Abstract
Astrocytes regulate brain‐wide functions and also show region‐specific differences, but little is known about how general and region‐specific functions are aligned at the single‐cell level. To explore this, we isolated adult mouse diencephalic astrocytes by ACSA‐2‐mediated magnetic‐activated cell sorting (MACS). Single‐cell RNA‐seq revealed 7 gene expression clusters of astrocytes, with 4 forming a supercluster. Within the supercluster, cells differed by gene expression related to ion homeostasis or metabolism, with the former sharing gene expression with other regions and the latter being restricted to specific regions. All clusters showed expression of proliferation‐related genes, and proliferation of diencephalic astrocytes was confirmed by immunostaining. Clonal analysis demonstrated low level of astrogenesis in the adult diencephalon, but not in cerebral cortex grey matter. This led to the identification of Smad4 as a key regulator of diencephalic astrocyte in vivo proliferation and in vitro neurosphere formation. Thus, astrocytes show diverse gene expression states related to distinct functions with some subsets being more widespread while others are more regionally restricted. However, all share low‐level proliferation revealing the novel concept of adult astrogenesis in the diencephalon.
Keywords: astrocytes, cerebral cortex, de novo astrocyte generation, proliferation, Smad4
Subject Categories: Neuroscience
Single cell RNA‐seq analysis identifies distinct subgroups of diencephalic astrocytes and finds that Smad4‐mediated astrogenesis occurs in the adult diencephalon.
Introduction
Parenchymal astrocytes have been found to be involved in ion and transmitter homeostasis, metabolic functions and participate in information processing by influencing synaptic functions in various regions of the mammalian brain (Lehre & Danbolt, 1998; Furness et al, 2008; Vandenberg & Ryan, 2013; Chung et al, 2015; Allen & Eroglu, 2017; Farmer & Murai, 2017; Mederos et al, 2018; Romanos et al, 2019). While ion and transmitter homeostasis is necessary for neuronal circuit physiology throughout the brain, metabolism and circuit functions may differ in different brain regions. Indeed, astrocytes have also been found to differ between brain regions (Ben Haim & Rowitch, 2017; Khakh & Deneen, 2019; Westergard & Rothstein, 2020), e.g. during development instructing different dendritic trees of neurons, or exhibiting region‐specific morphologies (Denis‐Donini & Estenoz, 1988; Emsley & Macklis, 2006). Accordingly, gene expression analysis of astrocytes from different brain regions highlighted specific expression patterns, sometimes even between subdivisions within the same brain region (Ben Haim & Rowitch, 2017; John Lin et al, 2017; Morel et al, 2017; Boisvert et al, 2018; Itoh et al, 2018; Batiuk et al, 2020; Bayraktar et al, 2020). An important question remaining, is, however, to which extent these region‐specific differences apply to all astrocytes in a given region, or only to subsets of astrocytes. In this regard, it is interesting to note that the deletion of pan‐astrocyte transcription factors (TFs), such as Sox9 and Nfia revealed region‐specific functions and requirements (Kang et al, 2012; Lozzi et al, 2020). This prompts the question of how regional diversity is aligned with pan‐astrocyte functions. Are there region‐specific subtypes of astrocytes performing common functions, or are there common astrocyte subtypes or states present throughout the brain performing the universal functions of transmitter up‐take or ion homeostasis, while others are more dedicated to region‐specific tasks?
To explore astrocyte regionalization, we chose the diencephalon (DIE) as this region is composed of two major functional subdivisions: the thalamic regions dedicated to sensory information processing and the hypothalamic regions involved in regulation of homeostasis and neuroendocrine functions regulating many physiological processes. Moreover, we choose the DIE as we had noted previously some similarity in gene expression between DIE astrocytes and the adult neural stem cells (NSCs; Beckervordersandforth et al, 2010; Gotz et al, 2015) which was less pronounced in comparison to cerebral cortex (CTX) grey matter (GM) astrocytes (Gotz et al, 2015; Sirko et al, 2015). As astrocytes in the intact brain normally lack progenitor or NSC hallmarks (Robel et al, 2011), we were interested to determine, if the DIE astrocytes were special in this regard. To explore these aspects at the single‐cell level, we set‐out to enrich DIE astrocytes by ACSA‐2‐mediated magnetic‐activated cell sorting (MACS) as previously described in CTX GM and hippocampus (Batiuk et al, 2017, 2020; Kantzer et al, 2017). After optimizing this method further to remove a considerable fraction of ependymal cells, we succeeded in performing a single‐cell RNA‐sequencing (scRNA‐seq) analysis on more than 20,000 astrocytes from the DIE. This revealed not only an unprecedented molecular diversity of astrocyte states or subtypes but also the novel concept of ongoing astrogenesis in the adult DIE.
Results
MACS isolation with ACSA‐2 enriches astrocytes and ependymal cells
To selectively isolate astrocytes in the DIE, we used Aldh1l1‐eGFP mice (Gong et al, 2003) that reliably label astrocytes (Fig 1A, Fig EV1A, Appendix Fig S1A–D). The parenchyma was dissected by broadly omitting the region lining the ventricle, where tanycytes and ependymal cells are located (Fig EV1A). To enrich astrocytes, we took advantage of the ACSA‐2 antibody following a recently described protocol for magnetic‐assisted cell sorting (MACS) (Batiuk et al, 2017, 2020). As expected, cells isolated by MACS were GFP+ as shown by immunostaining of acutely plated isolated cells (Appendix Fig S1E).
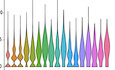
Figure 1. Astrocyte diversity in the diencephalon revealed by scRNA‐seq.
-
AScheme depicting the region (in turquoise green) dissected from 2‐ to 3‐mo‐old Aldh1l1‐eGFP mice (drawing left) removing any ependyma. Schemes to the right depicting the column‐based magnetic cell selection using first nano‐sized beads for cell depletion of myelin containing material followed by positive cell selection for ACSA‐2 cells. Cells isolated by ACSA‐2 magnetic‐associated cell separation were used for scRNA‐seq using 10×Genomics strategy.
-
B–Dt‐SNE projections of 25,761 single‐cell transcriptomes with clusters colour coded and annotated post hoc based on their transcriptional profile identities (B) and showing the expression of representative astrocyte‐ (C) and ependyma (D)‐specific marker genes (numbers reflect scaled log‐normalized read counts for the specified gene for each cell, levels indicated on the right; purple: high expression, grey: low to no expression). Cell numbers in the different clusters are: Cluster 0 = 6,564, Cluster 1 = 4,617, Cluster 2 = 4,147, Cluster 3 = 4,053, Cluster 4 = 1,375, Cluster 5 = 1,033, Cluster 6 = 991, Cluster 7 = 663, Cluster 8 = 633, Cluster 9 = 414, Cluster 10 = 333, Cluster 11 = 222, Cluster 12 = 207, Cluster 13 = 188, Cluster 14 = 154, Cluster 15 = 85, Cluster 16 = 82. For all differentially expressed genes per cluster of scRNA‐seq data from diencephalic astrocytes after ACSA‐2 MACS following an optimized dissection see Dataset EV1 for Fig 1B.
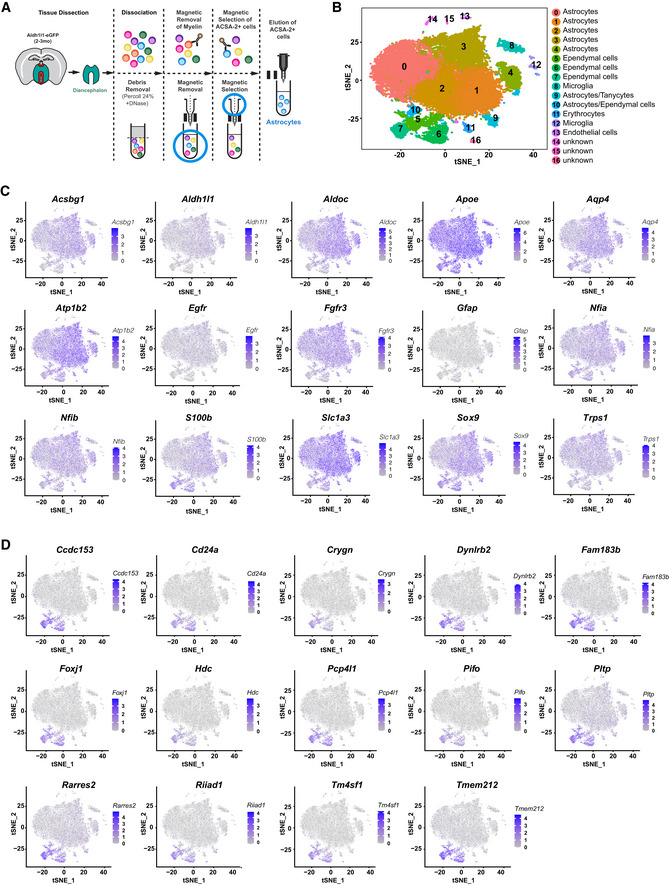
Figure EV1. Gene expression of astrocytes and ependymal cells in the diencephalon revealed by ACSA‐2 MACS followed by scRNA‐seq.
-
AScheme depicting dissected region (in turquoise green and dorsal part in red) from 2‐ to 3‐mo‐old Aldh1l1‐eGFP mice (drawing left) and isolation by ACSA‐2 magnetic‐associated cell separation (schemes to the right) followed by 10×Genomics strategy.
-
B–Et‐SNE projections of 4,651 single‐cell transcriptomes with clusters colour coded and annotated post hoc based on their transcriptional profile identities (B) and showing the expression of representative astrocyte‐specific marker genes (C–E; numbers reflect scaled log‐normalized read counts for the specified gene for each cell, levels indicated on the right; purple: high expression, grey: low to no expression) grouped in three categories (C: relatively equal amongst clusters, D: higher in clusters 1–4, E: higher in clusters 0 and 5). Violin plot in E showing log count values of Atp1b2 expression for each cluster. Colour‐coding corresponds to clusters shown in B. Cell numbers in the different clusters are: Cluster 0 = 1,042, Cluster 1 = 972, Cluster 2 = 779, Cluster 3 = 431, Cluster 4 = 414, Cluster 5 = 252, Cluster 6 = 219, Cluster 7 = 115, Cluster 8 = 85. For all differentially expressed genes per cluster of scRNA‐seq data from diencephalic astrocytes after ACSA‐2 MACS see Source Data for EV1B.
Source data are available online for this figure.
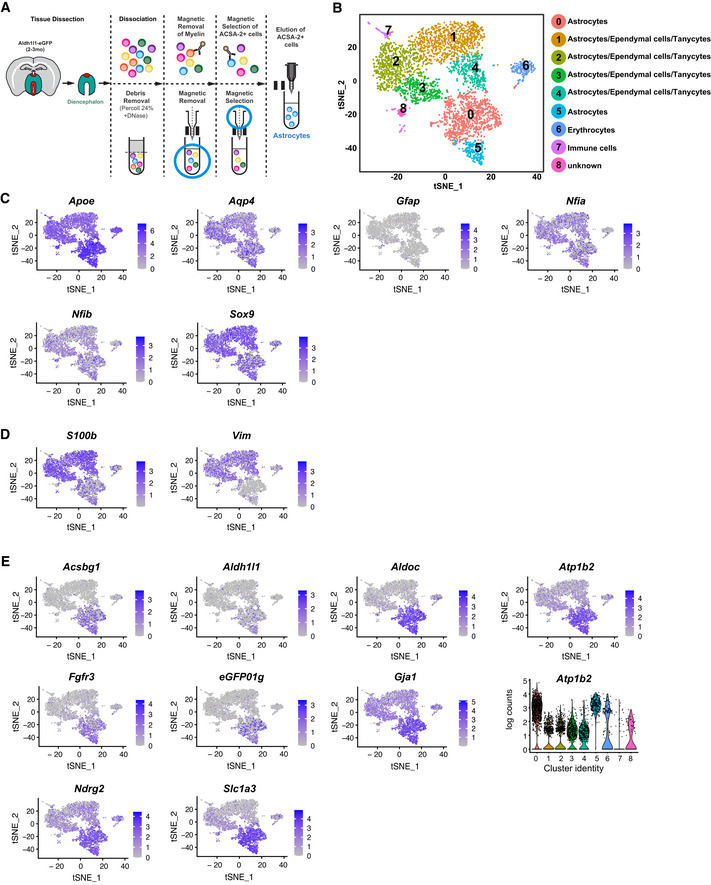
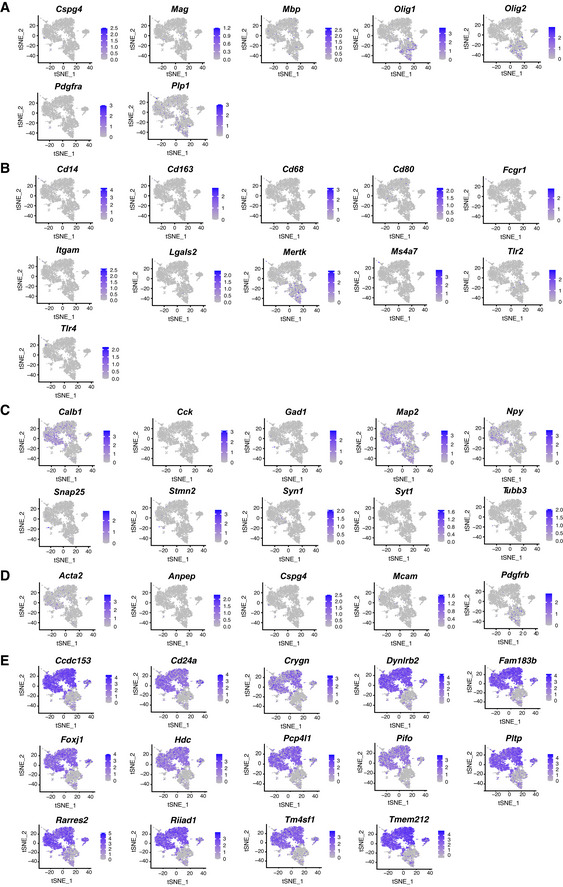
To study the heterogeneity of the isolated cells, we performed scRNA‐seq and confirmed that 90.28% of all cells expressed astrocyte markers (Fig EV1B–E). Shared neighbour modularity optimization clustering of scRNA‐seq data was based on 11 dimensions of PCA projection of 2,839 variable genes and resulted in 9 groups of cells (more details in methods section). We identified 6 of them as astrocyte‐like cells, two as erythrocytes and immune cells, and one small cluster remained not annotated (Fig EV1B). We based our assignments on high expression of astroglial genes (Fig EV1C–E) and low expression of genes typical for oligodendrocytes (Fig EV2A), microglia (Fig EV2B), neurons (Fig EV2C) and pericytes (Fig EV2D). All astrocyte‐like clusters expressed equal levels of well‐known astroglial genes (e.g. Aqp4, Sox9, Nfia, Fig EV1C). Other astrocyte markers, such as Slc1a3, Fgfr3, Aldoc, Ndrg2, Gja1, were highly expressed in clusters 0 and 5, while S100b and Vim expression was higher in clusters 1–4 (Fig EV1D and E, and Source data for Fig EV1). GFP expressed in the Adlh1l1‐eGFP mice was higher in clusters 0 and 5, but scattered cells were present in all clusters (Fig EV1E), consistent with the plating and immunostaining results (Appendix Fig S1E).
