Abstract
The S-M checkpoint is an intracellular signaling pathway that ensures that mitosis is not initiated in cells undergoing DNA replication. We identified cid1, a novel fission yeast gene, through its ability when overexpressed to confer specific resistance to a combination of hydroxyurea, which inhibits DNA replication, and caffeine, which overrides the S-M checkpoint. Cid1 overexpression also partially suppressed the hydroxyurea sensitivity characteristic of DNA polymerase δ mutants and mutants defective in the “checkpoint Rad” pathway. Cid1 is a member of a family of putative nucleotidyltransferases including budding yeast Trf4 and Trf5, and mutation of amino acid residues predicted to be essential for this activity resulted in loss of Cid1 function in vivo. Two additional Cid1-like proteins play similar but nonredundant checkpoint-signaling roles in fission yeast. Cells lacking Cid1 were found to be viable but specifically sensitive to the combination of hydroxyurea and caffeine and to be S-M checkpoint defective in the absence of Cds1. Genetic data suggest that Cid1 acts in association with Crb2/Rhp9 and through the checkpoint-signaling kinase Chk1 to inhibit unscheduled mitosis specifically when DNA polymerase δ or ɛ is inhibited.
Orderly progression through the eukaryotic cell cycle requires that mitosis be inhibited not only during normal, unperturbed DNA replication but also when cells are exposed to drugs, such as the ribonucleotide reductase inhibitor hydroxyurea (HU), that inhibit S-phase progression. This aspect of cell cycle regulation is performed by an intracellular signal transduction pathway termed the S-M checkpoint. In the fission yeast Schizosaccharomyces pombe, this pathway serves both to inhibit the activity of Cdc2, the key mitosis-promoting cyclin-dependent kinase and, separately, to promote recovery from S-phase arrest. DNA polymerase α (Pol α) and the products of the cdc18, cut5/rad4, and orp1/cdc30 genes are required for the generation of the S-M checkpoint signal as well as being essential for prereplication complex assembly or the initiation of DNA replication itself (12, 20, 23, 39). Fission yeast cells lacking any one of these essential gene products fail to enter S phase but also fail to inhibit entry into mitosis. In contrast, mutations in genes required either later in S phase, in G2, or in G1 result in cell cycle arrest without progression into unscheduled mitosis. These observations suggest that a major S-M checkpoint signal is established at an early stage during DNA replication and that generation of this signal requires assembly of the initiation complex itself.
In S. pombe, as in the budding yeast Saccharomyces cerevisiae and probably in other eukaryotes, Pol δ and ɛ have essential functions that are required, along with Pol α, for chromosomal DNA replication (8, 13, 18). Pol δ and ɛ are thought to be responsible for the elongation of primers generated by the Pol α-primase complex, although recent reports surprisingly conclude that the catalytic domain of Pol ɛ is nonessential (11, 24). Since Pol α continues to be required for lagging-strand synthesis, it could retain responsibility for generation of the S-M checkpoint signal throughout S phase. In the budding yeast S. cerevisiae, a related but distinct role may be played by Pol ɛ, mutation of which can allow cells to enter mitosis in the presence of HU (35). S. pombe or S. cerevisiae cells with a deletion of the gene encoding the catalytic subunit of Pol ɛ nonetheless arrest in early S phase without attempting to enter mitosis (13, 33); this is in sharp contrast to the loss of S-M checkpoint function in fission yeast cells lacking Pol α or containing a catalytically inactive form of the protein (6, 12).
Downstream from the essential, DNA replication-associated components of the S-M checkpoint, a number of nonessential signaling components have been identified. In fission yeast these include the “checkpoint Rad” proteins Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1, which are also required for cell cycle arrest following DNA damage (1, 2, 14, 37). Components involved in checkpoint signalling following HU treatment differ subtly from those involved following DNA polymerase inhibition, with Crb2/Rhp9 being required for the latter but not the former (21, 38, 45). The Cds1 protein kinase functions downstream from the checkpoint Rad proteins to promote cell survival after both forms of S-phase inhibition (34). Recent evidence has also suggested an S-M checkpoint-signaling role for the Chk1 protein kinase, which plays a role similar to that of Cds1 but is required for cell cycle arrest following DNA damage (42). Although cells lacking chk1 (chk1Δ), like those lacking cds1 (cds1Δ), arrest normally after exposure to HU, chk1+ function is required to prevent aberrant mitosis after temperature-sensitive (ts) Pol δ mutants are shifted to their restrictive temperature (16). Even in the presence of wild-type Pol δ, after protracted incubation in HU at 37°C, chk1Δ cells lose viability more rapidly than do wild-type controls and enter aberrant mitoses (17). In addition, cds1Δ chk1Δ cells are S-M checkpoint defective and lose viability more rapidly than do cds1 mutants (and as rapidly as checkpoint rad mutants) after exposure to HU at 30 to 32°C, the optimal temperature range for fission yeast growth (7, 26, 46). These findings suggest either that absence of Cds1 leads to the generation of DNA structures recognized as damage by a Chk1-dependent checkpoint pathway (26) or that Cds1 and Chk1 have a degree of functional overlap. The latter interpretation is supported by the observations that moderate Chk1 overexpression can suppress the HU sensitivity of cds1Δ cells and that Cds1 and Chk1 have very similar activities in vitro (46). On the other hand, unlike Cds1, Chk1 phosphorylation (and, by inference, activity) is not elevated after HU treatment, except in cells lacking Cds1 (26, 43).
