Abstract
Myelinating glia express high levels of a unique set of genes which code for structural proteins of the myelin sheath. Few transcription factors have so far been implicated in the regulation of any myelin gene. Here we show that the protein zero (P0) gene, a myelin gene exclusively expressed in the Schwann cell lineage of the peripheral nervous system, is controlled in its expression by the high-mobility-group domain protein Sox10 both in tissue culture and in vivo. Induction of wild-type Sox10, but not of other transcription factors or Sox10 mutants, strongly increased endogenous P0 expression in tissue culture. This activation was mediated by the P0 promoter, which was stimulated by Sox10 in transient transfections. Detailed analyses revealed the involvement of a proximal and a distal promoter region. The distal region functioned only in conjunction with the proximal one and contained a single Sox consensus binding site, which accounted for most of its activity. In contrast, the proximal region mediated Sox10 responsiveness on its own. It contained multiple binding sites for Sox proteins, with two high-affinity sites being the most significant. P0 expression also depended on Sox10 in vivo, as evident from the analysis of Schwann cell precursors in mouse embryos with Sox10 mutation at day 12.5 of embryogenesis. To our knowledge this is the most conclusive link to date between a glial transcription factor and cell-specific activation of myelin gene expression.
The nervous system contains two major types of cells, neurons and glia. The task of glial cells is to support development, survival, and functionality of neurons. Glial cells are found associated with neuronal cell bodies as well as with axons. Vertebrates have developed special types of glial cells that form multilamellar sheaths around axonal segments. These myelin sheaths act as electrical insulators and confine the spread of action potentials to the nodes of Ranvier which separate the myelinated segments. Myelinating glia are thus essential for the rapid saltatory conduction of nerve impulses that are characteristic of the vertebrate nervous system.
The myelin sheath is a specialized organelle that contains a small number of highly abundant proteins (25). These include myelin basic protein, proteolipid protein (PLP), protein zero (P0), and peripheral myelin protein 22 (PMP-22). Whereas myelin basic protein is an integral part of myelin in both the central and peripheral nervous systems, other myelin proteins are essentially confined to either peripheral or central nervous system. PLP, for instance, is only expressed at low levels in Schwann cells, which constitute the myelinating cells of the peripheral nervous system. Furthermore, PLP is not integrated into Schwann cell myelin (16). In the central nervous system, however, PLP accounts for approximately 40% of total myelin protein, and PLP transcripts are highly abundant in oligodendrocytes, which are the myelinating cells of the central nervous system.
For P0 and PMP-22, the situation is the exact reverse. Both are preferentially present in Schwann cells (24, 25). P0, in particular, seems to be expressed at significant levels in no cells other than those of the Schwann cell lineage. This transmembrane glycoprotein of the immunoglobulin superfamily is detected in neural crest cells committed to the glial lineage and continues to be present throughout development of the Schwann cell lineage at low levels (10, 21). Upon myelination, P0 expression is massively upregulated. As a consequence, P0 makes up more than 50% of the total myelin protein in mature Schwann cells, where it is directly involved in myelin compaction (1, 7, 26, 27, 42).
Its highly restricted expression has made P0 an attractive target for the analysis of cell-specific transcriptional regulation. Using cultures of rat Schwann cells and transgenic mice, it was shown that a 1.1-kb promoter region of the rat P0 promoter is sufficient to mediate Schwann cell-specific expression both in vitro and in vivo (27, 29, 30). Transient transfections have been used to analyze this region in further detail. These experiments revealed the presence of a minimal promoter responsible for basal levels of transcription, as well as a strongly activating proximal and a modulatory distal region within this fragment (6). Although DNase footprinting experiments have succeeded in identifying a number of cis-acting sequences within the P0 promoter, the relevant trans-acting factors are not known. This situation is symptomatic for all myelin gene promoters studied to date.
At the same time, a number of transcription factors which exhibit preferential expression in myelinating glia have been identified and found to be important for gliogenesis and maintenance of the glial phenotype (for reviews, see references 31, 40, and 50). However, the target genes through which these factors act are not known. One of these transcription factors is Sox10 (19). This transcription factor belongs to the group of Sox proteins which contain as their DNA binding domain a high-mobility-group (HMG) box with similarity to the one originally identified in the mammalian sex-determining factor Sry (35, 47). During development, Sox10 is first expressed widely in cells of the emerging neural crest (4, 19, 38, 46). Mutation of Sox10 therefore leads to a combination of neural crest defects that lead to embryonic lethality in homozygously affected mice and to pigmentation defects, deafness, and colonic aganglionosis in heterozygously affected mice and human patients suffering from combined Waardenburg-Hirschsprung syndrome (13, 36, 46). The neural crest-derived Schwann cell lineage also seemed to be affected in mice homozygous for the Sox10 mutation, arguing for a role of Sox10 in the early phases of Schwann cell development (13).
During late stages of embryogenesis and in the adult, Sox10 expression is primarily found in myelinating glia, including both Schwann cells and oligodendrocytes. This continued expression in myelinating glia clearly indicates that Sox10 has a function not only during early committment and determination but also during later stages of glial development. As for other glial transcription factors, no target gene which could help to explain Sox10 function in glial cells has so far been characterized.
Here we show that the P0 gene is a direct transcriptional target of Sox10, thus providing both one of the first examples of a target gene for a glial transcription factor and of a transcription factor intricately involved in the regulation of myelin-specific genes.
MATERIALS AND METHODS
Plasmids.
Plasmid pMPTRE was constructed by inserting a hygromycin resistance cassette into pUHD10-3, which carries a tetracycline-regulatable promoter (8). For inducible expression, rat cDNAs for several transcription factors were inserted into pMPTRE behind the tetracycline-regulatable promoter, using the following restriction sites: HindIII for Sox10 and Sox10dom (13), NotI/XhoI for Sox10059 (20), NotI for Sox11 (18), and EcoRI for Krox-20 (19).
The luciferase reporter plasmids containing either the rat P0 promoter from positions −915 to +48 (P0luc) or the β-globin TATA box (pTATAluc) were as described elsewhere (12, 41). The luciferase reporter carrying the mouse P0 promoter from positions −1300 to +48 (mP0luc) was constructed similarly to P0 luc, with mouse P0 sequences being generated by genomic PCR and inserted between XhoI and HindIII sites in front of luciferase. Deletion mutants of P0luc (see Fig. 4A) were generated by use of a BglII site at position −435 or a BamHI site at position −38 or by PCR-directed mutagenesis. The mutation of potential Sox binding sites within the P0 promoter (see Fig. 5A and 6B) was through the use of a Quick Change mutagenesis kit (Stratagene). A region corresponding to positions −229 to −116 of the rat P0 promoter was also inserted into pTATAluc in front of the TATA box, yielding pTATA+Proxluc. Plasmid pTKluc contained the full length murine thymidine kinase (TK) promoter in front of luciferase. For bacterial expression of rat Sox10, cDNA sequences corresponding to amino acids 1 to 189 and 101 to 180 were generated by PCR and inserted into pGEX vectors as BamHI/HindIII fragments.
FIG. 4.
Two regions of the P0 promoter mediate Sox10-dependent activation. (A) Schematic representation of rat P0 promoter regions used to drive luciferase reporter gene expression in panels B and C. Endpoints of each fragment are marked by their position relative to the transcriptional start site (+1). (B and C) Original Tet-On N2A cells (open bars) and derivatives capable of expressing Sox10 (filled bars) were transfected in quadruplicates with luciferase reporters driven by the β-globin minimal promoter (TATA) or various regions of the rat P0 promoter, shown in panel A. From each transfected quadruplicate, one duplicate was left untreated and the other was treated with doxycycline to induce transcription factor expression. Luciferase activities in extracts from transfected cells were determined in three independent experiments. Before doxycycline treatment, actual activities of luciferase reporters (light units per microgram of total protein) were 60 to 80 for TATA, 300 to 550 for P0, 200 to 300 for −558 P0, 350 to 450 for −435 P0, 600 to 850 for −295 P0, 600 to 700 for −155 P0, 30 to 40 for minP0, 80 to 100 for d1 P0, and 70 to 90 for d2 P0. Data were normalized and are presented as fold increase of luciferase activities in the presence of Sox10 expression relative to its absence.
FIG. 5.