Figure EV2. Expression of oligodendrocyte‐, microglia‐, neuron‐, pericyte‐ and ependyma cell‐specific genes in ACSA‐2 sorted cells from the DIE.
The lower level of several astrocyte marker genes in clusters 1–4 prompted us to explore also ependymal cell gene expression, as our previous analysis of subependymal zone (SEZ) cells had shown that NSCs and ependymal cells share marker gene expression with astrocytes and typically differ only in their expression levels (Beckervordersandforth et al, 2010). As we had removed most of the ventricular lining by dissection (except the dorsal part indicated in red in Fig 1A), and the first description of ACSA‐2 selection (Kantzer et al, 2017) had reported that ACSA‐2 would not select ependymal cells, we were surprised to find high expression of ependymal cell genes in clusters 1–4 (Fig EV2E). To examine the possible contamination of ependymal cells during the tissue dissection procedure, we labelled ependymal cells by injecting the CellTrace CFSE/FlashTag (FT) into the lateral ventricle 20 min before dissection (Fig EV3A and B). Indeed, we found a considerable fraction of FT+ cells (Fig EV3C and D) consistent with the possibility that ependymal cells were included in our dissection. In contrast, by omitting the dorsal part of the ventricular lining and the connection to the lateral ventricles, we found a drastic reduction of FT+ cells allowing us to remove most ependymal cells from our cell preparation (Fig EV3D).
Figure EV3. Optimization of ependymal cell removal using cell tracker/FlashTag.
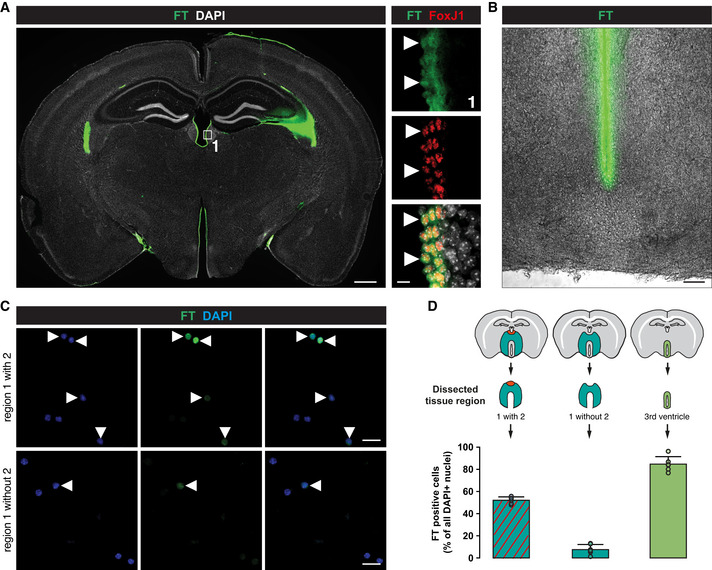
- Micrographs of coronal forebrain sections of 3‐mo‐old C57BL/6J mice stained as indicated showing that FlashTag (FT) labelled cells are all positive for the ependymal cell marker FoxJ1. Double‐positive cells are indicated by white arrowheads. Scale bars: 500 µm (overview) and 20 µm (magnification).
- Brightfield micrograph of a coronal forebrain section of 3‐mo‐old C57BL/6J mice, showing that FT‐positive cells are only lining around the lateral wall of the third ventricle and not penetrating within the parenchyma. Scale bar: 100 µm.
- Removing the dorsal part of the ventricle followed by ACSA‐2 MACS from FT labelled 3‐mo‐old C57BL/6J mice resulted in very little remaining FT‐positive cells (lower panel) compared to the tissue preparation where the dorsal part was included in the preparation (upper panel). FT labelled DAPI cells are highlighted by white arrowheads. Scale bars: 20 µm.
- Quantification of FT‐positive cells after ACSA‐2 MACS using different areas of dissection.
Data information: In D data are presented as mean ± SEM. Each dot represents one field per coverslip.
Using this improved dissection, we next dissociated cells from 8 Aldh1l1‐eGFP animals injected with FT followed by ACSA‐2 MACS (Fig 1A). ScRNA‐seq performed on these cells resulted in 25,911 cells, with a total of 23,337 genes detected and a median UMI count of 2,360 per cell. Gene selection was performed on the subset of 1,501 selected variable genes (mean expression value 0.0125–4, dispersion ≥ 0.5). Clustering on 15 dimensions of PCA data projection (for further details see Methods) revealed 17 clusters (Fig 1B, Dataset EV1 for Fig 1B). First, we examined astrocyte and ependymal cell gene expression and found that 83.5% cells expressed all astrocyte markers (clusters 0, 1, 2, 3, 4, 9 and 10, Figs 1C and EV4, Appendix Fig S2), and only a minority (10.4%, clusters 5, 6 and 7) expressed ependymal cell markers (Fig 1D). This method now provided us with an excellent dataset to explore astrocyte gene expression (Dataset EV1 for Fig 1B).
Figure EV4. Expression of oligodendrocyte‐, microglia‐, neuron‐, and pericyte cell‐specific genes in ACSA‐2 sorted DIE astrocytes using an optimized dissection.

Spatial mapping of cluster gene expression confirms cell identities and reveals differences in spatial mapping of astrocyte clusters
To confirm the existence of ependymal and other cell identities in our dataset, we mapped expression profiles of scRNA‐seq cells (see Methods for more details) to the spatial gene expression dataset of a coronal section of the mouse brain provided by 10xGenomics Version 1.1.0 (see Materials and Methods). This analysis revealed striking differences in predicted positions for each cluster identified by our scRNA‐seq (Fig 2A–R). Cluster 0 shared most gene expression with cells in the CTX (Fig 2A), while this was not the case for cluster 1 (Fig 2B). This may suggest that astrocytes from cluster 0 may be more similar to astrocytes in the CTX than those in cluster 1. Astrocytes in cluster 2 shared gene expression with cells in the thalamus and the upper cortical layers (Fig 2C).
Figure 2. Spatial gene expression mapping of diencephalic astrocyte clusters.
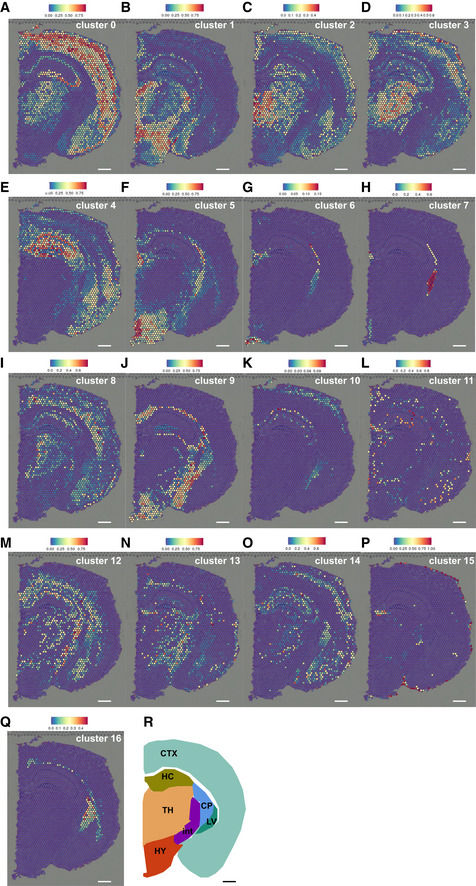
-
A–QMapping of the single‐cell data shows different distribution of genes expressed in the respective clusters. Astrocyte clusters 0–4, 9 and 10 are shown in panels A–E and J, K, respectively.
-
RSchematic of mouse coronal brain section. CP: caudatoputamen; CTX: cerebral cortex; HC: hippocamus; HY: hypothalamus; int: internal capsule; LV: lateral ventricle, TH: thalamus.
Data information: Colour‐coding bars represent degree of gene expression similarity between all cells from given cluster and mixture of all cells from given single pixel of Slide‐seq data. Scale bars: 300 µm
To evaluate our approach, we also explored the predicted positions of clusters with clear assignment of non‐astrocyte identity, such as ependymal cells, microglia, and erythrocytes. Indeed, genes expressed by the erythrocytes of cluster 11 were shown in small scatters equally distributed throughout the forebrain (Fig 2L). The ependymal cell clusters 6 and 7 showed a tight spatial restriction to ependymal lining of the lateral or third ventricles with no other localization detectable (Fig 2G and H). These data thus support the validity of the cluster annotation (Fig 1B) and validate the differential predictions observed in the spatial mapping analysis of the different astrocyte clusters.
Interestingly, ependymal cells from cluster 5 showed not only the predicted position at the ventricle where ependymal cells are located, but also distributing further into the hypothalamus parenchyma (Fig 2F). This could be indicative of some shared gene expression with parenchymal cells, which is not the case for the ependymal cells from cluster 6 and 7 (Fig 2G and H). Likewise, similarity to cluster 9 (annotated as astrocytes/tanycytes) is predicted to localize from the most ventral part of the third ventricle, where tanycytes are localized, to the hypothalamus parenchyma and is absent from the thalamus (Fig 2J). Cells of cluster 9 express many astrocyte genes consistent with the predicted parenchymal localization, as well as tanycyte genes consistent with the predicted localization at the ventral ventricular lining (Fig 1). To further probe the finding of shared gene expression between cells lining the third ventricle and astrocytes, we examined the protein S100a6 that has been detected in NSCs (Yamada & Jinno, 2014; Kjell et al, 2020) and is highly enriched in cluster 9 (Fig 3A). Immuno‐staining for S100a6 strongly labelled NSCs in the subependymal zone (SEZ) lining the lateral ventricles and white matter (WM, Fig 3B) as well as tanycytes of the third ventricle (Fig 3C). Additionally, some parenchymal astrocytes in the hypothalamus often in the vicinity to blood vessels were S100a6‐positive (Fig 3C). This is consistent with the predicted localization of the gene expression of clusters 6 and 9 (Fig 2G and J). Thus, immunostaining for S100a6 corroborates the spatial mapping of gene expression and suggests that the DIE parenchyma also harbours a small subset of S100a6 expressing astrocyte‐like cells that may possess NSC potential.
Figure 3. Diencephalic astrocyte subtypes share gene expression between ependyma and tanycytes.
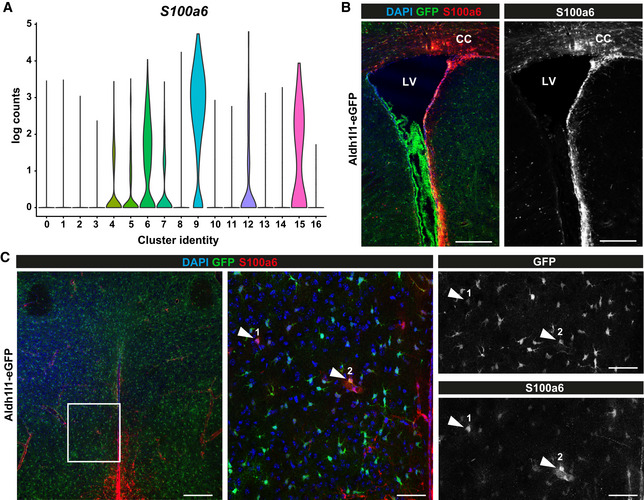
-
AViolin plot showing scaled log‐normalized read counts of S100a6 expression for each cluster. Colour‐coding corresponds to clusters shown in Fig 1B.
-
B, CIn vivo validation of S100a6 expressing DIE astrocytes in Aldh1l1‐eGFP mouse (3 months) with immunolabelling for S100a6 (B: in the SEZ, C: in the DIE). Inset in left panel of C is depicted in middle and right panels. S100a6/GFP double‐positive cells are indicated by arrowheads. LV: lateral ventricle, CC: corpus callosum. Scale bars: 200 µm (B and left panel C), 50 µm (middle and right panels C).
Diversity of gene expression of diencephalic astrocytes
The above analysis confirmed that the optimized dissection protocol resulted in isolation of 21,503 astrocytes that comprised 7 gene expression states or clusters (0, 1, 2, 3, 4, 9 and 10) based on high expression of almost all astrocyte marker genes (Fig 1C), and low expression of all other cell type markers (Fig EV4A–D), including ependymal cell genes (Fig 1D). Notably, clusters 4, 9 and 10 were not part of the large supercluster of astrocytes composed by clusters 0, 1, 2 and 3.
To get a first impression on how the clusters differ in gene expression, we performed Gene Ontology (GO) term analysis using Gorilla (Fig 4, Dataset EV2 for Fig 4). Many GO terms were related to well‐known astrocyte functions and hallmarks, but interestingly were significant in distinct clusters (Fig 4A–G). For example, GO terms related to ion regulation and sodium transport were significant in clusters 2 and 3, while significantly enriched GO terms related to potassium transport were restricted to cluster 3 (Fig 4C and D). Notably, gene expression of clusters 2 and 3 was predicted to map in a widespread manner throughout the forebrain including the CTX (Fig 2C and D). Conversely, GO terms related to mitochondrial function were significantly and selectively enriched in cluster 1 (Fig 4B), a cluster whose gene expression was predicted to be restricted to the entire DIE and not shared with the CTX (Fig 2B). This could mean that specific metabolic aspects of astrocyte gene expression may be adapted in a region‐specific manner to the neuronal and network requirements. Fatty acid metabolism was significantly enriched in GO terms of cluster 4 along with glutamate metabolism (Fig 4E, Dataset EV2 for Fig 4). Thus, one interpretation of the data could be that these astrocyte clusters may correspond to different states of astrocytes engaged in specific functions, or, that different subtypes of astrocytes may be more biased towards the metabolic support functions while others may engage more in the ion homeostasis. In either case, these data suggest that these well‐known functions are partitioned either temporally to astrocytes in different transcriptional states or more long‐term to astrocyte subtypes.
