Abstract
Non-small cell lung cancer (NSCLC) is one of the most malignant cancer types. N6-methyladenosine (m6A), an abundant eukaryotic mRNA modification, has been observed in multiple diseases, particularly cancer. Methyltransferase-like 14 (METTL14) is a central component of the m6A methyltransferase complex and has been reported to promote tumor development in several cancer types. The present study aimed to investigate the role of METTL14 in NSCLC. Relevant clinical and mRNA sequencing data for m6A-related genes were downloaded from The Cancer Genome Atlas database. R software was used to evaluate the expression of m6A regulators in NSCLC. The biological functions of METTL14 were evaluated using Cell Counting Kit-8, colony formation, Transwell migration and western blot analyses. The results demonstrated that METTL14 expression was upregulated in NSCLC tissues and cell lines, and its expression was high in cancer tissues from patients with NSCLC with all four stages (I, II, III and IV) of disease. METTL14 downregulation inhibited cell proliferation and migration in A549 and SK-MES-1 lung cancer cell lines. Knockdown of METTL14 in lung cancer cell lines increased E-cadherin expression and suppressed N-cadherin expression. Furthermore, METTL14 downregulation reduced the expression levels of the transcription factor Twist and the p-AKT/AKT ratio. In conclusion, the present findings revealed that silencing of METTL14 suppressed NSCLC malignancy by inhibiting Twist-mediated activation of AKT signaling. These data suggest that METTL14 may be a potential therapeutic target for NSCLC.
Keywords: non-small cell lung cancer, methyltransferase-like 14, epithelial-mesenchymal transition, Twist
Introduction
Lung cancer is the most common malignancy and cause of cancer-related mortality worldwide (1). Furthermore, ~85% of lung cancer cases are non-small cell lung cancer (NSCLC) cases (1). Despite advances in diagnosis and clinical therapy, the prognosis of NSCLC remains unfavorable, with an overall 5-year survival rate of 17% in 2016 in North America (2). The poor prognosis is mainly ascribed to cancer metastasis (3). Therefore, it is critical to determine the mechanisms underlying NSCLC progression and metastasis.
Epithelial-mesenchymal transition (EMT) is a reversible process by which epithelial cells acquire a migratory and invasive mesenchymal phenotype, which promotes metastasis (4,5). The process of EMT is generally accompanied by decreased expression levels of E-cadherin and increased expression levels of N-cadherin (5), which results in increased cellular invasiveness. Therefore, it is essential to explore the mechanisms underlying the EMT process involved in NSCLC metastasis.
N6-methyladenosine (m6A) is the most abundant chemical modification of RNA in eukaryotes (6). It is a dynamic and reversible process, which is mediated mainly by methyltransferase-like (METTL)3 (7) and 14 (8), and erased by the demethylases fat-mass and obesity-associated protein (9) and alkylation repair homolog protein 5 (ALKBH5) (10). The m6A modification is recognized by YTH domain-containing family proteins 1/2/3 (YTHDF1/2/3) (11). Recently, multiple studies have demonstrated that m6A modifications are associated with the development and progression of diverse cancer types, including cervical (12), prostate (13) and gastric (14) cancer, hepatocellular carcinoma (HCC) (15), colon cancer (16), and NSCLC (17). A number of m6A regulators are upregulated in NSCLC (18–20). For example, METTL3 is upregulated in NSCLC tissues, and METTL3 enhances NSCLC progression by modulating the levels of Bcl-2 through m6A modification (18). ALKBH5 is highly expressed in NSCLC tissues and cells, and increased ALKBH5 expression is closely associated with a poor prognosis (19). Functionally, ALKBH5 promotes the proliferation and represses the apoptosis of NSCLC cells in vitro, and silencing of ALKBH5 inhibits tumor growth in vivo (19). YTHDF1 is highly expressed in NSCLC cells, and knockdown of YTHDF1 reduces the proliferation, invasion and migration of NSCLC cells (20).
METTL14 serves as a central component of the m6A methyltransferase complex and has been found to participate in tumor development (21,22). For example, METTL14 enhances the proliferation and migration of pancreatic cancer cells (21). METTL14 overexpression results in a decrease in p53 effector related to peripheral myelin protein 22 mRNA levels in an m6A-dependent manner; thus, METTL14 is a potential target for the treatment of pancreatic cancer (21). Furthermore, METTL14 is markedly upregulated in breast cancer tissues (22). Upregulation of METTL14 may promote the migration and invasion of breast cancer cells by modulating hsa-microRNA-146a-5p expression (22). However, to the best of our knowledge, the expression patterns and pathophysiological role of METTL14 in NSCLC remain unclear. Therefore, the present study aimed to investigate the role of METTL14 in NSCLC.
Materials and methods
Public databases analysis
The Cancer Genome Atlas (TCGA)-NSCLC and all corresponding clinical data of 535 NSCLC samples and 88 normal samples were downloaded from TCGA data portal (http://gdc-portal.nci.nih.gov/). mRNA expression of METTL14 in an NSCLC dataset (GSE31210) (23) were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Samples with incomplete clinical characteristics were excluded. DepMap databases (https://depmap.org) were used to obtain the expression of m6A regulators in the cell lines.
Cell culture and treatment
The normal human bronchial epithelial cell lines (16HBE and BEAS-2B) and lung cancer cell lines (SK-MES-1, A549, H1299, H460 and H520) were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. All cell lines, except for the H520 cell line, were cultured in DMEM medium (Wisent Inc.). H520 cells were cultured in RPMI-1640 medium (Biological Industries). All cells were maintained in medium containing 10% fetal bovine serum (Biological Industries) and incubated at 37°C with 5% CO2.