Inhibition of mitosis in response to activation of the S-M checkpoint in fission yeast is achieved through inhibitory phosphorylation of Cdc2 at tyrosine residue 15 (Y15). Thus, cells that overproduce the Cdc25 protein phosphatase, which acts to remove Cdc2 Y15 phosphorylation, or that express mutant forms of Cdc2 that do not require activation by Cdc25 fail to inhibit mitosis when DNA replication is inhibited by HU (15). Mutants of this sort are defective only in the aspect of S-M checkpoint control that governs mitotic entry, and hence their loss of viability following exposure to HU is less dramatic than that seen with checkpoint rad mutants, which in addition lack the checkpoint function governing recovery from S-phase inhibition. In contrast, mitotic entry is inhibited following HU treatment of cds1 or rqh1 mutants, but these are HU sensitive, probably because they lack the ability to organize recovery from S-phase arrest (34). The mechanisms by which Cds1 and Chk1 could promote inhibitory phosphorylation of Cdc2 include phosphorylation-mediated inactivation of Cdc25, stabilization of the Mik1 protein kinase, which acts in concert with Wee1 to phosphorylate Cdc2 at Y15, and phosphorylation of Wee1 (7, 19, 36, 46).
In mammalian cells, many components of the checkpoint pathways outlined above are conserved, including analogues of several of the checkpoint Rad proteins and the Chk1 and Cds1 protein kinases. For some years it has been known that the S-M and G2 DNA damage checkpoints can be overridden by treatment of mammalian cells with a variety of structurally diverse drugs, including methylxanthines such as caffeine and several other inhibitors of protein kinases or protein phosphatases. We recently demonstrated that caffeine can also override the S-M checkpoint in fission yeast (44). Caffeine treatment of S. pombe cells arrested in S phase by HU leads to progression into unscheduled mitosis and rapid loss of cell viability, similar to that seen in a checkpoint rad mutant exposed to HU alone. The sensitivity of wild-type fission yeast cells to a combination of HU and caffeine is suppressed by overexpression of either Cds1 or Chk1. These data are consistent with the notion that caffeine acts by inhibition of the S-M checkpoint pathway upstream from these protein kinases, either at or close to the point of action of the checkpoint Rad proteins. By exploiting this toxicity of HU and caffeine, we were able to identify a novel gene (termed cid1, for “caffeine-induced death resistant”) that, when overexpressed, confers resistance specifically to this combination of drugs. Here we describe the results of a detailed analysis of cid1, which led us to conclude that the product of this gene, while not essential under normal circumstances, is a nucleotide transferase-like protein specifically required to inhibit mitosis and promote cell survival when DNA polymerase δ or ɛ is inhibited.
MATERIALS AND METHODS
Fission yeast strains and methods.
