ABSTRACT
Outbreaks of emerging viral pathogens like severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are a major medical challenge. There is a pressing need for antivirals that can be rapidly deployed to curb infection and dissemination. We determined the efficacy of interferon lambda-1 (IFN-λ) as a broad-spectrum antiviral agent to inhibit SARS-CoV-2 infection and reduce pathology in a mouse model of disease. IFN-λ significantly limited SARS-CoV-2 production in primary human bronchial epithelial cells in culture. Pretreatment of human lung cells with IFN-λ completely blocked infectious virus production, and treatment with IFN-λ at the time of infection inhibited virus production more than 10-fold. To interrogate the protective effects of IFN-λ in response to SARS-CoV-2 infection, transgenic mice expressing the human angiotensin-converting enzyme 2 (ACE-2) were tested. One dose of IFN-λ administered intranasally was found to reduce animal morbidity and mortality. Our study with SARS-CoV-2 also revealed a sex differential in disease outcome. Male mice had higher mortality, reflecting the more severe symptoms and mortality found in male patients infected with SARS-CoV-2. The results indicate that IFN-λ potentially can treat early stages of SARS-CoV-2 infection and decrease pathology, and this murine model can be used to investigate the sex differential documented in COVID-19.
KEYWORDS: COVID-19, SARS-CoV-2, antiviral, interferon, lung infection, murine model
INTRODUCTION
The ongoing severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic and the prior emergence of SARS-CoV in 2002 and the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 have underscored the urgent need for effective antiviral agents that can be deployed rapidly and can combat a diversity of respiratory viral pathogens. The development of vaccines for SARS-CoV-2 has been the defining factor in controlling the coronavirus disease 2019 (COVID-19) pandemic. However, specific vaccines take many months to generate and do not address viral variants that escape vaccine immunity or the emergence of distinct pathogenic viruses.
Interferons (IFNs) are natural antiviral cytokines and are essential for an effective early defense response. Pathogen recognition receptors such as Toll-like receptors and retinoic acid-inducible gene I-like receptors are activated in response to virus and initiate signaling cascades to induce gene expression of type I IFNs (alpha and beta) and type III IFNs (lambda) (1, 2). Type I and III IFNs are secreted from infected cells and bind distinct cell surface receptors. Receptor binding stimulates the phosphorylation and activation of signal transducers and activators of transcription (STATs) that induce IFN-stimulated genes (ISGs), the main effectors of antiviral protection (3, 4). SARS-CoV-2 has evolved multiple strategies to block the induction and action of IFNs (5–8). The ability of SARS-CoV-2 to antagonize IFN innate immunity is coincident with insufficient levels of IFN found in patients early during COVID-19 (9–11). The critical role of IFN for a successful response to SARS-CoV-2 is evident in severe COVID-19 patients who produce autoantibodies to IFN and in patients with life-threatening disease who have genetic deficiencies in IFN immunity (12, 13).
Although SARS-CoV-2 can suppress the IFN response, exogenous IFN treatment has been shown to inhibit viral production in cultured lung cells (14, 15). This has led to the proposition that the administration of IFN can protect against SARS-CoV-2 infection and lessen the severity of COVID-19. This is supported by an early exploratory study with type I IFN showing a reduced duration of viral infection in the respiratory tract (16). Type I IFN receptors are expressed on most cells of the body, including immune cells. For this reason, type I IFN stimulation can trigger systemic and local inflammation by immune cell activation and recruitment to the site of infection. High levels of type I IFN can contribute to the damaging cytokine storm evident in the severe respiratory inflammation caused by SARS-CoV-2 infection. In contrast, type III IFNs (IFN-λ) bind to a different receptor whose expression is limited primarily to epithelial cells and therefore is not associated with inflammation. The potential benefit of IFN-λ as an antiviral treatment for SARS-CoV-2 infection has been proposed, and clinical trials are ongoing with subcutaneous administration of pegylated-IFN-λ (17).
We used an experimental mouse model to investigate the impact of intranasal administration of IFN-λ on SARS-CoV-2 infection and disease outcome. Since SARS-CoV-2 binds specifically to the human angiotensin-converting enzyme 2 (ACE-2) for entry, transgenic mice were used that express human ACE2 in epithelial cells of the respiratory tract. We provide evidence that intranasal administration of IFN-λ reduces animal morbidity and mortality in response to SARS-CoV-2 at a viral dose close to the estimated infectious dose in patient populations (18). These results indicate that IFN-λ could be an effective preventive or therapeutic antiviral agent to treat human SARS-CoV-2 infection. This preclinical model of infection also revealed a sex differential in disease outcome. Male mice had a higher mortality rate in response to SARS-CoV-2 than female mice. This finding reflects the more severe illness and mortality that have been well documented in male versus female COVID-19 patients worldwide (19, 20) (https://globalhealth5050.org).
