Abstract
Adenosine 5'‐monophosphate (AMP)‐activated protein kinase (AMPK) is an important cellular metabolite‐sensing enzyme that can directly sense changes not only in ATP but also in metabolites associated with carbohydrates and fatty acids. However, less is known about whether and how AMPK senses variations in cellular amino acids. Here, we show that cysteine deficiency significantly triggers calcium/calmodulin‐dependent protein kinase kinase 2 (CaMKK2)‐mediated activation of AMPK. In addition, we found that CaMKK2 directly associates with cysteinyl‐tRNA synthetase (CARS), which then binds to AMPKγ2 under cysteine deficiency to activate AMPK. Interestingly, we discovered that cysteine inhibits the binding of CARS to AMPKγ2, and thus, under cysteine deficiency conditions wherein the inhibitory effect of cysteine is abrogated, CARS mediates the binding of AMPK to CaMKK2, resulting in the phosphorylation and activation of AMPK by CaMKK2. Importantly, we demonstrate that blocking AMPK activation leads to cell death under cysteine‐deficient conditions. In summary, our study is the first to show that CARS senses the absence of cysteine and activates AMPK through the cysteine–CARS–CaMKK2–AMPKγ2 axis, a novel adaptation strategy for cell survival under nutrient deprivation conditions.
Keywords: AMPK, CARS, cell survival, cyst(e)ine
Subject Categories: Metabolism, Signal Transduction
AMP‐activated protein kinase AMPK senses low amino acid levels through a cysteine‐CARS‐CaMKK2‐AMPKγ2 axis.
Introduction
Sensing changes in external nutrient levels is essential for cellular activities and survival (Efeyan et al, 2015). (AMP)‐activated protein kinase (AMPK) is one of the most important energy‐sensing enzymes in cells and is activated when energy levels are deficient (Hardie et al, 2012; Jeon, 2016; Herzig & Shaw, 2018). AMPK is a heterotrimer composed of α, β and γ subunits, and when activated, AMPK can promote cell survival by inhibiting anabolism, promoting catabolism, and reducing reactive oxygen species (ROS) levels (Jeon, 2016; Hayes et al, 2020). When the nutrient supply in the cells is restored, the ATP/AMP ratio increases to inhibit the activity of AMPK. With the identification of AMP‐binding sites on the AMPKγ subunit isoforms, researchers realized that AMPK activation is achieved upon the changes in the level of ATP, the energy currency of cells (Xiao et al, 2007; Gu et al, 2017). However, this simple model cannot explain how AMPK responds instantaneously to complex cellular metabolic processes and precisely regulates downstream genes. A recent study showed that glycogen inhibits AMPK activity by directly binding to the glycogen‐binding site on the AMPKβ subunit isoforms (McBride et al, 2009). Zhang et al found that AMPK can be activated by sensing a decrease in the glycolytic intermediate fructose‐1'6 diphosphate before the AMP/ATP ratio is changed upon induced glucose deficiency (Zhang et al, 2017a; Li et al, 2019). Long‐chain fatty acids bind to the ADaM site of the AMPKβ1 subunit, thereby allosterically activating AMPK (Pinkosky et al, 2020). Thus, in addition to sensing the AMP/ATP ratio, AMPK also has fine‐tuned functions for sensing variations in glucose‐ and fatty acid‐derived metabolites. It is currently unclear whether and how AMPK responds to variations in other nutrients or metabolites, such as amino acids. Hence, further identification of the molecular basis with respect to these factors is vitally important to help us gradually discover the precise regulatory mechanism of AMPK involved in different cellular metabolic processes.
Amino acids not only participate in the translation of proteins but also support various other metabolic processes, such as nucleotide synthesis and redox homeostasis (Wu, 2009). Therefore, cellular sensing of different amino acid levels is important for cell growth, and many intracellular amino acid sensors have been identified. For example, GCN2 indirectly senses all twenty kinds of amino acids by combining with free tRNA (Dong et al, 2000). SLC38A9 and CASTOR1 were identified as arginine sensors; Sestrin2 and LARS were identified as leucine sensors; and all these sensors eventually activate mTORC1 (Han et al, 2012; Wang et al, 2015; Chantranupong et al, 2016; Wolfson et al, 2016; Wyant et al, 2017). It has been reported that 20 aminoacyl tRNA synthase (AARS) enzymes can sense the concentration of all 20 amino acids in cells by recognizing aminoacylation modifications of lysine residues on the corresponding protein (He et al, 2018). Although amino acid‐sensing pathways have been described in general, for many amino acids, only some universal sensing pathways have been identified, which obviously cannot explain the specific metabolic activities of different amino acids. Therefore, the identification of different amino acid‐specific sensing molecules will help us understand complex amino acid metabolism pathways.
In this study, by subjecting cells to conditions in which a single amino acid is absent, we discovered that cystine starvation significantly activated AMPK. By promoting the binding of AMPKγ2 to CaMKK2 upon cystine deficiency, the cysteine‐specific sensor CARS is important for AMPK activation. These findings not only provide a better understanding of the regulatory role of AMPK in amino acid sensing but also suggest a combination strategy of cystine starvation and CaMKK2 inhibitor and/or AMPK inhibitor treatment for potential cancer therapy.
Results
Cystine deprivation activates AMPK through CaMKK2
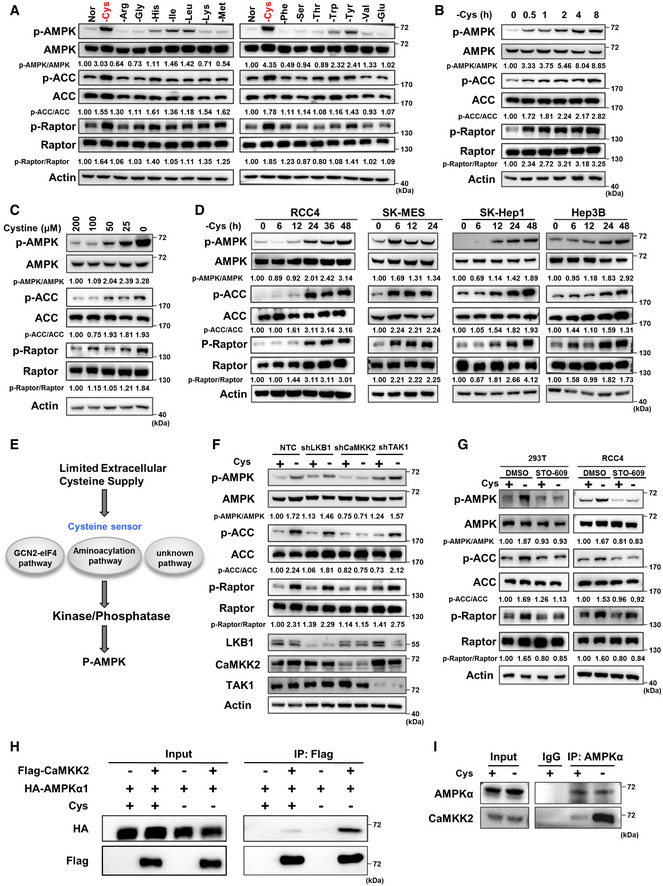
The AMPK pathway is activated by intermediate products of carbohydrate or fatty acid metabolism to promote cell survival (McBride et al, 2009; Zhang et al, 2017a; Pinkosky et al, 2020); however, it remains largely unclear whether AMPK directly senses changes in amino acid metabolites. To investigate the connection between the AMPK pathway and amino acid metabolism, we cultured cells in DMEM without selected individual amino acids. The results showed that, of the 15 amino acids tested, cystine deprivation markedly activated AMPK, as indicated by its phosphorylation, in both 293T cells and RCC4 cells, although tryptophan, tyrosine, isoleucine or leucine deprivation also activated AMPK (Figs 1A and EV1A). Thus, we focused on cystine deprivation and AMPK activation. It is worth mentioning that cysteine, a component of the culture medium, is easily oxidized, so cystine is added to form complete culture medium. After entering the cells, cystine will be decomposed into cysteine and participate in cell metabolism. Further experiments showed that cystine deprivation activated AMPK and its substrates including ACC and Raptor in a time‐dependent manner, starting 30 min after cystine was removed from the culture medium (Fig 1B). A cystine concentration gradient assay also showed that the gradual activation of AMPK and its substrates in 293T cells was correlated with cystine concentration in the culture medium (Fig 1C), confirming that cystine deprivation activates AMPK and AMPK‐mediated phosphorylation of its substrates including ACC and Raptor. Similar results were observed in other cell lines, including RCC4, SK‐MES, SK‐Hep1 and Hep3B cells (Fig 1D).
Figure 1. Cystine deprivation activates AMPK through CaMKK2.
-
A293T cells were cultured in complete medium made of dialysed serum for 24 h which was then replaced with medium in which one amino acid was eliminated and cultured for 8 h. Then, WB was used to measure p‐AMPK, p‐ACC, p‐Raptor and total AMPK, ACC, Raptor protein expression. “Nor” = normal medium. Actin served as the loading control. The red text "‐Cys" indicates that cystine deficiency most significantly activates AMPK and its substrates.
