Abstract
Polycomb response elements (PREs) are regulatory sites that mediate the silencing of homeotic and other genes. The bxd PRE region from the Drosophila Ultrabithorax gene can be subdivided into subfragments of 100 to 200 bp that retain different degrees of PRE activity in vivo. In vitro, embryonic nuclear extracts form complexes containing Polycomb group (PcG) proteins with these fragments. PcG binding to some fragments is dependent on consensus sequences for the GAGA factor. Other fragments lack GAGA binding sites but can still bind PcG complexes in vitro. We show that the GAGA factor is a component of at least some types of PcG complexes and may participate in the assembly of PcG complexes at PREs.
Polycomb group (PcG) proteins from complexes in vivo at regulatory sites, the Polycomb response elements (PREs), which mediate the silencing of neighboring genes. Transposons containing PREs generate new binding sites for PcG proteins on polytene chromosomes, indicating that PREs are the physical targets for PcG complex formation. Chromatin cross-linking experiments have also shown that PcG proteins are bound to and in the vicinity of known PRE sites (28, 35, 36). In these experiments, PcG proteins are found cross-linked over a few kilobases centered over fragments with known PRE activity, suggesting the possibility that a silencing complex initiated at a PRE involves at least a few kilobases either because of a cooperative spreading of the complex or because the PRE is in fact not a site but a region containing multiple sequences that interact with PcG proteins. None of the well-characterized PcG proteins can be shown to bind to DNA in vitro. One simple explanation for the fact that PcG complex formation appears to be specific for the PRE might be that it depends on other hitherto-unknown PcG proteins. One such candidate, the product of the pleiohomeotic (pho) gene, has recently been identified (3). However, it has not been shown yet that PHO interacts with other PcG proteins, and the presumptive consensus sequence for the binding of PHO is not always present in DNA fragments that have PRE activity, suggesting that other DNA-binding proteins might be involved. Other possibilities are suggested by the properties of PcG proteins and of PREs. Several of the PcG proteins can interact with one another, and experiments with Pc protein targeted to LexA or GAL4 binding sites indicate that a single PcG protein can recruit a silencing complex (26; S. Poux, D. McCabe, and V. Pirrotta, submitted for publication). Different genomic PcG sites show different degrees of dependence on different members of the PcG genes, both in the strength of the signal generated by immunostaining and the effect of different PcG mutations on the silencing of the accompanying genes. The silencing ability of a given PRE is strongly dependent on the genomic context in which it is introduced and on homologous pairing or the physical proximity to other PRE sequences in the nucleus (5, 10, 17, 34). All these observations suggest that the formation of a PcG complex at a PRE is normally dependent on multiple interactions and that PcG complex initiation is unlikely to be the result of a single sequence or a single recruiting protein.
PRE activity has generally been found in DNA fragments of several hundred to a few thousand base pairs. Such fragments often contain other activities which may be associated with PRE function. For example, the Fab-7 PRE region also contains a chromatin insulator or boundary element (14, 25, 38). The bxd PRE is flanked by embryonic enhancer elements (30). Both the bxd and the Fab-7 PREs are closely associated with target sites for the trithorax group (trxG) proteins TRX and the GAGA factor, which are usually thought to stimulate expression rather than silencing it (6, 7, 13). In this work, we have dissected the region containing the bxd PRE from the Ubx gene to show that residual PRE activity is associated with multiple smaller fragments and to determine if the different properties of the subfragments could help to identify functional components that contribute to the silencing function. Are different PcG proteins recruited to different parts of the PRE? Are sequence motifs repeated in different subfragments with PRE activity or does each fragment contribute distinct sequence elements that might be conserved in other known PREs? Finally, with smaller characterized PRE fragments, we hoped it might be possible to study the formation in vitro of minimal PcG complexes from embryonic nuclear extracts. The results presented here show that the bxd PRE is in fact a compound structure composed of sequences with different PRE-like activities and that many of its subfragments are able to interact in vitro with PcG complexes present in nuclear extracts. Surprisingly, the GAGA factor, often considered to be an activating protein and a member of the trxG, is a component of some PcG complexes and is important for their binding to PRE DNA in vitro.
MATERIALS AND METHODS
Fly strains and mutants.
All transgenic flies were produced using the Df(1)w67c23 strain, which is y− w−. The PcG mutations used were Pc3, Psc1, Su(z)21, and E(z)1. TrlR85 is a null mutation and is homozygously lethal, while Trl13C is a weaker allele that gives rise to some viable homozygous escapers (9). Both were provided by Gabriella Farkas. trxE2 is a strong, homozygously lethal allele with dominant phenotypes. For descriptions of the mutants, see reference 23.
Transposon constructs.
Most transposon constructs were assembled in the CaSpeR4 vector (29). Other vectors used were the S2 Ubx-lacZ construct (30), containing the S2 enhancer of the Ubx gene, and the YG CaSpeR construct in which the yellow gene is separated from the polylinker-miniwhite portion by a gypsy insulator element (34). In most cases, subfragments of the PRE region were oligomerized to produce 3, 4, or 6 tandem copies before being inserted into the transposon construct as indicated in Table 1. A transposon containing the LexA-PC gene (4) was constructed using the C4Y-hs vector (Poux et al., submitted). This vector uses the intronless yellow gene (12) as a marker and places the LexA-PC sequence under control of the hsp70 promoter (details available upon request).
TABLE 1.
PRE activities of subfragmentsa
| Constructb | Varc | No. of variegating lines/no. of lines tested with heterozygous mutationd:
|
Polytenese | Maintenancef | |||||
|---|---|---|---|---|---|---|---|---|---|
| Pc3 | Psc1 | Su(z)21 | E(z)1 | trxE2 | TrlR85 | ||||
| HA×4 | 8/22 | —g | — | — | — | — | — | — | |
| AB×6 | 11/27 | 3/6 | 1/6 | 3/6 | 5/6 | 5/6 | 0/5 | 0/2 | |
| BP×6 | 15/20 | 6/11 | 6/11 | 6/11 | 4/11 | 2/8 | 1/8 | 1/2 | |
| YGBP×6 | 19/20 | 3/6 | 4/6 | 4/6 | — | — | 3/6 | 2/3 | |
| YGBP×1 | 5/14 | — | — | — | — | — | — | — | |
| PF×4 | 22/31 | 3/12 | 7/12 | 8/13 | 2/12 | 3/9 | 0/9 | 3/5 | |
| HH2×4 | 4/12 | — | — | — | — | — | — | — | |
| HS×3 | 11/32 | — | — | — | — | — | — | — | |
| AB×6 S2 Ubx-lacZ | 4/8 | 0/6 | |||||||
| BP×6 S2 Ubx-lacZ | 5/13 | 5/13 | |||||||
| PF×4 S2 Ubx-lacZ | 4/8 | 0/5 | |||||||
For each fragment, the ability to induce variegated expression of the miniwhite gene in the CaSpeR4 vector was tested.
Oligomerization is indicated by the number following the multiplication sign.
Number of lines that variegate or are repressed when homozygous for the transposon/number of lines tested.
Mutations Pc3, Psc1, Su(z)21, and TrlR85 increased pigmentation, and mutations trxE2 and E(z)1 decreased it.
