Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common non- Hodgkin lymphoma and is characterized by a remarkable heterogeneity with diverse variants that can be identified histologically and molecularly. Large-scale gene expression profiling studies have identified the germinal center B-cell (GCB-) and activated B-cell (ABC-) subtypes. Standard chemo-immunotherapy remains standard front-line therapy, curing approximately two thirds of patients. Patients with refractory disease or those who relapse after salvage treatment have an overall poor prognosis highlighting the need for novel therapeutic strategies. Transducin b-like protein 1 (TBL1) is an exchange adaptor protein encoded by the TBL1X gene and known to function as a master regulator of the Wnt signaling pathway by binding to β-CATENIN and promoting its downstream transcriptional program. Here, we show that, unlike normal B cells, DLBCL cells express abundant levels of TBL1 and its overexpression correlates with poor clinical outcome regardless of DLBCL molecular subtype. Genetic deletion of TBL1 and pharmacological approach using tegavivint, a first-in-class small molecule targeting TBL1 (Iterion Therapeutics), promotes DLBCL cell death in vitro and in vivo. Through an integrated genomic, biochemical, and pharmacologic analyses, we characterized a novel, β-CATENIN independent, post-transcriptional oncogenic function of TBL1 in DLBCL where TBL1 modulates the stability of key oncogenic proteins such as PLK1, MYC, and the autophagy regulatory protein BECLIN-1 through its interaction with a SKP1-CUL1-F-box (SCF) protein supercomplex. Collectively, our data provide the rationale for targeting TBL1 as a novel therapeutic strategy in DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) comprises the majority of adult non-Hodgkin lymphoma cases worldwide and is traditionally categorized, based on the molecular profile, into two major subtypes, germinal center B-cell (GCB) and activated B-cell (ABC) lymphoma, mirroring the cell of origin as well as reflecting the differences in clinical outcomes.1-3 DLBCL is curable in 40-60% of the cases following standard front-line immuno-chemotherapy.4 However, patients with refractory disease, those who relapse after salvage chemotherapy and autologous stem cell transplant or chimeric antigen receptor T-cell therapy have an overall poor prognosis highlighting the need for novel therapeutic approaches. 4-6
Transducin b-like protein 1 (TBL1), encoded by the TBL1X gene, is an adaptor protein initially identified as a core component of the co-repressor silencing mediator for retinoid and thyroid hormone receptor (SMRT)– nuclear receptor co-repressor (N-COR) complex.7-9 SMRT/N-COR form a complex with TBL1, BCL6, and other proteins leading to transcriptional repression of target genes through HDAC3-mediated H3K9 deacetylation. 7,10 Subsequently, TBL1 was found to be a key player in enhancing the canonical Wnt signaling pathway by directly binding to β-CATENIN and recruiting it to the promoter of Wnt target genes (MYC, BIRC5, CCND1) to promote uncontrolled cell proliferation and survival.9,11,12 Previous work also showed that TBL1 binds to a S-phase kinase-associated protein 1 (SKP1)/Cullin-1(CUL1)/F-box protein complex (SCF complex) and protects b- CATENIN from proteasomal degradation.13,14 Tegavivint (Iterion Therapeutics) is a first-in-class small molecule compound that binds directly to the N-terminal domain of TBL1 and disrupts TBL1/β-CATENIN interaction leading to β-CATENIN degradation and subsequent downstream inhibition of the Wnt transcriptional program.15 Tegavivint has shown significant activity in preclinical models of acute myeloid leukemia (AML) and multiple myeloma (MM).15-17
Here, we report that, unlike normal B cells, DLBCL cells express abundant levels of TBL1 regardless of the molecular subtype and its overexpression correlates with poor clinical outcome. We demonstrate that both TBL1 genetic deletion and pharmacologic targeting with tegavivint induce DLBCL cell death in vitro and in vivo. More importantly, we show that β-CATENIN is dispensable for DLBCL cell survival and plays a minimal role in the tegavivint-mediated attenuation of known Wnt targets. Through an integrated genomic, biochemical, and pharmacologic approach we detail the crucial role played by TBL1 in controlling the stability of critical regulatory proteins involved in cell proliferation and autophagy induction. Our findings provide the rational for targeting TBL1 in DLBCL.
Methods
Cell lines and primary samples
DLBCL cell lines were purchased from ATCC and identity confirmed via short tandem repeats profiling (University of Arizona).18 Peripheral blood and tonsils from healthy donors and peripheral blood, bone marrow samples, and lymph nodes from DLBCL patients were obtained following written informed consent under a protocol approved by the Institutional Review Board of OSU in accordance with the Declaration of Helsinki.
Tissue microarray and immunohistochemistry
Details on tissue microarray (TMA) preparation and immunohistochemistry (IHC) are presented in the Online Supplementary Appendix.
Gene knock-down using short hairpin RNA and CRISPRCas9 system
TBL1X, CTNNB1, and CAND1 were genetically silenced using pLKO.1 short hairpin RNA (shRNA) vectors (Mission® shRNA, Sigma). Details are presented in the Online Supplementary Appendix.
Immunoblot and co-immunoprecipitation assay
Details are presented in the Online Supplementary Appendix (see Online Supplementary Table S4).
RNA extraction and real-time polymerase chain recation
Details are presented in the Online Supplementary Appendix (see Online Supplementary Table S5).
Chromatin-immunoprecipitation (ChIP)-sequencing and ChIP-polymerase chain reaction
For chromatin-immunoprecipitation-sequencing (ChIP-Seq) assays, 5x106 Pfeiffer and Riva cells treated with either vehicle control or tegavivint for 12 hours were processed according to Active Motif protocols.19 All raw sequence data are available in the Gene Expression Omnibus (GSE148232). ChIP-polymerase chain reaction (ChIP-PCR) was performed according to standard procedures. Details are presented in the Online Supplementary Appendix.
Confocal microscopy, proximity ligation assay, cytotoxicity assay and transmission electron microscopy
Details are presented in the Online Supplementary Appendix.
Animal studies
Animal studies were approved by the OSU Institutional Animal Care and Use Committee. Experimental details for the animal studies with tegavivint are included in the result section. For the in vivo TBL1X knock-down experiment, luciferase and Cas9 positive Riva cells were transduced with either doxycycline-inducible single guide RNA (sgRNA) targeting TBL1X or vector control only using a lentiviral luciferase vector previously described.20 20×106 of engineered Riva cells were engrafted into NSG mice via tail vein injection. On day 23, the tumor burden was visualized after intraperitoneal administration of 200 mL of D-luciferin potassium salt (Caliper Life Sciences) in phospate buffered saline using in vivo live-imaging system (IVIS) (Perkin Elmer Inc.). Additional details are presented in the Online Supplementary Appendix.
Statistical analysis
All the experiments include at least three independent replicates and results were expressed as the mean ± standard error of the mean (SEM). For correlated data such as patient cells treated with different conditions, paired t-tests for two group comparisons or linear mixed effects models for two or more group comparisons were used for analysis. Otherwise, for independent data, two-sample t-tests and analysis of variance (ANOVA) were used for comparisons between two groups and among more than two groups respectively. Survival analysis was performed using logrank test or Cox proportional hazard regression models. P-values <0.05 were considered significant after Holm’s procedure adjustment for multiple comparisons.