-
B–D293T cells were treated with cystine‐deficient medium for 0, 0.5, 1, 2, 4 or 8 h (B) or treated with medium containing 200, 100, 50, 25 or 0 µM cystine for 8 h (C); RCC4, SK‐MES, SK‐hep‐1 or Hep3B cells were treated with cystine‐deficient medium for indicated time (D). Protein levels of p‐AMPK, p‐ACC, p‐Raptor and total AMPK, ACC, Raptor were measured by WB, and actin served as the loading control.
-
EA schematic diagram of the hypothetical mechanism showing how cystine deprivation induces AMPK activation.
-
FWB analysis of p‐AMPK, p‐ACC, p‐Raptor and total AMPK, ACC, Raptor protein in 293T cells transfected with shRNAs targeting LKB1, CaMKK2, TAK1 or a non‐targeting control (NTC) that were treated with cystine‐deficient medium for 8 h. Actin served as the loading control.
-
GWB analysis of p‐AMPK, p‐ACC, p‐Raptor and total AMPK, ACC, Raptor protein in 293T cells and RCC4 cells treated with 1 µg/ml STO‐609 or DMSO for 8 h during cystine deprivation. Actin served as the loading control.
-
H293T cells were transfected with HA‐AMPKα1 alone or together with Flag‐CaMKK2 for 48 h and then cultured with cystine‐deficient medium or complete medium for 8 h. Cell lysates were immunoprecipitated with anti‐Flag, and then, WB analysis was performed.
-
I293T cells cultured with cystine‐deficient medium or complete medium for 8 h were harvested and subjected to immunoprecipitation with anti‐AMPKα, followed by WB analysis with anti‐AMPKα and anti‐CaMKK2.
Source data are available online for this figure.
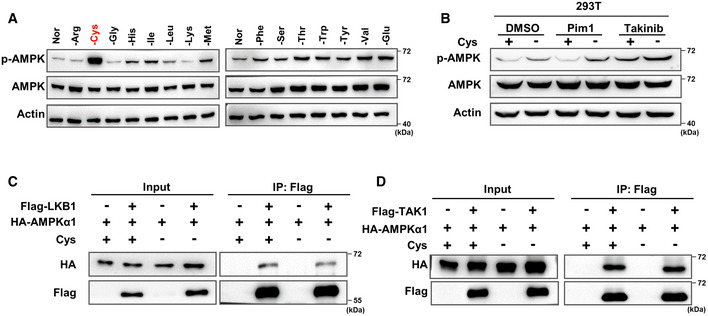
Figure EV1. Cystine starvation activates AMPK through CaMKK2 instead of LKB1 or TAK1.
-
ARCC4 cells were cultured in complete medium for 24 h and then replaced and cultured with the corresponding amino acid‐deficient medium for 24 h, followed by WB detection of p‐AMPK and total AMPK protein expression. Actin served as the loading control.
-
BWB analysis of p‐AMPK and total AMPK in 293T cells treated with DMSO, 1 µM Pim1 or 100 nM Takinib for 8 h under cystine deprivation. Actin served as the loading control.
-
C, D293T cells were transfected with HA‐AMPKα1 alone or with Flag‐LKB1 (C) or Flag‐TAK1 (D) alone for 48 h and then cultured with cystine‐deficient medium for 8 h. Cell lysates were immunoprecipitated with anti‐Flag antibody, and a WB analysis was performed.
Source data are available online for this figure.
Next, we sought to investigate the molecular mechanism by which cystine deprivation activates AMPK. We hypothesized that there might be at least two steps required to activate AMPK during cystine deprivation. First, cystine deficiency is sensed via a specific sensing mechanism, such as the known GCN2‐eIF4 pathway, aminoacylation pathway (Dong et al, 2000; He et al, 2018) or another unknown pathway(s), and second, the sensor protein transmits this deprivation signal to AMPK kinases or phosphatases, leading to the phosphorylation of AMPK (Fig 1E). To test this hypothesis, we first knocked down three AMPK kinases, LKB1, CaMKK2 and TAK1, by specific shRNAs. Interestingly, we observed that knocking down CaMKK2, but not LKB1 or TAK1, significantly inhibited the AMPK phosphorylation induced by cystine deprivation, and AMPK‐mediated ACC and Raptor phosphorylation (Fig 1F). Consistently, cystine deprivation‐induced AMPK phosphorylation and subsequent AMPK‐mediated ACC and Raptor phosphorylation were suppressed by a CaMKK2 inhibitor (STO‐609) but not by inhibitors of LKB1 (Pim1) or TAK1 (Takinib) (Figs 1G and EV1B), suggesting that CaMKK2 is involved in cystine deprivation‐induced AMPK activation. Since AMPK kinases directly bind to and phosphorylate the α subunit of AMPK, we first tested the binding of these three kinases to AMPKα1 during cystine starvation. Our results of the coimmunoprecipitation assay demonstrated that cystine deprivation enhanced the binding of CaMKK2 to AMPKα1 without affecting the association of LKB1 or TAK1 with AMPKα1 (Figs 1H and EV1EV1C and D). More importantly, our results also revealed that endogenous AMPKα1 interacted with CaMKK2 during cystine starvation (Fig 1I). These results indicate that the binding of CaMKK2 and AMPKα1 is enhanced under cystine deprivation conditions to facilitate AMPK and its substrate activation.
CARS is critical for CaMKK2‐mediated AMPK activation under cystine deprivation conditions
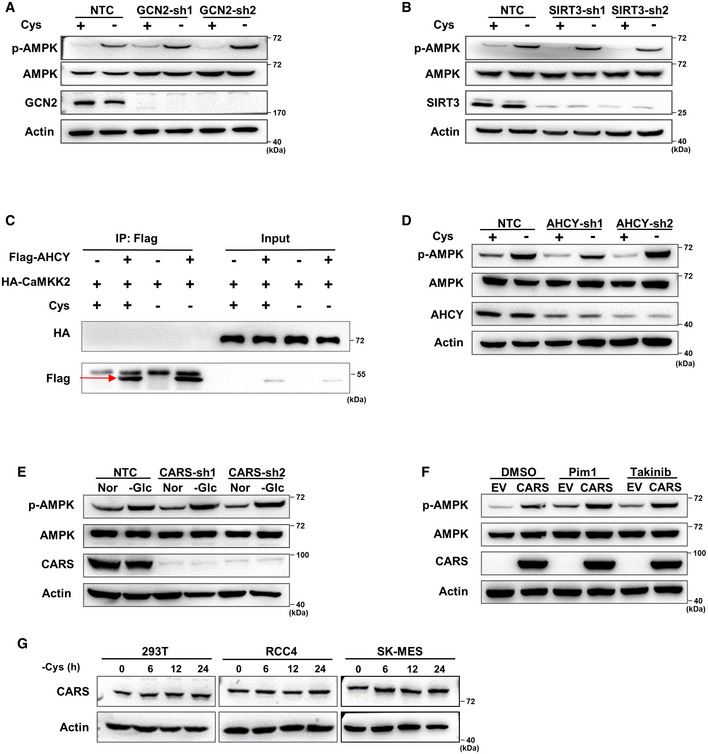
To explore whether the two known pathways are involved in the sensing of cystine starvation (Fig 1E), we next independently knocked down GCN2 and SIRT3 using specific shRNAs to block the GCN2‐eIF4 and aminoacylation pathways, respectively (Dong et al, 2000; He et al, 2018). The results showed that the suppression of GCN2 or SIRT3 led to no significant changes in AMPK phosphorylation (Fig EV2A and B), suggesting that neither the GCN2‐eIF4 nor aminoacylation pathways are involved in cystine deprivation‐induced AMPK activation. To identify the molecule that senses cystine changes, we speculated that the sensor is likely an upstream factor that binds to CaMKK2. Thus, we performed an immunoprecipitation assay with an anti‐CaMKK2 antibody followed by mass spectrometry analysis (IP‐MS) to identify the CaMKK2‐binding proteins under cystine deprivation conditions. By analysing the mass spectrometry results, we found that 238 proteins are potentially associated with CaMKK2, of which two proteins, AHCY and CARS, are known to be involved in cyst(e)ine metabolism (Fig 2A). Coimmunoprecipitation experiments revealed that CARS, but not AHCY, interacted with CaMKK2 in the presence or absence of cystine (Figs 2B and EV2C). Our further endogenous immunoprecipitation experiments revealed that CaMKK2 interacted with CARS independent of cystine concentration (Fig 2C). In addition, Flag pull‐down assays using prokaryotically expressed Flag‐CaMKK2 and His‐tagged CARS proteins revealed that CARS directly binds to CaMKK2 (Fig 2D). These results demonstrate the binding of CARS to CaMKK2, suggesting a novel function in the activation of AMPK during cystine deprivation.
Figure EV2. CARS is critical for AMPK activation under cystine starvation condition.
-
A, BWB analysis of p‐AMPK and total AMPK protein expression in 293T cells transfected with shRNAs targeting GCN2 (A) or Sirt3 (B) and further treated with cystine‐deficient medium for 8 h. Actin served as the loading control.
-
C293T cells were transfected with HA‐CaMKK2 alone or with Flag‐AHCY for 48 h and then cultured with cystine‐deficient medium or complete medium for 8 h. Cell lysates were immunoprecipitated with anti‐Flag, and a WB analysis was performed. The red arrow indicates the target band.
