Abstract
Somatic mutations in genes coding for splicing factors, e.g., SF3B1, U2AF1, SRSF2, and others are found in approximately 50% of patients with myelodysplastic syndromes (MDS). These mutations have been predicted to frequently occur early in the mutational hierarchy of the disease, therefore, making them particularly attractive potential therapeutic targets. Recent studies in cell lines engineered to carry splicing factor mutations have revealed a strong association with elevated levels of DNA:RNA intermediates (R-loops) and a dependency on proper ATR function. However, data confirming this hypothesis in a representative cohort of primary MDS patient samples have so far been missing. Using CD34+ cells isolated from MDS patients with and without splicing factor mutations as well as healthy controls we show that splicing factor mutation- associated R-loops lead to elevated levels of replication stress and ATR pathway activation. Moreover, splicing factor mutated CD34+ cells are more susceptible to pharmacological inhibition of ATR resulting in elevated levels of DNA damage, cell cycle blockade, and cell death. This can be enhanced by combination treatment with the low-dose splicing modulatory compound Pladienolide B. We further confirm the direct association between R-loops and ATR sensitivity and the presence of a splicing factor mutation using lentiviral overexpression of wild-type and mutant SRSF2 P95H in cord blood CD34+ cells. Collectively, our results from n=53 MDS patients identify replication stress and associated ATR signaling to be critical pathophysiological mechanisms in primary MDS CD34+ cells carrying splicing factor mutations, and provide a preclinical rationale for targeting ATR signaling in these patients.
Introduction
With a frequency of approximately 50%, somatically acquired mutations in genes coding for splicing factors such as SF3B1, U2AF1, SRSF2, and others are the most recurrent mutations in patients with myelodysplastic syndromes (MDS).1-3 Apart from DNMT3A, TET2, and ASXL1 mutations, splicing factor (sf) mutations are also frequently determined to be founder mutations in MDS.4-7 Therefore, sf mutations are particularly attractive targets for therapeutic intervention in MDS.
The precise mechanism of how sf mutations contribute to MDS pathophysiology is not fully understood. It has been shown that sf mutations perturb RNA splicing, e.g., by altering 3’-splice recognition sites, resulting in abnormal transcriptional programs in mutant cells.8-15 However, the overlap between the different sf mutations is limited16 raising the question how they can lead to a common myelodysplastic phenotype. Previous work has demonstrated that MDS-associated sf mutations induce increased formation of DNA:RNA intermediates (R-loops), suggesting elevated R-loops to be a novel common pathophysiological mechanism for sf mutated MDS.17,18 R-loops are transcription intermediates consisting of a DNA:RNA hybrids and a displaced single-stranded DNA (ssDNA).19 They are known to contribute to genome instability by exposing ssDNA and by blocking replication fork progression, causing replication stress and associated DNA damage.20
On the molecular level, perturbation in the dynamics of replication fork progression triggers phosphorylation of replication protein A (RPA) and activation of the ataxia telangiectasia and Rad3-related protein (ATR) pathway in order to repair replication stress-induced DNA damage.21 Chronically elevated R-loops as they occur in sf mutated cells make these cells dependent on proper ATR function in order to preserve the integrity of their genomes. This dependency represents a vulnerability towards pharmacological ATR inhibition that is specific to sf mutated MDS cells.22 A study on sf mutated cell lines has revealed that the R-loop-associated ATR activation seems to be independent of altered RNA processing.22 This opens the possibility for a potentially additive effect of the combination of ATR inhibition with splicing modulators, such as Pladienolide B (PladB), to selectively target sf mutated cells while sparing healthy non-mutated hematopoietic cells. PladB is a naturally occuring macrolide that targets the SF3b complex.23 PladB as well as its derivative E7107 have previously been shown to confer cytotoxic activity in several in vivo and in vitro models of sf mutated myeloid malignancies. 24,2 A next-generation splicing modulator, H3B- 8800, a derivative of PladB, is currently under investigation in a phase I clinical trial in MDS and related myeloid malignancies (clinicaltrials gov. Identifier: NCT02841540).26
In this work we investigated the impact of R-loop-accumulation and inhibition on ATR signaling in primary CD34+ cells isolated from the bone marrow (BM) of MDS patients. We show that sf mutated cells display elevated R-loop-formation as compared to non-sf mutated MDS and healthy cells, which results in increased replication stress and associated activation of the ATR signaling pathway. Pharmacological inhibition of ATR preferentially kills sf mutated cells by increasing DNA damage. We further show that this effect can be increased by the splicing modulator PladB specifically in sf mutated cells of MDS patients. Our work provides preclinical evidence in primary patient cells for targeting ATR as a novel therapeutic strategy in MDS.
Methods
Patient and healthy donor material
BM of MDS patients was obtained from residual diagnostic material. Healthy CD34+ cells were isolated from hip replacement surgery bone specimen (old CD34+ cells) or collected by iliac crest puncture of healthy volunteers (young CD34+ cells). For lentiviral transduction experiments, healthy CD34+ cells were isolated from cord blood. The study was approved by the Institutional Review Board of the Medical Faculty Mannheim, University of Heidelberg, Germany, and conducted in accordance with the declaration of Helsinki.
Sequencing
Sf mutations were either assessed by diagnostic myeloid panel sequencing (n=9) or by germline controlled whole exome sequencing (WES) (n=44).
Tissue culture and cell-based assays
Primary hematopoietic cells were cultured in StemSpan SFEM II supplemented with myeloid expansion supplement containing SCF, TPO, G-CSF, and GM-CSF (Stemcell Technologies). MOLM13 cells were obtained from the DSMZ (Germany) and cultured in RPMI-1614 supplemented with 20% heat-inactivated fetal calf serum.
Cell viability assay
Cell viability in response to treatment was determined by CellTiter-Glo® assay (Promega) according to the manufacturer’s recommendations.
Cell apoptosis assay
Apoptosis was determined by flow cytometry using Annexin V/ propidium iodide (PI) staining according to the manufacturer’s protocol (BD Bioscience) after treatment with AZD6738, PladB (alone and in combination), or vehicle for 24-48 hours.
Flow cytometry
Differential BM fractions were isolated from MNC stained with fluorochrome-conjugated antibodies against CD33, CD19, CD3, CD34 and CD38 by flow cytometry-based cell sorting (FACS). Cell isolation was performed on a BD FACSMelody Flow Cytometer using double sorting to ensure maximum purity.
