Gemtuzumab ozogamicin (GO) is a humanized anticluster of differentiation (CD) 33 monoclonal antibody linked to the cytotoxic agent calicheamicin.1-4 GO binds the CD33 antigen and is internalized; calicheamicin is released inside the blasts leading to DNA damage and death of leukemic cells.2,5,6
GO was approved for the treatment of patients with acute myeloid leukemia (AML) with expression of CD33 on blasts by the European Medicines Agency (here only for newly diagnosed patients in combination with intensive therapy) and the Food and Drug Administration based on positive results from the ALFA-0701 trial.1,7 However, a number of clinical trials with GO yielded partially conflicting results emphasizing the importance of a better understanding of factors influencing the clinical response to GO.6,7
Several factors can influence the response to GO. A meta-analysis by Hills et al. demonstrated a benefit of GO only in patients with Medical Research Council (MRC) favorable and intermediate cytogenetic risk, while GO did not improve outcome of patients with adverse cytogenetic risk.7 Another factor that might have an impact on the response to GO is the CD33-coding single nucleotide polymorphism (SNP) rs12459419 (NM_001772.4:c.41 C>T; Ala14Val in exon 2) as GO needs to bind to CD33.
CD33 consists of an amino-terminal V-set immunoglobulin (Ig)-like domain, coded by exon 2, and a C2-set Ig-like domain in its extracellular component. The presence of rs12459419 in exon 2 affects alternative splicing of CD33 resulting in the loss of exon 2 for the T allele which leads to a shorter isoform lacking the GO binding site. The C allele leads to the CD33 full-length isoform including the GO binding site.3,6 The genotype frequencies in the European population for the SNP are c.41C/C: 46.9%, c.41C/T: 44.1%, and c.41T/T: 8.9%.
Given the strong clinical need to clarify who could benefit from GO and the mechanisms responsible for a differential response to GO we studied the effect of rs12459419 in the AMLSG 09-09 phase III study (NCT00893399). This trial is a large, randomized study in adult AML patients who were eligible for intensive therapy and had a mutation in nucleophosmin1 (NPM1). According to MRC definitions, 8 99.5% of the patients in both treatment arms had intermediate-risk cytogenetics (Online Supplementary Table S1). Since the benefit of GO was only observed in favorable- and intermediate-risk groups,7 our cohort is well suited for analyzing the effect of the CD33 SNP. Of 588 patients included in this study, 545 patients had samples available for SNP analysis. Patients were randomly assigned (1:1) to receive 3 mg/m2 of GO (n=273, GO arm) versus no GO (n=272, standard arm) in combination with two cycles of induction chemotherapy.9
Samples were obtained from peripheral blood (n=459) and bone marrow (n=86); 507 samples were obtained at the time of diagnosis and 38 during follow-up. The SNP genotype was determined using the TaqMan SNP Genotyping Assay rs12459419 (Thermo Fisher Scientific, Waltham, MA, USA) on a 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). The allelic discrimination protocol was modified from that described by Easton et al.10 The Genotyping Assay revealed robust results for 540 out of 545 samples (call rate >99%). DNA from the remaining five patients was genotyped by Sanger sequencing as previously reported.11 In order to confirm the genotype we additionally analyzed three samples with a genotype known from the Genotyping Assay. Results from the Genotyping Assay and Sanger sequencing were concordant.
Genotype proportions were 45.2% c.41 C/C, 44.1% c.41 C/T and 10.7% c.41 T/T in the standard arm and 46.1% c.41 C/C, 45.3% c.41 C/T and 8.6% c.41 T/T in the GO arm.
We then analyzed whether rs12459419 genotypes influenced hematologic recovery times, response to therapy and clinical outcome in patients treated with or without GO.
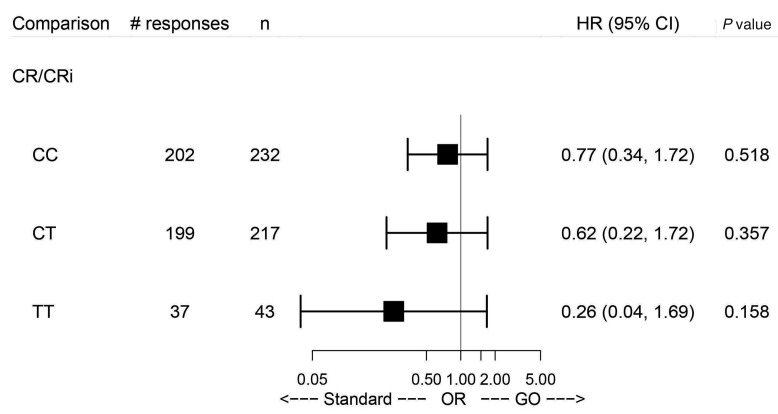
Figure 1.