The conditions for growth, maintenance, and genetic manipulation of fission yeast were as described previously (32). A complete list of the strains used in this study is given in Table 1. Except where otherwise stated, strains were grown at 30°C in yeast extract-peptone-dextrose (YPD) or Edinburgh minimal medium (EMM2) with appropriate supplements. Where necessary, gene expression from plasmids containing the nmt1 promoter (30) was repressed by the addition of 5 μM thiamine to the growth medium.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| HM123 | h− leu1-32 | Laboratory stock |
| 428/429 | h+/h− ade6-M210/ade6-M216 his7/his7 leu1-32/leu1-32 ura4-D18/ura4-D18 | Laboratory stock |
| cid1Δ | h− cid1::ura4+ leu1-32 ura4-D18 | This study |
| cid1Δ(LEU2) | h+ cid1::LEU2 leu1-32 ura4-D18 | This study |
| cid11Δ | h− cid11::ura4+ leu1-32 ura4-D18 | This study |
| cid12Δ | h− cid12::ura4+ leu1-32 ura4-D18 | This study |
| rad1Δ | h− rad1::ura4+ ade6-704 leu1-32 ura4-D18 | A. M. Carr |
| rad3Δ | h− rad3::ura4+ ade6-704 leu1-32 ura4-D18 | A. M. Carr |
| rad9Δ | h− rad9::ura4+ ade6-704 leu1-32 ura4-D18 | A. M. Carr |
| rad17Δ | h− rad17::ura4+ ade6-704 leu1-32 ura4-D18 | A. M. Carr |
| rad26Δ | h− rad26::ura4+ ade6-704 leu1-32 ura4-D18 | A. M. Carr |
| rqh1Δ | h− rqh1::ura4+ leu1-32 ura4-D18 | A. M. Carr |
| chk1Δ | h− chk1::ura4+ ade6-704 leu1-32 ura4-D18 | A. M. Carr |
| chk1 cid1Δ | h− cid1::LEU2 chk1::ura4 leu1-32 ura4-D18 | This study |
| cds1Δ | h− cds1::ura4+ leu1-32 ura4-D18 | A. M. Carr |
| cds1 cid1Δ | h− cid1::LEU2 cds1::ura4+ leu1-32 ura4-D18 | This study |
| crb2Δ | h− crb2::ura4+ leu1-32 ura4-D18 | P. Nurse |
| cdc1 | h− cdc1-P13 leu1-32 | P. A. Fantes |
| cdc1 cid1Δ | h− cdc1-P13 cid1::ura4+ ura4-D18 | This study |
| cdc6 | h− cdc6-121 | P. Nurse |
| cdc6 cid1Δ | h− cdc6-121 cid1::ura4+ ura4-D18 | This study |
| cdc17 | h+ cdc17-K42 | P. Nurse |
| cdc17 cid1Δ | h− cdc17-K42 cid1::ura4+ ura4-D18 | This study |
| cdc20 | h− cdc20-M10 leu1-32 | P. Nurse |
| cdc20 cid1Δ | h− cdc20-M10 leu1-32 cid1::ura4+ ura4-D18 | This study |
| cdc20-P7 | h− cdc20-P7 | P. Nurse |
| cdc20-P7 cid1Δ | h− cdc20-P7 cid1::ura4+ ura4-D18 | This study |
| cdc22 | h− cdc22-M45 | P. Nurse |
| cdc22 cid1Δ | h− cdc22-M45 cid1::ura4+ ura4-D18 | This study |
| cdc27 | h− cdc27-P11 leu1-32 | P. A. Fantes |
| cdc27 cid1Δ | h− cdc27-P11 cid1::ura4+ leu1-32 ura4-D18 | This study |
| cdc27 rad1Δ | h− cdc27-P11 rad1::ura4+ ura4-D18 | This study |
| cdc27 chk1Δ | h− cdc27-P11 chk1::ura4+ leu1-32 ura4-D18 | This study |
| cdc27 cds1Δ | h− cdc27-P11 cds1::ura4+ leu1-32 ura4-D18 | This study |
| cdc27 crb2Δ | h− cdc27-P11 crb2::ura4+ leu1-32 ura4-D18 | This study |
| cdc27 cid1Δ crb2Δ | h− cdc27-P11 cid1::LEU2 crb2::ura4+ leu1-32 ura4-D18 | This study |
| pol1 | h− swi7-H4 | H. Murakami |
| pol1 cid1Δ | h− swi7-H4 cid1::ura4+ ura4-D18 | This study |
| polδts1 | h− polδts1 | S. Francesconi |
| polδts1 cid1Δ | h− polδts1 cid1::ura4+ ura4-D18 | This study |
| polδts2 | h− polδts2 | S. Francesconi |
| polδts2 cid1Δ | h− polδts2 cid1::ura4+ ura4-D18 | This study |
| polδts3 | h− polδts3 | S. Francesconi |
| polδts3 cid1Δ | h− polδts3 cid1::ura4+ ura4-D18 | This study |
Plasmids and site-directed mutagenesis.