Immunofluorescence
R-loops and RPA were quantified by immunofluorescence analysis using anti-S9.6 (Kerafast) and anti-RPA (pSer33; Novus Biologicals) antibodies. Quantification of mean fluorescence intensity (MFI) was performed using the following formula: mean fluorescence of selected cell – (area of selected cell x mean fluorescence of background readings). Values are displayed as arbitrary units (A.U.).
Single-molecule DNA replication analyses
Replication track analyses were adapted from a published protocol. 27 In brief, CD34+ cells in culture were pulsed with 25 μM 5-chloro-2′-deoxyuridine (CldU) followed by 250 μM 5-iodo-2′- deoxyuridine (IdU), washed, and spotted onto SuperFrost® microscope slides for subsequent label detection.
Lentiviral transduction of CD34+ and MOLM13 cells
Lentiviral constructs expressing SRSF2 wild-type (WT) and SRSF2 P95H were transiently transfected into 293T cells together with a third-generation lentiviral packaging mix (AMS.P904, Amsbio). Lentiviral supernatants were concentrated (Lenti-X concentrator, Clontech) and used to infect CD34+ or MOLM13 cells by spin-infection (2,000xg, 33°C, 90 min).
RNA sequencing
For RNA sequencing the Illumina TruSeq Stranded mRNA kit (San Diego, USA) with 500 ng RNA as input was used. Sequencing was performed with 100 bp paired end on an Illumina NovaSeq device.
Statistical analysis
Data were analyzed using Prism 8 (GraphPad Software, La Jolla, CA) using t-test or one-way analysis of variance (ANOVA), or Mann-Whitney U test. P-value was considered significant at values less than 0.05 (n.s.: not significant, statistically significant *P<0.05, **P<0.01, ***P<0.001).
For detailed protocols, please see the Online Supplementary Methods.
Figure 1.
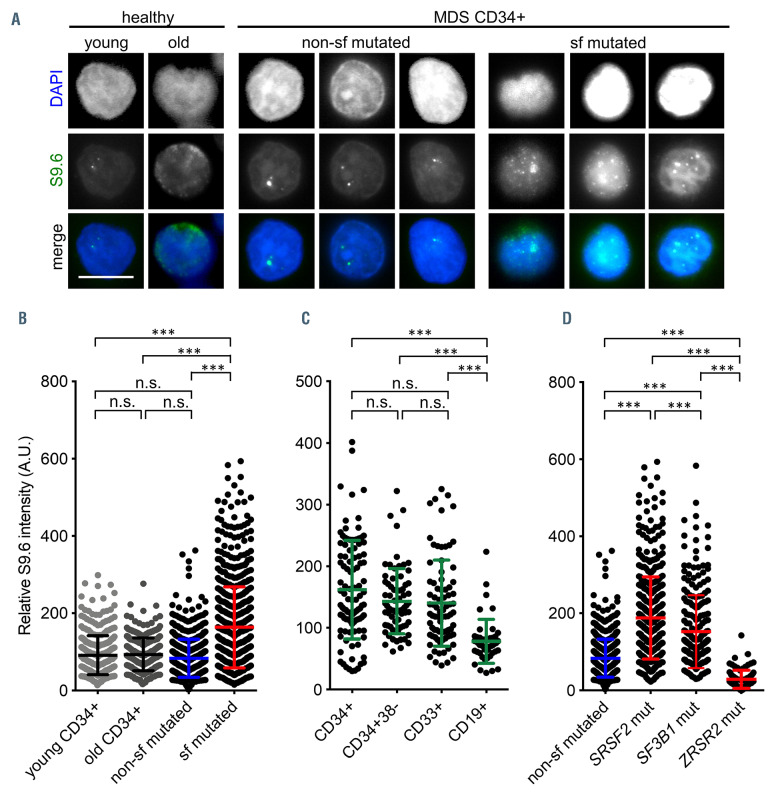
Splicing factor mutated myelodysplastic syndrome CD34+ cells exhibit elevated levels of R-loops. (A) Representative immunofluorescence images of R-loops using the antibody S9.6 (Kerafast, Boston) in CD34+ cells isolated from myelodysplastic syndrome (MDS) patient bone marrow with and without splicing factor (sf) mutations as well as young and old healthy controls. Scale bar, 5 μm. (B) Quantification of S9.6 mean fluorescence intensity (MFI); n=10 (658 cells, non-sf mutated), n=17 (809 cells, sf mutated), n=6 (363 cells, young CD34+), n=9 (301 cells, old CD34+). (C) Quantification of S9.6 MFI in fluorescence-activated cell sortingsorted hematopoietic subpopulations isolated from the bone marrow of three (n=1, SF3B1 K700 and n=2, SRSF2 P95) patients (CD34+: 103 cells; CD34+CD38–: 71 cells; CD33+: 81 cells; CD19+: 47 cells). (D) Quantification of S9.6 MFI in CD34+ cells carrying mutations in SRSF2, SF3B1, and ZRSR2; n=10 (658 cells, non-sf mutated), n=11 (484 cells, SRSF2 mutated), n=6 (325 cells, SF3B1 mutated); n=1 (88 cells, ZRSR2 mutated). Data are means +/-standard deviation. ***P≤0.001; n.s.: not significant; A.U: arbitrary units.
Results
Splicing factor mutated CD34+ cells from myelodysplastic syndrome patients have elevated levels of R-loops
Increased levels of R-loops have previously been associated with MDS-specific mutations in genes coding for splicing factors.17,22 Based on these cell-line data, we asked the question whether elevated R-loops would also represent a common vulnerability in sf mutated cells isolated from the BM of MDS patients. Patient characteristics are displayed in the Online Supplementary Table S1. Using an antibody against R-loops (S9.6, Kerafast), we evaluated levels of R-loops in CD34+ cells isolated from n=18 MDS patients carrying mutations in sf genes (SRSF2=10, SF3B1=7; ZRSR2=1) (sf mutated) and compared them to the levels of non-sf mutated CD34+ cells from MDS patients (n=10) (Figure 1A and B). We found a significant accumulation of R-loops in sf (SRSF2 and SF3B1) mutated cells (mean MFI [A.U.]: 83 vs. 163; P<0.001). In order to rule out that the accumulation of Rloops was solely an effect of aging we included CD34+ from healthy aged (>64 years of age; n=9) and young (<40 years of age, n=6) individuals as controls, which presented similar amounts of R-loops as non-mutated MDS cells (mean MFI [A.U.]: 91 [healthy young], 93 [healthy old]) (Figure 1A and B). It has previously been shown that the BM CD34+ fraction is enriched in MDS-initiating cells.4,28 In order to check whether certain cellular BM compartments were particularly prone to accumulate R-loops, we sorted several fractions, i.e., CD34+, CD34+CD38-, CD33+ (mature myeloid cells) and CD19– (B-lymphoid cells). As shown in Figure 1C, we detected similar levels of R-loops in CD34+ (mean MFI: 122), CD34+CD38– (mean MFI: 146), as well as in mature CD33+ (mean MFI: 126) myeloid BM cells. However, in accordance with the myeloid nature of MDS pathophysiology we did not find elevated levels of R-loops in CD19+ B cells (mean MFI: 68). We also confirmed the association of splicing perturbations induced by PladB with the accumulation of Rloops in non-sf mutated MOLM13 cell line cells (Online Supplementary Figure S1). Of note, the patients carrying a ZRSR2 mutation did not show increased levels of R-loops (Figure 1D).
