Abstract
The p53 tumor suppressor is activated by many diverse stress signals through mechanisms that result in stabilization and accumulation of the p53 protein. p53 is normally degraded through the proteasome following interaction with MDM2, which both functions as a ubiquitin ligase for p53 and shuttles to the cytoplasm, where p53 degradation occurs. Stabilization of p53 in response to stress is associated with inhibition of MDM2-mediated degradation, which has been associated with phosphorylation of p53 in response to DNA damage or activation of ARF. In this study we show distinct responses, as measured by phosphorylation, transcriptional activity, and subcellular localization, of p53 stabilized by different activating signals. Although normal cells and wild-type p53-expressing tumor cells showed similar responses to actinomycin D and camptothecin treatment, the transcriptional activity of stabilized p53 induced by deferoxamine mesylate, which mimics hypoxia, in normal cells was lost in all three tumor cell lines tested. Our results show that multiple pathways exist to stabilize p53 in response to different forms of stress, and they may involve down-regulation of MDM2 expression or regulation of the subcellular localization of p53 or MDM2. Loss of any one of these pathways may predispose cells to malignant transformation, although reactivation of p53 might be achieved through alternative pathways that remain functional in these tumor cells.
The p53 tumor suppressor protein functions to protect cells from malignant transformation, and the development of most tumors is associated with loss of p53 function (31). p53 has been shown to participate in the regulation of several processes which might inhibit tumor growth, including differentiation, senescence, and angiogenesis. However, central to the function of p53 appears to be the ability to induce both cell cycle arrest and apoptosis in stressed cells, at least in part by activating expression of p53-responsive target genes that mediate these responses (8).
p53 protein levels are usually maintained at low levels by rapid degradation through ubiquitin-dependent proteolysis (29), and p53 function is not essential for normal growth and development (14). Degradation of p53 is regulated by interaction with the MDM2 protein (19, 28), which both functions as a ubiquitin ligase (20) and shuttles from the nucleus to the cytoplasm, where degradation of p53 is thought to take place (30, 38, 48). MDM2 is itself transcriptionally regulated by p53 (6, 55), establishing a negative feedback loop where increased levels of p53 increase expression of MDM2, which targets p53 for degradation. The importance of regulation of p53 by MDM2 during normal development is dramatically illustrated by the complete rescue of the early embryonic lethality of MDM2-deficient mice by simultaneous deletion of p53 (24, 35).
Activation of p53 in response to potentially oncogenic signals, such as deregulated growth or DNA damage, depends to a large extent on the stabilization of the p53 protein, which rapidly accumulates in stressed cells. Stabilization of p53 is likely to reflect mechanisms that allow p53 to become resistant to MDM2-mediated degradation. Recent studies have shown that some DNA-damaging agents induce site-specific phosphorylation within the N terminus of p53, specifically at residues 15, 20, 33, 37, and 46 (5, 12, 13, 39, 44, 45), and this phosphorylation correlates well with stabilization of the p53 protein. Phosphorylation at serines 15 and 37 or at serine 20 was shown to reduce the interaction between p53 and MDM2 in vitro (43, 51), and replacement of both serines 15 and 37 with aspartic acid partially protected p53 from degradation by MDM2 (3). Taken together, these observations have led to the hypothesis that N-terminal phosphorylation at sites in or around the MDM2 binding region of p53 may regulate p53 stability. The most likely candidate kinases for phosphorylation of serine 15 are ATM and ATR (5, 13, 50), which both phosphorylate p53 at serine 15 in vitro. Inhibition of ATR function in cells leads to a reduction in serine 15 phosphorylation in response to ionizing radiation (IR) and UV radiation (50), and although loss of ATM substantially delays the stabilization of p53 in response to IR, phosphorylation of serine 15 is still detected in these cells (45). Assessing the importance of the phosphorylation events in a physiological context has proven rather difficult, and the observation that p53 proteins mutated in most known phosphorylation sites, including serines 15, 20, 33, and 37, can be stabilized in response to some DNA-damaging agents (3, 10) suggests the existence of phosphorylation-independent pathways leading to the stabilization of p53. More recently, a second mechanism for the stabilization of p53 has been described in the ARF protein (p14ARF in humans; p19ARF in mice) (42), which is activated in response to abnormal proliferative signals mediated by oncogene activation (36, 58) or aberrant E2F1 activity (7). ARF was shown to bind MDM2 and directly inhibit MDM2 activity without preventing the MDM2-p53 interaction (21, 25, 37, 47) or inducing phosphorylation of p53 (16). This showed that modifications of p53 that prevent binding to MDM2, by phosphorylation or other mechanisms, are not necessary for the stabilization of p53. Recent studies have shown that ARF relocalizes MDM2 to the nucleolus and that this activity is necessary for the inhibition of p53 degradation (49, 53). These results suggest that the separation of p53 and MDM2 to different subnuclear locations can also contribute to the inhibition of MDM2-mediated degradation of p53.
In this study, we have investigated the mechanisms underlying the stabilization of p53 in response to different cellular stresses. We have identified several mechanisms by which p53 could be stabilized, including specific down-regulation of MDM2 transcription and regulation of subcellular distribution of p53 and MDM2. Our results therefore suggest that each stress response utilizes a different pathway to induce a p53 response.
MATERIALS AND METHODS
Cell culture.
Four wild-type p53-expressing cell lines were used, the normal human fibrobast line MRC-5 and three tumor lines: the breast carcinoma cell line MCF-7, the colon cancer cell line RKO, and the osteosarcoma cell line U2OS. The cells were maintained in RPMI 1640 medium (MRC-5 cells) or Dulbecco modified Eagle medium (tumor cell lines) supplemented with 10% fetal calf serum at 37°C in an atmosphere of 10% CO2 in air.