Cycloheximide (CHX) was purchased from MedChemExpress
To explore the degradation of Twist, A549 cells were transfected with si-METTL14-1 for 48 h, as later described, and then treated with CHX (final concentration, 10 µg/ml) for 0, 1.5, 3, 4.5 and 6.0 h at 37°C. The expression levels of Twist were detected by western blot, as later described.
Cell transfection
Small interfering RNA (siRNA/si) against METTL14 (si-METTL14-1/si-ME-1 and si-METTL14-2/ si-ME-2) and negative control (si-NC) were obtained from Shanghai GenePharma Co., Ltd. The A549 and SK-MES-1 cells were cultured in plates and, when the cells reached 60–70% confluence, transient transfections with 100 pmol siRNA were performed using Lipofectamine 3000 (Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Next, the cells were cultured in serum-reduced medium and incubated at 37°C with 5% CO2 in incubator for 6 h, and then the medium was exchanged for complete culture medium. After 48 h, the cells were collected for the subsequent experiments. The sequences of the scrambled control group (si-NC) were as follows: Sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. The two pairs of synthesized siRNAs against METTL14 sequences were as follows: si-METTL14-1 forward, 5′-CCUAUUUCAGUGCUCCUAATT-3′ and reverse, 5′-UUAGGAGCACUGAAAUAGGTT-3′; siMETTL14-2 forward, 5′-GGACUUGGGAUGAUAUUAUTT-3′ and reverse, 5′-AUAAUAUCAUCCCAAGUCCTT.
Cell proliferation
Cell Counting Kit-8 (CCK-8) and colony formation assays were used to evaluate cell proliferation. For the CCK-8 assay, the cells were cultured in 96-well plates (A549 cells, 3,000 cells/well; SK-MES-1 cells, 6,000 cells/well). After being incubated for 0, 1, 2, 3 and 4 days at 37°C, the cells were treated with 10 µl CCK-8 reagent (Dojindo Laboratories, Inc.) at 37°C for 2 h. The absorbance at 450 nm was measured using a microplate reader.
For the colony information assay, cells (A549 cells, 300 cells/well; SK-MES-1 cells, 600 cells/well) were cultured in a 6-well plate for 2 weeks at 37°C to form colonies. Colonies were fixed with 1 ml 4% paraformaldehyde for 15 min and stained with 0.4% crystal violet for 15 min at room temperature. ImageJ software verion 1.48 (National Institutes of Health) was used to count the stained colonies (consisting of ≥50 cells).
Cell migration
The migration of the cells was evaluated using a Transwell® system (8 µm; Corning Inc.). The upper chambers of the 8-µm Transwell were filled with 200 µl of cells resuspended in serum-free medium (A549 cells, 2×104 cells/200 µl; SK-MES-1 cells, 4×104 cells/200 µl), and the lower chambers were filled with 600 µl medium supplemented with 20% FBS. Following incubation for 24 h at 37°C, the migrated cells on the lower side of the membranes were fixed with 4% paraformaldehyde for 15 min at room temperature and then stained with 0.4% crystal violet for 15 min at room temperature. Five random fields (magnification, ×200) were selected and images were captured using a light microscope. ImageJ software version 1.48h3 (National Institutes of Health) was used to count the migrated cells.
Western blotting
The cells were washed with PBS and lysed with RIPA Lysis Buffer (Beyotime Institute of Biotechnology). The cell lysates were centrifuged at 12,000 × g for 20 min at 4°C, and the supernatants were denatured for 10 min at 100°C. A BCA protein assay kit (Beyotime Institute of Biotechnology) was used to measure the protein concentration. Equal amounts of protein (30 µg/sample) were separated on 10% gels using SDS-PAGE, and then transferred onto 0.45-µm polyvinylidene difluoride membranes. Subsequently, the membranes were blocked with 5% skimmed dry milk for 2 h at room temperature and incubated with primary antibodies overnight at 4°C. Next, the membranes were washed and incubated with HRP-conjugated secondary antibody (dilution, 1:5,000) for 2 h at room temperature. Finally, the protein bands were visualized using chemiluminescence reagent (Pierce; Thermo Fisher Scientific, Inc.). Protein gray value detection was performed using ImageJ software (version 1.48h3; National Institutes of Health). Primary antibodies from Cell Signaling Technology, Inc. included those for E-cadherin (cat. no. 24E10), Snail (cat. no. C15D3), Slug (cat. no. C19G7), phosphorylated (p)-AKT (cat. no. D9E), p-GSK3β (cat. no. D3A4) and GAPDH (cat. no. D16H11). Primary antibodies from ProteinTech Group, Inc. included those for METTL14 (cat. no. 26158-1-AP), N-cadherin (cat. no. 22018-1-AP), zinc finger E-box binding homeobox 1 (ZEB1; cat. no. 21544-1-AP), AKT (cat. no. 10176-2-AP), GSK3β (cat. no. 22104-1-AP), β-actin (cat. no. 66009-1-Ig) and Twist (cat. no. 25465-1-AP). HRP-conjugated goat anti-mouse IgG (cat. no. SA00001-1) and HRP-conjugated goat anti-rabbit IgG (cat. no. SA00001-2) were purchased from Wuhan Sanying Biotechnology.