RESULTS
Interferon-lambda inhibits infection of human bronchial epithelial cells by SARS-CoV-2.
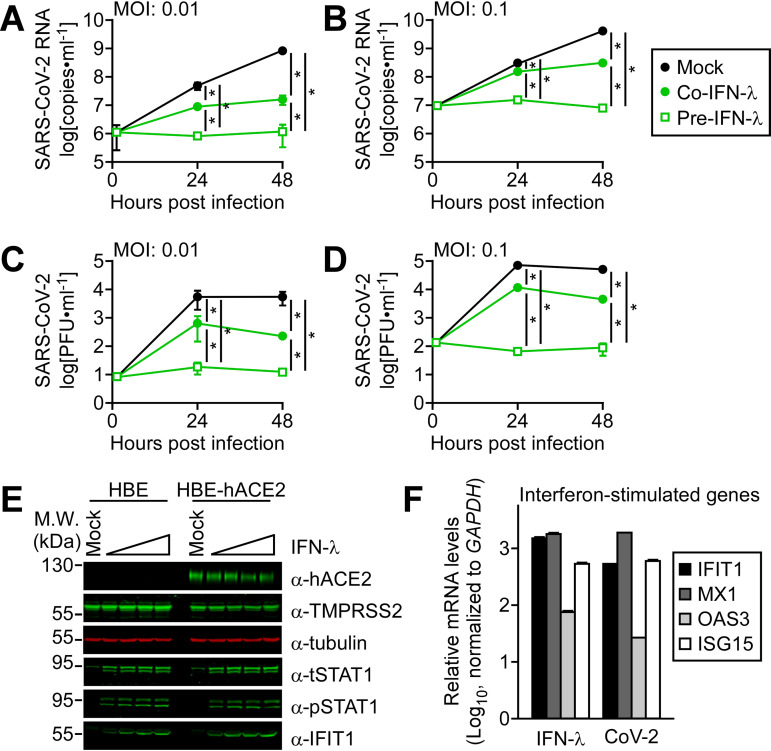
Coronaviruses are positive-sense RNA viruses with fast replication rates, producing virions within hours of infection. The innate immune response is therefore critical for host defense, but SARS-CoV-2 has evolved strategies to block the antiviral IFN responses. To determine the potential protective effects of IFN-λ administration, we evaluated its effects on virus production in cultured human lung cells. Infection is dependent on the presence of viral receptors and coreceptors, and the number of ACE2-expressing cells is greatest in the nasal epithelium and lower in the bronchi (21). We found that human bronchial epithelial cells immortalized with hTERT and CDK4 (HBE3-KT) were refractory to infection and expressed undetectable levels of hACE2, but they did express the TMPRSS2 cell surface protease that activates the SARS-CoV-2 spike glycoprotein. We therefore generated derivatives expressing hACE2 designated HBE-hACE2. The HBE-hACE2 cells were permissive for infection even at low multiplicities of infection (Fig. 1A to D). To investigate the effects of exogenous IFN-λ on infection by SARS-CoV-2, HBE-hACE2 cells were either pretreated with IFN-λ 24 h prior to SARS-CoV-2 infection or treated with IFN-λ at the same time of infection with a multiplicity of infection (MOI) of 0.1 or 0.01 PFU/cell. Viral RNA released in the culture media was measured by reverse transcriptase quantitative PCR (RT-qPCR) (Fig. 1A and B), and mature virus was measured by plaque assay (Fig. 1C and D). Results showed that addition of IFN-λ to the cultures during infection reduced the amount of viral RNA and virus produced by the HBE-hACE2 cells. Pretreatment with IFN-λ completely blocked the release of viral RNA and infectious virus from infected cells. Western blotting of proteins expressed in HBE and HBE-hACE2 cells showed that they responded to IFN-λ with tyrosine phosphorylation of the STAT1 transcription factor, increased steady-state levels of STAT1, and synthesis of the ISG protein IFIT1 (Fig. 1E). Analyses of RNA expression in the bronchial epithelial cells following treatment with IFN-λ showed an increase in ISG mRNA levels of IFIT1, MX1, OAS3, and ISG15 (Fig. 1F). ISG mRNA levels also increased with SARS-CoV-2 later in infection (24 h postinfection [hpi]). SARS-CoV-2 has been shown to suppresses ISG mRNA expression early in infection, and later in infection, there is an increase in ISG mRNA levels (5). The delayed ISG expression is proposed to provide a time frame for the virus to replicate unencumbered.
FIG 1.
IFN-λ inhibited SARS-CoV-2 replication in cultured human lung cells. Human bronchial epithelial cells expressing hACE2 (HBE-hACE2) were either mock treated (black-filled circles), pretreated for 24 h with IFN-λ (pre-IFN-λ) (green open squares), or treated with IFN-λ at the same time of infection (co-IFN-λ) (green-filled circles) with SARS-CoV-2. Culture medium was harvested at 0, 24, and 48 h postinfection, and viral RNA in culture medium was measured by RT-qPCR following infection at MOI of 0.01 PFU/cell (A) or MOI of 0.1 PFU/cell (B). Mature virus released into the culture media was quantified by plaque assay following infection at MOI of 0.01 PFU/cell (C) or MOI of 0.1 PFU/cell (D). Three independent experiments were performed with technical duplicates. *, P < 0.001 (two-way ANOVA). (E) Representative Western blots are shown of proteins expressed in the HBE and HBE-hACE2 cells following treatment with increasing amounts of IFN-λ (200 to 2,000 units/ml) for 24 h. (F) RT-qPCR measurements are shown for ISG mRNAs expressed in HBE-hACE2 cells relative to untreated controls in response to IFN-λ treatment (1,000 units/ml) or relative to mock treatment in response to a SARS-CoV-2 infection for 24 h.
Sex as a determinant of disease outcome following SARS-CoV-2 infection.
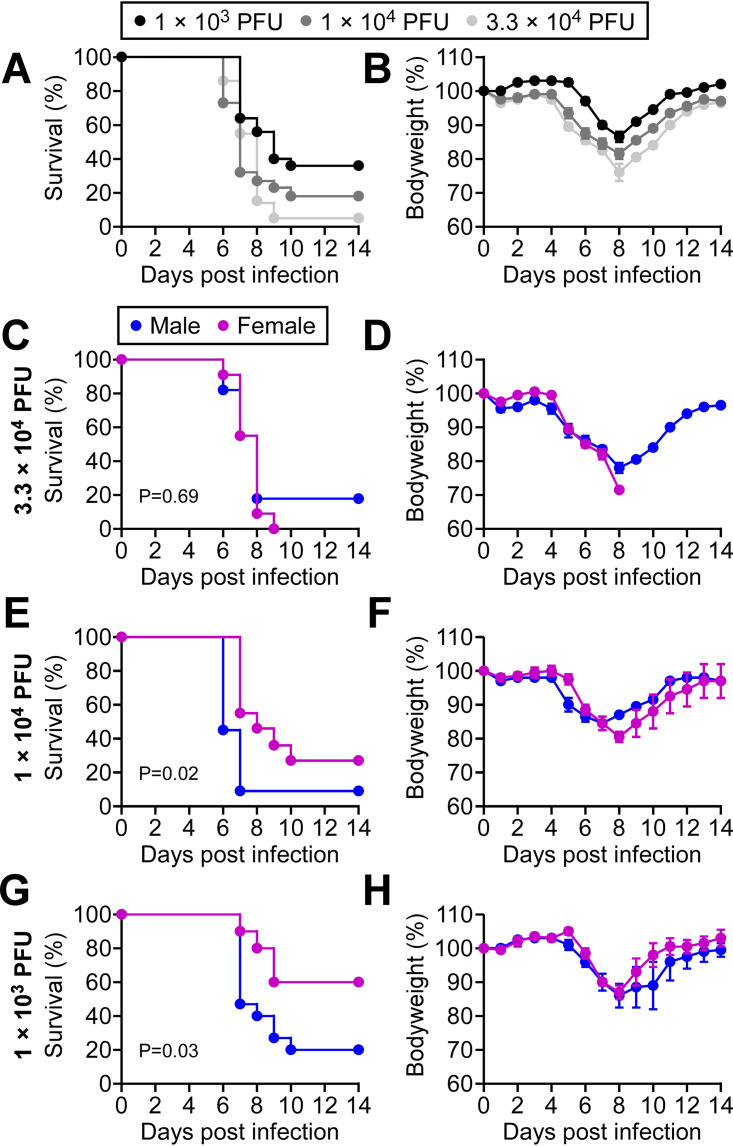
To interrogate COVID-19 pathology, we used a preclinical mouse model of disease and SARS-CoV-2 intranasal infection. Transgenic mice expressing human ACE2 in epithelial cells of the respiratory tract and other tissues (K18-hACE2) (22) were infected with increasing doses of SARS-CoV-2 from 1 × 103 PFU to 3.3 × 104 PFU (Fig. 2A). Survival decreased with a higher infectious dose; however, the animals that succumbed to disease all expired between 6 and 10 days following infection regardless of the dose. This time frame was accompanied by body weight reduction in response to infection in both survivors and nonsurvivors (Fig. 2B). We then evaluated whether there was a sex bias associated with a greater risk of death. It has been clearly documented worldwide that the severity and mortality due to COVID-19 are higher in human male patients than female patients (19, 23, 24). We found a sex differential was evident with lower viral doses in the mouse model. At the higher inoculum of 3.3 × 104 PFU, male and female mice succumbed to infection with similar kinetics and outcome (Fig. 2C and D). However, at 1 × 104 PFU and 1 × 103 PFU, a difference between the male and female response to infection was evident (Fig. 2E and G). Males had a significant increase in mortality compared to females. These findings indicate that male susceptibility is a physiological difference independent of lifestyle or comorbidity.