The isolation of pREP3Xcid1 was described previously (44). pREP1cid1 was generated by ligation of the cid1 cDNA insert from pREP3Xcid1 between the NdeI and BamHI sites of pREP1 (31). PCR using primers CID1MUTA and D10NOTI and primers CID1MUTB and D10OP5′ (Table 2) was used to generate the cid1 open reading frame in two fragments overlapping by 54 bp, with the region of overlap spanning codons 101 and 103, which were altered in the primer sequences to specify alanine rather than the aspartate residues specified by the wild-type gene at these positions. The resulting fragments were then mixed and used in a secondary PCR with primers D10OP5′ and D10NOTI. After digestion with NdeI and NotI, the final product was ligated into a derivative of pREP41 (31) containing a NotI site to generate pREP41cid1DADA. All plasmid constructions were confirmed by complete sequencing of the inserts using an ABI 377 sequencer and ABI PRISM dRhodamine reagents (Perkin-Elmer). Plasmids pREP1cds1 and pREP1chk1 were generously provided by Hiroshi Murakami (Imperial Cancer Research Fund, London, United Kingdom). In each of these plasmids the level of expression is attenuated by the presence of a CG tail in the 5′ untranslated region, resulting in cell cycle delay rather than the cell cycle arrest phenotype that results from the high-level expression of Cds1 or Chk1 in the absence of this element.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) |
|---|---|
| CID1A | TAATTAGCACACACATACAAAGAACGAATTTACCAGGCGACTGAGTCTTTCTTTCAAAAACCAAAATCCCCTCTAATAAAAAATCCCACTGGCTATATGT |
| CID1B | GAATTTTTATTGTAAACATTTCTTTTGGAATCATAAAAAAATTTGAGGCTACAAAAAGTAATTAGTCTTTATAAGTGTCTAATTCTAAATGCCTTCTGAC |
| CID11A | TTAGGTTATTAGGCGTTAATAAATCATCATTTAATATTTTTTAAGTTAATATTTTTATTTAGGAGGCTTAACTTACTATAAAATCCCACTGGCTATATGT |
| CID11B | TTAAATATTAATATTGCAAAGTTATCTCATTAATTTTACTTTGGCAATCTTTTTCCAATTTATTTATTTATTTCTTAATTAATTCTAAATGCCTTCTGAC |
| CID12A | TTACATATAATTACAAGGCACTCGCACGACCTCGTTATGTGCGAGGAGCCATGAAATTGAATCCATTGATATTAAAATTAAAATCCCACTGGCTATATGT |
| CID12B | ACCACATGCGGCAAGACAACTTAGGAATTGAAAAACAAATGTTTATTTAAACAGCGAGCATTATTTTTTAAATGCATTAAAATTCTAAATGCCTTCTGAC |
| 13cA | AGTACAGATGGGCGCTGGCTTATTTCCGGCGATGGAGGAGGCATGGTAAAGTATTTTGAACCGAATTTAAACAATGTCAAAAATCCCACTGGCTATATGT |
| 13cB | ACATTTTAACATATGCTCACGATGTTGACGAACCCCTTCATCAACTGATAATATGGTACTTAGGATACTAGGGAAATCTTAATTCTAAATGCCTTCTGAC |
| H9.01A | TCTTTCAAAGGTTTCGTTAATTAATGTTTCAATCGTTTAAAAGCGGCATACCCTTTATTTATTCTGTGATCCTTAATTAAAAATCCCACTGGCTATATGT |
| H9.01B | CTTTCGAAACTAATATCACCGGCCAACGGTATTTTGAAAGTGAGTCAGAGAGGGAAAAAACTGTTTTTTTCTGTTCTTATAATTCTAAATGCCTTCTGAC |
| CID1LEUA | TCAGCATTCTTTCTCTAAATAGGAATTTGTTACTTAATGGAGAAAAAAATGTTTCGATTTACCTAGTGTATTTGTTTGTATTATAGGATAATTATACTCT |
| CID1LEUB | CCAACCAAAAAAATTTTACATTAGTCTTTTTTTAATGCTGAGAAAGTCTTTGCTGATATGCCTTCCAACCAGCTTCTCTAATATAGTTTCGTCTACCCTA |
| CID1MUTA | TCTGGTTTAGCACTTAAAAATTCGGCTATGGCTTTGTGCGTGCTTATGGATTCG |
| CID1MUTB | CGAATCCATAAGCACGCACAAAGCCATAGCCGAATTTTTAAGTGCTAAACCAGA |
| D10OP5′ | TTTCATATGAACATTTCTTCTGCACAA |
| D10NOTI | TTTGCGGCCGCGCTCAGAATTGTCACC |
Gene disruption.
The one-step disruption method was used, following PCR-mediated generation of the entire ura4+ gene flanked by 80-bp segments from the 5′ and 3′ regions of the gene to be disrupted (5). Oligonucleotides used to generate ura4+ disruption cassettes for cid1, cid11, cid12, SPAC12G12.13c, and SPAC17H9.01 (CID1A and CID1B, CID11A and CID11B, CID12A and CID12B, 13cA and 13cB, and H9.01A and H9.01B, respectively) are listed in Table 2. Following transformation of strain 428/429, diploid ura+ progeny were screened for the desired integration pattern by diagnostic PCR amplifications using primer pairs spanning the presumptive recombination sites (details of the additional primers used for this purpose are available from the authors on request). Frequencies of homologous recombination (i.e., successful targeted gene disruption) ranged from 9 to 80%. Meiosis and sporulation were induced by plating onto malt extract agar, and tetrad dissection was performed with an MSM micromanipulator (Singer Instruments) as described by Moreno et al. (32). Construction of the cdc27 cid1Δ crb2Δ strain required the targeted disruption of cid1 using the S. cerevisiae LEU2 gene (which complements leu1-32), which was accomplished by an analogous method with a LEU2 cassette generated using primers CID1LEUA and CID1LEUB.