Statistical analysis
The R package ‘edgeR’ was used to analyze gene expression differences between NSCLC and normal tissues. The Kruskal-Wallis test with Steel-Dwass post hoc test was used to evaluate the association between METTL14 mRNA expression and tumor stages. The Mann-Whitney U test was used to evaluate the differences in tissue expression between non-metastatic patients and metastatic patients. A Cox proportional hazards model was used to perform univariate analysis and calculate the 95% CI. The R package ‘pheatmap’ was used to visualize the above results. R software (version 4.0.3; http://www.r-project.org/) was used for the aforementioned statistical analyses.
Each experiment was performed in triplicate
All experimental data are presented as the mean ± SD. Student's unpaired t-test was used to analyze differences between two groups, whereas one-way ANOVA followed by Tukey's post hoc test was used to analyze differences among multiple groups. GraphPad Prism 6.0 (GraphPad Software, Inc.) was used for statistical analyses of the experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
METTL14 is highly expressed in NSCLC tissues
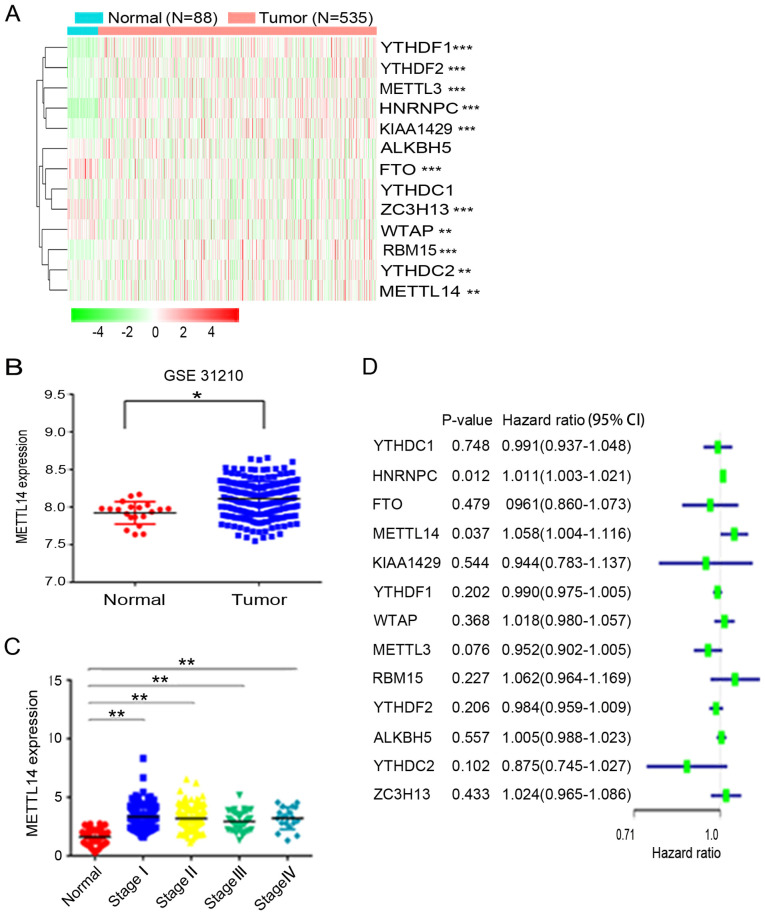
To evaluate the expression profile of m6A regulators in NSCLC, the present study analyzed the corresponding gene expression differences between 535 NSCLC tissues and 88 normal tissues based on data obtained from TCGA. The results revealed that the expression of YTHDF1, YTHDF2, METTL3, heterogeneous nuclear ribonucleoprotein C (HNRNPC), KIAA1429, FTO, vir like m6A methyltransferase associated (ZC3H13), WTAP, RNA binding motif protein 15 (RBM15), YTHDC2 and METTL14, but not that of ALKBH5 and YTH domain containing 1 (YTHDC1), was dysregulated in NSCLC (Fig. 1A). Among the aforementioned differentially expressed m6A-related genes, METTL14, METTL3, HNRNPC, ZC3H13, YTHDF1 and YTHDF2 were found to be highly expressed in NSCLC tissues. To further verify METTL14 expression, the GEO database was used and it was revealed that METTL14 expression was higher in NSCLC tissues than in normal tissues (Fig. 1B). Additionally, TGGA data demonstrated that METTL14 expression was significantly increased in patients with stage I–IV NSCLC compared with in patients in the normal control group (Fig. 1C). Furthermore, univariate analysis revealed that METTL14 is a risk gene of NSCLC. The hazard ratio of METTL14 was greater >1, indicating that the higher the expression of METTL14, the higher the risk of NSCLC (Fig. 1D).
Figure 1.
METTL14 is highly expressed in NSCLC tissues. (A) Expression of m6A regulators in NSCLC and normal tissues (red, upregulated; green, downregulated). The ‘edgeR’ R package was utilized to analyze the differences in gene expression profiles. (B) METTL14 expression in 246 NSCLC tissues and 20 normal tissues in the Gene Expression Omnibus database (dataset accession no. GSE31210). (C) Relative expression levels of METTL14 in patients with stage I, II, III and IV NSCLC based on TCGA database. Data were analyzed using the Kruskal-Wallis test with Steel-Dwass test. (D) Forest plot of the univariate Cox regression analysis. Univariate analysis was performed in m6A-related genes. All bars correspond to the 95% confidence intervals. *P<0.05, **P<0.01 and ***P<0.001. METTL14, methyltransferase-like 14; NSCLC, non-small cell lung cancer; TCGA, The Cancer Genome Atlas.