Induction of p53.
All cell types were plated at a density of 106 per 10-cm-diameter dish 24 h prior to treatment, with either actinomycin D (5 nM), deferoxamine mesylate (DFX; Sigma; 250 or 500 μM), camptothecin (CPT; 2 μM), or LLnL (calpain inhibitor I; Boehringer Mannheim; 10 μM).
Protein analysis.
To assay for p53 Ser15 or Ser20 phosphorylation, cells were washed three times in ice-cold phosphate-buffered saline (PBS) and lysed in 500 μl (per 10-cm-diameter dish) of NP-40 lysis buffer (100 mM NaCl, 100 mM Tris [pH 8.0], 1% NP-40) supplemented with proteasome inhibitors (Complete inhibitor mix; Boerhinger Mannheim) for 30 min at 4°C. p53 protein was immunoprecipitated overnight at 4°C by incubation with a 1:1 mixture of PAb1801-protein A and PAb421–protein A-Sepharose beads equilibrated in NP-40 lysis buffer. The immunoprecipitated protein was washed three times with NP-40 lysis buffer, and samples were resuspended in 50 μl of 2× sodium dodecyl sulfate (SDS) sample buffer, incubated at room temperature for 10 min, and then assayed by SDS–10% polyacrylamide gel electrophoresis. Western blots were probed with either anti-p53-P-Ser15 or anti-p53-P-Ser20 which had been preincubated with their respective unphosphorylated peptide, SVEPPLSQETFSD or LSQETFSDLWKLL (1 μg/ml), to reduce any cross-reactivity with p53 as previously described (43, 44). To assess immunoprecipitated p53, the blots were reprobed with the rabbit polyclonal antibody CM1 (34). Additionally, Western blot analysis to assess p53 protein levels in whole-cell extracts was performed in parallel using PAb1801.
To assess for MDM2 or p21Waf1/Cip1 protein levels in response to p53 induction, proteins from whole-cell extracts were separated by SDS–12% polyacrylamide gel electrophoresis as described above and analyzed by Western blotting with anti-human p21Waf1/Cip1 (Oncogene Science) or anti-human MDM2 (SMP14 or Ab-1; Oncogene Science) antibodies.
Northern blot analysis.
To assess MDM2 and p21Waf1/Cip1 mRNA induction, RKO cells (2 × 106) were plated on 15-cm2 dishes 24 h prior to treatment, as described above. The cells were washed three times with PBS and lysed in RNA lysis buffer, and total RNA was isolated by precipitation with sodium acetate, followed by phenol-chloroform extraction. RNA was assessed by Northern blot analysis and hybridized with a [32P]dCTP-labeled full-length human MDM2 cDNA probe or a full-length human p21Waf1/Cip1 cDNA probe. To assess RNA loading, the blots were stripped and then hybridized with a [32P]dCTP-labeled GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA probe (G3PDH; Clontech). The specificities of the probes were confirmed by showing loss of the ability to activate p21Waf1/Cip1 or MDM2 expression in RKO cells expressing E6, which fail to stabilize p53 in response to IR or actinomycin D.
Immunostaining.
MRC-5, MCF-7, or U2OS cells were plated on 10-cm2 dishes (106 cells) containing 1-cm-diameter sterile glass coverslips and treated as described above. The cells were washed three times with PBS and then fixed in ice-cold 4% paraformaldehyde in PBS for 10 min at room temperature. After fixation, the cells were permeabilized in cold PBS containing 0.2% Triton X-100 for 5 min. The cells were blocked in PBS containing 0.5% bovine serum albumin at room temperature for 30 min and then incubated overnight at 4°C with anti-p53 DO1 or CM1 or anti-MDM2 Ab-1 or SMP14 (Oncogene Science) in blocking solution. The cells were washed three times with PBS and incubated for 2 h at room temperature with a rabbit anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody (1:500; DAKO) or donkey anti-rabbit FITC-conjugated antibody (1:500; Amersham) in blocking solution containing 1 μg of DAPI (4′,6′-diamidino-2-phenylindole) (Sigma)/ml. The cells were washed three times with PBS, and slides were mounted with PBS-glycerol mount. For localization of the nucleolus, immunostaining with a goat anti-B23 antibody (Santa Cruz) was performed either in the presence or absence of anti-p53 or anti-MDM2 antibodies as described previously (57).
RESULTS
Phosphorylation of p53 on serine 15 or 20 in response to different activating signals.
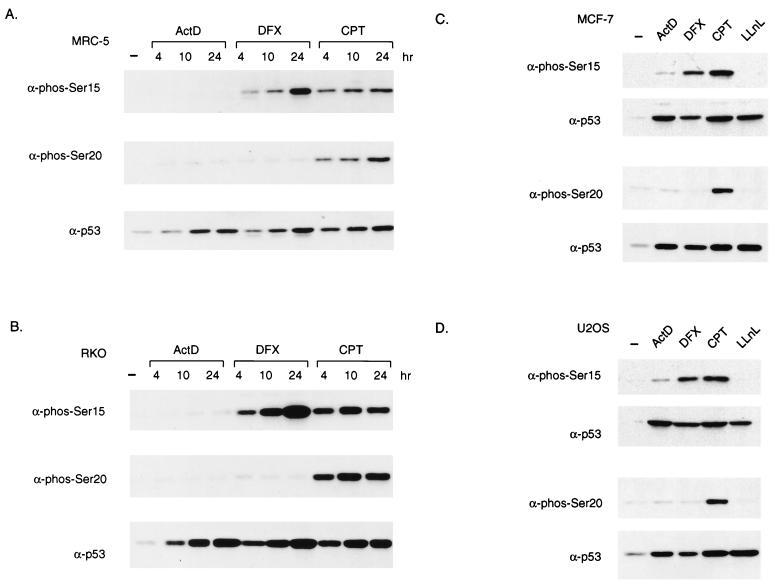
Previous studies have shown that many forms of stress lead to the stabilization of p53, and we have chosen to examine the response to actinomycin D, CPT, and DFX. Each of these p53-activating agents is likely to function through a different mechanism; inhibition of RNA polymerase II by actinomcyin D (32), topoisomerase I inhibition and induction of DNA strand breaks by CPT (22), and activation of hypoxia-inducible factor 1α by DFX (1). p53 was stabilized in response to each treatment with similar kinetics in both normal human fibroblasts (MRC-5 cells) and the colon cancer cell line RKO (Fig. 1A and B, lower blots).
