Efforts to combine anti-CD20 antibodies (such as rituximab or obinutuzumab) with BCR inhibitors or venetoclax lead to the necessity to better understand the largely unclear mechanisms of CD20 regulation and its function (s) (reviewed in Pavlasova and Mraz1). This is underscored by the observation that in chronic lymphocytic leukemia (CLL) the combination of ibrutinib with rituximab fails to provide a clinical benefit in comparison to ibrutinib alone2 likely as ibrutinib downmodulates CD20 levels.1,3-5 PI3Kδ inhibitor idelalisib has been approved in combination with rituximab or ofatumumab;6 however, it remains unclear if idelalisib affects CD20 levels or function (s). Here we show for the first time that single-agent idelalisib therapy in CLL leads to CD20 downmodulation in vivo by interfering with a previously unknown mechanism of CD20 transcriptional regulation via the IL4- STAT6 axis. We describe a novel mechanism of CD20 regulation in CLL B cells, which has implications for combinatorial therapy with PI3K inhibitors.
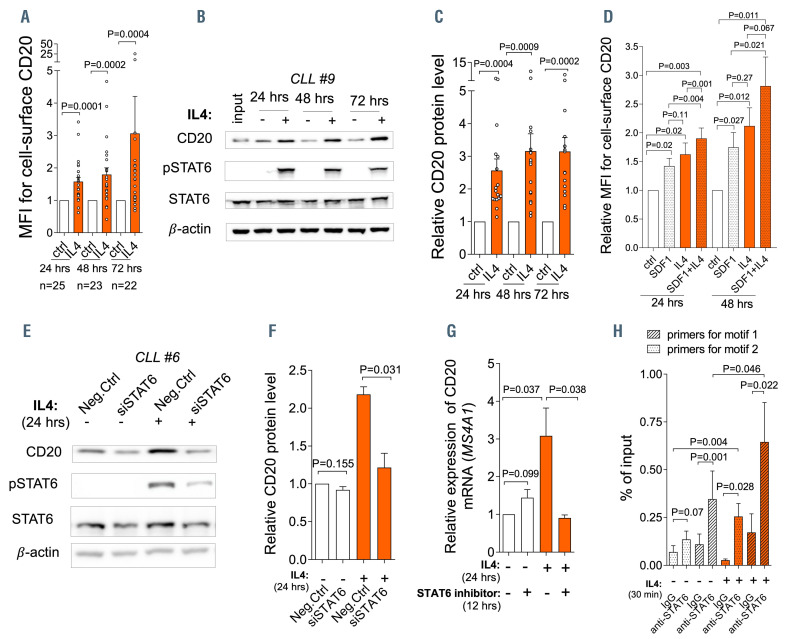
We have recently shown a concurrent upregulation of CD20 and cell surface immunoglobulin M (IgM) in CLL cells from immune niches and demonstrated that this functionally serves to increase BCR signaling propensity. 3,7 The SDF1 (CXCL12) chemokine induces approximately 30-50% upregulation of CD20,3 however, this cannot fully explain the ~2-fold higher CD20 levels in CLL cells from immune niches.3,7 Here we hypothesize that the same factor inducing cell surface IgM in the CLL microenvironment, namely interleukin 4 (IL4) produced by T cells,8 might also be inducing CD20. Indeed, stimulating primary CLL cells with IL4 led to a significant upregulation of CD20 (mean fold-change [FC] =1.6-3.1 [time span, 24-72 hrs], n=25; Figure 1A to C) and IgM on the cell-surface (Online Supplementary Figure S1A). IL4 also induced cell surface CD20 in normal B cells (~2-fold induction; Online Supplementary Figure S1B). This is in line with previous anecdotal observations suggesting a role for IL4 in CD20 regulation.9 We next compared the effects of SDF1 and IL4 on CD20 levels and noted that each factor independently induces CD20, and their combination has a more potent effect (Figure 1D). IgM induction was used as a control in this experiment since it is known to only be induced by IL48 and not SDF1 (Online Supplementary Figure S1C). The effect of IL4 on CD20 was transcriptional since CD20 mRNA (gene MS4A1) was induced similarly to its cell surface levels (Online Supplementary Figure S1D). The IL4 effect was observed irrespective of the immunoglobulin heavy-chain variable region (IGHV) status or the presence of chromosomal aberrations (Online Supplementary Figure S1E to J and data not shown). It is known that IL4 supports CLL cell viability, and to avoid any potential confounding effects from the different viability on CD20 levels, we gated on viable cells and we also separately analyzed CLL samples with comparable viability in control and IL4-treated cells (Online Supplementary Figure S1K to O). Altogether, the data show that IL4 (produced by T cells) induces CD20 in CLL cells.