Results
TBL1 is overexpressed and correlates with clinical outcome in diffuse large B-cell lymphoma regardless of its molecular subtype
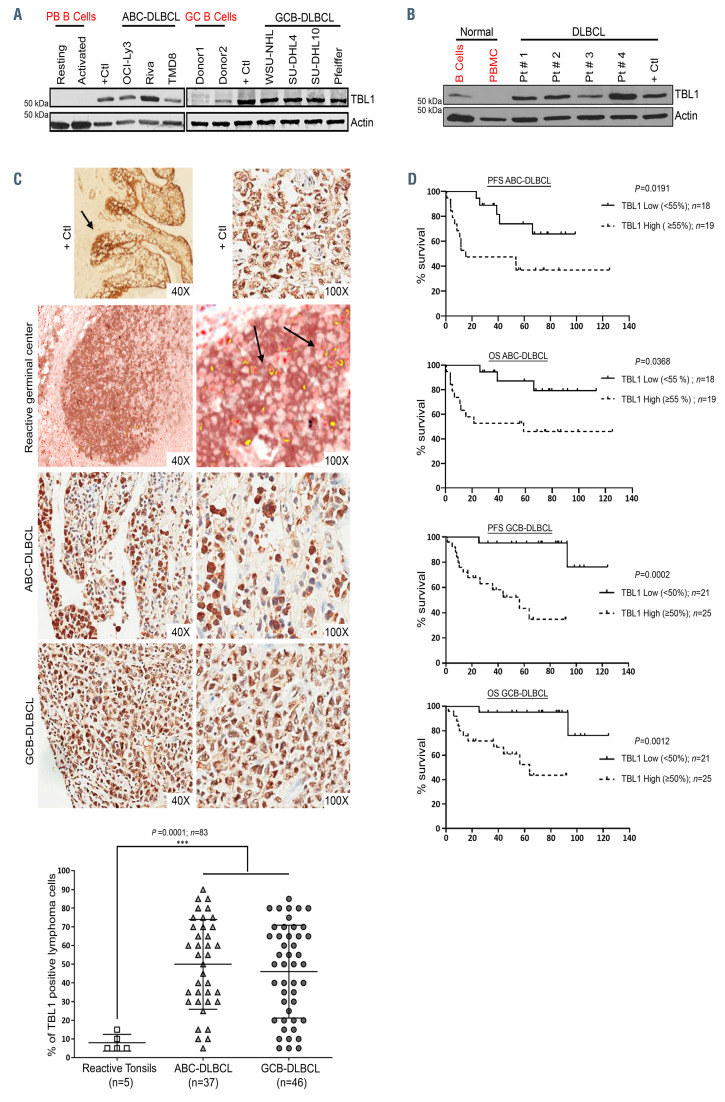
Immunoblot analysis showed abundant levels of TBL1 protein expression in seven DLBCL cell lines regardless of the molecular subtype (four GCB and three ABC, Figure 1A) and in primary DLBCL patient samples (n=4) (Figure 1B) (patient characteristics are listed in the Online Supplementary Table S1) compared to normal peripheral blood B cells and CD10+CD19+ sorted germinal center B cells from reactive tonsils (sorting strategy in the Online Supplementary Figure S1). In order to validate our findings, we performed IHC staining of TBL1 in tissue samples obtained from 83 primary de novo DLBCL patients uniformly treated with front-line R-CHOP (46 GCB- and 37 ABC-subtype, Figure 1C; Online Supplementary Figures S2 to S4; patient characteristics are listed in the Online Supplementary Tables S2 and S3). In our cohort, the percentage of DLBCL cells with strong (brown) TBL1 staining (similar to the intensity seen in the positive controls) ranged from 5% to 90% (GCB-DLBCL: mean percentage of positive cells 46%; range, 5-85%; ABC-DLBCL: mean percentage 49%; range, 5-90%) compared to 5% to 15% with a mean of 8% in tonsils with reactive lymphoid hyperplasia (n=5; P=0.0001). Normal GCB were identified by co-staining with CD10 (brown) and TBL1 (red) and coexpression showed in yellow (Figure 1C) or with TBL1 single staining (Online Supplementary Figure S2).21 No significant difference in the TBL1 expression patterns was seen between GCB- and ABC-DLBCL (dot plot in Figure 1C). Using the median percentage of TBL1 positive lymphoma cells (50% for GCB-DLBCL and 55% for ABC-DLBCL), patients were classified into high or low expressors. Importantly, a high percentage of TBL1 positive DLBCL cells was associated with significantly inferior progression free survival (PFS) and overall survival (OS) in both ABCand GCB-DLBCL (Figure 1D). However, the negative prognostic impact of TBL1 positivity on PFS/OS did not persist after adjusting for International Prognostic Index (IPI) and IPI factors in a Cox multivariate model.
TBL1 is critical for diffuse large B-cell lymphoma cell survival
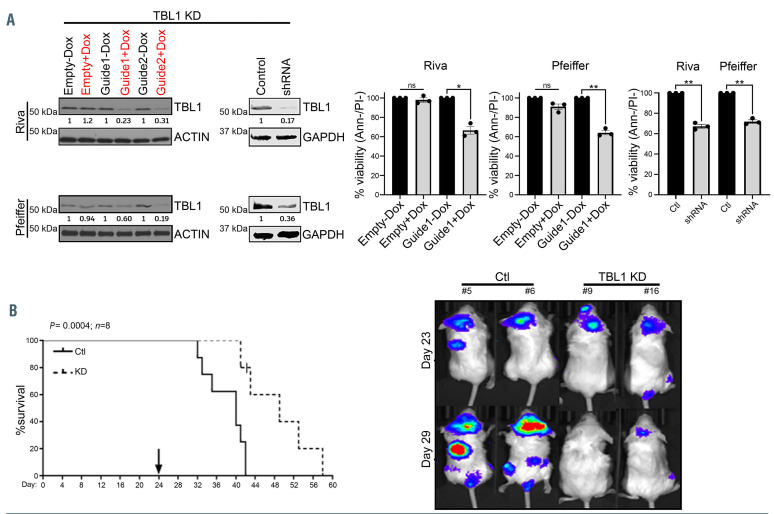
In vitro TBL1 knock-down using either a doxycyclineinducible CRISPR-Cas9 system or a TBL1X-specific shRNA was associated with significant DLBCL cell death regardless of the molecular subtype (Figure 2A) (P<0.05). Next, luciferase positive Riva cells were stably transduced with Cas9 and a doxycycline-inducible sgRNA targeting TBL1X exon 2 (guide 1). Prior to engraftment, efficient TBL1 knock-down after doxycycline induction was verified in vitro (Online Supplementary Figure S5). NSG mice (n=8 per group) received doxycycline (100 mg/kg) daily via oral gavage starting at day 24 after successful engraftment was verified by IVIS on day 23. TBL1 knock-down induced tumor regression after 5 days of doxycycline treatment as documented by in vivo imaging system (IVIS) and significantly prolonged the survival of these animals compared to control [median OS 48 days vs. 38 days, respectively, (P=0.0004)] (Figure 2B).
