and Evi9c (Fig. 3A). Evi9b is predicted to encode a 487-aa protein from an alternative translation initiation site. This protein is missing the first zinc finger motif and part of the proline-rich region of Evi9a (the full-length protein). Evi9c is predicted to encode a 239-aa protein that is missing all but the first zinc finger motif of Evi9a.
FIG. 3.
Three Evi9 isoforms are expressed in a tissue-specific fashion. (A) Protein structures of three Evi9 isoforms. Arrows indicate the locations of PCR primers used for RT-PCR. (B) Expression patterns of three Evi9 isoforms (upper panel) in various tissues and BXH2 leukemia cell lines were analyzed by RT-PCR. RT-PCR products were gel fractionated, transferred to a nylon membrane, and hybridized with probe AA53. Sizes of PCR products in kilobases are shown on the right. β-Actin (lower panel) was amplified from the same samples to check for RNA quality.
RT-PCR expression analysis showed that Evi9 isoform expression varies according to cell type (Fig. 3B). In the testis, kidney, and spleen, all three isoforms are expressed at similar relative levels. The same is true for BXH2 leukemia cell lines such as B119, B162, and B139. In other tissues like the cerebrum and intestine, expression of Evi9a is enriched, while in the cerebellum and thymus, expression of Evi9b is enriched. In other tissues, one or more isoforms appear to be missing. For example, only Evi9a appears to be expressed in the stomach, while Evi9b is expressed exclusively in the lung, being absent in bone marrow, heart, and brain stem. These results suggest that the Evi9 isoforms have cell-type-specific functions.
Evi9 transforms NIH 3T3 fibroblasts.
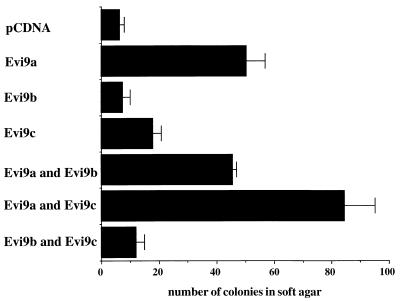
Since the enhanced expression of Evi9 by retroviral integration in BXH2 leukemias suggested that Evi9 might act as a dominant oncogene, the oncogenic potential of the various Evi9 isoforms was determined in an NIH 3T3 cell transformation assay. Each Evi9 isoform was inserted into a pCDNA3.1 expression vector and transfected into NIH 3T3 cells. NIH 3T3 cells transfected with Evi9a formed significantly increased anchorage-independent colonies in soft agar compared to cells transfected with vector alone (Fig. 4). In addition, Evi9c showed weak transforming activity for NIH 3T3 cells that was additive with the transforming activity of Evi9a. In contrast, Evi9b did not show transforming activity for NIH 3T3 cells when assayed alone or in combination with Evi9a or Evi9c. These results support the hypothesis that Evi9 functions as a dominant oncogene.
FIG. 4.
Anchorage-independent growth of NIH 3T3 cells expressing three different isoforms of Evi9. Numbers of colonies per 104 cells were determined after 3 weeks of growth in soft agar. Values are the means (±standard deviation) of three experiments.
Evi9 isoforms have different subcellular localization patterns.