Figure 1.
IL4 upregulates CD20 expression via STAT6. (A) Cell surface level of CD20 after interleukin 4 (IL4) treatment (20 ng/mL, PeproTech) for 24 hours (hrs) (n=25), 48 hrs (n=23), or 72 hrs (n=22) in comparison to untreated control cells (ctrl). (B) Representative immunoblot of CD20 and pSTAT6 (Tyr 641) protein levels in chronic lymphocytic leukemia (CLL) cells after IL4 stimulation (24-72 hrs). (C) Densitometric quantification of CD20 protein levels for independent replicates of the experiment described in (B) (n=17; 24-72 hrs). Untreated control (ctrl) without IL4 was set as 1 and compared to the other samples. (D) Normalized cell surface CD20 levels in primary CLL cells treated by SDF1a (100 ng/mL, PeproTech), IL4 (20 ng/mL), or their combination (SDF1a + IL4) for 24-48 hrs (n=7). The untreated ctrl was set as 1. (E) Peripheral blood CLL cells were electroporated with small interfering RNA (siRNA) against STAT6 (siSTAT6, 500 nM; Dharmacon) or negative control (Neg.Ctrl). IL4 (20 ng/ml) was added 48 hrs after transfection and cells were cultured for another 24 hrs. For the immunoblot, β-actin was used as a loading control and pSTAT6 (Tyr 641)/tSTAT6 levels were assessed. (F) Densitometric quantification of CD20 protein levels for independent replicates (n=4) of the experiment described in (E). Neg.Ctrl without IL4 was set as 1 and compared to the other samples. (G) Peripheral blood CLL cells were pretreated with pSTAT6 inhibitor (AS1517499, 1 mM, Selleckchem) for 12 hrs. Subsequently, IL4 (20 ng/mL) was added to the media, and cells were cultured for another 24 hrs. CD20 expression was determined by real-time quantitative polymerase chain reaction (TaqMan, ABI), and the expression of CD20 was normalized to endogenous control HPRT (n=6). (H) Chromatin immunoprecipitation analysis of samples that were immunoprecipitated with anti-STAT6 antibody in comparison with immunoglobulin G (IgG) antibody before (-) and after (+) IL4 stimulation (n=5; 40 ng/mL, 30 minutes). IgG antibody was used as an isotype control. For all in vitro experiments in Figure 1 and 2 CLL cells were purified by RosetteSep Human B Cell Enrichment Cocktail (Stemcell Technologies) to obtain purity ≥95% of CD5+CD19+ cells. For immunoblots, b-actin was used as a loading controls. In all experiments, the statistical difference was tested using a paired t-test, and the error bars indicate standard error of the mean.
Figure 2.