-
DWB analysis of p‐AMPK and total AMPK protein expression in 293T cells transfected with shRNAs targeting AHCY and further treated with cystine‐deficient medium for 8 h. Actin served as the loading control.
-
EWB analysis of p‐AMPK and total AMPK protein expression in 293T cells transfected with shRNAs targeting CARS and further treated with glucose‐deficient medium for 2 h. Actin served as the loading control.
-
FWB analysis of p‐AMPK and total AMPK protein expression in CARS‐overexpressing 293T cells treated with DMSO, pim1, or Takinib for 8 h. Actin served as the loading control.
-
GWB analysis of CARS protein expression in 293T, RCC4 and SK‐MES cells treated with cystine‐deficient medium for 0, 6, 12 or 24 h. Actin served as the loading control.
Source data are available online for this figure.
Figure 2. CARS is critical for AMPK activation under cystine deprivation conditions.
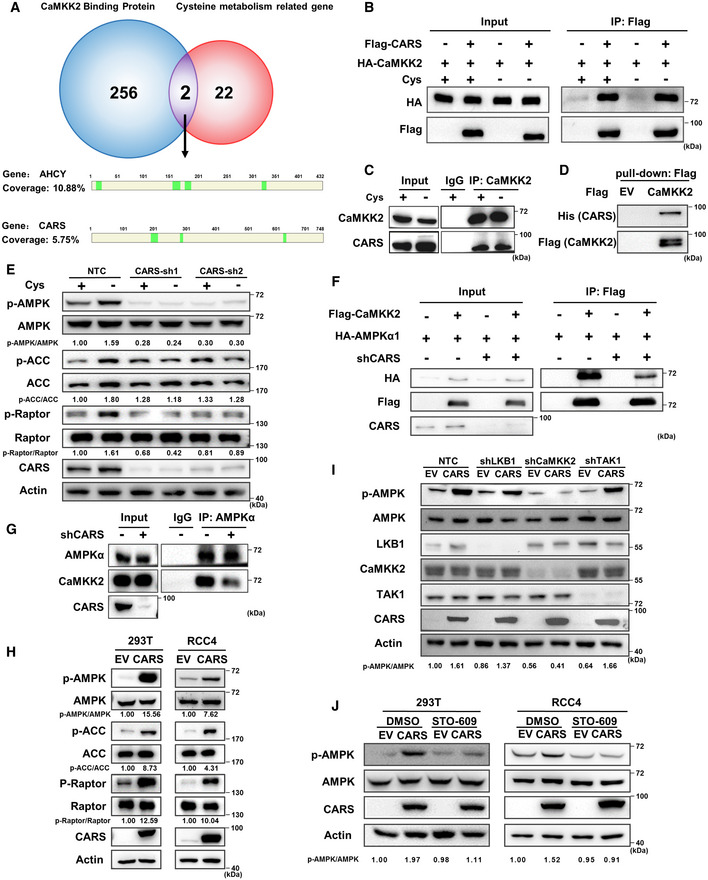
- Venn diagram showing the overlap of CaMKK2‐binding proteins during cystine deprivation and cyst(e)ine metabolism‐related proteins. The schematic diagram shows the peptides detected in the results of IP‐MS assays of the AHCY and CARS proteins. The CaMKK2‐binding proteins are listed in Table EV3, and the cyst(e)ine metabolism‐related proteins are listed in Table EV4.
- 293T cells were transfected with HA‐CaMKK2 alone or with Flag‐CARS for 48 h and then cultured with cystine‐deficient medium or complete medium for 8 h. Cell lysates were immunoprecipitated with anti‐Flag, and a WB analysis was performed.
- 293T cells cultured with cystine‐deficient medium or complete medium for 8 h were harvested and subjected to immunoprecipitation with anti‐CaMKK2, followed by WB analysis with anti‐CaMKK2 and anti‐CARS.
- Flag pull‐down assays were performed with Flag‐EV or Flag‐fused CaMKK2 protein and His‐CARS purified from E. coli.
- WB analysis of p‐AMPK, p‐ACC, p‐Raptor and total AMPK ACC, Raptor protein expression in 293T cells transfected with shRNAs targeting CARS that were further treated with cystine‐deficient medium for 8 h. Actin served as the loading control.
- 293T cells were first transfected with shRNAs targeting CARS or NTC, then transfected with HA‐AMPKα1 or HA‐AMPKα1 and Flag‐CaMKK2 for 48 h, and then, they were cultured with cystine‐deficient medium for 8 h. Cell lysates were immunoprecipitated with anti‐Flag and subjected to Western blot analysis.
- 293T cells were transfected with shRNAs targeting CARS or NTC; then, they were cultured with cystine‐deficient medium for 8 h. Cell lysates were immunoprecipitated with anti‐AMPKα and subjected to WB analysis with anti‐AMPKα and anti‐CaMKK2.
- WB analysis of p‐AMPK, p‐ACC, p‐Raptor and total AMPK, ACC, Raptor, CARS protein expression in 293T cells and RCC4 cells overexpressing Flag‐CARS. Actin served as the loading control.
- WB analysis of p‐AMPK and total AMPK protein expression in CARS‐overexpressing 293T cells were further transfected with shRNAs targeting LKB1, CaMKK2 or TAK1. Actin served as the loading control.
- WB analysis of p‐AMPK and total AMPK protein expression in CARS‐overexpressing 293T cells and RCC4 cells that were treated with 1 µg/ml STO‐609 or DMSO for 8 h. Actin served as the loading control.
Source data are available online for this figure.
To directly test the roles of CARS in AMPK activation induced by cystine deprivation, we knocked down CARS by shRNAs, and our results showed that AMPK and its substrate activation was dramatically abrogated by shCARS (Fig 2E). However, knocking down AHCY had no effect on cystine deprivation‐induced AMPK activation (Fig EV2D), which was consistent with our early observation that AHCY did not interact with CaMKK2 (Fig EV2C). Moreover, knockdown of CARS attenuated the binding of CaMKK2 to AMPKα1 under cystine deprivation conditions (Fig 2F), similar results were observed by endogenous immunoprecipitation experiments (Fig 2G). These results indicate that CARS is important for CaMKK2‐mediated AMPK and its substrate activation. Notably, the suppression of CARS had no effect on glucose deprivation‐induced AMPK activation (Fig EV2E), suggesting that CARS may specifically sense cystine deprivation to activate AMPK.
Moreover, we observed that overexpression of CARS significantly activated AMPK and its substrates in both 293T and RCC4 cells, and AMPK activation was attenuated by the suppression of CaMKK2 but not by suppression of the AMPK kinase LKB1 (Pim1) or TAK1 (Takinib) (Fig 2H and I). Consistently, CARS overexpression‐induced AMPK activation was abolished by the CaMKK2 inhibitor STO‐609 but not by inhibitors of LKB1 or TAK1 (Figs 2J and EV2F). We therefore concluded that, similar to cystine deprivation, overexpression of CARS activates AMPK and its substrates through CaMKK2. However, our Western blot results showed that cystine starvation had no effect on CARS protein expression, indicating that cystine deprivation‐induced AMPK activation is not caused by upregulation of the CARS protein (Fig EV2G). Taken together, these results provide evidence that CARS mediates AMPK activation induced by cystine deficiency.
CARS senses cystine starvation to activate AMPK by binding to AMPKγ2
At this point in our study, the data showed that CARS constitutively interacted with CaMKK2, regardless of cystine concentration (Fig 2B and C), while the binding of CaMKK2 and AMPKα1 was enhanced by cystine starvation and was dependent on CARS (Figs 1H and I, and 2F and G). Next, we sought to determine how CARS transmits the cystine deficiency signal to CaMKK2. Since the AMPK complex is composed of three subunits, α, β and γ, we first performed pull‐down assays using purified prokaryotically expressed His‐tagged CARS (Fig EV3A) and HA‐tagged AMPK subunit proteins. Our results indicated that only the HA‐tagged AMPKγ2 subunit was pulled down by the His‐tagged CARS protein (Fig 3A), suggesting that CARS directly binds to the AMPKγ2 subunit. Additional pull‐down experiments using GST‐tagged AMPKγ1 or AMPKγ2 protein confirmed the direct interaction of AMPKγ2 and CARS (Fig 3B). Since previous reports have suggested the functional implications of the homology differences in AMPKγ subunits and the tissue expression profile in organisms (Mahlapuu et al, 2004; Willows et al, 2017), we further compared the amino acid sequences of AMPKγ2 and AMPKγ1 (Fig EV3B) and found that the C‐terminus of AMPKγ2 (265–569 amino acids) is basically the same as the full‐length sequence of AMPKγ1, with the only difference in individual amino acids, while the N‐terminus (1–264 amino acids) of AMPKγ2 is unique. Therefore, we hypothesized that AMPKγ2 binds to CARS primarily through its N‐terminus. Pull‐down experiments with truncated AMPKγ2 protein and CARS protein purified from Escherichia coli revealed that AMPKγ2 binds to CARS via its N‐terminus (Fig 3C). On the other hand, based on the CARS protein structure analysis via the online website UniProt (http://www.ebi.ac.uk/interpro/), we constructed three vectors expressing the N‐terminus (N), middle fragment (M) and C‐terminus (C) of the CARS proteins, respectively, in which the N‐terminus fragment contains cysteine‐binding and catalytic domains, and the middle fragment contains tRNA‐binding domain. (Fig EV3C, upper panel). The results of Co‐IP experiments demonstrated that CARS binds to AMPKγ2 through its N‐terminus (Fig EV3C, bottom panel). Our data also showed that the AMP‐binding site mutant AMPKγ2‐R531G (Xiao et al, 2007) exhibited a similar effect as wild‐type AMPKγ2 on AMPK activation under cystine starvation conditions, suggesting that cystine starvation‐induced activation of AMPK is independent of intracellular ATP/AMP (Fig EV3D).