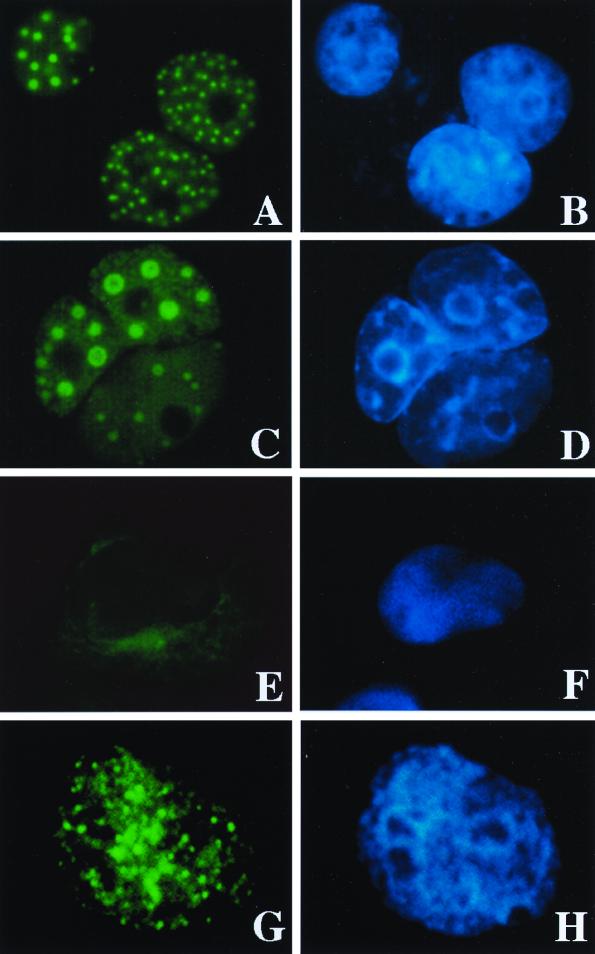
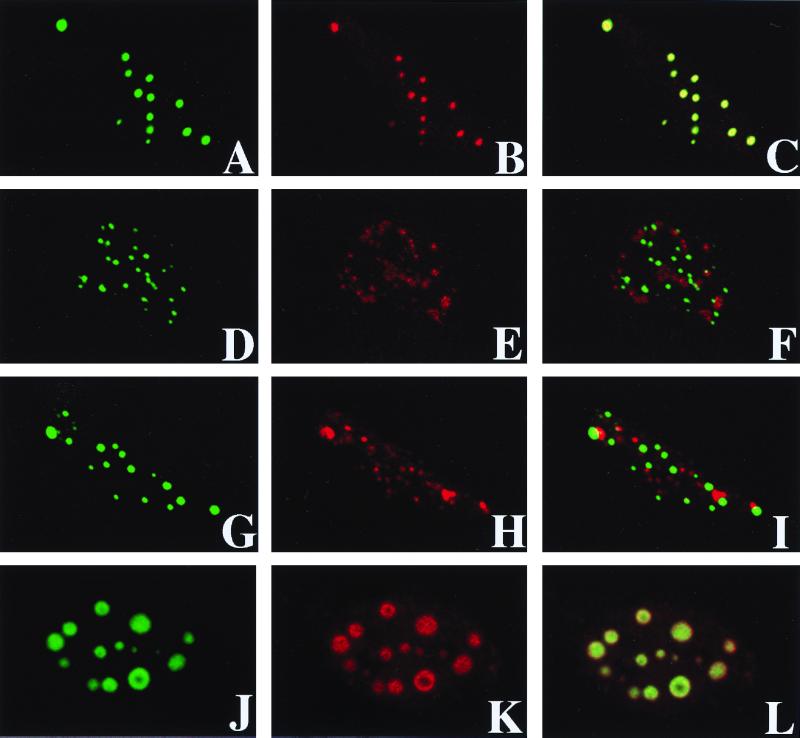
The subcellular localization of the different Evi9 isoforms was assessed by transient expression of each isoform followed by immunofluorescence using isoform-specific antibodies (Fig. 5). Evi9a and Evi9b, which are similar in their C-terminal regions, are located exclusively in the nucleus (Fig. 5A and C). In addition, both isoforms are distributed within spherical nuclear structures, 50 to 500 nm in diameter. In contrast, Evi9c, which lacks most of the C-terminal region of the protein, is localized predominantly in the cytoplasm (Fig. 5E). The nuclear localization of endogenous Evi9a and/or Evi9b proteins was also observed in B162 leukemia cells. The spherical nuclear localization was again noted, though the structures were more irregular in distribution and size (Fig. 5G).