Number of lines in which a PcG binding site on polytene chromosomes was created/number of lines tested. The determination was performed for selected lines in which the transposon was not inserted at an endogenous PcG site by staining with anti-PC antibodies and in some cases also with anti-PSC and anti-SU(Z)2 antibodies.
Number of lines in which repression was maintained/number of lines tested. The maintenance of repression was determined in embryos of lines containing the PRE fragment in front of an S2 Ubx-lacZ reporter gene.
—, not tested.
Histochemical staining.
Embryos were collected overnight, fixed, stained, and mounted as described previously (20). Rabbit anti-β-galactosidase antibody (Cappel) was preadsorbed with fixed wild-type embryos. A biotinylated goat anti-rabbit second antibody and a Vectastain ABC horseradish peroxidase kit (Vector Labs) were used to reveal the antibody complexes. The effect of Trl mutations was determined by crossing to TrlR85/TM3 hb-lacZ, where the balancer chromosome is marked with a lacZ inserted in the hb gene. Mutant embryos lack lacZ expression in the anterior region.
Antibodies.
Polyclonal antibodies against PcG proteins were raised in rabbits using glutathione S-transferase (GST)-PcG fusion proteins containing amino acids 191 to 354 of PC and amino acids 819 to 926 of PSC. The fusion proteins were expressed in BL21 bacteria and purified on glutathione-Sepharose as described by the manufacturer (Pharmacia). The rabbit sera were affinity purified by passing first through a GST-Sepharose column. The flowthrough was passed over the respective PcG-Sepharose columns, washed with phosphate-buffered saline (PBS) and eluted with 0.1 M glycine, pH 2.8. The antibodies were dialyzed overnight against PBS, aliquoted, and stored frozen at −20°C.
In vitro band shift and immunoprecipitation assays.
Embryonic nuclear extracts were prepared from overnight embryo collections. For LexA-PC extracts, overnight collections of embryos carrying the hsp70–LexA-PC gene were heat shocked at 37°C for 40 min. The isolation of nuclei and the preparation of the extracts were carried out essentially as described in reference 2. For the band shift assays, the PRE fragments were end-labeled with Klenow DNA polymerase. The binding reaction mixture, in a volume of 20 μl, contained 0.3 μg of nuclear extract, 1 to 2 fmol of end-labeled fragment, and 1.75 μg of poly(dI-dC) in band shift buffer (12 mM HEPES [pH 7.9], 4 mM Tris [pH 7.9], 60 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM dithiothreitol, 10% glycerol). After being incubating on ice for 15 min, the reaction mixture was analyzed by electrophoresis on a 4.5% nondenaturing acrylamide gel. For antibody supershift assays, 1 μl of the antibody was added to the reaction mixture. Extracts from bacteria containing a pET3-GAGA expression plasmid or the pET3 vector alone were made as described by reference 32.
For the immunoprecipitation assays, the binding reaction was scaled up to 100 μl and supplemented with 0.1% bovine serum albumin and 0.1% NP40. The binding reaction mixture was then incubated with protein A-Sepharose beads to which had been attached the appropriate antibody. After 2 h at 4°C, the beads were washed three times with 100 μl of bandshift buffer, pelleted, incubated for 1 h in 0.5% sodium dodecyl sulfate–0.2 μg of proteinase K/ml and extracted with phenol-chloroform before precipitation. The results were analyzed on a 5% nondenaturing acrylamide gel. As a control for nonspecific binding, the reaction mixtures contained an equimolar amount of one or more similarly sized fragments isolated from the Bluescript plasmid vector. For competition assays, a 150-fold molar excess of unlabeled competitor fragment was added to the binding reaction mixtures. The immunoprecipitation reactions involving a synthetic GAGA binding site were done using the sequence AAAGAGAGCCCGGGAGAGAGAAA, cloned in four copies in the SmaI site of the Bluescript vector and excised using EcoRI and EagI. For use as a competitor, the GAGA oligonucleotide was oligomerized with ligase and the ladder of products was used. For each binding reaction, the ratios between the test fragment and the control fragment in the input and bound fraction were determined by scanning the gel with a Bio-Rad Molecular Imager to determine the selectivity of the binding. The Ubx promoter fragment was a 561-bp fragment from position −201 to +360 from the transcription start and contained three GAGA binding sites. For the binding reaction, this fragment was cleaved with DdeI to generate three fragments. The hsp70 promoter fragment was a 456-bp XbaI-XmnI fragment starting 250 bp upstream of the transcription start site and containing five GAGAG sites. The LexA target fragment was made from an oligonucleotide with sequence ACTTGATACTGTATGAGCATACAGTATAACCA oligomerized in four copies.
Construction of the mutated BP fragment.
The GAGA site mutations indicated in Fig. 5 were introduced using two primers, M1 (CCGTAAAGCGCTAGCGATCCGA) and M2 (AACCGTATCTGGCCCTATTTCCGCAGTCG), each containing alterations in a GAGA consensus site. To introduce the mutations in the BP fragment, M2 was first used in a PCR with primer ACAGTTATGGCGACGGAGCTGCAG to generate a fragment containing the PstI end of the fragment, including 18 bp of the adjacent PF fragment. This fragment was then used together with the M1 oligonucleotide in a PCR that generated the PstI half of the BP fragment with both GAGA sites mutated. This fragment was used in turn for a third PCR in combination with primer CACGGAAGCCATAACGGCAGAAC from the BglI end of the fragment, including 7 bp from the AB region, just preceding the BglI site. The resulting fragment was cloned in the Bluescript vector and used to prepare the BP* mutant fragment for the binding assays. The two halves of the BP fragment were generated using the primer GCACCATAATGGCTGCG or its complement from the central part of the BP sequence (Fig. 5) in PCRs together with the primers from the PstI end and from the BglI end, respectively. The GAGA oligonucleotide, containing two consensus sequences was made by annealing AAAGAGAGCCCGGGAGAGAGAAA and its complement. To produce the mutant GAGA oligonucleotide, the first AGAGAG was replaced by TTCAAG, leaving a single GAGAG consensus.
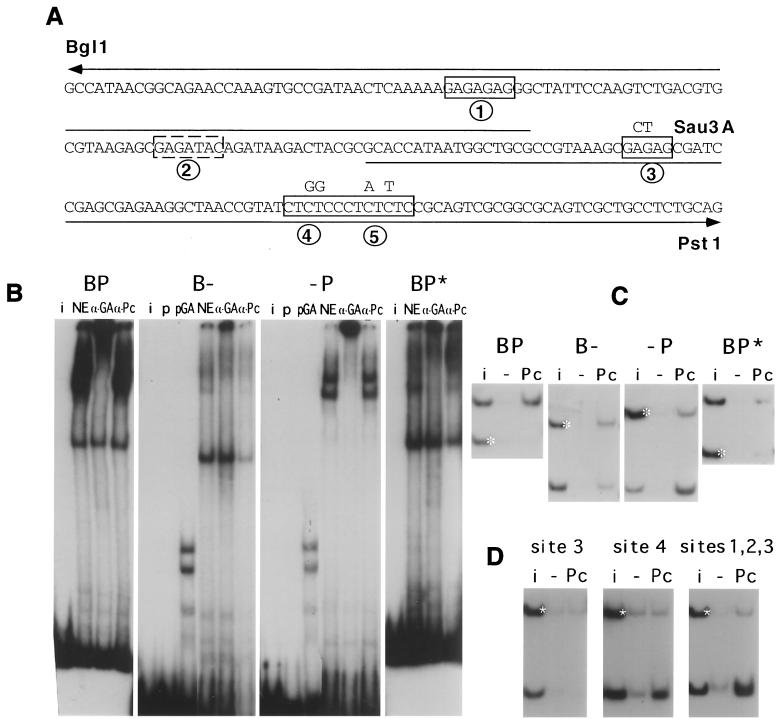
FIG. 5.