Figure EV3. CARS senses cystine starvation to activate AMPK through direct combination with AMPKγ2.
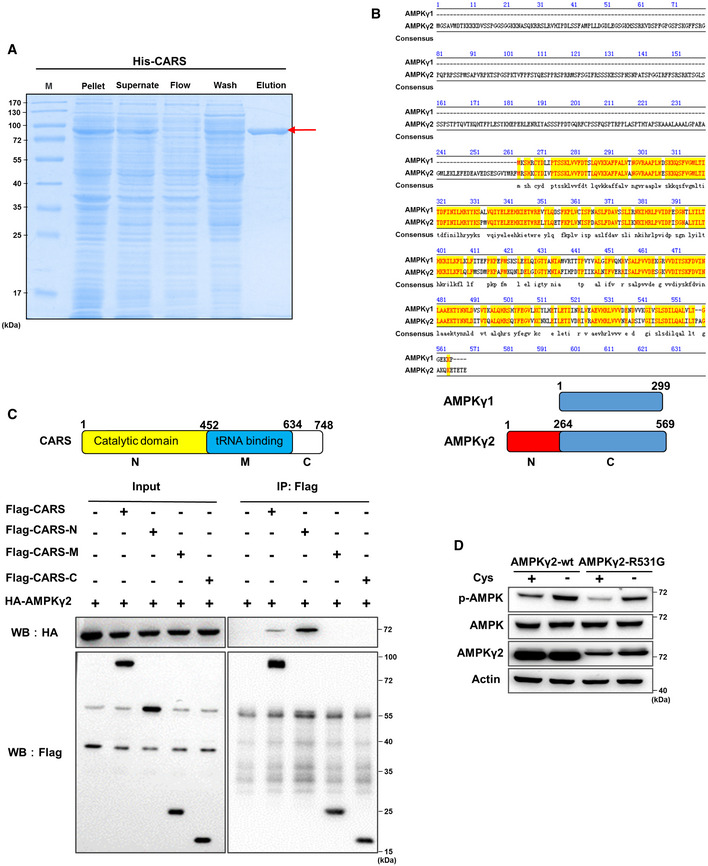
- Coomassie Blue staining of His‐CARS protein purified from E. coli. “M” = marker. The red arrow indicates the target band.
- Protein sequences of AMPKγ1 and AMPKγ2 were compared using Jellyfish software. The model below shows the segments of AMPKγ1 and AMPKγ2.
- 293T cells were transfected with HA‐AMPKγ2 alone or with Flag‐CARS, Flag‐CARS‐N‐terminus, Flag‐CARS‐M domain or Flag‐CARS‐C‐terminus for 48 h. Cell lysates were immunoprecipitated with anti‐Flag, and WB analysis was performed. The model above shows the segments of CARS.
- In 293T cells transfected with endogenous knocked down AMPKγ2, wild‐type AMPKγ2 or AMPKγ2R531G was treated with cystine‐deficient medium for 8 h, and WB was performed to detect of p‐AMPK, total AMPK and AMPKγ2 protein expression. Actin served as the loading control.
Source data are available online for this figure.
Figure 3. CARS activates AMPK by sensing cystine deficiency upon binding to AMPKγ2.
-
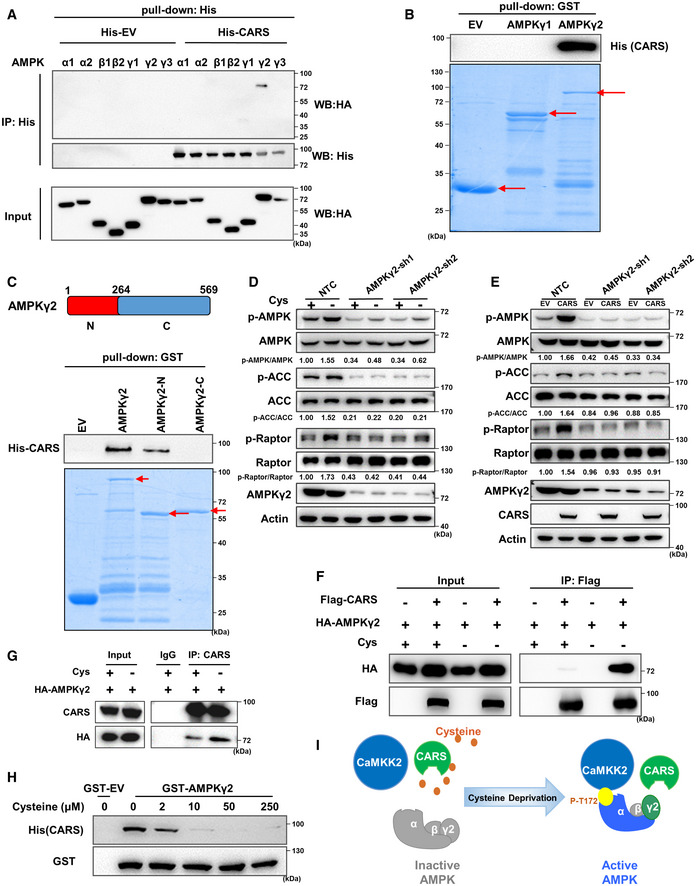
AHis pull‐down assay was performed with His‐EV‐ or His‐fused CARS protein and HA‐tagged AMPK subunit proteins overexpressed in 293T cells.
-
B, CGST pull‐down assay was performed with GST‐EV, GST‐AMPKγ1, GST‐AMPKγ2 (B) or GST‐EV, GST‐AMPKγ2, GST‐AMPKγ2‐N, GST‐AMPKγ2‐C (C) proteins and His‐tagged CARS protein purified from E. coli. The segments of AMPKγ2 are shown in the bar graph above (C). The red arrows indicate the target bands.
-
D, EWB analysis of p‐AMPK, p‐ACC, p‐Raptor and total AMPK, ACC, Raptor, AMPKγ2 protein expression in 293T cells transfected with shRNAs targeting AMPKγ2 that were further treated with cystine‐deficient medium for 8 h (D) or transfected with Flag‐CARS for 48 h (E). Actin served as the loading control.
-
F293T cells were transfected with HA‐AMPKγ2 alone or with Flag‐CARS for 48 h and then cultured with cystine‐deficient medium or complete medium for 8 h. Cell lysates were immunoprecipitated with anti‐Flag, and a WB analysis was performed.
-
G293T cells were transfected with HA‐AMPKγ2 for 48 h and then cultured with cystine‐deficient medium or complete medium for 8 h. Cell lysates were immunoprecipitated with anti‐CARS, followed by WB analysis with anti‐HA and anti‐CARS.
-
HGST pull‐down of His‐CARS by GST‐AMPKγ2 using protein purified from E. coli, followed by WB analysis. Cysteine (0, 2, 10, 50 and 250 μM) was added to the experimental buffer.
-
IThe schematic diagram shows that cystine deficiency‐induced AMPK activation is mediated by CaMKK2, which is recruited by CARS and the AMPKγ2 complex.
Source data are available online for this figure.
Moreover, we found that suppression of AMPKγ2 by shRNAs abolished cystine deprivation‐ or CARS overexpression‐induced AMPK and its substrate activation (Fig 3D and E), demonstrating that AMPKγ2 is essential for cystine deprivation‐ or CARS overexpression‐induced AMPK activation. More interestingly, we found that cystine deficiency promoted the binding of CARS and AMPKγ2 (Fig 3F and G), and with increasing cystine concentration, the binding of CARS and AMPKγ2 decreased gradually (Fig 3H). These results suggest that cysteine competitively inhibits the binding of CARS and AMPKγ2. In the presence of cyst(e)ine, cysteine occupied CARS at the N‐terminus, which blocked the interaction of CARS with AMPKγ2. However, when cystine was eliminated from the medium, CARS proteins were able to sense intracellular cyst(e)ine deficiency and bind to AMPKγ2, thus recruiting CaMKK2 to the AMPK complex and leading to phosphorylation and activation of AMPK (Fig 3I).