FIG. 5.
Subcellular localization of Evi9 isoforms. Evi9a (A and B), Evi9b (C and D), and Evi9c (E and F) mammalian expression vectors were introduced into COS7 cells, and the protein was detected using polyclonal Evi9 antibody Ab1 (Evi9a and Evi9c) or Ab2 (Evi9a and Evi9b). Endogenous Evi9 protein (Evi9a and Evi9b) was also detected in B162 leukemia cells using the Ab2 antibody (G and H). Primary antibodies were visualized with fluorescein isothiocyanate-labeled anti-rabbit immunoglobulin G (A, C, E, and G). DNA was stained with Hoechst 33342 (B, D, F, and H).
Evi9a colocalizes with BCL6.
The speckled hybridization pattern of Evi9a and Evi9b suggested that they are located within nuclear bodies or other subnuclear compartments (8, 11, 20, 33). Many proteins are known to localize to these nuclear compartments. Among these proteins, three—PML, Sp100, and BCL6—are involved in hematopoietic disease (7, 8, 36). To determine whether any of these proteins colocalize with Evi9a or Evi9b, expression constructs of these three proteins were cotransfected into HeLa cells along with a GFP-tagged Evi9a expression construct (Fig. 6). The nuclear localization of GFP-tagged Evi9a was identical to that of native Evi9a (indicating that the GFP tag did not cause Evi9a to become mislocalized [Fig. 5A and 6A]) as well as to BCL6 (Fig. 6A to C) but not to PML or Sp100 (Fig. 6D to I). Evi9a, without a GFP tag, also colocalized with BCL6, indicating that Evi9a binds to BCL6 in its native state (Fig. 6J to L). These results show that Evi9a (and Evi9b) and BCL6 colocalize within the nucleus.
FIG. 6.
Evi9 colocalizes with BCL6 but not with PML or Sp100. GFP-tagged Evi9a was cotransfected along with expression constructs of BCL6 (A to C), PML (D to F), or Sp100 (G to I) into HeLa cells. Evi9 localization was detected as GFP signals in panels A, D, and G. Polyclonal antisera against BCL6 (B), PML (E), or Sp100 (H) were also used, and the signals were detected using rhodamine-conjugated anti-rabbit immunoglobulin G. Colocalization was evaluated by merging the different images (C, F, and I). Colocalization of Evi9a and BCL6 was also confirmed by using nontagged Evi9a and Myc-tagged BCL6 constructs with anti-Evi9 Ab1 antisera (J) and anti-Myc epitope antibody (K). Colocalization was noted by merging the images (L).
Evi9a and BCL6 directly interact.
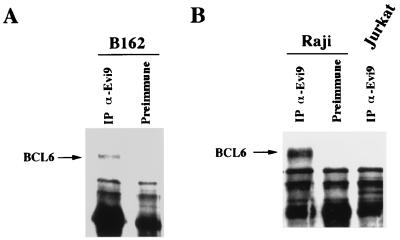
To test for a direct interaction between Evi9 and BCL6, cellular extracts from B162 cells, in which Evi9 is upregulated by retroviral integration, were subjected to immunoprecipitation using an anti-Evi9 antibody. Northern blot analysis showed that BCL6 is expressed in B162 cells and in the majority of BXH2 leukemia cell lines examined (data not shown). The immunoprecipitated material was then immunoblotted with an anti-BCL6 antibody. These experiments clearly showed that Evi9a is coimmunoprecipitated along with BCL6 (Fig. 7A). The interaction between Evi9a and BCL6 was also observed in the Raji B-lymphocytic cell line (Fig. 7B); however, no interaction was detected in the Jurkat T-cell lymphocytic cell line, which does not express BCL6 (Fig. 7B).
FIG. 7.