FIG. 1.
p53 is stabilized and differentially phosphorylated in response to different stress signals. (A and B) Western blot analysis of p53 protein immunoprecipitated from MRC-5 (A) or RKO (B) cells harvested at the indicated time points after treatment with actinomycin D (ActD; 5 nM), CPT (2 μM), or DFX (500 or 250 μM, respectively). The blots were probed with phosphoserine 15- or phosphoserine 20-specific antibodies (α-phos-Ser15 or -20) or PAb1801 (α-p53) to detect total p53 levels. (B) Western blot analysis of p53 protein immunoprecipitated from RKO cells harvested at the indicated time points after treatment with actinomycin D (5 nM), CPT (2 μM), or DFX (250 μM). The blots were probed with phosphoserine 15- or phosphoserine 20-specific antibodies or PAb1801 to detect total p53 levels. (C and D) Western blot analysis of Ser15 and Ser20 phosphorylation of immunoprecipitated p53 protein from MCF-7 (C) or U2OS (D) cell harvested 24 h after treatment with actinomycin D (5 nM), CPT (2 μM), DFX (250 μM), or LLnL (10 μM). The blots were probed with antibodies as described for panel A. −, no treatment.
DNA-damaging events that lead to the stabilization of p53 have also been shown to induce phosphorylation of residues within the N terminus of p53. Particular interest has recently focused on serine 15 and serine 20, which are phosphorylated following the exposure of cells to UV light or IR in vivo (44, 45). These residues lie in or close to the MDM2 binding site on p53, and there is evidence that phosphorylation of these sites reduces the binding of p53 to MDM2 and could therefore lead directly to the stabilization of the p53 protein (43, 51). Our previous studies indicated that N-terminal phosphorylation of p53 was not necessary for stabilization in response to actinomycin D (3), and we therefore examined phosphorylation of these sites in response to each treatment using phosphospecific antibodies (43, 44) (Fig. 1A and B). The pattern of phosphorylation was the same in both cell types; treatment with CPT induced phosphorylation at both serine 15 and serine 20, DFX treatment resulted in only serine 15 phosphorylation, and no phosphorylation of either site was detected in response to actinomycin D. No evidence of serine 20 phosphorylation in response to DFX or serine 15 or 20 phosphorylation in response to actinomycin D was seen at any time point, indicating that transient phosphorylation of these sites does not occur. To extend these observations, we analyzed two more wild-type p53-expressing cell lines that are widely used to study p53 activation and function, the breast carcinoma cell line MCF-7 (Fig. 1C) and the osteosarcoma cell line U2OS (Fig. 1D). Identical results were obtained using these lines, in which each treatment stabilized p53 efficiently. Only CPT treatment induced phosphorylation of both serines 15 and 20; DFX treatment resulted in phosphorylation of serine 15 but not serine 20, and actinomycin D did not induce significant phosphorylation of either site. As shown before, inhibition of p53 degradation with the proteasome inhibitor LLnL stabilized p53 without phosphorylation of serine 15 or 20. These observations are consistent with previous observations that phosphorylation of p53 N-terminal sites is not an essential step for stabilization of the protein in response to actinomycin D treatment (3).
Activation of p21Waf1/Cip1 expression by stabilized p53.