Figure 4. GO term enrichment analysis of astrocyte clusters.
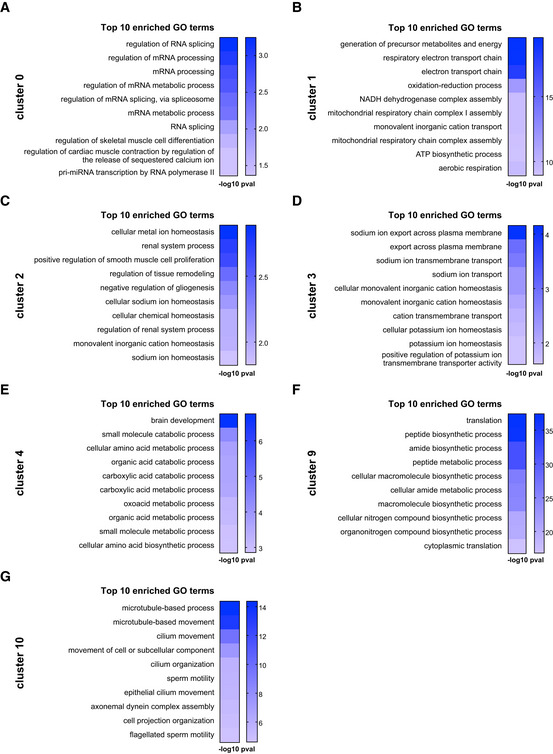
-
A–GGraphs showing top 10 enriched GO terms for each astrocyte cluster annotated in Fig 1B. For all significantly enriched GO terms (analysed by Gorilla) see Dataset EV2 for Fig 4. (P‐value threshold was set to 0.01, see http://cbl‐gorilla.cs.technion.ac.il).
This analysis also highlighted more unexpected GO terms specifically enriched in some of the astrocyte clusters. For example, cluster 0 is characterized almost exclusively by GO terms related to RNA processing and splicing (Fig 4A). Surprisingly, clusters 2 and 4 had significantly enriched GO terms related to the regulation of proliferation (Dataset EV2 for Fig 4). Cluster 2 also comprised GO terms on glial development and differentiation (Fig 4C), while cluster 4 contained many significantly enriched GO terms referring to development, proliferation and neurogenesis, suggestive of certain NSC hallmarks (Dataset EV2 for Fig 4). Interestingly, cells in this cluster also expressed Mlc1 (megalencephalic leukoencephalopathy associated with cysts 1) that is associated with astrocytes located at peri/juxtavascular positions (Teijido et al, 2004, 2007; Gilbert et al, 2019), which we identified as prone to proliferation after injury in the CTX (Bardehle et al, 2013; Sirko et al, 2013; Gotz et al, 2021). Cluster 9 also shares many astrocyte markers, but also expresses NSC markers, such as S100a6 (Fig 3) and tanycyte markers, such as Ptn (pleiotrophin), Thrsp (thyroid hormone responsive), Crym (crystallin), Slc16a2 (solute carrier family 16 member 2) and Col23a1 (collagen, type XXIII alpha 1) (Campbell et al, 2017; Chen et al, 2017) (for all genes expressed in each cluster see Dataset EV1 for Fig 1B).
Given the GO terms related to proliferation in some of the astrocyte clusters, we examined a proliferation index by compiling an index of 9 genes related to proliferation (Fig 5A, Dataset EV3 for Fig 5A and B). This showed that proliferation genes are widely expressed throughout all astrocyte populations with similar levels to the tanycyte/ependymal cell populations (Fig 5B). To explore further if cells are in a stable state or are dynamically changing (differentiation, proliferation), we performed RNA velocity analysis which compares levels of unspliced and spliced mRNAs in and between cells to derive activity measure and to determine the expected direction of the ongoing change (La Manno et al, 2018). In the adult brain, differences in the RNA velocity between clusters may be informative suggesting if and how some cells may convert into others or show transcriptional dynamics. It is noteworthy here that the 10xGenomics data have a general 3` bias. To mitigate this issue, we used the velocyto algorithm (La Manno et al, 2018) to calculate velocity per cell taking all detectable genes into account and subsequently average it per cell group as described in the Method section.
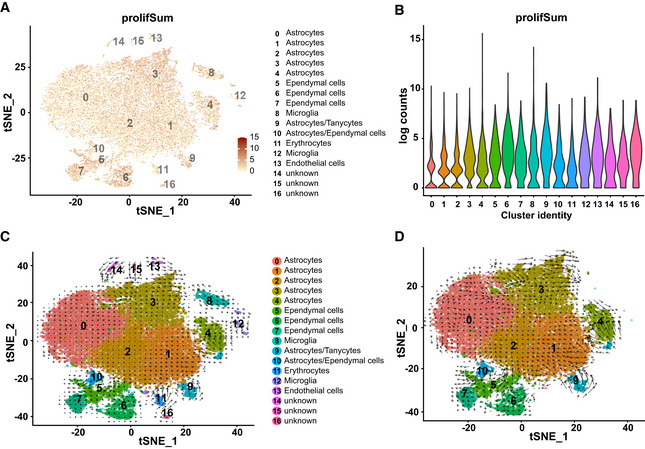
Figure 5. Widespread expression of proliferation‐associated genes in diencephalic astrocytes.
-
A, Bt‐Distributed stochastic neighbour embedding visualization of ACSA‐2‐MACSed cell populations of representative well‐known proliferation genes (A; numbers reflect scaled log‐normalized read counts for each cell, Dataset EV3 for Fig 5 A and B) and associated violin plot (B) showing distribution of proliferative genes across all clusters (cluster colour coding and annotation see in Fig 5C) as well as value per each single cell (A).
-
C, DVelocity analysis based on differences between spliced and unspliced transcripts for all cell clusters (C) and astrocyte clusters (D).
Interestingly, this analysis showed particularly high velocity in astrocyte clusters 4 and 9, with a direction towards the main astrocyte cloud (Fig 5C). This would be consistent with the generation of astrocytes from some of the tanycyte/ependymal cell populations, as previously suggested by GLASTCreERT2 fate mapping (Robins et al, 2013). Interestingly, also within the big astrocyte cloud velocity was high in clusters 1 and 3, with the former pointing towards cluster 2 and the latter showing an attractant within its own cluster at one end (upper left, Fig 5D). There was also notable dynamics visible at the island of cluster 4 (cells located between clusters 0 and 3). Taken together, both the proliferative index as well as the velocity analysis suggested the possibility of astrocyte proliferation, which we set‐out to examine further by monitoring DNA synthesis, expression of proliferation markers and clonal analysis.
Low‐level astrogenesis in the diencephalon persisting in 8‐month‐old mice
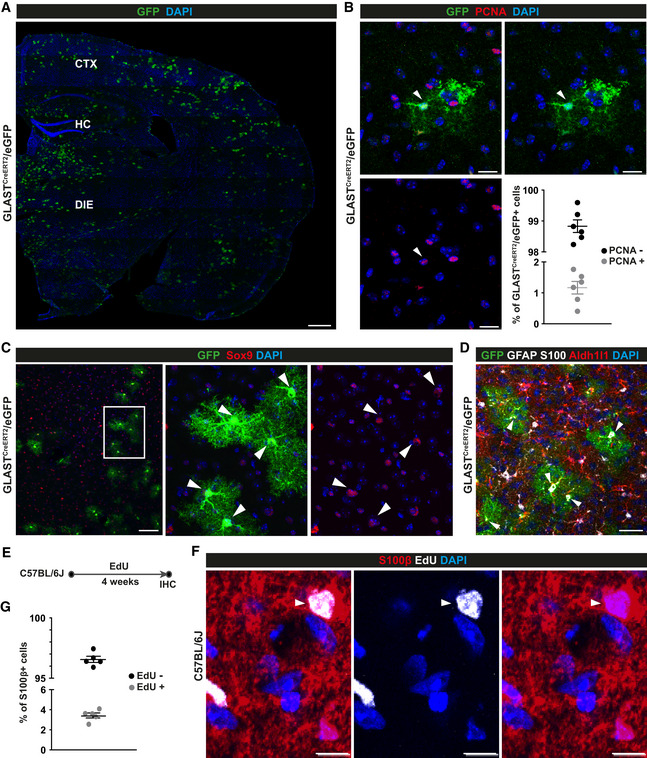
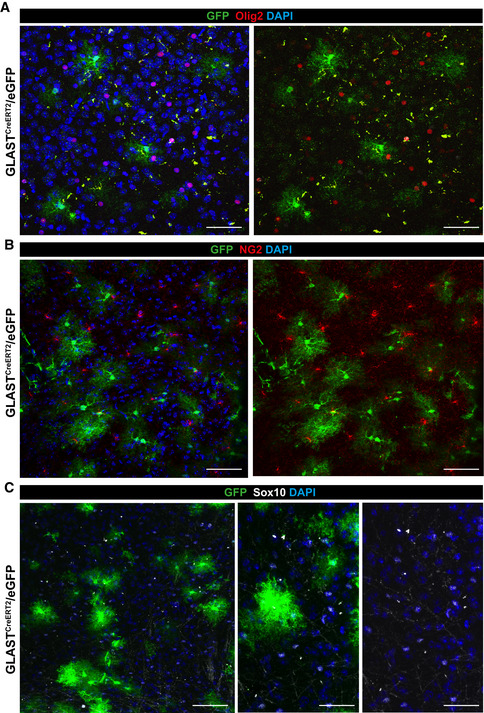
Given the above results, we explored if astrocyte proliferation occurs in the adult uninjured parenchyma of the DIE, i.e. beyond the age of 3 weeks when proliferating astrocytes were observed previously (Shoneye et al, 2020). Brain sections from GLASTCreERT2 mice (Mori et al, 2006) crossed to eGFP reporter mice (Nakamura et al, 2006) were immunostained for the proliferating cell nuclear antigen (PCNA) and GFP at 3 weeks after tamoxifen treatment (Fig 6A and B). As demonstrated before (Buffo et al, 2008; Bardehle et al, 2013; Sirko et al, 2015; Lange Canhos et al, 2021), cells labelled by this eGFP reporter line and this recombination protocol were almost all astrocytes, double‐positive for Sox9, S100, GFAP and Aldh1l1 (Fig 6C and D). Conversely, oligodendrocyte progenitors/NG2 glia (Psachoulia et al, 2009) stained for Olig2, NG2 and Sox10 were not labelled by GLASTCreERT2‐mediated recombination and hence we found no GFP double‐positive cells in the DIE (Fig EV5A–C). PCNA immunoreactivity was detected in a mean of 1.2% of the GFP‐labelled astrocytes in the DIE parenchyma (Fig 6B). As this is a small population and PCNA levels were relatively low, we next examined DNA synthesis during S‐phase by providing the thymidine analogue 5‐ethynyl‐2’‐deoxyuridine (EdU) for 4 weeks in the drinking water (Psachoulia et al, 2009) (Fig 6E). This resulted in 3.4% of S100β+ astrocytes that had incorporated EdU (Fig 6F and G), thereby further corroborating that some astrocytes are in S‐phase in the adult murine DIE. Slowly proliferating cells, such as also the oligodendrocyte progenitor cells known to proliferate in the adult brain, typically have a long G1 phase, e.g. up to 40 days at this age (Psachoulia et al, 2009). The small number of double‐positive astrocytes would thus be consistent with a slow cell cycle progression of many astrocytes expressing proliferation genes with a long G1 phase and hence only few of them in cell cycle at any given time. The alternative model of only a small subset of proliferating astrocytes and most being postmitotic is less consistent with the widespread expression of proliferation genes (Fig 5A).
Figure 6. Proliferation of diencephalic astrocytes in the adult mouse brain.
-
A–GMicrographs of maximum intensity projections of forebrain sections from 3‐mo‐old GLASTCreERT2/eGFP mice 21 days after tamoxifen treatment (A–D) and 3‐mo‐old C57BL/6J mice after EdU application for 4 weeks in drinking water (E, F) immunostained as indicated on top of the panels. Double‐positive cells are indicated by arrowheads. CTX: cerebral cortex; DIE: diencephalon; HC: hippocampus. Scale bars: 300 µm (A), 20 µm (B), 75 µm (C), 30 µm (D), 10 µm (F). (B, G) Histograms showing quantifications for B and F (B, n = 6 animals; 5 region of interest (ROI) covering the diencephalon on a total of 3 slides analysed per animal; G, n = 5 animals; 5 ROI covering the diencephalon on a total of 2 slides analysed per animal).
Data information: In B and G data are presented as mean ± SEM. Each data point represents one animal.
Figure EV5. Specificity for astrocyte labelling in GLASTCreERT2/eGFP mice.
-
A–CMaximum intensity projections showing immunostainings of brain sections of 3‐mo‐old GLASTCreERT2/eGFP mice for GFP and Olig2 (A) or NG2 (B) or Sox10 (C). Note that GFP+ cell population do not comprise cells of the oligodendroglial lineage. Scale bars: 50 µm (A, middle and right panels C), 75 µm (B and left panel C).