Microscopy.
Cells fixed in 70% ethanol were rehydrated and stained with 4′,6-diamidino-2-phenylindole (DAPI) before being examined by fluorescence microscopy (Zeiss Axioskop). Images were acquired using a Hamamatsu cooled charge-coupled device camera and Kromascan software (Kinetic Imaging) and were assembled using Adobe Photoshop.
Database searches and protein structure prediction.
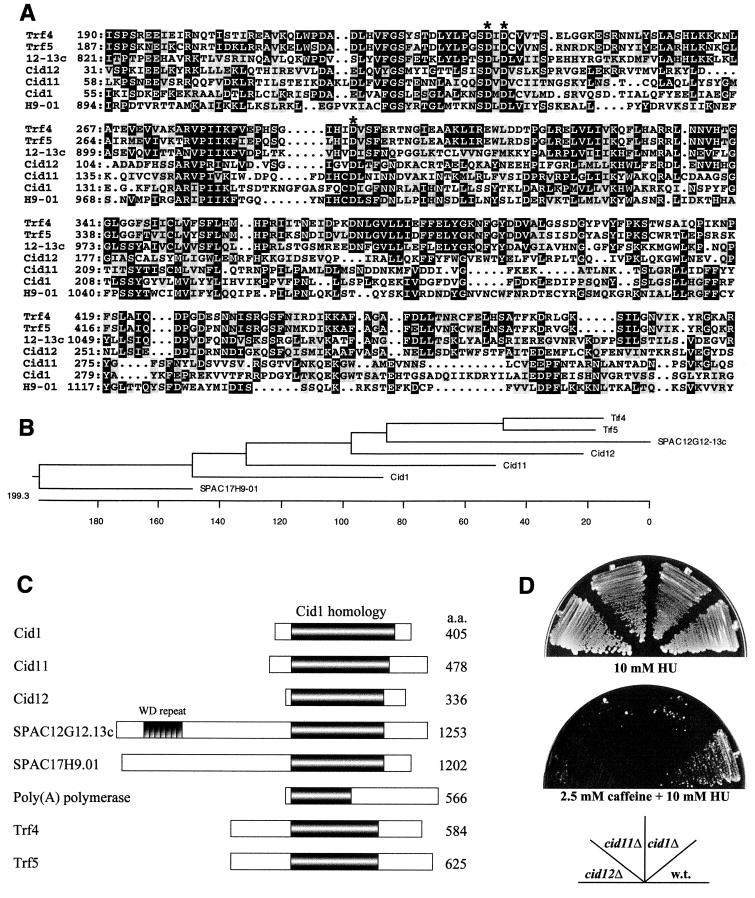
Database searches to identify Cid1-related sequences in S. pombe were performed using the Sanger Centre server (http://www.sanger.ac.uk/Projects/S_pombe/blast_server.shtml). Ψ-BLAST (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-psi_blast) searches were used to identify similarities between Cid1 and proteins in the SWISSPROT database. Three consecutive iterations of the algorithm were used to generate matches with the ‘expect’ numbers quoted in the text. A secondary-structure prediction for Cid1 and subsequent comparison with known protein crystal structures were performed using 3D-PSSM (L. Kelley, R. MacCallum, and M. Sternberg, unpublished data) (http://www.bmm.icnet.uk/servers/3dpssm). Multiple-sequence alignments were created using PILEUP (Genetics Computer Group, University of Wisconsin) and MacBoxshade (Michael D. Baron, Biotechnology and Biological Sciences Research Council). The cladogram shown in Fig. 6B was generated using MegAlign (DNASTAR, Inc.).
FIG. 6.