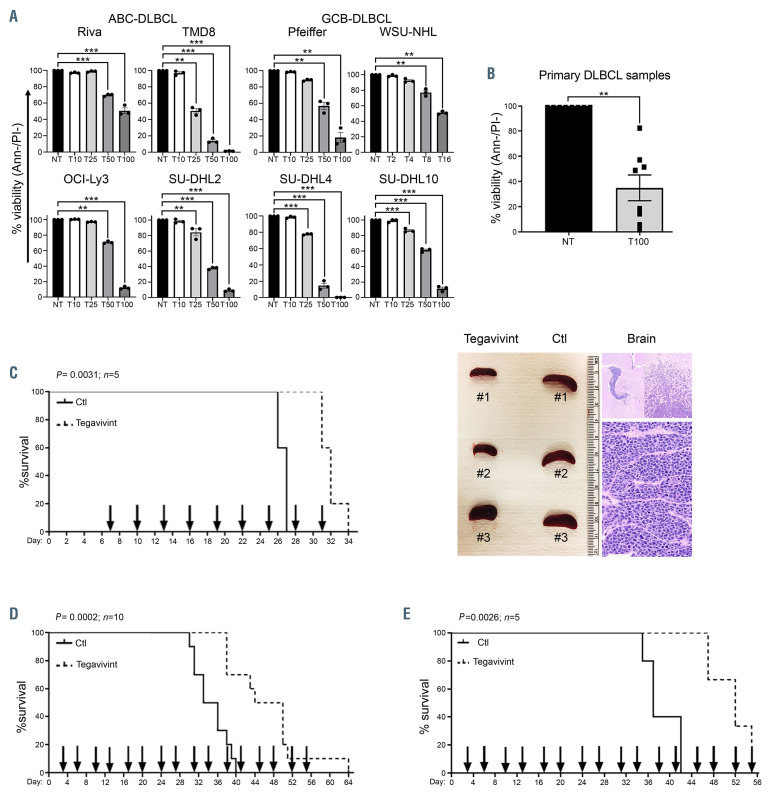
Next, we tested the in vitro cytotoxicity of tegavivint. As shown in Figure 3A and B, 24 hours treatment with tegavivint (2-100 nM) induced significant DLBCL cell death in eight DLBCL cell lines (four GCB and four ABC) and primary DLBCL samples (n=8; patient characteristics are listed in the Online Supplementary Table S1) compared to the vehicle control (P<0.001). Importantly, cytotoxic studies on normal resting and activated immune cell subsets from the peripheral blood of healthy donors (n=3) incubated for 24 hours with the highest concentration of tegavivint (100 nM) showed no significant toxicity on B and natural killer (NK) cells while marginal toxicity (<10% decrease in viability) was noted in activated (P=0.0287) but not resting T cells, consistent with their low TBL1 expression (Online Supplementary Figure S6A and B). In support of a selective TBL1-mediated DLBCL cell death, tegavivint lost the majority of its cytotoxic activity in DLBCL cells when TBL1 was efficiently knocked-down via shRNA (Online Supplementary Figure S7).
In order to evaluate the activity of tegavivint in vivo, we employed three murine models representative of disseminated ABC- and GCB-DLBCL. i) Seven-week-old NSG mice were engrafted with 20×106 Riva cells (ABC-DLBCL) via tail vein injection. Mice in groups of five were then randomized to receive either vehicle control or tegavivint at 25 mg/kg every 3 days via tail vein injection starting at day 7 post engraftment. Tegavivint significantly prolonged survival in the treated group (median OS of tegavivint vs. vehicle control mice 33 vs. 27 days, P=0.0031) (Figure 3C). Notably, 4 of 5 tegavivint-treated mice died of central nervous system (CNS) involvement with minimal systemic disease evaluated by the spleen size (Figure 3C) consistent with the known limited CNS penetration property of tegavivint. ii) Seven-week-old NSG mice were engrafted with 40×106 SU-DHL10 cells (GCB-DLBCL) via tail vein injection. Mice in groups of ten were then randomized to receive either vehicle control or intravenous tegavivint at 25 mg/kg starting at day 3 post engraftment on a twice weekly schedule (Monday-Thursday). Tegavivint significantly prolonged survival in the treated group (median OS of tegavivint vs. vehicle control mice 47 vs. 34.5 days, P=0.0002) (Figure 3D). ii) Seven-week-old NSG mice were tail vein engrafted with 1×106 of passage 3 DFBL-18689 cells from an ABC-DLBCL patient derived xenograft (PDX) mouse model obtained from the public repository of xenografts (Proxe).22 Mice in groups of five were then randomized to receive either vehicle control or tegavivint at 25 mg/kg via tail vein injection on a twice weekly schedule (Monday-Thursday) starting at day 3 post engraftment. Tegavivint significantly prolonged the survival of the treated animals (median OS of tegavivint vs. vehicle control mice 55 vs. 37 days, P=0.0026) (Figure 3E; Online Supplementary Figure S8).
Collectively, these data demonstrate that TBL1 is a novel therapeutic target in DLBCL and that tegavivint induces the desired pharmacologic effect by exhibiting selective TBL1-mediated lymphoma cell death with very limited toxicity to normal immune cells.
TBL1 modulates Wnt targets in a novel, post-transcriptional, β-CATENIN independent manner
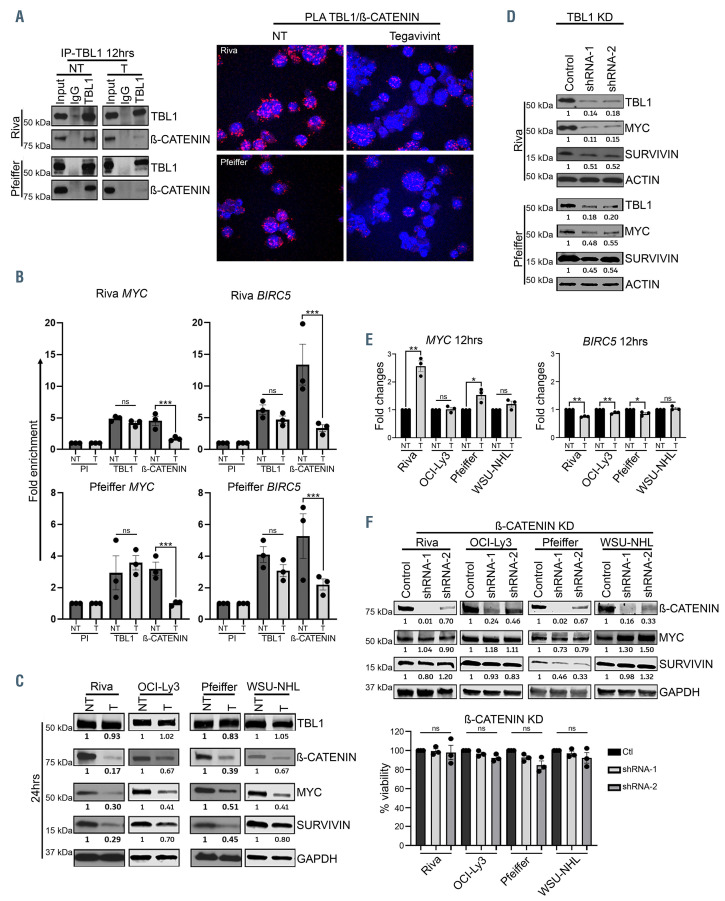
Co-immunoprecipitation (Co-IP) experiments in Riva and Pfeiffer cells showed that treatment with tegavivint for 12 hours led to the disruption of TBL1/β-CATENIN interaction (Figure 4A) which was further confirmed via PLA (Figure 4A; Online Supplementary Figure S9A and B). ChIP-seq analysis in the same cell lines revealed that genome-wide TBL1 promoter occupancy was not significantly affected by tegavivint treatment for 12 hours (Online Supplementry Figure S10A) while ChIP-PCR validation of selected Wnt/β-CATENIN target genes showed selective loss of β-CATENIN recruitment to the promoters of MYC and BIRC5 (Figure 4B) upon treatment (P<0.0005), TBL1 recruitment was unaffected. Treatment of four DLBCL cell lines with tegavivint for 24 hours resulted in decreased protein levels of β-CATENIN, MYC and SURVIVIN with no effect on TBL1 expression when compared to the untreated control (Figure 4C). Similarly, shRNA-mediated TBL1 knock-down led to significant decrease in MYC, and SURVIVIN expression compared to controls (Figure 4D). Despite the loss of β-CATENIN recruitment to the promoter regions of MYC and BIRC5 following treatment with tegavivint, quantitative realtime PCR (qRT-PCR) showed significant increase in MYC transcript in Riva (P<0.005) and Pfeiffer (P<0.05) and no changes in OCI-Ly3 and WSU-NHL (Figure 4E). A similar pattern was observed with the transcript of two other well-known β-CATENIN target genes, WISP1 and AXIN2 (Online Supplementary Figure S10B) while treatment with tegavivint resulted in a minimal but significant decrease in BIRC5 transcript in three out of four DLBCL cell lines tested (Figure 4E). Lastly, efficient shRNA knock-down of β-CATENIN did not significantly affect MYC or SURVIVIN protein expression and had minimal effect on cell viability in all the tested DLBCL cell lines (Figure 4F). In further support of a TBL1-mediated and β-CATENIN independent process, tegavivint maintained both its cytotoxic activity and its effect on MYC and SURVIVIN (protein and transcript) in the context of efficient β-CATENIN knock-down (Online Supplementary Figure S11).
