Abstract
Human ZNF648 is a novel poly C-terminal C2H2 zinc finger (ZnF) protein identified amongst the most dysregulated proteins in erythroid cells differentiated from induced pluripotent stem cells. Its nuclear localization and structure indicate it is likely a DNA-binding protein. Using a combination of ZNF648 overexpression in an induced pluripotent stem cells line and primary adult erythroid cells, ZNF648 knockdown in primary adult erythroid cells and megakaryocytes, comparative proteomics and transcriptomics we show that ZNF648 is required for both erythroid and megakaryocyte differentiation. Orthologues of ZNF648 were detected across Mammals, Reptilia, Actinopterygii, in some Aves, Amphibia and Coelacanthiformes suggesting the gene originated in the common ancestor of Osteichthyes (Euteleostomi or bony fish). Conservation of the C-terminal ZnF domain is higher, with some variation in ZnF number but a core of at least six ZnF conserved across all groups, with the N-terminus recognisably similar within but not between major lineages. This suggests the N-terminus of ZNF648 evolves faster than the C-terminus, however this is not due to exon-shuffling as the entire coding region of ZNF648 is within a single exon. As for other such transcription factors, the N-terminus likely carries out regulatory functions, but showed no sequence similarity to any known domains. The greater functional constraint on the ZnF domain suggests ZNF648 binds at least some similar regions of DNA in the different organisms. However, divergence of the N-terminal region may enable differential expression, allowing adaptation of function in the different organisms.
Introduction
The C2H2 zinc finger (ZnF) proteins represent one of the largest families of regulatory proteins in humans, involved in a variety of cellular activities including development, cell differentiation, genome integrity and tumour suppression (reviewed by Iuchi, 2001).1 The C2H2 motif represents the classical ZnF DNA-binding domain (reviewed by 2), with 675-700 C2H2 ZnF genes identified in the human genome, a large proportion of which are transcription factors.3 Such C2H2 ZnF transcription factors, including GATA 1 and 2, FOG and KLF1, play important roles in differentiation and development of red blood cells (reviewed by Kim and Bresnick4 and Siatecka and Bieker)5.
The generation of red blood cells in vitro as an alternative transfusion product is a goal for research groups and blood services globally. In particular, for patients with rare blood group phenotypes for who matched donor blood is difficult to source, and for those requiring regular transfusions who are at risk of alloimmunisation. Various types of stem cells have been used for in vitro erythroid culture systems. Of these, adult peripheral blood (PB) and umbilical cord blood (CB) CD34+ cells differentiate efficiently along the erythroid pathway with high rates of enucleation (60-95%).6-8 However, the erythroblasts have restricted expansion potential using current systems and are thus not a sustainable system, with repeat collections of stem cells required for successive cultures. In contrast, embryonic stem cells (ESC) and induced pluripotent stem cells (iPSC) have the potential to provide an inexhaustible source of progenitors for the generation of large numbers of erythroid cells. Of these, iPSC are particularly attractive as they can be derived from easily accessible adult cells, and without the associated ethical issues of ESC. However, iPSC-derived erythroid cells have historically exhibited terminal differentiation defects and severely impaired enucleation,9-12 although more recent advancements in culture techniques have demonstrated degrees of improvements.13-15
We originally sought to identify proteins potentially involved in the terminal differentiation defect of human iPSC-derived erythroid cells, comparing the proteome of erythroid cells differentiated from three iPSC lines, originating from different cellular origins, with those differentiated from adult PB CD34+ cells.12 The most differentially expressed proteins were -, - and -globin, as expected due to the known differences in the globin expression profiles of these cells. However, further interrogation of datasets revealed a novel, previously unstudied ZnF protein, ZNF648, amongst the most differentially expressed proteins, >20-fold lower in both early and late iPSC-derived erythroid cells compared to respective adult erythroid cells.
Human ZNF648 is a poly C2H2 ZnF protein which we show is essential for both erythroid and megakaryocyte differentiation. As conservation of protein sequences across species can indicate functional importance, we also explored the conservation and profile of ZNF648 through evolution. ZNF648 originated in the common ancestor of Osteichthyes (Euteleostomi or bony fish). However, conservation of the C-terminal ZnF domain is higher, with some variation in ZnF number but a core of at least six ZnF conserved across all groups, with the Nterminus recognizably similar within, but not between, major lineages. However, unlike the evolution of many C2H2 ZnF transcription factors, this is not due to exonshuffling, as the entire coding region of ZNF648 is within a single exon. If, as with other transcription factors, the N-terminal region of ZNF648 contains regulatory domains, these are potentially novel as no sequence similarity to any known domains was found. The greater functional constraint on the ZnF domain suggests ZNF648 binds at least some similar regions of DNA in the different organisms. However, divergence of the N-terminal region may enable differential expression and hence control of gene expression required for the different environmental conditions of the various organisms.
Methods
Cell culture and transduction
Adult CD34+ cells were isolated from Leukocyte Reduction System (LRS) cones, with informed consent from all donors, and used in accordance with the Declaration of Helsinki and approved by the National Health Service National Research Ethics Committee (reference number 08/H0102/26) and the Bristol Research Ethics Committee (reference 12/SW/0199). Adult CD34+ erythroid and megakaryocyte, K562 and HiDEP-1 cultures were performed as described previously.6,16,17 Lentiviral transduction for protein overexpression was performed with pXLG3 construct as described previously.18 Knockdown studies using pLKO.1 short hairpin RNA (shRNA) plasmid TRCN0000107710 (ZNF648 shRNA), TRCN0000107714 (ZNF648 shRNA2) or a scrambled control (Scr) shRNA were as described previously18 (all designed by the Broad Institute and purchased from Open Biosystems, GE Dharmacon, Lafayette, CO, USA). Dead cell removal was carried out with a dead cell removal kit (Miltenyi Biotech Ltd).
Megakaryocyte ploidy
DNA content was measured on day 14 cultured cells. Cells were incubated with anti-CD41 antibody for 30 min at 4°C, washed, fixed in 75% EtOH, stained with propidium iodide and measured using the MACSquant VYB Analyser.
Quantitative polymerase chain reaction analysis of ZNF648 transcript levels
Total RNA from cell pellets was extracted using the RNeasy Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). RNA quantity and purity were determined using NanoDrop Lite (NanoDrop Technologies, Wilmington, DE, USA), and RNA integrity was assessed by determining the RNA 28S/18S ratio. RNA (500 ng) was reverse-transcribed into cDNA using Oligo(dt)18 primers (Thermo Scientific, Vilnus, Lithuania) and SuperScript IV (Invitrogen, Vilnus, Lithuania). The cDNA products were amplified by quantitative polymerase chain reaction (qPCR) using the SYBR select master mix (Applied Biosystems, Vilnus, Lithuania). All reactions were carried out in triplicate. Real-time qPCR (RT-qPCR) was run in QuantStudio™ 3 Real-Time PCR System (Applied Biosystems). Primers used to detect ectopic ZNF648-GFP: forward 5’-GTGGAAATGTCTGGGAAAGC, reverse 5’-CAATTTGTGTGCGAGACCAC; primers used to detect endogenous ZNF648: forward 5’- AGCGTGAGAGACAGAGACACC, reverse 5’-GGATACCTGGGAAATGCAGA. Primers for PABPC1: forward 5’- AGCTGTTCCCAACCCTGTAATC, reverse 5’-GGATAGTATGCAGCACGGTTCTG. All primers were synthesized by Sigma-Aldrich. Results were normalized to PABPC1 levels. The threshold cycle (Ct) was determined and the relative gene expression was expressed using the 2-Ct method.
Transcriptomics and proteomics
Transcriptomics using Human Genome U133 Plus 2.0 arrays and tandem mass tag (TMT) proteomics were performed as described previously.12,19,20
Sequence retrieval and analysis
Annotated ZNF648 (ENSG00000179930) sequences were retrieved from the Ensembl database,21 with BLASTP and TBLASTN searches used to identify other ZNF648 homologues from RefSeq22 including sequences for which the protein had not been annotated from the genome. In order to identify more divergent ZNF648 homologues and trace the evolution of the family, more sensitive Hidden Markov Model (HMM) searches were used against a local database of metazoan proteomes using HMMER3. ExPASY prosite23 was used to find where the C-terminal cluster of C2H2 motifs started and separated the N-terminal region and C-terminal region. All-versus-all BLASTP searches24 were used to determine pairwise percentage identities of both the N and C terminal regions. These results were visualized using heatmaps generated in R25 with ggplot26 and Viridis (https://github.com/sjmgarnier/viridis).
Phylogenetics
Sequences were aligned with Mafft using the most accurate l- INS-i mode.27 We inferred a maximum likelihood phylogeny of ZNF648 (949 aligned sites/1134 positions/42 taxa) using the best-fitting LG+C60+G+F28,29 model in IQ-Tree 1.6.10. We used 1,000 ultrafast bootstraps30,31 to assess support.