Dissection of the BP fragment. (A) The sequence from the BglI site to the PstI site is shown with the GAGA consensus sites boxed. Site 2, which is not a canonical consensus, is boxed by a dashed line. The nucleotides mutated in the BP∗ fragment are shown above the sequence and the extents of the two half fragments are indicated by the line above the sequence (B-) and below the sequence (-P). (B) Band shift analysis of BP, its two halves, and the mutant BP∗ fragment. NE, nuclear extract alone; α-GA, NE plus anti-GAGA; α-Pc, NE plus anti-PC; pGA, binding to extracts of bacteria containing pET-GAGA; p, extracts containing the pET vector alone; i, input mixture. (C) Immunoprecipitation analysis of BP, B-, P-, and BP∗ fragments. White asterisk, control fragment i, input; −, no antibody; Pc, bound with anti-PC. (D) Binding of fragments containing only GAGA site 3, site 4 (+5), or sites 1 to 3, obtained by cleavage at the Sau3A site between GAGA sites 3 and 4.
Protein immunoprecipitation.
Antibodies were attached to protein A-Sepharose beads and cross-linked with dimethyl pimelimidate as described in reference 15. The beads were then washed once before use with 1 ml of interaction buffer (12 mM HEPES [pH 7.9], 4 mM Tris [pH 7.9], 5 mM MgCl2, 60 mM KCl, 0.1 mM EDTA, 0.5 mM dithiothreitol, 0.1 mg of bovine serum albumin/ml, 0.1% NP-40) and incubated with embryonic nuclear extract containing 200 μg of protein in 100 μl of interaction buffer for 2 h at 4°C. The supernatant was removed, and the beads were washed three times with 100 μl of interaction buffer. The bound protein was released by incubation with 0.1 M glycine, pH 2.8, and analyzed by Western blotting with the appropriate antibody, together with 10 μg of input extract.
RESULTS
PRE fragments variegate miniwhite.
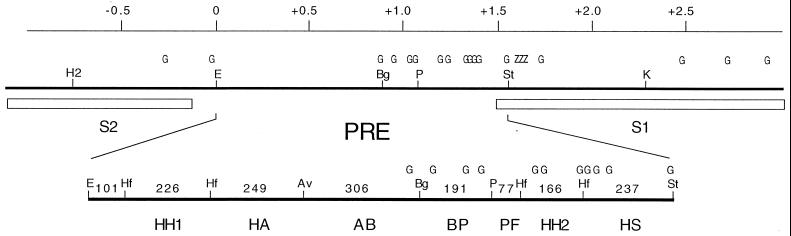
The bxd PRE activity was initially found in 6.5-kb HindIII fragment 2212H6.5 located about 20 kb from the transcription start site of the Ubx gene, in a region that contains several parasegmental enhancers (5). Previous mapping had shown that the principal PRE activity is contained in the 1.5-kb interval between the StyI and the EcoRI sites in Fig. 1. Fragments to the left or to the right of this interval did not by themselves produce either variegation of miniwhite gene expression or maintenance of the anterior repression of a Ubx-lacZ construct, though they are likely to contribute to silencing when part of a larger fragment (see, e.g., reference 37). The 1.5-kb EcoRI-StyI fragment induces variegation of the miniwhite gene in more than 60% of the lines; it maintains repression of a Ubx-lacZ construct in embryos and creates a new site of PcG protein binding on polytene chromosomes. To determine whether different parts of this sequence could make independent contributions to PcG silencing, we subdivided it into smaller fragments and tested each for its ability to induce variegation of the miniwhite gene. Considering that the smaller fragments might be less efficient in establishing the repressive complex, we oligomerized them and tested arrays of 3 to 6 copies. The different fragments have recognizably different effects on the expression of the miniwhite gene (Fig. 1 and Table 1). The strongest activity in terms of strength of variegation and frequency of lines displaying it was found in the two central fragments: a 180-bp BglI-PstI fragment (BP) and the adjacent 78-bp PstI-HinfI fragment (PF). The nonuniform pigmentation induced by the AvaII-BglI fragment (AB) was not of the usual highly variable mosaic type but generally in the form of an anterior-posterior gradient (Fig. 2A). A significant frequency of variegation was also obtained with the HA, HH2, and HS fragments, although more often in the form of a more general partial repression with faint spots of stronger pigmentation. A 400-bp fragment from the EcoRI end that includes the HH1 subfragment gave a frequency of variegation of marginal significance (4 of 30 lines). For comparison, the PRE fragment found most important for effective silencing in a recent paper by Tillib et al. (37), their fragment C, is delimited by nucleotide positions 218835 to 219249 of the BX-D sequence (24) and contains part of AB (positions 218822 to 219127) and part of BP (positions 219128 to 219318). Two other fragments found important in that study, fragments B and D, lie within the S2 fragment and the S1 fragment, respectively. In our experiments, neither S2 nor S1 had detectable PRE activity by itself.
FIG. 1.
Map of PRE region and subfragments. The bxd PRE region is shown with a scale marked in kilobases centered at the EcoRI site which corresponds to position 218241 in the BX-C sequence (24), while the StyI site is at position 219797. The Ubx promoter in this scale would lie at position +24. The positions of the embryonic enhancers S1 and S2 are indicated by boxes below the line. The map also shows the approximate positions of GAGAG sequences (G) and of Zeste binding sites (Z). The subfragments, whose sizes in base pairs are indicated, were prepared using restriction enzymes EcoRI (E), HinfI (Hf), AvaII (Av), BglI (Bg), PstI (P), StyI (St), and KpnI (K).
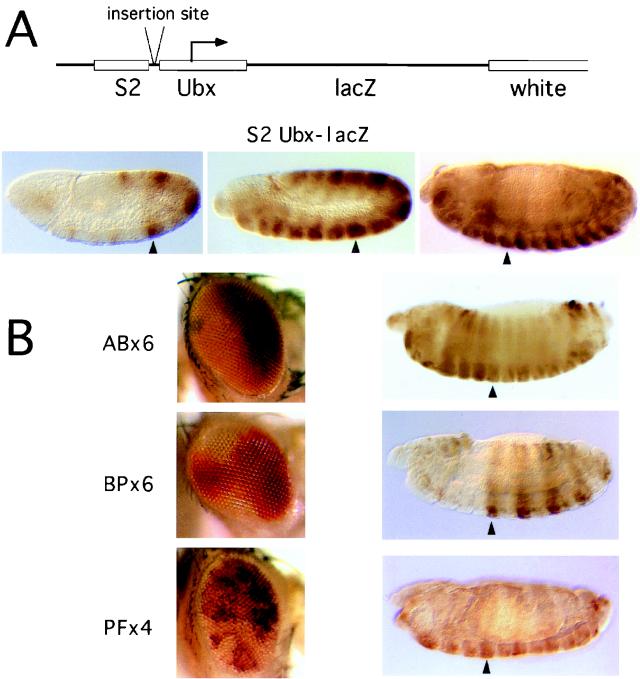
FIG. 2.