Figure 1.
TBL1 is abundantly expressed and correlates with clinical outcome in diffuse large B-cell lymphoma. (A) Immunoblot showing abundant TBL1 expression in whole cell lysates from the indicated germinal center B-cell (GCB)- and activated B-cell (ABC)-diffuse large B-cell lymphoma (DLBCL) cell lines compared to normal resting and activated peripheral blood (PB) and germinal center (GC) B cells (n=3). (B) Immunoblot showing abundant TBL1 expression in whole cell lysates from four primary DLBCL patient (Pt) samples compared to normal B-cells and peripheral blood mononuclear cells (PBMC). ACTIN was used as loading control; mantle cell lymphoma cell line (Jeko) was used as positive control (+Ctl) for (A)-(B). (C) Immunohistochemical (IHC) staining of lymph nodes involved by ABC- and GCB-DLBCL (at 40x and 100x) showing strong (dark) nuclear and cytoplasmic positivity for TBL1. As comparison, tonsillar germinal center were co-stained with CD10 (brown) and TBL1 (red). TBL1-CD10 co-expression at 40x and 100x is shown in yellow (black arrows) using the Nuance Multispectral Imaging System (model 3.0.2) and colocalization tool which allows to select any color for co-localization evaluation. Positive controls (tonsillar epithelium [left] and breast ductal carcinoma [right]) are both shown at 40x and 100x. Dot plot showing percentage of TBL1 positive lymphoma cells by IHC staining in DLBCL patient samples (n=83 total with 46 GCBDLBCL and 37 (ABC-DLBCL) to reactive tonsillar germinal center controls (n=5). Data represent median percentage of TBL1 positive cells ± standard deviation. ***P<0.0005 by ANOVA. (D) Percentage of TBL1 positive lymphoma cells (≥55% for ABC-DLBCL and ≥50% for GCB-DLBCL) inversely correlated with progression free survival (PFS) and overall survival (OS) in de novo GCB- and ABC-DLBCL patients treated with front-line R-CHOP chemotherapy. P-values were determined using the log-rank tests.
Figure 2.
TBL1 genetic knock-down results in significant diffuse large B-cell lymphoma cell death in vitro and in vivo. (A) TBL1 was knocked-down (KD) using either a doxycycline-inducible CRISPR-Cas9 system (two different guides: one and two targeting exon 2 and 3, respectively) or a TBL1-specific short hairpin RNA (shRNA) construct (n=3). 72 hours after transduction, efficient TBL1 knock-down was verified via immunoblot and viability determined by annexin/propidium (Ann-/PI-) staining and flow cytometry. Data represent means ± standard error of the mean. ns: P>0.05, *P<0.05, **P<0.005, by paired t-test. Empty vector and scramble were used as controls. +Dox/-Dox: with (red) or without (black) doxycycline 0.75 mg/mL. Numbers below the immunoblots reflect normalized value of quantified protein bands relative to the controls. (B) Kaplan–Meier curve showing overall survival (OS) of NSG mice engrafted with Cas9+ Riva cells transduced with either a doxycycline- inducible TBL1-targeting single guide RNA (sgRNA) (n=8) or empty vector control (n=8). Daily doxycycline induction was started on day 24, in vivo imaging system (IVIS) images were obtained on day 23 and day 29 post engraftment. Median OS was 38 days for the empty vector controls (Ctl) versus 48 days for the TBL1 KD group (P=0.0004 using the log-rank test).
Figure 3.
Tegavivint shows significant activity in in vitro and in vivo models of diffuse large B-cell lymphoma. (A-B) Cytotoxicity assay in the indicated diffuse large B-cell lymphoma (DLBCL) cell lines (n=3) and DLBCL patient samples (n=8: peripheral blood [n=4], lymph node [n=3] and bone marrow [n=1]) treated with the indicated concentration of tegavivint (T) in nM for 24 hours. Viability was determined by annexinV/propidium (Ann-/PI-) staining and flow cytometry. Data represent means ± standard error of the mean. *P<0.05, **P<0.005, ***P<0.0005 by linear mixed effects models with adjustment for multiple dose comparisons in (A) and with two sample t-test in (B). (C) Kaplan–Meier curve showing overall survival (OS) of NSG mice engrafted with Riva (activated B-cell [ABC]-DLBCL) and randomized to receive either vehicle control (n=5) or tegavivint (n=5) given at 25 mg/kg via tail vein injection every 3 days starting at day 7 post engraftment. Median OS was 27 days for the controls (Ctl) versus 33 days for the treated group (P=0.0031 using the log-rank test). Representative picture of the spleen size (left image) and hematoxylin and eosin stain showing brain involvement by DLBCL cells (right image). (D) Kaplan–Meier curve showing OS of NSG mice engrafted with SU-DHL10 (germinal center B-cell [GCB]-DLBCL) and randomized to receive either vehicle control (n=10) or tegavivint (n=10) given at 25 mg/kg via tail vein injection twice weekly starting at day 3 post engraftment. Median OS was 34.5 days for the controls (Ctl) versus 47 days for the treated group (P=0.0002 using the log-rank test). (E) Using an adaptive transfer model of DFBL-18689, recipient NSG mice (n=5/group) were randomized to receive either vehicle control or tegavivint at 25 mg/kg via tail vein injection on a twice weekly schedule (Monday-Thursday) starting at day 3 post engraftment. Kaplan–Meier curve showing OS (P=0.0026 using the logrank test). Median OS: 55 days (tegavivint) and 37 days (control [Ctl]). Two mice in the treated group were censored due to engraftment failure assessed by flow cytometry for circulating lymphoma cells and magnetic resonance imaging for spleen volume.
Figure 4.
TBL1 modulates MYC levels in a post-transcriptional/β-CATENIN independent manner. (A) Diffuse large B-cell lymphoma (DLBCL) cell lines were treated with either dimethylsulphoxide (DMSO) control (NT) or tegavivint (T) for 12 hours. Lysates were immunoprecipitated with anti-TBL1 or anti-IgG and then immunoblotted with the indicated antibodies. Confocal images of proximity ligation assay (PLA) (60x) using anti-TBL1 and anti-β-CATENIN antibodies after treatment of DLBCL cells with either DMSO control (NT) or tegavivint (T) for 12 hours. Red indicates PLA signal, blue indicates cell nuclei (Dapi) (n=3). (B) Chromatin immunoprecipitation (ChIP) assay on crosslinked chromatin from DLBCL cells after treatment with either DMSO control (NT) or tegavivint (T) for 12 hours. Either preimmune (PI) or the indicated immune antibodies were used and the retained DNA was amplified using MYC or BIRC5 promoter-specific primers. Fold enrichment with each antibody was calculated relative to the PI. n=3; data represent means ± standard error of the mean (SEM). ns: P>0.05, *P<0.05, ***P<0.0005 by linear mixed effects models. (C) Immunoblot showing TBL1, β-CATENIN, MYC and SURVIVIN expression after treatment of DLBCL cells with either DMSO control (NT) or tegavivint (T) for 24 hours (n= 3). (D) Immunoblot showing MYC and SURVIVIN protein levels after TBL1 knock-down (KD) using two TBL1 specific shRNA constructs (n=3). Lysates are the same used in Figure 2A however probed for different proteins. (E) Quantitative real-time polymerase chain reaction (qRT-PCR) showing the mRNA fold changes of the indicated oncogenes after treating DLBCL cells with tegavivint (T) for 12 hours relative to the untreated control (NT). n=3, data represent means ± SEM. ns: P>0.05, *P<0.05, **P<0.005 by paired t-test. (F) Immunoblot showing MYC and SURVIVIN expressions after efficient β-CATENIN KD using two β-CATENIN specific short hairpin RNA (shRNA) constructs. Viability obtained by trypan blue exclusion 72 hours post-transduction. n=3, data represent means ± SEM. ns: P>0.05 by paired t-test (ns: not significant). Tegavivint (T): Riva, Pfeiffer: 70 nM; OCI-ly3: 50 nM; WSU-NHL: 15 nM (for all experiments).