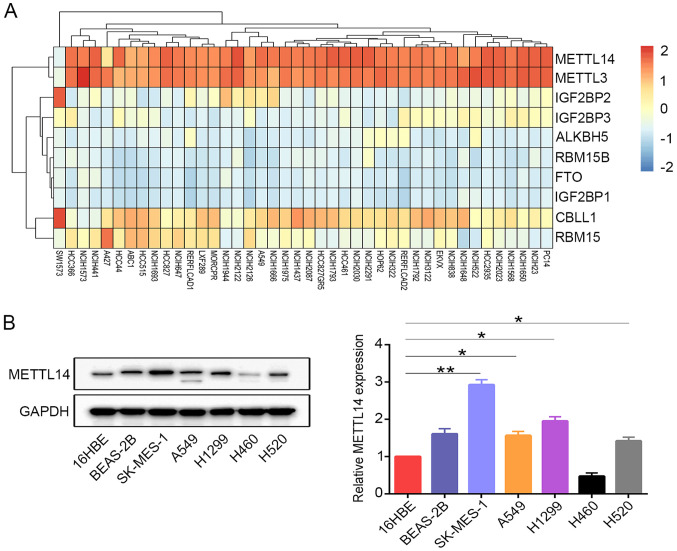
METTL14 is highly expressed in SK-MES-1 cells, A549 cells, H1299 cells and H520 cells
The present study investigated the expression levels of several m6A regulators in various NSCLC cell lines. The results revealed that only METTL3 and METTL14 were highly expressed in the majority of the NSCLC cell lines (Fig. 2A). METTL14 protein expression was further examined in the cell lines using western blotting. Consistently, it was observed that METTL14 expression in the lung cancer cell lines, with the exception of H460 cells, was increased compared with that in the normal human bronchial epithelial cells (16HBE) (Fig. 2B). SK-MES-1 cells were selected as ‘loss-of-function’ models, as they expressed high levels of METTL14 protein. A549 cells were selected for use, as METTL14 expression was consistent compared with other NSCLC cell lines (except SK-MES-1 cells).
Figure 2.
METTL14 is highly expressed in NSCLC cell lines. (A) Dependency heatmap of m6A regulators in different NSCLC cell lines (red, upregulated; blue, downregulated).. (B) Protein levels of METTL14 in the normal human bronchial epithelial cell lines (16HBE and BEAS-2B) and lung cancer cell lines (SK-MES-1, A549, H1299, H460 and H520) were examined by western blotting. *P<0.05 and **P<0.01 (one-way ANOVA with Tukey's post hoc test). METTL14, methyltransferase-like 14; NSCLC, non-small cell lung cancer.
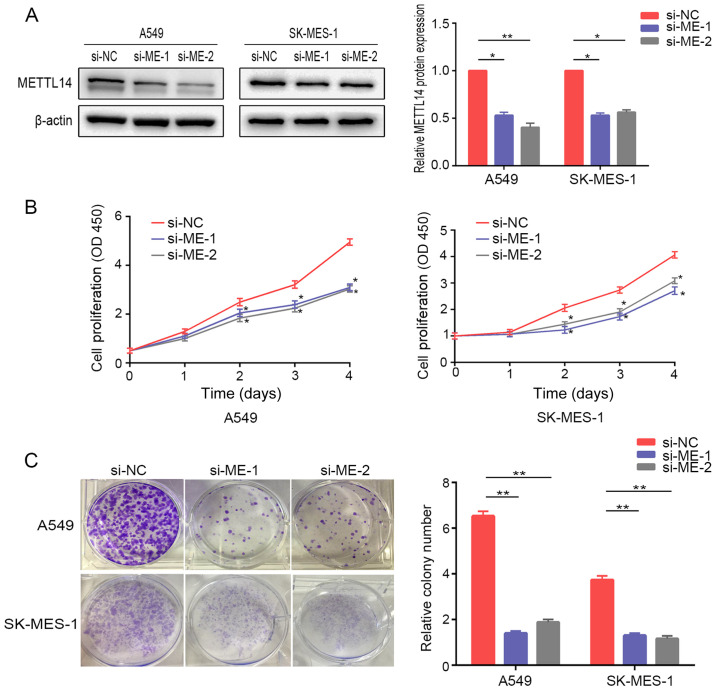
Knockdown of METTL14 inhibits NSCLC cell proliferation
To explore the potential effects of METTL14 on cell proliferation, METTL14 expression was knocked down in A549 and SK-MES-1 cells. Western blot analysis demonstrated that METTL14 siRNAs efficiently reduced METTL14 protein expression (Fig. 3A). The CCK-8 assay revealed that proliferation of A549 and SK-MES-1 cells was significantly reduced after suppression of METTL14 at days 2–4 (Fig. 3B). Consistently, colony formation assays demonstrated that knockdown of METTL14 was associated with a marked reduction in number of A549 and SK-MES-1 cell colonies (Fig. 3C).
Figure 3.
METTL14 knockdown inhibits non-small cell lung cancer cell proliferation. (A) Interference efficiency of two different METTL14 siRNAs was detected in A549 and SK-MES-1 cells. (B) Proliferation of A549 and SK-MES-1 cells following METTL14 knockdown was determined using Cell Counting Kit-8 assays. (C) A colony formation assay was performed for 2 weeks in A549 and SK-MES-1 cells after knockdown of METTL14. Representative images (left) and the relative number of colonies (right) are shown. *P<0.05 and **P<0.01 vs. si-NC (one-way ANOVA with Tukey's post hoc test). METTL14/ME, methyltransferase-like 14; NC, negative control; OD, optical density; siRNA/si, small interfering RNA.