Variegation and maintenance activities. (A) Top, S2 Ubx-lacZ construct and the insertion site for PRE fragments; bottom, expression patterns of the S2 enhancer construct in embryos at (from left to right) early extension, extended, and retracted germ band stages. Arrowheads indicate PS6. (B) Representative eye variegation patterns of constructs AB×6, BP×6, and PF×4 are shown together with embryos containing the corresponding S2 Ubx-lacZ reporter constructs. The embryos are shown at the late germ band retraction stage. Maintenance of early repression was obtained only with the BP×6 construct, in which expression continued to be repressed anterior of parasegment 6 (arrowhead) and in which the pattern was arrested at the pair rule stage.
If a fragment has high PRE activity, multiple copies might completely silence the miniwhite marker gene, preventing the recovery of many transgenic fly lines. To evaluate the importance of this effect, we constructed a transposon (YGBP×6) containing six copies of the BP fragment and including, as an independent marker for transformation, the yellow gene, protected against PRE silencing by a gypsy insulator element which, when interposed between the PRE and the marker gene, has been shown to block silencing very efficiently (34). YGBP×6 lines show an even higher frequency of repression of the miniwhite gene. Six copies of BP are in fact able to repress completely the expression of the miniwhite gene in 12 of 23 lines heterozygous for the transposon. This means that half of the lines would not have been recovered in the absence of the yellow marker. For comparison, an analogous transposon (YGBP) containing a single copy of BP induces variegation at a much lower frequency (Table 1).
Genetic interactions.
Might different PRE subfragments interact preferentially with some subset of PcG proteins? When we tested the dominant effects of different PcG mutations on the variegation induced by the different subfragments, we did not observe such specificities. For all constructs, some lines are affected by a given PcG mutation and some are not (Table 1). A line affected by mutations in one PcG gene might not respond to mutations in another PcG gene. In no case was the variegation dependent on Suvar(2)5, indicating that the tandem repeats contained in these transposons do not induce the heterochromatin-like silencing observed by Dorer and Henikoff (8) with arrays of white transposons.
While the PRE region has an overall silencing activity, it also contains targets for some trxG proteins. A 400-bp region immediately upstream of the PstI site and corresponding to our fragment AB plus BP has been reported to interact with trx (6, 7); the central part of the PRE is extremely rich in consensus binding sequences for the GAGA factor; a set of three zeste binding sites is immediately adjacent to the PRE core fragments. We tested a selection of the lines of the different PRE fragments for the effect of mutations in these three factors. Loss-of-function mutations in the zeste gene (za) or the z1 mutation have no effect on transposons that do not include the zeste binding sites (EcoRI-StyI or smaller PRE fragments). However, single copies of transposons that include the PRE Zeste sites (EcoRI-KpnI or larger) become more strongly repressed in the presence of either of the zeste mutations (not shown). In contrast, when Zeste is overexpressed from a heat shock promoter, miniwhite gene expression from these transposons is strongly stimulated (not shown). The presence of the rest of the PRE region is not necessary since transposons containing the S1 fragment, which contains the Zeste binding sites but no PRE activity, also respond to zeste (not shown). The interaction with Zeste is independent of silencing and does not require pairing since it affects flies heterozygous for the transposon.
Heterozygous mutations in the trx gene decreased the expression of the white gene in several AB lines and, to a lesser extent, in BP lines, indicating that trx function stimulates expression in a dosage-dependent way. As with the PcG mutations, both the presence and the strength of the effect are strongly dependent on the site of insertion. Although the bxd PRE contains clusters of GAGA sites, mutations in the Trl gene, encoding the GAGA factor, had little effect in the majority of our lines. Surprisingly, however, in contrast with the generally positive effect of the GAGA factor on gene expression, a few lines displayed a detectable increase in eye pigmentation when heterozygous for the null mutation TrlR85 (9), suggesting that it contributes to repression. This was seen in one of eight BP lines and in three of six YGBP×6 lines tested, indicating that, in some cases, a decrease in GAGA factor reduces the silencing effect of the PRE fragment. A similar involvement of GAGA factor in promoting effective PcG silencing has been shown for the Fab-7 PRE (13).
Immunochemical staining of polytene chromosomes provides a direct test for the ability of a transposon construct to recruit a given PcG protein. Polytene chromosome binding at the insertion site was observed with BP and PF constructs but not with AB. The ability to induce detectable PcG protein binding at the site of insertion depended on the site: it was visible in some BP and PF lines but not in others. In summary, then, we could detect no differential interactions of individual PcG proteins with any one fragment. Although a given line might interact with one PcG mutation but not another or show binding to one protein but not another, these specificities were site dependent and not fragment dependent, suggesting that at different chromosomal sites different PcG proteins make different contributions to the formation of the complex.
Maintenance of repression in embryos.
The strict functional test of bxd PRE activity is its ability to maintain the pattern of repression of a Ubx promoter construct in the embryo. To test this, we assembled three constructs, placing the AB6, BP6, or PF4 fragment oligomers in front of a Ubx-lacZ reporter gene under the control of the Ubx S2 enhancer (Fig. 2A) (5, 30). This enhancer has a pair rule pattern of expression in the even-numbered parasegments when it is first activated at the syncytial blastoderm but shifts to an all-parasegment pattern in the course of germ band extension (Fig. 2B). Five of 13 BP6 lines tested showed effective maintenance of the S2 pattern of expression: the even-numbered PS pattern persists in the late embryo and expression anterior of PS6 never develops. In contrast, maintenance was not seen in five PF4 lines and six AB6 lines tested, although in most cases the adults showed variegated or nonuniform eye coloration.
The ability to maintain repression conferred by the BP6 oligomer was lost in embryos homozygous for a Pc mutation, resulting in the appearance of expression in the thoracic segments at the end of germ band extension. A much weaker and less consistent derepressing effect was observed in embryos homozygous for the TrlR85 null mutation or in embryos homozygous for the weaker Trl13C mutation, obtained by crossing males containing the reporter transposon with female escapers homozygous for the Trl13C mutation to minimize the maternal Trl contribution (1). However, since some embryos show incomplete maintenance even in a Trl+ background and since the degree of maintenance varies from one experiment to another, we could not consider the effect significant.
Immunoprecipitation of PcG complexes.