Figure 5.
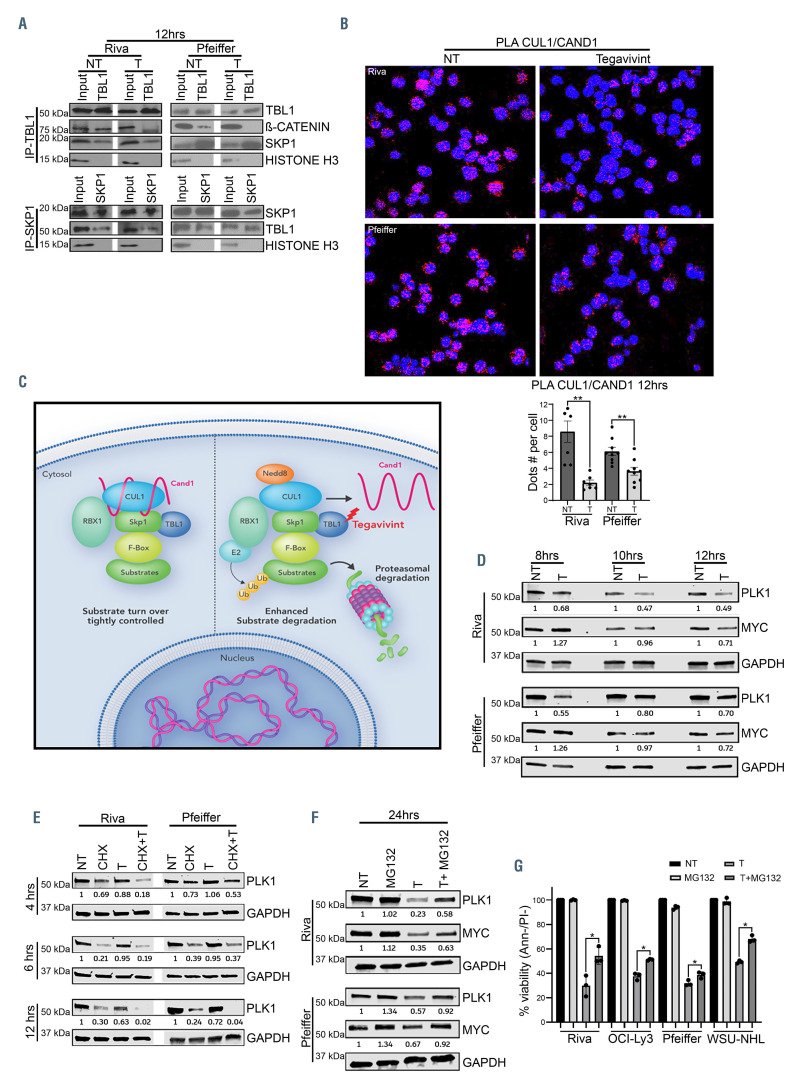
TBL1 is a critical component of a SCF complex in diffuse large B-cell lymphoma through which it modulates MYC stability. (A) The indicated diffuse large B-cell lymphoma (DLBCL) cell lines were treated with either dimethylsulfoxide (DMSO) control (NT) or tegavivint (T) for 12 hours, immunoprecipitated with either an anti-TBL1 or anti-SKP1, and then immunoblotted with the indicated antibodies. β-CATENIN and histone H3 were used as positive and negative controls, respectively. (B) Confocal images (60x) of proximity ligation assay (PLA) showing the CAND1-CUL1 interaction in the indicated DLBCL cells lines after treatment with either DMSO control (NT) or tegavivint (T) for 12 hours. Red indicates PLA signal, blue indicates cell nuclei (Dapi). Graph represents the mean value of PLA signal from no less than 300 cells (n=3). **P<0.005 by paired t-test. (C) Proposed model of the mechanism of tegavivint-induced activation of the SCF complex in DLBCL. (D) Immunoblot showing PLK1 and MYC expression levels after treatment with either DMSO control (NT) or tegavivint (T) at the indicated time points. (E) Immunoblot showing PLK1 level after treatment with either the translation inhibitor cycloheximide (CHX), tegavivint (T) or the combination (CHX+T) for the indicated time points. (F) Immunoblot showing PLK1 and MYC expression after treatment with either DMSO control (NT), tegavivint (T), the proteasome inhibitor MG132 or the combination (T+MG132) for 24 hours. (G) Cytotoxicity assay in DLBCL cell lines treated with either DMSO control (NT) or tegavivint (T), MG132 or the combination (T+MG132). Viability was determined by annexinV/propidium (Ann-/PI-) staining and flow cytometry at 24 hours. n= 3, data represent means ± SEM. *P<0.05, **P<0.005 by linear mixed effects models including interaction test between T and MG132. CHX: 70 mg/mL added for 1 hour and then washed before adding tegavivint treatment. MG132 (Riva, OCI-Ly3 and WSU-NHL: 0.5 uM and Pfeiffer: 0.3 uM) was added 6 hours after tegavivint treatment was started.
Collectively our data provide evidence that b- CATENIN contributes minimally to the transcriptional regulation of critical Wnt targets and to DLBCL cell survival and that TBL1 regulates the stability of MYC in a b- CATENIN independent fashion.
TBL1 is a critical component of the SCF complex in diffuse large B-cell lymphoma
The SCF complex controls the ubiquitination and degradation of a large number of proteins through the specificity-conferring F-box proteins such as bTRCP and FBW7.23,24 Its activity is regulated by the interaction of CUL1 with two proteins: i) the cullin associated and neddylation dissociated 1 (CAND1) and ii) NEDD8. CAND1 functions as an inhibitor of the SCF complex while NEDD8 binding to CUL1 promotes the targeted substrate ubiquitination and degradation.23 We hypothesized that TBL1 modulates target protein stability through the SCF complex and that targeting TBL1 with tegavivint enhances SCF complex activity leading to increased downstream degradation of key regulatory proteins. Co- IP and PLA in Riva and Pfeiffer cells showed TBL1-SKP1- CUL1 association which was unaffected by tegavivint (Figure 5A; Online Supplementary Figure S12). Importantly, tegavivint treatment of Riva and Pfeiffer induced significant decrease of CAND1-CUL1 association by PLA (P<0.005) (Figure 5B; Online Supplementary Figure S13) with no effect on the expression of these two proteins (Online Supplementary Figure S14A).
These results suggest a model in which TBL1, as a critical component of an SCF complex, modulates the stability of critical regulatory proteins (Figure 5C).