Results
Structure of ZNF648
The gene for human ZNF648 is mapped to chromosome 1q25.3. It has 2 exons and 1 intron with the entire open reading frame located in the first half of exon 2. The gene codes for a protein of 568 amino acids with 10 C2H2 ZnF arranged adjacent to each other at the C-terminus, a typical architecture for a human ZnF protein. NCBI BLAST (Basic Local Alignment Search Tool; https://blast.ncbi.nlm.nih.gov/) search revealed no homology of ZNF648 with other human transcription factors.
About 40% of human ZnF superfamily members have an N-terminal KRAB domain (Krüppel-Associated Box), which can confer transcriptional repression by recruiting KAP-1 (reviewed by Emerson and Thomas32). However homology modeling indicated ZNF648 does not contain this domain, a BTB/POZ effector33 or SCAN domain.34
In considering a role for ZNF648 as a DNA binding protein, tandemly arranged C2H2 ZnF are often connected by short linker regions which play an important role in conferring high-affinity binding to the DNA.35 These linker regions are made up of seven to eight amino acids, with the consensus sequence TGEKP identified in 50% of C2H2 ZnF proteins35 with variations of this sequence also verified. 36,37 The presence of this region is often used as a predictor for the DNA binding property of a protein. Interrogation of the ZNF648 sequence revealed the consensus linker sequence between zinc fingers 2 and 3, with alternatively recognised motifs between ZnFs 1 and 2, 5 and 6, 6 and 7, 7 and 8 (Online Supplementary Figure S1). The position of these linkers suggests two potential DNA-binding clusters, firstly ZnF 1, 2 and 3 and secondly ZnF 5, 6, 7 and 8. It is therefore predicted that ZNF648 binds to DNA.
Expression profile of ZNF648 during erythropoiesis
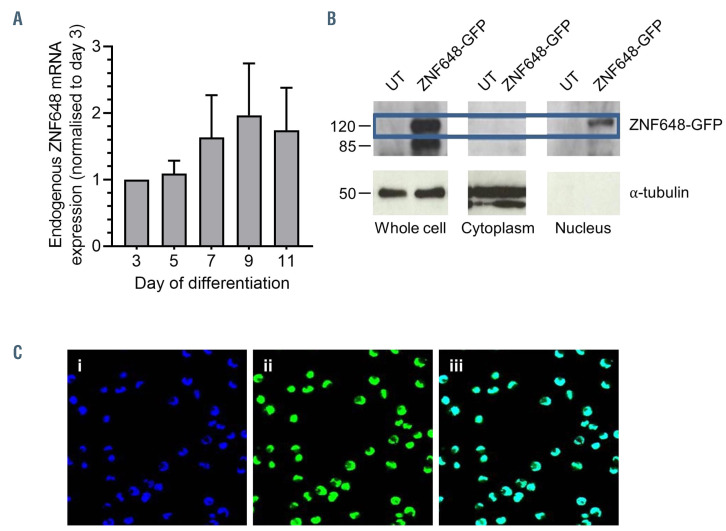
Adult PB CD34+ cells were cultured in our erythroid culture system which supports efficient erythroid differentiation, with yields of up to 95% reticulocytes on day 19.6,8 Analysis of ZNF648 transcripts throughout differentiation by qPCR showed a slight upward trend, but levels did not reach significance at any time point (Figure 1A). Endogenous ZNF648 protein was below the level of detection by immunoblot. ZNF648 antibodies (sc-249727; Santa Cruz and ab170269; Abcam) were therefore validated by transduction of cells with ZNF648 and ZNF648- GFP, with bands of the expected size detected by both antibodies, but no protein detected in control cells (Online Supplementary Figure S2).
Overexpression of ZNF648 in K562 cells showed ZNF648 exclusively localized to the nucleus by both confocal microscopy and immunoblot of cell fractions (Figure 1B and C), supporting the hypothesis that ZNF648 is a DNA-binding protein.
Overexpression of ZNF648 in erythroid cells reduces proliferation and advances differentiation
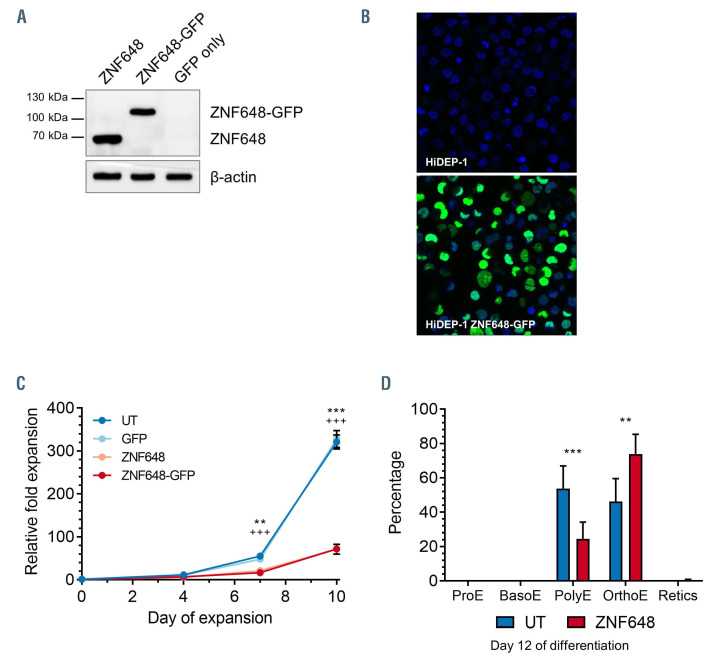
In order to begin to explore the role of ZNF648 in erythropoiesis we initially expressed exogenous ZNF648 in HiDEP-1 cells, an erythroid cell line created by immortalising erythroid cells differentiated from iPSC.38 HiDEP-1 cells exhibit many of the same properties as iPSCderived erythroid cells, e.g., impaired erythroid differentiation, defective enucleation and fetal/embryonic globin expression.17,38 As with iPSC-derived erythroid cells, the level of ZNF648 protein is substantially lower (approximately 20-fold) in HiDEP-1 cells compared to adult erythroid cells (Frayne unpublished comparative proteomic data for HiDEP-1 vs. primary adult erythroid cells; ZNF648 quantified from 33 peptide spectra matches [PSM]).
HiDEP-1 cells were transduced with ZNF648, ZNF648- GFP or with a green fluorescent protein (GFP) control construct. Transduction efficiency was over 90%. Expression was confirmed by immunoblot (Figure 2A), with the expected size bands for the untagged and GFPtagged proteins observed. Quantification from abundance values following TMT labeling of tryptic peptides and analysis by nano-LC MS/MS gave an approximately 10-fold increase in ZNF648 (from 23 unique peptides and 75 PSM). Confocal analysis of the ZNF648-GFP transduced cells showed exclusive nuclear localisation (Figure 2B), as for the transduced K562 cells. Cells transduced with ZNF648 and ZNF648-GFP had equivalent viability, but a reduced proliferation rate (∼80% reduction by day 10, P<0.01 for both controls vs. ZNF648 and vs. ZNF848- GFP; Figure 2C) and accelerated differentiation; at day 12 significantly fewer polychromatic normoblasts (P<0.001) and significantly more orthochromatic normoblasts (P<0.01) compared to controls (Figure 2D). Enucleation rates were not increased.
We also analyzed the effect of ZNF648 overexpression on cultured adult erythroid cells. Erythroid cells differentiated from adult PB CD34+ cells at day 3 in culture were transduced with ZNF648-GFP. GFP+ cells were isolated on day 8 and maintained in our erythroid culture system thereafter. As with HiDEP-1, overexpression of ZNF648 reduced the proliferation rate (e.g., by ∼50% on day 15; Online Supplementary Figure S3A), without affecting viability, and advanced the rate of differentiation, with significantly fewer basophilic normoblasts (P<0.05) and significantly more orthochromatic normoblasts and reticulocytes (both P<0.05) on day 13 of culture compared to controls (Online Supplementary Figure S3B).
However, a major issue with such ectopic expression studies is the non-physiological levels of ZNF648 achieved. As the expression of endogenous ZNF648 is below the level of detection by antibody-dependant assays, this likely masks the true role of ZNF648 in erythropoiesis. We therefore took the alternative approach of knocking down ZNF648 in adult erythroid cells.
Figure 1.
Expression and localisation of ZNF648 during erythroid differentiation. (A) Expression of ZNF648 transcript levels at days 3, 5, 7, 9 and 11 in culture analysed by quantitative polymerase chain reaction (qPCR), n=2. (B) Western blot of whole cell lysate, cytoplasmic fraction, and nuclear fraction of control K562 cells and K562 cells transduced with ZNF648-GFP probed with antibody to green fluorescent protein (GFP). Antibody to -tubulin was used as a control for cytoplasmic fraction protein loading. Molecular weight (MW) markers shown on left hand side. (C) images of K562 cells transduced with ZNF648-GFP (i) DAPI staining (ii) ZNF648-GFP fluorescence (iii) Merged image of panels i and ii. Images obtained using a Leica SP5 confocal microscope. Magnification 400x.