Two consensus Sox binding sites are present within the P0 promoter. (A) Purified GST-Sox10 protein (amino acids 101 to 180) was analyzed in electrophoretic mobility shift assays for its ability to bind to radiolabeled oligonucleotides containing consensus Sox binding sites from the P0 promoter (B and F; for sequence, see Fig. 6B) or mutant versions thereof (mtB and mtF; for sequence, see Fig. 6B). The relative localization of these sites in the P0 promoter is shown at the bottom. −, no protein added; +, GST-Sox10 added. (B) Tet-On N2A cells capable of expressing Sox10 were transfected in quadruplicates with luciferase reporters driven by the β-globin minimal promoter (TATA) or the rat P0 promoter. In addition to the wild-type P0 promoter (P0), versions were used in which site B (P0mtB) or site F (P0mtF) were mutated. minP0 denotes the minimal P0 promoter from positions −38 to +48. From each transfected quadruplicate, one duplicate was left untreated and the other was treated with doxycycline to induce transcription factor expression. Luciferase activities in extracts from transfected cells were determined in three independent experiments. Before doxycycline treatment, actual activities of luciferase reporters (light units per micrograms of total protein) were 60 to 80 for TATA, 300 to 550 for P0, 30 to 40 for minP0, 500 to 600 for P0mtB, and 550 to 670 for P0mtF. Data are presented after normalization as fold increase of luciferase activities in the presence of Sox10 expression relative to its absence.
FIG. 6.
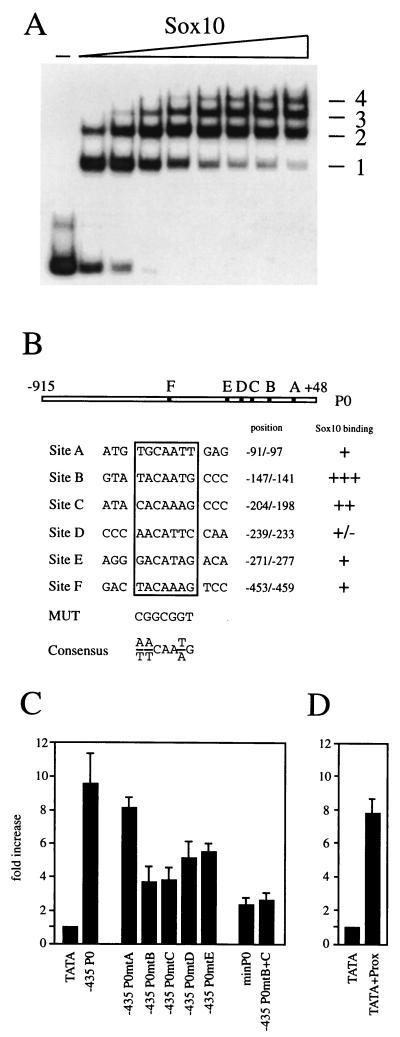
The proximal region of the P0 promoter contains multiple Sox binding sites. (A) Increasing amounts of purified GST-Sox10 protein (amino acids 101 to 180) were analyzed in electrophoretic mobility shift assays for the ability to bind to a radiolabeled fragment spanning positions −295 to −38 of the rat P0 promoter. Protein-DNA complexes are numbered consecutively. −, no protein added. (B) Summary of potential Sox binding sites (A to F) and their relative localization in the rat P0 promoter. The consensus Sox binding site (Consensus) and sequence to which binding sites within the P0 promoter were mutated (MUT) are shown. DNA binding of GST-Sox10 to all sites was determined and rated from strong (+++) to weak (±). (C and D) Tet-On N2A cells capable of expressing Sox10 were transfected in quadruplicates with luciferase reporters driven by the β-globin minimal promoter (TATA), a combination of rat P0 sequences from positions −229 to −116, and β-globin minimal promoter (TATA+Prox) or shortened versions of the rat P0 promoter (−435 P0 and minP0; see also Fig. 4). In addition to the wild-type −435 P0 luciferase construct, versions were used in which site A (−435 P0mtA), B (−435 P0mtB), C (−435 P0mtC), D (−435 P0mtD), E (−435 P0mtE), or B and C (−435 P0mtB+C) were mutated as indicated in panel B. From each transfected quadruplicate, one duplicate was left untreated and the other was treated with doxycycline to induce transcription factor expression. Luciferase activities in extracts from transfected cells were determined in three independent experiments. Before doxycycline treatment, actual activities of luciferase reporters (light units per microgram of total protein) were 60 to 80 for TATA, 350 to 450 for −435 P0, 120 to 150 for −435 P0mtA, 380 to 460 for −435 P0mtB, 430 to 530 for −435 P0mtC, 380 to 450 for −435 P0mtD, 530 to 630 for −435 P0mtE, 630 to 710 for −435 P0mtB+C, and 180 to 230 for TATA+Prox. Data were normalized and are presented as fold increase of luciferase activities in the presence of Sox10 expression relative to its absence.
Cell culture, transfections, and luciferase assays.
G418-resistant N2A neuroblastoma cells expressing the reverse tetracycline-controlled transactivator (rtTA) were a gift from E.-M. Mandelkow (EMBL Outstation, Hamburg, Germany). Stable transfection was performed using calcium phosphate precipitates and hygromycin selection. The resulting transfectants were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, G418 (400 μg/ml; Gibco BRL), and hygromycin (150 μg/ml; Roche Diagnostics). For Western blot analysis, cells were harvested after 24 h in the presence or absence of doxycycline (2.5 μg/ml; Sigma).
For luciferase assays, cells were transfected in quadruplicates on 35-mm-diameter plates with 2 μg of luciferase reporter plasmid per plate, using Superfect reagent as instructed by the manufacturer (Qiagen). After transfection, cells were returned to Dulbecco's modified Eagle medium containing 10% fetal calf serum. To induce expression of the effector, doxycycline was added at a concentration of 2.5 μg/ml to half of the plates. Cells were harvested 62 h posttransfection, and extracts were assayed for luciferase activity (48). To compare induction rates between single experiments, doxycycline-dependent activation of a particular luciferase reporter was normalized to the doxycycline-dependent activation of pTATAluc. For normalization to an internal control, 2 μg of plasmid pCH110 (Clontech) was cotransfected, and aliquots of the extracts were analyzed in liquid β-galactosidase assays according to standard protocols.
RNA preparation, reverse transcription, PCR, and RNase protection.
Total RNA was isolated from N2A stable transfectants and from embryonic day 12.5 (E12.5) mouse embryos, using TRIZOL reagent (Gibco BRL). Two micrograms of each RNA sample was reverse transcribed into cDNA for 1 h at 42°C, using 400 U of Moloney murine leukemia virus reverse transcriptase, 45 pmol of oligo(dT) primer, and 0.5 mM deoxynucleoside triphosphate in 30 μl of 50 mM Tris-HCl (pH 8.3)–75 mM KCl–3 mM MgCl2–1 mM dithiothreitol.
For quantitation, 2 μl of cDNA was amplified with primer pairs specific for P0, Sox10, Sox11, Krox-20, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The following primer pairs were used: P0 (5′-GCCCTGCTCTTCTCTTCTTT-3′ and 5′-CCAACACCACCCCATACCTA-3′, yielding a 0.4-kb product), Sox10 (5′-GAGGAGGTGGGCGTTGGGCTCTTC-3′ and 5′-AGCTCTGTCTTTGGGGTGGTTGGA-3′, yielding a 0.8-kb product), Sox11 (5′-CGTTGGAAGATGCTGAAGGACA-3′ and 5′-CCGCTGGATGAGGAGGTGGACA-3′, yielding a 0.65-kb product), Krox-20 (5′-CACCACTTCCACCTCCTCTC-3′ and 5′-CTCACCACCTCCACTTGCTC-3′, yielding a 0.3-kb product), and GAPDH (5′-GCCATCAA(C/T)GACCCCTTCATT-3′ and 5′-CGCCTGCTTCACCACCTTCTT-3′, yielding a 0.7-kb product). In addition to 40 pmol of each primer in a selected pair, PCR mixtures contained 0.2 mM deoxynucleoside triphosphate, 0.4 μCi of [32P]dCTP, and 1 U of Taq DNA polymerase in 40 μl of 10 mM Tris-HCl (pH 8.3)–10% (vol/vol) dimethyl sulfoxide–50 mM KCl–2 mM MgCl2. After denaturation (1 min at 94°C), repeated cycling was performed; each cycle consisted of denaturation (30 s at 94°C), primer-specific annealing (30 s at 56°C for P0, 58°C for Krox-20 and GAPDH, and 62°C for Sox10 and Sox11), and an elongation step (45 s at 72°C). Amplification products obtained after 20, 23, and 26 cycles were separated for each gene on 4% polyacrylamide gels and analyzed by autoradiography. RNase protection experiments were carried out with a HybSpeed RPA kit (Ambion) using 15 μg of total RNA hybridized to a 456-bp antisense probe specific for mouse P0.
Proteins, cell extracts, and Western blots.
Regions corresponding to amino acids 1 to 189 or 101 to 180 of rat Sox10 were produced in bacteria as glutathione S-transferase (GST) fusion proteins and purified according to standard procedures (39). In some experiments, the GST moiety was removed by thrombin cleavage.