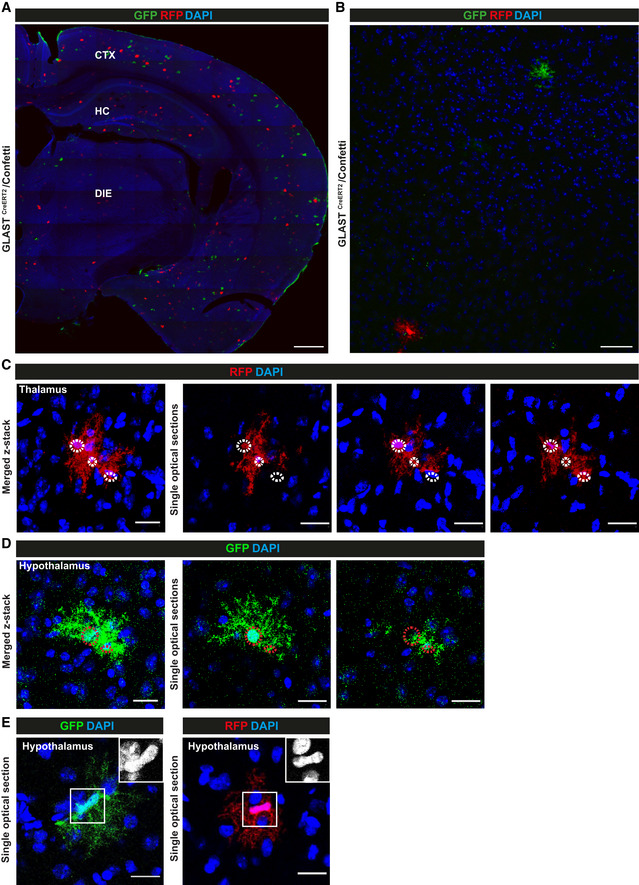
In order to determine if astrocytes were capable to complete the cell cycle and generate daughter cells, we used the previously established protocol for sparse recombination allowing clonal analysis (Bardehle et al, 2013; Lange Canhos et al, 2021) in GLASTCreERT2 mice crossed to the multicolour Confetti reporter mice (Snippert et al, 2010). Cells labelled by RFP or GFP antibody staining (the latter comprising cytoplasmic YFP and membrane CFP as the nuclear GFP is virtually not expressed (see (Calzolari et al, 2015)) were mostly single astrocytes distant from each other in both regions (Fig 7A and B and Appendix Fig S3A–C). Interestingly, however, clusters of 2–3 astrocytes labelled by the same fluorescent protein were observed in close vicinity (< 10 µm) in the DIE at 21 days post‐tamoxifen (dpt) (Fig 7C and D). As these clusters were not present at 3 dpt, they should have been generated by cell division. Notably, no such multi‐cell clones were detectable in the GM of the CTX consistent with previous data showing that astrocytes do not proliferate in the intact, uninjured CTX GM (Buffo et al, 2008; Sirko et al, 2009, 2013; Bardehle et al, 2013; Heimann et al, 2017). In the DIE, however, around 2% of clones had more than 1 cell, consistent with the above‐described detection of PCNA+ or EdU‐incorporating diencephalic astrocytes (Fig 6). Thus, astrocytes continue to proliferate generating new astrocytes, albeit at low level, in the intact DIE of 3‐month‐old mice.
Figure 7. Diencephalic astrocytes give rise to progeny in vivo .
-
A–EMicrographs of maximum intensity projections (A–D) or single optical section (E) of forebrain sections stained as indicated for fluorescent proteins in astrocytes labelled in 8 months (A, B, E) and 3 months (C, D) GLASTCreERT2/Confetti mice at 21 dpt. Note that clusters of duplets (D) and triplets (C) labelled in the same colour can be found only in the DIE. Also note that astrocyte somata are often very close together, as also observed after live in vivo imaging of dividing astrocytes (Bardehle et al, 2013). Somata of single cells are highlighted in circles. For the number and proportion of cell clones amongst all recombined astrocytes see Source data for Fig 7. For an example of a cell clone generated from a single RFP positive astrocyte in the DIE see Movie EV1.
Data information: CTX: cerebral cortex; DIE: diencephalon; HC: hippocampus. Scale bars: 300 µm (A), 100 µm (B), 50 µm (C, D), 20 µm (E).
Source data are available online for this figure.
Next, we asked if astrocytes may generate more than 1 daughter cell, as recently observed after repetitive injury for a few astrocyte clones in the CTX GM (Lange Canhos et al, 2021). However, even 2 months post‐tamoxifen clones observed in the DIE contained only 2 cells. To determine if new astrocytes are still generated in older mice, recombination was induced in 8‐month‐old mice. Indeed, also at this stage we observed 2‐cell clones (Fig 7E), suggesting that astrogenesis is still present in 8‐month‐old mice. Notably, no spatial bias was observed when analysing the distribution of clones with more than 1 cell (n = 40) with almost exactly 50% in thalamic or hypothalamic regions, respectively. Taken together, these data support a model of slow astrocyte proliferation driven by the widespread expression of proliferation genes in astrocytes leading to low‐level astrogenesis as shown by clonal analysis.
Smad4 is required for proliferation and neurosphere formation of diencephalic astrocytes
To identify possible molecular regulators of DIE astrocyte proliferation, we used genome‐wide expression data from population analysis of GLASTCreERT2/eGFP labelled astrocytes from the intact CTX GM where astrocytes do not proliferate (Buffo et al, 2008; Bardehle et al, 2013) and the DIE where astrocytes express proliferation genes and proliferate at low levels. As we had seen the proliferation in GLASTCreERT2/eGFP labelled astrocytes, we used this line crossed to the B6N.129Rpl22 tm1.1Psam/J (RiboTag) mice (Sanz et al, 2009) to express the Hemagglutinin (HA)‐tagged ribosomal protein Rpl22 upon Cre‐mediated recombination (Fig 8A–C, Appendix Fig S4A). Immunostaining for the HA‐tag showed a good recombination rate and astrocyte specificity in GLASTCreERT2 mice (see also Buffo et al, 2008) as virtually all HA‐labelled cells had astrocyte morphology in both regions (Fig 8B) and co‐localized with S100β (Fig 8C) and Sox9 (Appendix Fig S4B).
Figure 8. RNA‐seq of astrocytes from adult mouse cerebral cortex grey matter and diencephalon.
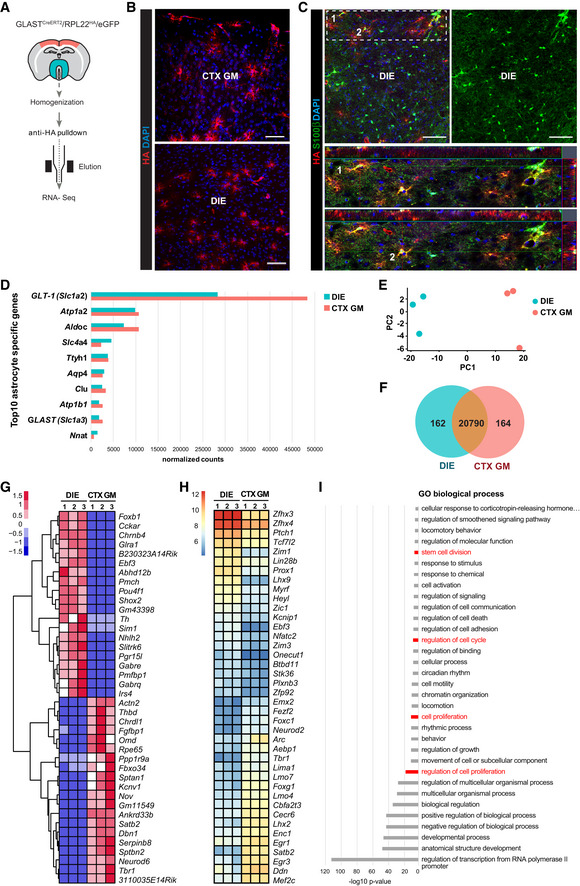
-
ASchematic drawing of RiboTag‐assisted mRNA isolation from astrocytes isolated from regions dissected as indicated in red (CTX GM) and turquoise green (DIE).
-
BMicrographs of forebrain sections of 3‐mo‐old GLASTCreERT2/RPL22HA/eGFP 21 dpt mice stained for HA show maximum intensity projections of HA+ expressing cells with astroglial morphology. CTX GM: cerebral cortex grey matter; DIE: diencephalon. Scale bars: 100 µm.
-
CMicrographs of forebrain sections of 3‐mo‐old GLASTCreERT2/RPL22HA/eGFP mice 21dpt stained for HA and S100β showing the specificity of the mouse line with HA detected only in S100β+ astrocytes. DIE: diencephalon. Scale bar: 70 μm.
-
DHistogram showing expression levels of the top 10 astrocyte‐specific markers genes in DIE (blue bars) and CTX GM (red bars) samples.
-
EPrincipal Component Analysis (PCA) from DeSeq2 expression data shows distinct clusters of samples from 3 DIE (blue) and 3 CTX GM (red) astrocyte sample preparations.
- F
-
G, HHeatmaps depicting top 40 differentially expressed genes (G, row scaled log‐normalized read counts) or transcription factors (H, Library size normalized log2 transformed read counts; from DIE and CTX GM astrocytes. Transcription factors were identified according to the content of AnimalTFDB database (Zhang et al, 2012).
-
IBar chart depicting significantly enriched GO terms (analysed first by Gorilla followed by Revigo) based on the 379 differentially expressed transcription factor genes (DeSeq2, cutoff pAdj0.01) comparing DIE and CTX GM astrocytes. Terms related to cell division, cell proliferation etc. are highlighted in red. (P‐value threshold was set to 0.01 http://cbl‐gorilla.cs.technion.ac.il).
Data information: CTX GM: cerebral cortex grey matter; DIE: diencephalon; n = 3 biological replicates, tissue from 3 mice was pooled
The DIE and CTX GM were dissected from 9 mice (Fig 8A), and 3 samples each were pooled for ribosome‐associated and HA‐purified RNA‐seq. The top 10 astrocyte‐specific genes based on previous analyses (Cahoy et al, 2008; Sharma et al, 2015; Zhang et al, 2016) were all highly expressed in these samples (Fig 8D), while neuronal, oligodendroglial, endothelial and microglial marker gene expression was either hardly detectable or much lower compared to other astrocyte‐specific marker genes (Appendix Fig S4C). Thus, consistent with previous data (Boisvert et al, 2018; Itoh et al, 2018), this protocol allows assessing the translatome of astrocytes in the regions of interests.
Region‐specific differences were revealed by principal component analysis (PCA) as all biological replicates from the same brain region cluster together (Fig 8E). Differential gene expression analysis using log(fc) ≥ 2 and P‐value threshold of 0.005 or lower identified 326 differentially expressed genes (Fig 8F) which amount to < 1.5% of all genes with detectable expression levels (21,116, Dataset EV4 for Fig 8). Thus, astrocytes from DIE and CTX GM share most of their gene expression and differ in a small proportion of their transcriptome. Amongst the top 20 regionally enriched genes were several region‐specific TFs, such as Tbr1, Neurod6 and Satb2 enriched in the CTX GM astrocytes, while Foxb1, Ebf3, Pou4f1 were enriched in DIE astrocytes (Fig 8G, Appendix Fig S4D). Interestingly, patterning TFs were mostly expressed in cluster 4 of the scRNA‐seq analysis (Dataset EV1 for Fig 1B). To explore the landscape of differentially expressed TFs further, we plotted r‐log‐normalized expressions for the top 20 TFs in both CTX GM and DIE (Fig 8H) and performed GO term analysis (Fig 8I). Interestingly, this analysis further corroborated our findings above with terms related to stem cell division, cell proliferation etc.
In our search for molecular regulators of astrocyte proliferation in the DIE, we noted inhibitors of BMP and Wnt signalling, such as Chrdl1, Nov (also known as Ccn3, an inhibitor of BMP2 and Notch signalling (Sakamoto et al, 2017)), Dkk3 and others amongst the highest differentially expressed genes in CTX GM astrocytes (Dataset EV4 for Fig 8). Notably, Chrdl1 has been detected in mostly upper layer CTX GM astrocytes as key regulator of synapse maturation (Blanco‐Suarez et al, 2018; Bayraktar et al, 2020). Conversely, we observed BMP‐responsive gene expression, such as Aqp4, AldoC, F3, Hopx (see (Chen et al, 2018) in the scRNA‐seq data of the DIE astrocytes. An unbiased search for signalling pathways using the Reactome pathway database (Reactome Version 70) (Fabregat et al, 2018) contained the term “SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription” (Fig 9A) as significantly upregulated in DIE astrocytes, with Sp1, Smad7, Ccnt2 showing higher expression in these astrocytes. Smads are TFs acting as down‐stream effectors in TGFβ or/and BMP signalling (Luo, 2017; Ampuja & Kallioniemi, 2018; Dituri et al, 2019), and both of these signalling pathways regulate NSC properties, including quiescence (Mira et al, 2010) as well as astrocyte differentiation during development (Mabie et al, 1997; Stipursky et al, 2014). Smad4, a common TGFβ/BMP signalling mediator, is expressed across all DIE astrocyte clusters (Fig 9B). After confirming that Smad4 is also detectable at protein level in DIE astrocytes (Fig 9C) and showed higher expression levels in DIE compared to CTX GM astrocytes isolated by MACS (Appendix Fig S4E), we proceeded to explore its functional role.
Figure 9. Smad4 regulates diencephalic astrocyte proliferation and neurosphere formation.
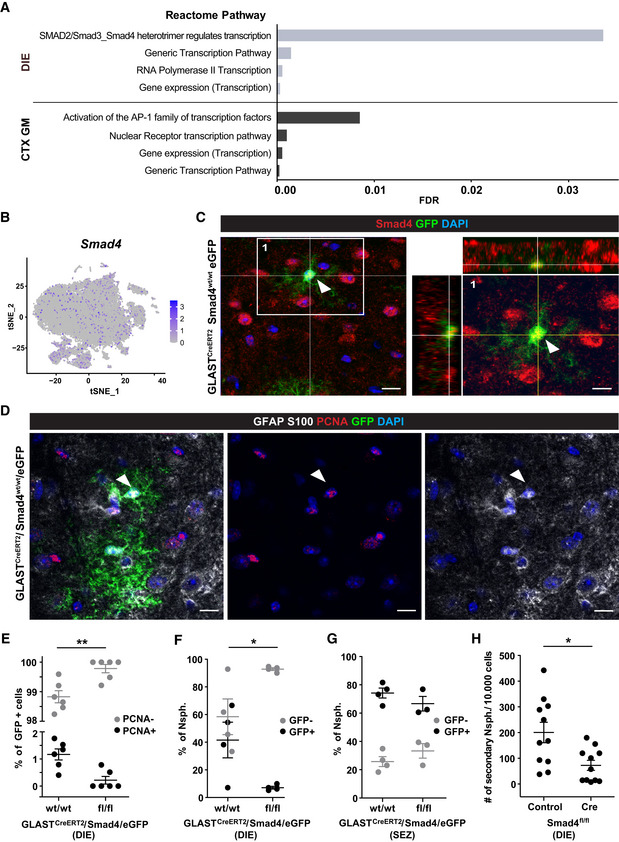
-
AReactome pathway analysis for all 174 transcription factor genes with higher expression in DIE (upper) and all 205 genes higher in CTX GM (lower) astrocytes.
-
Bt‐SNE visualization of DIE astrocytes isolated by ACSA‐2 MACS (Fig 1A) showing Smad4 expression (scaled log‐normalized read counts).