Figure 2.
Exogenous expression of ZNF648 in the induced pluripotent stem cells line HiDEP-1 impedes proliferation and advances differentiation. HiDEP-1 cells were transduced with ZNF648, ZNF648-GFP or a control green fluorescent protein (GFP) construct. (A) Western blot of HiDEP-1 cells probed with ZNF648 antibody (Abcam). Molecular wieght (MW) markers shown on left hand side (B) Images of control and HiDEP-1 cells transduced with ZNF648-GFP; DAPI nuclear staining in blue. (C) Relative fold expansion of ZNF648 and ZNF648-GFP expressing HiDEP-1 cells compared to untransduced (UT) and GFP control cells. *GFP vs. ZNF648, +GFP vs. ZNF648-GFP. **/++ P<0.01; ***/+++ P<0.001, n=2 each for ZNF648 and ZNF648-GFP ± standard deviation, t-test. (D) proportion of HiDEP-1 cells at each stage of differentiation present at day 12 of culture. ProE: proerythroblast; BasoE: basophilic erythroblast; PolyE: polychromatic erythroblast; OrthoE: orthochromatic erythroblast; Retics: reticulocytes. **P<0.01, ***P<0.001, 2-way ANOVA. N=3 ± standard deviation.
ZNF648 knockdown impedes erythroid cell differentiation
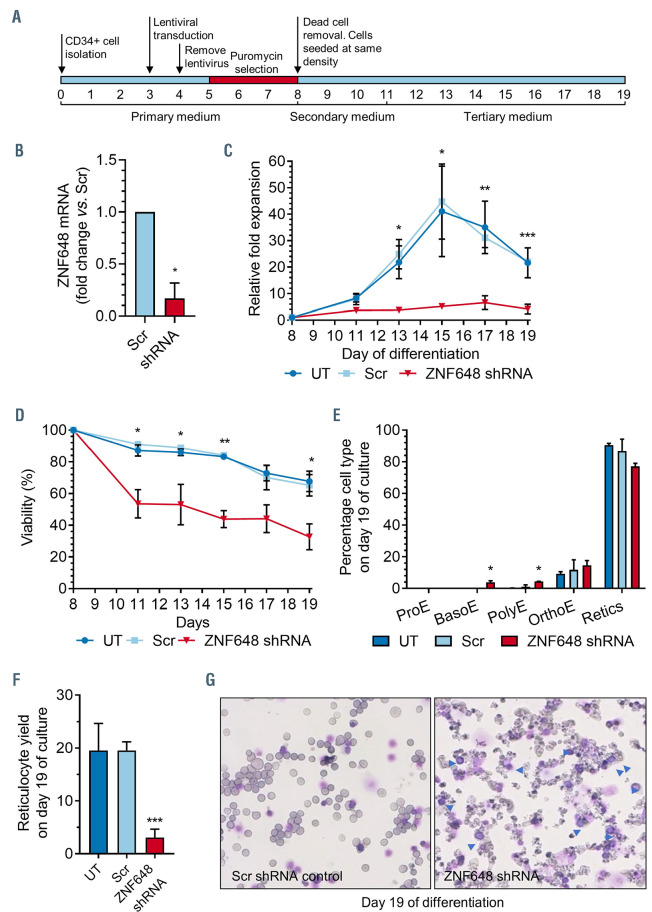
Erythroid cells differentiated from adult PB CD34+ cells were transduced with a ZNF648 shRNA or with a Scr control shRNA at day 3 in culture. Following puromycin selection, dead cells were removed, and surviving cells seeded at the same number and density (schematic of protocol shown in Figure 3A). Knockdown (KD) of ZNF648 was verified by qPCR which showed an average of ∼85% reduction in transcripts compared to control (Figure 3B). We also obtained ZNF648 protein abundance values from TMT labeled tryptic peptides analyzed by nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC MS/MS) (see below). ZNF648 protein level was reduced by ∼70% (data from 11 PSM).
Morphological analysis showed no difference in the population of seeded cells between ZNF648 KD and Scr control cultures, with the majority of cells having the appearance of pro-erythroblasts (Online Supplementary Figure S4). Cells were maintained in our erythroid culture system with counts performed and samples taken for analysis throughout differentiation (day 8, 11, 13, 15, 17 and 19 of culture).
Figure 3.
ZNF648 knockdown impedes erythroid differentiation. Erythroblasts differentiated from adult peripheral blood (PB) CD34+ cells at day 3 in culture were transduced with scrambled (Scr) short hairpin RNA (shRNA) as control or ZNF648 shRNA, puromycin selected and seeded at same cell density along with untransduced cells (UT) that served as a further control. (A) Schematic of experimental design. (B) quantitative polymerase chain reaction (qPCR) of ZNF648 transcript levels normalised to Scr control. (C) Relative fold expansion during differentiation compared to cell number at day 8 in culture. (D) Viability of cells during differentiation assessed by trypan blue exclusion. (E) Proportion of cells at different stages of differentiation at day 19 in culture. ProE: proerythroblast; BasoE: basophilic erythroblast; PolyE: polychromatic erythroblast; OrthoE: orthochromatic erythroblast; Retics: reticulocytes. (F) Reticulocyte yield at day 19 of culture. (G) Morphology of cells at day 19 of culture stained with May- Grünwald-Giemsa, representative images of 3 independent cultures. *P<0.05, **P<0.01, ***P<0.001, ttest, n=3 ± standard devation.
ZNF648 KD had a striking, negative effect on the expansion and viability of cells, with significantly fewer cells recorded at all time points from day 13 (P<0.05 to P<0.001, n=3), due to both decreased cell expansion rate and significantly increased cell death (P<0.05 to P<0.01, n=3; Figure 3C and D). However, a proportion of cells do survive, possibly due to higher levels of remaining ZNF648, but their differentiation was impaired, with significantly more (P<0.05, n=3) early erythroid cells (basophilic and polychromatic normoblasts) still present at day 19 of culture compared to control cultures (Figure 3E). In addition, although the percentage of orthochromatic normoblasts appears similar between the controls and ZNF648 KD cultures at day 19 this is due to a combination of more control cells having already enucleated (with ∼30% more reticulocytes; Figure 3E) and significantly more ZNF648 KD cells dying (Figure 3D; P<0.05, n=3), possibly due to impeded enucleation. The reticulocyte yield, calculated as percentage of enucleation multiplied by the cumulative fold expansion,39 is also significantly decreased (P<0.001, n=3) in the ZNF648 KD cultures (Figure 3F). High levels of debris were clearly visible in the ZNF648 KD cultures due to the increased cell death (Figure 3G).
We also analyzed four other ZNF648 shRNA. Unlike the above ZNF648 shRNA which targets the 3’ non-coding region of ZNF648, these shRNA all have complementary sequences within the ZNF648 coding region. We were therefore able to determine their efficiency by evaluating reduction in the level of ectopically expressed ZNF648 on immunoblot (Supplementary Figure 5A and B). Three of the shRNA did not reduce ZNF648 levels. However, ZNF648 shRNA2 reduced ectopic ZNF648 protein ∼2-fold (also see Online Supplementary Figure 6A). In order to compare endogenous protein levels, we again used abundance values from mass spectrometry (TMT labelled tryptic peptides analysed by nano-LC MS/MS) of adult erythroid cells transduced with Scr or ZNF648 shRNA2, which showed ∼20% reduction in endogenous ZNF648. Hence, ZNF648 shRNA2 reduces ZNF648 levels, but not to the same magnitude as the above ZNF648 shRNA.
We therefore used ZNF648 shRNA2 in our KD protocol. Expansion of cells transduced with ZNF648 shRNA2 was reduced (Online Supplementary Figure 6B), however, the magnitude decrease was less (e.g., at day 15, 4.5-fold less) than for ZNF648 shRNA, in line with the less efficient KD of ZNF648 by ZNF648 shRNA2. Terminal differentiation was also impaired, with significantly more orthochromatic normoblasts (P<0.05, n=3; Online Supplementary Figure S6C) still present at day 19 of cultures compared to controls in which significantly more cells had already enucleated (P<0.05, n=3; Online Supplementary Figure S6C). The reticulocyte yield of ZNF648 shRNA2 cultures at day 19 was also significantly lower (P<0.05) than control cultures (Online Supplementary Figure S6D). The effect of ZNF648 shRNA2 therefore parallels that of the first ZNF648 shRNA but the effects are less pronounced due to the smaller magnitude decrease in ZNF648 levels.