Extracts from N2A neuroblastoma cells were prepared and analyzed by Western blotting and enhanced chemiluminescence ECL detection as described elsewhere (43). Polyclonal rabbit antisera directed against Sox10, Sox11, or Krox-20 (1:3,000 dilution) served as primary antibodies; horseradish peroxidase-coupled protein A served as the secondary detection reagent (19).
Electrophoretic mobility shift assay.
In general, 0.5 ng of 32P-labeled probe (for sequences, see Fig. 6B and 7A) were incubated with recombinant protein for 20 min on ice in a 20-μl reaction mixture containing 10 mM HEPES (pH 8.0), 5% glycerol, 50 mM NaCl, 5 mM MgCl2, 2 mM dithiothreitol, 0.1 mM EDTA, 4 μg of bovine serum albumin, and 2 μg of poly(dG-dC) as nonspecific competitor. Samples were loaded onto native 4% polyacrylamide gels and electrophoresed in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA [pH 8.3]) at 120 V for 1.5 h. Gels were dried and exposed for autoradiography.
FIG. 7.
The high-affinity binding site C in the proximal region of the P0 promoter binds Sox10 cooperatively. (A) Schematic representation of the DNA fragments used in panel B. Fragment endpoints are defined by their position relative to the transcription start site of the P0 gene. Intact binding sites are shown as open boxes; mutations are indicated by crossed-out boxes. (B) Purified Sox10 protein (amino acids 1 to 189) was analyzed in electrophoretic mobility shift assays for its ability to bind to a radiolabeled fragment spanning positions −229 to −116 of the rat P0 promoter. In addition to the wild-type fragment (wt), versions were used in which site B (mutB), C (mutC), or both (mutB+C) were mutated. −, no protein added; +, Sox10 protein added (C) Purified Sox10 protein (amino acids 1 to 189) was analyzed in electrophoretic mobility shift assays for its ability to bind to a radiolabeled fragment spanning positions −206 to −185 of the rat P0 promoter. In addition to the wild-type fragment (C/C′), versions were used in which C (mutC/C′), or C′ (C/mutC′) were mutated. −, no protein added; +, Sox10 protein added.
In situ hybridization, histology, and microscopy.
Mouse embryos at E12.5 were fixed overnight at 4°C in 4% paraformaldehyde, dehydrated, bleached, and embedded in 20% gelatin. After overnight fixation in 4% paraformaldehyde, blocks were vibratome sectioned (100 μm). Digoxigenin (DIG)-labeled riboprobes were produced with a DIG-RNA labeling kit (Roche Diagnostics). Whole-mount in situ hybridization on vibratome slices was performed essentially as described elsewhere (15, 21) with hybridization temperature raised to 60°C for P0 and kept at 65°C for cadherin-6.
For histological analysis, E12.5 mouse embryos were fixed overnight at 4°C in 4% paraformaldehyde, dehydrated, and embedded in Technovit 7100 resin (Kulzer); 6-μm sections were stained with toluidine blue.
RESULTS
Inducible expression of Schwann cell transcription factors.
During development of the Schwann cell lineage, P0 is expressed in an overlapping pattern with several transcription factors that are preferentially present in these cells, including the POU protein Tst-1/Oct6/SCIP, the zinc finger protein Krox-20, and the HMG domain protein Sox10 (3, 19). Krox-20 and Sox10, in particular, are both strongly expressed in mature Schwann cells which produce large amounts of P0.
To test whether P0 expression is regulated by any of these transcription factors, we generated cell lines that allowed their inducible expression. The system chosen was the Tet-On system, where rapid induction of a gene under the control of a tetracycline-responsive promoter (TRE) is achieved by addition of tetracycline or its derivative doxycycline to cells which express the rtTA transactivator (9). rtTA-positive N2A neuroblastoma were selected as starting material because of their neural origin and the absence of endogenous Tst-1/Oct6/SCIP and Sox10 (19). Only Krox-20 was present in low amounts (data not shown).
Clonal and polyclonal lines were established by stable transfection using TRE–Tst-1, TRE–Krox-20, and TRE-Sox10 constructs. Both polyclonal and several clonal lines exhibited tightly regulated expression of these transcription factors with low background levels under normal culture conditions and high induction rates after addition of doxycycline (Fig. 1A). Upon long exposure of Western blots, minimal amounts of each transcription factor were detectable in the uninduced state, as were low levels of transcripts by a sensitive reverse transcription-PCR (RT-PCR) approach (Fig. 1B). Residual expression in the uninduced state, however, did not impair the usefulness of this system. Experiments described below were performed with the polyclonal and various clonal lines with essentially identical results, thus ruling out that the observed effects are integration or selection artifacts caused by the process of stable transfection.
FIG. 1.
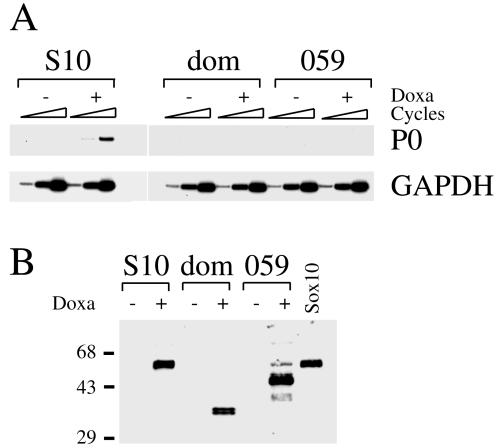
Sox10 selectively increases endogenous P0 expression in stable N2A transfectants. (A) Western blot analyses of extracts prepared from N2A Tet-On cell lines generated by stable transfection and kept for 24 h in the absence (−) or presence (+) of doxycycline (Doxa). Expression of Sox10 (S10), Sox11 (S11), and Krox-20 (K20) proteins was detected using polyclonal antisera raised against the proteins. Numbers on the right indicate sizes of molecular weight markers in kilodaltons. (B) RT-PCR analysis of cDNA obtained from N2A Tet-On cell lines capable of inducibly expressing Sox10 (S10), Sox11 (S11), or Krox-20 (K20). Transcription factor induction (+) was through treatment with doxycycline (Doxa). cDNAs from uninduced cells (−) served as a control. P0 transcript levels were compared in the various transfectants by semiquantitative PCR using increasing numbers of amplification cycles. PCRs with primers specific for GAPDH, Sox10, Sox11, and Krox-20 were performed to monitor the experiment.
Activation of P0 gene expression by induction of Sox10.
To identify potential activators of P0 gene expression, we screened for variations in the levels of endogenous P0 transcripts coincident with induction of Tst-1/Oct6/SCIP, Krox-20, or Sox10 in each stable cell line. Doxycycline treatment of mock-transfected Tet-On N2A cells did not cause activation of endogenous P0 expression (data not shown). Similarly, induction of Krox-20 or Tst-1/Oct6/SCIP did not lead to any detectable increase in endogenous P0 message, as judged by semiquantitative RT-PCR analysis (Fig. 1B and data not shown). Among the cell lines tested, those that inducibly expressed Sox10 were the only ones for which significant alterations in the amounts of endogenous P0 message could be detected. Upon Sox10 induction, we detected on average a greater than 10-fold increase in the intensity of the P0-specific signal in semiquantitative RT-PCRs (Fig. 1B).
To analyze whether this effect was specific to Sox10 or could be obtained with other Sox proteins, we generated another set of N2A lines which inducibly expressed Sox11 (Fig. 1). This Sox protein was chosen because it is present in cells of various neural lineages and because it behaves as a very potent transcriptional activator on artificial promoter constructs with binding sites for Sox proteins (18). Sox10 and Sox11 belong to different subgroups of Sox proteins (47), significant (59%) amino acid identities between them being restricted to their HMG domains. After induction of Sox11, only a slight activation of P0 expression was observed, clearly pointing to the specificity of P0 gene induction by Sox10.
Consequences of Sox10 mutations on P0 gene induction.
Several mutations that lead to functional inactivation of Sox10 have been found in mice and Waardenburg-Hirschsprung patients (5, 13, 20, 36). We generated stable N2A lines which inducibly expressed naturally occurring Sox10 mutations (Fig. 2B). The Sox10Dom mutation was originally identified as the gene defect in the spontaneous mouse mutant Dominant megacolon (Dom) (13, 46), while the Sox10059 mutation derives from a Waardenburg-Hirschsprung patient (36). Both mutations consist of frameshifts that lead to Sox10 proteins with carboxy-terminal truncations varying in length. Sox10Dom consists of only the first 193 residues of Sox10 followed by 99 unrelated amino acids; Sox10059, on the other hand, contains the first 360 residues of Sox10 followed by 40 unrelated amino acids (Fig. 2B). Both proteins still have an intact HMG domain and are capable of DNA binding (20). When endogenous P0 expression was quantified in these stable cell lines in the absence or presence of doxycycline, no significant difference was detected (Fig. 2A). Unlike the case for wild-type Sox10, induction of neither mutant led to an increase of endogenous P0 expression. Thus, naturally occurring Sox10 mutations are defective for P0 activation in the N2A Tet-On system.
FIG. 2.
Sox10 mutants fail to increase endogenous P0 expression in stable N2A transfectants. (A) RT-PCR analysis on cDNA obtained from N2A Tet-On cell lines before (−) and after (+) induction of Sox10 proteins by treatment with doxycycline (Doxa). P0 transcript levels were compared in transfectants capable of inducibly expressing Sox10 (S10) or the Sox10 mutants Sox10Dom (dom) and Sox10059 (059) by semiquantitative PCR using increasing numbers of amplification cycles. PCRs with primers specific for GAPDH were performed as control. (B) Western blot analysis of extracts prepared from stable transfectants kept for 24 h in the absence (−) or presence (+) of doxycycline. Sox10 (S10) and its two mutant forms Sox10Dom (dom) and Sox10059 (059) were detected using a polyclonal antiserum raised against Sox10. As a positive control, Sox10 from transiently transfected COS cells (Sox10) is shown. Numbers on left indicate sizes of molecular weight markers in kilodaltons.
Activation of the P0 promoter by Sox10.
The region responsible for glia-specific expression of P0 has been mapped to a 1.1-kb promoter region (27, 29), the most important determinants being present between positions −350 to +45 relative to the transcriptional start site (6). Therefore, it was of interest to determine whether the Sox10-dependent increase of P0 expression was mediated by the promoter region. This question was addressed in transient transfection experiments using a luciferase reporter under the control of rat P0 promoter sequences from positions −915 to +48 (Fig. 3A). The P0 luciferase reporter was transfected into the stable Tet-On N2A lines, and transcription factor expression was activated by treatment with doxycycline. In general, activation of reporter gene expression was measured as the ratio of luciferase activity in the presence and absence of doxycycline relative to the β-globin minimal promoter. In several experiments, activation rates were additionally determined relative to the full-length TK promoter or the simian virus 40 (SV40) early promoter, with reference plasmids being transfected either in separate plates (TK-driven reporter) or as internal control in the same plates as the P0 luciferase reporter (SV40-based reporter). As shown paradigmatically in Fig. 3B and C, results were very similar under all conditions.
FIG. 3.
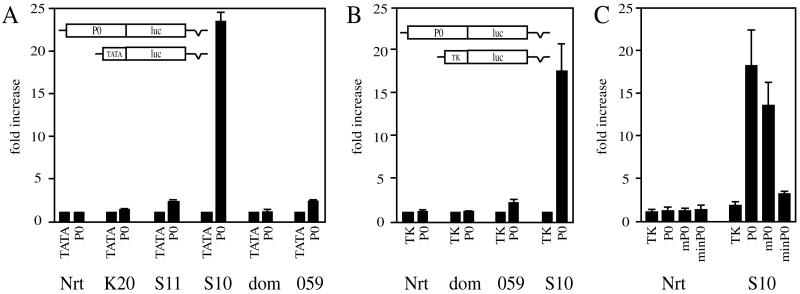
Sox10 increases activity of the P0 promoter. (A) Tet-On N2A cells (Nrt) or various derivatives capable of expressing Krox-20 (K20), Sox11 (S11), Sox10 (S10), Sox10Dom (dom) or Sox10059 (059) were transfected in quadruplicates with luciferase (luc) reporters driven by the β-globin minimal promoter (TATA) or the rat P0 promoter (P0). From each transfected quadruplicate, one duplicate was left untreated and the other was treated with doxycycline to induce transcription factor expression. Luciferase activities in extracts from transfected cells were determined in three independent experiments. Data were normalized to pTATAluc as described in Materials and Methods and are presented as fold increase of luciferase activities in the presence of transcription factor expression relative to its absence. (B and C) For select transfections, data were normalized to pTKluc, a luciferase reporter driven by the full-length TK promoter (B), or to pCH110, an internal control plasmid which carries lacZ under the control of the SV40 early promoter (C). Transfections in panel C were also carried out with the mouse P0 promoter (mP0) and the rat minimal P0 promoter (minP0; see also Fig. 4). Measured activities (light units per microgram of total protein) for the rat P0 promoter before doxycycline treatment were in the range of 150 to 290 in Nrt, 90 to 110 in K20, 110 to 180 in S11, 300 to 550 in S10, 120 to 200 in dom, and 380 to 450 in 059.
There was no significant stimulation of the P0 promoter in the original N2A Tet-On cells or in the stable Krox-20 cell lines after transcription factor induction (Fig. 3A). The induction of Sox11 led to a twofold stimulation of the P0 promoter, which is in good agreement with the already described low-level Sox11-dependent induction of endogenous P0 (Fig. 1B). In parallel experiments, we observed a 23-fold increase in reporter gene expression upon induction of Sox10. When wild-type Sox10 was replaced by the Sox10059 mutant or the Sox10Dom mutant, stimulation of the P0 promoter was severely reduced or abolished (Fig. 3A).
Induction of Sox10 also activated transcription from a mouse P0 promoter spanning positions −1300 to +48 (Fig. 3C). Stimulation rates were on average 75% of those observed for the rat promoter fragment. The Sox10-dependent induction of the endogenous P0 is thus faithfully mimicked in transient transfections by luciferase reporters under the control of the P0 promoter.
Sox10-responsive regions in the P0 promoter.
To determine which part of the P0 promoter mediates Sox10-dependent activation, we generated shortened versions of the P0 promoter by successively removing sequences from the distal end (Fig. 4A). The resulting promoter constructs were then analyzed for the ability to be activated in a Sox10-dependent manner by transfection into the stable S10 cell line (Fig. 4B). Control transfections in the original N2A Tet-On cells confirmed that none of the P0 promoter constructs was activated upon addition of doxycycline in the absence of Sox10 (Fig. 4B).
Deletion of the distal-most 357 bp left inducibility unaltered (compare the 17-fold stimulation for P0 to the 16-fold stimulation for −558 P0 in Fig. 4B), arguing against a role of the distal portion in mediating Sox10-dependent stimulation of the P0 promoter. This conclusion was corroborated in the complementary experiment. A compound construct consisting of the distal-most 357 bp and the minimal P0 promoter was not activated by Sox10 (d1 P0 in Fig. 4C). However, further truncation of the P0 promoter to position −435 led to a significant reduction in Sox10 responsiveness, with stimulation rates dropping from 16-fold to 6-fold (Fig. 4B). Thus, there is an important determinant of Sox10 responsiveness between positions −558 and −435. However, this region failed to confer Sox10-dependent stimulation onto the minimal P0 promoter when artificially placed next to it (d2 P0 in Fig. 4B). Therefore, region −558 to −435 is not sufficient to confer Sox10 responsiveness to the P0 promoter. Additional determinants must be present in the proximal region.
Deletion of an additional 140 bp from positions −435 to −295 did not further reduce the rate of Sox10-dependent stimulation (Fig. 4B). The remaining Sox10-dependent increase of P0 promoter activity was lost only upon gradual removal of sequences from position −295 via −155 to −38. We conclude that Sox10 increases P0 promoter activity through two regions, with positions −295 to −38 mediating the basic effect and −558 to −435 strongly augmenting it.
Sox10-responsive elements in the P0 promoter.
Although suggestive, deletion analysis of the P0 promoter does not prove that the effect of Sox10 on the P0 promoter is direct. To evaluate this possibility, we searched for potential Sox10 binding sites in the P0 promoter, in particular within those two regions shown to mediate the Sox10-dependent promoter stimulation. The published consensus sequences for Sox binding sites consist of a core binding element of 7 bp and allow both adenosine and thymidine at three out of seven positions (11, 28) (Fig. 6B). We identified two sites in the P0 promoter which fully conformed to this consensus, site B (positions −147 to −141) and site F (positions −453 to −459). Interestingly, these sites are localized in the two regions identified as mediating Sox10-dependent stimulation (Fig. 5). In electrophoretic mobility shift assays, both sites were bound by the bacterially expressed HMG domain of Sox10, with site B showing higher affinity than site F (Fig. 5A). Both sites were mutagenized such that the Sox consensus was replaced by a GC-rich element (MUT in Fig. 6B). As a consequence, Sox10 binding was obliterated (Fig. 5A).
These same mutations were also introduced into the context of the rat P0 promoter, and the mutant promoters were analyzed for their responsiveness to Sox10 (Fig. 5B). Mutation of site F reduced Sox10-dependent stimulation from 23-fold to 8-fold, an effect very similar to the one observed after truncation of the P0 promoter to position −435. Thus, the action of site F is sufficient to explain the activity of the more distal of the two Sox10-responsive regions within the P0 promoter.
Mutation of site B also led to a severe decrease in Sox10-dependent stimulation, lowering activation rates from 23-fold to 5-fold (Fig. 5B). Combination of both mutations, however, did not further reduce activation rates (data not shown).