-
CExample of a Smad4/GFP double‐positive astrocyte (indicated by arrowhead) in a single optical section of DIE from 3‐mo‐old GLASTCreERT2/eGFP mice 21 dpt immunostained as indicated on top of the panel. Orthogonal projections of the area highlighted in left panel of C shown at higher magnification in the right panel. Scale bars: 15 µm (left panel) and 10 µm (right panel).
-
DExample of a proliferating (PCNA+, arrowhead) astrocyte in a single optical section of DIE from 3‐mo‐old GLASTCreERT2/eGFP mice 21 dpt immunostained as indicated on top with single channels shown in middle and right panels. Scale bars: 15 µm.
-
EHistogram depicting percentage of PCNA‐positive cells of all GFP‐positive cells in the DIE of the genotype indicated on the x‐axis (n = 6 animals per genotype; 5 ROI covering the diencephalon on a total of 3 slides were analysed per animal; PCNA‐negative and PCNA‐positive cells comparing wt/wt vs. fl/fl animals, adjusted P‐value = 0.002).
-
F, GHistograms depicting percentage of the neurosphere‐generating cells obtained from the DIE (F) or SEZ (G) of GLASTCreERT2/Smad4fl/fl/eGFP or GLASTCreERT2/Smad4WT/WT/eGFP, as indicated on the x‐axis (n = 3–4 animals for SEZ, 4 to 8 technical replicates analysed per animal; 4 animals for DIE, 8 technical replicates analysed per animal).
-
HHistogram depicting the number of secondary neurospheres generated from 10,000 Smad4f/f neurosphere cells at 7 days after infection with MLV‐based retrovirus containing either CAG‐NLS‐Cre or only CAG‐GFP (n = 11 cultures/animals, 3 technical replicates per condition analysed; control vs. Cre, P‐value = 0.0149). Note the decrease in DIE astrocyte proliferation in vivo (E) and in neurosphere formation upon Smad4 deletion (F, H).
Data information: In E, F, G and H data are presented as mean ± SEM. F: GFP‐negative and GFP‐positive cells comparing wt/wt vs fl/fl animals, adjusted P‐value = 0.0408; G: GFP‐negative and GFP‐positive cells comparing wt/wt vs fl/fl animals, ns). Each dot represents one n. *P < 0.05, **P < 0.01 (E, F, G: nonparametric 2way ANOVA followed by Sidak's multiple comparisons test, H: nonparametric Mann–Whitney test).
To do so, genetic deletion was induced in Smad4fl/fl mice (Yang et al, 2002) crossed to GLASTCreERT2 and eGFP reporter mice mentioned above, as previously described (Colak et al, 2008). The number of proliferating PCNA+ and GFP+ astrocytes was examined at 21 dpt (example shown in Fig 9D) and found to be significantly reduced in the DIE of GLASTCreERT2/Smad4fl/fl/eGFP compared to control tamoxifen‐treated GLASTCreERT2/Smad4WT/WT/eGFP mice (Fig 9E). As astrocyte proliferation in vivo is often accompanied by neurosphere‐forming potential of some of these cells (Buffo et al, 2008; Sirko et al, 2009, 2013; Bardehle et al, 2013; Heimann et al, 2017), we asked if we could find neurosphere‐forming activity from DIE cells and if they were affected by Smad4 deletion. To do so, we dissociated cells at 21 dpt. Indeed, in control mice we observed neurospheres derived from recombined eGFP+ cells of the DIE of GLASTCreERT2/eGFP mice that were significantly reduced from GLASTCreERT2/Smad4fl/fl/eGFP mice (Fig 9F). Notably, neurosphere numbers from GFP‐negative cells were not affected (Fig 9F). In contrast, GFP+ neurospheres from SEZ cells did not differ between genotypes (Fig 9G), consistent with our previous data that Smad4 regulates other fate decisions in the SEZ (Colak et al, 2008). To avoid potential issues with biased selection of cells in neurosphere culture condition after Smad4 deletion in vivo, we deleted Smad4 acutely in the cultured primary neurosphere cells in vitro by infection with an MLV‐based retrovirus containing CAG‐NLS‐Cre. This reduced the number of secondary neurospheres to less than half compared to control CAG‐GFP infected cultures 6 days after infection (Fig 9H). Taken together, these results support the idea that Smad4 regulates proliferation of DIE astrocytes in vivo and neurosphere formation in vitro.
Discussion
Comparison of diencephalic astrocytes and ependymal cells gene expression
Previous characterization of ACSA‐2 MACS isolation suggested contamination by some other cell types, but ependymal cells would not express the ACSA‐2 antigen ATP1B2 (Batiuk et al, 2017; Kantzer et al, 2017). However, labelling ependymal cells specifically by CellTrace dye injection and removing this population by sub‐dissection suggests that ACSA‐2 also isolates ependymal cells (Figs EV1, EV2E and Fig 1). Indeed, clusters 1–4 in the analysis without further sub‐dissection expressed many ependymal marker genes (Chen et al, 2017; Shah et al, 2018; Fig EV2E) including expression of transcription factor Foxj1, the master regulator of ciliogenesis (Yu et al, 2008), reflecting the multi‐ciliated nature of ependymal cells. Thus, we propose that these represent ependymal cells that indeed also share expression of some astrocyte genes (Fig EV1) as also noted before (Tavazoie et al, 2008; Beckervordersandforth et al, 2010; Shah et al, 2018). Differentiating the gene expression between clusters of ependymal cells, we also noted enrichment of some genes involved in promoting or inhibiting proliferation, Mns1 or Ecrg4 (1500015O10Rik), respectively, shared between cluster 1 and 4, while Nnat that is highly expressed in development but also in hypothalamus signalling (Joseph et al, 1995; Ivanova & Kelsey, 2011; Kanno et al, 2016, 2019) was shared between clusters 2 and 3. Thus, these data provide information about third ventricle ependymal cells, allowing comparisons to other ependymal cells isolated from the lateral ventricles (Shah et al, 2018) or spinal cord (Llorens‐Bobadilla et al, 2020).
However, it is also important to note that some genes expressed in ependymal cells and tanycytes are shared with astrocytes, and can indeed be detected in parenchymal astrocytes. We demonstrate this here for S100a6, with high signal in tanycytes, SEZ and DG cells (Yamada & Jinno, 2014; Kjell et al, 2020), as well as in the parenchymal astrocyte subsets in the hypothalamus (Fig 3, highest in cluster 9), but not thalamic regions, consistent with the spatial mapping shown in Fig 2J. Thus, some parenchymal subsets of astrocyte‐like cells share expression with tanycytes and other NSCs. Given the expression of S100a6 in NSCs, these parenchymal hypothalamic astrocytes could also be the origin of neurosphere‐forming cells obtained from the DIE, but as dissection cannot remove all ependymal cells, our gene expression analysis can now be used to identify novel, more specific markers for this population of astrocytes (e.g. by overlapping cluster 1 and cluster 9 gene expression for common markers).
Beyond S100a6 and NSC‐like hallmarks, shared gene expression between ependyma/tanycytes and hypothalamic astrocytes is important as hypothalamic astrocytes have been implicated in special functions, such as glucose sensing and the regulation of hunger and satiety as well as hormonal regulatory loops (Chowen et al, 2019; MacDonald et al, 2020). Indeed, cluster 5, 6, 7 and 10 cells express highest levels of Slc2a1, the glucose transporter 1, suggesting shared metabolic functions. Especially, cluster 9 shares GO terms of translation with the astrocyte cluster 1 (Dataset EV1 for Fig 1B and Dataset EV2 for Fig 4) highlighting the expression of Ntrk2, a neurotrophin receptor that has been described in astrocytes (Holt et al, 2019). Likewise, expression of TFs regulated by Ca2+, such as Aebp1 (see Dataset EV1 for Fig 1B), is shared between clusters of ependymal cells and those expanding to parenchymal astrocytes, as Foxj1. FoxJ1 has been described in both, astrocytes and NSCs (Jacquet et al, 2009, 2011; Devaraju et al, 2013). Thus, our data now allow further dissecting shared and distinct gene expression and function of ependymal cells and astrocytes, and point to regional differences in these similarities within the DIE.
Unprecedented astrocyte heterogeneity in the diencephalon
To tackle astrocyte heterogeneity, it was important to examine a larger number of astrocyte cells by lowering the ependymal cell contamination, now reaching an unprecedented number of astrocytes above 21,000 from one brain region, while only about 5,000 astrocytes were contained in the experiment comprising the high ependymal cell fraction. This allowed identifying a large diversity in gene expression with 7 astrocyte clusters from the DIE, identifying either distinct states or subtype identities. Notably, 4 of these clusters form a big supercluster (0–3) and comprise the vast majority of astrocytes (more than 19,000 cells), while clusters 4, 9 and 10 comprise only < 2,000 cells and hence represent more rare subtypes or states.
Previous scRNA‐seq of murine brain cells described 6–7 astrocyte subtypes/gene expression clusters in different brain regions according to developmental boundaries (Zeisel et al, 2018; Wheeler et al, 2020). So far in a single adult brain region, only astrocytes from the cerebral CTX GM and hippocampus were examined using the same MACS protocol for isolation (Batiuk et al, 2020; Bayraktar et al, 2020) reporting a lower number of clusters of astrocytes with distinct gene expression hallmarks. This shows the importance to collect many astrocytes from one region as done here to achieve sufficient resolution for detecting further differences in gene expression, even though the sequencing methods are not directly comparable. To which extent cell clustering in scRNA‐seq expression analysis represent indeed distinct subtypes or different states of cells is an ongoing issue of debate. This is impossible to discern without live imaging of cells expressing reporter of genes discriminating different cluster expression to observe if these change dynamically or rather remain stable. Here we used spatial mapping of the expression of all genes of a given cluster as a further approach to monitor where cells with a similar gene expression profile may be located (Fig 2). Indeed, this shows highly specific localization patterns for cells with a distinct identity, e.g. ependymal cells, microglia, endothelial cells, thereby verifying the reliability of this approach. It is possible, that not only astrocyte gene expression contributes to the similarities of gene expression explored by spatial mapping. We aimed to minimize this possibility by including the entire set of genes significantly enriched in cells of a given cluster for mapping. Moreover, several of these genes, e.g. the glutamate transporters and glutamine synthase, enriched in the respective clusters are highly selective for astrocytes and hence unlikely to include other cell types.
Interestingly, this analysis shows different spatial distribution also for the different astrocyte clusters, with a largely DIE restricted pattern for cluster 1, enrichment in thalamus and CTX for clusters 0, 2 and 3 and lateral DIE and hippocampus for cluster 4. This is intriguing as it highlights that the gene expression of some astrocyte states or subsets is shared with other cells more widespread throughout the forebrain (clusters 0, 2 and 3), while others are more region‐specific (clusters 1 and 4). This fits well with the expression of patterning TFs, such as Lhx2, Emx2 and Foxg1 in astrocytes of cluster 4, that are not detectable in clusters 0–3. These data could suggest that the region‐specific gene expression observed previously (John Lin et al, 2017; Morel et al, 2017; Itoh et al, 2018; Lozzi et al, 2020) and in this study in population RNA‐seq of astrocytes may at least partially be due to astrocyte subtypes or states. In this regard, it is notable that both clusters 4 and 9 are smaller, while the main astrocyte cloud comprised of clusters 0–3 express no or only common forebrain patterning factors, such as Foxg1. Conversely, clusters 2 and 3 gene expression have significant GO term enrichment all related to ion transport and ion homeostasis, highlighting pan‐astrocyte functions that may indeed be shared across regions. Interestingly, astrocyte gene expression clusters with significant GO terms related to mitochondrial functions, like cluster 1, show more restricted mapping, prompting the idea that metabolic adaptations may be more region‐specific and depending on the respective neuronal networks. However, this may also be clearly subject to different states of the animals depending on sensory inputs, arousal etc. Importantly, however, our work revealed for the first time gene expression states or subtypes of astrocytes that are more shared across the forebrain, while others are more restricted, at a given time in these animals from which cells were collected. It will be fascinating to unravel if these are stable over time or dynamic and if the latter in which time scale.
Continued low‐level astrogenesis in the diencephalon
To explore the dynamic gene expression of these astrocyte clusters, we performed velocity analysis based on differences between spliced and unspliced transcripts (La Manno et al, 2018). This showed higher values in clusters 3, 4, 9 as well as 0 and 1 (Fig 5C and D), i.e. dynamic transcription in several of the astrocyte gene expression clusters. Velocity arrows were sometimes directed between clusters, e.g. from cluster 9 to cluster 1. As cluster 9 also expresses tanycyte cell markers, this is consistent with previous fate mapping analysis suggesting that these may generate parenchymal astrocytes (Robins et al, 2013). Proliferation is one known contributing factor to high velocity scores and GO term analysis highlighted neural precursor cell proliferation in cluster 4 that had particularly high velocity scores. However, expression of proliferation‐associated genes was not restricted to these cell clusters, but rather widespread with little detectable differences between the clusters (Fig 5A and B). Consistent with this, we observed astrocytes labelled by Sox9 and S100β in the DIE of Aldh1l1‐eGFP mice or GLASTCreERT2 fate mapping to start proliferation, as shown by EdU incorporation and PCNA immunoreactivity (Fig 6 and Appendix Fig S1B), and some also completing it as shown by clonal analysis (Fig 7). This discriminates the astrocytes in the DIE from those in the cerebral CTX GM where no such clones could be observed (Bardehle et al, 2013; Lange Canhos et al, 2021). Importantly, this is not limited to adolescent stages (Shoneye et al, 2020), as we also found 2‐cell clones of astrocytes when recombination was induced in 8‐month‐old mice. The only other region where astrocytes proliferate in the absence of injury known so far is the hilus of the DG (Cosgrave et al, 2010; Nigussie et al, 2016; Garcia‐Martinez et al, 2020), but it is not clear if these derive from the subgranular NSCs or are indeed a proliferative astrocyte population. To our knowledge, the proliferation of DIE astrocytes is therefore the first evidence for continued astrocyte proliferation in the adult brain parenchyma.