Cid1 belongs to a novel protein family in S. pombe. (A) Alignment of the predicted protein sequences of Cid1 and related proteins in S. pombe and S. cerevisiae. Only the region of significant similarity to Cid1 (approximately 300 amino acid residues) is shown in each case, with amino acid residue numbers given on the left. 12-13c denotes the predicted product of SPAC12G12-13c, and H9-01 denotes the predicted product of SPAC17H9-01. Amino acid residues found at the same position in three or more of the aligned sequences are shaded in black, and conservative substitutions are highlighted in grey. The conserved aspartate triad residues are indicated by asterisks. (B) Cladogram showing the relationship between Cid1 family members in S. pombe and the Trf4 and Trf5 proteins of S. cerevisiae. The length of each pair of branches represents the distance between sequence pairs. Units indicate the number of substitution events. (C) Schematic representation of the overall structural similarity between Cid1, Cid11, Cid12, SPAC12G12.13c, SPAC17H9.01, and poly(A) polymerase from S. pombe and Trf4 and Trf5 from S. cerevisiae. The extent of the region of significant similarity between these proteins is indicated by the shaded area, and the location of the seven tandem WD repeats in SPAC12G12.13c is also shown. (D) Deletion of any one of the smaller cid1-related genes results in sensitivity to HU in the presence of low-dose caffeine. Strains HM123 (wild type [w.t.]), cid1Δ, cid11Δ, and cid12Δ were streaked as indicated onto YPD agar containing 10 mM HU or 2.5 mM caffeine plus 10 mM HU. The plates were photographed after 7 days of incubation at 30°C.
RESULTS
The cid1 deletion confers sensitivity to the combination of HU and caffeine.
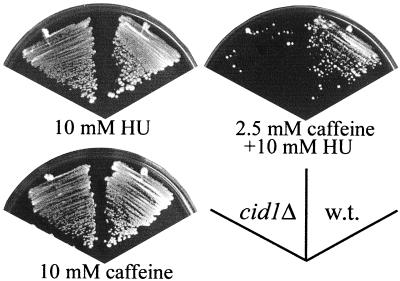
Targeted integration of a DNA fragment consisting of the ura4+ selectable marker flanked by 80-bp sequences derived from the 5′ and 3′ regions of the genomic cid1 sequence was used to delete one cid1 allele in a diploid S. pombe strain. After induction of meiosis, sporulation, and tetrad dissection, ura+ (and therefore cid1-deleted) progeny were found to be viable. The sensitivities of the cid1 deletion strain (cid1Δ) to HU and caffeine were indistinguishable from those of a wild-type strain when each drug was administered singly (Fig. 1), in marked contrast to checkpoint rad, cds1, and rqh1 mutants, which are unusually HU sensitive. The cid1Δ strain was nonetheless specifically sensitive to a combination of HU and low-dose caffeine that allowed growth of wild-type cells. The lack of sensitivity of the cid1Δ strain to individual drugs is consistent with the observation that Cid1 overexpression confers resistance specifically to the checkpoint-overriding activity of caffeine rather than conferring drug resistance in a more general sense.
FIG. 1.
Deletion of cid1 confers sensitivity specifically to the combination of HU and low-dose caffeine. Fission yeast strains HM123 (wild type [w.t.]) and cid1Δ were streaked onto YPD agar plates containing 10 mM HU, 10 mM HU plus 2.5 mM caffeine, or 10 mM caffeine, as indicated. The plates were photographed after 5 to 7 days of incubation at 30°C.
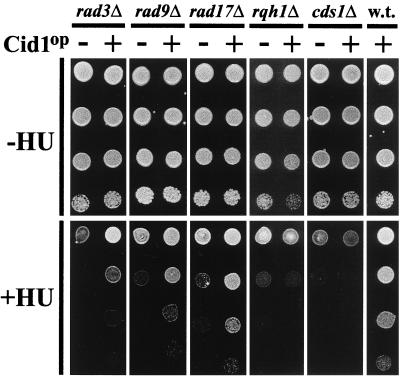
Cid1 overexpression partially suppresses the HU sensitivity of checkpoint rad mutants.
Cid1 overexpression confers specific resistance to a combination of HU and low-dose caffeine (44). If reinforcement of S-M checkpoint signaling explains this resistance, it might be expected that Cid1 overexpression would also suppress S-M checkpoint defects in mutants lacking known components of this pathway. To test this hypothesis, the effect of Cid1 overexpression on the HU sensitivity of a variety of HU-sensitive mutants was determined (Fig. 2). Overexpression of Cid1 in the checkpoint rad mutants rad3Δ, rad9Δ, and rad17Δ clearly suppressed the toxicity of HU, although growth was not completely restored to wild-type levels. Similar results were obtained for rad1, rad26, and hus1 mutants (data not shown). In contrast, the HU sensitivities of the rqh1Δ and cds1Δ strains were unaffected by Cid1 overexpression. In the absence of HU, Cid1 overexpression had no perceptible effect on cell cycle progression in any of these strains. Thus, Cid1 can function to reinforce the S-M checkpoint signal when one of the checkpoint Rad proteins is absent, but cannot suppress the HU sensitivity of rqh1Δ or cds1Δ.