Figure 2.
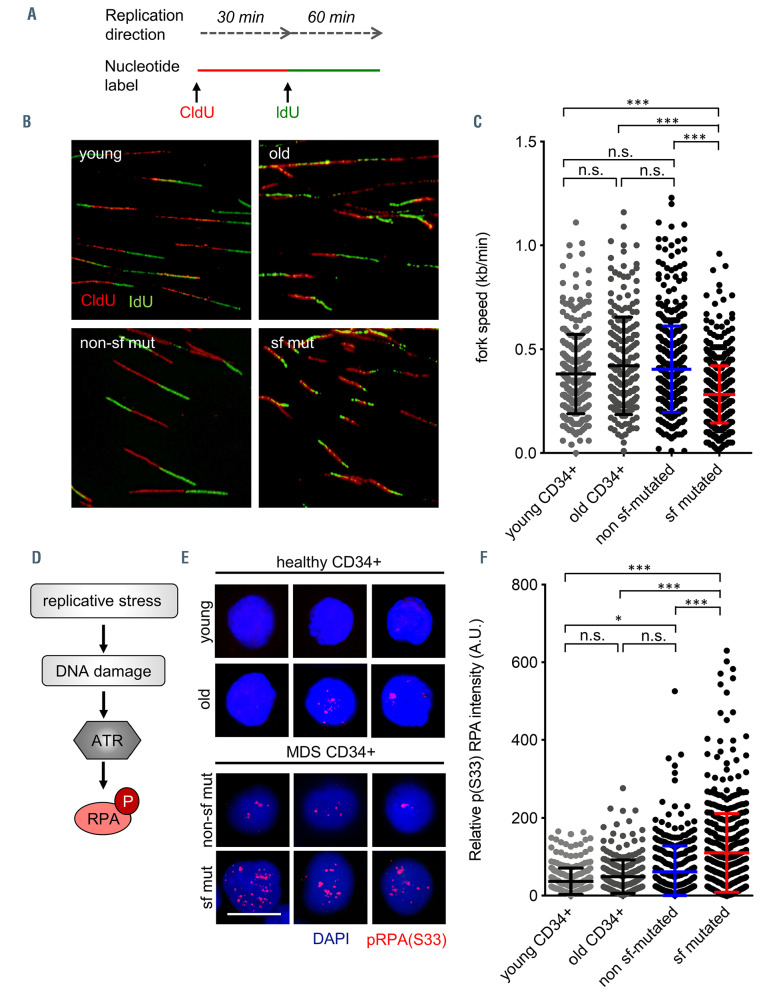
Elevated levels of R-loops induce replication stress in splicing factor mutated myelodysplastic syndrome CD34+ cells. (A) Experimental procedure and representative images of CldU/IdU labeled DNA fibers. (B) Quantification of DNA fork velocity in CD34+ cells isolated from myelodysplastic syndrome (MDS) patient bone marrow with and without splicing factor (sf) mutations as well as healthy controls (young and old). n=441 (non-sf mutated), n=400 (sf mutated), n=279 (young CD34+), n=290 (old CD34+). (C) Schematic illustration of the DNA damage response pathway activated in response to replication stress and representative immunofluorescence images of pRPA (S33) in sf mutated and non-sf mutated MDS CD34+ cells. Scale bar, 5 μm. (D) Schematic illustration of the DNA damage response pathway activated in response to replication stress. (E) Representative immunofluorescence images of pRPA (S33) in healthy young, healthy old, MDS sf mutated, and MDS non-sf mutated CD34+ cells. Scale bar, 5 μm. (F) Quantification of pRPA (S33) mean fluorescence intensity (MFI). n=7 (373 cells, non-sf mutated), n= 13 (522 cells, sf mutated), n=4 (279 cells, young CD34+), n=6 (290 cells, old CD34+). Data are means +/- standard deviation. *P≤ 0.05; *** P≤0.001; n.s.: not significant; CldU: 5-chloro-2′-deoxyuridine; IdU: 5-iodo-2′-deoxyuridine (IdU); A.U: arbitrary units.
Figure 3.
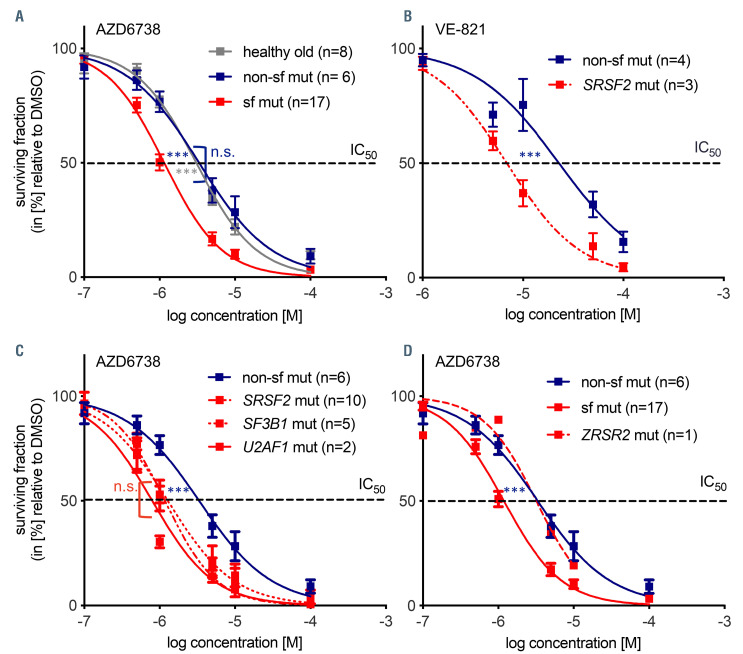
Splicing factor mutated myelodysplastic syndrome CD34+ cells are hypersensitive to ATR inhibition. Dose-response experiments measuring cell viability with Celltiter-Glo® at 48 hours after continuous in vitro drug exposure with ATR inhibitors AZD6738 (n=8 [healthy old], n=6 [non-sf mutated], n=17 [sf mutated]) myelodysplastic syndrome (MDS) patients (A) and VE-821 (n=4 [non-sf mutated], n=3 [sf mutated MDS patients]) (B). (C) Sensitivity of MDS CD34+ cells carrying mutations in different sf genes towards AZD6738; n=6 (non-sf mutated patients), n=10 patients (SRSF2 mutated patients), n=5 (SF3B1 mutated patients) and n=2 (U1AF1 mutated patients). (D) Dose-response experiments measuring cell viability by CellTiter-Glo® 48 hours after continuous drug exposure with ATR inhibitor AZD6738 in an MDS patient cells carrying a ZRSR2 mutation; n=6 (non-sf mutated patients), n=15 (SRSF2, SF3B1, and U2AF1 mutated patients), n=1 (ZRSR2 mutated patient UPN18). IC50: 3.3 μM (non-sf mutated) versus 1.2 μM (sf mutated) versus 3.2 μM (ZRSR2 mutated). *P≤ 0.05; ***P≤0.001; n.s.: not significant; sf: splicing factor.