However, one may argue that DIE astrogenesis may not matter due to its low degree with about 2% of all labelled astrocytes. However, this may be a regulated process that provides new astrocytes where and when they may be critical. For example, adult neurogenesis works in a “turn‐over” mode (Weinandy et al, 2011) with few of the newly generated neurons surviving long‐term, but providing transiently more excitable young neurons as a particularly plastic neuronal population to the neuronal circuitry (Sailor et al, 2017). Indeed, the proportion of 2‐cell clones was lower after longer post‐tamoxifen intervals as if only some of the newly generated astrocytes survive, similar to adult neurogenesis where only a minority of newly generated neurons persist (Weinandy et al, 2011; Calzolari et al, 2015; Goodman & Hajihosseini, 2015; Ryu et al, 2016; Pfisterer & Khodosevich, 2017). Especially in the hypothalamus, astrocyte plasticity is critical for the neuronal circuitry mediating metabolic control and homeostasis (Kim et al, 2014; Garcia‐Caceres et al, 2016, 2019; Argente‐Arizon et al, 2017; Zhou, 2018). Generation of new astrocytes may provide a further level of plasticity and be subject to regulation by metabolic changes, a fascinating hypothesis to pursue further. Thus, this first comprehensive analysis of thalamic and hypothalamic astrocyte heterogeneity opens many fascinating avenues towards a better understanding of the distinct region‐specific astrocyte functions.
Materials and Methods
Experimental animals
C57BL/6J mice (Charles River Laboratories; Sulzfeld, Germany) were used as controls. Smad4 floxed/floxed mice (Jackson Laboratories; SMAD4tm2.1CXD/J, loxP sites flanking exon 8) (Yang et al, 2002), R26R‐Confetti reporter mice (Jackson Laboratories; Gt(ROSA)26Sortm1(CAG‐Brainbow2.1)Cle/J) (Snippert et al, 2010) and RiboTag mice (Jackson Laboratories; B6N.129‐Rpl22tm1.1Psam/J) (Sanz et al, 2009) were maintained on a C57BL/6J background. For selective deletion of Smad4 in astrocytes, mice homozygous for the Smad4 floxed alleles were crossed with GLASTCreERT2 mice (Slc1a3tm1(cre/ERT2)Mgoe) (Mori et al, 2006) and the CAG‐eGFP reporter line (FVB.B6‐Tg(CAG‐cat,‐EGFP)1Rbns/KrnzJ) (Nakamura et al, 2006). Recombination was induced by administration of Tamoxifen at the age of 9 weeks, both sexes were used. Tamoxifen (Sigma‐Aldrich) was dissolved in corn oil (Sigma‐Aldrich) to a final concentration of 40 mg/ml and delivered by oral gavage (0.4 mg per gram body weight, gbw) 3 times with 48 h interval. GLASTCreERT2/Smad4wt/wt/eGFP mice exposed to Tamoxifen in the same way were used as controls and for astrocyte labelling. For selective induction of HA labelling in astrocytes, homozygous mice carrying Rpl22HA floxed/floxed were crossed with GLASTCreERT2 mice. Recombination analysis was done as described above. For clonal analysis, adult mice at 2–3 months and 8 months of age obtained from crosses between GLASTCreERT2 and R26R‐Confetti reporter mice (GLASTCreERT2/Confetti) were induced with a single intraperitoneal injection of 100 μg Tamoxifen/gbw. Aldh1l1‐eGFP mice (Tg(Aldh1l1‐EGFP)OFC789Gsat/Mmucd) (Gensat Project) (Gong et al, 2003) were maintained on the FVB/N background and used for ACSA‐2 based sorting. Both sexes were used. Proliferating cells were labelled with EdU (Thermo Fisher Scientific; 0.2 mg/ml water containing 1% sucrose) in drinking water for 4 consecutive weeks.
Genotypes were determined by PCR using following primers: Aldh1l1‐eGFP (forward: TTC ACC TTG ATG CCG TTC T reverse: GCC GCT ACC CCG ACC AC), CAG‐eGFP (forward: CTG CTA ACC ATG TTC ATG CC reverse: GGT ACA TTG AGC AAC TGA CTG), Confetti (1) (forward: GAA TTA ATT CCG GTA TAA CTT CG reverse: AGA GTA TAA AAC TCG GGT GAG C), Confetti (2) (forward: CTC CTG GCT TCT GAG GAC C reverse: CCA GAT GAC TAC CTA TCC TC), GLASTCreERT2 (1) (forward: GAG GCA CTT GGC TAG GCT CTG AGG A reverse: GAG GAG ATC CTG ACC GAT CAG TTG G), GLASTCreERT2 (2) (reverse: GGT GTA CGG TCA GTA AAT TGG ACA T), RPL22HA (forward: GGG AGG CTT GCT GGA TAT G reverse: TTT CCA GAC ACA GGC TAA GTA CAC), Smad4fl/fl (forward: TAA GAG CCA CAG GGT CAA GC reverse: TTC CAG GAA AAA CAG GGC TA).
All mice were kept under specified pathogen‐free conditions and housed in groups of 2–3 animals in filter top cages and a 12‐h/12‐h light/dark cycle. Mice had free access to water (acidified and desalinated) and standard rodent chow (Altromin, 1310M). All experimental procedures were performed in accordance with animal welfare policies and approved by the Government of Upper Bavaria (Germany).
Tissue immunohistochemistry
Mice were anaesthetized by intraperitoneal injection of ketamine (100 mg/kgbw) and xylazine (10 mg/kgbw) and after transcardial perfusions with 4% paraformaldehyde (PFA, vol/vol; Roth) in phosphate‐buffered saline (PBS) for 20 min, brains were postfixed overnight and cryoprotected for further analysis. For immunohistology, sections (30 µm, for clonal analysis 60 µm) were pre‐incubated for 30 min in blocking solution (10% normal goat serum (Invitrogen), 0.5% Triton X‐100 (Sigma) in PBS). The following primary antibodies were diluted in blocking solution and incubated with the sections for 24 h at 4°C: chick antibody to GFP (1:500, Aves Lab, GFP‐1020), mouse antibody to GFAP (1:500, Sigma, G3893), mouse antibody to S100β (1:500, Sigma, S2532), rabbit antibody to S100β (1:500, Abcam, ab52642), rabbit antibody to GFAP (1:250, Dako, Z0334), rabbit antibody to S100 (1:250, Sigma, S2644), mouse antibody to PCNA (1:200, Dako, M0879), rabbit antibody to S100a6 (1:500, Abcam, ab181975), rabbit antibody to RFP (1:500, Rockland, 600‐401‐379), rabbit antibody to Sox9 (1:1,500, Merck/Millipore, AB 5535), mouse antibody to Olig2 (1:250, Merck/Millipore, MABN50), goat antibody to Sox10 (1:50, Santa Cruz, sc‐17342), rabbit antibody to NG2 (1:400, Merck/Millipore, AB 5320), mouse antibody to HA (1:1,000, Abcam, ab130275), rabbit antibody to Smad4 (1:100, LIFE Technologies, PA534806), mouse antibody to Foxj1 (1:500, Invitrogen, 14‐9965‐82) and mouse antibody to Aldh1l1 (1:100, Merck/Millipore, MABN 495). After washing in PBS, secondary antibodies were diluted in blocking solution and incubated at room temperature for 2 h using fluorophore‐coupled (1:500) antibody to chick Alexa488 (Invitrogen, A11039), antibody to mouse Cy3 (Dianova, 115‐165‐003), antibody to rabbit Cy3 (Dianova, 711‐165‐152), antibody to mouse A555 (Invitrogen, A21127), antibody to mouse A647 (Invitrogen, A21240), antibody to goat A647 (Dianova, 705‐605‐003) and antibody to rabbit A647 (Dianova, 111‐605‐144) as secondary antibodies. Nuclei were stained with DAPI (1:1,000, final concentration of 0.1 µg/ml, Sigma, D9564) for 5 min at RT. PCNA staining was performed in free‐floating sections pretreated with 0.01 M Sodium citrate (pH 6) in 0.05% Tween20 at 96°C for 20 min. EdU incorporation was detected by Click‐iT™ EdU Alexa Fluor™ 647 Imaging Kit (Thermo Fisher Scientific) according to the manufacturer instructions.
Neurosphere culture
Neurosphere cultures were prepared as previously described (Buffo et al, 2008) using tissue from the superficial SEZ layer (Fischer et al, 2011) and the area of the DIE, sparing the region around the third ventricle (Fig EV1A). Cells were plated at a density of 5 cells/µl in 600 µl of neurosphere medium (DMEM/F12, Gibco; with 1% of PenStrep (vol/vol), 1% B27 (vol/vol) and 2 mM l‐Glutamine, all Invitrogen) supplemented with fibroblast growth factor 2 (FGF2) and epidermal growth factor (EGF) (both at 20 ng/ml, Invitrogen) in a single well of a 24 well plate (Eppendorf). For in vitro deletion of Smad4, neurospheres were generated from 2‐ to 3–mo‐old Smad4fl/fl mice. 12 h after passaging the primary generated spheres by dissociation with 0.05% trypsin/EDTA (Invitrogen) for 12 min followed by inhibition with Ovomucoid (Roche), cells were infected with a MLV‐based retrovirus containing either CAG‐NLS‐Cre or only CAG‐GFP and neurosphere formation was assayed 6 days after infection (Colak et al, 2008).
MACS purification of astrocytes and Single‐Cell RNA Sequencing
Diencephalic cells were first isolated from nine 3‐mo‐old Aldh1l1‐eGFP mice of both sexes by ACSA‐2 (Anti‐ACSA‐2 MicroBead Kit, mouse; Miltenyi Biotec) magnetic‐associated cell separation as described before resulting in the dataset provided as Source data for Fig EV1 (Batiuk et al, 2017). In a second set of experiments, cells were isolated from eight 3‐mo‐old Aldh1l1‐eGFP mice after dissection under visual inspection for any remaining ependymal cells. Briefly, an intraventricular injection of 500 µl carboxyfluorescein esters (CFSEs/CellTrace™, Thermo Fisher Scientific) was performed to label ependymal cells 20 min before the dissection with the following coordinates from Bregma: RC: −0.2 mm; ML: −1.0 mm; DV: −2.0 mm. The final concentration of 10 mM working solution was obtained by adding 8 µl of DMSO (Invitrogen) and 1 µl of Fast Green (Sigma‐Aldrich) to one vial of CellTrace CFSE as previously described (Govindan et al, 2018). Visual inspection of remaining FT labelling was carried out during dissection to exclude ependymal cells from the final sample. At this stage, the Aldh1l1‐eGFP signal is not visible and therefore does not interfere with FT detection. MACS procedures were carried out following a validated protocol resulting in the data provided as Dataset EV1 for Fig 1B (Batiuk et al, 2017). For both experiments, cells were eluted and resuspended in PBS to reach a final concentration of 1,000 cells/µl. 17,000 cells were then processed using the Single‐Cell 3’ Reagent Kits v2 (for Source data for Fig EV1) or v3.1 (for the 10x dataset after optimized dissection, Dataset EV1 for Fig 1B) from 10xGenomics according to the manufacturer instructions. This was followed by GEM generation and barcoding, post‐GEM‐RT cleanup, cDNA amplification and library construction. Illumina sequencing libraries were sequenced on one lane with HiSeq 4000 system (100 bp sequencing, 50 k row reads per cell, Source data for Fig EV1) or with a NovaSeq 6000 (NovaSeq Flow cell Type SP, 20 K row reads per cell for the second 10× dataset, Dataset EV1 for Fig 1B) after quality assessment with the Bioanalyzer (Agilent).
Real‐time PCR
RNA was isolated directly after cell sorting and centrifugation with the Arcturus Picopure Kit (Thermo Fisher Scientific) then cDNA was synthesized with the Maxima First‐Strand cDNA Synthesis Kit (Life Technologies) and qPCR was performed on a LightCycler480 instrument (Roche) using LightCycler Probe Master kit (Roche) and Monocolor Hydrolysis Probe (Roche) according to manufacturer’s instructions (20 μl final volume). The expression of each gene was analysed in triplicates. Data were processed with the ΔΔC t method (Livak & Schmittgen, 2001). Expression levels were normalized to GAPDH. Primers used for qPCR are listed below. Smad4 (forward: GAGAACATTGGATGGACGACT, reverse: CACAGACGGGCATAGATCAC); GAPDH (forward: TTCACCACCATGGAGAAGG, reverse: CACACCCATCACAAACATGG). For each sample, we analysed three technical replicates.
Cell immunocytochemistry
Cells after MACS isolation were fixed in 4% PFA for 10 min at RT after 20 min adhesion time on Poly‐D‐Lysine (PDL) coated coverslips. After washing in PBS and blocking for 30 min in 2% BSA (Sigma) in PBS, cells were incubated for 24 h at 4°C with the primary antibody (chicken antibody to GFP, 1:500, Aves Lab, GFP‐1020). Nuclei were stained with DAPI (1:10,000, Sigma, D9564). Secondary antibodies were used as described above.
RiboTag‐assisted isolation of ribosomal‐associated mRNA of adult astrocytes and transcriptome analysis
The same area of dissection as used for scRNA‐seq (Figure EV1A turquoise green and red) was used from GLASTCreERT2/RPL22HA/eGFP mice. Per replicate material from 3 mice was pooled. mRNA was isolated as described before (Sanz et al, 2009), purified using RNeasy Micro Kit (Qiagen 74004) and cDNA was prepared using Ovation RNA‐seq System v2 (NuGEN 7102). Amplified cDNA was fragmented, and libraries were prepared using IonXpress plus gDNA & Amplificon Library Preparation kit (4471269, 4471252, 4471250). Samples were barcoded during the library preparation with sample‐specific indices. Equimolar ratio of each sample was then pooled and sequenced on an Ion Proton Sequencer.
The raw reads were separated based on the barcode sequences and the barcodes/adapters were trimmed. The pre‐processed reads were checked for quality and then aligned using STAR software 2.4.2a to Mus Musculus genome (mm10) and were quantified against mm10 ensemble release 89 annotations. The gene counts normalization and differential gene expression analysis was performed using DESeq2 (Love et al, 2014) and R (R Foundation for Statistical Computing, Vienna, Austria, http://www.R‐project.org/). Genes with fold change > 2 and adjusted P‐value < 0.005 were considered as differentially regulated.