Interaction of Evi9 and BCL6 in leukemia cell lines. (A) Cellular extracts of B162 leukemia cells were immunoprecipitated (IP) with anti-Evi9 antibody Ab1 or preimmune serum followed by immunoblotting using anti-BCL6 antibody. (B) A similar experiment showed interaction of Evi9 and BCL6 in the B-lymphocytic cell line Raji. No interaction was observed in the T-lymphocytic cell line Jurkat, which does not express BCL6.
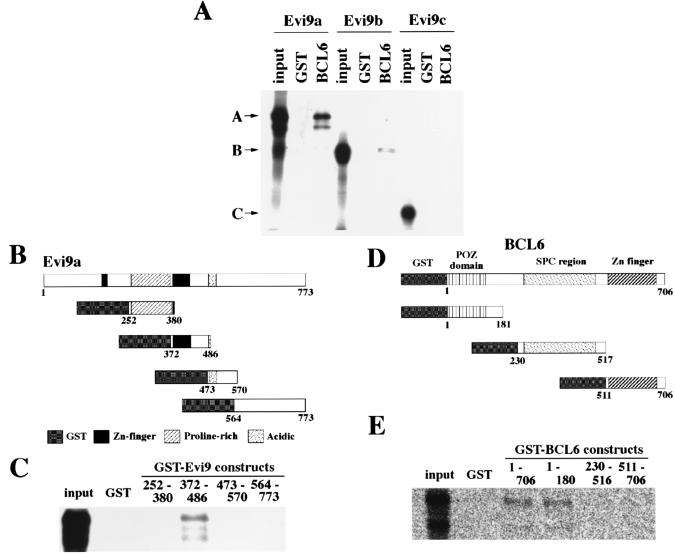
GST pull-down assays were used to confirm that these two proteins directly interact. Purified GST-tagged BCL6 protein, or GST alone, was immobilized on glutathione-Sepharose beads and incubated with in vitro-translated Evi9a, Evi9b, or Evi9c, and the bound proteins were analyzed by SDS-PAGE. Consistent with immunolocalization and immunoprecipitation studies, Evi9a and Evi9b were found to physically interact with BCL6 (Fig. 8A). On the other hand, Evi9c, which is found in the cytoplasm, did not bind to BCL6 (Fig. 8A).
FIG. 8.
Direct interaction of Evi9 and BCL6 in vitro. (A) GST pull-down assays were performed using in vitro-translated Evi9 isoforms and the full-length GST-BCL6 fusion protein. Evi9a and Evi9b show binding with BCL6, whereas Evi9c does not. (B) GST fusion constructs of Evi9. (C) The Evi9 fusion construct 372–486, which contains the second zinc finger motif, shows interaction with in vitro-translated BCL6 protein. (D) GST fusion constructs of BCL6. (E) The BCL6 fusion constructs 1–706 (full-length protein) and 1–180 show interaction with Evi9.
Deletion mutagenesis was carried out to map the protein interaction domains. Among the Evi9a deletion mutants tested for the ability to bind to BCL6, only the mutant containing the second zinc finger motif (aa 372 to 486) bound to BCL6 (Fig. 8B and C). This suggested that the BCL6 interaction domain is located in, or very near, the second zinc finger motif of Evi9. Likewise, the Evi9 interaction domain was localized within, or very near, the POZ domain of BCL6 (Fig. 8D and E).
DISCUSSION
We show here that the Evi9 common retroviral integration site in BXH2 myeloid leukemias encodes three isoforms (Evi9a, Evi9b, and Evi9c) of a novel C2H2-zinc finger protein that does not show homology with any protein in current databases. Retroviral integration at Evi9 leads to increased Evi9 expression without altering the size of Evi9 transcripts, suggesting that retroviral integration increases Evi9 expression by an enhancer activation mechanism (15).