TBL1 modulates MYC stability through the SCF complex
Polo-like kinase-1 (PLK1), an essential regulator of mitosis and a novel target in lymphoma, is degraded by SCFbTrCP.25 Importantly, PLK1 plays a critical protective role in MYC stability by preventing its SCFFbw7-mediated proteasomal degradation.26-28 We hypothesized that targeting TBL1 with tegavivint may enhance the proteasomalmediated degradation of MYC directly by SCFFbw7 and indirectly by enhancing SCFbTrCP–mediated PLK1 degradation. Time course analysis showed that treatment of four DLBCL cell lines with tegavivint led to PLK1 downregulation while leaving its transcript unaffected (Figure 5D; Online Supplementary Figure S14B and C), decreased MYC levels with concomitant increase in its phosphorylation on Ser62 and Thr58 residues at 10 hours, consistent with enhanced MYC proteasomal degradation (Online Supplementary Figure S14D). Adding the translation inhibitor cycloheximide (CHX) to tegavivint significantly shortened the half-life of PLK1 protein compared to CHX alone (Figure 5E; Online Supplementary Figure S14E). Consistent with this, the proteasomal inhibitor MG132 partially restored PLK1 and MYC protein levels as well as cell viability when combined with tegavivint versus tegavivint alone (P<0.05) (Figure 5F and G). In further support of an SCF/TBL1-modulated process, efficient shRNA knock-down of CAND1 (and TBL1) in Riva and Pfeiffer cells led to significant decrease in viability as well as downregulation of PLK1 and MYC protein expression (Online Supplementary Figure 14F and G).
Altogether, our data support a novel model in which TBL1 regulates MYC stability directly through SCFFbw7 and indirectly by enhancing SCFbTrCP-mediated PLK1 degradation.
TBL1 is involved in the modulation of cytoprotective autophagy
Autophagy is a catabolic pathway that sustains metabolism by recycling cytoplasmic contents to support macromolecular synthesis during periods of cellular stress with accumulating evidence supporting its pro-survival role in lymphoma.29,3
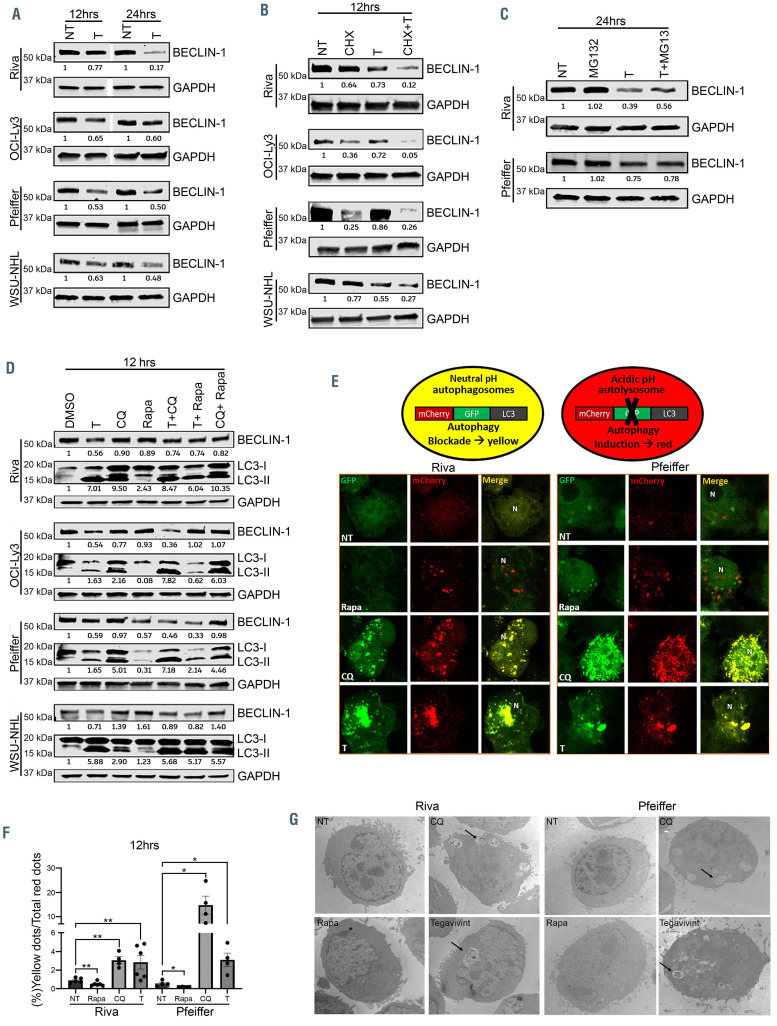
As SCF complexes regulate the stability of key autophagy inducers such as BECLIN-1 and MYC,31-33 we hypothesized that tegavivint impairs the autophagy flux by affecting these key autophagy inducers. Immune blot analysis showed downregulation of BECLIN-1 protein level in four DLBCL cell lines at 12 and 24 hours (Figure 6A) whereas BECN1 transcript level was not affected by tegavivint (Online Supplementary Figure S15). Co-treatment with CHX and tegavivint significantly shortened the half-life of BECLIN-1 compared to either CHX or tegavivint alone (Figure 6B) and incubation with MG132 partially restored BECLIN-1 protein level when combined with tegavivint compared to tegavivint alone (Figure 6C). In order to evaluate the effect of tegavivint on the autophagy flux, four DLBCL cell lines were treated with either dimethylsulfoxide (DMSO) control, chloroquine (autophagy blocker), rapamycin (autophagy inducer), tegavivint or combinations. The increase in the amount of LC3-II observed in chloroquine-treated cells represents the amount of LC3 delivered to the lysosome for degradation and is a well-established measure of autophagic flux.34 As shown in Figure 6D, treatment with tegavivint led to an increase in the levels of LC3-II which could be due to autophagy induction or inhibition. Co-treatment with chloroquine led to further accumulation of LC3-II, suggesting autophagy induction in two of the four cell lines tested. However, treatment with rapamycin in combination with either tegavivint or chloroquine resulted in an increase of LC3-II compared with rapamycin alone across the four cell lines tested, suggesting that tegavivint may impair autophagic flux. In order to clarify this discordance, we transfected Riva and Pfeiffer cells with a GFP– mCherry-LC3 construct. This fluorescence reporter monitors LC3 flux by relying on both green-fluorescence protein (GFP) sensitivity and mCherry resistance to the acidic/proteolytic environment in the lysosomes. With autophagy blockade, both mCherry (red fluorescence protein) and GFP co-localize in a vesicular compartment that has not fused with lysosomes (autophagosomes) resulting in yellow puncta, whereas autophagy induction leads to fusion of these compartments (autolysosomes) and subsequent quenching of GFP signal resulting in singly mCherry red puncta.35 As shown in Figure 6E, treatment of these cells with either chloroquine or tegavivint increased yellow puncta assessed by confocal microscopy whereas rapamycin treatment was associated with increased red puncta solely. By calculating the ratio between yellow and red puncta, we found that the yellow signal was significantly increased in chloroquine and tegavivint-treated DLBCL cells (P<0.05) compared to controls, suggesting autophagosome accumulation (Figure 6E and F). Ultrastructurally, Riva and Pfeiffer treated with chloroquine or tegavivint for 12 hours showed accumulation of enlarged vesicles with characteristic enclosed cytoplasmic ultrastructures, supporting the notion that tegavivint treatment impairs the terminal stages of autophagy (Figure 6G).
Figure 6.