FIG 2.
Male sex is a risk factor for death in a preclinical model of SARS-CoV-2 infection. Male and female K18-hACE2 transgenic mice were infected intranasally with three different doses of SARS-CoV-2. (A) Kaplan-Meier survival curves are presented from pooled data of three independent experiments in panels C, E, and G. (B) Averaged body weight changes are presented for each viral dose of mice in A. (C and D) Data for survival and body weight changes for 11 male and 12 female mice infected with 3.3 × 104 PFU of SARS-CoV-2. Data are stratified for male sex (blue line) and female sex (pink line). (E and F) Data for survival and body weight changes for 11 male and 10 female mice infected with 1.0 × 104 PFU of SARS-CoV-2. Data are stratified for male sex (blue line) and female sex (pink line) (G and H) Data for survival and body weight changes for 15 male and 10 female mice infected with 1.0 × 103 PFU of SARS-CoV-2. Data are stratified for male sex (blue line) and female sex (pink line). Results represent pooled data from three independent experiments. P values are provided in the figure.
Interferon lambda administration promotes survival in a mouse model of SARS-CoV-2 infection.
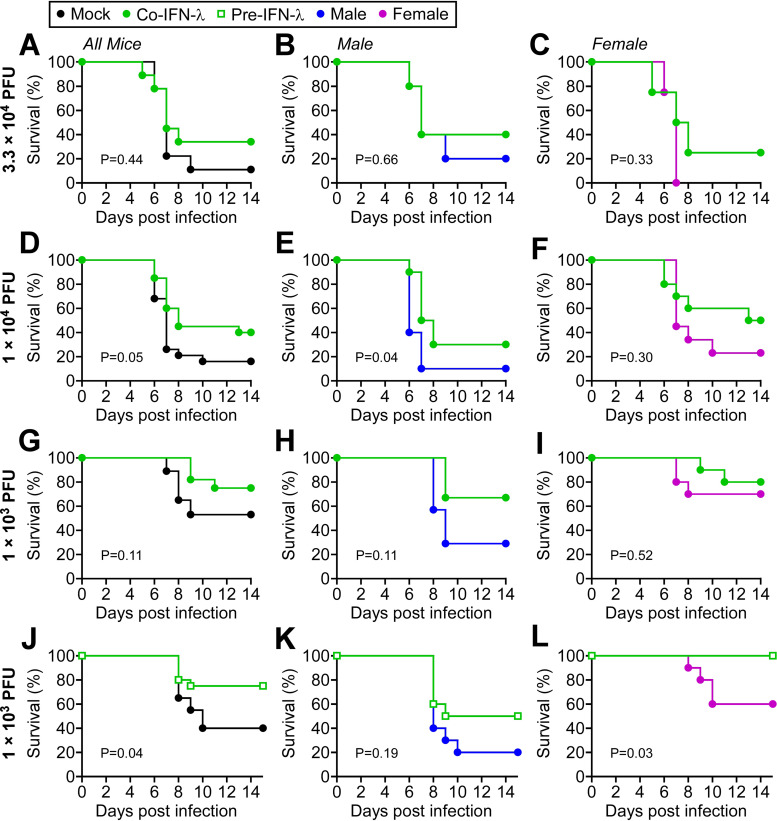
Since IFN-λ inhibited SARS-CoV-2 infection of cultured human bronchial epithelial cells, we examined the protective effects of IFN-λ in the K18-hACE2 preclinical mouse model. Kaplan-Meier curves are shown representing the results of infection with three different doses of SARS-CoV-2 (3.3 × 104 PFU, 1 × 104 PFU, and 1 × 103 PFU) either without administration of IFN-λ or with intranasal administration of IFN-λ at the time of infection (closed green circles) (Fig. 3A, D, and G). Corresponding body weight changes are presented in Fig. S1 in the supplemental material. Results indicate increased survival of SARS-CoV-2 infection with IFN-λ administration at lower doses of virus. Although IFN-λ afforded protection from lower inoculums of SARS-CoV-2, stratification of the results for male and female mice did not show a sex difference in IFN-λ protection (Fig. 3B, C, E, F, H, and I). The expression of ISG mRNAs in lung homogenates of mice treated with IFN-λ was similar in males and females 24 h following administration (Fig. S2). Changes in ISG expression were not detectable in whole-lung homogenates with this SARS-CoV-2 low inoculum of 1 × 103 PFU (Fig. S2).
FIG 3.
IFN-λ administration reduced mortality in a mouse model of SARS-CoV-2 infection. Male and female K18-hACE2 transgenic mice were infected with the specified dose of SARS-CoV-2, and survival is displayed as Kaplan-Meier curves. The left panels for each set of infections represent both male and female mice, and the right panels are stratified for male and female mice. (A to C) Data for survival of mice infected with 3.3 × 104 PFU virus (5 male mice, 5 female mice; black closed circle) and mice treated with IFN-λ at the time of infection (5 male, 4 female; green closed circle). (D to F) Data for survival of mice infected with 1.0 × 104 PFU virus (10 male, 9 female; black closed circle) and mice treated with IFN-λ at the time of infection (10 male, 10 female; green closed circle). (G to I) Data for survival of mice infected with 1.0 × 103 PFU virus (7 male, 10 female; black closed circle) and mice treated with IFN-λ at the time of infection (6 male, 10 female; green closed circle). (J to L) Data for survival of mice infected with 1.0 × 103 PFU virus (10 male, 10 female; black closed circles) and mice pretreated with IFN-λ for 24 h prior to infection (10 male, 10 female; green open squares). Results represent pooled data from two independent experiments. P values are provided in the figure.