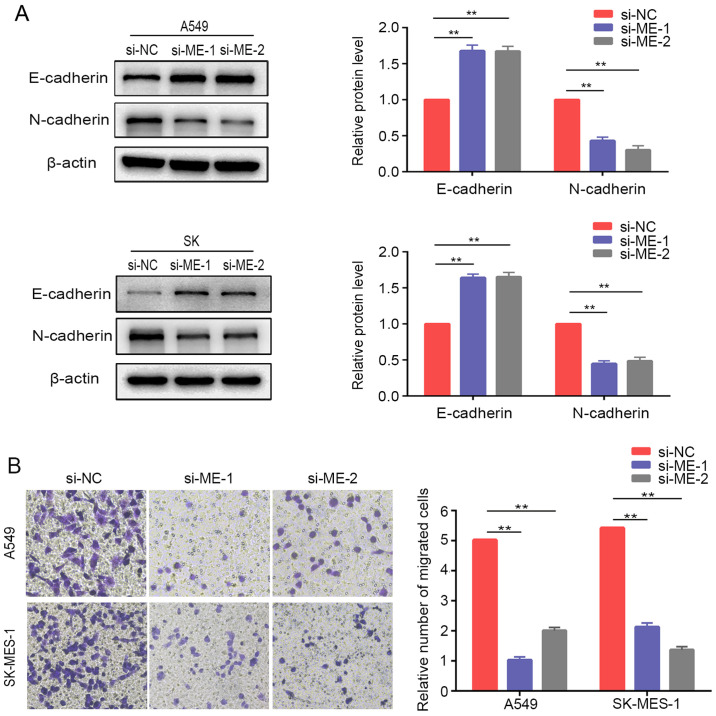
Knockdown of METTL14 inhibits EMT in NSCLC cells
The present study further examined the relative protein expression of EMT markers in lung cancer cells after METTL14 inhibition using siRNAs. The results demonstrated that E-cadherin was upregulated, while N-cadherin was downregulated compared with the expression levels of the control cells (Fig. 4A). Similarly, Transwell® assays demonstrated that downregulation of METTL14 markedly inhibited the migration of A549 and SK-MES-1 cells (Fig. 4B). Overall, these findings suggested that METTL14 served a crucial role in regulating the EMT process and promoting migration of lung cancer cells. However, the differences in tissue expression of METTL14 between non-metastatic and metastatic patients in TCGA database were not statistically significant (Fig. S1).
Figure 4.
METTL14 knockdown inhibits non-small cell lung cancer cell migration and epithelial-mesenchymal transition. (A) E-cadherin and N-cadherin expression was evaluated by western blotting in A549 cells and SK-MES-1 cells after METTL14 knockdown. (B) Migration of A549 and SK-MES-1 cells following METTL14 knockdown was measured using Transwell migration assays (magnification, ×200 times). **P<0.01 (one-way ANOVA with Tukey's post hoc test). METTL14/ME, methyltransferase-like 14; NC, negative control; si, small interfering RNA.
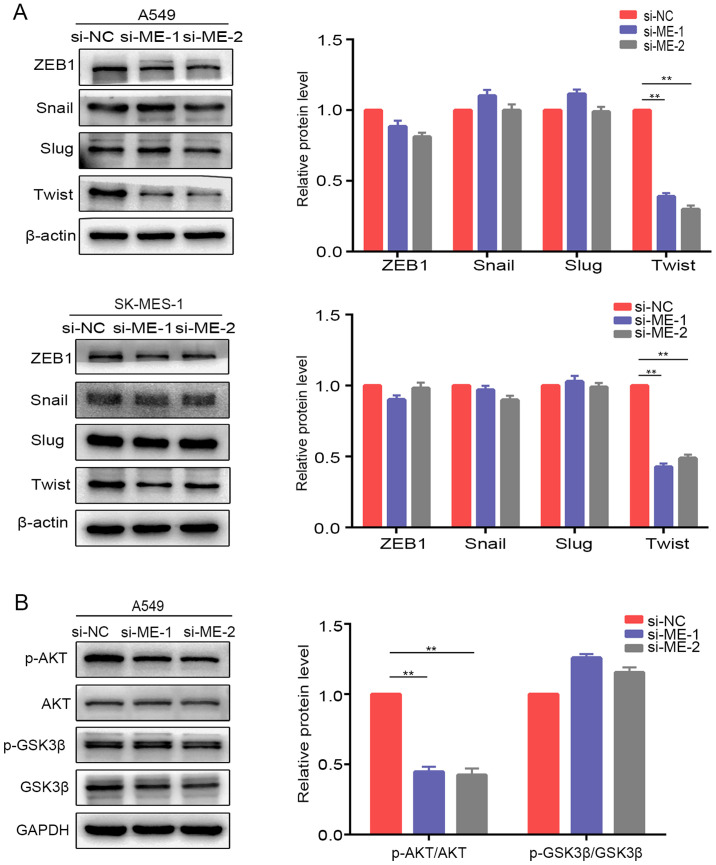
Silencing of METTL14 inhibits the activation of AKT signaling by downregulating Twist expression in NSCLC cells
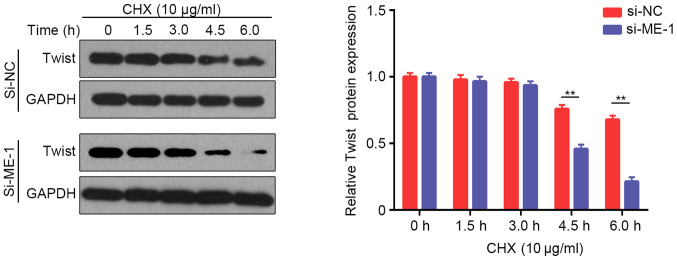
Considering that certain transcription factors (ZEB1, Snail, Slug and Twist) are involved in EMT regulation (24), the present study examined the protein expression of these factors after knockdown of METTL14 in A549 and SK-MES-1 cells. Western blotting revealed that Twist expression was downregulated after knockdown of METTL14 in A549 and SK-MES-1 cells, whereas there were no obvious alterations in ZEB1, Snail or Slug expression (Fig. 5A). These results suggested that METTL14 may promote EMT in NSCLC by regulating Twist expression. A previous study revealed that Twist can enhance cell invasion via the PI3K/AKT/GSK3β signaling pathway (25). Therefore, the present study examined the expression of proteins in this pathway. The levels of p-AKT/AKT were decreased in A549 cells after transfection with si-ME-1 (Fig. 5B). However, the levels of p-GSK3β/GSK3β exhibited no visible changes. To further evaluate whether METTL14 influenced the levels of p-AKT by regulating the stabilization of Twist, the present study analyzed the changes in Twist degradation after treatment with CHX, a protein synthesis inhibitor. The expression levels of Twist were examined at different time points (0, 1.5, 3, 4.5 and 6 h) after treatment with CHX. Inhibition of METTL14 using siRNA-1 accelerated Twist degradation (Fig. 6), which indicated that the stabilization of Twist was attenuated after METTL14 knockdown. Overall, these findings suggested that METTL14 knockdown may decrease the stability of Twist by affecting the levels of p-AKT.
Figure 5.
Silencing of METTL14 inhibits Twist expression via inactivation of AKT signaling in non-small cell lung cancer cells. (A) Expression levels of epithelial-mesenchymal transition-related proteins (ZEB1, Snail, Slug and Twist) in A549 and SK-MES-1 cells were detected by western blotting following knockdown of METTL14. (B) Levels of p-AKT, AKT, p-GSK3β and GSK3β were evaluated in A549 cells after knockdown of METTL14 **P<0.01 (one-way ANOVA with Tukey's post hoc test). METTL14/ME, methyltransferase-like 14; NC, negative control; p-, phosphorylated; si, small interfering RNA; ZEB1, zinc finger E-box binding homeobox 1.