TBL1 modulates cytoprotective autophagy through the SCF complex. (A) Immunoblot showing BECLIN-1 expression in the indicated diffuse large B-cell lymphoma (DLBCL) cell lines after treatment with either dimethylsulfoxide (DMSO) control (NT) or tegavivint (T) for 12 hours and 24 hours (n=3). (B) Immunoblot showing BECLIN-1 expression in the indicated DLBCL cell lines after treatment with either cycloheximide (CHX), tegavivint (T) or the combination (CHX+T) (n=3). (C) Immunoblot showing BECLIN-1 expression in the indicated DLBCL cell lines after treatment with either DMSO control (NT), tegavivint (T), MG132 or the combination (T+MG132) at 24 hours (n=3). (D) Immunoblot showing BECLIN-1 and microtubule-associated protein light-chain 3 (LC3-I and LC3-II) expression in the indicated DLBCL cell lines after treatment with either DMSO control (NT), tegavivint (T), chloroquine (CQ), rapamycin (Rapa) or combinations at 12 hours and 24 hours (n=3). (E) Top: schematic depicting tandem LC3 reporter. Bottom: confocal microscopy images (120x) of the indicated DLBCL cell lines transduced with a lentivirus expressing a tandem LC3 plasmid (mCherry-eGFP-LC3) and subsequently treated with either DMSO control (NT), tegavivint (T), chloroquine (CQ) or rapamycin (Rapa) for 12 hours. (F) Histograms represent quantification of yellow (GFP/mCherry) puncta/total red (mCherry) puncta (%) of no less than 100 cells per condition. n=4, data represent means ± standard error of the mean (SEM). *P<0.05, **P<0.005 by a linear mixed effects model with Holm’s adjustment for multiple comparisons for each cell line. (G) Representative transmission electron microscopy images showing ultrastructural changes in the indicated DLBCL cell lines after treatment with either DMSO control (NT), tegavivint, chloroquine (CQ) or rapamycin (Rapa) at 12 hours. The arrows indicate accumulation of autophagic vacuoles containing cytoplasmic material after exposure to tegavivint or chloroquine (n=3). Chloroquine: 50uM and rapamycin: 10uM for all cell lines.
Collectively, these results indicate that tegavivint impairs cytoprotective autophagy by promoting SCFmediated degradation of key autophagy inducers such as MYC and BECLIN-1.
Discussion
In the work presented here, we showed that TBL1 encoded by the TBL1X gene is abundantly expressed in DLBCL cells regardless of the molecular subtype. While further studies are required to validate TBL1 on DLBCL samples using IHC, and for this to predict relapse in a prospective manner, in this initial evaluation we show that TBL1 overexpression correlates with poor survival in de novo DLBCL patients uniformly treated with standard front-line immuno-chemotherapy.
Genetic deletion of TBL1X or pharmacologic treatment with tegavivint significantly affected DLBCL cell viability in vitro while minimally toxic against normal immune cells and significantly prolonged the survival in four animal models of disseminated DLBCL, supporting the therapeutic potential of targeting TBL1 in this disease.
While tegavivint has been shown to mediate AML and MM cell death by disrupting the TBL1/β-CATENIN interaction followed by proteasomal-mediated degradation of β-CATENIN and inhibition of the Wnt/transcriptional program,16,17 our findings are novel and provide insight into a previously undiscovered post-transcriptional, b- CATENIN-independent oncogenic network modulated by TBL1 in DLBCL. Despite β-CATENIN expression in a subset of DLBCL cases,36,37 our data minimize the transcriptional relevance of β-CATENIN in promoting Wnt signaling and DLBCL cell survival while suggesting a possible role as a direct or indirect transcriptional repressor for some of the Wnt downstream targets (MYC, WISP1 and AXIN2) as previously shown in melanoma.38 However, this scenario may differ in a setting such as the tumor microenvironment where cytokine-induced Wnt signaling is active.39
The SCF-type of E3 ubiquitin ligase is a multi-protein complex composed of three static subunits RBX1, CUL1, SKP1 and variable F-box subunits which confer substrate selectivity to the complex.23,24,40-42 Here we showed that TBL1 modulates the stability of MYC, PLK1 and BECLIN- 1 through its direct interaction with the SCF complex. Mechanistically, our data support the hypothesis that tegavivint binding to TBL1 leads to conformational changes in the SCF complex resulting in CAND1-CUL1 dissociation followed by enhanced proteasomal degradation of these critical regulatory/pro-survival proteins. In further support of these findings, we showed that efficient shRNA knock-down of CAND1 led to significant decrease in DLBCL viability as well as downregulation of PLK1 and MYC protein expression. CAND1 functions as an inhibitor of the SCF complex by preventing the access of SKP1 and F-box proteins to the CUL1 catalytic core.23 In agreement with our work, C60, a small molecule inhibitor of CAND1, has been shown to destabilize CAND1-CUL1 interaction while increasing global ubiquitination in Epstein-Barr virus-positive lymphoma cell lines.43
Interestingly, TBL1 itself has an F-box motif and a WD40 domain for substrate recognition42 suggesting the possibility that TBL1 directly interacts with specific targets to modulate their stability. In support of this hypothesis which warrants further investigation, data produced in human embryonic kidney 293T cells showed that TBL1 directly interacts with and protects β-CATENIN from Siah-mediated ubiquitination and degradation as part of an SCFTBL1 complex.13,14
MYC rearrangement is identified in a subset of DLBCL cases (5-15%), MYC protein over-expression is seen more commonly (30–50% of DLBCL).44 Regardless of the genetic mechanism, MYC over-expression is associated with a more aggressive clinical behavior and inferior outcome in DLBCL.45 In addition, it has been recently showed that PLK1 signaling promotes MYC protein stability, and in turn, MYC directly induces PLK1 transcription, establishing PLK1 as a therapeutic vulnerability in high grade B-cell lymphomas.28 Our findings are of particular importance because they indicate that tegavivint enhances the proteasomal-mediated degradation of MYC directly by SCFFbw7 and indirectly by enhancing SCFbTrCP– mediated PLK1 degradation supporting the notion that TBL1 holds promise as a therapeutic target particularly in MYC overexpressing DLBCL.
Cytoprotective autophagy plays an important role in promoting lymphoma cell survival in adverse conditions and adequate levels of BECLIN-1 are necessary to maintain the autophagy flux as supported by the fact that BECLIN-1+/- mice have defective autophagy.29,30,33,46 Consistent with this published work and with that on MYC as a potent inducer of autophagy,31 our data indicate that tegavivint impairs pro-survival autophagy by promoting the SCF-mediated proteasomal degradation of both BECLIN-1 and MYC.
Although we have identified TBL1 to play a central role in modulating the turnover of PLK1, MYC, and BECLIN- 1 through the SCF complex, there are a number of other potential SCF targets that will need to be explored to further define the role of TBL1 in the context of the SCF complex and to fully characterize the mechanism of action of tegavivint. For example, in addition to b- CATENIN and PLK1, the TBL1 interacting SCF complex (SCFbTrCP) targets multiple proteins regulating mTOR signaling including the Ras homolog endriched in brain protein (RHEB), an upstream direct mTORC1 activator.47 Constitutive activation of mTOR signaling is critical for cellular growth and metabolism in both ABC- and GCBDLBCL48 and mTORC1 inhibition is known to lead to lymphoma cell cycle arrest and compensatory autophagy induction.49 While TBL1 targeting represents an attractive approach to simultaneously affect multiple prosurvival signaling pathways leading to a more profound lymphoma cell death than that achievable with agents targeting each of these pathways individually, this approach, given the complexity of the prosurvival pathways regulated by TBL1, may induce upregulation of compensatory mechanisms ultimately leading to drug resistance and providing the rational for combination strategies.
While ongoing efforts are focused on studying the functional role of nuclear TBL1, the findings presented here demonstrate that TBL1 serves as a master regulator of a post-transcriptional oncogenic network and provide a rationale for TBL1 targeting in DLBCL with tegavivint which is currently in a phase-I clinical trial for patients with primary or recurrent desmoid tumors (clinicaltrials gov. Identifier: NCT03459469).
Supplementary Material
Acknowledgments
We are grateful to the patients and healthy volunteers who provided tissue samples for these studies, to the OSU Comprehensive Cancer Center Leukemia Tissue Bank Shared Resource (supported by NCIP30 CA016058) for sample procurement, to the OSU Comparative Pathology and Mouse Phenotyping Shared Resource (CPMPSR,supported in part by NCIP30 CA016058) for excellent histotechnologic support (Ms. Brenda Wilson and Ms. Tessa VerStraete), to the OSU Campus Microscopy and Imaging Facility (CMIF supported in part by NIH Grant P30 CA016058) for excellent support with electron and confocal microscopy, to Somersault18:24 for the professional illustration of the proposed model of TBL1 posttranscriptional role within the SCF complex. The authors would also like to acknowledge Dr. Aharon Freud (OSU Department of Pathology) for providing normal pediatric tonsil controls.