Rescue of ZNF648 knockdown improves phenotype
We also rescued ZNF648 KD cells by co-transduction of ZNF648 shRNA and ZNF648 coding region tagged with GFP (Online Supplementary Figure S7); as shown above (Figure 2) ZNF648-GFP and untagged ZNF648 have the same effect on proliferation and viability. Co-transduction was required due to death of cells immediately following ZNF648 KD. As ZNF648 shRNA targets the 3’ non-coding region of ZNF648 it cannot target the ZNF648 transgene. The increase in ZNF648 transcript levels following transduction is shown in the Online Supplementary Figure S7C (day 8 panel), with densitometry values from ZNF648 western blot showing >50-fold increase in ZNF648 protein level. A schematic of the protocol is shown in the Online Supplementary Figure S7A, with GFP+ cells isolated from the co-transduced population by fluorescence- activated cell sorting (FACS) and cells from all cultures seeded at the same density in erythroid culture.
The viability of the ZNF648 KD population again declined to <50% of control cells by day 13 of culture (Online Supplementary Figure S7B). In contrast, viability of the rescued population mirrored that of the Scr shRNA control for the first 3 days, but declined to a greater extent thereafter, although was higher than that of the ZNF648 KD population to day 19. The lower viability of the rescued culture seen by day 13 compared to control may be due to a more rapid loss of ZNF648. As the ZNF648 transgene lacks a 3’ non-coding region it is less stable than endogenous transcripts, decreasing its half-life. This is exacerbated as differentiation continues, as the nuclei condense, and transcriptional activity reduces. A reduced level of ZNF648-GFP mRNA and protein is indeed seen by day 13 of culture (Online Supplementary Figure S7C and D).
ZNF648 rescue also significantly improved the differentiation potential of the cells (Online Supplementary Figure S7E and F), with no basophilic or polychromatic normoblasts remaining at day 19 in culture, in contrast to the ZNF648 KD cultures, along with ~30% more reticulocytes. There was also notably less debris in the rescue than ZNF648 KD cultures, more clearly seen at day 11 (Online Supplementary Figure S7F). However, the enucleation rate of the rescued population was not fully recovered, being still ~35% lower than the control. Again, this is likely due to physiological levels of ZNF648 not being achieved. Nevertheless, the data clearly supports that the observed phenotype following ZNF648 KD is due to reduced levels of ZNF648.
Perturbation of the transcriptome and proteome following ZNF648 knockdown.
Analysis of transcriptome
In order to determine the mechanism by which loss of ZNF648 impedes erythroid differentiation, and results in reduced viability, we initially looked at transcript levels for key erythroid transcription factors and proteins following ZNF648 KD in adult erythroid cells on day 8 of culture (5 days post transduction with ZNF648 shRNA) by PCR. Expression of transcription factors GATA1, FOG1, KLF1 and BCL11A-XL were unchanged, as was AHSP. However, there was a decrease in expression of HBB (-globin), SCL4A1 (Band 3), GYPA (glycophorin A) and CA1 (carbonic anhydrase 1) (Online Supplementary Figure S8), possibly indicating some defect or delay in differentiation even in these early cells, despite no obvious morphological differences compared to control cells. There was also reduced expression of E2F2, consistent with decreased cell expansion rate.40
For a more in-depth and global analysis of the effect of ZNF648 KD, comparative transcriptomic analysis was performed. Following transduction of adult erythroid cells with Scr or ZNF648 shRNA, total RNA was extracted at day 8 from three independent cultures and used to screen human genome arrays. Following statistical analysis, 299 probe ID (representing 208 unique genes) were down-regulated ≥2-fold and 124 probe ID (representing 84 unique genes) were up-regulated ≥2-fold upon ZNF648 KD (Online Supplementary Table S1).
All genes with increased or decreased expression ≥2- fold were entered into the Reactome Knowledgebase (https://reactome.org) to reveal significantly over-represented pathways in the datasets.41,42 Identified pathways, sorted by P-value, are provided unless too few proteins (<5) were assigned or pathway association was not significant (P>0.05).
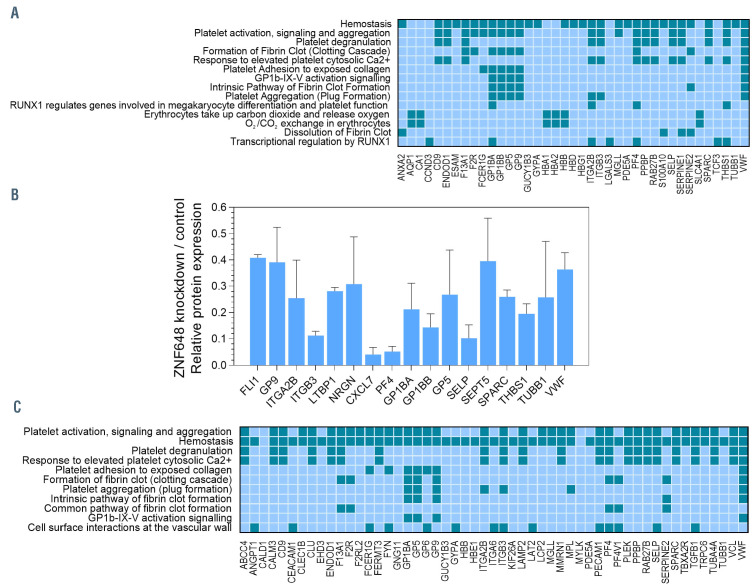
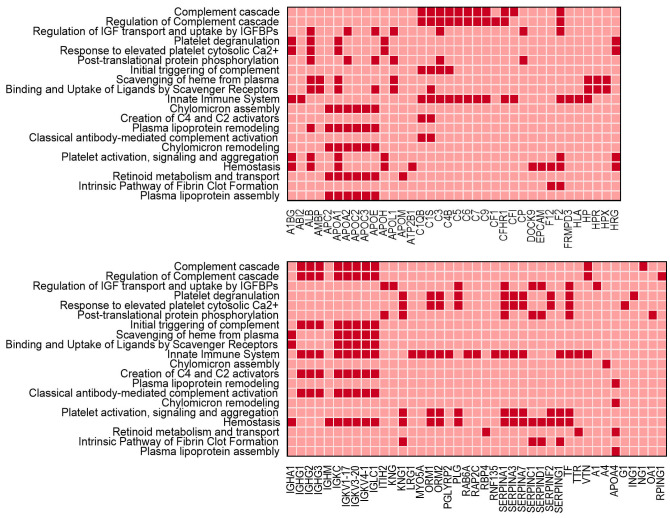
Of the genes with decreased expression, 153 were found in Reactome with the data demonstrating significant over-representation of megakaryocyte genes, with almost all identified pathways associated with megakaryocyte or platelet regulation and function (Figure 4A). Of the genes with increased expression, 47 were found in Reactome but no pathways were significantly over-represented.
Analysis of proteome
Transcript abundance does not directly correlate with protein abundance, due to substantial post-transcriptional, translational and protein degradation regulation.43 We therefore investigated the change in proteome following ZNF648 KD to further, and more directly inform how ZNF648 KD disrupts erythropoiesis, helping delineate its role in this process.
TMT comparative proteomics was performed on adult erythroid cells on day 8 in culture following transduction with Scr or ZNF648 shRNA, as for the transcriptomic analysis above. A total of 7,868 unique proteins were quantified, with 123 unique proteins decreased in level by ≥2-fold following ZNF648 KD (Online Supplementary Table S2A). In keeping with the transcriptomic data, interrogation of the proteomic dataset revealed a striking number of these 123 proteins are known megakaryocyte proteins (14%; Figure 4B). Following analysis with Reactome, almost all assigned pathways were again associated with megakaryocyte or platelet regulation and function (Figure 4C; 86 of the 123 proteins found in Reactome). Of note, six of the ten proteins with greatest magnitude decrease in level were megakaryocyte-associated proteins (Online Supplementary Figure S9). These proteins were all found to decrease in level during normal erythropoiesis (Online Supplementary Figure S10), hence their decrease on ZNF648 KD is not due to delayed differentiation. Megakaryocyte proteins in Figure 4B were also decreased on transduction of cells with ZNF648 shRNA2 but by 20-30% compared to 50-70% with ZNF648 shRNA, in line with the magnitude of ZNF648 KD by the two shRNA.
Figure 4.