FIG. 2.
Overexpression of Cid1 partially suppresses the HU sensitivity of checkpoint rad mutants. Cells of strains rad3Δ, rad9Δ, rad17Δ, rqh1Δ, cds1Δ, and HM123 (wild type [w.t.]) transformed with either pREP1 (−) or pREP1cid1 (+) were plated at 10-fold serial dilutions either onto minimal agar supplemented with adenine (−HU) or onto the same agar additionally supplemented with 2 mM (rad3Δ, rad9Δ and rad17Δ) or 5 mM (rqh1Δ and cds1Δ) HU (+HU). The plates were photographed after 5 days of incubation at 30°C.
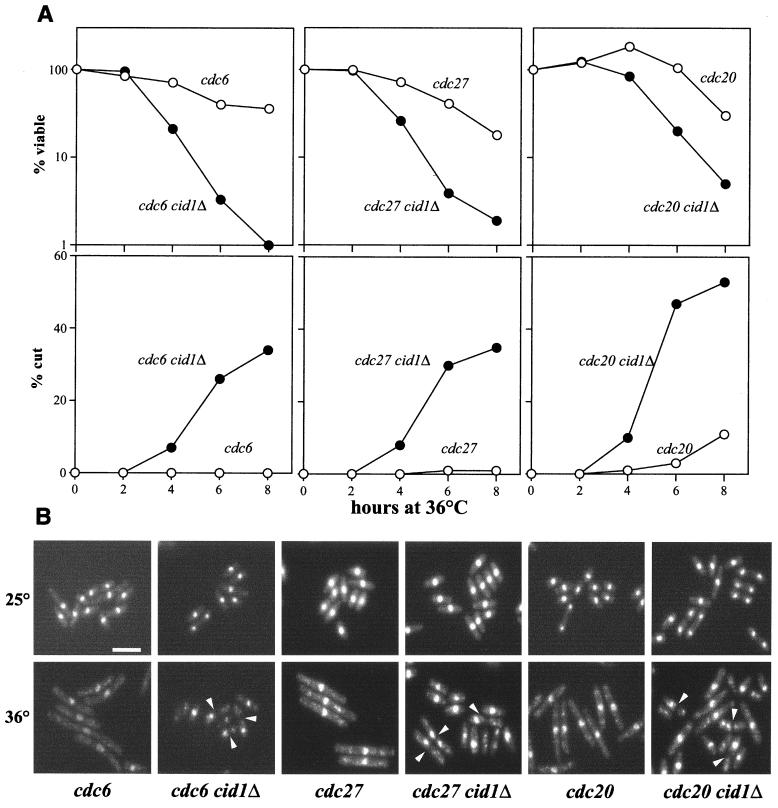
Deletion of cid1 leads to loss of checkpoint control when Pol δ or ɛ is inhibited.
Although the cid1Δ strain was not checkpoint defective upon HU treatment, earlier studies have concluded that fission yeast checkpoint components responding to ribonucleotide reductase inhibition are distinct from those responding to other aspects of inhibition of DNA synthesis (38). To learn more about the function of cid1, genetic interactions with genes that control various aspects of S-phase progression were sought. No synthetic phenotype was seen when cid1Δ was combined with cdc22-M45, which encodes a ts ribonucleotide reductase subunit, in line with the lack of HU sensitivity of the cid1Δ strain. After shifting to the restrictive temperature of 36°C, the cid1Δ cdc22-M45 strain, like the parental cdc22-M45 strain or the cid1Δ strain treated with HU, arrested with the cdc (for “cell division cycle”) phenotype, i.e., as elongated cells each with a single nucleus. Similarly, no synthetic genetic interactions were seen between cid1Δ and the following genes: cut5/rad4, chk1, swi7/pol1 (which encodes Pol α), cdc17 (DNA ligase I), or cdc1 (a subunit of Pol δ). In contrast, mutations in pol3/cdc6 or cdc27, which encode other Pol δ subunits, or in cdc20, which encodes Pol ɛ, exhibited genetic interactions with cid1Δ, some of which were allele specific. In each case, the single parental cdc mutant arrested with the characteristic phenotype and substantial retention of cell viability after the shift to the restrictive temperature (Fig. 3). The cid1Δ strain itself displayed no loss of viability after the shift to 36°C (data not shown). Strains carrying the cid1 deletion in combination with cdc6-121, polδts1, polδts2, cdc27-P11, or cdc20-P7 (but not polδts3 or cdc20-M10) failed to arrest with the Cdc phenotype, however, and displayed substantial loss of viability within 6 h after the shift to the restrictive temperature. This loss of viability was correlated with the appearance of cells with the “cut” phenotype, in which septation (and, by inference, mitosis) is executed without nuclear division. Significantly elevated levels of cut cells were seen by 4 h after the temperature shift, at which time all of the cdc20-P7 cells were in G1 or S phase (reference 13 and data not shown). No significant numbers of cut cells were seen in the parental cid1+ cdc and cid1Δ strains (Fig. 3 and data not shown). Thus, the S-M checkpoint, which is normally intact in cdc20-P7 cells, can be disrupted by deletion of cid1. The cell cycle position from which the cid1Δ strains containing ts pol3/cdc6 or cdc27 alleles enter mitosis is less clear, since these cdc strains fail to arrest homogeneously in early S phase. It is nonetheless likely that at least some of these cells acquire the cut phenotype as a result of mitotic entry before completion of bulk DNA synthesis.