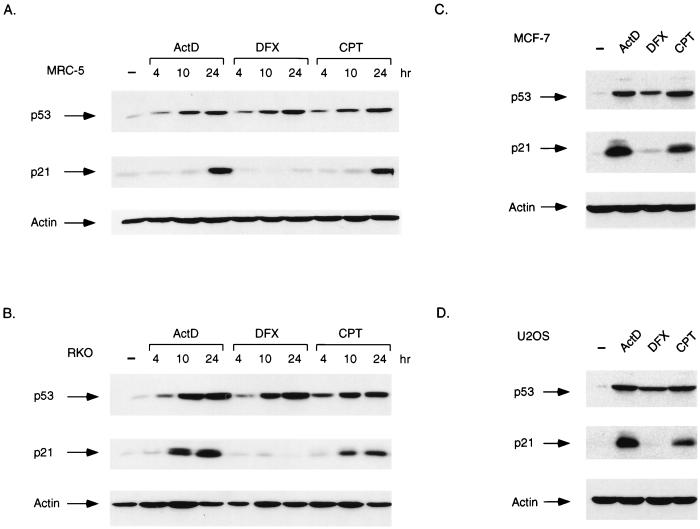
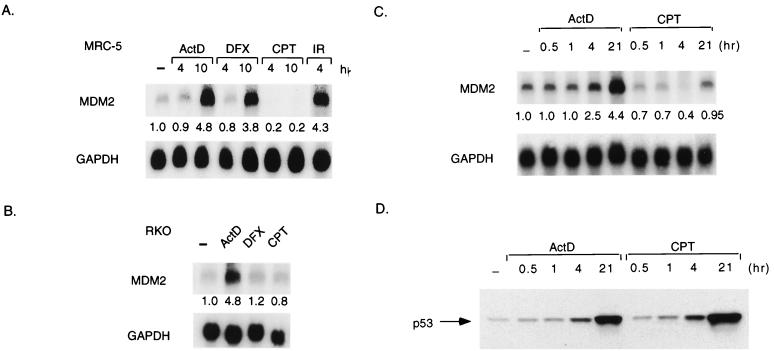
The ability of p53 to inhibit cell growth is closely related to the function of p53 as a transcription factor, and we therefore examined the expression of a p53-responsive gene encoding the cyclin-dependent kinase inhibitor p21Waf1/Cip1, a protein that plays an important role in establishing p53-dependent cell cycle arrest (11, 15, 52). Both actinomycin D and CPT treatment led to a time-dependent increase in p21Waf1/Cip1 protein levels in MRC-5 and RKO cells, indicating that the p53 protein stabilized in response to these drugs is transcriptionally active (Fig. 2A and B). However, despite a robust elevation of p53 levels in response to DFX, no evidence for an increase in p21Waf1/Cip1 protein levels could be detected in either cell line. Analysis of MCF-7 cells (Fig. 2C) and U2OS cells (Fig. 2D) confirmed the elevation of p21Waf1/Cip1 protein expression in response to actinomycin D and CPT but not DFX. p21Waf1/Cip1 is one of the principal mediators of p53-dependent cell cycle arrest, and flow cytometric analysis of U2OS and RKO cells treated with DFX confirmed that this treatment did not result in a G1 or G2 arrest (data not shown).
FIG. 2.
Activation of p21Waf1/Cip1 protein expression. (A) Western blot analysis of p53 and p21Waf1/Cip1 protein levels in MRC-5 cells harvested at the indicated times after treatment with actinomycin D (ActD; 5 nM), DFX (500 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. (B) Western blot analysis of p53 and p21Waf1/Cip1 protein levels in RKO cells harvested at the indicated times after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. (C) Western blot analysis of p53 and p21Waf1/Cip1 protein levels in MCF-7 cells harvested 24 h after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. (D) Western blot analysis of p53 and p21Waf1/Cip1 protein levels in U2OS cells harvested 24 h after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. −, no treatment.
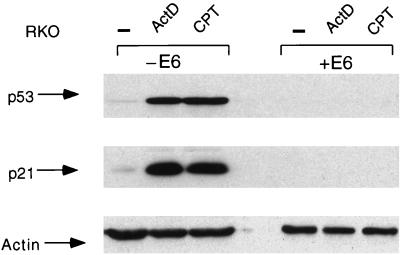
Expression of p21Waf1/Cip1 can be regulated by several p53-independent mechanisms, and in order to confirm that the elevation of p21Waf1/Cip1 seen in response to actinomycin D and CPT was dependent on p53, we repeated the analysis using RKO cells stably expressing the human papillomavirus type 16 E6 protein. E6 efficiently targets p53 for degradation through an MDM2-independent mechanism, and the E6-expressing cells are unable to stabilize p53 in response to any treatment, making them functionally p53 null (27). Treatment of RKO+E6 cells with either actinomycin D or CPT failed to stabilize p53, and neither treatment led to the enhanced expression of p21Waf1/Cip1 (Fig. 3).
FIG. 3.
Activation of p21Waf1/Cip1 protein expression in response to CPT is p53 dependent. Western blot analysis of p53 (upper blot) and p21Waf1/Cip1 (middle blot) protein levels in RKO cells (−E6) and RKO cells stably expressing human papillomavirus E6 (+E6) harvested 24 h after treatment with or without actinomycin D (ActD; 5 nM) or CPT (2 μM). To assess protein loading, the Western blots were reprobed with antiactin antibody (lower blot). −, no treatment.
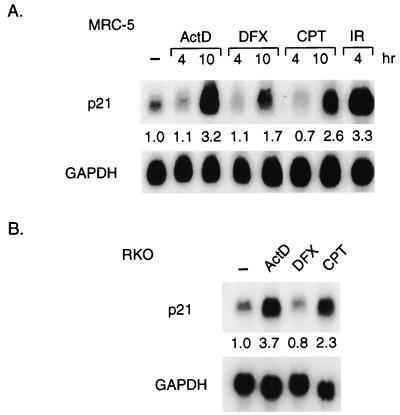
To confirm that changes in p21Waf1/Cip1 protein levels in response to different treatments were a reflection of specific changes in transcriptional activity, we analyzed mRNA expression in normal human MRC-5 cells by Northern blotting (Fig. 4A). Treatment with actinomycin D and CPT led to increased p21Waf1/Cip1 mRNA expression with slightly slower kinetics than that induced by IR, consistent with increased transcriptional activity of the stabilized p53. Treatment with DFX also led to a slight increase in p21Waf1/Cip1 mRNA levels, although it was not sufficient to result in an increase in protein. Similarly, clear elevation of p21Waf1/Cip1 mRNA was induced by actinomycin D and CPT treatments in RKO cells, although in these cells no increase of p21Waf1/Cip1 mRNA was seen in response to DFX (Fig. 4B).
FIG. 4.
Activation of p21Waf1/Cip1 transcription. (A) Northern blot analysis of p21Waf1/Cip1 mRNA levels in MRC-5 cells harvested at the indicated times after treatment with actinomycin D (ActD; 5 nM), DFX (500 μM), CPT (2 μM), or IR (10 Gy). The blots were reprobed for GAPDH expression as a loading control. Quantification of the signal relative to the GAPDH control was expressed as a ratio of the signal in untreated cells (shown under each lane). (B) Northern blot analysis of p21Waf1/Cip1 mRNA levels in RKO cells harvested 21 h after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for GAPDH expression as a loading control. Quantification of the signal relative to the GAPDH control was expressed as a ratio of the signal in untreated cells (shown under each lane). −, no treatment.