Funding Statement
Funding: This work was supported by the NCI (K08 CA226352 to LA; R35 CA198183 to JCB) and by a Research Scholar grant from the American Society of Hematology (LA)
References
- 1.Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin. 2016;66(6):443-459. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937-1947. [DOI] [PubMed] [Google Scholar]
- 4.Sehn LH, Donaldson J, Chhanabhai M, et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J Clin Oncol. 2005;23(22):5027-5033. [DOI] [PubMed] [Google Scholar]
- 5.Chow VA, Shadman M, Gopal AK. Translating anti-CD19 CAR T-cell therapy into clinical practice for relapsed/refractory diffuse large B-cell lymphoma. Blood. 2018;132(8):777-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkozy C, Sehn LH. Management of relapsed/refractory DLBCL. Best Pract Res Clin Haematol. 2018;31(3):209-216. [DOI] [PubMed] [Google Scholar]
- 7.Yoon HG, Chan DW, Huang ZQ, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22(6):1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Gene Dev. 2000;14(9):1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 9.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116(4):511-526. [DOI] [PubMed] [Google Scholar]
- 10.Hatzi K, Jiang Y, Huang C, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4(3):578-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Wang C-Y. TBL1–TBLR1 and β-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat Cell Biol. 2008;10(2):160-169. [DOI] [PubMed] [Google Scholar]
- 12.Perissi V, Scafoglio C, Zhang J, et al. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell. 2008;29(6):755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimitrova YN, Li J, Lee Y-T, et al. Direct ubiquitination of β-CATENIN by Siah-1 and regulation by the exchange factor TBL1. J Biol Chem. 2010;285(18):13507-13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Stevens J, Rote CA, et al. Siah-1 mediates a novel β-CATENIN degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell. 2001;7(5):927-936. [DOI] [PubMed] [Google Scholar]
- 15.Soldi R, Horrigan SK, Cholody MW, et al. Design, synthesis, and biological evaluation of a series of anthracene-9, 10-dione dioxime β-CATENIN pathway inhibitors. J Med Chem. 2015;58(15):5854-5862. [DOI] [PubMed] [Google Scholar]
- 16.Fiskus W, Sharma S, Saha S, et al. Pre-clinical efficacy of combined therapy with novel β-CATENIN antagonist BC2059 and histone deacetylase inhibitor against AML cells. Leukemia. 2015;29(6):1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savvidou I, Khong T, Cuddihy A, McLean C, Horrigan S, Spencer A. β-CATENIN inhibitor BC2059 is efficacious as monotherapy or in combination with proteasome inhibitor bortezomib in multiple myeloma. Mol Cancer Ther. 2017;16(9): 1765-1778. [DOI] [PubMed] [Google Scholar]
- 18.ANSI/ATCC ASN-0002-2011. Authentica - tion of human cell lines: standardization of STR profiling. https://webstore.ansi.org/standards/atcc/ansiatccasn00022011. [Google Scholar]
- 19.de Almeida Nagata DE, Chiang EY, Jhunjhunwala S, et al. Regulation of tumorassociated myeloid cell activity by CBP/EP300 bromodomain modulation of H3K27 acetylation. Cell Rep. 2019;27(1): 269-281. [DOI] [PubMed] [Google Scholar]
- 20.Aubrey BJ, Kelly GL, Kueh AJ, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10(8):1422-1432. [DOI] [PubMed] [Google Scholar]
- 21.Fiore C, Bailey D, Conlon N, et al. Utility of multispectral imaging in automated quantitative scoring of immunohistochemistry. J Clin Pathol. 2012;65(6):496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Townsend EC, Murakami MA, Christodoulou A, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell. 2016;29(4):574-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldenberg SJ, Cascio TC, Shumway SD, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullindependent ubiquitin ligases. Cell. 2004;119(4):517-528. [DOI] [PubMed] [Google Scholar]
- 24.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5(9):739-751. [DOI] [PubMed] [Google Scholar]
- 25.Giráldez S, Galindo-Moreno M, Limón-Mortés MC, et al. G1/S phase progression is regulated by PLK1 degradation through the CDK1/bTrCP axis. FASEB J. 2017;31(7): 2925-2936. [DOI] [PubMed] [Google Scholar]
- 26.Popov N, Schülein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF b-TrCP antagonizes SCF Fbw7-mediated turnover. Nat Cell Biol. 2010;12(10):973-981. [DOI] [PubMed] [Google Scholar]
- 27.Xiao D, Yue M, Su H, et al. Polo-like kinase-1 regulates Myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol Cell. 2016;64(3): 493-506. [DOI] [PubMed] [Google Scholar]
- 28.Ren Y, Bi C, Zhao X, et al. PLK1 stabilizes a MYC-dependent kinase network in aggressive B cell lymphomas. J Clin Invest. 2018;128(12):5517-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alinari L. Toward autophagy-targeted therapy in lymphoma. Blood. 2017;129(13): 1740-1742. [DOI] [PubMed] [Google Scholar]
- 30.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7(12):961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart LS, Cunningham JT, Datta T, et al. ER stress–mediated autophagy promotes Mycdependent transformation and tumor growth. J Clin Invest. 2012;122(12):4621-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui D, Xiong X, Zhao Y. Cullin-RING ligases in regulation of autophagy. Cell Div. 2016;11(1):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang R, Zeh H, Lotze M, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4): 571-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent- tagged LC3. Autophagy. 2007;3(5): 452-460. [DOI] [PubMed] [Google Scholar]
- 36.Ge X, Lv X, Feng L, Liu X, Wang X. High expression and nuclear localization of bcatenin in diffuse large B-cell lymphoma. Mol Med Rep. 2012;5(6):1433-1437. [DOI] [PubMed] [Google Scholar]
- 37.Bognar M, Vincendeau M, Erdmann T, et al. Oncogenic CARMA1 couples NF-κB and bcatenin signaling in diffuse large B-cell lymphomas. Oncogene. 2016;35(32):4269-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-CATENIN signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231-235. [DOI] [PubMed] [Google Scholar]
- 39.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11): 1461-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciechanover A. The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J. 1998;17(24):7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hussain M, Lu Y, Liu Y-Q, et al. Skp1: implications in cancer and SCF-oriented anti-cancer drug discovery. Pharmacol Res. 2016;111:34-42. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14(4):233-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tikhmyanova N, Tutton S, Martin KA, et al. Small molecule perturbation of the CAND1-Cullin1-ubiquitin cycle stabilizes p53 and triggers Epstein-Barr virus reactivation. PLoS Pathog. 2017;13(7):e1006517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114(17):3533-3537. [DOI] [PubMed] [Google Scholar]
- 45.Valera A, López-Guillermo A, Cardesa-Salzmann T, et al. MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica. 2013;98(10):1554-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077-15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harraz MM, Tyagi R, Cortés P, Snyder SH. Antidepressant action of ketamine via mTOR is mediated by inhibition of nitrergic Rheb degradation. Mol Psychiatry. 2016;21(3):313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2015;125(1):22-32. [DOI] [PubMed] [Google Scholar]
- 49.Wanner K, Hipp S, Oelsner M, et al. Mammalian target of rapamycin inhibition induces cell cycle arrest in diffuse large B cell lymphoma (DLBCL) cells and sensitises DLBCL cells to rituximab. Br J Haematol. 2006;134(5):475-484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.