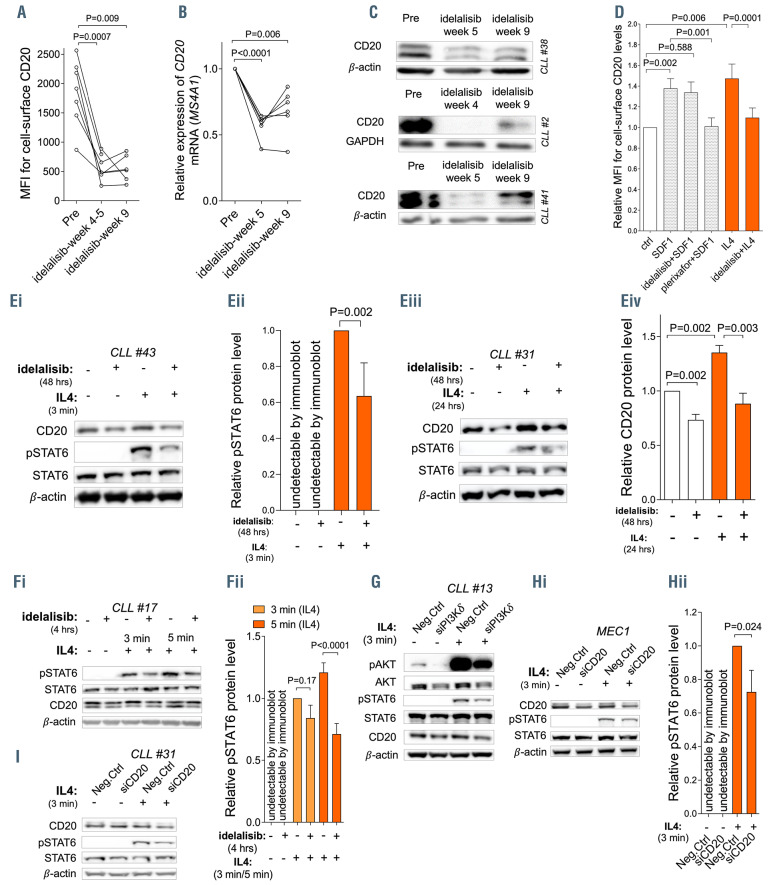
CD20 is downmodulated by idelalisib and embedded in the IL4 pathway. (A) Cell-surface CD20 levels and (B) relative expression of CD20 mRNA in paired samples before (Pre) and after 5 weeks (idelalisib-week 5) and 9 weeks (idelalisib-week 9) of idelalisib therapy in vivo (cell surface CD20: n=1 week 4, n=6 week 5, n=6 week 9; CD20 mRNA: n=6 week 5 and week 9). Chronic lymphocytic leukemia (CLL) cells were isolated by density centrifugation (Ficoll-Paque) followed by magnetic anti-CD3 MicroBeads separation (Miltenyi Biotec) or in some cases negative selection with RosetteSep Human B Cell Enrichment Cocktail (Stemcell Technologies) was used to obtain purity of ≥95% of CD5+19+ cells. (C) Representative examples (n=3) of CD20 protein levels in CLL cells obtained before (Pre) and during idelalisib therapy in vivo (week 4/5 and 9). (D) CLL cells (purity ≥95%) were pretreated with idelalisib (2 mM, Selleckchem) for 4 hours (hrs) or plerixafor (5 mg/mL, Selleckchem) for 4 hrs and then SDF1 (100 ng/mL, CXCR4 ligand) or interleukin 4 (IL4) (20 ng/mL) were added into the media for 24 hrs (for the result of the 48 hrs treatment with SDF1/IL4 see the Online Supplementary Figure S3A). Cell surface CD20 levels were measured and the results are visualized as a fold-change to untreated control (ctrl) (n=12). Viable CLL cells were gated for assessment of cell surface CD20 levels. The pretreatment of CLL cells by idelalisib or plerixafor (CXCR4 inhibitor) for 4 hrs was performed to ensure a full inhibition of the pathways before exposure to the receptor ligands. (Ei) Representative immunoblot of CLL cells treated in vitro with idelalisib (2 mM; 48 hrs) and subsequently stimulated by IL4 (40 ng/mL; 3 minutes [min]). (Eii) Densitometric quantification of pSTAT6 (Tyr 641) protein levels for independent replicates (n=7) of the experiment described in (Ei). (Eiii) Representative immunoblot of CLL cells treated in vitro with idelalisib (2 mM; 48 hrs) then washed twice with clean culture media and stimulated by IL4 in full media (20 ng/mL; 24 hrs). (Eiv) Densitometric quantification of CD20 protein levels for independent replicates of the experiment described in (Eiii) (n=7). (Fi) Representative immunoblot of pSTAT6 (Tyr 641) downmodulation after pretreatment of CLL cells with idelalisib (2 mM; 4 hrs) followed by IL4 stimulation (40 ng/mL; 3 and 5 min). (Fii) Densitometric quantification of pSTAT6 (Tyr 641) protein level for independent replicates (n=5) of the experiment described in (Fi). (G) Representative immunoblot of CLL cells transfected with small interfering RNA (siRNA) against the PI3Kδ isoform (siPI3Kδ, 500 nM, Dharmacon) or a negative control (Neg.Ctrl). Seventy-two hrs after transfection, cells were stimulated by IL4 (40 ng/mL; 3 min) and STAT6 phosphorylation (Tyr 641) was assessed. Phosphorylation of AKT (Ser 473) was used as a surrogated marker for PI3Kδ downmodulation after siPI3Kδ since we were not able to detect PI3Kδ protein due to issues with anti- PI3Kδ primary antibody. (Hi) Representative immunoblot of MEC1 cells transfected with siRNA against CD20 (siCD20, 500 nM, Thermo Fisher Scientific) or negative control (Neg.Ctrl). Forty-eight hrs after the transfection, cells were stimulated by IL4 (40 ng/mL; 3 min). (Hii) Densitometric quantification of pSTAT6 (Tyr 641) protein level for independent replicates (n=4) of the experiment described in (Hi). Neg.Ctrl without IL4 was set as 1 and compared to the other samples. (I) Representative immunoblot of primary CLL cells transfected with siRNA against CD20 (siCD20) or negative control (Neg.Ctrl), cultured for 48 hrs, and then stimulated by IL4 (40 ng/mL; 3 min). For immunoblots, β-actin or GAPDH were used as a loading controls. In all experiments, the statistical difference was tested using a paired t-test, and the error bars indicate standard error of the mean.
We next hypothesized that CD20 induction by IL4 might be mediated by STAT6 as it is a well-known key IL4 signaling effector. Indeed, STAT6 silencing by small interfering RNA (siRNA) or a specific STAT6 inhibitor (AS1517499) impaired the IL4-induced CD20 expression (Figure 1E to G). siRNA against STAT6 or STAT6 inhibitor did not have any effect on cell viability (Online Supplementary Figure S2A, and data not shown). Chromatin immunoprecipitation revealed that STAT6 binds to the CD20 promotor in CLL cells (-2 nucleotides from transcription start-site [TSS]), and the occupancy of this novel site by STAT6 was significantly increased (~2-fold) immediately after IL4 treatment (30 minutes, Figure 1H). We also observed a weaker STAT6 binding at another putative binding site (-197 nucleotides from TSS; Figure 1H; Online Supplementary Figure S2B and C). We also noted higher STAT6 phosphorylation in freshly obtained unstimulated CXCR4dim CD5bright CLL subpopulation in comparison to CXCR4bright CD5dim cells using flow cytometry (Online Supplementary Figure S2D and E). The CXCR4dimCD5bright cells are regarded as an intraclonal CLL cell subpopulation that has recently exited immune niches versus resting CXCR4brightCD5dim cells.3,7 However, we could not reliably detect phosphorylated STAT6 in CLL cells by a less sensitive immunoblotting technique suggesting that its levels were very low in peripheral blood in general. Altogether, these data demonstrate a novel direct role for STAT6 in transcriptional CD20 regulation upon IL4 stimulation. This is potentially a mechanism coupling the regulation of two molecules (IgM and CD20) which are required for BCR signaling3,7,8 and which are both induced by IL4 produced in the immune microenvironment. This mechanism might be a part of the pathway crosstalk by which IL4 alternates the BCR pathway, a phenomenon described by others.10