The upregulated expression of Evi9 in myeloid leukemogenesis suggests that Evi9 might act as a dominant oncogene. This possibility was confirmed using an NIH 3T3 transformation assay, which showed that Evi9a and Evi9c, but not Evi9b, have transforming potential. These findings also suggested that the N-terminal region of Evi9, including a single zinc finger motif that is deleted in Evi9b, may be important for the oncogenic potential of Evi9. While Evi9a is predominantly localized in the nucleus, Evi9c is cytoplasmic. It is not clear at this point how Evi9c transforms NIH 3T3 cells, given its cytoplasmic localization. Perhaps the nucleus contains a small amount of Evi9c which is not detectable by immunohistochemistry, and it is this Evi9c that is important for cellular transformation. Alternatively, Evi9c may function in the cytoplasm by an as yet undefined mechanism.
The nucleus is not an amorphous structure but has many subnuclear functional domains (37). The speckled nuclear localization pattern of the larger of the two Evi9 isoforms (Evi9b and Evi9c), which contain the C-terminal region, suggested that Evi9 might interact with subnuclear structures such as nuclear bodies. In an attempt to identify interacting partners for Evi9, we tested PML (9, 18, 38), Sp100 (37), and BCL6/LAZ3 (16, 23, 40) for the ability to interact with Evi9 since all three protein are involved in hematopoietic disease and are located in subnuclear structures. PML and Sp100 are located within what is called the PML oncogenic domain (POD), which seems to play some role in transcriptional regulation (11). Our findings, however, showed that Evi9 is not localized to PODs, and it is thus very likely that Evi9 nuclear speckles belong to a functionally distinct subdomain. Instead, we find that BCL6, which is known to show POD-independent nuclear dots (7), localizes and physically interacts with Evi9.
The BCL6 gene is located at human chromosome 3q27 and was found to be involved in diffuse large B-cell lymphoma (16, 23, 40). BCL6 encodes a zinc finger protein which functions as a sequence-specific transcriptional repressor (3, 31), and recent studies have suggested that the repressor function of BCL6 might result from its interaction with components of a corepressor complex containing N-CoR, SMRT, and histone deacetylase via its N-terminal POZ domain (14, 39). Similar interactions have been found between corepressors and PLZF, another POZ/zinc finger protein which is fused to retinoic acid receptor in acute promyelocytic leukemia with a t(11;17) chromosome translocation (6, 12, 39). Unlike the case for BCL6, very weak expression of PLZF is found in most BXH2 leukemia, with or without retroviral integrations at Evi9 (data not shown), suggesting that PLZF is not a major partner for Evi9 in BXH2 leukemias.
The interaction of transcription factors with corepressor complexes results in target gene repression through histone deacetylation and modulation of chromatin assembly (22). Our preliminary experiment show that Evi9 exhibits repressional potential for transcription (Y. Saiki et al., unpublished data), suggesting that the association of Evi9 and BCL6 may lead to recruitment of BCL6 and a corepressor complex to Evi9 target genes. Alternatively, Evi9 may bind the DNA target cooperatively with POZ/zinc finger proteins, and Evi9 may be recruited to the corepressor complexes by these partner proteins. Some cofactors may be required to increase the DNA binding specificity of Evi9, since it is likely that Evi9 can bind only a hexanucleotide sequence with its two C-terminal zinc fingers (28).
The biological significance of Evi9 and BCL6 interactions in leukemogenesis remains to be clarified. BCL6 is only weakly expressed in NIH 3T3 cells (data not shown), and coexpression of BCL6 did not alter the transforming activity of Evi9 against NIH 3T3 cells (data not shown). In addition, the shortest isoform of Evi9 (Evi9c), which does not interact with BCL6, also shows transforming activity for NIH 3T3 cells. These data suggest that there are two major possibilities. The first is that Evi9 and BCL6 interaction is not functionally important in leukemia cells; the second is that the interaction is important in leukemia cells but not in NIH 3T3 cells. Given the facts that Evi9 is expressed in the nucleus and is associated with BCL6 in leukemia cells, the latter possibility is more feasible. Experiments to test the functional importance of this interaction in leukemia cells are under way.