The CARS–CaMKK2–AMPK axis promotes cell survival when cystine is eliminated
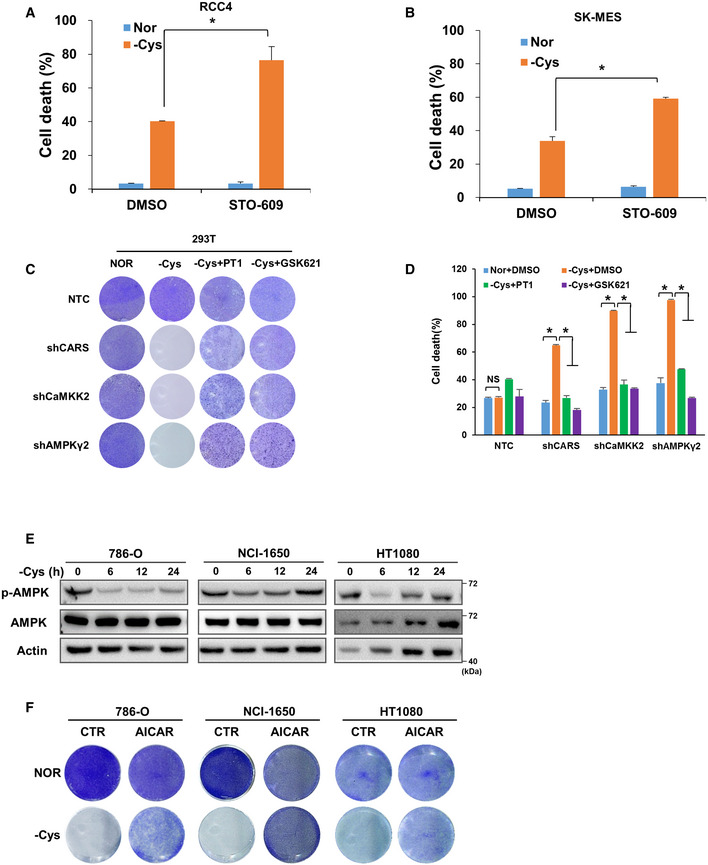
To investigate the effect of AMPK activation on cell survival under cystine deprivation conditions, we added the CaMKK2 inhibitor STO‐609 to block AMPK activation induced by cystine deficiency in 293T, RCC4 and SK‐MES cells and evaluated cell survival with crystal violet staining experiments. Our results showed that cystine starvation alone had a marginal effect on cell survival; however, when cystine‐starved cells were treated with STO‐609, the majority of the cells died (Fig 4A). Flow cytometry analysis confirmed that the combination of cystine deprivation and STO‐609 treatment triggered dramatic cell death compared with cystine deprivation alone (Figs 4B and EV4A and B). Moreover, knocking down CARS, CaMKK2 or AMPKγ2 markedly induced cell death under cystine deprivation conditions, which was attenuated by adding the AMPK activator AICAR, PT1 and GSK621 (Figs 4C and EV4C), although the mechanism of PT1’s role in AMPK activation remains controversial (Jensen et al, 2015). Similar results were observed when cell death was detected by flow cytometry (Figs 4D and EV4D). These results demonstrate that AMPK activation by the CARS‐CaMKK2 axis is important for cell survival when cystine is absent.
Figure 4. The CARS‐CaMKK2‐AMPK axis promotes cell survival when cystine is eliminated.
-
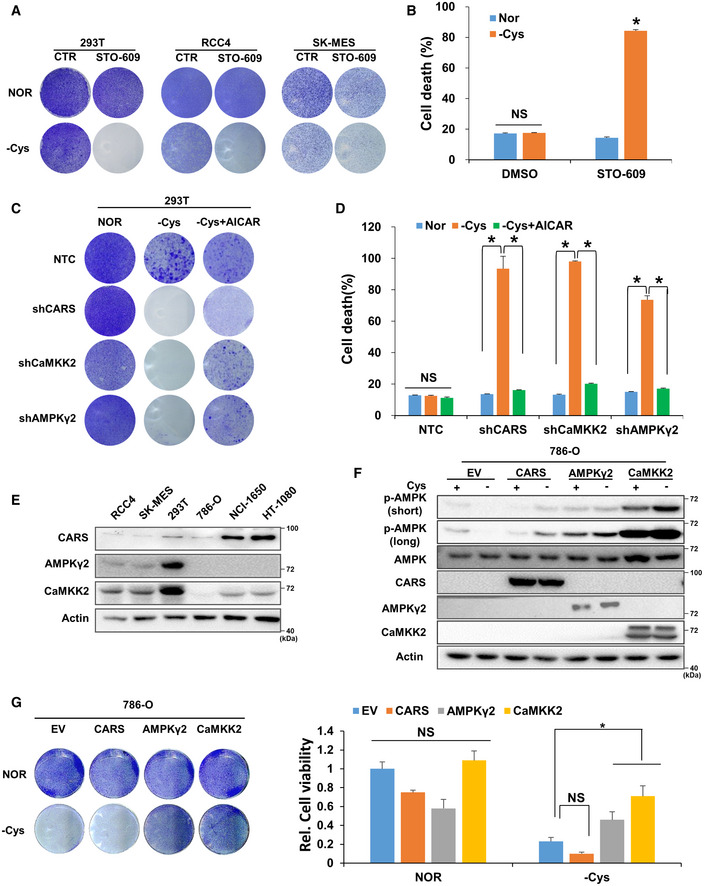
AThe crystal violet assay was performed in cystine‐deficient medium‐cultured 293T, RCC4 and SK‐MES cells with or without 1 µg/ml STO‐609 for 24 h.
-
BThe cell death level was determined in 293T cells with the same treatment conditions as in Fig 4A. Data are presented as the mean (± SD) of three independent experiments. NS, not significant; *P < 0.05 [two‐tailed Student’s t‐test], compared with the indicated groups.
-
C, DThe crystal violet assay (C) and apoptosis rate assay (D) were performed in 293T cells transfected with shRNAs targeting NTC, CARS, CaMKK2 or AMPKγ2 and further treated with cystine‐deficient medium for 24 h with or without 1 mM AICAR. Data are presented as the mean (± SD) of three independent experiments. NS, not significant; *P < 0.05 [two‐tailed Student’s t‐test], compared with the indicated groups.
-
EWB analysis of CARS, CaMKK2 and AMPKγ2 protein expression in RCC4, SK‐MES, 293T, 786‐O, NCI‐1650 and HT1080 cells. Actin served as the loading control.
-
F, GWB analysis of p‐AMPK, total AMPK, CARS, AMPKγ2 and CaMKK2 protein expression (F) and a crystal violet assay were performed (G, left) with 786‐O cells transfected with Flag‐EV, Flag‐CARS, Flag‐CaMKK2 or Flag‐AMPKγ2 and further treated with cystine‐deficient medium for 8 h. Actin served as the loading control. After scanning, 20% glacial acetic acid was added to dissolve the crystal violet, and the absorbance of each well was measured at 570 nm (OD570) to calculate relative cell viability (G, right). Data are presented as the mean (± SD) of three independent experiments. NS, not significant; *P < 0.05 [two‐tailed Student’s t‐test], compared with the indicated groups.
Source data are available online for this figure.
Figure EV4. The cysteine–CARS–AMPK pathway promotes cell survival.
-
A, BCell death levels were determined in cystine‐deficient medium‐cultured RCC4 or SK‐MES cells with or without 1 µg/ml STO‐609 for 24 h. Data are presented as the mean (± SD) of three independent experiments. “Nor” = normal medium; “‐Cys” = cystine‐deficient medium. *P < 0.05 [two‐tailed Student’s t‐test], compared with the indicated groups.
-
C, DThe crystal violet assay (C) and apoptosis rate assay (D) were performed in 293T cells transfected with shRNAs targeting NTC, CARS, CaMKK2 or AMPKγ2 and further treated with cystine‐deficient medium for 24 h with or without 100 μM PT1 or 30 μM GSK621. Data are presented as the mean (± SD) of three independent experiments. NS, not significant; *P < 0.05 [two‐tailed Student’s t‐test], compared with the indicated groups.
-
E786‐O, NCI‐1650 and HT1080 cells were treated with cystine‐deficient medium for 0, 6, 12 and 24 h, respectively. The protein levels of p‐AMPK and total AMPK were measured by WB, and actin served as the loading control.
-
FThe crystal violet assay was performed in cystine‐deficient medium‐cultured 786‐O, NCI‐1650 and HT1080 cells with or without 1 mM AICAR.
Source data are available online for this figure.
In addition to 293T, RCC4 and SK‐MES cells that can activate AMPK when cystine is deficient, we also identified several other cell lines, such as 786‐O, NCI‐1650 and HT1080, wherein AMPK was not activated even when cystine was eliminated from the culture system (Fig EV4E). Consistent with these results, cystine deprivation alone resulted in extensive cell death, which was attenuated by the addition of AICAR (Fig EV4F). However, our Western blot data further showed that AMPKγ2 and CaMKK2 were significantly lower in cystine‐nonresponsive 786‐O, NCI‐1650 and HT1080 cells than in cystine‐responsive 293T, RCC4 and SK‐MES cells (Fig 4E). These results further underline the importance of the CARS–CaMKK2–AMPK axis in protecting cells under cystine‐deficient conditions. By overexpressing CARS, AMPKγ2 and CaMKK2 in 786‐O cells, we found that AMPK was more notably activated when cystine was deficient (Fig 4F). Further, crystal violet staining experiments showed that overexpression of AMPKγ2 and CaMKK2 significantly suppressed the cell death of 786‐O cells induced by cystine deprivation (Fig 4G). These results indicate that the differential expression of AMPKγ2 and CaMKK2 in different cells may determine their sensitivity to cystine deficiency and the subsequent AMPK activation and cell survival.