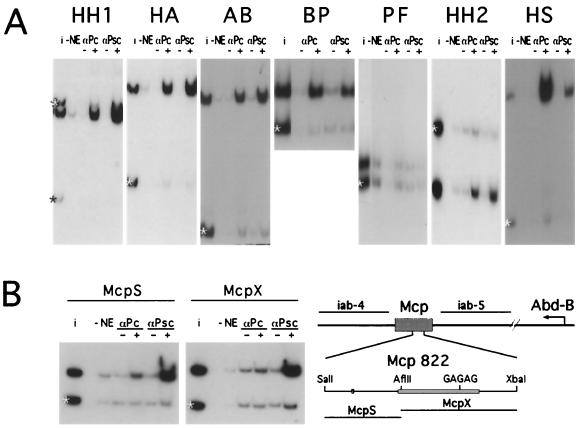
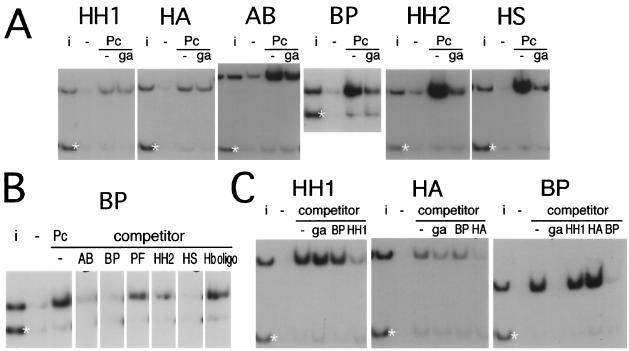
The in vivo experiments show that the PRE region is composed of multiple fragments with silencing activities that appear to differ both quantitatively and qualitatively. We next tested the different fragments for their ability to bind in vitro to PcG proteins present in embryonic nuclear extracts. All the PRE fragments shown in Fig. 1 interact with the nuclear extracts, resulting in a low-mobility complex when tested in gel mobility shift assays (not shown). To determine whether PcG proteins are present in these complexes, we used immunoprecipitation. For comparison, the immunoprecipitation reaction mixtures contained one or more reference DNA fragments from the plasmid vector or from other Ubx regions, and in all experiments the results were scanned and quantitated to determine the degree of selective enrichment in the immunoprecipitate relative to the input. Figure 3A shows that most of the subfragments from the EcoRI-StyI region are selectively immunoprecipitated both by anti-PC and by anti-PSC antibody, albeit with different efficiencies. Fragments BP, HH2, and HS, which contain GAGA sites, are more efficiently precipitated by anti-PC, while HH1, HA, and AB, which lack multiple GAGA sites, are more efficiently precipitated by anti-PSC. The only fragment that clearly fails to immunoprecipitate with either of the two antibodies is PF, although in vivo it has strong PRE activity as determined by the ability to induce PcG-dependent variegation of the miniwhite gene. Although the PF fragment is only 78 bp long, the failure to immunoprecipitate is not due to its small size since smaller fragments can be efficiently precipitated in this assay (see experiments below).
FIG. 3.
Immunoprecipitation binding assay. (A) For each fragment, a mixture including one or more control fragments (∗) was incubated with nuclear extract (NE) and with beads without antibody (−) or with anti-PC antibody (αPc) or anti-PSC antibody (αPsc). The radioactivity bound to the beads was analyzed by gel electrophoresis together with an aliquot of the input mixture (i). (B) Binding to the Mcp PRE. The locations of the Mcp PRE in the bithorax complex and the region containing the major DNase hypersensitive sites (16) are shown. The two Mcp fragments indicated were incubated with nuclear extract and immunoprecipitated with anti-PC or anti-PSC.
We also examined the interactions of the S1 and S2 fragments that flank the PRE region and that contain embryonic enhancer activities to see if they might also contribute to the assembly of PcG complexes. These fragments were split into two at the KpnI and HincII sites, respectively (Fig. 1). Both halves of S2 show significant immunoprecipitation with both PC and PSC antibodies. For S1, a weak interaction with the SK fragment but not with the KS fragment was found (not shown). For comparison, we tested the binding to another PRE from the bithorax complex, the Mcp PRE (Fig. 3B). The 822-bp SalI-XbaI Mcp fragment, which contains only a single GAGA consensus sequence (16), was cut with AflII into two fragments, both of which are efficiently immunoprecipitated by anti-PSC but only very poorly by anti-PC, resembling in this respect the behavior of the GAGA-less fragments of the bxd PRE. In contrast, no immunoprecipitation was observed with the Ubx PBX, BXD, or BX enhancers (30–32) (results not shown).
Involvement of GAGA sequences.
As expected from the presence of multiple GAGA sites, the BP fragment also forms complexes that are efficiently immunoprecipitated by anti-GAGA antibody (not shown) and competed by excess unlabeled GAGA oligonucleotide. In the course of these experiments we found that, surprisingly, the interaction of the BP fragment with PC or with PSC is also efficiently competed by an oligonucleotide containing GAGA consensus sequences, suggesting that it might be mediated by a factor that binds to this sequence. Three other PRE fragments contain GAGA sites and interact with the bacterially produced GAGA factor in band shift assays (not shown), while two others have neither property. We tested the ability of the GAGA oligonucleotide to compete for the immunoprecipitation of each of the PRE fragments with anti-PC antibody. Figure 4A shows that the oligonucleotide efficiently competes for the binding of the GAGA-containing fragments but has little or no effect with the HH1 and HA fragments, which have no GAGA sites, and has only a slight effect with AB, which has only one GAGA site. These results suggest that at least two kinds of PcG complexes are detected by the immunoprecipitation assay: one dependent on the GAGA factor and one independent.
FIG. 4.
Binding competition. (A) The binding detected by the anti-PC antibody (Pc) was competed by the presence of an unlabeled oligonucleotide containing the GAGA consensus (ga). Strong competition was observed with the BH, HH2, and HS fragments, weak competition was observed with AB, and none was observed with HH1, HA, or AB. (B) The immunoprecipitation of the BP fragment with anti-PC was carried out in the presence of unlabeled DNA of the various other fragments as the competitor or of an unlabeled oligonucleotide containing the Hunchback consensus binding sequence (Hb oligo). (C) BP fragment does not compete with the binding of HH1 or HA. No competitor (−) or GAGA oligonucleotide (ga), BP, HH1, or HA unlabeled competitors were added as indicated.
To verify the existence of two mechanisms of interaction with DNA, we tested the ability of different fragments to compete with one another. We first examined the effect on the immunoprecipitation of BP of unlabeled competing DNA from the other fragments (Fig. 4B). The GAGA-containing fragments HS, HH2, and BP themselves are good competitors. The AB fragment, containing a single GAGA site, competes weakly. No competition was observed with an oligonucleotide containing a Hunchback (HB) consensus binding sequence. We then looked in more detail into the competition between BP and the non-GAGA-containing fragments HH1 and HA (Fig. 4C). The immunoprecipitation of HH1 or HA is not competed either by GAGA oligonucleotide or BP DNA, while the BP complex is inhibited by both of these but not by either HH1 or HA competitor DNA. HH1 and HA do not compete with BP, but they do compete with each other (not shown). These results confirm the existence of at least two modes for the binding of PcG complexes to DNA in vitro.
Mutated GAGA sites abolish binding.
Footprinting experiments show that BP contains five sites that can interact with the bacterially produced GAGA factor in vitro (not shown). Sites 1, 3, 4, and 5 (Fig. 5A) are typical consensus sequences containing a GAGAG core; site 2 is noncanonical. When probes corresponding to the two halves of BP are incubated with nuclear extract and tested for immunoprecipitation with anti-PC, the right half (-P) shows good binding, though it is apparently weaker than that shown by the intact BP fragment, while the left half (B-) does not interact visibly (Fig. 5C). Band shift and supershift experiments (Fig. 5B) show that -P still forms low-mobility complexes that are supershifted by anti-GAGA antibody. The B- fragment is responsible for the higher-mobility complex seen already with BP, but this complex contains no GAGA protein and is not supershifted by anti-PC antibody. The B- fragment still interacts well with bacterially produced GAGA protein but only very weakly with GAGA in the nuclear extract. In fact, a very faint, low-mobility band that is supershifted by anti-GAGA antibody is still visible.