Next we analyzed the consequence of site B mutation in the context of the −435 P0 luciferase construct. In this set of experiments, Sox10-dependent stimulation dropped from 9.5-fold to 4-fold in the absence of a functional site B (Fig. 6C). This corroborates that site B is responsible for a significant fraction of the Sox10 responsiveness. At the same time, it indicates that there must be additional Sox10-responsive elements in the proximal P0 promoter.
Activation of the type II collagen gene in chondrocytes by Sox9, a protein closely related to Sox10, had previously been shown to depend on an enhancer with multiple functional Sox binding sites (2, 49). In addition, these sites only loosely conformed to the consensus DNA binding motif for Sox proteins. To investigate whether the proximal region of the P0 promoter also contains multiple binding sites for Sox10, we performed electrophoretic mobility shift analyses with a DNA fragment encompassing positions −295 to −38 of the P0 promoter (Fig. 6A). Using increasing amounts of a fusion protein between GST and the Sox10 HMG domain, we obtained a ladder of regularly spaced protein-DNA complexes. Any given complex differs from the one with the next-higher mobility by possession of one additional HMG domain molecule. The presence of multiple HMG domain molecules in these complexes could be due to interactions of each HMG domain with the DNA fragment or to protein-protein interactions between the proteins themselves. Because we were unable to detect protein-protein interactions between Sox10 HMG domains in pull-down experiments (data not shown), we favor the hypothesis that the number of complexes is a direct indicator of the number of independent Sox10 binding sites within this region. In these experiments, we were able to resolve at least four separate complexes, thus revealing the presence of multiple Sox10 binding sites in this region.
By allowing mismatches, we identified several candidate binding sites for Sox proteins in addition to the already described site B. Electrophoretic mobility shift analysis with these sites revealed that additional sites, termed A, C, D, and E, were bound by the HMG domain of Sox10 (Fig. 6B). Only site C showed an affinity approaching that of site B. Sites A, D and E were significantly weaker binding sites when assayed on their own (Fig. 6B).
Each of these sites was mutated in the context of the −435 P0 luciferase reporter to the same GC-rich element previously employed (MUT in Fig. 6B). The resulting P0 promoter mutants were analyzed for their response to Sox10 (Fig. 6C). Mutation of site A had the least effect, with Sox10-dependent stimulation dropping from 9.5-fold to 8-fold. The strongest reductions were observed for P0 promoters that lacked a functional site B or C. In each of these cases, stimulation decreased from 9.5-fold to 4-fold. Mutations of sites D and E were of intermediate consequence, with activation rates dropping from 9.5-fold to 5- to 5.5-fold. Combination of site B and site C mutations curtailed Sox10 responsiveness to levels comparable to those obtained with the minimal promoter. Combination of site B mutations with mutations of any other site, on the other hand, did not lead to a reduction in Sox10 responsiveness significantly below the level observed for site B mutation alone (data not shown). The proximal Sox10-responsive region therefore contains multiple Sox10 binding sites as its functional elements, with sites B and C being the most important ones. Corroborating the importance of these sites, a fragment corresponding to positions −229 to −116 of the P0 promoter and encompassing sites B and C was sufficient to confer Sox10 responsiveness to a luciferase reporter under the control of the β-globin minimal promoter (Fig. 6D).
To analyze whether Sox10 binds cooperatively to sites B and C, we performed electrophoretic mobility shift assays with DNA fragments corresponding to positions −229 to −116 of the P0 promoter. These fragments contained either wild-type or mutant versions of sites B and C in all possible combinations (Fig. 7A). Using recombinant protein corresponding to amino acids 1 to 189 of Sox10, we obtained three complexes of different mobilities with a probe that contained wild-type sequences for both site B and site C (wt in Fig. 7B). This indicates that there are three Sox10 molecules bound to the probe instead of the expected two. When we used a probe that carried a mutation in site B, the complex with the lowest mobility was lost and the strength of the complex with the highest mobility was significantly reduced. Only the complex with intermediate mobility remained almost unaltered (mutB in Fig. 7B). When we used a probe with mutation in site C, the complex with the lowest mobility was again lost. Now there was a strong reduction of the complex with the intermediate mobility, and the high-mobility complex remained unaffected (mutC in Fig. 7B). When both sites were mutated, binding was abolished completely (mutB+C in Fig. 7B). Together, these results indicate that site B binds a single Sox10 molecule, whereas site C is bound by two Sox10 molecules. Thus, while we failed to detect cooperativity between sites B and C, there was cooperative binding at site C.
We had previously noticed a potential Sox10 binding site with one mismatch in close proximity to site C. This site (C′ in Fig. 7A), however, failed to bind Sox10 by itself (data not shown). When we mutated C′ in the context of an oligonucleotide probe that contained both C and C′, we observed an increase in the mobility of the Sox10-DNA complex, indicating that only one Sox10 molecule was bound to this probe (C/mutC′ in Fig. 7B), whereas two molecules are bound to the wild type (C/C′ in Fig. 7B). When site C was mutated in the context of this probe, binding was almost completely abolished (mutC/C′ in Fig. 7B). Thus, C′ can be occupied at significant levels only in the presence of site C, as expected in case of cooperativity.
P0 expression in Sox10 mutant mice.
Our experiments show that in the N2A Tet-On system, Sox10 regulates expression of the P0 gene by directly binding to its promoter. To obtain in vivo evidence for these tissue culture findings, we used Dom mice. These mice express a truncated and functionally inactive version of the Sox10 gene (13, 46). Homozygous Dom embryos exhibit severe defects early in development. On average they die at E13.5 for unknown reasons. Therefore, we concentrated our analyses on E12.5 embryos.
First we analyzed expression of P0 and Sox10 in total embryos by semiquantitative RT-PCR (Fig. 8A). In the heterozygotes, there was a dramatic reduction of P0 expression. This decrease was even more pronounced in homozygous Dom embryos, with P0 transcripts being almost undetectable. These RT-PCR data were independently confirmed by RNase protection analyses (Fig. 8B). A parallel analysis of Sox10 expression revealed that message levels in heterozygotes and wild-type embryos were comparable. This was expected, as inactivation of Sox10 in the Dom mouse does not lead to a loss of Sox10 transcripts (13). However, we detected a significant reduction of Sox10 expression in the homozygous embryos, indicating that at this age there is already a significant loss of Sox10-expressing cells throughout the embryo. Therefore, it is impossible to distinguish by this assay whether the strong decrease of P0 expression in homozygous Dom embryos is due to a loss of P0-expressing cells, to a loss of P0 expression within still existing cells, or to a combination of both. The reduced P0 expression in heterozygotes, however, is clearly not due to significant cell loss, but rather points to a direct involvement of Sox10 in the regulation of P0 expression.
FIG. 8.
Reduction of P0 expression is observed in Dom embryos at E12.5. (A) RT-PCR analysis on cDNA obtained from wildtype (+/+), heterozygous (Dom/+), and homozygous (Dom/Dom) embryos at E12.5. P0 transcript levels were compared in the various genotypes by semiquantitative PCR using increasing numbers of amplification cycles. PCRs with specific primers for GAPDH and Sox10 were performed to monitor the experiment. −RT, PCR on all three genotypes without prior reverse transcription of RNA. Only the results obtained with the highest number of cycles are shown. (B) RNase protection analysis on RNA from wild-type (+/+), heterozygous (Dom/+), and homozygous (Dom/Dom) embryos at E12.5. Yeast RNA (yRNA) served as control. Antisense RNA probes for P0 and β-actin (open arrowheads) and protected fragments (filled arrowheads) are marked.