Increased R-loops cause replication stress in splicing factor mutated myelodysplastic syndrome CD34+ cells
In order to interrogate the hypothesis that elevated levels of R-loops cause replicative stress, we performed fiber assays. This assay evaluates alterations in DNA replication fork dynamics, a known complication of stalled or collapsed forks. As expected, the presence of sf mutations and associated elevated levels of R-loops reduced the rate of DNA replication from 0.404 (range, 0.385-0.423) kb/min to 0.282 (range, 0.272-0.293) kb/min (P<0.001), strongly implying the presence of fork stalling and associated replication stress (Figure 2A and B). In comparison, healthy young CD34+ cells showed a median fork rate of 0.381 (range, 0.359-0.403) kb/min, and a median fork rate of 0.4206 (range, 0.388-0.453) was determined in healthy old (median age: 81 years, range, 76-88) CD34+ cells (Figure 2A to C).
Elevated R-loops cause ATR pathway activation
As a consequence of replication stress and fork stalling, sf mutated myelodysplastic syndrome CD34+ cells showed increased activation of the ATR pathway as demonstrated by increased staining with pRPA (S33), a well-established downstream target of ATR kinase (Figure 2D to F) (mean MFI [A.U.]: 62 [non-sf mutated] vs. 110 [sf mutated]; P<0.001). This dependence on proper ATR function to repair replication stress-induced DNA damage represents a vulnerability presented primarily by sf mutated cells in a synthetic lethal fashion. ATR inhibitors, particularly AZD6738, are tested in early phase clinical trials, primarily for the treatment of solid cancers. For this reason, we next investigated whether inhibition of ATR in MDS patients could constitute a possible therapeutic target to selectively affect sf mutated CD34+ cells.
Splicing factor mutated myelodysplastic syndrome CD34+ cells are hypersensitive towards ATR inhibition
In order to determine the sensitivity of MDS CD34+ cells to ATR inhibitors we treated MDS CD34+ cells with two different ATR inhibitors (VE-821 and AZD6738, respectively) for 48 hours before cell viability assessment by CellTiter-Glo® assay. The presence of a sf mutation resulted in enhanced sensitivity associated with a reduced half maximal inhibitory concentration IC50 in MDS patient CD34+ cells (IC50=3.1 [healthy old] vs. 3.3 μM [MDS non-sf mutated] vs. 1.2 μM [MDS sf mutated]) (Figure 3A and B). We did not detect significant differences in IC50 values between the three most common sf hotspot mutations (IC50 SF3B1=1.2 [n=5] vs. SRSF2=1.2 [n=10], vs. U2AF1=0.8 μM [n=2]) (Figure 3C). Interestingly, the IC50 was unchanged in one MDS patient (UPN18) with a ZRSR2 mutation (3.3 [non-sf mutated] vs. 3.2 μM [ZRSR2]) (Figure 3D). This is consistent with the absence of increased levels of R-loops in this patient (Figure 1D).
Figure 4.
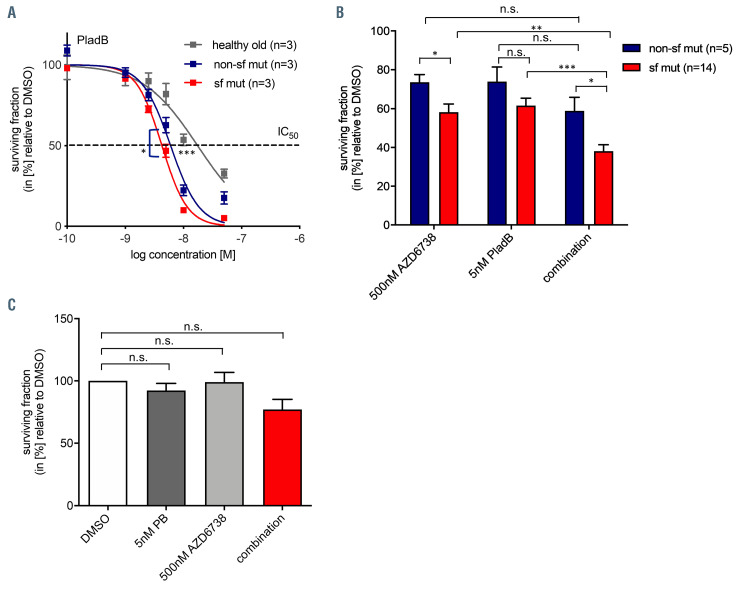
Combinatorial effect of ATR inhibition and splicing modulation on splicing factor mutated and non-mutated myelodysplastic syndrome CD 34+ cells. (A) Dose-response experiment measuring cell viability at 48 hour after continuous drug exposure towards Pladienolide B; n=3 (healthy old), n=3 (nonsf mutated), n=3 (sf mutated) myelodysplastic syndrome (MDS) patients. (B) Cells were exposed to 500 nM AZD6738, 5 nM Pladienolide B, or a combination of both, and viability was determined by CellTiter-Glo® 48 hours after continuous drug exposure in vitro; n=5 (non-sf mutated patients), n=14 (sf mutated patients). (C) Combinatorial effect of ATR inhibition and splicing modulation on ZRSR2 mutated MDS CD34+ cells (UPN48) (n=3 technical replicates of one patient). Data are means +/- standard deviation. *P≤0.05; ** P≤0.01; ***P≤0.001; n.s.: not significant; sf: splicing factor; PladB: Pladienolide B; UPN: unique patient number.