Currently we are investigating the possible involvement of EVI9 in human hematopoietic disease. The human homologue of Evi9 is located at chromosome 2p13 (T. Nakamura and M. Yoshida, unpublished data), where chromosomal abnormalities associated with human malignant lymphoma have been found (13). We are also making a targeted mutation of Evi9 using embryonic cell knockout technology in order to better understand the biological relevance of the Evi9 gene.
ACKNOWLEDGMENTS
This research was sponsored in part by Grant-in-Aid for Scientific Research on Priority Areas (A) from the Ministry of Education, Science, Sports and Culture, Japan, and by the National Cancer Institute under contract with ABL.
We thank Tohru Miki for providing the BCL6 expression plasmid, Thomas Sternsdorf for expression plasmids and antibodies for human PML and Sp100, Mitsuyasu Kato for help in using confocal laser microscopy, and Ryoko Iwata for technical assistance.
REFERENCES
- 1.Bardwell V J, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 2.Bedigian H G, Johnson D A, Jenkins N A, Copeland N G. Spontaneous and induced leukemias of myeloid origin in recombinant inbred BXH mice. J Virol. 1984;51:586–594. doi: 10.1128/jvi.51.3.586-594.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang C-C, Ye B H, Chaganti R S K, Dalla-Favera R. BCL6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;77:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copeland N G, Jenkins N A. Myeloid leukemias: disease genes and mouse models. In: Hiai H, Hino O, editors. Animal models for cancer predisposition syndromes. S. Basel, Switzerland: Karger; 1999. pp. 53–63. [DOI] [PubMed] [Google Scholar]
- 6.David G, Alland L, Hong S-H, Wong C-W, DePinho A R, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 7.Dhordain P, Albagli O, Ansieau S, Koken M H M, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert J P, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- 8.Doucas V, Evans R M. The PML nuclear compartment and cancer. Biochim Biophys Acta. 1996;1288:M25–M29. doi: 10.1016/s0304-419x(96)00028-5. [DOI] [PubMed] [Google Scholar]
- 9.Dyck J A, Maul G G, Miller W H, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 10.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X P, Neilson E G, Rauscher F J., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 11.Hodges M, Tissot C, Howe K, Grimwade D, Freemont P S. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am J Hum Genet. 1998;63:297–304. doi: 10.1086/301991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong S-H, David G, Wong C-W, Dejean A, Privalsky M L. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houldsworth J, Mathew S, Rao P H, Dyomina K, Louie D, Parsa N, Offit K, Chaganti R S. REL proto-oncogene is frequently amplified in extranodal diffuse large cell lymphoma. Blood. 1996;87:25–29. [PubMed] [Google Scholar]
- 14.Huynh K D, Bardwell V J. The BCL6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- 15.Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 16.Kerckaert J P, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- 17.Knochel W, Poting A, Koster M, el Baradi T, Nietfeld W, Bouwmeester T, Pieler T. Evolutionary conserved modules associated with zinc fingers in Xenopus laevis. Proc Natl Acad Sci USA. 1989;86:6097–6100. doi: 10.1073/pnas.86.16.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koken M H M, Puvio-Dutilleul F, Guillemin M C, Viron A, Cruz-Linares G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de The H. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;14:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Largaespada D A, Shaughnessy J D, Jenkins N A, Copeland N G. Retroviral insertion at the Evi-2 locus in BXH2 myeloid leukemia cell lines disrupts Nf1 expression without changes in steady-state Ras-GTP levels. J Virol. 1995;69:5095–5102. doi: 10.1128/jvi.69.8.5095-5102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeBrun D P, Matthews B P, Feldman B J, Cleary M L. The chimeric oncoproteins E2A-PBX1 and E2A-HLF are concentrated within spherical nuclear domains. Oncogene. 1997;15:2059–2067. doi: 10.1038/sj.onc.1201367. [DOI] [PubMed] [Google Scholar]