To determine whether the GAGA consensus sequences on the right half of the fragment are responsible for the interactions, we mutated them (Fig. 5A) and tested the mutated BP fragment (BP*). The BP* fragment, still containing one canonical GAGAG and one noncanonical site, is no longer immunoprecipitated by anti-PC antibody (Fig. 5C), and the band shift experiment reveals only a weak residual affinity for GAGA-containing complexes (Fig. 5C). Cleavage with Sau3A, which separates site 3 from sites 4 and 5, shows that while sites 4 and 5 suffice for binding to PC-containing complexes, site 3 alone does not. However, a fragment containing sites 1, 2, and 3 can bind (Fig. 5D). These results suggest that at least two consensus GAGAG sequences are necessary for interaction with either the endogenous GAGA factor or with a PC-containing complex. The mutated BP* fragment does not bind in vitro to a PcG complex either in single copy or when oligomerized.
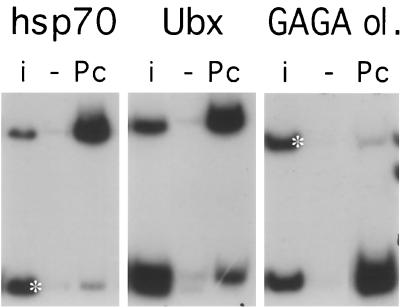
If GAGA binding sites are the targets of PcG complexes in vitro, are they sufficient? We examined this question by testing other DNA fragments known to interact with the GAGA factor in vivo. The hsp70 promoter contains prominent GAGA sites that are important for its heat shock-inducible promoter activity (22). When an hsp70 promoter fragment was tested in our assay, it was efficiently immunoprecipitated by anti-PC antibody (Fig. 6). The Ubx promoter contains a set of GAGA sites that are important for its promoter activity both in vivo and in vitro (2, 19). A DNA fragment from the Ubx promoter containing these sites is also efficiently immunoprecipitated in our assay. Note, however, that another fragment, just downstream of the Ubx transcription start site, also binds in this assay although it contains no GAGAG sequence. Even an oligomer consisting of four copies of a synthetic oligonucleotide containing two GAGA binding sites is strongly immunoprecipitated (Fig. 6). This oligonucleotide binds well also as a monomer, but the introduction of two nucleotide changes in one of the GAGA consensus sites destroys the binding activity. These results show that two nearby consensus sequences are necessary and sufficient in vitro to interact with PcG complexes present in nuclear extracts, though they are not sufficient to recruit a PcG complex in vivo. Two or more GAGA sites separated by the entire length of the fragment, as in BP* oligomers, do not support in vitro binding (not shown).
FIG. 6.
Binding to other GAGA-containing sequences. Shown are anti-PC immunoprecipitation reactions with fragments containing the hsp70 promoter, the Ubx promoter, and a synthetic GAGA consensus oligonucleotide oligomerized to four copies (GAGA ol.). Note that the Ubx promoter fragment (nucleotides −201 to +360 surrounding the transcription start) was cut into three fragments with DdeI. The largest fragment, containing three GAGAG sites, binds strongly, but another fragment, downstream of the transcription start, also immunoprecipitates though it contains no GAGAG. A control fragment, where included, is marked with a white asterisk. i, input mixture; Pc, anti-PC antibody.
When the BP* mutant fragment, oligomerized in six copies, was incorporated in a reporter construct containing the Ubx S2 enhancer and Ubx-lacZ gene, only 1 of 9 transgenic lines was able to partly maintain the repressed pattern of S2 expression, compared to 5 of 13 maintaining lines found for the wild-type BP construct. These numbers are too low to be statistically significant. That the mutations at best only reduce the frequency of maintaining lines might be explained if BP* still contains other determinants for PRE activity and suggests that, while the GAGA sites may contribute, they are not the only sequences involved in recruiting a PcG complex in vivo.
GAGA factor is a constituent of a PcG complex.
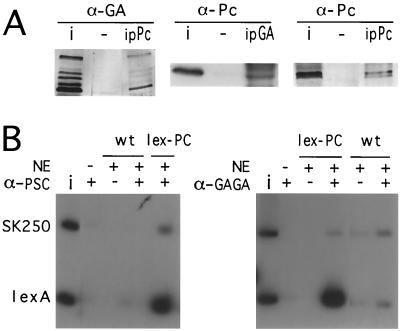
The preceding experiments show that PcG complexes bind in vitro to sites containing GAGA consensus sequences and are able to bind the GAGA factor. Is the in vitro binding of PcG complexes to these sites mediated by the GAGA factor itself or does some other PcG component recognize the same GAGA consensus sequence? We used two approaches to ask if the GAGA factor was in fact associated with the multiprotein PcG complexes present in the nuclear extract. Anti-PC antibody immunoprecipitates the GAGA protein from Drosophila embryonic extracts (Fig. 7A) but does not interact with the GAGA protein expressed in bacteria (not shown). Conversely, an anti-GAGA antibody immunoprecipitates the PC protein from the embryonic extracts.
FIG. 7.
Coimmunoprecipitation of PC and the GAGA factor. (A) Nuclear extract proteins were immunoprecipitated with beads containing no antibody (−), with anti-PC beads (ipPc), or with anti-GAGA beads (ipGA). The material recovered from the beads was analyzed by Western blotting; blots were developed with anti-PC (α-Pc) or with anti-GAGA (α-GA). Additional bands in the anti-PC blot (middle) are due to the immunoglobulins present in the immunoprecipitate. i, input nuclear extract. (B) Nuclear extracts (NE) from embryos expressing LexA-PC cause binding of a PSC-containing complex to a DNA fragment containing four LexA binding sites (LexA), while wild-type extracts do not bind. Immunoprecipitation with anti-GAGA shows that the GAGA factor is also contained in this complex. SK250, a control fragment from the Bluescript vector; i, input.
To confirm the participation of the GAGA factor in PcG complexes, we used another approach. A construct expressing a chimeric PC protein fused to the LexA DNA-binding domain was expressed in flies under control of the hsp70 promoter. The LexA-PC protein is functional in vivo and participates in PcG complexes, as shown by immunostaining of polytene chromosomes and by its ability to repress in vivo a reporter construct containing LexA binding sites (Poux et al., submitted). When nuclear extracts from embryos expressing LexA-PC are used in the binding reactions, anti-PC or anti-PSC antibodies can precipitate a DNA probe containing four LexA binding sites, while no binding occurs with wild-type extracts (Fig. 7B), showing that the LexA-PC protein can recruit a PcG complex to the LexA target sequence. In a parallel experiment, anti-GAGA antibodies immunoprecipitated the LexA probe in the presence of the LexA-PC extract but not in the presence of wild-type extract. These experiments confirm that the GAGA factor is a constituent of at least some PcG complexes formed in vivo.
DISCUSSION
PRE activity in vivo.