Activation of MDM2 expression by stabilized p53.
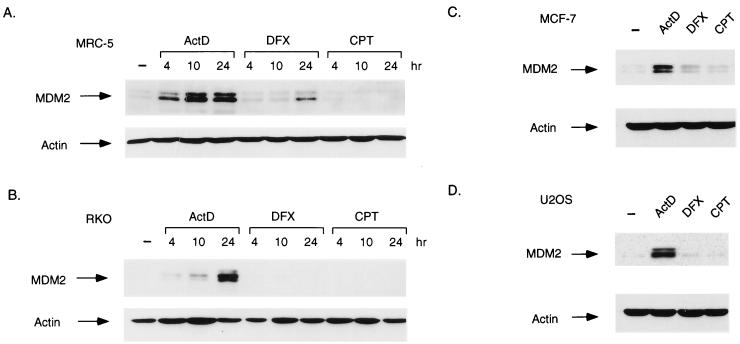
The failure of p53 stabilized in response to DFX to activate expression of p21Waf1/Cip1 prompted us to look at the expression of MDM2, also the product of a p53-inducible gene. Analysis of MRC-5 cells (Fig. 5A) showed that MDM2 protein expression was elevated following treatment of the cells with actinomycin D; a weaker activation of MDM2 protein expression was seen in response to DFX, and no increase in MDM2 protein was detected in response to CPT, despite the presence of significant levels of p53 and activation of p21Waf1/Cip1 following CPT treatment (Fig. 2A). Examination of the response to each treatment in RKO cells (Fig. 5B) also showed an increase in MDM2 protein following actinomycin D treatment, but no elevation of MDM2 in response to either CPT or DFX was observed. An identical pattern of MDM2 expression was seen in the two other tumor cell lines, MCF-7 (Fig. 5C) and U2OS (Fig. 5D).
FIG. 5.
Activation of MDM2 protein expression. (A) Western blot analysis of MDM2 protein levels in MRC-5 cells harvested at the indicated times after treatment with actinomycin D (ActD; 5 nM), DFX (500 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. (B) Western blot analysis of MDM2 protein levels in RKO cells harvested at the indicated times after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. (C) Western blot analysis of MDM2 protein levels in MCF-7 cells harvested 24 h after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. (D) Western blot analysis of MDM2 protein levels in U2OS cells harvested 24 h after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for actin expression as a loading control. −, no treatment.
Northern blot analysis confirmed that the pattern of MDM2 protein expression in these cell lines was a reflection of transcriptional activation. In MRC-5 cells (Fig. 6A), treatment with both actinomycin D and DFX led to the increased expression of MDM2 mRNA, while CPT treatment led to a reduction rather than an increase in MDM2 mRNA levels. In RKO cells (Fig. 6B), actinomycin D treatment increased MDM2 mRNA levels while DFX and CPT treatments failed to induce MDM2 mRNA. These results show that although the normal and tumor lines responded similarly to actinomycin D and CPT treatments, a difference was seen in response to DFX. Treatment with DFX stabilized p53 in both normal and tumor cells, but only the normal cells showed evidence of a transcriptional activity of this stabilized p53, with loss of this response in the tumor cell lines.
FIG. 6.
Activation of MDM2 mRNA expression. (A) Northern blot analysis of MDM2 mRNA levels in MRC-5 cells harvested at the indicated times after treatment with actinomycin D (ActD; 5 nM), DFX (500 μM), CPT (2 μM), or IR (10 Gy). The blots were reprobed for GAPDH expression as a loading control. Quantification of the signal relative to the control is shown under each lane. (B) Northern blot analysis of MDM2 mRNA levels in RKO cells harvested 21 h after treatment with actinomycin D (ActD; 5 nM), DFX (250 μM), or CPT (2 μM). The blots were reprobed for GAPDH expression as a loading control. Quantification of the signal relative to the GAPDH control was expressed as a ratio of the signal in untreated cells (shown under each lane). (C) Northern blot analysis of human MDM2 mRNA from RKO cells harvested after treatment with or without actinomycin D (5 nM) or CPT (2 μM) at the times indicated. The Northern blots were stripped and then hybridized with a human GAPDH cDNA probe to assess loading. Quantification of the signal relative to the GAPDH control was expressed as a ratio of the signal in untreated cells (shown under each lane). (D) Western blot analysis of p53 protein from RKO cells after treatment as described for panel C. −, no treatment.
Intriguingly, these data also suggested that treatment of cells with CPT resulted in elevation of p21Waf1/Cip1, but not MDM2, levels. To ascertain whether CPT treatment prevented the elevation of MDM2 in response to p53 or reduced basal levels of MDM2 expression (as suggested in the analysis of MRC5 cells shown in Fig. 6A), we carried out Northern blot analyses of a time course following actinomycin D and CPT treatment in RKO cells (Fig. 6C). These results showed that although actinomycin D treatment led to a time-dependent increase in MDM2 mRNA expression which correlated well with elevation in p53 protein levels (Fig. 6D), treatment with CPT led to a transient drop in MDM2 mRNA, followed by a return to basal levels. No increase in MDM2 transcription in response to p53 induction was seen at any time point.
Subcellular localization of p53 and MDM2.