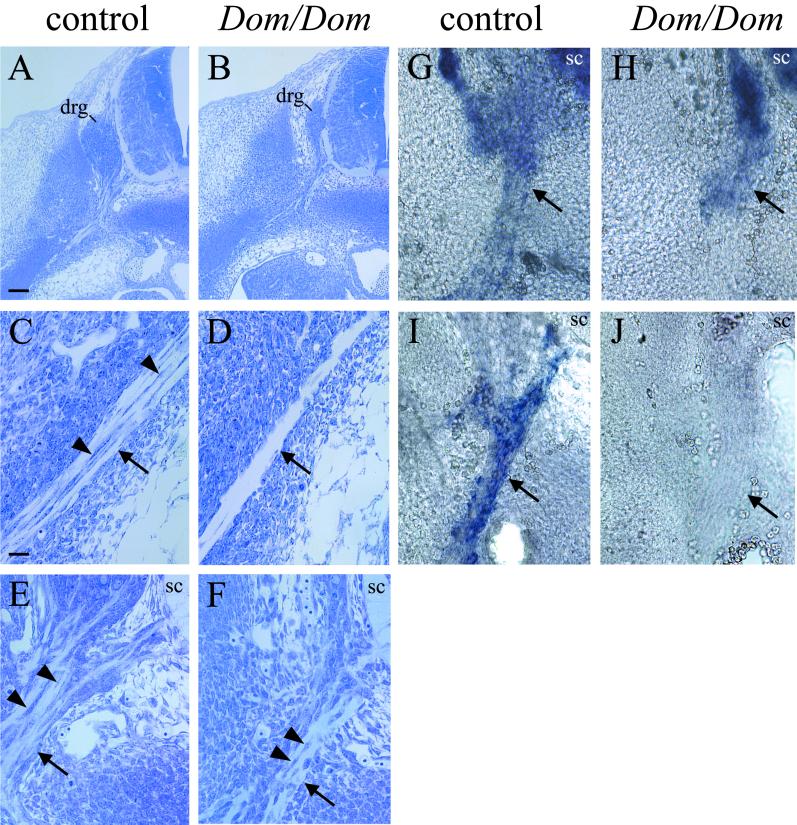
To determine the underlying cause of decreased P0 expression in homozygous Dom mice, we next performed histology and in situ hybridization on E12.5 embryos. Histological analysis of homozygous Dom embryos demonstrated a severe reduction in the size of dorsal root ganglia (Fig. 9A and B). Furthermore, most peripheral nerves appeared thinned and lacked Schwann cell precursors (Fig. 9C and D). This defect in Schwann cell development will be described elsewhere in greater detail (D.E.G. and M.W., unpublished data). Nevertheless, it is clear that the loss of Schwann cell precursors contributes strongly to the decrease in P0 expression observed in homozygous Dom embryos by RT-PCR.
FIG. 9.
P0 is not expressed in spinal nerves of homozygous Dom embryos at E12.5. (A to F) Histology of control (A, C, E) and Dom/Dom (B, D, F) embryos on E 12.5. (C and D) Distal parts of the spinal nerve that are not visible in the overview (A and B). (E and F) Proximal spinal nerves. In situ analysis of vibratome sections shows the proximal spinal nerves of control (G and I) and Dom/Dom (H and J) E12.5 embryos with probes specific for cadherin-6 (G and H) and P0 (I and J). Dorsal root ganglia (drg) and spinal cord (sc) are indicated; arrows point toward the spinal nerves, and arrowheads point toward Schwann cell precursors. Magnifications in A and B and in C to H are identical; scale bars are 100 and 25 μm, respectively.
However, in the vicinity of the dorsal root ganglia and the ventral root, spinal nerves still contain significant numbers of neural crest-derived cells associated with the nerve fibers (Fig. 9E). Although these cells show a slightly altered morphology compared to control cells (Fig. 9F), their location and Sox10 expression identify them as Schwann cell precursors (Fig. 9E and data not shown). In agreement, these cells, like their wild-type counterparts were found by in situ hybridization to express cadherin-6 (Fig. 9G and H). At E12.5, cadherin-6 expression is largely restricted to motor neurons within and Schwann cell precursors outside the central nervous system and thus represents one of the few available markers for cells of the Schwann cell lineage at this time of development (15).
When spinal nerves of wild-type mouse embryos were analyzed by in situ hybridization with a P0 probe, P0 expression was readily observed in cells lining these nerves (Fig. 9I). In contrast, nerves of homozygous Dom embryos were completely devoid of P0 even in the region close to dorsal root ganglia and ventral roots where cadherin-6-positive cells were still present (Fig. 9J). Reduced RT-PCR detection of P0 in homozygous embryos is, therefore, also due to loss of P0 expression in existing glial progenitor cells. This result shows that Sox10 is needed in these cells for P0 expression.
DISCUSSION
In this study we have examined several transcription factors which are preferentially expressed in cells of the Schwann cell lineage for the ability to regulate P0 expression using a Tet-On cell culture system in which expression of these transcription factors could be induced. We had to choose a cell line with low or missing expression of the transcription factors to be analyzed. In addition, we assumed that a cell of neural origin would provide a better environment for the function of these transcription factors than a nonneural cell. N2A neuroblastoma fulfilled these criteria. However, it has to be kept in mind that N2A cells do not express glial genes. This prevents a priori the detection of inhibitory effects on glial target genes. Thus, our system is unable to address the previously reported repression of P0 gene expression by Tst-1/Oct6/SCIP (32). In addition, it cannot be excluded that N2A cells lack some glia-specific transcription factors or coactivators that are needed for the function of Sox10, Krox-20, or Tst-1/Oct6/SCIP in myelinating glia. As a consequence, it might be possible to detect only a subset of effector-target gene relationships.
Despite these limitations, we were able to show Sox10-dependent induction of endogenous P0 expression in these cells. This effect was not observed for Krox-20, for Tst-1/Oct6/SCIP, or for naturally occurring Sox10 mutants previously shown to be functionally inactive (13, 20). Sox11 induced only low levels of endogenous P0 expression, despite the fact that Sox11 behaves as a very strong transcriptional activator of minimal promoters with Sox binding sites (18). The stimulatory effect on P0 gene expression is therefore specific for Sox10.
Sox10-dependent stimulation was also observed for a luciferase reporter plasmid under control of the P0 promoter. Within the P0 promoter, Sox10 responsiveness was mapped to two regions, each of which contained binding sites for Sox proteins. These binding sites were directly involved in mediating the stimulatory activity of Sox10 as indicated by mutagenesis. Taken together, these results strongly suggest that the observed stimulation of P0 gene expression in the N2A system results from direct activation of the P0 promoter by bound Sox10 molecules.
P0 expression is confined to cells of the Schwann cell lineage. Given the findings that Sox10 is the major Sox protein in Schwann cells (19) and that Sox10-dependent activation of P0 gene expression is conferred by the same regions previously shown to contain all necessary determinants for Schwann cell-specific expression both in vivo (29) and in vitro (6, 27), it seems likely that Sox10 is involved in controlling P0 gene expression in vivo in cells of the Schwann cell lineage.
This conclusion is strongly supported by experimental evidence. During embryogenesis, P0 is expressed at low levels first in a subpopulation of neural crest cells which give rise to peripheral glia and later in cells of the Schwann cell lineage (10, 21). These cells are also positive for Sox10 (19). At E12.5, they are already strongly reduced in mouse embryos homozygous for the Dom mutation of Sox10, indicating that Sox10 expression is essential for early Schwann cell development. However, residual Schwann cell precursors are still present in these embryos in the vicinity of dorsal root ganglia and ventral roots. These cells still express the glial marker cadherin-6 but fail to express P0, whereas their counterparts in control embryos are positive for both. This proves that P0 expression in these cells depends on the presence of functional Sox10 protein. A second evidence for the in vivo regulation of P0 expression by Sox10 is provided by the observation that P0 expression is strongly decreased in heterozygous Dom mice despite the fact that there is no detectable reduction in the number of Schwann cell precursors. At this developmental stage, a single wild-type Sox10 allele seems insufficient for maintaining normal levels of P0 expression.
Expression patterns of Sox10 and P0 are not completely congruous. There are a number of cells which express Sox10 but do not contain detectable levels of P0. These include early neural crest cell populations, melanoblasts, cells of the oligodendrocyte lineage, as well as nonmyelinating Schwann cells (19, 46). Thus, while there is good evidence for a role of Sox10 in the regulation of P0 expression, it is unlikely that Sox10 alone is responsible for this effect. It is much more likely that Sox10 has to cooperate with other factors. This is a reasonable assumption, as Sox proteins generally tend to exert their numerous functions in conjunction with other proteins (for reviews, see references 35 and 47) and as Sox10 has been shown on synthetic promoter constructs to be a weak transcriptional activator on its own but a strong activator in combination with other transcription factors such as Tst-1/Oct6/SCIP and Pax-3 (19, 20). This feature has been attributed to a proposed role of Sox proteins in determining higher-order structure of protein-DNA complexes by both bending DNA and providing interfaces for protein-protein interactions (37). According to this hypothesis, cells which express P0 should contain a factor that is capable of synergistic interaction with Sox10 and that is absent from those Sox10 positive cells which do not express P0.
One of the proteins that would have been a good candidate for a Sox10 cooperation partner with regard to regulation of P0 gene expression is Krox-20. In the Schwann cell lineage, Krox-20 expression starts after transition from Schwann cell precursors to embryonic Schwann cells and from late embryogenesis onward is restricted to myelinating Schwann cells (33, 51). Thus, Krox-20 occurrence parallels P0 expression in the Schwann cell lineage from the embryonic Schwann cell stage onward. In addition, both Krox-20 and P0 are absent from oligodendrocytes, which are a main site of Sox10 expression in the adult. Finally, there is a GC-rich element in the P0 promoter that could potentially function as a binding site for Krox-20 (6).