Dissection of the PRE reveals that it is a compound region containing several sequences that are able to different extents to induce variegated expression of the miniwhite gene, respond to PcG mutations, and create new binding sites for PcG proteins on polytene chromosomes. The separate fragments are definitely weaker in activity than the whole. A single copy of a fragment containing BP, AB, and part of HA silences very effectively (34), indicating that the different sequences normally cooperate to achieve more-complete silencing to a degree that is not attained by multiple tandem copies of one fragment. The different subfragments most likely contribute complementary functions, but it has not been possible to demonstrate that different PcG proteins interact with different subfragments. As with the entire PRE, the response to different PcG mutations depends strongly on the site of insertion of the transposon construct. The genomic context makes therefore a strong contribution not only to the strength of the silencing but also to the relative importance of the different PcG components of the silencing complex. The activity of PRE-containing transposons inserted at different sites suggests that this contribution is due not only to sequences flanking the insertion site but also to the interaction in trans with other genomic PRE sites (34).
Only one of the three subfragments tested in embryos, BP, was able to maintain repression of the Ubx-lacZ reporter gene. This could be due simply to the relative PRE strengths of the different fragments. That is, increasing the number of copies of the other fragments might achieve the same silencing strength. Another possibility is that the complex formed at the BP fragment is qualitatively different from that recruited by the other fragments; for example, it might be able to recruit PcG proteins sufficiently early in embryonic development to have an effect on the Ubx-lacZ gene, while other PRE fragments might be able to institute silencing only at later stages. Different affinities for PcG complexes could also account for the different abilities to create binding sites for PcG proteins on polytene chromosomes. However, the fact that the PF fragment, though able to induce variegation at a high rate and to bind PcG proteins on polytenes, failed to show any detectable PcG complex formation in the immunoprecipitation assays suggests that the nature and composition of the complexes and/or the mode and timing of their recruitment are likely to differ for the different fragments.
The role of the GAGA factor.
The in vitro experiments show that GAGAG-containing sequences are binding sites for PcG complexes and that the GAGA factor is associated with PcG complexes present in the nuclear extracts. Ion exchange chromatography of nuclear extracts confirms that, while PcG proteins elute over a broad range of salt concentrations, the in vitro binding activity constitutes a small minority and copurifies with the GAGA factor (B. Horard and V. Pirrotta, unpublished experiments). The multiplicity and heterogeneity of PcG complexes present in nuclear extracts would not be detected in affinity-based purification schemes such as that used by Shao et al. (33), who did not find the GAGA factor to be a constituent of their PRC1 PcG complex. In contrast, Hodgson and Brock (submitted for publication) find the GAGA factor, along with PH, in a multiprotein complex that binds in vitro to PRE regions corresponding to ours. We cannot exclude the possibility that some other PcG protein also recognizes the GAGA consensus sequence, but the association of the GAGA factor with PcG complexes shows that it is most likely involved in at least one mode of PcG binding to PRE DNA. Does this reflect a role for the GAGA factor in PcG silencing in vivo? The GAGA factor was originally identified as a transcription-stimulating factor both in vivo and in vitro and was classified as a trxG protein because it stimulated the activity of homeotic genes while its mutants had phenotypes indicative of homeotic insufficiency (2, 9). However, some evidence suggests that it can also be associated with repressive functions. The GAGA factor, together with another activator, NTF-1, also binds to an 11-bp element required for the repression of tailless by the torso-dependent pathway (22). Evidence that it might be involved in PRE function was presented by Hagstrom et al. (13), who found that GAGA mutations decrease the silencing effected by the Fab-7 PRE. In the Ubx gene, the bxd PRE region contains the largest concentration of GAGA binding sites. If we take each continuous G(AG)n stretch as one binding site, the 1-kb interval containing the core of the PRE contains 13 sites while the next highest concentration (8 sites) is found in a 1-kb region containing the bx PRE (not to be confused with the BX enhancer). Chromatin cross-linking and immunoprecipitation experiments confirm that these regions bind the GAGA factor in vivo (34). Our results suggest that at these sites the GAGA factor is not an antagonist of silencing and is not simply an accessory or a facilitator of PcG complex formation but may, in concert with other factors, contribute to targeting PcG complexes.
It was surprising, in view of our in vitro results, that the effects of Trl mutations on either miniwhite variegation or the silencing of the Ubx-lacZ reporter were sporadic and strongly dependent on the insertion site. One possible explanation is that, in vivo, the GAGA factor is only one of a set of DNA-binding recruiting proteins and that, while it contributes to, it is not essential for, the assembly of PcG complexes. Chromatographic fractionation of nuclear extracts indicates in fact that only a fraction of the PcG complexes present in embryonic extracts are associated with the GAGA factor (B. Horard and V. Pirrotta, unpublished results). Furthermore, embryos contain an important maternal supply of the GAGA factor, which would mask the effect of a reduced zygotic contribution. Later, other recruiting factors might be involved. Finally, our results cannot exclude the possibility that, although the GAGA factor is a component of PcG complexes, can target their binding in vitro, and is apparently important for the function of the Fab-7 PRE (13), it is not primarily involved in recruitment at the bxd PRE. Its role might be instead primarily architectural. Katsani et al. (18) have shown that the GAGA factor binds to DNA as a multimer that recognizes clustered GAGA consensus sequences, and they argue that such binding would be expected to bend DNA in a way incompatible with nucleosome assembly. GAGA binding would then clear the PRE core of nucleosomes and bend it to facilitate interactions among other DNA-binding components.
PcG complexes with other GAGA binding sites.
The presence of GAGA binding sites alone appears to be sufficient in vitro to bind a PcG complex since not only the PRE fragments but also the Ubx promoter and the hsp70 promoter bind in our experiments though they have no known PcG silencing activity in vivo. In addition, a GAGA-containing oligonucleotide also binds efficiently to PcG complexes. Nevertheless, GAGA protein binding to a DNA sequence is not sufficient to recruit PcG complexes in vivo. Clearly the in vitro binding reaction does not reflect the in vivo activity. The most probable explanation of this discrepancy is that the binding detected in vitro is due to complexes that are preassembled in vivo and are then dissociated from the chromatin during the preparation of nuclear extracts. If the nature and composition of PcG complexes are templated by the PREs at which they are assembled, GAGA-containing PcG complexes would be efficiently targeted to GAGA binding sites in vitro while, in vivo, complex formation would require the de novo recruitment and assembly of PcG complexes, involving other DNA binding components or cofactors. We favor this interpretation because it would also explain the variable compositions of PcG complexes detected at different chromosomal sites. In vivo, the large majority of GAGA binding sites visible on polytene chromosomes are not associated with PcG binding, suggesting that only a small fraction of the GAGA protein is involved in PcG complexes. This interpretation also accounts for the fact that the LexA-GAGA protein cannot recruit PcG complexes to LexA binding sites (Poux et al., submitted). We note also that the target of PcG complexes in vivo is chromatin, not naked DNA. The presence of nucleosomes might normally increase the selectivity, allowing PcG complexes to assemble only at sites where other recruiting or architectural proteins are also bound.
In view of these results, the existence of GAGA sites at the Ubx promoter raises other possibilities. In the presence of a PRE, a GAGA factor bound at the Ubx promoter might participate in the silencing activity by interacting with GAGA-containing PcG complexes recruited at the PRE, mediating or contributing to promoter silencing. Both the hsp70 and hsp26 promoters are efficiently repressed by the presence of a PRE in the same transposon construct (V. Pirrotta, unpublished results). The GAGA factor might contribute to silencing in these cases also. The miniwhite gene, which is also silenced by the PRE, does not contain typical clustered GAGA sites in its promoter region but only a few scattered sites in the transcribed region. The expression of the miniwhite gene is strongly dependent on the site of insertion and on distant enhancers within or outside of the transposon construct. The silencing of these enhancers might be in part responsible for the effect of the PRE on miniwhite expression. Alternatively, other proteins binding to the miniwhite promoter region might interact with PcG complexes.