Subcellular localization of p53 and MDM2 has recently been shown to play a critical role in controlling p53 function and stability. p53 contains three nuclear localization sequences (33, 40, 41), and failure to localize to the nucleus results in an inability of p53 to activate transcription of target genes like p21Waf1/Cip1 and MDM2. p53 and MDM2 also contain nuclear export signals (38, 46), and the ability of MDM2 to shuttle from the nucleus to the cytoplasm appears to be important for the degradation of p53 (18, 30, 48).
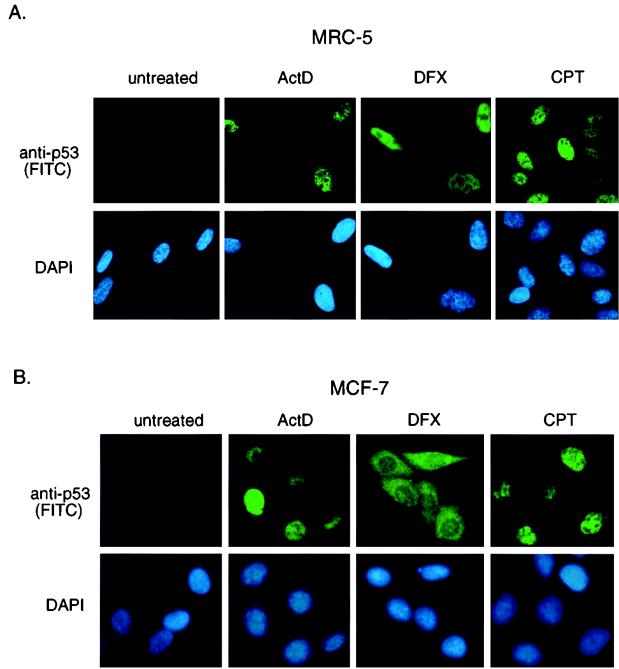
We examined the localization of the p53 protein that was expressed in response to different treatments in MRC-5 (Fig. 7A), MCF-7 (Fig. 7B), and U2OS cells (data not shown). Essentially identical results were obtained in MCF-7 cells and U2OS cells. Treatment of each cell line with actinomycin D or CPT resulted in the accumulation of nuclear p53, consistent with the retention of at least some transcriptional activity of this stabilized p53. In contrast, although p53 protein stabilized following DFX was localized mainly to the nucleus in MRC-5 cells (Fig. 7A), both nuclear and cytoplasmic accumulation of p53 was seen in MCF-7 and U2OS cells (Fig. 7B and data not shown).
FIG. 7.
Subcellular localization of p53. (A) Immunofluorescence staining of MRC-5 cells at 10 h after treatment with actinomycin D (ActD; 5 nM), DFX (500 μM), or CPT (2 μM). Localization of human p53 protein was assessed using DO1 and visualized using an FITC-conjugated secondary antibody. Cells were counterstained with DAPI to localize the nucleus. (B) Immunofluorescence staining of MCF-7 cells at 10 h after treatment with actinomycin D (5 nM), DFX (250 μM), or CPT (2 μM). Localization of human p53 protein was assessed as described for panel A.
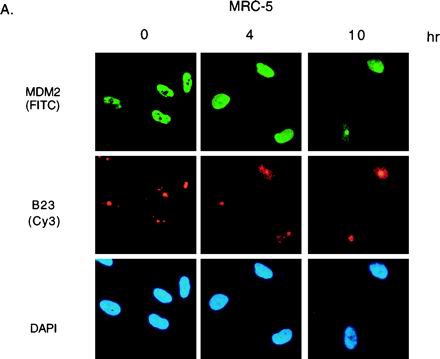
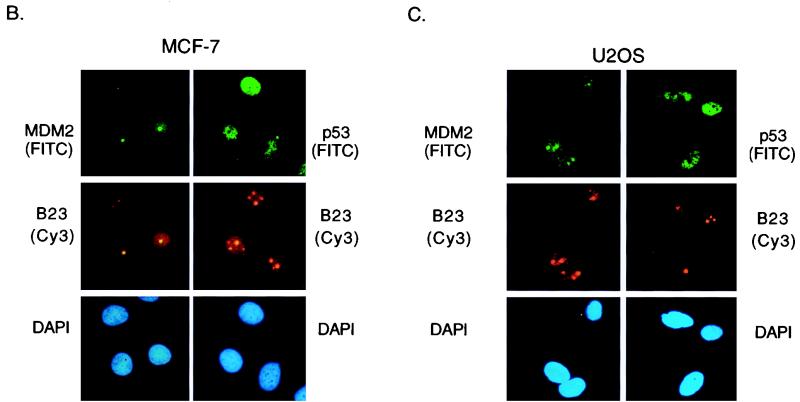
Treatment of cells with actinomycin D resulted in elevation of p53 protein which was not phosphorylated at serine 15 or 20 (Fig. 1) and retained the ability to bind to MDM2 in coprecipitation assays (data not shown). In order to determine whether p53 and MDM2 were colocalized in cells treated with actinomycin D, we examined the subcellular localization of MDM2 in actinomycin D-treated cells. The overall level of MDM2 protein in MRC-5 cells was first elevated by inhibition of the proteasome, using LLnL, before treatment with actinomycin D (Fig. 8A). As described previously, MDM2 stabilized in untreated cells localized to the nucleus, with exclusion from the nucleoli, as shown by costaining with the nucleolar protein B23. In response to actinomycin D treatment, the localization of MDM2 changes, occupying the whole nucleus after 4 h and showing association with B23 staining by 10 h posttreatment. Similar patterns of staining were seen in both MCF-7 and U2OS cells in response to actinomycin D treatment (Fig. 8B and C). Previous studies have shown that treatment of cells with 80 to 200 nM actinomycin D results in a dose-dependent and reversible relocation of nucleolar proteins to the nucleoplasm (56). Although our studies involved treatment of cells with only 5 nM actinomycin D, it is possible that our results indicate either association of MDM2 with nucleolar proteins such as B23 released from the nucleolus following actinomycin D treatment or a relocation of MDM2 to the nucleolus. In either case, p53 and MDM2 proteins showed distinct subnuclear localization patterns following actinomycin D treatment (Fig. 8).