However, previous protein-DNA interaction studies on Schwann cell nuclear extracts failed to reveal Krox-20 binding. Instead, Sp1 binding was detected (6). In agreement, we were unable to obtain a significant increase of endogenous P0 expression or P0 promoter activity in N2A cells upon induction of Krox-20. Importantly, stable N2A clones that coexpressed Sox10 and Krox-20 in an inducible manner failed to show stimulation of endogenous P0 expression and P0 promoter activity above levels obtained for Sox10 alone (data not shown). Thus, we have no evidence implicating Krox-20 in the regulation of P0 expression either alone or in conjunction with Sox10. Recently, Zorick et al. (52) reported a threefold increase in expression of a luciferase reporter under the control of the rat P0 promoter by Krox-20 in Schwann cells. In light of all available data, this result is best explained as an indirect effect, with Krox-20 activating other transcription factors that in turn stimulate P0 expression.
The proposed role of Sox10 as both an accessory and an architectural factor in P0 gene expression is also compatible with its binding to multiple sites within the proximal P0 promoter. Given the ability of Sox10 to bend DNA significantly (R. I. Peirano and M. Wegner, unpublished data), the combined interaction of Sox10 with all identified sites could lead to dramatic changes in promoter topology. Two of these sites (B and C) have high affinity for Sox10, and their mutation has the strongest influence on the ability of Sox10 to activate the P0 proximal promoter. Site B exactly matches the Sox consensus site, whereas site C does not. Nevertheless, site C displays high affinity because it allows cooperative binding of two Sox10 molecules. It is likely that sites B and C will be occupied first. Additional binding of the other sites might vary during different phases of Schwann cell development and might be a means of regulating promoter activity by altering promoter topology and allowing different sets of transcription factors to bind.
Sox proteins are known to interact functionally with other transcription factors (for a review, see reference 47). This might also be important in the context of the P0 promoter. That other proteins do indeed bind to the P0 promoter has become evident in DNase footprint analysis with Schwann cell extracts (6). These experiments were carried out with the promoter-proximal region because this region had been found to confer most of the Schwann cell-specific activity of the P0 promoter. This is the same region that also contains most of the Sox10 binding sites identified in this study. With the exception of site A, Sox10 binding sites do not overlap with the previously mapped protein interaction sites. Instead they are interspersed. While contradictory at first glance, these results fit together well. The use of dI-dC as competitor in the DNase footprinting experiments (6) very likely prevented detection of Sox binding sites, because dI-dC is a very efficient competitor for proteins binding to the minor grove of AT-rich DNA such as Sox proteins (44). Thus, both studies taken together would have to be interpreted in such a way that Sox10 binds between sites occupied by other proteins, possibly altering overall promoter topology by DNA bending and by allowing additional protein-protein interactions.
Binding and activation characteristics of Sox10 on the P0 promoter are also reminiscent of Sox9 function on the type-II collagen gene (col2a1). Col2a1 is the major extracellular matrix component of cartilage, and Sox9 is very strongly expressed in the cartilage forming chondrocytes. It has been shown that chondrocyte-specific expression of the col2a1 gene is mediated by a chondrocyte-specific enhancer present in intron 1 of the gene. This enhancer contains as functional elements multiple binding sites for Sox9, just as the P0 promoter contains multiple Sox10 binding sites (2, 22, 34). Mutation of these binding sites severely disturbed the chondrocyte-specific activity of this enhancer in vitro and in vivo. Additionally, expression of a Sox9 transgene under the control of the Hoxb2 promoter induced ectopic expression of both the endogenous col2a1 gene and a col2a1-lacZ transgene, clearly proving that chondrocyte-specific function of the enhancer required Sox9 (2). However, col2a1 expression was not observed in all regions and cells that exhibited ectopic Sox9 expression, just as type II collagen is not produced by all cells which endogenously express Sox9 during development, thus, providing further analogy to the situation described in this study for Sox10 and P0.
Recent results on the col2a1 enhancer have also shown that Sox9 functions in concert with Sox6 and L-Sox5, two other Sox proteins only distantly related to Sox9 (23). It will be interesting to determine whether such a cooperation with other Sox proteins can also be detected for Sox10 on the P0 promoter. Sox6 and L-Sox-5 are, however, unlikely to be these proteins, as neither of them is expressed at significant levels in Schwann cells (Peirano and Wegner, unpublished data).
Regulation of P0 expression by Sox10 might also help to explain the pathophysiology of Sox10 mutations. Heterozygous Sox10 mutations in humans primarily result in Waardenburg-Hirschsprung syndrome (5, 36, 45). Recently, however, a patient with a novel heterozygous Sox10 mutation that leads to a carboxy-terminal 82-amino-acid extension of the open reading frame has been described. This patient not only exhibited the classical symptoms of Waardenburg-Hirschsprung but in addition showed myelin abnormalities in both central and peripheral nervous systems which are characteristic of Pelizaeus-Merzbacher disease and Charcot-Marie-Tooth disease type 1, respectively (14); the latter is usually caused by mutations in the genes coding for structural myelin proteins such as connexin-32, PMP-22, and P0 (for a review, see reference 17). Similar phenotypic manifestations of Sox10 and P0 mutations may be explained by the fact that P0 expression is under the control of Sox10. Sox10 and P0 mutation would then both lead to comparable loss of functional P0 protein.
In conclusion, we have shown both in vivo and in a tissue culture model that Sox10 regulates P0 expression by directly acting on its promoter. This finding has important implications for our understanding of Schwann cell biology and human disease.
ACKNOWLEDGMENTS
This work was supported by grant We1326/7-1 from the Deutsche Forschungsgemeinschaft to M.W.
E.-M. Mandelkow and M. Goossens are acknowledged for the gift of the N2A neuroblastoma expressing the rtTA transactivator and Dom mice, respectively.
REFERENCES
- 1.Baron P, Shy M, Honda H, Sessa M, Kamholz J, Pleasure D. Developmental expression of P0 mRNA and P0 protein in the sciatic nerve and the spinal nerve roots of the rat. J Neurocytol. 1994;23:249–257. doi: 10.1007/BF01275529. [DOI] [PubMed] [Google Scholar]
- 2.Bell D M, Leung K K H, Wheatley S C, Ng L J, Zhou S, Ling K W, Sham M H, Koopman P, Tam P P L, Cheah K S E. Sox9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard A D, Sinanan A, Parmantier E, Zwart R, Broos L, Meijer D, Meier C, Jessen K R, Mirsky R. Oct-6 (SCIP/Tst-1) is expressed in Schwann cell precursors, embryonic Schwann cells, and postnatal myelinating Schwann cells: comparison with Oct-1, Krox-20, and Pax-3. J Neurosci Res. 1996;46:630–640. doi: 10.1002/(SICI)1097-4547(19961201)46:5<630::AID-JNR11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 4.Bondurand N, Kobetz A, Pingault V, Lemort N, Encha-Razavi F, Couly G, Goerich D E, Wegner M, Abitbol M, Goossens M. Expression of the SOX10 gene during human development. FEBS Lett. 1998;432:168–172. doi: 10.1016/s0014-5793(98)00843-6. [DOI] [PubMed] [Google Scholar]
- 5.Bondurand N, Kuhlbrodt K, Pingault V, Enderich J, Sajus M, Tommerup N, Warburg M, Hennekam R C, Read A P, Wegner M, Goossens M. A molecular analysis of the Yemenite deaf-blind hypopigmentation syndrome: SOX10 dysfunction causes different neurocristopathies. Hum Mol Genet. 1999;8:1785–1789. doi: 10.1093/hmg/8.9.1785. [DOI] [PubMed] [Google Scholar]
- 6.Brown A M, Lemke G. Multiple regulatory elements control transcription of the peripheral myelin protein zero gene. J Biol Chem. 1997;272:28939–28947. doi: 10.1074/jbc.272.46.28939. [DOI] [PubMed] [Google Scholar]
- 7.Giese K P, Martini R, Lemke G, Soriano P, Schachner M. Mouse P0 gene disruption leads to hypomyelination, abnormal expression of recognition molecules, and degeneration of myelin and axons. Cell. 1992;71:565–576. doi: 10.1016/0092-8674(92)90591-y. [DOI] [PubMed] [Google Scholar]