In summary, our results demonstrate that cystine deficiency promotes the binding of CARS and AMPKγ2, whereby CARS recruits its partner CaMKK2 to activate AMPK, leading to cell survival under nutrient stress conditions (Fig 5).
Figure 5. Working model: CARS‐CaMKK2‐AMPKγ2 pathway activates AMPK to promote cell survival under cystine starvation condition.
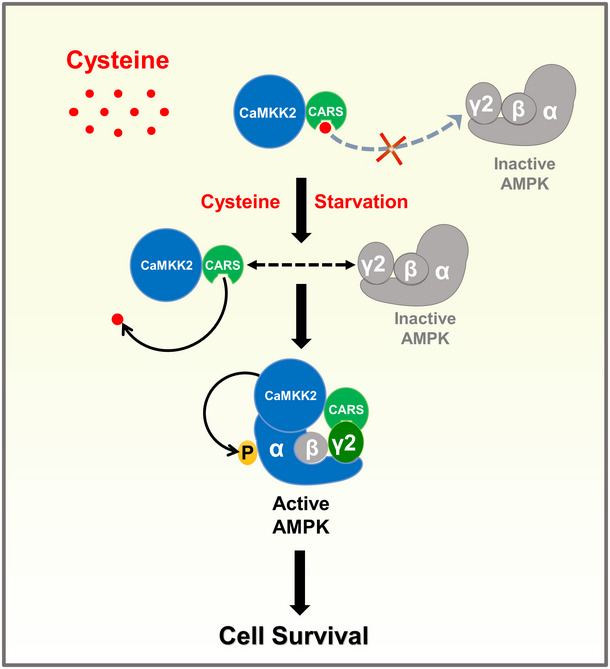
In the presence of cyst(e)ine, cysteine blocked the interaction of CARS with AMPKγ2, the AMPK complex is in an inactive state. In the deficiency of cystine, CARS is able to bind to AMPKγ2, thus recruiting CaMKK2 to the AMPK complex and leading to phosphorylation and activation of AMPK, which ultimately promotes cell survival.
Discussion
(AMP)‐activated protein kinase directly senses metabolic intermediates, which has recently attracted increased attention (Steinberg & Kemp, 2009; Hardie et al, 2012; Hardie, 2014; Zhang et al, 2014). In this study, we report, for the first time, that CARS senses cysteine deprivation conditions and activates AMPK to promote cell survival. We discovered a fine‐tuned sensing mechanism of AMPK to cyst(e)ine variation. Under cystine deficiency conditions, the CARS protein fails to bind cysteine, and this dissociation can release the inhibitory effect of cysteine on the binding of CARS to AMPKγ2. CARS then recruits the AMPK complex to the vicinity of CARS‐bound CaMKK2, causing CaMKK2‐induced AMPK phosphorylation and activation, thereby promoting the survival of cells under cystine deprivation stress. Hence, we identified a novel mechanism whereby AMPK is activated to trigger cell survival when the amino acid cystine is depleted.
(AMP)‐activated protein kinase can sense intracellular glucose and lipids (McBride et al, 2009; Zhang et al, 2017a; Li et al, 2019; Pinkosky et al, 2020), but surprisingly, little is known about the relationship between AMPK and its ability to sense amino acids. A previous study showed that withdrawal of essential amino acids induced the activation of AMPK through CaMKK2 (Ghislat et al, 2012), but Dalle Pezze et al found that AMPK activation by amino acid replenishment is mediated by CaMKK2 (Dalle Pezze et al, 2016). These reports indicate that there is still considerable confusion in this field, and that, to date, no data linking AMPK to the deficiency of a single specific amino acid have been documented. In this study, we explored, for the first time, the connection between the availability of each amino acid and AMPK activation and found that cystine deficiency alone can notably activate AMPK (Figs 1A–D and EV1A). As a heterotrimer, AMPK has two α subunits, two β subunits and three γ subunit forms, suggesting 12 possible combinations, which clearly indicate functional redundancy (Ross et al, 2016). Therefore, different combinations of AMPK complexes may correspond to different functions under different physiological conditions, and the three kinases of AMPK may also be potentially critical for sensing differences in nutrient levels (Hawley et al, 2003; Hong et al, 2003; Woods et al, 2003, 2005; Mahlapuu et al, 2004; Momcilovic et al, 2006; Koay et al, 2010; Oakhill et al, 2010; Kim et al, 2012; Ross et al, 2016) In this regard, intriguingly, we discovered that AMPK activation induced by cystine deprivation is specifically dependent on the AMPKγ2 subunit (Figs 3D and E, and EV3D), and this activation can be mediated only by CaMKK2 (Fig 1F and G).
In recent years, aminoacyl tRNA synthase (AARS), a sensor of amino acid sensors, has increasingly piqued the interest of scientists (Hountondji et al, 2000; Yanagisawa et al, 2010; Han et al, 2012; He et al, 2018). The traditional function of AARS is linking amino acids to corresponding tRNAs to form aminoacyl tRNAs during protein translation. AARS in most eukaryotes has a novel domain that does not participate in aminoacylation, suggesting that AARS may be involved in other biological processes (Guo & Schimmel, 2013). Recently, many nontranslational functions of AARS have been discovered, most notably the sensing of metabolites (Ko et al, 2000, 2001; Yakobov et al, 2017; Gupta & Laxman, 2019). As a metabolic sensing molecule, AARS also conforms to the principle of making the best use of biological evolution. For instance, AARS must first be able to bind directly to a sensed metabolite; second, the molecule then must transmit a signal. Our results show that CARS, an AARS, can sense cyst(e)ine deprivation and enhance its binding with AMPKγ2 (Fig 3F–G), ultimately activating AMPK by binding its kinase CaMKK2 (Fig 2A–D). These results substantially extend our understanding of the metabolic sensing function of AARS.
As cysteine is the only reducing amino acid in cells, an increasing number of reports in recent years have shown that cysteine plays an important role in the occurrence and development of tumours. Ioannis et al found that lung cancer with EGFR mutations is particularly sensitive to cysteine loss. Eliminating cysteine in mice can inhibit the growth of lung cancer cell‐transplanted tumours (Poursaitidis et al, 2017); Shira et al found that clearing cysteine from mouse serum can significantly inhibit the development of prostate cancer and breast cancer, as well as increases the average survival time of lymphoma mice by twofold (Cramer et al, 2016); Michael A et al found cysteine deprivation can induce the specific ferroptosis of mouse pancreatic cancer cells and then inhibit the development of pancreatic cancer (Badgley et al, 2020). Collectively, these studies showed that the elimination of cysteine to inhibit the occurrence and development of tumours has broad application prospects. However, any therapy for tumour treatment must consider the scope of the application and possible cell resistance. In our study, we found that some tumour cells prevent cell death by activating AMPK under cystine deprivation, but the addition of the CaMKK2 inhibitor STO‐609 inhibited this phenomenon, suggesting that blocking AMPK activation may be a possible therapeutic option when cells are insensitive to cystine elimination. Additionally, considering the importance of AMPKγ for cell survival under cysteine deprivation conditions (Fig 4D), and the differences in the expression of AMPKγ subunits in different tissues (Mahlapuu et al, 2004), treating cancer cells with low AMPKγ2 expression in combination with cysteine starvation might be a potential cancer targeting strategy.
In this study, we show that AMPK is activated by cystine deprivation. In addition, we found that CARS, the sensor of cysteine, can bind to AMPKγ2 when cystine is deficient and then recruit the AMPK complex to CARS‐bound CaMKK2, causing AMPK phosphorylation and activation by CaMKK2. In addition, we observed that AMPK activation in the absence of cystine is closely related to the survival of cells. Hyemin et al (Lee et al, 2020) found that AMPK activation can inhibit ferroptosis, which again illustrates the importance of AMPK activation when cells face oxidative stress. In addition to cystine, there are several amino acids whose deficiency activates AMPK. Determination of the amino acids accurately sensed to activate AMPK, the sensors involved and the physiological significance of this activation on cellular fate and function warrants further study. We found that CARS functions as a sensor of cysteine, but whether other AARS can also sense specific amino acid content to regulate molecular pathways in cells or whether they can sense the corresponding tRNA instead of amino acids remains to be further explored.
Materials and Methods
Cell culture and reagents
All cells were purchased from ATCC and cultured in Dulbecco’s modified essential medium (DMEM) containing 10% FBS and 1% penicillin–streptomycin. All cells were cultured in a humidified incubator at 37°C with 5% CO2. Cystine and other amino acid deprivation experiments were carried out as described previously (Zhang et al, 2017b). In brief, cells were cultured in DMEM deprived of a selected nutrient. Amino acid deprivation studies of Figs 1A and I, 2C and G and 3G used dialysed serum (Biological Industries; Cat# O4‐011‐1), while normal serum was used for the rest of the experiments. Takinib (HY‐103490), Pim1 (HY‐10371) and GSK621 (HY‐100548) were purchased from MCE, and AICAR (A8184), ATO‐609 (B6787) and PT1 (B7572) were purchased from APEXBIO.