FIG. 8.
Subnuclear localization of MDM2. (A) Immunofluorescence staining of MRC-5 cells, pretreated for 3 h with LLnL to stabilize MDM2 and harvested at the indicated times after treatment with 5 nM actinomycin D. Colocalization of MDM2 (AB-1) visualized using an FITC-conjugated secondary antibody with B23 protein visualized using a Cy3-conjugated secondary antibody is shown; the cells were counterstained with DAPI to localize the nucleus. (B) Immunofluorescence staining of MCF-7 cells 10 h after treatment with 5 nM actinomycin D. Colocalization of MDM2 (AB-1) or p53 (DO1) with B23 protein is shown. The cells were counterstained with DAPI to localize the nucleus. (C) Immunofluorescence staining of U2OS cells 16 h after treatment with 5 nM actinomycin D. Colocalization of MDM2 (AB-1) or p53 (DO1) with B23 protein is shown. The cells were counterstained with DAPI to localize the nucleus.
DISCUSSION
The ability to activate p53 in response to various types of potentially oncogenic stress plays an important role in preventing malignant progression. Many forms of stress have now been shown to activate p53, including DNA damage, activation of oncogenes, and hypoxia (4). Despite the diversity of these signals, a point of convergence in the stress responses appears to be the inhibition of p53 degradation by MDM2 and subsequent stabilization of the p53 protein. Two independent mechanisms to disrupt MDM2 function have been described; phosphorylation of p53 to prevent MDM2 binding and activation of expression of ARF.
In this study we show that different p53-activating signals, such as CPT, actinomycin D, and DFX, appear to utilize different mechanisms to prevent MDM2-mediated degradation of p53 (summarized in Table 1). Although activation of p53 in response to CPT leads to phosphorylation of p53 at both serines 15 and 20, which would inhibit the interaction of p53 with MDM2, CPT treatment is not accompanied by elevation of MDM2 levels, and the stabilization of p53 is likely to reflect, at least in part, inhibition of MDM2 expression. The p53 stabilized following CPT treatment resulted in enhanced expression of p21Waf1/Cip1, and it seems likely that treatment with CPT specifically down-regulated expression of MDM2. Inhibition of MDM2 expression may be a common mechanism to stabilize p53. Previous reports have indicated that both UV irradiation (54) and treatment with the topoisomerase II inhibitor etoposide also inhibits expression of MDM2 (2). Treatment with approximately 10-fold-higher concentrations of actinomycin D than those used in this study has also been shown to reduce MDM2 expression in a lymphoblast cell line (9).
TABLE 1.
Summary of p53 responses to actinomycin D (Act-D), DFX, and CPT treatment in normal (MRC-5) and tumor (RKO, MCF-7, and U2OS) cells
| Parameter | Valuea
|
|||||
|---|---|---|---|---|---|---|
| Act-D
|
DFX
|
CPT
|
||||
| Normal | Tumor | Normal | Tumor | Normal | Tumor | |
| Serine 15 phosphorylation | − | − | + | + | + | + |
| Serine 20 phosphorylation | − | − | − | − | + | + |
| p21 expression | + | + | ± | − | + | + |
| MDM2 expression | + | + | + | − | − | − |
| Cytoplasmic p53 | − | − | ± | + | − | − |
+, present; −, absent; ±, present at low levels.
In contrast to CPT treatment, DFX stabilized p53 that was phosphorylated on serine 15 but not serine 20 and was unable to activate transcription of p21Waf1/Cip1 or MDM2 in three tumor cell lines but retained some transcriptional activity in normal MRC-5 cells. Interestingly, even in normal cells the transcriptional activity of p53 in response to DFX was clearly much weaker than that seen for comparable levels of p53 induced by other treatments, such as actinomycin D. Activation of p53 has been shown to depend on both elevation of the protein levels and a shift from the latent to the DNA binding form of the protein itself (23). Our results suggest that DFX, which mimics hypoxia, may only partially activate stabilized, but latent, p53 in normal cells and that this response is entirely lost during tumor progression. Interestingly, the loss of transcriptional activity in the tumor cell lines correlated with an accumulation of p53 in the cytoplasm, potentially indicating a defect in pathways that allow nuclear localization of p53.
Treatment of cells with actinomycin D induced p53 that is stable and transcriptionally functional, despite the presence of high levels of MDM2. p53 stabilized in response to actinomycin D was not phosphorylated at either serine 15 or 20, and as expected, this unphosphorylated p53 retained the ability to form an interaction with MDM2 in coprecipitation assays. However, our results suggest that following actinomycin D treatment the two proteins can occupy distinct nuclear locations that might prevent or reduce complex formation in the intact cell, explaining the potential paradox of how p53 can be transcriptionally active in a cell containing high levels MDM2 without modifications that prevent binding. These results show some similarity to the stabilization of unphosphorylated p53 by nucleolar localization of MDM2 following interaction with ARF (49, 53), although previous studies showing that neither MCF-7 nor U2OS cells express ARF (47) suggest that ARF is not directly involved here. MDM2 has been shown to bind ribosomal proteins and RNA (17), and recently, a nucleolar localization signal has been identified in the C terminus of MDM2 (32a). It will be of interest to determine whether actinomycin D leads to nucleolar localization of MDM2 or to nucleoplasmic association of MDM2 with components of the nucleolus released in response to actinomycin D treatment.