Megakaryocyte-associated proteins and pathway classification of transcripts and proteins decreased in level following ZNF648 knockdown. Total RNA and whole cell protein lysates were prepared from erythroid cells differentiated from adult peripheral blood (PB) CD34+ cells transduced with scrambled (Scr) or ZNF648 short hairpin RNA (shRNA), at day 8 in culture. RNA was used to screen human genome arrays, with transcripts decreased in level by ≥2-fold following ZNF648 knockdown (KD) entered into Reactome. Data from three independent KD experiments (A). Tryptic peptides were prepared from cell lysates and labeled with Tandem Mass Tags followed by nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC MS/MS) with (B) relative decrease in level of megakaryocyte associated proteins following ZNF648 KD, identified amongst the 123 unique proteins decreased by ≥2-fold. Data from two independent KD experiments ± standard deviation, and (C) Reactome analysis of the proteins decreased in level. In (A) and (C) dark shades indicate genes/proteins apportioned to the pathway shown on the left. Pathways shown with minimum probability of over representation in the dataset, corrected for false discovery rate of P<0.006 for transcripts and P<1.42x10-5 for proteins.
175 unique proteins were found to be increased in level following ZNF648 KD (Online Supplementary Table 2B), 119 of which were found in Reactome. Interrogation of the pathways assigned (Figure 5) revealed proteins typically expressed by cells of the monocyte lineage (e.g., apolipoproteins, complement cascade and regulatory components, SERPINS) as well as immunoglobulin chains normally synthesized by B cells, although some of the above are also expressed by granulocytes and megakaryocytes.
Consistent with the PCR data, no change in the level of GATA1, FOG1, KLF1, BCL11A-XL and AHSP, as well as key erythroid proteins ITGA4, CD71, CD44, -Spectrin or -Spectrin was detected in the transcriptomic or proteomic datasets. A decreased level of -globin and glycophorin A was detected in both datasets, and of Band 3 and CA1 in the proteomic dataset, in line with the PCR data. The raw data for the transcriptomic analysis did also show a decrease for E2F2, SCL4A1 and CA1 but levels were variable and therefore did not pass the statistical cut-offs.
Overall, the data demonstrate a distorted genetic readout on ZNF648 KD, with altered expression of genes normally associated with cells of other haematopoietic lineages. In particular, there was a striking effect on proteins associated with megakaryocyte or platelet function.
Figure 5.
Pathway classification of proteins increased in level following ZNF648 knockdown. Tandem Mass Tag labeling and nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC MS/MS) was performed on whole cell lysates from erythroid cells transduced with ZNF648 or scrambled (Scr) (control) short hairpin RNA (shRNA) at day 8 in culture. Proteins increased in level following ZNF648 knockdown were entered into Reactome. Dark red indicates proteins apportioned to the pathway shown on the left. Top 20 pathways shown with minimum probability of over representation in the dataset, corrected for false discovery rate, of P<2.11x10-5.
Proteins increased in level following ZNF648 knockdown are also increased in erythroid cells differentiated from three induced pluripotent stem cell lines
As ZNF648 is reduced in level in erythroid cells differentiated from our three iPSC lines, we cross referenced the most abundantly upregulated proteins (≥4-fold; 112 proteins) following ZNF648 KD with our previously published proteomic dataset of erythroid cells differentiated from the iPSC lines compared to differentiation stage matched normal adult erythroid cells.12 Of the 112 proteins, 69 were found in the iPSC proteomic dataset, and of these 46 (67%; Online Supplementary Table S3) were also increased in the iPSC-derived erythroid cells, with the same pathways assigned by Reactome (Online Supplementary Figure S11). Proteins that increased in level by <4-fold did not show a correlative increase in the iPSC-derived erythroid cells.
Analysis of all proteins increased in the iPSC-derived erythroid cells (≥2-fold; see Trakarnsanga et al.12) for datasets) highlighted dysregulation of the same proteins and pathways as above, but also proteins associated with other pathways, showing greater dysregulation in these cells (Online Supplementary Figure S12). However, there was no correlation between proteins decreased in level following ZNF648 KD and those decreased in iPSCderived versus adult erythroid cells, suggesting differences in the regulation or programming of iPSC-derived erythroid cells compared to adult cells.
ZNF648 knockdown impedes megakaryocyte differentiation
We compared the abundance of ZNF648, as well as abundance of the megakaryocyte proteins that decreased in level in erythroid cells following ZNF648 KD, in erythroblasts and megakaryoblasts. For this we took advantage of available RNA sequencing (RNAseq) data for normal erythroblasts and megakaryoblasts differentiated from CD34+ cells accessible via the Bloodspot website.44,45 The expression of ZNF648 was an order of magnitude lower in erythroblasts compared to megakaryoblasts (see also PCR analysis of ZNF648 transcript levels in erythroblasts and megakaryoblasts, Online Supplementary Figure S13). Similarly, the expression of all 17 megakaryocyte proteins shown in Figure 4B were between 2- to 8-fold lower in erythroid cells. We confirmed expression of some by PCR (Online Supplementary Figure S13). Higher levels of ITGA2B, PF4, GP9 and ITGB3 in megakaryocytes compared to erythroblasts have been reported previously. 46 As a control, we looked at the expression of erythroid markers Band 3, glycophorin A, ITGA4, CD36, CD44, -Spectrin and -Spectrin in the Bloodspot dataset, all of which were at a higher level in erythroblasts than megakaryoblasts as expected.
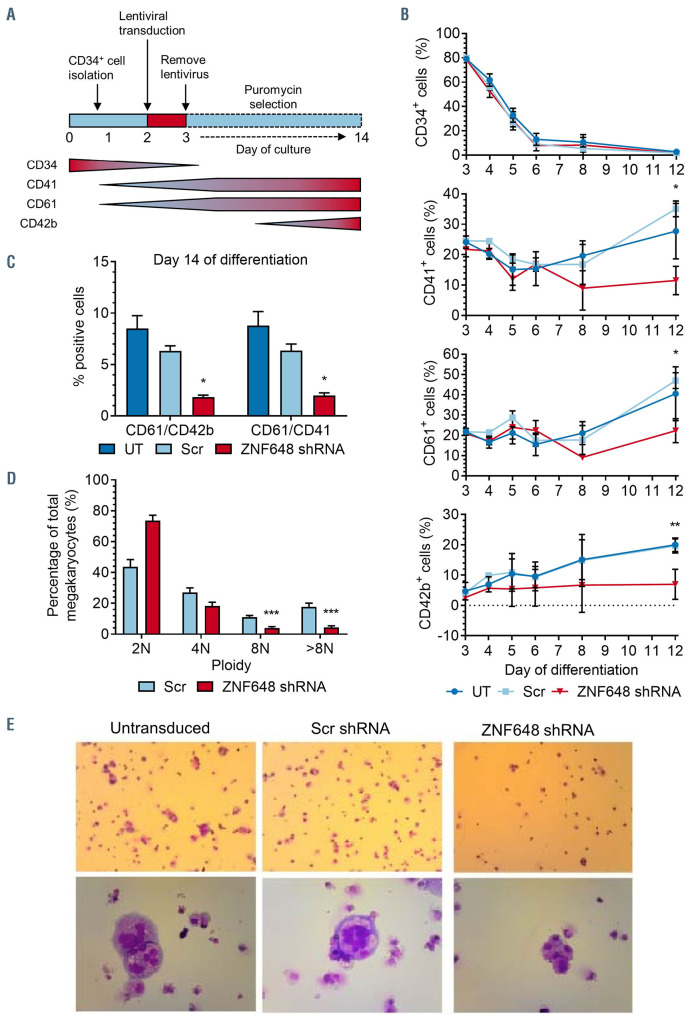
Finally, using the same ZNF648 shRNA, we knocked down ZNF648 in megakaryoblasts to determine if ZNF648 also plays a regulatory role in this lineage.
Adult CD34+ cells transferred to megakaryocyte culture medium were transduced on day 1 with ZNF648 shRNA or with the control Scr shRNA, followed by puromycin selection. Untransduced cells served as a further control (schematic of experimental design shown in Figure 6A). As expected, there was similar loss of CD34 expression with time in all cultures (Figure 6B). In order to determine the effect of ZNF648 KD on megakaryopoiesis, megakaryocyte membrane protein marker expression was assessed during differentiation. There was no significant difference between control and ZNF648 KD cultures from day 3 through to day 8, although the level of CD42b was consistently lower in the latter. However, on day 12 the levels of CD61, CD41 and CD42b were significantly lower in the ZNF648 KD cultures, indicating a defect in megakaryocyte differentiation (Figure 6B). In order to further assess megakaryocyte differentiation, we measured the levels of CD61 and CD41 as indicators of commitment to the megakaryocyte lineage, and CD61 and CD42b as indicators of mature megakaryocytes on day 14 of culture. As seen in Figure 6C both megakaryocyte commitment and maturity were significantly reduced following ZNF648 KD. Finally, a unique characteristic of megakaryocytes is their capacity to become polyploid. We therefore measured the ploidy status of CD61+ cells. There was a significant shift to lower ploidy status for the ZNF648 KD cells (Figure 6D). This is also seen in the cell images where fewer large cells are present in ZNF648 KD cultures (Fig. 6E, upper panel) and those detected have less nuclear content (Figure 6E, lower panel), indicating the cells are not able to mature properly to large polyploid megakaryocytes. Overall, the data support a role for ZNF648 in megakaryocyte differentiation.