Simultaneous ATR and splicing modulation show augmented efficacy in splicing factor mutant myelodysplastic syndrome CD34+ cells
Splicing modulators, such as PladB or derivates such as H3B-8800 are under investigation for the treatment of sf mutated MDS.26,29 We confirmed that PladB as a single substance had increased efficacy in MDS cells as compared to age-matched healthy CD34+ cells and that there was a slight, but significant increase in sensitivity in sf mutated versus non-sf mutated MDS CD34+ cells (IC50= 17.5 [healthy old] vs. 6.9 [non-sf mutated] vs. 4.3 μM [MDS sf mutated] [MDS sf mutated vs. non-sf mutated; P=0.0175], [healthy old vs. MDS; P<0.001]) (Figure 4A). Based on previous studies showing enhanced efficacy of splicing modulators in sf mutated cells of hematologic malignancies,26 we evaluated this concept, also in combination with ATR inhibition. We tested treatment with PladB in combination with AZD6738 in n=19 primary MDS samples (Figure 4B). Despite high inter-patient variability, we observed augmented activity of the combination of 500 nM AZD6738 and 5 nM PladB as compared to the single substances when analysing cytotoxicity in CellTiter-Glo® assays (Figure 4B). However, calculation of combination indexes by CompuSyn software (version 1.0; ComboSyn, Inc. Paramus, NJ, USA) did not result in significant additive or synergistic effects. Of note, combination of ATR inhibitor AZD6738 with an established standard therapy such as 5-Azacytitidine did not reveal additive or synergistic effects either. Again, a tendency for relatively lower cell viability was observed in the sf mutated group compared to the non-sf mutated group (P=0.20) (Online Supplementary Figure 2A to C). Interestingly, CD34+ cells of one available ZRSR2 mutated patient (UPN48) did neither respond to AZD6738 treatment alone nor to AZD6738 treatment in combination with PladB (Figure 4C).
Figure 5.
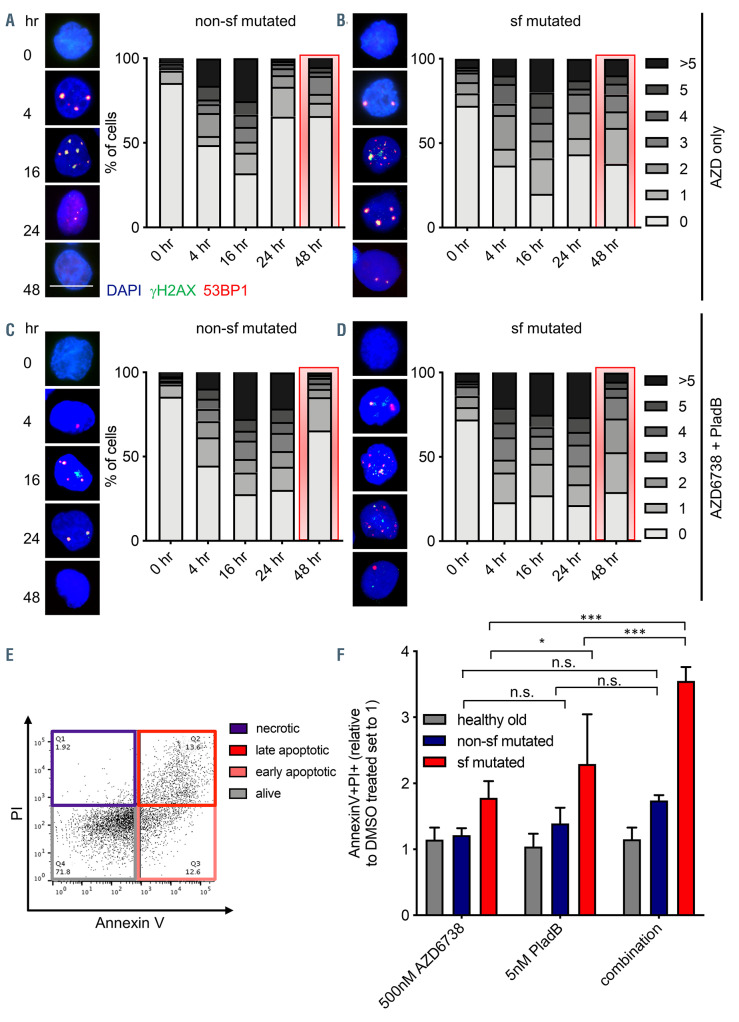
Unresolved DNA damage leads to elevated cytotoxicity in splicing factor mutated myelodysplastic syndrome CD34+ cells treated with AZD6738 and Pladienolide B. (A to D) Representative immunofluorescence images of DNA repair kinetics following exposure to 500 nM AZD6738, alone or in combination with 5 nM Pladienolide B in cultured splicing factor (sf) not mutated (non-sf mutated) (A and B) or mutated (C and D) myelodysplastic syndrome (MDS) CD34+ cells (n=2 [non-sf mutated] and n=3 [sf mutated] independent experiments, respectively; 25-130 cells were analyzed per timepoint and experiment). For comparison reasons, the 48-hour timepoints are highlighted in red. Scale bar, 5 μm. 0, 1, 2, 3, 4, 5, >5 indicate the number of H2AX/53BP1 foci. (E) Representative flow cytometry result to quantify apoptosis rates using AnnexinV/propidium iodide (PI). (F) Quantification of AnnexinV+ /PI+ cells in response to in vitro exposure to 500 nM AZD6738, 5 nM Pladienolide B (PladB), or a combination of both in sf mutated and non-sf mutated MDS CD34+ cells (n=2-7 independent experiments). Data are means +/- standard deviation. *P≤0.05; ***P≤0.001; n.s.= not significant; hr: hour.
Figure 6.