Figure 6.
A549 cells were transfected with si-ME-1, and then treated with 10 µg/ml CHX for 0, 1.5, 3.0, 4.5 and 6.0 h. The expression levels of Twist were evaluated using western blotting. **P<0.01 (Student's unpaired t-test). CHX, cycloheximide; METTL14/ME, methyltransferase-like 14; NC, negative control; si, small interfering RNA.
Discussion
Due to the high mortality rate (accounting for 18.4% of global cancer-associated deaths in 2018), NSCLC remains a serious public health problem worldwide, and the prognosis of most patients remains poor due to tumor metastasis (26). Accumulating evidence has demonstrated that m6A regulators contribute to cancer progression and metastasis (27). However, limited studies have focused on the tumorigenic effects of m6A regulators in NSCLC. To the best of our knowledge, METTL14 has not been reported previously as a regulator of NSCLC progression, and the underlying mechanisms remain elusive. Therefore, clarification of the involvement of METTL14 in NSCLC carcinogenesis will be beneficial for improving the treatment of NSCLC.
METTL14 is an important subunit of the methyltransferase complex and serves a structural role in substrate recognition (28). METTL14 has been proposed as an oncogene in various tumors. For instance, METTL14 promotes the proliferation of Epstein Barr virus (EBV)-transformed cells and tumor growth in vivo and is considered to be a novel target for the treatment of EBV-associated tumors (29). METTL14 is highly expressed in acute myeloid leukemia cells and exerts its oncogenic role by modulating MYB proto-oncogene, transcription factor and MYC expression via m6A modification (30). A previous study also demonstrated that long intergenic non-protein coding RNA 942 promotes the proliferation of breast cancer cells by elevating the expression levels of METTL14 and its downstream targets, including C-X-C motif chemokine receptor 4 and cytochrome P450 family 1 subfamily B member 1 (31). The present study revealed that METTL14 expression was upregulated in NSCLC, and its expression was higher in patients with NSCLC with stage I, II, III and IV disease compared with that in patients in the control group. Additionally, the present study demonstrated that METTL14 knockdown inhibited cell proliferation and migration of lung cancer lines in vitro. Therefore, to the best of our knowledge, the present study was the first to demonstrate that METTL14 acted as an oncogene and promoted the growth of NSCLC. To further verify our conclusion, GSE31210 data were analyzed and it was revealed that METTL14 expression was higher in NSCLC tissues than in normal tissues. However, other conflicting studies have reported that the expression levels of METTL14 in lung adenocarcinoma and lung squamous cell carcinoma are downregulated (32,33). The discrepancies in METTL14 expression in NSCLC may be due to the different cell lines used. Downregulation of METTL14 has also been demonstrated in gastric (34), liver (35) and colon (36) cancer. Furthermore, there have been controversial reports regarding the role of METTL14 in liver cancer. One study revealed a decrease in m6A modification levels in human HCC, and depletion of METTL14 was an adverse prognostic factor for recurrence-free survival and promoted HCC metastasis (35). By contrast, another study reported that downregulation of METTL14 reduced proliferation and migration of liver cancer cells (37). The discrepancies in the findings of the aforementioned studies highlight that the expression levels and role of METTL14 may differ in different types of tumors. Further investigations will be required to address this controversy.