Body weight changes of infected mice. Body weights of SARS-CoV-2-infected mice corresponding to the results in Fig. 3. The averages of bodyweights are shown for infected mice without IFN treatment (mock) (black-filled circles), with IFN-λ treatment at the time of infection (green-filled circles), or with IFN-λ treatment 24 h prior to infection (green open squares). The specific viral doses and body weights are presented for all mice (left panels) and are stratified into survivors and nonsurvivors. Ten mice were infected with 3.3 × 104 PFU virus, and 9 mice were cotreated with IFN-λ. Nineteen mice were infected with 1.0 × 104 PFU virus, and 20 mice were cotreated with IFN-λ. Seventeen mice were infected with 1.0 × 103 PFU virus, and 16 mice were cotreated with IFN-λ. Twenty mice were pretreated with IFN-λ and infected with 1.0 × 103 PFU virus. Download FIG S1, PDF file, 2.6 MB (2.6MB, pdf) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ISG mRNA expression in the lungs of mice. Mice were treated with control PBS, treated with IFN-λ (2 μg), or infected with SARS-CoV-2 (1 × 103 PFU) for 24 h, and lungs were harvested. Lung homogenates were prepared for RT-qPCR, and the levels of specific ISG mRNAs were measured with primers shown in Table S1. mRNA levels are shown for IFN-λ treatment or SARS-CoV-2 infection relative to the PBS control. Three mice were used for each treatment, and the mean ± SEM is presented. Download FIG S2, PDF file, 0.5 MB (468.9KB, pdf) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence of oligonucleotide primers used in the study. Download Table S1, DOCX file, 0.01 MB (15.2KB, docx) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SARS-CoV-2 has developed multiple strategies to evade the IFN defense response (5–8). For this reason, administration of IFN-λ prior to infection is predicted to provide the most significant protection. To address this possibility, mice were administered IFN-λ intranasally 24 h before infection with 1 × 103 PFU SARS-CoV-2, and disease progression was measured (Fig. 3J) (open green squares). IFN-λ administration had a clear preventative effect on mortality. Stratifying the IFN-λ-treated animals by sex showed that all deaths occurred in male mice (Fig. 3L). Together, the results indicate that intranasal IFN-λ administration is an effective preventative therapeutic.
Viral clearance by IFN-λ administration in a SARS-CoV-2 mouse model of infection.
To determine if the antiviral effect of IFN-λ is the primary mechanism by which IFN-λ protects animals from SARS-CoV-2 pathology, we measured virus production in IFN-λ-treated and untreated infected mice (Fig. 4). Mice were either infected intranasally with 1 × 103 PFU SARS-CoV-2 or were pretreated with IFN-λ 24 h prior to the infection. The lungs and brains of infected mice were harvested 2 days postinfection, and infectious virus was measured in organ homogenates by plaque assays. There was no evidence of virus in the brains of infected mice at the limit of detection, 120 PFU/g tissue. However, the lungs from eight mice infected with virus produced titers between 1.1 × 105 and 1.5 × 106 PFU/g tissue. In stark contrast, seven mice that received IFN-λ before infection had no detectable virus in the lungs, and one animal had only 630 PFU/g tissue. These results indicate that the primary mode of action of IFN-λ in promoting animal survival is suppression of virus production.
FIG 4.
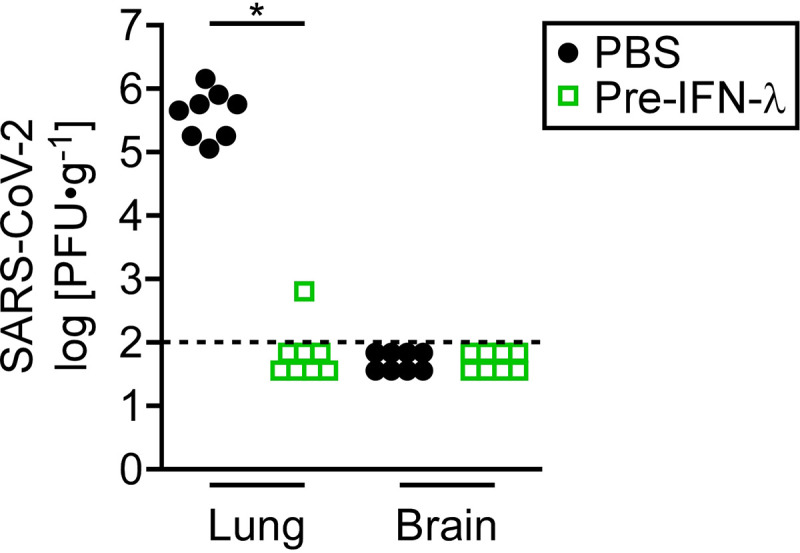
IFN-λ acts to inhibit SARS-CoV-2 replication in the lung. Male and female K18-hACE2 transgenic mice were either mock treated (black closed circles) or pretreated with intranasal IFN-λ for 24 h (green open squares) prior to infection with 1 × 103 PFU SARS-CoV-2. Viral titers were measured by plaque assay in lungs and brains harvested from eight mice 2 days postinfection. The dotted horizontal line indicates the limit of detection. Circles and boxes represent individual mice. Results represent pooled data from two independent experiments. *, P = 0.0002 (Mann-Whitney).
Histology reflected increased pathology in SARS-CoV-2-infected male mice.
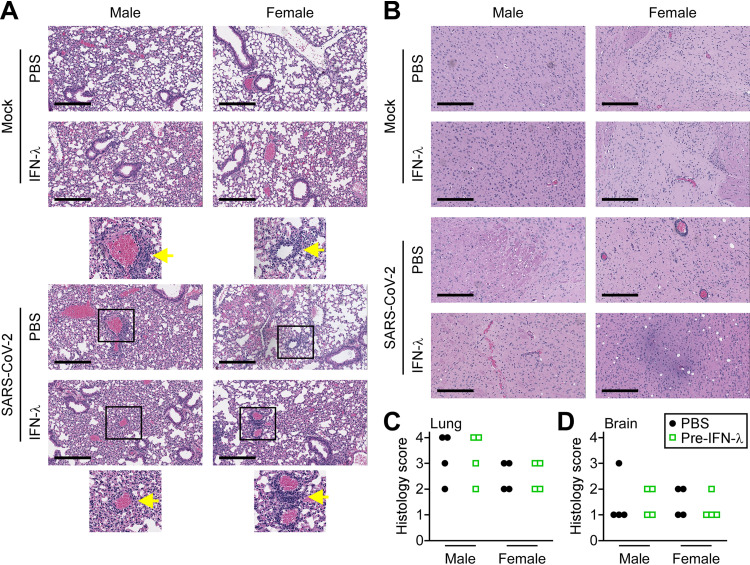
The pathology in mouse brains and lungs caused by virus infection was assessed by histology. Histology scoring parameters are provided in Table S2. Lungs and brains of mice were harvested 5 days following intranasal infection and evaluated for inflammation and hemorrhage (Fig. 5). Mock phosphate-buffered saline (PBS)-treated mice showed normal lung architecture with thin alveolar walls and clear air space. Following SARS-CoV-2 infection, a greater number of male mice showed moderate to severe bronchitis and perivascular lung inflammatory cells in comparison to less severe pathology in infected female mice. The difference is consistent with the sex differential in survival data (Fig. 2 and 3). Administration of IFN-λ alone had a minor inflammatory effect in the lungs. There were no obvious changes in the brains of male or female mice following infection with SARS-CoV-2 (Fig. 5B and D).
FIG 5.
Lung and brain histology following SARS-CoV-2 infection. (A) Representative hematoxylin and eosin (H&E) staining of mouse lung tissue sections. Top panels show images of lungs from uninfected mice (mock) treated with PBS or IFN-λ for 5 days. Bottom panels show representative lung sections from mice that were pretreated with PBS or IFN-λ and infected with 1 × 103 PFU SARS-CoV-2 for 5 days. Scale bar, 300 μm. Magnified images are shown with arrows identifying perivascular immune cell infiltration. (B) H&E-stained sections of brain tissue from mice treated as in panel A. Quantitative histology scoring for lungs (C) and brains (D) of male and female mice is described in Table S2 in the supplemental material.