Plasmids and established stable cells
Lentiviral shRNAs targeting the LKB1, CaMKK2, TAK1, CARS, AMPKγ2, GCN2 and Sirt3 plasmids were purchased from Sigma. The sequences of shRNAs used in this study are listed in Table EV1. pSin‐3XFlag‐CaMKK2, pSin‐3XFlag‐LKB1, pSin‐3XFlag‐TAK1, pSin‐3XFlag‐AHCY, pSin‐3XFlag‐CARS‐wt, pSin‐3XFlag‐CARS‐mut1, pSin‐3XFlag‐mut2, pSin‐3XFlag‐AMPKγ2‐wt and pSin‐3XFlag‐AMPKγ2‐R531G vectors were used to construct a Flag‐overexpressing plasmid, and pSin‐HA‐AMPKα1, pSin‐HA‐AMPKα2, pSin‐HA‐AMPKβ1, pSin‐HA‐AMPKβ2, pSin‐HA‐AMPKγ1, pSin‐HA‐AMPKγ2, pSin‐HA‐AMPKγ3 and pSin‐HA‐CaMKK2 vectors were used to construct a HA‐expressing plasmid. All plasmids targeting specific genes were cotransfected with plasmids carrying Δ8.9 and VSVG into HEK293T cells using PEI (Invitrogen) for 48 h, and then, the viruses were collected by filtering the supernatant of the HEK293T cells. The collected viruses were used to infect cells in the presence of 8 μg/ml polybrene (Sigma‐Aldrich). The transduced cells were selected by puromycin.
Western blot analysis
Cells were harvested as described previously (Zhang et al, 2017b). Protein concentration was measured using a Bradford Assay Kit (Sangon Biotech). Equal amounts of proteins were fractionated by 6‐10% SDS–PAGE. The following primary antibodies were used: anti‐p‐AMPK (CST; 2535; 1:1,000), anti‐AMPK (CST; 253; 1:750), anti‐p‐ACC‐S79 (CST; 3661; 1:1,000), anti‐p‐Raptor‐S792 (CST; 2083; 1:1,000), anti‐AMPKγ2 (CST; 2536s; 1:1,000), anti‐ACC (PTG; 67373‐1‐Ig; 1:1,000), anti‐Raptor (PTG; 20984‐1‐AP; 1:1,000), anti‐CARS (PTG; 15296‐1‐AP; 1:1,000), anti‐CaMKK2 (PTG; 11549‐1‐AP; 1:1,000), anti‐LKB1 (PTG; 10746‐1‐AP; 1:1,000), anti‐TAK1 (PTG: 12330‐2‐AP; 1:1,000), anti‐HA‐HRP(CST; 2999; 1:5,000), anti‐His‐Tag (Biotechnology; D110002; 1:2,000) and anti‐GST‐Tag (PTG; 10000‐0‐AP; 1:1,000). HRP‐conjugated anti‐rabbit and anti‐mouse (Bio‐Rad) secondary antibodies were used. Signals were detected using a Western ECL substrate (Bio‐Rad). Detailed information for all antibodies used is provided in Table EV2.
Immunoprecipitation
Immunoprecipitation was carried out as described previously (Wu et al, 2017). In brief, 293T cells were collected and lysed in IP buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1.5 mM MgCl2, 0.5% NP‐40) for 1–2 h, and the supernatant was transferred to a 20‐μl solution containing protein A/G beads and precleared for 1 h. After protein quantification, the corresponding primary antibody was added and incubated overnight. Next, the protein A/G beads were incubated for 2 h and washed four times with IP buffer. Protein loading was added, and the beads were placed in a 100°C metal bath to fully denature the protein. Western blot analysis was used to detect protein binding.
LC‐MS and data analysis
The sample was prepared according to a preparation kit (Thermo: 90057), and LC‐MS was carried out as described previously (Jiang et al, 2019). A Q‐Exactive Plus mass spectrometer (Thermo, USA) coupled to an EASY‐nLC 1200 HPLC system (Thermo, USA) with a nanoelectrospray ion source and operated in data‐dependent mode was used to analyse the samples. Raw LC‐MS data were further analysed using proteomics discovery software (version 2.1, Thermo Fisher Scientific) against the human UniProt database (version 20140922, 20,193 sequences). A result is considered positive when the peptide of detected constitutes more than 5% of the total peptides in the protein.
Fusion protein pull‐down experiment
The PET‐22b‐CARS plasmid was transfected into Escherichia coli (BL21), and the expression of the His‐CARS tag protein was induced with IPTG. The bacterial cells were collected, and the proteins were subjected to by ultrasonic lysis. The His‐CARS protein was selected on a nickel column, which was rinsed with imidazole, by elution with solutions of imidazole at different concentrations, which was used to obtain purified His protein. The PET‐22b‐Flag‐CaMKK2 plasmid was transfected into E. coli (BL21), and the expression of the Flag‐CaMKK2 tag protein was induced using IPTG. Pierce™ anti‐DYKDDDDK magnetic agarose (Thermo; A36797) was used to purify Flag‐tagged protein. PGEX‐4T1‐EV, PGEX‐4T1‐AMPKγ1, PGEX‐4T1‐AMPKγ2, PGEX‐4T1‐AMPKγ2‐N and PGEX‐4T1‐AMPKγ2‐C plasmids were transfected into E. coli (BL21). IPTG was used to induce the expression of GST‐tagged proteins, and then, GSH beads were used to adsorb the GST‐tagged proteins. Next, we used PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.1% Tween 20) buffer to rinse and purify the GST‐tagged proteins. Finally, the purified Flag‐tagged protein or GST‐tagged protein and His‐CARS protein were subjected to a pull‐down experiment. The proteins were detected by WB and Coomassie Blue staining.
Crystal violet staining
After cells were treated as indicated, the culture medium was discarded, and the cells were carefully washed twice with PBS to remove the residual culture medium. The cells were fixed with methanol for 30 min and stained with 0.5% crystal violet for 30 min, which was washed away with PBS. After natural drying, the plates were scanned with a scanner. To quantify the cells stained with crystal violet, 20% glacial acetic acid was added to each well and mixed by pipetting. The absorbance at 570 nm (OD570) of each well was measured by a microplate reader, and relative cell viability was calculated according to the following formula: [OD570 (treated group) − OD570 (blank)]/[OD570 (control) − OD570 (blank)] × 100%.
Flow cytometry
Cultured cells were collected from the 6‐well plate and washed twice with PBS to remove the residual culture medium. For the cell death analysis, the collected cells were stained with 5 μg/ml propidium iodide (PI), and the percentage of PI‐positive dead cells was recorded by flow cytometry (BD FACS Celesta). The data were further analysed by FlowJo software.
Statistical analysis
Data are presented as the mean (± SD) of three independent experiments unless otherwise noted. Group differences (P < 0.05) were analysed by two‐tailed Student’s t‐test.
Author contributions
PG, HZ and LS conceived and supervised this study. PG, HZ, LS and MY designed the experiments. MY, RY, YZ, YQ, ZJ, HL, LS and YW executed the experiments. PG, HZ, LS, MY, RY and YZ wrote the manuscript. All the authors read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Source Data for Expanded View
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Acknowledgements
This work is supported in part by National Key R&D Program of China (2018YFA0800300, 2018YFA0107103), National Natural Science Foundation of China (91957203, 81930083, 82130087, 81874060), the Chinese Academy of Sciences (XDB39020100) and the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S054).
The EMBO Journal (2021) 40: e108028.
See also: KK Brown (November 2021)
Contributor Information
Linchong Sun, Email: sunlc@scut.edu.cn.
Huafeng Zhang, Email: hzhang22@ustc.edu.cn.
Ping Gao, Email: pgao2@ustc.edu.cn.
Data availability
The immunoprecipitation mass spectrometry data of CaMKK2‐binding proteins relating to Fig 2A have been deposited at PRIDE (Perez‐Riverol et al, 2019) hosted at the EBI (https://www.ebi.ac.uk/pride/archive/projects/PXD027017/). Data are available via ProteomeXchange with identifier PXD027017.