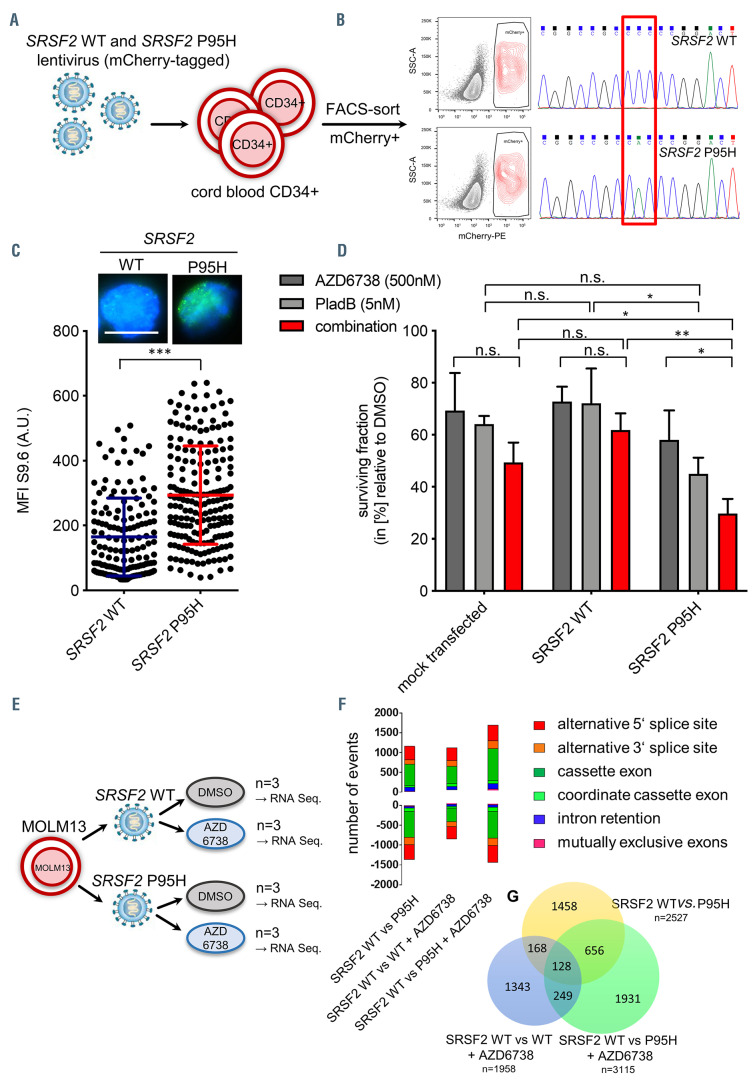
Mechanistic validation that hypersensitivity towards ATR inhibition is driven by the presence of the splicing factor mutations and increased induction of aberrant splicing events. (A) Experimental procedure of lentivirus-mediated overexpression of SRSF2 wild-type (WT) and SRSF2 P95H in CD34+ isolated form cord blood. (B) Sequencing of the SRSF2 locus in transduced and sorted mCherry+ cells. (C) Representative immunofluorescence images and quantification of R-loops using antibody S9.6 (Kerafast) in mCherry+ cells (n=2 independent experiments; cells analyzed =150 [SRSF2 WT]/ 193 [SRSF2 P95]). (D) Cytotoxicity tests of AZD6738 alone and in combination with Pladienolide B (PladB) in transduced and sorted mCherry+ cells. Cell viability was determined by Celltiter-Glo® after 48 hours (n=3 independent experiments). (E) Experimental procedure of RNA sequencing experiment in order to determine aberrant splicing events in SRSF2 WT or P95H MOLM13 cells in the presence or absence of AZD6738. (F) Number of aberrantly spliced events decreased in SRSF2 WT MOLM13 cells treated with AZD6738. (G) Venn diagram demonstrating total numbers of splicing events in SRSF2 WT in comparison to SRSF2 P95H treated with dimethyl sulfoxide (DMSO), SRSF2 WT treated with AZD6738, and SRSF2 P95H treated with AZD6738. Data are means +/- standard deviation. *P≤0.05; ***P≤0.001; n.s.: not significant; A.U: arbitrary units.
ATR inhibition causes elevated levels of DNA damage associated with cell cycle arrest and cell death
Our data demonstrated that increasing concentrations of ATR inhibitors led to a loss in cell viability likely due to the fact that replication stress-induced DNA damage is no longer efficiently repaired. In order to gain a deeper understanding of the underlying molecular mechanisms we treated non-sf mutated and sf mutated MDS CD34+ cells with 500 nM AZD6738 alone or in combination with 5 nM PladB and followed the DNA damage response dynamics over a period of 48 hours (h). As shown in Figure 5, treatment with AZD6738 led to the accumulation of H2AX/53BP1 foci within 4-16 h, which were then largely repaired within 24-48 h post-exposure. However, in contrast to non-sf mutated CD34+ cells (Figure 5A), sf mutated MDS CD34+ not only showed delayed repair kinetics but they also retained levels of persistent DNA damage after 48 h (Figure 5B, 48-h bar graphs are highlighted in red). This was particularly evident in sf mutated cells when treated with both AZD6738 and PladB (Figure 5C and D). The inability to cope with increased levels of DNA damage upon both AZD6738 and PladB treatment was further reflected in increased apoptosis rates (Figure 5E and F) specifically in sf mutated cells. Results for vehicle and PladB only treated sf mutated and non-sf mutated cells are depicted in the Online Supplementary Figure S3A to D and show that even at baseline, sf mutated CD34+ cells have slightly elevated numbers of γH2AX/53BP1 foci as compared to non-sf mutated cells in line with previous publications. 17,18
Mechanistic validation that hypersensitivity towards ATR inhibition is driven by the presence of the splicing factor mutations and increased induction of aberrant splicing events
The patient-derived CD34+ cells that we used in our study not only contained sf mutations but also other MDS typical recurrent mutations (Online Supplementary Table S1). In order to rule out confounding effects of other genetic alterations on ATR inhibitor sensitivity, we overexpressed WT and mutant SRSF2 in CD34+ cord blood cells, which we assumed not to carry any MDS-associated mutations (Figure 6A and B). As shown in Figure 6C we confirmed that increased levels of R-loops were directly associated with the presence of the sf mutation (mean MFI= 164.5. [SRSF2 WT] vs. 193.8 [SRSF2 P95]; P<0.001). In addition, SRSF2 P95H expressing CD34+ cells exhibited a reduced sensitivity to ATR inhibition alone and in combination with PladB (Figure 6D) similar to the levels seen in sf mutated MDS patient cells. This confirmed the direct association of the presence of sf mutations and elevated levels of R-loops and ATR hypersensitivity. As the presence of sf mutations has been shown to enhance alternative splicing,8-15 we evaluated whether ATR inhibition affects such alternative splicing events by performing RNA sequnecing. MOLM13 cells were transduced with and SRSF2 WT and P95H- expressing lentiviral particles and treated with AZD6738 or or dimethyl sulfoxide (DMSO) (Figure 6E; Online Supplementary Figure S4A). Of the n=4 experimental groups, SRSF2 WT (+ DMSO) was taken as reference and tested against SRSF2 WT + AZD6738, SRSF2 P95H + DMSO and SRSF2 P95H + AZD6738. The stability of the vector integration was confirmed within the RNA sequencing experiment (Online Supplemental Figure S4F). We found that n=1958 events in SRSF2 WT + AZD6738, n=2527 events in SRSF2 P95H, and n=3115 events in the SRSF2 P95H + AZD6738 group were differentially spliced (Figure 6F and G). As expected, the presence of SRSF2 P95H significantly increased differential splicing events. However, treatment of SRSF2 WT cells with ATR inhibitor AZD6738 also resulted in increased numbers of differential splicing events. SRSF2 P95H treated with AZD6738 showed the highest number of alternative splicing changes. Furthermore, we observed that the number of increased and decreased splicing events was approximately the same in all three experimental groups (Figure 6F) as were the patterns of splicing alterations (e.g., cassette exon, alternative 5’/3’ splice site) (Online Supplementary Figure S4G and I). Of note, only approximatly one third (45% and 33%, repectively) of events were shared between SRSF2 P95H and SRSF2 P95H + AZD6738 (Figure 6G), suggesting additional effects to aberrant splicing, beyond the presence of the sf mutation (Online Supplemental Figure S4I). Overall, the results suggest that ATR inhibition in combination with an sf mutation enhance differential splicing even further.