ZNF648 sequence conservation and evolution origins
Conservation of protein sequences across species can indicate functional importance. Having established a role for ZNF648 in humans, we next investigated the conservation and profile of ZNF648 through evolution to gain further information on this novel protein.
We used BLAST and HMMER sequence similarity searches to identify homologues of ZNF648 across eukaryotes, then inferred phylogenetic trees to determine orthology relationships and pinpoint the evolutionary origin of the ZNF648 gene.
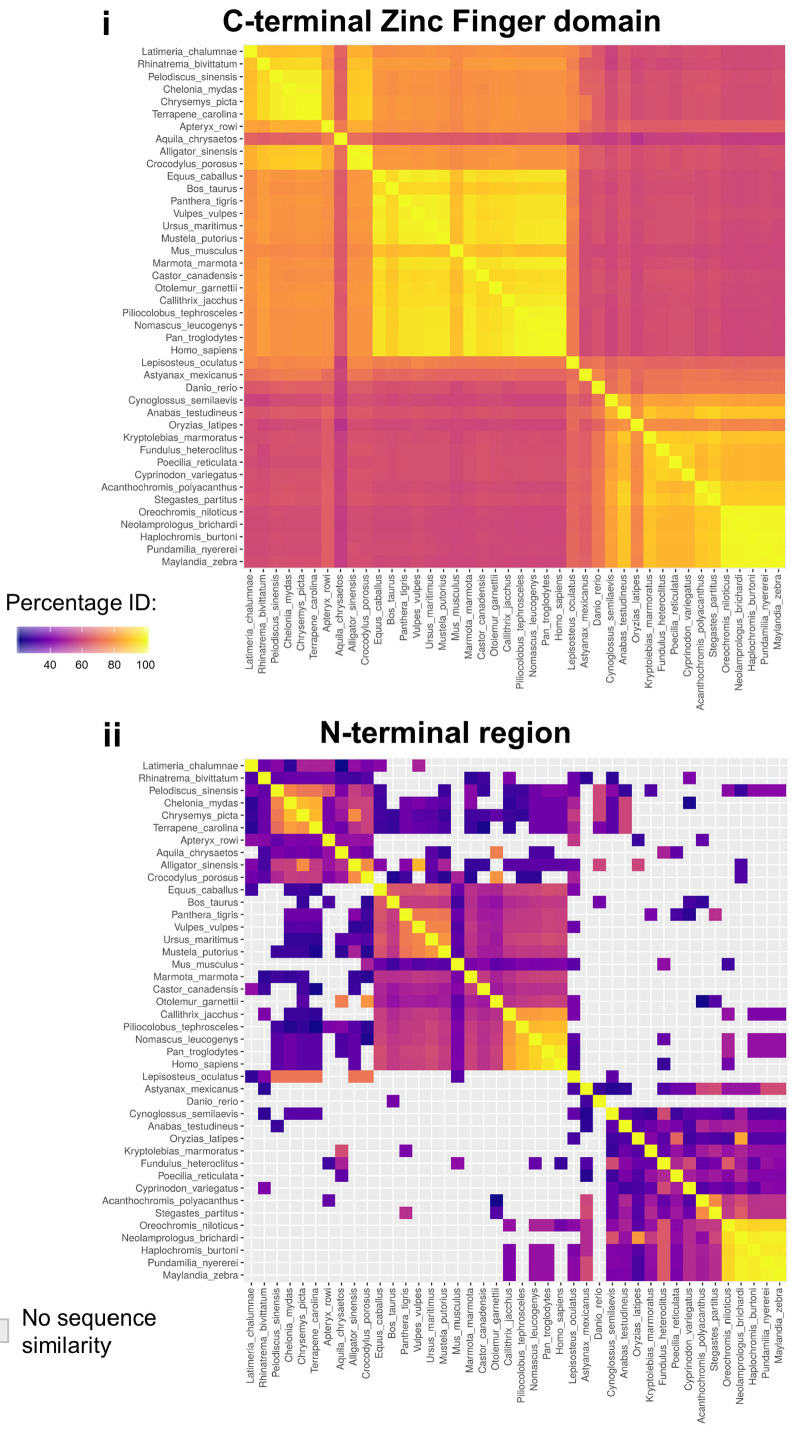
Among mammals, ZNF648 is conserved (Online Supplementary Table S4). However, the C-terminus, which contains ten tandemly arranged C2H2 ZnF motifs in all, is more highly conserved than the N-terminus (94-100% amino acid sequence identity in the C-terminus, human residues 279-568, compared to 50-98% N-terminal sequence identity, residues 1-278). The N-terminal region was however conserved between closely related family members (Online Supplementary Table S5).
Broadening our search beyond mammals, we detected orthologues of ZNF648 across Reptilia, in some Amphibia and Aves, in Coalacanthiformes and across the bony fish (Actinopterygii) but not outside this group, suggesting that the gene originated in the common ancestor of Osteichthyes (Euteleostomi) after their evolutionary divergence from gnathostomes. As in mammals, ZNF648 C-terminal conservation (28-100%) is higher than N-terminal conservation (21-97%), with the N-terminus recognisably similar within, but not between, major lineages (Figure 7). This suggests that the N-terminus of ZNF648 evolves faster than the C-terminus, perhaps due to reduced or lineage-specific functional constraints. We hypothesize that the C-terminus (containing the ZnF motifs) evolves slowly because it performs the same function, binding to a DNA target motif, while the N-terminus may vary in function across lineages. We used sensitive profile-profile sequence searches but were unable to detect any significant similarity between the N-terminus of the ZNF648 protein family and any characterized database sequences, suggesting that this represents a novel functional domain; in other ZnF proteins, the non-ZnF region of the protein acts as an effector domain, for example binding to other proteins to bring about transcription repression.32
Figure 6.
ZNF648 knockdown impedes megakaryocyte differentiation. Isolated adult peripheral blood CD34+ cells were transferred to megakaryocyte differentiation medium and transduced with scrambled (Scr) short hairpin RNA (shRNA) as control or ZNF648 shRNA followed by puromycin selection. Untransduced cells served as a further control. (A) Schematic of experimental design and expression profile of CD34, CD41, CD61 and CD42b during normal megakaryopoiesis, (B) expression of membrane markers CD34, CD41 (platelet glycoprotein IIb), CD61 (platelet glycoproteins IIIa) and CD42b (GPIb ) analysed by flow cytometry with antibodies CD41- PE, CD61-APC (both from Biolegend), CD34-VB BD and CD42b-PE BD (both from Pharming). Data was acquired with a MacsQuant VYB Analyser using a plate reader, n=6. (C) Proportion of CD41/CD61 and CD61/CD42b positive cells on day 14 of culture, n=6 ± standard error of the mean. (D) Ploidy status of cells at day 14 of culture, n=3. (E) Morphology of cells at day 14 of culture stained with May-Grünwald-Giemsa. Upper panel 100x magnification, lower panel 400x magnification. Images representative of three independent cultures. *P<0.05, **P<0.01, ***P<0.001, Student’s t-test.
Figure 7.
Pairwise sequence identity and phylogeny of representative ZNF648 orthologues across Osteichthyes. (A) (i) C-terminus (human residues 279-568;), (ii) N-terminus (human residues 1-278). Warm colors indicate higher pairwise percentage identity. The C-terminus (containing the ZnF motifs) is conserved across Osteichthyes, but the N-terminus is much less conserved, showing high identity only within major lineages.
Notwithstanding, proteins or regions with low sequence similarity can still have homologous function via adoption of a similar structure47 so we analysed the Nterminal half of ZNF648 from a range of species to see if residues shared a common fold. All sequences were aligned using Clustal Omega48 and a secondary structure prediction carried out by PsiPred.49 From this secondary structure alignment, confidence that the N-terminal half of these proteins share a common fold is low. However, there are two significant patches of sequence with shared charge similarity between all mammalian sequences but also found in reptilia and coelacanth (Online Supplementary Figure S14), which suggests that even if they do not share the same fold they may have targeting or protein-protein interaction sites in common.
In the C-terminus, the number of detected C2H2 zinc finger motifs varies among taxa, from e.g., 11 in Reptilia, ten in mammals to seven in Aves (see schematic in the Online Supplementary Figure S15). All outside the mammalian clade have an additional conserved C2H2 ZnF at the N-terminal end of the ZnF domain, lost in mammals. Of potential interest only Reptilia and Coelacanth have all 10 conserved mammalian ZnF, which along with the conserved patches of charge similarity in their N-terminal region suggests a possible closer functional relationship between ZNF648 in these species.
Figure 8.