Ibrutinib leads to a significant reduction of CD20 levels in CLL by interfering with SDF1 signaling.1,3,4 Notably, in vivo therapy with idelalisib as a single agent prominently reduced CD20 mRNA and protein levels within several weeks (Figure 2A to C). Next, we tested if idelalisib interferes with the IL4- or SDF1-dependent induction of CD20. Idelalisib clearly inhibited CD20 induction by IL4, but not by SDF1 (Figure 2D; Online Supplementary Figure S3A). The repression of CD20 induction by idelalisib was observed with doses ≥0.5 mM (Online Supplementary Figure S3B). Therefore, we hypothesized that PI3K is involved in STAT6 activation, while the CXCR4-SDF1 axis does not include STAT6. Indeed, CXCR4 signaling does not induce any STAT6 phosphorylation (Online Supplementary Figure S3C), while idelalisib treatment in vitro (48 hours) significantly impaired STAT6 phosphorylation and CD20 induction by IL4 (Figure 2Ei to Eiv). In order to exclude secondary effects of idelalisib or decreased cell viability, we also performed a short pretreatment of cells by idelalisib (4 hours) and observed an identical decrease in pSTAT6 levels (Figure 2F, and data not shown; idelalisib had no effect on cell viability at 4 hours and a minor effect at 48 hours). In vitro, the treatment of CLL cells with idelalisib (1 or 2 mM) also led to a reduction of CD20 levels prior to IL4 exposure (Online Supplementary Figure S3D; Figure 2Eiii to Eiv, and data not shown), however, this was less pronounced than during IL4 stimulation. This suggests that idelalisib might affect other CD20 regulators besides STAT6 or low-level basal STAT6 activity influences also “basal” CD20 transcription. Indeed, we detected some STAT6 phosphorylation in unstimulated CLL cells (see above).
The silencing of PI3Kδ by siRNA decreased the STAT6 phosphorylation after IL4 (Figure 2G; Online Supplementary Figure S3E), indicating a direct role of the PI3Kδ isoform in IL4-induced CD20 expression and a specific on-target effect of idelalisib leading to CD20 downmodulation. This is in line with studies describing the involvement of PI3Kδ in IL4 signaling of normal B cells.11 However, idelalisib does not impair IL4-induced increase in CLL cell viability in vitro,12 suggesting that the pro-survival effect of IL4 is independent of STAT6 or that a weaker STAT6 phosphorylation is sufficient to provide a pro-survival signal. Altogether, our data demonstrate that PI3Kδ inhibition interferes with CD20‘s transcriptional activation by the IL4-STAT6 axis.
Besides the effects of idelalisib on STAT6 phosphorylation and CD20 levels, we also noticed a minor decrease in cell surface IL4 receptor (IL4Ra) levels after 48 hours of idelalisib treatment in vitro (Online Supplementary Figure S3F). However, this decrease is not responsible for the reduced responsiveness to IL4, since incubation of CLL cells with idelalisib for 4 hours also impaired STAT6 phosphorylation after IL4 (Figure 2F), but IL4Ra or CD20 levels remained unchanged during these short-term experiments (Online Supplementary Figure S3G and H). Moreover, we did not observe a significant downmodulation of IL4Ra levels during idelalisib therapy in vivo (Online Supplementary Figure S3I). This supports a direct role of PI3K in STAT6 signaling and the on-target effect of idelalisib.
Figure 3.