Single‐cell and spatial transcriptomics
ScRNA‐seq reads from two diencephalon preparations differing by exclusion of the dorsal part of the ventricular lining and connections to the lateral ventricle in the second sample were aligned against mm10 mouse genome (builds versions 1.2.0 and 2020A from 10xGenomics) using CellRanger software versions 3.0.2 and 6.0.1 with default settings. Subsequent single‐cell expression analysis and clustering was facilitated by Seurat version 2.3.4 (Butler et al, 2018) on R version 3.5.3 (R Foundation for Statistical Computing, Vienna, Austria, http://www.R‐project.org/.). We obtained 4,651 and 25,911 cells with UMI counts of at least 500 and mitochondrial gene counts lower than 30% and 40%, respectively. Data were log‐normalized to 10,000 counts per cell and scaled to regress out UMI counts and percentage of mitochondrial counts. Genes with variance to mean ratio of logged values lower than 0.5 and average expression lower than 0.0125 or bigger than 4 were removed leaving us with 2,839 and 1,501 variable genes. The respective dimensions of PCA projection were used to perform clustering by shared nearest neighbour modularity optimization algorithm with resolution of 0.6 and 10 iterations of Louvain algorithm (Waltman & van Eck, 2013). Same number of PCA dimensions were used to represent data by 2D t‐SNE projections. Differential expression analysis of genes specific for each cluster was done comparing each cluster to remaining data with Wilcox rank sum algorithm providing us with log fold change (fc), adjusted P‐value and coverage over the cells. To calculate single‐cell velocity of gene expression from exonic and intronic reads, we used Velocyto algorithm (Svensson & Pachter, 2018) with projection time eq. 1, gamma fit on 2% quantiles of expression values and slope calculations smoothed over 25 nearest cells. Velocity values, logarithmic scaled and multiplied by to highlight interesting features were smoothed over 200 cells and overlaid as arrows onto t‐SNE projection of clustered scRNA‐seq data. The spatial gene expression dataset of coronal section of a mouse brain was provided by 10xGenomics (https://support.10xgenomics.com/spatial‐gene‐expression/datasets/1.1.0/V1_Adult_Mouse_Brain). The dataset has sequencing depth of 115,569 read pairs, 6,018 median genes and 28,944 median UMI counts per spot with 2,702 spots cover by tissue (spot size 55 μm). 10 µm thick slice of the coronal plane from fresh frozen embedded and cryosectioned (Visium Spatial Protocols‐Tissue Preparation Guide CG000240) mouse brain tissue (Strain C57BL/6) from BioIVT Asterand were placed on Visium Gene Expression Slides. Microscopic picture was recorded over 2 ms with Nikon Ti2‐E microscope using 10× objective and 4.5× gain. The library (T1T2‐F3) was prepared with Visium Spatial Gene Expression Reagent Kits CG000239 according to manufacturer protocol and sequenced on Illumina NovaSeq 6000. Data was mapped against mouse genome (mm10) with Space Ranger 1.1.0. Spatial dataset was regularized by negative binomial regression to normalize UMI counts. Subsequently, clusters from our scRNA‐seq diencephalon data were projected into Visium spatial dataset by finding similarities between them with anchors transfer algorithm with SCT normalization of query data and log2 normalization of PCA cells embeddings of reference data, and 5 neighbours to define anchors (Seurat version 3.2; Stuart et al, 2019).
Quantification and statistical analysis
Confocal laser scanning (Zeiss LSM710) or epifluorescence (Zeiss, Axiophot) microscopes were used to quantify immunopositive cells in sections or cell culture. Quantification of immunostaining was performed on ≥ 3 sections per animal from each experimental group (≥ 3 animals) on multi‐channel, confocal 3D stacks, using Zeiss ZEN 2010 software. The levels of individual immunostaining were adjusted independently for optimal visualization. Sections stained for antibodies mentioned above were documented using constant exposure settings.
Statistical analysis of data was performed using R (R Foundation for Statistical Computing, Vienna, Austria, http://www.R‐project.org/.) and PRISM (GraphPad, v 7.03). Sample numbers and experimental repeats are indicated in figure legends. As listed below, statistical tests were chosen dependent on sample size, normality of distribution and number of comparisons. As indicated in figure legends, quantitative data are presented as mean ± standard error of the mean (SEM) or median ± interquartile range (IQR) with 25–75% range (borders) and 10–90% range (whiskers) of data distribution. Normality of data distribution was tested using the Kolmogorov–Smirnov test (Fig 9E–H) or Shapiro–Wilk test (Appendix Fig S4E). All tests used in this study are only composed of nonparametric tests. For multiple comparisons we used Sidak test as alpha adjustment is part of this method. P‐values were calculated by nonparametric 2way ANOVA followed by Sidak's multiple comparisons test (Fig 9E: wt/wt vs fl/fl, P‐value = 0.0020; Fig 9F: wt/wt vs fl/fl, P‐value = 0.0408; Fig 9G: wt/wt vs fl/fl, not significant (ns)) or nonparametric Mann‐Whitney test (Fig 9H: one‐tailed, P‐value = 0.0149; Appendix Fig S4E: two‐tailed, ns). The minimum level of significance was defined as P < 0.05 and is indicated based on the P‐value (*P < 0.05; **P < 0.01; ***P < 0.001).
Author contributions
SO, SC, TS‐E, MT, SS, RB and JF‐S performed and designed experiments. SO analysed the data in Figs 4 and 6, 7, 8, 9 with M.T. contributing to experiments and analysis for Fig 7. SC performed statistics for Fig 9E–H. SC performed experiments and analysed data shown in Fig 3B and C. SS with MT performed experiments and analysed data shown in Figs 6A,C,D, Figs 7A and B, and EV5, Appendix Fig S1A–D, Appendix Fig S3 and Appendix Fig S4A. PS analysed data in Figs 1, 2, 3A, 4, and 5 with input from JF‐S and MG, in Figs EV1 and EV2 with input from JF‐S., MG and SC and Fig EV4 with input from JF‐S and MG. The protocol for scRNA‐seq of astrocytes was established by SC, TS‐E and JF‐S who also performed the MACS isolation and library preparations. NK and MR carried out the bulk RNA‐seq analysis, analysed the data shown in Fig 8 and Appendix Fig S4C and PS performed the transcriptional regulator analysis in Fig 8H and Appendix Fig S4D. RB had the idea of how to label ependymal cells, performed and analysed the experiments in Fig EV3. MT, SC, SO and SU established, maintained and performed experiments with transgenic mouse lines. SS, SO and SC prepared figures and graphs. SS initially established the idea of the project and provided input during the project. MG designed the project and experiments, discussed the results, wrote the manuscript with input from all co‐authors, coordinated, directed and financed the project.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Movie EV1
Source Data for Figure 7
Source Data for Figure EV1B
Acknowledgements
We would like to thank Jaime Eugenin von Bernhardi and Leda Dimou for initial help with long‐term BrdU‐labelling experiments to detect proliferating cells in the diencephalon, Luisa Lange Canhos for providing expertise in clonal labelling and initial samples of GLASTCreERT2/Confetti, Detlef Franzen for excellent technical assistance and Jovica Ninkovic for excellent comments on the manuscript. We thank the Next‐Generation Sequencing Core Facility (Helmholtz Zentrum Muenchen) for sequencing. This work was funded by the DFG, the German Research Foundation, within the Priority Program 1757 to MG and MR, and to MG: the Collaborative Research Center 870, European Research Council (advanced ERC grant ChroNeuroRepair GA Nr. 340793), the EU consortium NSC Reconstruct (Nr. 874758) and SyNergy (EXC 2145 / Projekt‐ID 390857198). Open Access funding enabledand organized by Projekt DEAL.
The EMBO Journal (2021) 40: e107532.
Contributor Information
Swetlana Sirko, Email: swetlana.sirko@med.uni-muenchen.de.
Pawel Smialowski, Email: pawel.smialowski@helmholtz-muenchen.de.
Judith Fischer‐Sternjak, Email: judith.fischer@helmholtz-muenchen.de.
Magdalena Götz, Email: magdalena.goetz@helmholtz-muenchen.de.
Data availability
The datasets produced in this study are available in the following database: 10×Genomics single‐cell RNA‐seq and bulk RNA‐seq: Gene Expression Omnibus Accession ID: GSE149115.
References
- Allen NJ, Eroglu C (2017) Cell biology of astrocyte‐synapse interactions. Neuron 96: 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampuja M, Kallioniemi A (2018) Transcription factors‐Intricate players of the bone morphogenetic protein signaling pathway. Genes Chromosomes Cancer 57: 3–11 [DOI] [PubMed] [Google Scholar]
- Argente‐Arizon P, Guerra‐Cantera S, Garcia‐Segura LM, Argente J, Chowen JA (2017) Glial cells and energy balance. J Mol Endocrinol 58: R59–R71 [DOI] [PubMed] [Google Scholar]
- Bardehle S, Krüger M, Buggenthin F, Schwausch J, Ninkovic J, Clevers H, Snippert HJ, Theis FJ, Meyer‐Luehmann M, Bechmann I et al (2013) Live imaging of astrocyte responses to acute injury reveals selective juxtavascular proliferation. Nat Neurosci 16: 580–586 [DOI] [PubMed] [Google Scholar]
- Batiuk MY, Martirosyan A, Wahis J, de Vin F, Marneffe C, Kusserow C, Koeppen J, Viana JF, Oliveira JF, Voet T et al (2020) Identification of region‐specific astrocyte subtypes at single cell resolution. Nat Commun 11: 1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiuk MY, de Vin F, Duque SI, Li C, Saito T, Saido T, Fiers M, Belgard TG, Holt MG (2017) An immunoaffinity‐based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA‐2 antibody. J Biol Chem 292: 8874–8891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Bartels T, Holmqvist S, Kleshchevnikov V, Martirosyan A, Polioudakis D, Ben Haim L, Young AMH, Batiuk MY, Prakash K et al (2020) Astrocyte layers in the mammalian cerebral cortex revealed by a single‐cell in situ transcriptomic map. Nat Neurosci 23: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, Kirchhoff F, Hirrlinger J, Haslinger A, Lie DC et al (2010) In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 7: 744–758 [DOI] [PubMed] [Google Scholar]
- Ben Haim L, Rowitch DH (2017) Functional diversity of astrocytes in neural circuit regulation. Nat Rev Neurosci 18: 31–41 [DOI] [PubMed] [Google Scholar]
- Blanco‐Suarez E, Liu TF, Kopelevich A, Allen NJ (2018) Astrocyte‐Secreted Chordin‐like 1 Drives Synapse Maturation and Limits Plasticity by Increasing Synaptic GluA2 AMPA Receptors. Neuron 100: 1116–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert MM, Erikson GA, Shokhirev MN, Allen NJ (2018) The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep 22: 269–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo A, Rite I, Tripathi P, Lepier A, Colak D, Horn AP, Mori T, Gotz M (2008) Origin and progeny of reactive gliosis: a source of multipotent cells in the injured brain. Proc Natl Acad Sci USA 105: 3581–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA et al (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28: 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari F, Michel J, Baumgart EV, Theis F, Gotz M, Ninkovic J (2015) Fast clonal expansion and limited neural stem cell self‐renewal in the adult subependymal zone. Nat Neurosci 18: 490–492 [DOI] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AMJ, Resch JM, McCarroll SA et al (2017) A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci 20: 484–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Van Gulden S, McGuire TL, Fleming AC, Oka C, Kessler JA, Peng CY (2018) BMP‐responsive protease HtrA1 Is differentially expressed in astrocytes and regulates astrocytic development and injury response. J Neurosci 38: 3840–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Wu X, Jiang L, Zhang Y (2017) Single‐cell RNA‐Seq reveals hypothalamic cell diversity. Cell Rep 18: 3227–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowen JA, Frago LM, Fernandez‐Alfonso MS (2019) Physiological and pathophysiological roles of hypothalamic astrocytes in metabolism. J Neuroendocrinol 31: e12671 [DOI] [PubMed] [Google Scholar]
- Chung WS, Allen NJ, Eroglu C (2015) Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb Perspect Biol 7: a020370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D, Mori T, Brill MS, Pfeifer A, Falk S, Deng C, Monteiro R, Mummery C, Sommer L, Gotz M (2008) Adult neurogenesis requires Smad4‐mediated bone morphogenic protein signaling in stem cells. J Neurosci 28: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrave AS, McKay JS, Morris R, Quinn JP, Thippeswamy T (2010) The effects of nitric oxide inhibition prior to kainic acid treatment on neuro‐ and gliogenesis in the rat dentate gyrus in vivo and in vitro. Histol Histopathol 25: 841–856 [DOI] [PubMed] [Google Scholar]
- Denis‐Donini S, Estenoz M (1988) Interneurons versus efferent neurons: heterogeneity in their neurite outgrowth response to glia from several brain regions. Dev Biol 130: 237–249 [DOI] [PubMed] [Google Scholar]
- Devaraju K, Barnabe‐Heider F, Kokaia Z, Lindvall O (2013) FoxJ1‐expressing cells contribute to neurogenesis in forebrain of adult rats: evidence from in vivo electroporation combined with piggyBac transposon. Exp Cell Res 319: 2790–2800 [DOI] [PubMed] [Google Scholar]
- Dituri F, Cossu C, Mancarella S, Giannelli G (2019) The interactivity between TGFbeta and BMP signaling in organogenesis, fibrosis, and cancer. Cells 8: 1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley JG, Macklis JD (2006) Astroglial heterogeneity closely reflects the neuronal‐defined anatomy of the adult murine CNS. Neuron Glia Biol 2: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B et al (2018) The reactome pathway knowledgebase. Nucleic Acids Res 46: D649–D655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WT, Murai K (2017) Resolving astrocyte heterogeneity in the CNS. Front Cell Neurosci 11: 300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Beckervordersandforth R, Tripathi P, Steiner‐Mezzadri A, Ninkovic J, Gotz M (2011) Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nat Protoc 6: 1981–1989 [DOI] [PubMed] [Google Scholar]
- Furness DN, Dehnes Y, Akhtar AQ, Rossi DJ, Hamann M, Grutle NJ, Gundersen V, Holmseth S, Lehre KP, Ullensvang K et al (2008) A quantitative assessment of glutamate uptake into hippocampal synaptic terminals and astrocytes: new insights into a neuronal role for excitatory amino acid transporter 2 (EAAT2). Neuroscience 157: 80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Cáceres C, Balland E, Prevot V, Luquet S, Woods SC, Koch M, Horvath TL, Yi C‐X, Chowen JA, Verkhratsky A et al (2019) Role of astrocytes, microglia, and tanycytes in brain control of systemic metabolism. Nat Neurosci 22: 7–14 [DOI] [PubMed] [Google Scholar]