Criteria used to score histopathology of mouse lungs in Fig. 5. Download Table S2, DOCX file, 0.01 MB (7.6KB, docx) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
SARS-CoV-2 is a positive-strand RNA virus with rapid replication kinetics and an estimated doubling time of 6 h (25). Therefore, an effective defense response is dependent on the swift action of innate immunity. Both type I IFN (IFNs-α/β) and type III IFN (IFN-λ) are critical components of the first line of innate defense. They bind distinct cell surface receptors to induce a similar set of IFN-stimulated genes (ISGs) that function in antiviral protection. IFNs-α/β receptors are expressed on all somatic cells, including immune cells; however, IFN-λ receptors are limited principally to epithelial cells (26). IFN-λ does not activate macrophages, T cells, or other cells of the immune system responsible for the cytokine storm and inflammation that are coincident with SARS-CoV-2 acute respiratory syndrome. Consequently, IFN-λ plays a protective role, primarily at mucosal sites such as the respiratory tract. For this reason, we investigated the potential therapeutic value of intranasal administration of IFN-λ as an antiviral in response to SARS-CoV-2 infection. The effects of IFN-λ on SARS-CoV-2 infection were first evaluated in cultured human lung cells. Our results and those of others demonstrated that IFN-λ inhibits SARS-CoV-2 production in cultured human lung cells (Fig. 1) (14, 15). IFN-λ addition at the time of infection reduced viral titers, and pretreatment of cells with IFN-λ completely blocked SARS-CoV-2 from establishing an infection. This differential effect is not unexpected since hours are needed for host cells to express antiviral ISG proteins in response to IFN-λ.
The human ACE2 metalloprotease has been identified as the primary functional receptor for SARS-CoV and for SARS-CoV-2 (27, 28). Originally, ACE2 was reported to be an ISG, raising concerns for the efficacy of IFN-based therapies for COVID-19 (29). However, it was later shown that IFN only induces expression of a truncated product of ACE2 that is not able to bind SARS-CoV-2 spike protein (30–32). Because the SARS-CoV-2 spike protein binds poorly to murine ACE2, we tested the prophylactic and therapeutic effects of IFN-λ in infected mice expressing the human ACE2 transgene driven by the cytokeratin-18 promoter (K18-hACE2) (33, 34).
Our studies with intranasal infection with a low dose of SARS-CoV-2 revealed a sex difference in disease outcome (Fig. 2). The precise infectious dose of SARS-CoV-2 during human respiratory transmission remains to be determined, but a computational estimate is approximately 300 virus particles (18). In our study, a high viral dose (3.3 × 104 PFU) produced a rapidly fatal disease in both male and female mice. However, as the inoculum was reduced to 1 × 103 PFU, male mice were found to have a higher mortality rate. This sex differential is reflective of the more severe symptoms and mortality that are well documented in male patients that suffer from COVID-19 (19, 23, 35).
Since the hACE2 gene is located on the X chromosome, it has been speculated that variants of hACE2 could be more detrimental in men and contribute to the sex differential. However, hACE2 in the transgenic mice is independent of X chromosome location, encodes the same gene in male and female mice, and is regulated by the same promoter. It has also been proposed that contributing factors of smoking or comorbid conditions of diabetes or heart disease may impact the sex differential, but these are not factors with the transgenic mice. A known difference between males and females is the increased responsiveness of the RNA-sensing Toll-like receptor 7 in females that leads to increased IFN expression (36, 37). Infection of a low number of lung cells with 1 × 103 PFU SARS-CoV-2 did not detect changes in ISG mRNA expression in lysates of male or female lungs (Fig. S2 in the supplemental material). Understanding the sex bias in response to SARS-CoV-2 infection is critical in considering therapeutic intervention and prediction of patient outcome. The transgenic model system will be used in the future to investigate the mechanisms that lead to worsened outcome in males following SARS-CoV-2 infection.
IFNs have been approved clinically for the treatment of several cancers, autoimmune diseases, and viral infections (38). The potential therapeutic value of IFN-λ for COVID-19 has been examined in clinical trials with a single subcutaneous injection of a pegylated form of IFN-λ (pegIFN-λ) either 3 days following diagnosis of mild to moderate COVID-19 (39) or within 7 days of symptoms or diagnosis (40). The former trial found no harmful effects of IFN-λ but no improvement in symptoms, whereas the latter trial found an improvement in viral clearance and respiratory symptoms. Experiments with a mouse-adapted SARS-CoV-2 indicated subcutaneous injection of pegIFN-λ reduced mortality in mice, although the animals were only evaluated for 5 days (41). Because SARS-CoV-2 infection commonly leads to an acute respiratory syndrome, intranasal administration or inhalation may be more therapeutic than subcutaneous injections. For this reason, we evaluated the protective effects of intranasal administration of unmodified IFN-λ in the K18-hACE2 transgenic mice with different doses of SARS-CoV-2. Overall, IFN-λ treatment at the time of viral infection was found to maintain body weight of survivors and reduce mortality (Fig. 3; Fig. S1). IFN-λ had even greater protection if administered 1 day prior to SARS-CoV-2 infection. In fact, all female mice survived infection with 1 × 103 PFU with IFN-λ pretreatment. The preventative value of IFN-λ is not unexpected since the antiviral ISGs are expressed before the SARS-CoV-2 products can inhibit IFN signaling and ISG expression (5–8).
To investigate the mechanism by which IFN-λ promotes survival following SARS-CoV-2 infection, we measured viral titers in lungs and brains of mice 2 days postinfection. The lungs of infected mice showed substantial viral production, whereas there was nearly undetectable virus in mice that received intranasal administration of IFN-λ prior to infection (Fig. 4). The results indicate that the protective effects of IFN-λ against SARS-CoV-2 infection in this model are mediated, in large part, by the ability of IFN-λ to block infectious virus production. However, since not all of the mice survive, it is possible that multiple doses of IFN-λ are needed to completely eliminate the virus. Alternatively, infection may trigger an irreversible inflammatory response leading to the death of a mouse.
Our preclinical model showed that administration of IFN-λ can reduce morbidity and mortality caused by SARS-CoV-2 infection and that IFN-λ can be effective as a preventative therapy. Previously, type I IFN-α was used in nasal drops of healthy medical staff in the COVID-19 epidemic area of Hubei Province in China, and the results indicated protection (42). The findings in our study indicate that intranasal administration of the less inflammatory IFN-λ can reduce replication of virus in the lungs of mice and improve survival. Our findings are particularly relevant for the use of broad-spectrum antivirals against newly emergent pathogens and for disease prevention in emergency responders and health care workers.
MATERIALS AND METHODS
Biocontainment.