Most human cancers show loss of normal p53 function, although this is not always reflected by mutational inactivation of the p53 protein itself. Disruption of the pathways which allow activation of p53 in response to stress could also contribute to tumor development, and many cancer cells that retain wild-type p53 show loss of the ARF protein which is necessary for the stabilization of p53 in response to abnormal proliferation (47). The observation that many different pathways lead to p53 stabilization presents the possibility that tumors arising following inactivation of one pathway (such as loss of ARF) may retain the ability to activate p53 in response to signals that utilize different pathways. Tumor cells lacking ARF, for example, show normal p53 activation in response to DNA-damaging signals (25, 26). Tumors that arise following loss of kinases, such as ATM, that phosphorylate p53 at serine residue 15 in response to UV irradiation may retain the ability to activate p53 in phosphorylation-independent pathways, such as that engaged by actinomycin D. An understanding of the pathways utilized by different drugs to activate p53 could be used to enhance and complement therapeutic options.
ACKNOWLEDGMENTS
We thank Marion Lohrum for critical review of the manuscript and all other members of the Vousden lab for advice and encouragement. We are also extremely grateful to Chuck Sherr for pointing out the consequences of actinomycin D treatment for the nucleolus.
This work was sponsored by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.An W G, Kanekal M, Simon M C, Maltepe E, Blagosklonny M V, Neckers L M. Stabilization of wild-type by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 2.Arriola E L, Rodriguez Lopez A, Chresta C M. Differential regulation of p21waf-1/cip-1 and Mdm2 by etoposide: etoposide inhibits the p53-Mdm2 autoregulatory loop. Oncogene. 1999;18:1081–1091. doi: 10.1038/sj.onc.1202391. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft M, Kubbutat M H, Vousden K H. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashcroft M, Vousden K H. Regulation of p53 stability. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 5.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 6.Barak Y, Juven T, Haffner R, Oren M. Mdm-2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates S, Phillips A C, Clarke P A, Stott F, Peters G, Ludwig R L, Vousden K H. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 8.Bates S, Vousden K H. p53 in signalling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:1–7. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 9.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blattner C, Tobiasch E, Litfen M, Rahmsdorf H J, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 11.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–556. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 12.Bulavin D, Saito S, Hollander M C, Sakaguchi K, Anderson C W, Appella E, Fornace A J., Jr Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999;18:6845–6854. doi: 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canman C E, Lim D-S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 14.Choi J, Donehower L A. p53 in embryonic development: maintaining a fine balance. Cell Mol Life Sci. 1999;55:38–47. doi: 10.1007/s000180050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 16.de Stanchina E, McCurrach M E, Zindy F, Shieh S Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine A J. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med. 1996;2:439–451. [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 20.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 21.Honda R, Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsiang Y H, Hertzberg R, Hecht S, Liu L F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 23.Hupp T R, Lane D P. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 24.Jones S N, Roe A E, Donehower L A, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 25.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 27.Kessis T D, Slebos R J, Nelson W G, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 29.Kubbutat M H G, Vousden K H. Keeping an old friend under control: regulation of p53 stability. Mol Med Today. 1998;4:250–256. doi: 10.1016/s1357-4310(98)01260-x. [DOI] [PubMed] [Google Scholar]
- 30.Lain S, Midgley C, Sparks A, Lane E B, Lane D P. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODS. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 31.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Ljungman M, Zhang F, Chen F, Rainbow A J, McKay B C. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 32a.Lohrum M A E, Ashcroft M, Kubbutat M H G, Vousden K H. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- 33.Middeler G, Zerf K, Jenovai S, Thulig A, Tschodrich-Rotter M, Kubitscheck U, Peters R. The tumor suppressor p53 is subject to both nuclear import and export, and both are fast, energy-dependent and lectin-inhibited. Oncogene. 1997;14:1407–1417. doi: 10.1038/sj.onc.1200949. [DOI] [PubMed] [Google Scholar]
- 34.Midgley C A, Fisher C J, Bartek J, Vojtesek B, Lane D P, Barnes D M. Analysis of p53 expression in human tumours: an antibody raised against human p53 expressed in E. coli. J Cell Sci. 1992;101:183–189. doi: 10.1242/jcs.101.1.183. [DOI] [PubMed] [Google Scholar]
- 35.Montes de Oca Luna R, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 36.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 37.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 38.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaulsky G, Goldfinger N, Ben-Ze'ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10:6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaulsky G, Goldfinger N, Tosky M S, Levine A, Rotter V. Nuclear localization is essential for the activity of p53 protein. Oncogene. 1991;6:2055–2065. [PubMed] [Google Scholar]
- 42.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 43.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 44.Shieh S Y, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stott F, Bates S A, James M, McConnell B B, Starborg M, Brookes S, Palmero I, Hara E, Ryan K M, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao W, Levine A J. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao W, Levine A J. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 53.Weber J D, Taylor L J, Roussel M F, Sherr C J, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 54.Wu L, Levine A J. Differential regulation of the p21/WAF-1 and mdm2 genes after high dose UV irradiation: p53-dependent and p53-independent regulation of the mdm2 gene. Mol Med. 1997;3:441–451. [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X W, Bayle J H, Olson D, Levine A J. The p53 mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 56.Yokoyama Y, Niwa K, Tamaya T. Scattering of the silver-stained proteins of nucleolar organizer regions in Ishikawa cells by actinomycin D. Exp Cell Res. 1992;202:77–86. doi: 10.1016/0014-4827(92)90406-x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of Mdm2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- 58.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]