References
- Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee H‐J, Purohit V, Sagalovskiy IR, Ma A, Kapilian J, Firl CEM et al (2020) Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 368: 85–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Scaria Sonia M, Saxton Robert A, Gygi Melanie P, Shen K, Wyant Gregory A, Wang T, Harper JW, Gygi Steven P, Sabatini David M (2016) The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer SL, Saha A, Liu J, Tadi S, Tiziani S, Yan W, Triplett K, Lamb C, Alters SE, Rowlinson S et al (2016) Systemic depletion of L‐cyst(e)ine with cyst(e)inase increases reactive oxygen species and suppresses tumor growth. Nat Med 23: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Pezze P, Ruf S, Sonntag AG, Langelaar‐Makkinje M, Hall P, Heberle AM, Razquin Navas P, van Eunen K, Tölle RC, Schwarz JJ et al (2016) A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat Commun 7: 13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia‐Barrio M, Anderson J, Hinnebusch AG (2000) Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA‐binding domain. Mol Cell 6: 269–279 [DOI] [PubMed] [Google Scholar]
- Efeyan A, Comb WC, Sabatini DM (2015) Nutrient‐sensing mechanisms and pathways. Nature 517: 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislat G, Patron M, Rizzuto R, Knecht E (2012) Withdrawal of essential amino acids increases autophagy by a pathway involving Ca2+/calmodulin‐dependent kinase kinase‐beta (CaMKK‐beta). J Biol Chem 287: 38625–38636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Yan Y, Novick SJ, Kovach A, Goswami D, Ke J, Tan MHE, Wang L, Li X, de Waal PW et al (2017) Deconvoluting AMP‐activated protein kinase (AMPK) adenine nucleotide binding and sensing. J Biol Chem 292: 12653–12666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Schimmel P (2013) Essential nontranslational functions of tRNA synthetases. Nat Chem Biol 9: 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Laxman S (2019) tRNA wobble‐uridine modifications as amino acid sensors and regulators of cellular metabolic state. Curr Genet 66: 475–480. [DOI] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S (2012) Leucyl‐tRNA synthetase is an intracellular leucine sensor for the mTORC1‐signaling pathway. Cell 149: 410–424 [DOI] [PubMed] [Google Scholar]
- Hardie DG (2014) AMPK–sensing energy while talking to other signaling pathways. Cell Metab 20: 939–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Mäkelä TP, Alessi DR, Hardie DG (2003) Complexes between the LKB1 tumor suppressor, STRAD/and MO25/ are upstream kinases in the AMP‐activated protein kinase cascade. J Biol Chem 2: 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Dinkova‐Kostova AT, Tew KD (2020) Oxidative stress in cancer. Cancer Cell 38: 167–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XD, Gong W, Zhang JN, Nie J, Yao CF, Guo FS, Lin Y, Wu XH, Li F, Li J et al (2018) Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab 27: 151–166 [DOI] [PubMed] [Google Scholar]
- Herzig S, Shaw RJ (2018) AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19: 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M (2003) Activation of yeast Snf1 and mammalian AMP‐activated protein kinase by upstream kinases. Proc Natl Acad Sci USA 100: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hountondji C, Beauvallet C, Pernollet JC, Blanquet S (2000) Enzyme‐induced covalent modification of methionyl‐tRNA synthetase from bacillus stearothermophilus by methionyladenylate: identification of the labeled amino acid residues by matrix‐assisted laser desorption‐ionization mass spectrometry. J Protein Chem 19: 563–568 [DOI] [PubMed] [Google Scholar]
- Jensen TE, Ross FA, Kleinert M, Sylow L, Knudsen JR, Gowans GJ, Hardie DG, Richter EA (2015) PT‐1 selectively activates AMPK‐gamma1 complexes in mouse skeletal muscle, but activates all three gamma subunit complexes in cultured human cells by inhibiting the respiratory chain. Biochem J 467: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM (2016) Regulation and function of AMPK in physiology and diseases. Exp Mol Med 48: e245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, Xing B, Sun W, Ren L, Hu BO et al (2019) Proteomics identifies new therapeutic targets of early‐stage hepatocellular carcinoma. Nature 567: 257–261 [DOI] [PubMed] [Google Scholar]
- Kim E, Lee SH, Lee KS, Cheong H‐K, Namkoong K, Hong CH, Oh BH (2012) AMPK γ2 subunit gene PRKAG2 polymorphism associated with cognitive impairment as well as diabetes in old age. Psychoneuroendocrinology 37: 358–365 [DOI] [PubMed] [Google Scholar]
- Ko YG, Kang YS, Kim EK, Park SG, Kim S (2000) Nucleolar localization of human methionyl‐tRNA synthetase and its role in ribosomal RNA synthesis. J Cell Biol 149: 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko Y‐G, Kim E‐K, Kim T, Park H, Park H‐S, Choi E‐J, Kim S (2001) Glutamine‐dependent antiapoptotic interaction of human glutaminyl‐tRNA synthetase with apoptosis signal‐regulating kinase 1. J Biol Chem 276: 6030–6036 [DOI] [PubMed] [Google Scholar]
- Koay A, Woodcroft B, Petrie EJ, Yue H, Emanuelle S, Bieri M, Bailey MF, Hargreaves M, Park J‐T, Park K‐H et al (2010) AMPK beta subunits display isoform specific affinities for carbohydrates. FEBS Lett 584: 3499–3503 [DOI] [PubMed] [Google Scholar]
- Lee H, Zandkarimi F, Zhang Y, Meena JK, Kim J, Zhuang LI, Tyagi S, Ma LI, Westbrook TF, Steinberg GR et al (2020) Energy‐stress‐mediated AMPK activation inhibits ferroptosis. Nat Cell Biol 22: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang C‐S, Zong Y, Feng J‐W, Ma T, Hu M, Lin Z, Li X, Xie C, Wu Y et al (2019) Transient receptor potential V channels are essential for glucose sensing by aldolase and AMPK. Cell Metab 30: 508–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Johansson C, Lindgren K, Hjälm G, Barnes BR, Krook A, Zierath JR, Andersson L, Marklund S (2004) Expression profiling of the gamma‐subunit isoforms of AMP‐activated protein kinase suggests a major role for gamma3 in white skeletal muscle. Am J Physiol Endocrinol Metab 286: E194–E200 [DOI] [PubMed] [Google Scholar]
- McBride A, Ghilagaber S, Nikolaev A, Hardie DG (2009) The glycogen‐binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metab 9: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic M, Hong SP, Carlson M (2006) Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP‐activated protein kinase in vitro . J Biol Chem 281: 25336–25343 [DOI] [PubMed] [Google Scholar]
- Oakhill JS, Chen ZP, Scott JW, Steel R, Castelli LA, Ling N, Macaulay SL, Kemp BE (2010) β‐Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP‐activated protein kinase (AMPK). Proc Natl Acad Sci USA 107: 19237–19241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Riverol Y, Csordas A, Bai J, Bernal‐Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkosky SL, Scott JW, Desjardins EM, Smith BK, Day EA, Ford RJ, Langendorf CG, Ling NXY, Nero TL, Loh K et al (2020) Long‐chain fatty acyl‐CoA esters regulate metabolism via allosteric control of AMPK beta1 isoforms. Nat Metab 2: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poursaitidis I, Wang X, Crighton T, Labuschagne C, Mason D, Cramer SL, Triplett K, Roy R, Pardo OE, Seckl MJ et al (2017) Oncogene‐selective sensitivity to synchronous cell death following modulation of the amino acid nutrient cystine. Cell Rep 18: 2547–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FA, Jensen TE, Hardie DG (2016) Differential regulation by AMP and ADP of AMPK complexes containing different gamma subunit isoforms. Biochem J 473: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg GR, Kemp BE (2009) AMPK in health and disease. Physiol Rev 89: 1025–1078 [DOI] [PubMed] [Google Scholar]
- Wang S, Tsun Z‐Y, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W et al (2015) Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347: 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willows R, Navaratnam N, Lima A, Read J, Carling D (2017) Effect of different gamma‐subunit isoforms on the regulation of AMPK. Biochem J 474: 1741–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM (2016) Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D (2005) Ca2+/calmodulin‐dependent protein kinase kinase‐beta acts upstream of AMP‐activated protein kinase in mammalian cells. Cell Metab 2: 21–33 [DOI] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D (2003) LKB1 is the upstream kinase in the AMP‐activated protein kinase cascade. Curr Biol 13: 2004–2008 [DOI] [PubMed] [Google Scholar]
- Wu G (2009) Amino acids: metabolism, functions, and nutrition. Amino Acids 37: 1–17 [DOI] [PubMed] [Google Scholar]
- Wu G, Yuan M, Shen S, Ma X, Fang J, Zhu L, Sun L, Liu Z, He X, Huang DE et al (2017) Menin enhances c‐Myc‐mediated transcription to promote cancer progression. Nat Commun 8: 15278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant GA, Abu‐Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, Sabatini DM (2017) mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 171: 642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT et al (2007) Structural basis for AMP binding to mammalian AMP‐activated protein kinase. Nature 449: 496–500 [DOI] [PubMed] [Google Scholar]
- Yakobov N, Debard S, Fischer F, Senger B, Becker HD (2017) Cytosolic aminoacyl‐tRNA synthetases: unanticipated relocations for unexpected functions. Biochem Biophys Acta 1861: 387–400 [DOI] [PubMed] [Google Scholar]
- Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S (2010) A paralog of lysyl‐tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol 17: 1136–1143 [DOI] [PubMed] [Google Scholar]
- Zhang C‐S, Hawley SA, Zong Y, Li M, Wang Z, Gray A, Ma T, Cui J, Feng J‐W, Zhu M et al (2017a) Fructose‐1,6‐bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C‐S, Jiang B, Li M, Zhu M, Peng Y, Zhang Y‐L, Wu Y‐Q, Li T, Liang YU, Lu Z et al (2014) The lysosomal v‐ATPase‐Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 20: 526–540 [DOI] [PubMed] [Google Scholar]
- Zhang CS, Li M, Lin SC (2017b) Methods to study lysosomal AMPK activation. Methods Enzymol 587: 465–480 [DOI] [PubMed] [Google Scholar]