Analysis comparing gemtuzumab ozogamicin versus standard treatment in the three subgroups defined by the genotypes with regard to complete remission/complete remission with incomplete hematologic recovery. HR: hazard ratio; 95% CI: 95% confidence interval; CR: complete remission; CRi: complete remission with incomplete hematologic recovery; OR: odds ratio; GO: gemtuzumab ozogamicin.
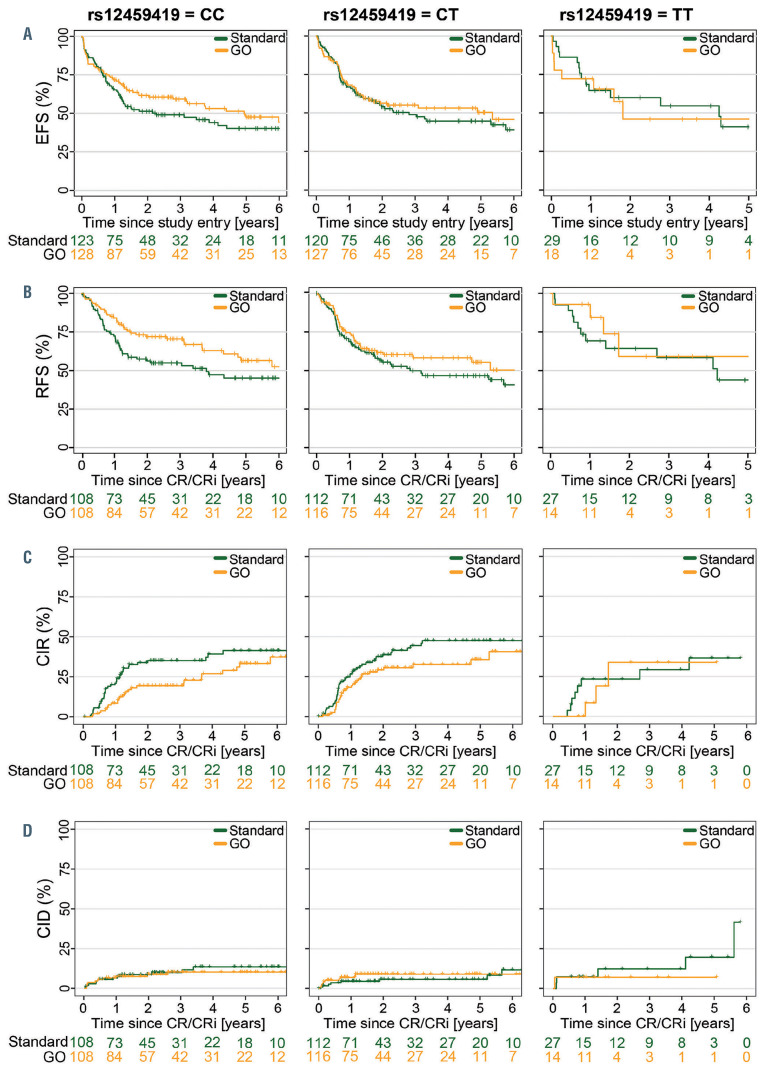
Figure 2.
Association of rs12459419 genotypes with clinical outcome by treatment arm. (A) Event-free survival. (B) Relapse-free survival. (C) Cumulative incidence of relapse. (D) Cumulative incidence of death in remission. EFS: event-free survival; GO: gemtuzumab ozogamicin; RFS: relapse-free survival; CR: complete remission; CRi: complete remission with incomplete hematologic recovery; CIR: cumulative incidence of relapse; CID: cumulative incidence of death in remission.
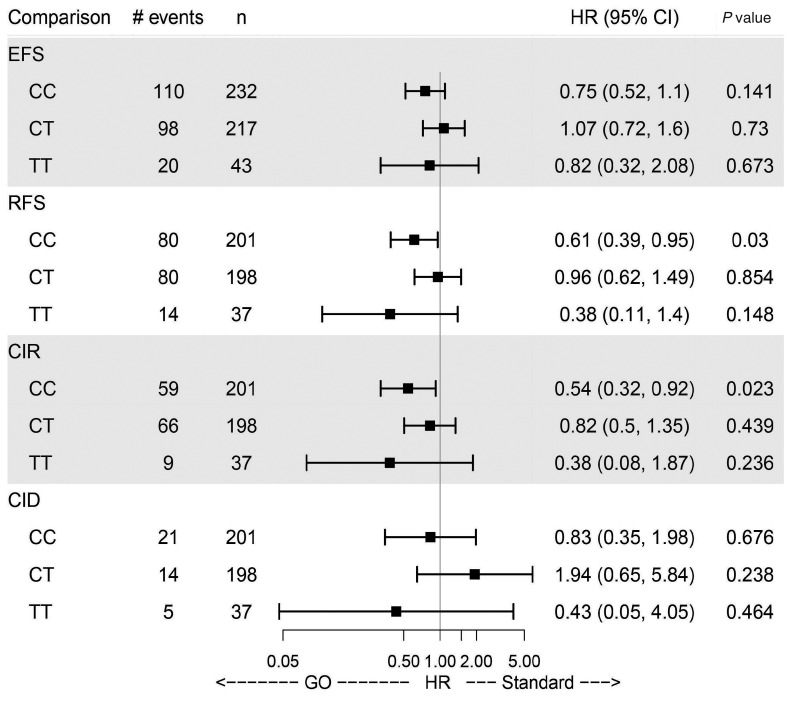
Figure 3.