- 8.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 10.Hagedorn L, Suter U, Sommer L. P0 and PMP22 mark a multipotent neural crest-derived cell type that displays community effects in response to TGF-(beta) family factors. Development. 1999;126:3781–3794. doi: 10.1242/dev.126.17.3781. [DOI] [PubMed] [Google Scholar]
- 11.Harley V R, Lovell-Badge R, Goodfellow P N. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, Gerrero R, Simmons D M, Park R E, R. L C, Swanson L W, Rosenfeld M G. Tst-1, a member of the POU domain gene family, binds the promoter of the gene encoding the cell surface adhesion molecule Po. Mol Cell Biol. 1991;11:1739–1744. doi: 10.1128/mcb.11.3.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbarth B, Pingault V, Bondurand N, Kuhlbrodt K, Hermans-Borgmeyer I, Puliti A, Lemort N, Goossens M, Wegner M. Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc Natl Acad Sci USA. 1998;95:5161–5165. doi: 10.1073/pnas.95.9.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue K, Tanabe Y, Lupski J R. Myelin deficiencies in both the central and peripheral nervous system associated with a SOX10 mutation. Ann Neurol. 1999;46:313–318. doi: 10.1002/1531-8249(199909)46:3<313::aid-ana6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Inoue T, Chisaka O, Matsunami H, Takeichi M. Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Dev Biol. 1997;183:183–194. doi: 10.1006/dbio.1996.8501. [DOI] [PubMed] [Google Scholar]
- 16.Kamholz J, Sessa M, Scherer S, Vogelbacker H, Mokuno K, Baron P, Wrabetz L, Shy M, Pleasure D. Structure and expression of proteolipid protein in the peripheral nervous system. J Neurosci Res. 1992;31:231–244. doi: 10.1002/jnr.490310204. [DOI] [PubMed] [Google Scholar]
- 17.Keller M P, Chance P F. Inherited neuropathies: from gene to disease. Brain Pathol. 1999;9:327–341. doi: 10.1111/j.1750-3639.1999.tb00230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and Sox proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 19.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhlbrodt K, Schmidt C, Sock E, Pingault V, Bondurand N, Goossens M, Wegner M. Functional analysis of Sox10 mutations found in human Waardenburg-Hirschsprung patients. J Biol Chem. 1998;273:23033–23038. doi: 10.1074/jbc.273.36.23033. [DOI] [PubMed] [Google Scholar]
- 21.Lee M-J, Brennan A, Blanchard A, Zoidl G, Dong Z, Tabernero A, Zoidl C, Dent M A R, Jessen K R, Mirsky R. P0 is constitutively expressed in the rat neural crest and embryonic nerves and is negatively and positively regulated by axons to generate non-myelin-forming and myelin-forming Schwann cells, respectively. Mol Cell Neurosci. 1997;8:336–350. doi: 10.1006/mcne.1996.0589. [DOI] [PubMed] [Google Scholar]
- 22.Lefebvre V, Huang W, Harley V R, Goodfellow P N, DeCrombrugghe B. Sox9 is a potent activator of the chondrocyte-specific enhancer of the proα1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemke G. The molecular genetics of myelination: an update. Glia. 1993;7:263–271. doi: 10.1002/glia.440070402. [DOI] [PubMed] [Google Scholar]
- 25.Lemke G. Unwrapping the genes of myelin. Neuron. 1988;1:535–543. doi: 10.1016/0896-6273(88)90103-1. [DOI] [PubMed] [Google Scholar]
- 26.Lemke G, Axel R. Isolation and sequence of a cDNA encoding the major structural protein of peripheral myelin. Cell. 1985;40:501–508. doi: 10.1016/0092-8674(85)90198-9. [DOI] [PubMed] [Google Scholar]
- 27.Lemke G, Lamar E, Patterson J. Isolation and analysis of the gene encoding peripheral myelin protein zero. Neuron. 1988;1:73–83. doi: 10.1016/0896-6273(88)90211-5. [DOI] [PubMed] [Google Scholar]
- 28.Mertin S, McDowall S G, Harley V R. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messing A, Behringer R R, Hammang J P, Palmiter R D, Brinster R L, Lemke G. P0 promoter directs expression of reporter and toxin genes to Schwann cells of transgenic mice. Neuron. 1992;8:507–520. doi: 10.1016/0896-6273(92)90279-m. [DOI] [PubMed] [Google Scholar]
- 30.Messing A, Behringer R R, Wrabetz L, Hammang J P, Lemke G, Palmiter R D, Brinster R L. Hypomyelinating peripheral neuropathies and schwannomas in transgenic mice expressing SV40 T-antigen. J Neurosci. 1994;14:3533–3539. doi: 10.1523/JNEUROSCI.14-06-03533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirsky R, Jessen K R. The neurobiology of Schwann cells. Brain Pathol. 1999;9:293–311. doi: 10.1111/j.1750-3639.1999.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monuki E S, Kuhn R, Weinmaster G, Trapp B, Lemke G. Expression and activity of the POU transcription factor SCIP. Science. 1990;249:1300–1303. doi: 10.1126/science.1975954. [DOI] [PubMed] [Google Scholar]
- 33.Murphy P, Topilko P, Schneider-Maunoury S, Seitanidou T, Baron-van Evercooren A, Charnay P. The regulation of Krox-20 expression reveals important steps in the control of peripheral glial cell development. Development. 1996;122:2847–2857. doi: 10.1242/dev.122.9.2847. [DOI] [PubMed] [Google Scholar]
- 34.Ng L-J, Wheatley S, Muscat G E O, Conway-Campbell J, Bowles J, Wright E, Bell D M, Tam P P L, Cheah K S E, Koopman P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 35.Pevny L H, Lovell-Badge R. Sox genes find their feet. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 36.Pingault V, Bondurand N, Kuhlbrodt K, Goerich D E, Prehu M-O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith J C, Read A P, Wegner M, Goossens M. Sox10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 37.Prior H M, Walter M A. Sox genes: architects of development. Mol Med. 1996;2:405–412. [PMC free article] [PubMed] [Google Scholar]
- 38.Pusch C, Hustert E, Pfeifer D, Südbeck P, Kist R, Roe B, Wang Z, Balling R, Blin N, Scherer G. The SOX10/Sox10 gene from human and mouse: sequence, expression, and transactivation by the encoded HMG domain transcription factor. Hum Genet. 1998;103:115–123. doi: 10.1007/s004390050793. [DOI] [PubMed] [Google Scholar]
- 39.Renner K, Leger H, Wegner M. The POU-domain protein Tst-1 and papovaviral T-antigen function synergistically to stimulate glia-specific gene expression of JC virus. Proc Natl Acad Sci USA. 1994;91:6433–6437. doi: 10.1073/pnas.91.14.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherer S. The biology and pathobiology of Schwann cells. Curr Opin Neurol. 1997;10:386–397. doi: 10.1097/00019052-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber J, Sock E, Wegner M. The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain. Proc Natl Acad Sci USA. 1997;94:4739–4744. doi: 10.1073/pnas.94.9.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro L, Doyle J P, Hensley P, Colman D R, Hendrickson W A. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17:435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 43.Sock E, Enderich J, Rosenfeld M G, Wegner M. Identification of the nuclear localization signal of the POU domain protein Tst-1/Oct6. J Biol Chem. 1996;271:17512–17518. doi: 10.1074/jbc.271.29.17512. [DOI] [PubMed] [Google Scholar]
- 44.Solomon M J, Strauss F, Varshavsky A. A mammalian high mobility group protein recognizes any stretch of six A.T base pairs in duplex DNA. Proc Natl Acad Sci USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southard-Smith E M, Angrist M, Ellison J S, Agarwala R, Baxevanis A D, Chakravarti A, Pavan W J. The Sox10(Dom) mouse: modeling the genetic variation of Waardenburg-Shah (WS4) syndrome. Genome Res. 1999;9:215–225. [PubMed] [Google Scholar]
- 46.Southard-Smith E M, Kos L, Pavan W J. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 47.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wegner M, Drolet D W, Rosenfeld M G. Regulation of JC virus by the POU-domain transcription factor Tst-1, implications for progressive multifocal leukoencephalopathy. Proc Natl Acad Sci USA. 1993;90:4743–4747. doi: 10.1073/pnas.90.10.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou G, Lefebvre V, Zhang Z, Eberspaecher H, de Crombrugghe B. Three high-mobility group-like sequences within a 48-base pair enhancer of the Col2a1 gene are required for cartilage-specific expression in vivo. J Biol Chem. 1998;273:14989–14997. doi: 10.1074/jbc.273.24.14989. [DOI] [PubMed] [Google Scholar]
- 50.Zorick T S, Lemke G. Schwann cell differentiation. Curr Opin Cell Biol. 1996;8:870–876. doi: 10.1016/s0955-0674(96)80090-1. [DOI] [PubMed] [Google Scholar]
- 51.Zorick T S, Syroid D E, Arroyo E, Scherer S S, Lemke G. The transcription factors SCIP and Krox-20 mark distinct stages and cell fates in Schwann cell differentiation. Mol Cell Neurosci. 1996;8:129–145. doi: 10.1006/mcne.1996.0052. [DOI] [PubMed] [Google Scholar]
- 52.Zorick T S, Syroid D E, Brown A, Gridley T, Lemke G. Krox-20 controls SCIP expression, cell cycle exit and susceptibility to apoptosis in developing myelinating Schwann cells. Development. 1999;126:1397–1406. doi: 10.1242/dev.126.7.1397. [DOI] [PubMed] [Google Scholar]