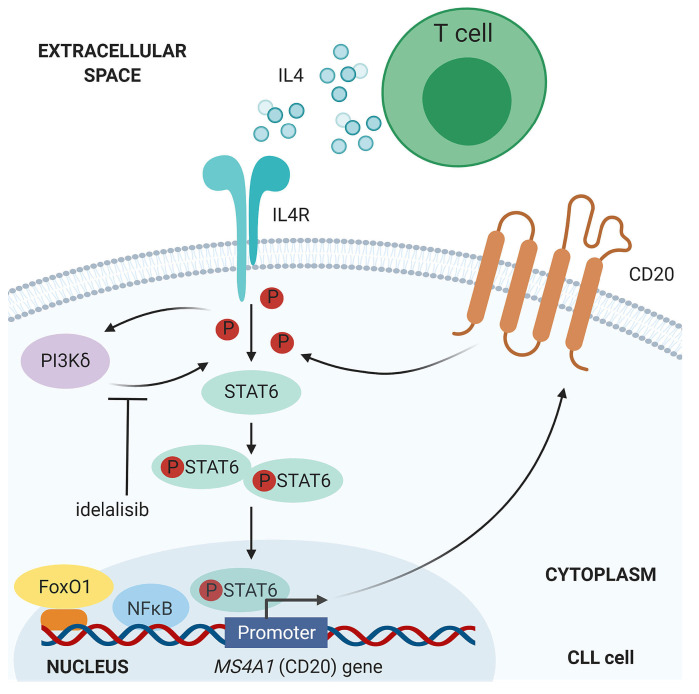
Schematic overview for CD20 downmodulation by idelalisib via impaired IL4-STAT6 axis. In chronic lymphocytic leukemia (CLL), IL4-STAT6 axis upregulates CD20 gene expression through the STAT6 phosphorylation and its direct binding to CD20 (MS4A1) promoter. The IL4-STAT6-CD20 axis is inhibited by PI3Kδ inhibitor idelalisib. NFκB and FoxO1 represent other two known regulators of CD20 transcription in CLL (NFκB is a positive regulator, and FoxO1 is an indirect negative regulator).1,4,5 This figure was created with tools at BioRender.com.
We next asked if CD20 downmodulation by idelalisib might affect the function(s) of CD20 in some signaling pathways other than BCR. To our surprise, CD20 silencing by siRNA significantly impaired the response to IL4 in MEC1 and primary CLL cells (Figure 2H and I; Online Supplementary Figure S3J), and had a minor effect on the phosphorylation of IKK after CpG or CD40L (data not shown). This suggests that CD20 is embedded in several receptor-associated pathways, including the regulation of IL4 signaling propensity. IL4 maintains CD20 levels via STAT6 activation, and CD20 increases responsiveness of CLL cells to IL4 via a still unclear mechanism. We noted that IL4 treatment in CLL cells does not lead to internalization of the IL4 receptor (data not shown), allowing cells to respond to IL4 continuously. These observations of the role of CD20 in T-cell interactions are in line with studies of CD20 in normal B cells, since CD20 knockout in mice or CD20 loss in humans leads to defects in T-cell dependent immunity.1,13 However, full understanding of this phenome will require further insight into CD20 functions and interaction partners, which is a long-standing question in the field.1
Altogether, here we describe a novel CD20 regulatory axis and reveal for the first time that T- cell interactions via IL4 induce CD20 transcription via STAT6 binding to its promotor (summarized in Figure 3). The PI3Kδ is involved in CD20 induction by the IL4-STAT6 axis, and consequently, idelalisib therapy represses CD20 in CLL. Idelalisib has been approved in combination with anti- CD20 antibodies without a comparison to the adminstration of single agent idelalisib,6 and is currently mainly used in therapy of relapsed/refractory disease and/or in cases of a BTK inhibitor intolerance. Downmodulation of CD20 by idelalisib likely reduces the rituximab-induced apoptosis and CDC, since complement fragment deposition is closely dependent on cell surface CD20 levels,14 and might impair ADCC/ADCP since these are also facilitated by opsonization of target cells with complement.15 Indeed, idelalisib inhibited in vitro the immune cell-mediated mechanisms induced by anti-CD20 antibodies,16 but this requires further investigation and might also include the effect of idelalisib on effector cells. Based on our data the benefit of rituximab addition should be tested in a clinical trial since this might fail to show improvement of progression-free/ overall survival, similarly to the lack of benefit for ibrutinib plus rituximab combination versus ibrutinib alone.2 It would be interesting to also test the combination of idelalisib/PI3Kδ inhibitor with anti-CD20 antibodies whose efficacy is less dependent on CD20 levels such as obinutuzumab. In conclusion, any clinicallyused inhibitor blocking PI3Kδ or interfering with the IL4- STAT6 pathway will reduce CD20 expression with potential consequences for combinatorial therapy.