All experiments conducted with SARS-CoV-2 were conducted either in a biosafety level 3 laboratory located in the Center for Infectious Disease (CID) or an animal biosafety level 3 laboratory located in the Laboratory of Comparative Medicine, Division of Laboratory Animal Resources. All personnel completed safety training and used personal protective equipment that included powered air-purifying respirators, Tyvek coveralls and booties, and double gloves. All protocols involving infectious virus and protocols for the inactivation of virus in samples prepared for export to a biosafety level 2 laboratory were approved by the University CID Safety Committee.
Mouse model and infections.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocol was approved by the Institutional Animal Care and Use Committee of Stony Brook University (IACUC no. 1596316). The Division of Animal Laboratory Research at Stony Brook University operates in accordance with the American Association for Laboratory Animal Science (AALAS), the American College of Laboratory Animal Medicine (ACLAM), and Animal Welfare Assurance ID D16-00006 (A3011-01) of the NIH. Mice were housed in either standard filter-topped shoebox microisolator cages or filter-ventilated cages. Rooms had 10 to 15 air changes per hour and were maintained at 70 to 72°F. Pelleted irradiated Purina mouse chow was provided ad libitum, and hyperfiltered water (2 μm) was provided via water bottles ad libitum. All mice were provided with Enviro-Dri nesting material. Mice were euthanized with 3 liter/min carbon dioxide inhalation, consistent with the recommendations of the panel on euthanasia of the American Veterinary Medical Association (AVMA guidelines) and Stony Brook University IACUC.
K18-hACE2 transgenic mice [Jackson Laboratory, B6.Cg-Tg(K18-ACE2)2Prlman/J, 9 to 10 weeks old] were anesthetized via intraperitoneal injection of 100 mg·ml−1 ketamine and 20 mg·ml−1 xylazine per kilogram of body weight. Mice were infected by intranasal inoculation of 1 × 103, 1 × 104, or 3.3 × 104 PFU SARS-CoV-2 passage 4 stock in 20 μl PBS containing 0.1% bovine serum albumin (BSA) and monitored daily for signs of disease and weight loss. For IFN-λ pretreatment, anesthetized animals received intranasal inoculation of 2 μg IFN-λ (30,000 units) in 20 μl PBS with 0.1% BSA. For IFN-λ treatment at the time of SARS-CoV-2 infection, anesthetized animals received SARS-CoV-2 mixed with 2 μg IFN-λ (30,000 units) IFN-λ in 20 μl PBS with 0.1% BSA. On day 2 following infection, mice were euthanized, lungs and brains were removed and homogenized in 2 ml Dulbecco’s modified Eagle medium (DMEM) with 10% FBS, and the viral titers in tissues were determined by plaque assay. For histopathology analysis, lungs and brains were harvested on day 5 postinfection and fixed in 10% formalin for 7 days at room temperature. Tissues were embedded in paraffin, thin sectioned, stained with hematoxylin-eosin, and inspected by light microscopy.
Cells, virus, and IFN-λ.
Human bronchial epithelial (HBE) 3-KT cells were obtained from American Type Culture Collection (ATCC) and cultured in airway epithelial cell basal medium supplemented with the bronchial epithelial cell growth kit (ATCC). HEK293-FT cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and penicillin-streptomycin. Vero E6 cells were maintained in DMEM supplemented with 10% HyClone iron-supplemented bovine calf serum (Cytiva). All cells were cultured at 37°C in a humidified atmosphere with 5% CO2. The SARS-CoV-2 strain (SARS-CoV-2/USA-WA1/2020) was obtained from Biodefense and Emerging Infections Research Resources Repository (NR no. 52281; BEI Resources, VA, USA). Virus stocks were prepared by passage of the seed virus (passage 1 [p1]) at an MOI of 0.01 per cell, and the infected cell culture supernatant was collected at 72 hpi, clarified by centrifugation at 400 × g for 10 min, and then stored at −70°C. Human IFN-λ1 (IL-29) was purchased from PBL Assay Science and diluted in PBS with 0.1% BSA.
Generation of HBE-hACE2 cells.
The human ACE2 cDNA was amplified from pcDNA3.1-hACE2-C9 (Addgene plasmid no. 1786) (27) by PCR using the primers listed in Table S1 in the supplemental material, and the SpeI/XhoI-digested fragment was inserted into corresponding sites of pLenti6/v5-D-TOPO (Life Technologies; kindly provided by Thomas Stamminger, Ulm University). HEK293-FT cells were cotransfected with pLenti6-hACE2, pLP1, pLP2, and pVSV-G using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Twenty hours later, cells were washed with PBS and incubated with fresh medium for 2 days. The supernatant was collected and added to HBEC3-KT cells with 5 μg polybrene/ml. Forty-eight hours postransduction, cells were treated with 10 μg blasticidin S/ml (Thermo Fisher), and resistant cells were evaluated.
Plaque assay.
Vero E6 cells were seeded in 6-well cell culture plates at 4.8 × 105 cells per well and incubated for 2 days to obtain confluent monolayers. Cells were infected with 0.4 ml of sample diluted in growth medium for 1 h at 37°C with occasional rocking. The virus inoculum was removed and replaced with 2.8 ml of an overlay medium consisting of DMEM, 2.5% bovine calf serum, and 0.76% gum tragacanth. Plaques were allowed to develop by incubating for 44 to 48 h, at which time the overlay medium was aspirated, and the cells were fixed and stained for 15 min with 0.5% crystal violet, 50% methanol, and 0.8% glutaraldehyde.
Western blot analyses.
Total cell extracts were prepared in sample buffer and subjected to SDS-PAGE. Separated proteins were transferred to nitrocellulose membranes and blocked with Tris-buffered saline containing 0.1% Tween 20 (TBST) and 3% BSA. The following primary antibodies were used: ACE2 (catalog no. 10108-T24; Sino Biological; 1:2,500), TMPRSS2 (catalog no. 14437-1-AP; Proteintech; 1:2,000), STAT1 (catalog no. 9172; Cell Signaling Technology; 1:2,000), phospho-STAT1 (Y701) (catalog no. 9167; Cell Signaling Technology; 1:2,000), IFIT1 (catalog no. 23247-1-AP; Proteintech; 1:2,000), and α-tubulin (catalog no. T5182; MilliporeSigma; 1:5,000). The IRDye 800CW-conjugated goat anti-rabbit IgG (Li-COR; 1:5,000) and the IRDye 680CW-conjugated goat anti-mouse IgG (Li-COR, 1:5,000) were used as secondary antibodies. The Western blot images were acquired on an Odyssey CLx imaging system (Li-COR) and analyzed using Image Studio software (Li-COR).
Quantification of SARS-CoV2 RNA and cellular mRNA by RT-qPCR.
The SARS-CoV2 RNA in the cell culture media was isolated using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's protocol. RNA levels were determined by RT-qPCR using the primers listed in Table S1 and a TaqPath 1-Step RT-qPCR master mix (Thermo Fisher) on a StepOne Plus real-time PCR system (Applied Biosystems). To determine the viral RNA copy numbers, a standard curve was generated by using SARS-CoV-2 nucleocapsid RNA. Briefly, a SARS-CoV2 nucleocapsid plasmid (Addgene; plasmid no. 153201) was linearized by digestion with XbaI and purified using a PCR purification kit (Qiagen). The nucleocapsid gene was in vitro transcribed using a MAXIscript SP6 kit (Thermo Fisher), resulting in a 1,533-base RNA fragment. DNA in the reaction mixture was removed by incubation with Turbo DNase (Thermo Fisher), and RNA was further purified using an RNeasy minikit (Qiagen).