- 21.Liao X, Du Y, Morse H C, Jenkins N A, Copeland N G. Proviral integrations at the Evi5 locus disrupt a novel 90 kDa protein with homology to the Tre2 oncogene and cell-cycle regulatory proteins. Oncogene. 1997;14:1023–1029. doi: 10.1038/sj.onc.1200929. [DOI] [PubMed] [Google Scholar]
- 22.Lin R J, Egan D A, Evans R M. Molecular genetics of acute promyelocytic leukemia. Trends Genet. 1999;15:179–184. doi: 10.1016/s0168-9525(99)01710-2. [DOI] [PubMed] [Google Scholar]
- 23.Miki T, Kawamata N, Hirosawa S, Aoki N. Gene involved in the 3q27 translocation associated with B-cell lymphoma, BCL-5, encodes a kruppel-like zinc-finger protein. Blood. 1994;83:26–32. [PubMed] [Google Scholar]
- 24.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 25.Moriyama M, Yamochi T, Semba K, Akiyama T, Mori S. BCL6 is phosphorylated at multiple sites in its serine- and proline-clustered region by mitogen-activated protein kinase (MAPK) in vivo. Oncogene. 1997;14:2465–2474. doi: 10.1038/sj.onc.1201084. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura T, Largaespada D A, Shaughnessy Jr J D, Jenkins N A, Copeland N G. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 27.Onizuka T, Moriyama M, Yamochi T, Kuroda T, Kazama A, Kanazawa N, Sato K, Kato T, Ota H, Mori S. BCL6 gene product, a 92- to 98-kD nuclear phosphoprotein, is highly expressed in germinal center B cells and their neoplastic counterparts. Blood. 1995;86:28–37. [PubMed] [Google Scholar]
- 28.Pavletich N P, Pabo C O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 29.Pengue G, Lania L. Kruppel-associated box-mediated repression of RNA polymerase II promoters is influenced by the arrangement of basal promoter elements. Proc Natl Acad Sci USA. 1996;93:1015–1020. doi: 10.1073/pnas.93.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts T, Chernova O, Cowell J K. NB4S, a member of the TBC1 domain family of genes, is truncated as a result of a constitutional t(1;10)(p22;q21) chromosome translocation in a patient with stage 4S neuroblastoma. Hum Mol Genet. 1998;7:1169–1178. doi: 10.1093/hmg/7.7.1169. [DOI] [PubMed] [Google Scholar]
- 31.Seyfert V L, Allman D, He Y, Staudt L M. Transcriptional repression by the proto-oncogene BCL6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 32.Shaughnessy J D, Jr, Largaespada D A, Tian E, Fletcher C F, Cho B C, Vyas P, Jenkins N A, Copeland N G. Mrvi1, a common MRV integration site in BXH2 myeloid leukemias, encodes a protein with homology to a lymphoid-restricted membrane protein Jaw1. Oncogene. 1999;18:2069–2084. doi: 10.1038/sj.onc.1202419. [DOI] [PubMed] [Google Scholar]
- 33.Skinner P J, Koshy B T, Cummings C J, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–978. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 34.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 35.Spector D. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 36.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szostecki C, Guldner H H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 38.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmos-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARa in acute promyelocytic leukemia cells. Cell. 1994;76:345–358. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 39.Wong C-W, Privalsky M L. Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARα, and BCL6. J Biol Chem. 1998;273:27695–27702. doi: 10.1074/jbc.273.42.27695. [DOI] [PubMed] [Google Scholar]
- 40.Ye B H, Lista F, Lo Coco F, Knowles D M, Offit K, Chaganti R S K, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 41.Zollman S, Godt D, Prive G G, Couderc J L, Laski F A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]