Supplementary Material
Funding Statement
Funding: this work was supported by the Ministry of Health of the Czech Republic, grant no. NU20-03-00292. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (grant agreement no. 802644). All rights reserved.
References
- 1.Pavlasova G, Mraz M. The regulation and function of CD20: an "enigma" of B-cell biology and targeted therapy. Haematologica. 2020;105(6):1494-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burger JA, Sivina M, Jain N, et al. Randomized trial of ibrutinib vs ibrutinib plus rituximab in patients with chronic lymphocytic leukemia. Blood. 2019;133(10):1011-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlasova G, Borsky M, Seda V, et al. Ibrutinib inhibits CD20 upregulation on CLL B cells mediated by the CXCR4/SDF-1 axis. Blood. 2016;128(12):1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skarzynski M, Niemann CU, Lee YS, et al. Interactions between ibrutinib and anti-CD20 Antibodies: competing effects on the outcome of combination therapy. Clin Cancer Res. 2016;22(1):86-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pyrzynska B, Dwojak M, Zerrouqi A, et al. FOXO1 promotes resistance of non-Hodgkin lymphomas to anti-CD20-based therapy. Oncoimmunology. 2018;7(5):e1423183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furman RR, Sharman JP, Coutre SE, et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370(11):997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlasova G, Borsky M, Svobodova V, et al. Rituximab primarily targets an intra-clonal BCR signaling proficient CLL subpopulation characterized by high CD20 levels. Leukemia. 2018;32(9):2028-2031. [DOI] [PubMed] [Google Scholar]
- 8.Guo BC, Zhang L, Chiorazzi N, Rothstein TL. IL-4 rescues surface IgM expression in chronic lymphocytic leukemia. Blood. 2016; 128(4):553-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venugopal P, Sivaraman S, Huang XK, Nayini J, Gregory SA, Preisler HD. Effects of cytokines on CD20 antigen expression on tumor cells from patients with chronic lymphocytic leukemia. Leuk Res. 2000; 24(5):411-415. [DOI] [PubMed] [Google Scholar]
- 10.Guo BC, Rothstein TL. B cell receptor (BCR) cross-talk: IL-4 creates an alternate pathway for BCR-induced ERK activation that is phosphatidylinositol 3-kinase independent. J Immunol. 2005; 174(9): 5375-5381. [DOI] [PubMed] [Google Scholar]
- 11.Bilancio A, Okkenhaug K, Camps M, et al. Key role of the p110 delta isoforrn of PI3K in B-cell antigen and IL-4 receptor signaling: comparative analysis of genetic and pharmacologic interference with p110 delta function in B cells. Blood. 2006;107(2):642-650. [DOI] [PubMed] [Google Scholar]
- 12.Herman SEM, Gordon AL, Wagner AJ, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116(12):2078-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morsy DE, Sanyal R, Zaiss AK, Deo R, Muruve DA, Deans JP. Reduced T-Dependent Humoral Immunity in CD20-Deficient Mice. J Immunol. 2013;191(6):3112-3118. [DOI] [PubMed] [Google Scholar]
- 14.Golay J, Lazzari M, Facchinetti V, et al. CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood. 2001;98(12):3383-3389. [DOI] [PubMed] [Google Scholar]
- 15.Campagne MV, Wiesmann C, Brown EJ. Macrophage complement receptors and pathogen clearance. Cell Microbiol. 2007;9(9):2095-2102. [DOI] [PubMed] [Google Scholar]
- 16.Da Roit F, Engelberts PJ, Taylor RP, et al. Ibrutinib interferes with the cell-mediated anti-tumor activities of therapeutic CD20 antibodies: implications for combination therapy. Haematologica. 2015; 100(1): 77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.