Cell culture lysates and mouse lung and brain homogenates were prepared by disruption in 2 ml buffer RLT (Qiagen) supplemented with 1% 2-mercaptoethanol. Total RNA was purified with an RNeasy Plus minikit (Qiagen) followed by reverse transcription using EcoDry premix (TaKaRa). cDNA levels were measured by qPCR using primers listed in Table S1 and a DyNAmo HS SYBR green master mix (Thermo Fisher). The fold change in gene expression was determined by the threshold cycle (ΔΔCT) method using a differential with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and comparison with the untreated control.
Statistical analyses.
Statistical analyses were performed with GraphPad Prism 9. The proportion of surviving animals was analyzed using the two-tailed log-rank test. The organ viral burden was analyzed by the Mann-Whitney test. Two-way analysis of variance (ANOVA) with Turkey’s multiple comparisons was performed to calculate log-transformed in vitro data.
ACKNOWLEDGMENTS
We thank Hong Qin for technical assistance and Jeronimo Cello, the Laboratory Safety and Security Officer. We acknowledge the technical support provided by the Research Histology Core Laboratory, Department of Pathology, and the support of the Division of Animal Laboratory Resources at Stony Brook University.
This study was supported by funding received from the G. Harold and Leila Y. Mathers Foundation, the Stony Brook University School of Medicine Infrastructure Awards for COVID-19, the SUNY Research Seed Grant Program for COVID-19, and the Stony Brook University Office of the Vice President for Research COVID-19 Seed Grant Program.
Footnotes
This article is a direct contribution from Nancy C. Reich, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Eleanor Fish, Toronto General Research Institute, University Health Network & Department of Immunology, University of Toronto, Toronto, Ontario, Canada, and Peter Sarnow, Stanford University School of Medicine.
Citation Sohn S-Y, Hearing J, Mugavero J, Kirillov V, Gorbunova E, Helminiak L, Mishra S, Mackow E, Hearing P, Reich NC, Kim HK. 2021. Interferon-lambda intranasal protection and differential sex pathology in a murine model of SARS-CoV-2 infection. mBio 12:e02756-21. https://doi.org/10.1128/mBio.02756-21.
Contributor Information
Nancy C. Reich, Email: nancy.reich@stonybrook.edu.
Hwan Keun Kim, Email: hwan.kim@stonybrook.edu.
Stacy M. Horner, Duke University Medical Center
REFERENCES
- 1.Lazear HM, Schoggins JW, Diamond MS. 2019. Shared and distinct functions of type I and type III interferons. Immunity 50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. 2015. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stark GR, Darnell JE, Jr.. 2012. The JAK-STAT pathway at twenty. Immunity 36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, Guo F, Zhao Z, Zhou Z, Xiang Z, Wang J. 2020. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sa Ribero M, Jouvenet N, Dreux M, Nisole S. 2020. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog 16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, Danziger O, White KM, Rathnasinghe R, Uccellini M, Gao S, Aydillo T, Mena I, Yin X, Martin-Sancho L, Krogan NJ, Chanda SK, Schotsaert M, Wozniak RW, Ren Y, Rosenberg BR, Fontoura BMA, Garcia-Sastre A. 2020. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci USA 117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Pere H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pene F, Marin N, Roche N, Szwebel TA, Merkling SH, Treluyer JM, Veyer D, Mouthon L, Blanc C, Tharaux PL, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kerneis S, Terrier B. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultze JL, Aschenbrenner AC. 2021. COVID-19 and the human innate immune system. Cell 184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastard P, Orlova E, Sozaeva L, Levy R, James A, Schmitt MM, Ochoa S, Kareva M, Rodina Y, Gervais A, Le Voyer T, Rosain J, Philippot Q, Neehus AL, Shaw E, Migaud M, Bizien L, Ekwall O, Berg S, Beccuti G, Ghizzoni L, Thiriez G, Pavot A, Goujard C, Fremond ML, Carter E, Rothenbuhler A, Linglart A, Mignot B, Comte A, Cheikh N, Hermine O, Breivik L, Husebye ES, Humbert S, Rohrlich P, Coaquette A, Vuoto F, Faure K, Mahlaoui N, Kotnik P, Battelino T, Trebusak Podkrajsek K, Kisand K, Ferre EMN, DiMaggio T, Rosen LB, Burbelo PD, McIntyre M, Kann NY, et al. 2021. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med 218:e20210554. doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, Rosain J, Bilguvar K, Ye J, Bolze A, Bigio B, Yang R, Arias AA, Zhou Q, Zhang Y, Onodi F, Korniotis S, Karpf L, Philippot Q, Chbihi M, Bonnet-Madin L, Dorgham K, Smith N, Schneider WM, Razooky BS, Hoffmann HH, Michailidis E, Moens L, Han JE, Lorenzo L, Bizien L, Meade P, Neehus AL, Ugurbil AC, Corneau A, Kerner G, Zhang P, Rapaport F, Seeleuthner Y, Manry J, Masson C, Schmitt Y, Schluter A, Le Voyer T, Khan T, Li J, COVID-STORM Clinicians, et al. 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, Aoued H, Tharp GM, Pellegrini KL, Manfredi C, Sorscher E, Mainou B, Lobby JL, Kohlmeier JE, Lowen AC, Shi PY, Menachery VD, Anderson LJ, Grakoui A, Bosinger SE, Suthar MS. 2020. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J Virol 94:e00958-20. doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, Drosten C, Weber F. 2020. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem 295:13958–13964. doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Chen V, Shannon CP, Wei XS, Xiang X, Wang X, Wang ZH, Tebbutt SJ, Kollmann TR, Fish EN. 2020. Interferon-alpha2b treatment for COVID-19. Front Immunol 11:1061. doi: 10.3389/fimmu.2020.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prokunina-Olsson L, Alphonse N, Dickenson RE, Durbin JE, Glenn JS, Hartmann R, Kotenko SV, Lazear HM, O'Brien TR, Odendall C, Onabajo OO, Piontkivska H, Santer DM, Reich NC, Wack A, Zanoni I. 2020. COVID-19 and emerging viral infections: the case for interferon lambda. J Exp Med 217:e20200653. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu S. 2021. Computational characterization of inhaled droplet transport to the nasopharynx. Sci Rep 11:6652. doi: 10.1038/s41598-021-85765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peckham H, de Gruijter NM, Raine C, Radziszewska A, Ciurtin C, Wedderburn LR, Rosser EC, Webb K, Deakin CT. 2020. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun 11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vahidy FS, Pan AP, Ahnstedt H, Munshi Y, Choi HA, Tiruneh Y, Nasir K, Kash BA, Andrieni JD, McCullough LD. 2021. Sex differences in susceptibility, severity, and outcomes of coronavirus disease 2019: cross-sectional analysis from a diverse US metropolitan area. PLoS One 16:e0245556. doi: 10.1371/journal.pone.0245556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, III, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schafer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi-Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O'Neal WK, Randell SH, Boucher RC, Baric RS. 