Discussion
Somatic heterozygous mutations in SF genes such as U2AF1, SRSF2, and SF3B1 are the most frequently occurring mutations in MDS. Their high allele frequencies in clinical samples suggest that they often affect the founding or dominating clones of the disease and implicate RNA splicing alterations to be critical in disease initiation and progression. Due to their high frequency in MDS and dominating variant allele frequencies they are attractive targets for the development of new molecularly driven therapeutic strategies. Elevated levels of R-loops have emerged as a possible unifying pathomechanism common to U2AF1 S34 and SRFS2 P95 mutations in MDS.17
The association of sf mutations particularly in SRSF2 and U2AF1 with increased R-loop formation had previously been demonstrated in cell line models.17,22 Furthermore, a recent study has demonstrated increased R-loop accumulation in induced pluripotent stem cells and n=3 primary MDS patient samples carrying an SF3B1 mutation.18 Our study for the first time confirms this connection in a large representative clinical cohort of primary cells of MDS patients and therewith takes this finding into a pre-clinical setting. We screened a total of 28 MDS patient samples (ten non-sf mutated, 18 sf mutated [SRSF2=10, SF3B1=7; ZRSR2=1]) for their levels of Rloops in purified CD34+ BM cells, a cell population known to be enriched for MDS disease initiating cells. These results corroborate the hypothesis that accumulation of R-loops might indeed constitute a common pathophysiological mechanism in MDS patients with sf mutations.
In line with previous studies on cell lines and as a consequence of elevated levels of R-loops we found highly activated replication stress signaling in sf mutated MDS patient CD34+ cells. This let us hypothesize that sf mutated BM cells from MDS patients may be dependent on proper ATR function in order to counteract replication stress-induced DNA damage. As a consequence, interfering with ATR-mediated DNA repair by ATR inhibitors VE-821 and AZD6738 causes mutated cells to no longer be able to tolerate R-loops driven replication stress causing them to cease their cell cycle and undergo apoptosis. This synthetic lethal connection between increased levels of R-loops and ATR inhibition makes therapeutic treatment possible without interfering too much with healthy hematopoiesis. While high doses of ATR inhibitors also impaired viability of non-sf mutated MDS CD34+ cells, careful titration allowed us to identify doses of ATR inhibitors that were still tolerated by CD34+ cells of nonsf mutated MDS patients and cells from healthy individuals. This suggests that at least in vitro there seems to be a therapeutic window for mutation-specific therapeutic targeting. Our in vitro experimental results support this hypothesis in an elaborate pre-clinical patient sample cohort of n=24, showing that hematopoietic cells with sf mutations become sensitive to even low doses of ATR inhibitor. It remains to be evaluated how this concept can be applied in the in vivo situation and ultimately in humans. We specifically chose AZD6738 as this substance has already moved into clinical trials. In addition, certain co-occuring mutations may affect response to ATR inhibition. Mutations interfering with the DNA damage response such as those affecting TP53, for instance, may predict resistance. Given the fact that ATR inhibition affects the DNA damage response we investigated the influence of TP53 mutational status on the response towards AZD6738 (Online Supplemental Figure S5A). We did not detect statistically significant effects of the presence of mutated TP53 on the cells’ sensitivity towards AZD6738 in either the sf mutated or the non-sf mutated cohort. However, there was only one sample with a low level mutation of TP53 in the sf mutated group. Since the role of TP53 in the DNA damage response is primarily downstream of ATM signaling its exact impact on ATR inhibitor response needs to be carefully investigated in further studies. In order to assess any confounding effects of co-occurring mutations, our sample cohort was either analyzed by NGS panel sequencing (n=9) of recurrently mutated genes in myeloid diseases or by germline controlled WES (n=44). However, we did not discover molecular markers alleviating the sf mutation-mediated sensitivity against ATR inhibition. Mutations in ZRSR2 might be an exception among the sf mutations since the two patients carrying ZRSR2 mutations, which we were able to analyze, did neither display elevated levels of R-loops nor did they respond to ATR inhibition with the same sensitivity as the other sf mutations. The pathophysiological mechanism underlying ZRSR2 mutations in MDS may be different compared to common mutations in the other sf (SF3B1, U2AF1, and SRSF2) and possibly related to its aberrant retention of U12-type introns30 rather than the major U2-dependent spliceosome.
Another confounding factor in our study could be due to an enrichment of higher risk IPSS-R categories in the non-sf mutated group mediating cell-intrinsic or, -extrinsic effects on ATR inhibitor responsiveness. The reason for this imbalance may be a natural bias in the distribution of sf mutations to be enriched in IPSS-R31 lower risk MDS categories. For SF3B1 this has been shown multiple times32,33 and is, last but not least, reflected in the WHO 2016 classification of myeloid neoplasms. Since our study includes a total of 12 patients with SF3B1 mutations, this per se accounts for an enrichment of lower risk categories in the sf mutated group. The absence of a sf mutation probably vice versa leads to an enrichment of higher risk categories in the non-sf mutated group.32 We did not have enough samples to artificially equalize this natural bias in differential mutational profiles among MDS risk groups. However, we have attempted to perform a subgroup analysis with matched IPSS-R risk status (n=4/4; Online Supplementary Figure S5B and C). In this analysis, the sf mediated hypersensitivity against ATR inhibition was retained.