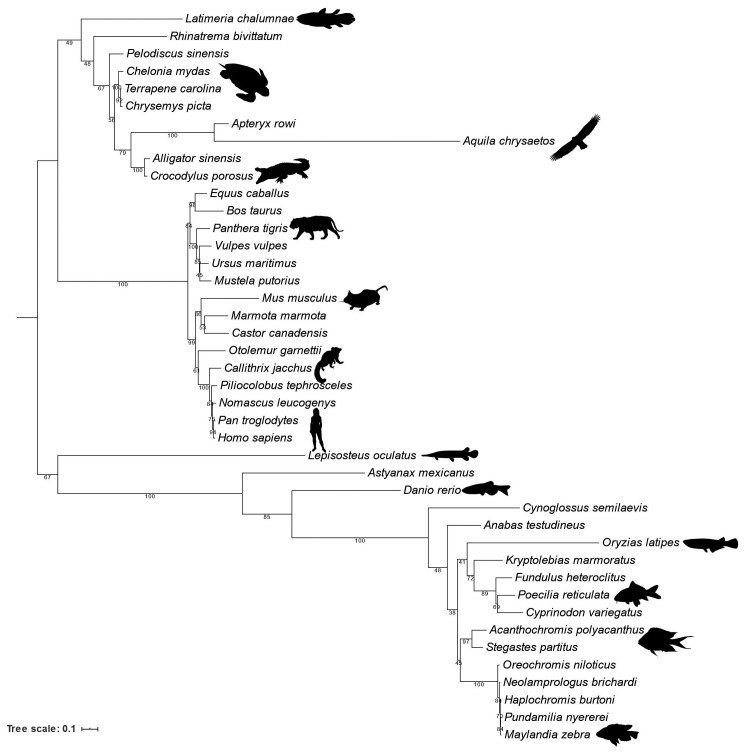
ZNF648 phylogenetic tree. A maximum likelihood phylogeny of ZNF648 was inferred (949 aligned sites/1134 positions/42 taxa) using the best-fitting LG+C60+G+F model in IQ-Tree 1.6.10. Branch supports are 1,000 ultrafast bootstraps. ZNF648 is conserved across the major lineages of Osteichthyes (Euteleostomi), but several lineage-specific losses have occurred. The topology of the tree is congruent with our understanding of species relationships, suggesting that the gene traces back to the common ancestor of Osteichthyes.
While the ZNF648 gene has a broad phylogenetic distribution in bony fishes, it has been lost repeatedly and independently in a number of descendant lineages, particularly in many lineages of birds, and in the common ancestor of all Lepidosaurians (snakes, lizards and tuatara). The patchy distribution of ZNF648 orthologues across these groups appears to be the result of independent losses rather than gains (for example, by de novo origin of a new C2H2 ZnF-motif containing sequence) because the retained sequences follow the species tree (Figure 8). Phylogenetic trees inferred separately for the C-terminal ZnF domain and N-terminal region (Online Supplementary Figure S16A and B) are consistent with the hypothesis that both domains have evolved on the same underlying tree, indicating that while N-terminal sequence conservation is low the two domains are likely to have evolved together as a unit since the common ancestor of bony fish.
Discussion
Our data show that ZNF648 is required for both erythroid and megakaryocyte differentiation in humans, with the order of magnitude higher level of ZNF648 in megakaryoblasts suggesting differential levels of ZNF648 may be required by the two lineages. In addition, the striking reduction in abundance of proteins associated with the megakaryocyte lineage following ZNF648 KD in erythroid cells supports a function for these proteins in erythropoiesis.
Of interest, the level of transcription factor Fli1 was found to be decreased in erythroid cells in both the transcriptomic and proteomic datasets following ZNF648 KD. Fli1 is critical for megakaryopoiesis,50 with cross-antagonism between FLI-1 and KLF1, a transcription factor essential for erythroid differentiation,5 implicated in the control of erythroid versus megakaryocyte lineage differentiation. 51 However, Fli1 mutant mice have aberrant red blood cell development, as well as severely impeded megakaryopoiesis, 52 suggesting a requirement for Fli1 or its relative level also in erythropoiesis. Fli1 directly regulates the expression of many megakaryocyte proteins, including ITGA2B, GP1BA, GP9 and PF4,53,54 all decreased in level on ZNF648 KD in erythroid cells. Therefore, ZNF648 may, at least in part, exert its effect upstream of Fli1.
Furthermore, ZNF648 may suppress differentiation to other haematopoietic lineages, as the level of proteins normally expressed by e.g., monocytes is increased following its KD, contributing to the distorted genetic readout, defective phenotypes and cell death observed. Interrogation of RNAseq datasets via the Bloodspot website44,45 showed ZNF648 is also expressed by neutrophils, monocytes, CD8+ T cells and hematopoietic stem cells as well as megakaryocytes and erythroid cells. Megakaryocytes have the highest level of expression, with the other cell types having similar levels to erythroid cells.
We do not yet know how ZNF648 functions, but its specific nuclear localisation and presence of canonical linker sequences between ZnFs 1-3 and 5-8 support it as a DNAbinding transcriptional regulator. It is known that binding of adjacent poly C2H2 ZnF proteins to DNA is carried out by 24–75% of the ZnF, with two to three successive fingers responsible for the specific DNA motif interaction, with other fingers facilitating binding. For example, TFIIIA with nine ZnF binds its DNA recognition motif via fingers 1-3, with finger 5 and 7-9 also interacting with the DNA. Similarly, analysis of the linker sequences between ZnF in ZNF648 suggest specific DNA recognition by fingers 1-3 as the consensus linker sequence, inducing highest binding affinity, is found between fingers 2 and 3, but that fingers 5-8 also interact. The remaining ZnF in such proteins may interact with other molecules, including RNA and proteins (reviewed by Iuchi1). Notwithstanding, it has also been suggested that different groupings of adjacent ZnF in poly-ZnF proteins bind different specific sites in the genome, thus independently regulating a transcriptional programme.32 Alternatively, ZNF648 could interact with DNA indirectly via interaction with other factors, act as a scaffold or modulate the binding of other DNA-binding proteins. Future work elucidating the target DNA-binding site(s) and interacting partners for ZNF648 will help reveal the specific function of this protein.
In order to further determine the functional significance of ZNF648 we explored its sequence conservation across a wide range of species and through evolution, identifying a clear common ancient ancestor, that of Osteichthyes (Euteleostomi/bony fish). This is in contrast to the large number of poly-ZnF proteins encoded in the human genome which are postulated to be a recent invention, the ancestral size of this gene family being small with rapid expansion on the primate lineage; a substantial proportion of the proteins having no mouse ortholog.32 The biological function of a large majority of these proteins is still unknown. Instead ZNF648 may have occurred with a much earlier expansion of C2H2-ZnF proteins in metazoans. 55
There has been clear differential divergence within the ZNF648 sequence through evolution. The C-terminal sequential C2H2 ZnF domain is highly conserved across mammals, reptilians, actinopterygii, in a member of the Sarcopterygii order Coelacanth and the amphibian Caecilian. In contrast the remaining N-terminal region of the protein is much less conserved; it is recognizably similar within each of the major groups surveyed (mammals, reptiles, actinopterygians), with greater conservation between more closely related members, but similarity between groups is low. ZNF648 orthologues were clearly distinguishable from other C2H2 ZnF proteins in our phylogenetic analyses, indicating that the conserved sequences we detected across Osteichthyes do not represent other, more distantly related ZnF families.
Rapid evolution of the N-terminal regions of poly C2H2 ZnF proteins, often involving loss or gain of regulatory KRAB and SCAN domain, has been observed previously for mammalian poly C2H2 ZnF proteins and predicted to be due to an exon-shuffling mechanism.56 In such proteins the ZnF domain is often located within a single exon, with the N-terminal domain on separate exon(s) and separated from the ZnF exon often by a large intron.57,58 However, analysis of the intron/exon organisation of ZNF648 in a range of organisms (mammals, reptilians and actinopterygii) showed the entire coding sequence is within a single exon. It is therefore likely that the common ancestor had a “complete” ZNF648 gene (i.e., with both the N-terminal region and C-terminal ZnF domain), but that the N-terminal region has evolved more quickly than the C-terminal domain. Greater functional constraint on the DNA interacting ZnF domain suggests ZNF648 binds at least some similar regions of DNA in the different organisms. However, divergence of the N-terminal region, which likely contains binding sites for proteins that regulate ZNF648 function but has a potentially novel functional domain, having no sequence similarity to known regulatory domains, may enable differential expression of genes in the different organisms. This could essentially create control points connecting cellular requirements to DNA-binding, and hence control of gene expression required for the different environmental conditions of the various organisms.
Finally, ZNF648 in humans is also expressed in other non-hematopoietic cell types (Online Supplementary Figure S17) indicating a broader regulatory role. Hence, although all organisms with a ZNF648 gene have red blood cells and platelets (or the more primitive thrombocytes in lower vertebrates) with ZNF648 possibly involved in regulating the differentiation of these cells throughout evolution, it may additionally or alternatively be involved in other processes.