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J, Wong LR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB, Jr., Perlman S. 2021. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL. 2020. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol 20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradhan A, Olsson PE. 2020. Sex differences in severity and mortality from COVID-19: are males more vulnerable? Biol Sex Differ 11:53. doi: 10.1186/s13293-020-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheemarla NR, Watkins TA, Mihaylova VT, Wang B, Zhao D, Wang G, Landry ML, Foxman EF. 2021. Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J Exp Med 218:e20210583. doi: 10.1084/jem.20210583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donnelly RP, Kotenko SV. 2010. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res 30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH, II, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, HCA Lung Biological Network, et al. 2020. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng KW, Attig J, Bolland W, Young GR, Major J, Wrobel AG, Gamblin S, Wack A, Kassiotis G. 2020. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat Genet 52:1294–1302. doi: 10.1038/s41588-020-00732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onabajo OO, Banday AR, Yan W, Obajemu A, Stanifer ML, Santer DM, Florez-Vargas O, Piontkivska H, Vargas J, Kee C, Tyrrell DLJ, Mendoza JL, Boulant S, Prokunina-Olsson L. 2020. Interferons and viruses induce a novel primate-specific isoform dACE2 and not the SARS-CoV-2 receptor ACE2. bioRxiv doi: 10.1101/2020.07.19.210955. [DOI] [PMC free article] [PubMed]
- 32.Blume C, Jackson CL, Spalluto CM, Legebeke J, Nazlamova L, Conforti F, Perotin JM, Frank M, Butler J, Crispin M, Coles J, Thompson J, Ridley RA, Dean LSN, Loxham M, Reikine S, Azim A, Tariq K, Johnston DA, Skipp PJ, Djukanovic R, Baralle D, McCormick CJ, Davies DE, Lucas JS, Wheway G, Mennella V. 2021. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat Genet 53:205–214. doi: 10.1038/s41588-020-00759-x. [DOI] [PubMed] [Google Scholar]
- 33.McCray PB, Jr., Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang LI, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J, Silva J, Mao T, Oh JE, Tokuyama M, Lu P, Venkataraman A, Park A, Liu F, Meir A, Sun J, Wang EY, Casanovas-Massana A, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Yale IRT, Shaw A, Fournier JB, Odio CD, Farhadian S, Dela Cruz C, Grubaugh ND, Schulz WL, Ring AM, Ko AI, Omer SB, Iwasaki A, Yale IMPACT Research Team . 2020. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berghofer B, Frommer T, Haley G, Fink L, Bein G, Hackstein H. 2006. TLR7 ligands induce higher IFN-alpha production in females. J Immunol 177:2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- 37.Gupta S, Nakabo S, Blanco LP, O'Neil LJ, Wigerblad G, Goel RR, Mistry P, Jiang K, Carmona-Rivera C, Chan DW, Wang X, Pedersen HL, Gadkari M, Howe KN, Naz F, Dell'Orso S, Hasni SA, Dempsey C, Buscetta A, Frischmeyer-Guerrerio PA, Kruszka P, Muenke M, Franco LM, Sun HW, Kaplan MJ. 2020. Sex differences in neutrophil biology modulate response to type I interferons and immunometabolism. Proc Natl Acad Sci USA 117:16481–16491. doi: 10.1073/pnas.2003603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin FC, Young HA. 2014. Interferons: success in anti-viral immunotherapy. Cytokine Growth Factor Rev 25:369–376. doi: 10.1016/j.cytogfr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jagannathan P, Andrews JR, Bonilla H, Hedlin H, Jacobson KB, Balasubramanian V, Purington N, Kamble S, de Vries CR, Quintero O, Feng K, Ley C, Winslow D, Newberry J, Edwards K, Hislop C, Choong I, Maldonado Y, Glenn J, Bhatt A, Blish C, Wang T, Khosla C, Pinsky BA, Desai M, Parsonnet J, Singh U. 2021. Peginterferon lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat Commun 12:1967. doi: 10.1038/s41467-021-22177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feld JJ, Kandel C, Biondi MJ, Kozak RA, Zahoor MA, Lemieux C, Borgia SM, Boggild AK, Powis J, McCready J, Tan DHS, Chan T, Coburn B, Kumar D, Humar A, Chan A, O'Neil B, Noureldin S, Booth J, Hong R, Smookler D, Aleyadeh W, Patel A, Barber B, Casey J, Hiebert R, Mistry H, Choong I, Hislop C, Santer DM, Lorne Tyrrell D, Glenn JS, Gehring AJ, Janssen HLA, Hansen BE. 2021. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir Med 9:498–510. doi: 10.1016/S2213-2600(20)30566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinnon KH, III, Leist SR, Schafer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Jr., Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Z, Wang T, Chen L, Chen X, Li L, Qin X, Li H, Luo J. 2021. The effect of recombinant human interferon alpha nasal drops to prevent COVID-19 pneumonia for medical staff in an epidemic area. Curr Top Med Chem 21:920–927. doi: 10.2174/1568026621666210429083050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Body weight changes of infected mice. Body weights of SARS-CoV-2-infected mice corresponding to the results in Fig. 3. The averages of bodyweights are shown for infected mice without IFN treatment (mock) (black-filled circles), with IFN-λ treatment at the time of infection (green-filled circles), or with IFN-λ treatment 24 h prior to infection (green open squares). The specific viral doses and body weights are presented for all mice (left panels) and are stratified into survivors and nonsurvivors. Ten mice were infected with 3.3 × 104 PFU virus, and 9 mice were cotreated with IFN-λ. Nineteen mice were infected with 1.0 × 104 PFU virus, and 20 mice were cotreated with IFN-λ. Seventeen mice were infected with 1.0 × 103 PFU virus, and 16 mice were cotreated with IFN-λ. Twenty mice were pretreated with IFN-λ and infected with 1.0 × 103 PFU virus. Download FIG S1, PDF file, 2.6 MB (2.6MB, pdf) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ISG mRNA expression in the lungs of mice. Mice were treated with control PBS, treated with IFN-λ (2 μg), or infected with SARS-CoV-2 (1 × 103 PFU) for 24 h, and lungs were harvested. Lung homogenates were prepared for RT-qPCR, and the levels of specific ISG mRNAs were measured with primers shown in Table S1. mRNA levels are shown for IFN-λ treatment or SARS-CoV-2 infection relative to the PBS control. Three mice were used for each treatment, and the mean ± SEM is presented. Download FIG S2, PDF file, 0.5 MB (468.9KB, pdf) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence of oligonucleotide primers used in the study. Download Table S1, DOCX file, 0.01 MB (15.2KB, docx) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Criteria used to score histopathology of mouse lungs in Fig. 5. Download Table S2, DOCX file, 0.01 MB (7.6KB, docx) .
Copyright © 2021 Sohn et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.