Analysis comparing gemtuzumab ozogamicin versus standard treatment in the three subgroups defined by the genotypes with regard to clinical endpoints. Effect estimates are taken from multivariate Cox models and are therefore adjusted for age, gender, European LeukemiaNet 2010 risk class and white blood cell count. HR: hazard ratio; 95% CI: 95% confidence interval; EFS: eventfree survival; RFS: relapsefree survival; CR: complete remission; CIR: cumulative incidence of relapse; CID: cumulative incidence of death in remission: GO: gemtuzumab ozogamicin.
Time to hematologic recovery from the start of the first and second induction cycles was not influenced by the SNP genotype in the GO arm (Online Supplementary Figure S1).
With respect to the clinical endpoints complete remission (CR) or CR with incomplete hematologic recovery (CRi) as well as event-free survival, relapse-free survival, cumulative incidence of relapse and cumulative incidence of death in remission (defined according to European LeukemiaNet 2017 criteria12), multivariate regression models (logistic regression and Cox proportional hazards models) were used to evaluate whether the effect of GO varies with the SNP genotype. More explicitly, the likelihood ratio test was used to test for the presence of an interaction between treatment with GO and SNP genotype while other prognostic factors (age, gender, European LeukemiaNet 2010 risk class13 and white blood cell count) were accounted for by including them as additional covariates. Of 485 patients who achieved CR/CRi, 157 patients experienced a relapse and 46 patients died in CR. No significant differences for CR/CRi could be observed between the genotypes in the treatment arms (Figure 1).
For event-free survival, no significant difference in the treatment effect of GO compared to the standard arm was observed across the SNP genotypes (Figures 2A and 3). Specifically, patients in the GO arm with the c.41 C/C genotype did not have a superior event-free survival compared to those with other genotypes. However, relapsefree survival was significantly improved for patients with the c.41 C/C genotype in the GO arm (P=0.03) compared to those in the standard arm (Figures 2B and 3). It is important to state that the early death rate during induction was significantly higher (10.3%) in the GO arm than in the standard arm (5.7%) (P=0.05) but not influenced by the different SNP genotypes (data not shown). However, the higher early death rate in the GO arm may explain why we see an effect of the CD33 SNP on relapse-free survival but not on event-free survival.
Furthermore, patients with c.41 C/C genotype had a significantly reduced cumulative incidence of relapse when treated with GO (P=0.023) (Figures 2C and 3). Patients with c.41 C/T or c.41 T/T genotype did not benefit from GO with regard to relapse-free survival and cumulative incidence of relapse (Figures 2B, C and 3). There were no significant differences in cumulative incidence of death in remission in relation to genotypes between the treatment arms or between the genotypes within a treatment arm (Figures 2D and 3).
Because subgroup analysis of the AMLSG 09-09 trial revealed a significant prognostic advantage in female patients, patients <70 years and FLT3-ITD-negative patients when receiving GO, we analyzed whether these observations could be explained by different distributions of the SNP genotypes. We found no significant association of the SNP with mutations in FLT3-ITD, FLT3-TKD or DNMT3A (Online Supplementary Table S1). For separate analysis of female and male patients see Online Supplementary Figure S2A, B.
Additionally, we separately analyzed the influence of rs12459419 on the effect of GO in younger patients (≤40 years) versus older ones (>40 years). Due to low event numbers (especially in the group of younger patients) additional covariate adjustment was omitted for these subgroup analyses. Here, younger patients with the c.41 C/C genotype had no extra benefit from GO compared to younger patients with other SNP genotypes (data not shown).