Other recruiting proteins.
The immunoprecipitation experiments also detected binding that is not competed by GAGA oligonucleotides with PRE fragments that do not contain consensus GAGA binding sites. This implies that other recognition sequences and other DNA-binding proteins are involved in these cases. The recent discovery that PHO, a Drosophila PcG protein homologue of the mammalian YY-1 factor, binds to DNA suggests that it might be one such recruiter of PcG complexes (3). There are in fact a number of putative PHO binding sites with the minimal consensus GCCAT in the PRE region: one in AB, two in BP (a third site is destroyed by the BglI cleavage), and three in the PF fragment. These bind PHO protein in vitro and are important for PRE activity in vivo (11). However, none are found in the HH or HA fragments; hence these presumably depend on other recruiting proteins. The PF fragment, on the other hand, though it contains three putative PHO sites, is conspicuous for its inability to bind PcG complexes in our extracts, suggesting that PHO is either not present in the complex containing PC and PSC or does not interact directly with it. The fact that the mammalian PHO homologue YY-1 causes sharp bends in the DNA (27) raises the possibility that PHO too might serve a primarily architectural role without necessarily interacting directly with PcG complexes.
Although PF does not contain GAGA sites, it is almost as effective in inducing PcG-dependent variegation of the miniwhite gene as the BP fragment and it can generate new PcG binding sites at the site of insertion on polytene chromosomes. Yet PF cannot maintain repression of the Ubx-lacZ reporter gene in embryos. One possible explanation for these results is that PF is the target for yet another PcG recruiting mechanism that either functions poorly under our in vitro binding conditions or depends on proteins that are not present in the embryonic extracts. The fact that the PF fragment can recruit silencing complexes in larval cells but cannot maintain repression in the embryo would be consistent with a requirement for proteins present only at later developmental stages. Another possible explanation is that PF does interact with certain PcG complexes which do not include PC or PSC and hence escaped our detection.
The picture of the PRE that emerges from these experiments is that of a mosaic of multiple interaction sites which may require different DNA-binding proteins to recruit PcG components. A similar conclusion was reached by Tillib et al. (37), based on deletions that abolish the activity of the bxd PRE and by Hodgson and Brock (submitted), who used an in vitro binding approach similar to ours. GAGA sites are associated with some PREs but not others (e.g., the Mcp PRE). If the GAGA factor acts as a recruiting protein, it is most likely only one of many possible recruiters. Different recruiters might interact specifically with different PcG proteins, accounting for the fact that the binding sites for different PcG proteins on polytene chromosomes do not completely coincide. Nevertheless, the ability of PcG proteins to interact with one another or to enter into a chain of recruitment (26) means that, in most cases, strong binding sites for one PcG protein will be able to recruit at least to some degree the other PcG proteins. The difference between direct and indirect recruitment may be responsible for the fact that a strong chromosomal binding site for one PcG protein is sometimes a weak binding site for another PcG protein.
ACKNOWLEDGMENTS
B.H. and C.T. contributed equally to this work.
We are grateful to Peter Becker and members of his laboratory for the use of his fly cages and help in preparing the wild-type embryonic extract. Thanks are due to Carl Wu and Peter Harte for repeated gifts of antibody, to Gabriella Farkas for the Trl mutants, to Bob Kingston for the LexA-PC gene and to Ivan Dellino for constructing the BP* mutant. We thank Hugh Brock for valuable discussions and for sharing results prior to publication.
This work was supported by a grant from the Swiss National Science Foundation, by the Human Frontiers Science Program, and by a contribution from the Georges and Antoine Claraz Donation.
REFERENCES
- 1.Bhat K M, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 2.Biggin M D, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 3.Brown J L, Mucci D, Whiteley M, Dirksen M, Kassis J A. The Drosophila polycomb group gene pleiohomeotic encodes a sequence-specific DNA binding protein with homology to the multifunctional mammalian transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 4.Bunker C A, Kingston R E. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol Cell Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan C S, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y-L, King B O, O'Connor M, Mazo A, Huang D-H. Functional reconstruction of trans regulation of the Ultrabithorax promoter by the products of two antagonistic genes, trithorax and Polycomb. Mol Cell Biol. 1995;15:6601–6612. doi: 10.1128/mcb.15.12.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinwalla V, Jane E P, Harte P J. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorer D R, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 9.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 10.Fauvarque M-O, Dura J-M. polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 1993;7:1508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- 11.Fritsch C, Brown J, Kassis J, Müller J. The DNA-binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- 12.Geyer P K, Corces V G. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- 13.Hagstrom K, Müller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 15.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 522–523. [Google Scholar]
- 16.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassis J A. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6 kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsani K, Hajibagheri M, Verrijzer C. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laney J D, Biggin M D. zeste, a nonessential gene, potently activates Ultrabithorax transcription in the Drosophila embryo. Genes Dev. 1992;6:1531–1541. doi: 10.1101/gad.6.8.1531. [DOI] [PubMed] [Google Scholar]
- 20.Lawrence P A, Johnston P. Analysis of function of the pair-rule genes hairy, even-skipped and fushi tarazu in mosaic Drosophila embryos. Development. 1989;107:847–854. doi: 10.1242/dev.107.4.847. [DOI] [PubMed] [Google Scholar]
- 21.Lee H-S, Kraus K W, Wolfner M F, Lis J T. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- 22.Liaw G-J, Rudolph K M, Huang J-D, Dubnicoff T, Courey A J, Lengyel J A. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 1995;9:3163–3173. doi: 10.1101/gad.9.24.3163. [DOI] [PubMed] [Google Scholar]
- 23.Lindsley D L, Zimm G G. The genome of Drosophila melanogaster. San Diego, Calif: Academic Press; 1992. [Google Scholar]
- 24.Martin C H, Mayeda C A, Davis C A, Ericsson C L, Knafels J D, Mathog D R, Celniker S E, Lewis E B, Palazzolo M J. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci USA. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124:1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 26.Müller J. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 1995;14:1209–1220. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natesan S, Gilman M Z. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 28.Orlando V, Jane E P, Chinwalla V, Harte P J, Paro R. Binding of Trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriquez R L, Denhardt D T, editors. Vectors. A survey of molecular cloning vectors and their uses. Boston, Mass: Butterworths; 1988. pp. 437–445. [Google Scholar]
- 30.Pirrotta V, Chan C S, McCabe D, Qian S. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics. 1995;141:1439–1450. doi: 10.1093/genetics/141.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poux S, Kostic C, Pirrotta V. Hunchback-independent silencing of late Ubx enhancers by a Polycomb group response element. EMBO J. 1996;15:4713–4722. [PMC free article] [PubMed] [Google Scholar]
- 32.Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao Z, Raible F, Mollaaghababa R, Guyon J, Wu C, Bender W, Kingston R. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 34.Sigrist C J A, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila Polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strutt H, Paro R. The Polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]