- García‐Cáceres C, Quarta C, Varela L, Gao Y, Gruber T, Legutko B, Jastroch M, Johansson P, Ninkovic J, Yi C‐X et al (2016) Astrocytic insulin signaling couples brain glucose uptake with nutrient availability. Cell 166: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Martinez Y, Sanchez‐Huerta KB, Pacheco‐Rosado J (2020) Quantitative characterization of proliferative cells subpopulations in the hilus of the hippocampus of adult Wistar rats: an integrative study. J Mol Histol 51: 437–453 [DOI] [PubMed] [Google Scholar]
- Gilbert A, Vidal XE, Estevez R, Cohen‐Salmon M, Boulay AC (2019) Postnatal development of the astrocyte perivascular MLC1/GlialCAM complex defines a temporal window for the gliovascular unit maturation. Brain Struct Funct 224: 1267–1278 [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME et al (2003) A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925 [DOI] [PubMed] [Google Scholar]
- Goodman T, Hajihosseini MK (2015) Hypothalamic tanycytes‐masters and servants of metabolic, neuroendocrine, and neurogenic functions. Front Neurosci 9: 387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Sirko S, Beckers J, Irmler M (2015) Reactive astrocytes as neural stem or progenitor cells: In vivo lineage, In vitro potential, and Genome‐wide expression analysis. Glia 63: 1452–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S, Bribian A, Lopez‐Mascaraque L, Gotz M, Grothe B, Kunz L (2021) Heterogeneity of astrocytes: electrophysiological properties of juxtavascular astrocytes before and after brain injury. Glia 69: 346–361 [DOI] [PubMed] [Google Scholar]
- Govindan S, Oberst P, Jabaudon D (2018) In vivo pulse labeling of isochronic cohorts of cells in the central nervous system using FlashTag. Nat Protoc 13: 2297–2311 [DOI] [PubMed] [Google Scholar]
- Heimann G, Canhos LL, Frik J, Jager G, Lepko T, Ninkovic J, Gotz M, Sirko S (2017) Changes in the proliferative program limit astrocyte homeostasis in the aged post‐traumatic murine cerebral cortex. Cereb Cortex 27: 4213–4228 [DOI] [PubMed] [Google Scholar]
- Holt LM, Hernandez RD, Pacheco NL, Torres Ceja B, Hossain M, Olsen ML (2019) Astrocyte morphogenesis is dependent on BDNF signaling via astrocytic TrkB.T1. eLife 8: e44667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Itoh Y, Tassoni A, Ren E, Kaito M, Ohno AI, Ao Y, Farkhondeh V, Johnsonbaugh H, Burda J et al (2018) Cell‐specific and region‐specific transcriptomics in the multiple sclerosis model: Focus on astrocytes. Proc Natl Acad Sci USA 115: E302–E309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E, Kelsey G (2011) Imprinted genes and hypothalamic function. J Mol Endocrinol 47: R67–R74 [DOI] [PubMed] [Google Scholar]
- Jacquet BV, Muthusamy N, Sommerville LJ, Xiao G, Liang H, Zhang Y, Holtzman MJ, Ghashghaei HT (2011) Specification of a Foxj1‐dependent lineage in the forebrain is required for embryonic‐to‐postnatal transition of neurogenesis in the olfactory bulb. J Neurosci 31: 9368–9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet BV, Salinas‐Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT (2009) FoxJ1‐dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 136: 4021–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Lin C‐C, Yu K, Hatcher A, Huang T‐W, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W et al (2017) Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci 20: 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Dou D, Tsang W (1995) Neuronatin mRNA: alternatively spliced forms of a novel brain‐specific mammalian developmental gene. Brain Res 690: 92–98 [DOI] [PubMed] [Google Scholar]
- Kang P, Lee H, Glasgow S, Finley M, Donti T, Gaber Z, Graham B, Foster A, Novitch B, Gronostajski R et al (2012) Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74: 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno N, Fujiwara K, Yoshida S, Kato T, Kato Y (2019) Dynamic changes in the localization of neuronatin‐positive cells during neurogenesis in the embryonic rat brain. Cells Tissues Organs 207: 127–137 [DOI] [PubMed] [Google Scholar]
- Kanno N, Higuchi M, Yoshida S, Yako H, Chen M, Ueharu H, Nishimura N, Kato T, Kato Y (2016) Expression studies of neuronatin in prenatal and postnatal rat pituitary. Cell Tissue Res 364: 273–288 [DOI] [PubMed] [Google Scholar]
- Kantzer CG, Boutin C, Herzig ID, Wittwer C, Reiß S, Tiveron MC, Drewes J, Rockel TD, Ohlig S, Ninkovic J et al (2017) Anti‐ACSA‐2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia 65: 990–1004 [DOI] [PubMed] [Google Scholar]
- Khakh BS, Deneen B (2019) The emerging nature of astrocyte diversity. Annu Rev Neurosci 42: 187–207 [DOI] [PubMed] [Google Scholar]
- Kim JG, Suyama S, Koch M, Jin S, Argente‐Arizon P, Argente J, Liu Z‐W, Zimmer MR, Jeong JK, Szigeti‐Buck K et al (2014) Leptin signaling in astrocytes regulates hypothalamic neuronal circuits and feeding. Nat Neurosci 17: 908–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjell J, Fischer‐Sternjak J, Thompson AJ, Friess C, Sticco MJ, Salinas F, Cox J, Martinelli DC, Ninkovic J, Franze K et al (2020) Defining the adult neural stem cell niche proteome identifies key regulators of adult neurogenesis. Cell Stem Cell 26: 277–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A et al (2018) RNA velocity of single cells. Nature 560: 494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Canhos L, Chen M, Falk S, Popper B, Straub T, Gotz M, Sirko S (2021) Repetitive injury and absence of monocytes promote astrocyte self‐renewal and neurological recovery. Glia 69: 165–181 [DOI] [PubMed] [Google Scholar]
- Lehre KP, Danbolt NC (1998) The number of glutamate transporter subtype molecules at glutamatergic synapses: chemical and stereological quantification in young adult rat brain. J Neurosci 18: 8751–8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Llorens‐Bobadilla E, Chell JM, Le Merre P, Wu Y, Zamboni M, Bergenstrahle J, Stenudd M, Sopova E, Lundeberg J, Shupliakov O et al (2020) A latent lineage potential in resident neural stem cells enables spinal cord repair. Science 370: eabb8795 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozzi B, Huang TW, Sardar D, Huang AY, Deneen B (2020) Regionally distinct astrocytes display unique transcription factor profiles in the adult brain. Front Neurosci 14: 61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K (2017) Signaling cross talk between TGF‐beta/Smad and other signaling pathways. Cold Spring Harb Perspect Biol 9: a022137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabie PC, Mehler MF, Marmur R, Papavasiliou A, Song Q, Kessler JA (1997) Bone morphogenetic proteins induce astroglial differentiation of oligodendroglial‐astroglial progenitor cells. J Neurosci 17: 4112–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald AJ, Holmes FE, Beall C, Pickering AE, Ellacott KLJ (2020) Regulation of food intake by astrocytes in the brainstem dorsal vagal complex. Glia 68: 1241–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederos S, Gonzalez‐Arias C, Perea G (2018) Astrocyte‐neuron networks: a multilane highway of signaling for homeostatic brain function. Front Synaptic Neurosci 10: 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortigüela R, Marqués‐Torrejón MÁ, Nakashima K et al (2010) Signaling through BMPR‐IA regulates quiescence and long‐term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7: 78–89 [DOI] [PubMed] [Google Scholar]
- Morel L, Chiang MSR, Higashimori H, Shoneye T, Iyer LK, Yelick J, Tai A, Yang Y (2017) Molecular and functional properties of regional astrocytes in the adult brain. J Neurosci 37: 8706–8717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Tanaka K, Buffo A, Wurst W, Kuhn R, Gotz M (2006) Inducible gene deletion in astroglia and radial glia–a valuable tool for functional and lineage analysis. Glia 54: 21–34 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Colbert MC, Robbins J (2006) Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res 98: 1547–1554 [DOI] [PubMed] [Google Scholar]
- Nigussie F, Huang PS, Lukauskis K, Bawa B, Moussa E, Abbott LC (2016) Neural cell proliferation and survival in the hippocampus of adult CaV 2.1 calcium ion channel mutant mice. Brain Res 1650: 162–171 [DOI] [PubMed] [Google Scholar]
- Pfisterer U, Khodosevich K (2017) Neuronal survival in the brain: neuron type‐specific mechanisms. Cell Death Dis 8: e2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psachoulia K, Jamen F, Young KM, Richardson WD (2009) Cell cycle dynamics of NG2 cells in the postnatal and ageing brain. Neuron Glia Biol 5: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robel S, Berninger B, Gotz M (2011) The stem cell potential of glia: lessons from reactive gliosis. Nat Rev Neurosci 12: 88–104 [DOI] [PubMed] [Google Scholar]
- Robins SC, Stewart I, McNay DE, Taylor V, Giachino C, Goetz M, Ninkovic J, Briancon N, Maratos‐Flier E, Flier JS et al (2013) alpha‐Tanycytes of the adult hypothalamic third ventricle include distinct populations of FGF‐responsive neural progenitors. Nat Commun 4: 2049 [DOI] [PubMed] [Google Scholar]
- Romanos J, Benke D, Saab AS, Zeilhofer HU, Santello M (2019) Differences in glutamate uptake between cortical regions impact neuronal NMDA receptor activation. Commun Biol 2: 127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JR, Hong CJ, Kim JY, Kim EK, Sun W, Yu SW (2016) Control of adult neurogenesis by programmed cell death in the mammalian brain. Mol Brain 9: 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailor KA, Schinder AF, Lledo PM (2017) Adult neurogenesis beyond the niche: its potential for driving brain plasticity. Curr Opin Neurobiol 42: 111–117 [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Matsushita Y, Minamizato T, Katsuki Y, Katsube KI, Yamaguchi A (2017) The bone regeneration model and primary osteoblastic cell culture used in the analysis of Ccn3 transgenic and knockout mice. Methods Mol Biol 1489: 309–324 [DOI] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS (2009) Cell‐type‐specific isolation of ribosome‐associated mRNA from complex tissues. Proc Natl Acad Sci USA 106: 13939–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PT, Stratton JA, Stykel MG, Abbasi S, Sharma S, Mayr KA, Koblinger K, Whelan PJ, Biernaskie J (2018) Single‐cell transcriptomics and fate mapping of ependymal cells reveals an absence of neural stem cell function. Cell 173: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique‐Hoyos N, Kongi K, Cantuti L, Hanisch U‐K, Philips M‐A et al (2015) Cell type‐ and brain region‐resolved mouse brain proteome. Nat Neurosci 18: 1819–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoneye T, Orrego AT, Jarvis R, Men Y, Chiang MSR, Yang Y (2020) Differential proliferation and maturation of subcortical astrocytes during postnatal development. Front Neurosci 14: 435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirko S, Behrendt G, Johansson P, Tripathi P, Costa MR, Bek S, Heinrich C, Tiedt S, Colak D, Dichgans M et al (2013) Reactive glia in the injured brain acquire stem cell properties in response to sonic hedgehog. [corrected]. Cell Stem Cell 12: 426–439 [DOI] [PubMed] [Google Scholar]
- Sirko S, Irmler M, Gascón S, Bek S, Schneider S, Dimou L, Obermann J, De Souza Paiva D, Poirier F, Beckers J et al (2015) Astrocyte reactivity after brain injury‐: The role of galectins 1 and 3. Glia 63: 2340–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirko S, Neitz A, Mittmann T, Horvat‐Brocker A, von Holst A, Eysel UT, Faissner A (2009) Focal laser‐lesions activate an endogenous population of neural stem/progenitor cells in the adult visual cortex. Brain 132: 2252–2264 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon‐Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD et al (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144 [DOI] [PubMed] [Google Scholar]
- Stipursky J, Francis D, Dezonne RS, Bergamo de Araujo AP, Souza L, Moraes CA, Alcantara Gomes FC (2014) TGF‐beta1 promotes cerebral cortex radial glia‐astrocyte differentiation in vivo . Front Cell Neurosci 8: 393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive integration of single‐cell data. Cell 177: 1888–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V, Pachter L (2018) RNA velocity: molecular kinetics from single‐cell RNA‐Seq. Mol Cell 72: 7–9 [DOI] [PubMed] [Google Scholar]
- Tavazoie M, Van der Veken L, Silva‐Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia‐Verdugo JM, Doetsch F (2008) A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijido O, Casaroli‐Marano R, Kharkovets T, Aguado F, Zorzano A, Palacin M, Soriano E, Martinez A, Estevez R (2007) Expression patterns of MLC1 protein in the central and peripheral nervous systems. Neurobiol Dis 26: 532–545 [DOI] [PubMed] [Google Scholar]
- Teijido O, Martinez A, Pusch M, Zorzano A, Soriano E, Del Rio JA, Palacin M, Estevez R (2004) Localization and functional analyses of the MLC1 protein involved in megalencephalic leukoencephalopathy with subcortical cysts. Hum Mol Genet 13: 2581–2594 [DOI] [PubMed] [Google Scholar]
- Vandenberg RJ, Ryan RM (2013) Mechanisms of glutamate transport. Physiol Rev 93: 1621–1657 [DOI] [PubMed] [Google Scholar]
- Waltman L, van Eck NJ (2013) A smart local moving algorithm for large‐scale modularity‐based community detection. Eur Phys J B 86: 471 [Google Scholar]
- Weinandy F, Ninkovic J, Gotz M (2011) Restrictions in time and space–new insights into generation of specific neuronal subtypes in the adult mammalian brain. Eur J Neurosci 33: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Westergard T, Rothstein JD (2020) Astrocyte diversity: current insights and future directions. Neurochem Res 45: 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MA, Clark IC, Tjon EC, Li Z, Zandee SEJ, Couturier CP, Watson BR, Scalisi G, Alkwai S, Rothhammer V et al (2020) MAFG‐driven astrocytes promote CNS inflammation. Nature 578: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J, Jinno S (2014) S100A6 (calcyclin) is a novel marker of neural stem cells and astrocyte precursors in the subgranular zone of the adult mouse hippocampus. Hippocampus 24: 89–101 [DOI] [PubMed] [Google Scholar]
- Yang X, Li C, Herrera PL, Deng CX (2002) Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32: 80–81 [DOI] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, Roy S (2008) Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 40: 1445–1453 [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G et al (2018) Molecular architecture of the mouse nervous system. Cell 174: 999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H‐M, Chen H, Liu W, Liu H, Gong J, Wang H, Guo A‐Y (2012) AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Research 40: D144–D149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Sloan S, Clarke L, Caneda C, Plaza C, Blumenthal P, Vogel H, Steinberg G, Edwards M, Li G et al (2016) Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89: 37–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YD (2018) Glial regulation of energy metabolism. Adv Exp Med Biol 1090: 105–121 [DOI] [PubMed] [Google Scholar]