In conclusion, in humans the novel C2H2 ZnF protein ZNF648 is involved in the regulation of both erythroid and megakaryocyte differentiation. Functional importance of ZNF648 is further implied by its maintenance through evolution from the common ancestor of Osteichthyes, with conservation within the ZnF domain but divergence of the N-terminal region allowing adaptation of function in the different organisms.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Kate Heesom, Director of the Bristol University Proteomic Facility, UK for performing Mass Spectrometry, the University of Bristol Genomics Facility, Dr Lee Carpenter, Oxford for providing the iPSC, Profs Yukio Nakamura and Ryo Kurita, RIKEN BioResource Research Center, Japan for the HiDEP-1 cells.
Funding Statement
Funding: The work was funded by the Government of Brunei Darussalam via an In-Service Training Scholarship to JHM, The Wellcome Trust (grant numbers 087430/Z/08 and 102610), BrisSynBio a BBSRC/EPSRC Synthetic Biology Research Centre (BB/L01386X/1) and NIHR Blood and Transplant Research Unit (NIHR BTRU) in Red Cell Products (IS-BTU-1214-10032). This manuscript presents independent research funded by the National Institute for Health Research (NIHR). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. T.A.W. is funded by a Royal Society University Research Fellowship (UF140626). ERR is funded by a Royal Society Fellows Enhancement Award to TAW (RGF\EA\180199)
References
- 1.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58(4):625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43(1):1-21. [DOI] [PubMed] [Google Scholar]
- 3.Vaquerizas JM, Kummerfeld SK, Teichmann SA, et al. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252-263. [DOI] [PubMed] [Google Scholar]
- 4.Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26(47):6777-6794. [DOI] [PubMed] [Google Scholar]
- 5.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118(8):2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffiths RE, Kupzig S, Cogan N, Mankelow TJ, et al. Maturing reticulocytes internalize plasma membrane in glycophorin A-containing vesicles that fuse with autophagosomes before exocytosis. Blood. 2012;119(26):6296-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmins NE, Athanasas S, Gunther M, et al. Ultra-high-yield manufacture of red blood cells from hematopoietic stem cells. Tissue Eng Part C Methods. 2011;17(11):1131-1137. [DOI] [PubMed] [Google Scholar]
- 8.Kupzig S, Parsons SF, Curnow E, et al. Superior survival of ex vivo cultured human reticulocytes following transfusion into mice. Haematologica. 2017;102(3):476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias J, Gumenyuk M, Kang H, et al. Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev. 2011;20(9):1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn I, Klich K, Arauzo-Bravo MJ, et al. Erythroid differentiation of human induced pluripotent stem cells is independent of donor cell type of origin. Haematologica. 2015;100(1):32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapillonne H, Kobari L, Mazurier C, et al. Red blood cell generation from human induced pluripotent stem cells: perspectives for transfusion medicine. Haematologica. 2010;95(10):1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trakarnsanga K, Wilson MC, Griffiths RE, et al. Qualitative and quantitative comparison of the proteome of erythroid cells differentiated from human iPSCs and adult erythroid cells by multiplex TMT labelling and nanoLC-MS/MS. PLoS One. 2014;9(7):e100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen M, Varga E, Aarts C, et al. Efficient production of erythroid, megakaryocytic and myeloid cells, using single cell-derived iPSC colony differentiation. Stem Cell Res. 2018;29:232-244. [DOI] [PubMed] [Google Scholar]
- 14.Olivier EN, Zhang S, Yan Z, et al. PSC-RED and MNC-RED: albumin-free and lowtransferrin robust erythroid differentiation protocols to produce human enucleated red blood cells. Exp Hematol. 2019;75:31-52. [DOI] [PubMed] [Google Scholar]
- 15.Razaq MA, Taylor S, Roberts DJ, et al. A molecular roadmap of definitive erythropoiesis from human induced pluripotent stem cells. Br J Haematol. 2017;176(6):971-983. [DOI] [PubMed] [Google Scholar]
- 16.Salunkhe V, Papadopoulos P, Gutierrez L. Culture of megakaryocytes from human peripheral blood mononuclear cells. Bioprotocol. 2015;5(21):e1639. [Google Scholar]
- 17.Trakarnsanga K, Wilson MC, Lau W, et al. Induction of adult levels of beta-globin in human erythroid cells that intrinsically express embryonic or fetal globin by transduction with KLF1 and BCL11A-XL. Haematologica. 2014;99(11):1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satchwell TJ, Hawley BR, Bell AJ, et al. The cytoskeletal binding domain of band 3 is required for multiprotein complex formation and retention during erythropoiesis. Haematologica. 2015;100(1):133-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singleton BK, Burton NM, Green C, et al. Mutations in EKLF/KLF1 form the molecular basis of the rare blood group In(Lu) phenotype. Blood. 2008;112(5):2081-2088. [DOI] [PubMed] [Google Scholar]
- 20.Trakarnsanga K, Griffiths RE, Wilson MC, et al. An immortalized adult human erythroid line facilitates sustainable and scalable generation of functional red cells. Nat Commun. 2017;8:14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham F, Achuthan P, Akanni W, et al. Ensembl 2019. Nucleic Acids Res. 2019;47(D1):D745-D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Castro E, Sigrist CJ, Gattiker A, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Web Server issue):W362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Team RC. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2017. [Google Scholar]
- 26.Wickham H. ggplot2. Wiley interdisciplinary reviews: computational statistics. 2011;3(2):180-185. [Google Scholar]
- 27.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21(6):1095-1109. [DOI] [PubMed] [Google Scholar]
- 29.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307-1320. [DOI] [PubMed] [Google Scholar]
- 30.Hoang DT, Chernomor O, von Haeseler A, et al. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35(2):518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoang DT, Vinh LS, Flouri T, et al. MPBoot: fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC Evol Biol. 2018;18(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5(1):e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albagli O, Dhordain P, Deweindt C, et al. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ.1995;6(9):1193-1198. [PubMed] [Google Scholar]
- 34.Edelstein LC, Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1-17. [DOI] [PubMed] [Google Scholar]
- 35.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183-212. [DOI] [PubMed] [Google Scholar]
- 36.Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50(3):111-131. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q, Segal DJ, Ghiara JB, et al. , 3rd. Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A. 1997;94(11):5525-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurita R, Suda N, Sudo K, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8(3):e59890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daniels DE, Downes DJ, Ferrer-Vicens I, et al. Comparing the two leading erythroid lines BEL-A and HUDEP-2. Haematologica. 2020;105(8):e389-e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilon AM, Arcasoy MO, Dressman HK, et al. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Biol. 2008;28(24):7394-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabregat A, Korninger F, Viteri G, et al. Reactome graph database: efficient access to complex pathway data. PLoS Comput Biol. 2018;14(1):e1005968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabregat A, Sidiropoulos K, Viteri G, et al. Reactome diagram viewer: data structures and strategies to boost performance. Bioinformatics. 2018;34(7):1208-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4):227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagger FO, Kinalis S, Rapin N. BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 2019;47(D1):D881-D885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Ge B, Casale FP, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167(5):1398-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macaulay IC, Tijssen MR, Thijssen-Timmer DC, et al. Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins. Blood. 2007;109(8):3260-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koonin EV, Galperin MY. Sequence - evolution - function: computational approaches in Comparative Genomics. Boston: Kluwer Academic; 2003. [PubMed] [Google Scholar]
- 48.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195-202. [DOI] [PubMed] [Google Scholar]
- 50.Jackers P, Szalai G, Moussa O, et al. Etsdependent regulation of target gene expression during megakaryopoiesis. J Biol Chem. 2004;279(50):52183-52190. [DOI] [PubMed] [Google Scholar]
- 51.Starck J, Cohet N, Gonnet C, Sarrazin S, et al. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol. 2003;23(4):1390-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawada H, Ito T, Pharr PN, et al. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int J Hematol. 2001;73(4):463-468. [DOI] [PubMed] [Google Scholar]
- 53.Pang L, Xue HH, Szalai G, et al. Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood. 2006;108(7):2198-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Crispino JD, Letting DL, et al. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 2002;21(19):5225-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Najafabadi HS, Garton M, Weirauch MT, et al. Non-base-contacting residues enable kaleidoscopic evolution of metazoan C2H2 zinc finger DNA binding. Genome Biol. 2017;18(1):167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tadepally HD, Burger G, Aubry M. Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains. BMC Evol Biol. 2008;8:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellefroid EJ, Marine JC, Ried T, et al. Clustered organization of homologous KRAB zinc-finger genes with enhanced expression in human T lymphoid cells. EMBO J. 1993;12(4):1363-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huntley S, Baggott DM, Hamilton AT, et al. A comprehensive catalog of human KRABassociated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16(5):669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.