Various studies have demonstrated that EMT is associated with NSCLC progression (38–40). Interestingly, previous studies have demonstrated that m6A regulators exert crucial roles in the regulation of EMT (41,42). METTL3 has been demonstrated to be an essential protein in the development of ovarian cancer by regulating AXL receptor tyrosine kinase translation and EMT (41). YTHDF2 has been reported to promote carcinogenesis of pancreatic cancer via EMT (42). Consistently, the present study demonstrated that silencing METTL14 in NSCLC cells significantly increased E-cadherin expression and suppressed N-cadherin expression. These results suggested that METTL14 may participate in the EMT process in NSCLC. However, the present study compared the expression difference of METTL14 in the tissues of non-metastatic patients and metastatic patients based on TCGA, and the results demonstrated that the differences in tissue expression between non-metastatic patients and metastatic patients were not statistically significant. The limited samples may contribute to this result; a larger sample size will be collected and used in future work.
EMT is a process that is precisely regulated by several major transcription factors, including Twist, Snail, Slug and ZEB1 (24). Twist, acting as a crucial transcription factor of EMT, is considered to be involved in tumorigenesis and metastasis (43). In the present study, Twist expression was reduced after downregulation of METTL14 in NSCLC cells. Therefore, it was deduced that lower METTL14 expression may inhibit the EMT process by repressing Twist expression. Twist can induce EMT changes via the activation of the PI3K/AKT signaling pathway (44). Therefore, the present study explored the role of METTL14/Twist in the activation of AKT signaling pathways. Consistently, it was revealed that METTL14 downregulation suppressed the levels of p-AKT/AKT; however, the levels of p-GSK3β/GSK3β exhibited no visible changes. It was also found that Twist expression was attenuated in siRNA-METTL14-transfected lung cancer cells after treatment with CHX. Therefore, it was deduced that METTL14 knockdown decreased the stability of Twist by affecting the levels of p-AKT and, ultimately, inhibited the EMT process in NSCLC cells. However, determining the relationship between AKT signaling and Twist stability and function still requires further investigation.
More in vivo and in vitro experiments need to be performed to determine the role of METTL14 in NSCLC and its underlying molecular mechanism, which will provide a basis for the treatment of NSCLC.
In conclusion, the present study demonstrated that METTL14 expression was upregulated in NSCLC. In addition, knockdown of METTL14 inhibited Twist, restored E-cadherin expression by inactivating AKT signaling, and decreased the migration of NSCLC cells. To the best of our knowledge, the present study was the first to demonstrate that METTL14 expression was upregulated in NSCLC, and METTL14 can promote NSCLC progression via EMT. Taken together, these results indicated that METTL14 may modulate the metastatic potential of NSCLC cells.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was funded by the Key Research and Development Projects in Anhui Province (grant no. 201904a07020082).
Funding
The present study was funded by the Key Research and Development Projects in Anhui Province (grant no. 201904a07020082).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YQL conceived and designed the experiments. FY, WQY and JL performed the experiments and confirm the authenticity of all the raw data. FY and WQY analyzed the data and wrote the paper. YQL revised the paper. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Roche J. The epithelial-to-mesenchymal transition in cancer. Cancers (Basel) 2018;10:52. doi: 10.3390/cancers10020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Liu J, Chen WJ, Ye QQ, Chen WT, Li CL, Wu HT. Regulation of N6-methyladenosine in the differentiation of cancer stem cells and their fate. Front Cell Dev Biol. 2020;8:561703. doi: 10.3389/fcell.2020.561703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Zhuo L, Wang J, Zhang Q, Li Q, Li G, Yan L, Jin T, Pan T, Sui X, et al. METTL3 plays multiple functions in biological processes. Am J Cancer Res. 2020;10:1631–1646. [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian R, Zhang S, Sun D, Bei C, Li D, Zheng C, Song X, Chen M, Tan S, Zhu X, et al. M6A Demethylase FTO plays a tumor suppressor role in thyroid cancer. DNA Cell Biol. 2020;39:2184–2193. doi: 10.1089/dna.2020.5956. [DOI] [PubMed] [Google Scholar]
- 10.Guo T, Liu DF, Peng SH, Xu AM. ALKBH5 promotes colon cancer progression by decreasing methylation of the lncRNA NEAT1. Am J Transl Res. 2020;12:4542–4549. [PMC free article] [PubMed] [Google Scholar]
- 11.Lasman L, Krupalnik V, Viukov S, Mor N, Aguilera-Castrejon A, Schneir D, Bayerl J, Mizrahi O, Peles S, Tawil S, et al. Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev. 2020;34:1373–1391. doi: 10.1101/gad.340695.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Wan Y, Zhang Z, Jiang Y, Lang J, Cheng W, Zhu L. FTO demethylates m6A modifications in HOXB13 mRNA and promotes endometrial cancer metastasis by activating the WNT signaling pathway. RNA Biol. 2021;18:1265–1278. doi: 10.1080/15476286.2020.1841458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma XX, Cao ZG, Zhao SL. m6A methyltransferase METTL3 promotes the progression of prostate cancer via m6A-modified LEF1. Eur Rev Med Pharmacol Sci. 2020;24:3565–3571. doi: 10.26355/eurrev_202004_20817. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Jiang X, Li D, Jiang X. HBXIP promotes gastric cancer via METTL3-mediated MYC mRNA m6A modification. Aging (Albany NY) 2020;12:24967–24982. doi: 10.18632/aging.103767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu N, Qin S, Zhang X, Bo X, Liu Z, Tan C, Wen G, Jiang H. Multiple m6A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer. 2020;20:165. doi: 10.1186/s12885-020-6638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Kang M, Zhang B, Meng F, Song J, Kaneko H, Shimamoto F, Tang B. m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer. 2019;18:185. doi: 10.1186/s12943-019-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, Zheng H, Li B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512:479–485. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Liu S, Zhao T, Dang C. METTL3 mediated m6A modification of Bcl-2 mRNA promotes non small cell lung cancer progression. Oncol Rep. 2021;46:163. doi: 10.3892/or.2021.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Z, Qian Q, Zhao X, Ma L, Chen P. N6-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene. 2020;731:144348. doi: 10.1016/j.gene.2020.144348. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Xiao D, Qiu T, Li J, Liu Z. Loading MicroRNA-376c in extracellular vesicles inhibits properties of non-small cell lung cancer cells by targeting YTHDF1. Technol Cancer Res Treat. 2020 Dec 7; doi: 10.1177/1533033820977525. (Epub ahead of print). doi: 10.1177/1533033820977525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, Li X, Xu S, Miao J, Guo J, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N6 adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19:130. doi: 10.1186/s12943-020-01249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi D, Wang R, Shi X, Xu L, Yilihamu Y, Sang J. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6 methyladenosine and hsa miR 146a 5p expression. Oncol Rep. 2020;43:1375–1386. doi: 10.3892/or.2020.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 24.Ahmadiankia N, Khosravi A. Significance of epithelial-to-mesenchymal transition inducing transcription factors in predicting distance metastasis and survival in patients with colorectal cancer: A systematic review and meta-analysis. J Res Med Sci. 2020;25:60. doi: 10.4103/jrms.JRMS_174_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng L, Li N, Guo F, Jian XC, Jiang CH, Yin P, Min AJ, Huang L. Twist-related protein 1 enhances oral tongue squamous cell carcinoma cell invasion through β-catenin signaling. Mol Med Rep. 2015;11:2255–2261. doi: 10.3892/mmr.2014.2904. [DOI] [PubMed] [Google Scholar]
- 26.Ni L, Li Z, Shi X, Yao C, Sun J, Ai M, Li S, Liu Y, Xu F, Zhou Y, et al. Rosthorin A inhibits non-small cell lung cancer cell growth and metastasis through repressing epithelial-mesenchymal transition via downregulating Slug. Anticancer Drugs. 2020;31:997–1003. doi: 10.1097/CAD.0000000000000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, Yang D, Zheng ZC, Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 29.Lang F, Singh RK, Pei Y. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 2019;15:e1007796. doi: 10.1371/journal.ppat.1007796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205.e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, Lu S, Xu D, Wu Y, Chen Q, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39:5358–5372. doi: 10.1038/s41388-020-1338-9. [DOI] [PubMed] [Google Scholar]
- 32.Li F, Wang H, Huang H, Zhang L, Wang D, Wan Y. m6A RNA Methylation regulators participate in the malignant progression and have clinical prognostic value in lung adenocarcinoma. Front Genet. 2020;11:994. doi: 10.3389/fgene.2020.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Xiao M, Zhang L, Li L, Zhu G, Shen E, Lv M, Lu X, Sun Z. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. 2021;35:e23655. doi: 10.1002/jcla.23655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Zhang S, He C, Xue P, Zhang L, He Z, Zang L, Feng B, Sun J, Zheng M, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Yang S. PRDX2 promotes the proliferation and metastasis of non-small cell lung cancer in vitro and in vivo. Biomed Res Int. 2020 Aug 27; doi: 10.1155/2020/8359860. (Epub ahead of print). doi: 10.1155/2020/8359860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L, Chen Y, Jing L, Nie K, Xiao Y, Yu H. MicroRNA-629 promotes the tumorigenesis of non-small-cell lung cancer by targeting FOXO1 and activating PI3K/AKT pathway. Cancer Biomark. 2020;29:347–357. doi: 10.3233/CBM-201685. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Y, Ye X, Chen R, Gao Q, Zhao D, Ling C, Qian Y, Xu C, Tao M, Xie Y. Sirtuin 7 promotes non small cell lung cancer progression by facilitating G1/S phase and epithelial mesenchymal transition and activating AKT and ERK1/2 signaling. Oncol Rep. 2020;44:959–972. doi: 10.3892/or.2020.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, Duan P. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151:356–365. doi: 10.1016/j.ygyno.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, Lu Y, Zeng J, Du F, Gong A, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16:2259–2271. doi: 10.1080/15384101.2017.1380125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, Zhang B, Xu N, Wang MJ, Liu Q. miR-326 regulates EMT and metastasis of endometrial cancer through targeting TWIST1. Eur Rev Med Pharmacol Sci. 2017;21:3787–3793. [PubMed] [Google Scholar]
- 44.Yu X, Zheng Y, Zhu X, Gao X, Wang C, Sheng Y, Cheng W, Qin L, Ren N, Jia H, et al. Osteopontin promotes hepatocellular carcinoma progression via the PI3K/AKT/Twist signaling pathway. Oncol Lett. 2018;16:5299–5308. doi: 10.3892/ol.2018.9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.