In a separate set of experiments we evaluated the effect of ATR inhibition on alternative splicing, which is known to be increased in sf mutated cells.8-15 Interestingly, treatment of non-sf mutated cells with AZD6738 resulted in elevated numbers of differentially spliced events to a similar extent as seen by ectopically expressed SRSF2 P95. The exact mechanism for this is unknown, however, splicing is inherently linked to transcription, 34 a process known to be intertwined with DNA repair.35,36 As such, increased DNA damage induces alterations in RNA splicing of transcripts involved in genome stability maintenance.37,38 Furthermore, we found that the combination of ATR inhibitor treatment of SRSF2 mutated cells resulted in the highest number and unique differential splicing alterations. We hypothesize that the elevated levels of R-loops induced by the mutation leads to further shifts in splicing factors and RNA binding proteins likely to prevent RNA-induced genome instability.39,40
Given the fact that splicing modulators have been shown to confer therapeutic activity on sf mutated cells,26 we hypothesized that combining low-dose AZD6738 with such a compound may further enhance the therapeutic window in sf mutated cells. We therefore performed experiments with PladB, which augmented the sensitivity of mutated cells towards ATR inhibitors. However, the combination failed to reach additive or synergistic effects. This could be a reflection of a possible lack of efficacy of splicesome modulators in MDS, which was also observed in the first clinical evaluation of H3B-8800 in myeloid malignancies.29 In conclusion, our study shows increased levels of R-loops in a large cohort of primary MDS CD34+ cells that carry sf mutations (SRSF2, SF3B1, and U2AF1), which results in high sensitivity towards pharmacologic ATR inhibition. Our data strengthen the preclinical rationale for this novel therapeutic strategy in MDS. A recently initiated single-arm phase Ib clinical trial is assessing safety and seeking preliminary evidence for efficacy for AZD6738 in patients with MDS and CMML, who have failed to respond to first-line treatments (clinicaltrials gov. Identifier: NCT03770429). R-loops could be potential novel biomarkers for response to this treatment. Future efforts should focus on combination strategies including ATR inhibition.
Supplementary Material
Funding Statement
Funding: This work was supported by funds from the Deutsche Forschungsgemeinschaft (DFG, project FL1054/1-1 and NO817/5-2), funds from the “Deutsche Krebshilfe” (Project 70113953), funds from the Gutermuth Foundation, funds from the HW & J. Hector fund (Project M83), Baden Wuerttemberg, and the Dr. Rolf M. Schwiete Fund (Project 20/2016), Mannheim. DN is an endowed Professor of the German José- Carreras-Stiftung (DJCLS H 03/01).
References
- 1.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578-3584. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 3.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mian SA, Rouault-Pierre K, Smith AE, et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat Commun. 2015;6:10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mossner M, Jann JC, Wittig J, et al. Mutational hierarchies in myelodysplastic syndromes dynamically adapt and evolve upon therapy response and failure. Blood. 2016;128(9):1246-1259. [DOI] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616-3627; quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pellagatti A, Roy S, Di Genua C, et al. Targeted resequencing analysis of 31 genes commonly mutated in myeloid disorders in serial samples from myelodysplastic syndrome patients showing disease progression. Leukemia. 2016;30(1):247-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiozawa Y, Malcovati L, Galli A, et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun. 2018;9(1):3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilagan JO, Ramakrishnan A, Hayes B, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25(1):14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim E, Ilagan JO, Liang Y, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komeno Y, Huang YJ, Qiu J, et al. SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative pre-mRNA splicing. Mol Cell Biol. 2015;35(17):3071-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Przychodzen B, Jerez A, Guinta K, et al. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122(6):999-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Lieu YK, Ali AM, et al. Diseaseassociated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015; 112(34):E4726-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yip BH, Steeples V, Repapi E, et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J Clin Invest. 2017;127(9):3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellagatti A, Armstrong RN, Steeples V, et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood. 2018;132(12):1225-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi P, Halene S, Abdel-Wahab O. How do messenger RNA splicing alterations drive myelodysplasia? Blood. 2017;129(18):2465-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Chen JY, Huang YJ, et al. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol Cell. 2018;69(3):412-425.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh S, Ahmed D, Dolatshad H, et al. SF3B1 mutations induce R-loop accumulation and DNA damage in MDS and leukemia cells with therapeutic implications. Leukemia. 2020;34(9):2525-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16(10):583-597. [DOI] [PubMed] [Google Scholar]
- 20.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46(2):115-124. [DOI] [PubMed] [Google Scholar]
- 21.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16(1):2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen HD, Leong WY, Li W, et al. Spliceosome mutations induce R loop-associated sensitivity to ATR inhibition in myelodysplastic syndromes. Cancer Res. 2018;78(18):5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11(11): 847-859. [DOI] [PubMed] [Google Scholar]
- 24.Lee SC, Dvinge H, Kim E, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obeng EA, Chappell RJ, Seiler M, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016; 30(3):404-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seiler M, Yoshimi A, Darman R, et al. H3B- 8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat Med. 2018;24(4):497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klusmann I, Rodewald S, Muller L, et al. p53 activity results in DNA replication fork processivity. Cell Rep. 2016;17(7):1845-1857. [DOI] [PubMed] [Google Scholar]
- 28.Woll PS, Kjallquist U, Chowdhury O, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25(6):794-808. [DOI] [PubMed] [Google Scholar]
- 29.Steensma DP, Wermke M, Klimek VM, et al. Results of a clinical trial of H3B-8800, a splicing modulator, in patients with myelodysplastic syndromes (MDS), acute myeloid leukemia (AML) or chronic myelomonocytic leukemia (CMML). Blood. 2019;134(Suppl 1):S673-673. [Google Scholar]
- 30.Madan V, Kanojia D, Li J, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makishima H, Yoshizato T, Yoshida K, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017; 49(2):204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malcovati L, Karimi M, Papaemmanuil E, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126(2):233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14(3):153-165. [DOI] [PubMed] [Google Scholar]
- 35.Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell. 2013;52(3):291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci. 2010;35(6):333-338. [DOI] [PubMed] [Google Scholar]
- 37.Cloutier A, Shkreta L, Toutant J, Durand M, Thibault P, Chabot B. hnRNP A1/A2 and Sam68 collaborate with SRSF10 to control the alternative splicing response to oxaliplatin- mediated DNA damage. Sci Rep. 2018;8(1):2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shkreta L, Toutant J, Durand M, Manley JL, Chabot B. SRSF10 connects DNA damage to the alternative splicing of transcripts encoding apoptosis, cell-cycle control, and DNA repair factors. Cell Rep. 2016;17(8):1990-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018;23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.