Published data about whether CD33 SNP rs12459419 can influence the response to GO have been conflicting. The first study to analyze the effect of the rs12459419 on the response to GO was the AAML0531 trial, a randomized phase III study in pediatric and adolescent AML patients (aged 0-29 years).3 In that study, Lamba et al. showed that the T allele was significantly associated with higher levels of the CD33 isoform lacking exon 2. Patients harboring the c.41 C/C genotype treated with GO had a significantly reduced cumulative incidence of relapse and higher relapse-free survival than patients in the non-GO arm. Importantly, this positive effect was restricted to patients with favorable and intermediate cytogenetic risk. The clinical benefit of GO was not visible in patients with the c.41 C/T or c.41 T/T genotype of SNP rs12459419.3
In contrast, in the UK MRC/NCRI AML 15 (ISRCTN17161961) and AML 17 (ISRCTN55675535) trials, GO was given to adult AML patients aged 13 to 69 years. Here, the SNP genotype had no impact on outcome, even if only patients with favorable-risk cytogenetics were considered.14 Of note, the trial included multiple randomization steps with different chemotherapy regimens.14
Moreover, Short et al. could not show a significant impact of the SNP on overall survival or relapse-free survival in patients (n=113) treated with decitabine plus GO.15 However, this study was performed in a population of patients unlikely to benefit from GO as it included patients with high-risk myelodysplastic syndrome and AML patients with unfavorable risk features.15
Our results show a similar signal as the data from Lamba et al. with regard to an improved relapse-free survival and reduced cumulative incidence of relapse for patients with the c.41 C/C genotype treated with GO. It is unlikely that the discrepancies between the results of the studies by Gale et al. and Lamba et al. are only related to the difference in age of the patients as we showed an impact of the SNP in adult patients.
In summary, our study suggests that CD33 SNP rs12459419 is one of the predictive factors that affects the rate of relapse in patients with NPM1-mutated AML receiving GO. As testing for the SNP is technically simple, it can be easily included in routine diagnostics. In order to support our findings a prospective study in a larger cohort is desirable.
Supplementary Material
Funding Statement
Funding: this study was supported by grant DJCLS R13/14 from the Deutsche José Carreras Leukämie-Stiftung e.V., grant 01GM1909A from the German Federal Ministry of Education and Research and by Sonderforschungsbereich SFB 1074, project B3 (to KD) from the Deutsche Forschungsgemeinschaft.
References
- 1.Lambert J, Pautas C, Terré C, et al. Gemtuzumab ozogamicin for de novo acute myeloid leukemia: final efficacy and safety updates from the open-label, phase III ALFA-0701 trial. Haematologica. 2019;104(1):113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ricart AD. Antibody-drug conjugates of calicheamicin derivative: gemtuzumab ozogamicin and inotuzumab ozogamicin. Clin Cancer Res. 2011;17(20):6417-6427. [DOI] [PubMed] [Google Scholar]
- 3.Lamba JK, Chauhan L, Shin M, et al. CD33 splicing polymorphism determines gemtuzumab ozogamicin response in de novo acute myeloid leukemia: report from randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol. 2017;35(23):2674-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thol F, Schlenk RF. Gemtuzumab ozogamicin in acute myeloid leukemia revisited. Expert Opin Biol Ther. 2014;14(8):1185-1195. [DOI] [PubMed] [Google Scholar]
- 5.Sievers EL. Efficacy and safety of gemtuzumab ozogamicin in patients with CD33-positive acute myeloid leukaemia in first relapse. Expert Opin Biol Ther. 2001;1(5):893-901. [DOI] [PubMed] [Google Scholar]
- 6.Godwin CD, Gale RP, Walter RB. Gemtuzumab ozogamicin in acute myeloid leukemia. Leukemia. 2017;31(9):1855-1868. [DOI] [PubMed] [Google Scholar]
- 7.Hills RK, Castaigne S, Appelbaum FR, et al. The addition of gemtuzumab ozogamicin to induction chemotherapy in acute myeloid leukaemia: an individual patient data meta-analysis of randomised trials in adults. Lancet Oncol. 2014;15(9):986-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kühnl A, Grimwade D. Molecular markers in acute myeloid leukaemia. Int J Hematol. 2012;96(2):153-163. [DOI] [PubMed] [Google Scholar]
- 9.Schlenk RF, Paschka P, Krzykalla J, et al. Gemtuzumab ozogamicin in NPM1 -mutated acute myeloid leukemia: early results from the prospective randomized AMLSG 09-09 phase III study. J Clin Oncol. 2020;38(6):623-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easton EF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thol F, Damm F, Lüdeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889-2896. [DOI] [PubMed] [Google Scholar]
- 12.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453-474. [DOI] [PubMed] [Google Scholar]
- 14.Gale RE, Popa T, Wright M, et al. No evidence that CD33 splicing SNP impacts the response to GO in younger adults with AML treated on UK MRC/NCRI trials. Blood. 2018;131(4):468-471. [DOI] [PubMed] [Google Scholar]
- 15.Short NJ, Richard-Carpentier G, Kanagal-Shamanna R, et al. Impact of CD33 and ABCB1 single nucleotide polymorphisms in patients with acute myeloid leukemia and advanced myeloid malignancies treated with decitabine plus gemtuzumab ozogamicin. Am J Hematol. 2020;95(9):E225-E228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.