FIG. 3.
Deletion of cid1 causes loss of checkpoint integrity when Pol δ or ɛ is inhibited. t.s. Pol δ (cdc6, cdc27) and Pol ɛ (cdc20) strains and the respective double mutants with cid1Δ, as indicated, were grown in liquid culture to mid-logarithmic phase at 25°C and shifted to 36°C, the restrictive temperature. (A) Samples of 500 cells taken at the indicated times after the shift to 36°C were plated in duplicate onto YPD agar and incubated at 25°C. After 4 days of growth, viability (top panels) was scored as a percentage of the number of colonies formed by the sample taken at time zero. Samples taken at the same time points were fixed, DAPI stained, and examined by fluorescence microscopy. The percentage of each sample exhibiting the cut phenotype (bottom panels) was scored by counting a total of at least 200 cells for each time point. (B) Representative fields of DAPI-stained cells of the indicated strains grown at 25°C (top panels) or 6 h after the shift to 36°C (bottom panels). Cut cells are indicated (arrowheads). Bar, 10 μm.
Cid1 overexpression suppresses the HU sensitivity of Pol δ mutants.
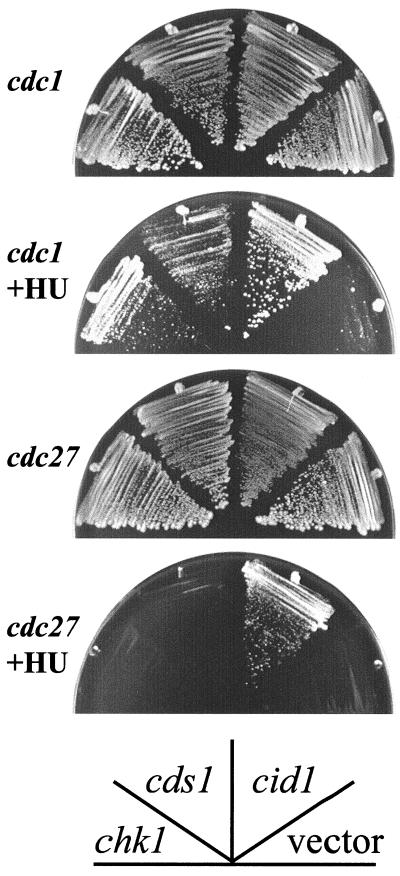
Strains carrying the cdc1-P13 or cdc27-P11 alleles encoding ts Pol δ subunits were found previously to be unusually sensitive to low-dose HU (27). Given the genetic interaction between cid1 deletion and genes encoding various components of the Pol δ holoenzyme, the effect of Cid1 overexpression on the HU sensitivity of cdc1-P13 and cdc27-P11 strains was tested (Fig. 4). Expression of cid1 from the nmt1 promoter allowed cdc1-P13 and cdc27-P11 to grow at concentrations of HU (5 and 10 mM, respectively) that did not allow colony formation in the respective control strains transformed with an “empty” vector. These data therefore provide a second independent strand of genetic evidence linking cid1 with Pol δ function. Moderate overexpression of Cds1 or Chk1 was also able to suppress the HU sensitivity of cdc1-P13 but not that of cdc27-P11 (Fig. 4).
FIG. 4.
Cid1 overexpression partially suppresses the HU sensitivity of cdc1-P13 and cdc27-P11 mutants. cdc1 or cdc27 strains transformed with pREP1cid1, pREP1cds1, pREPchk1, or an “empty” vector (pREP1) as indicated were streaked onto YPD plates or plates containing 5 mM (cdc1) or 10 mM (cdc27) HU. The plates were photographed after 5 days of growth at 30°C.
cid1 and crb2/rhp9 contribute to checkpoint integrity in an additive fashion.
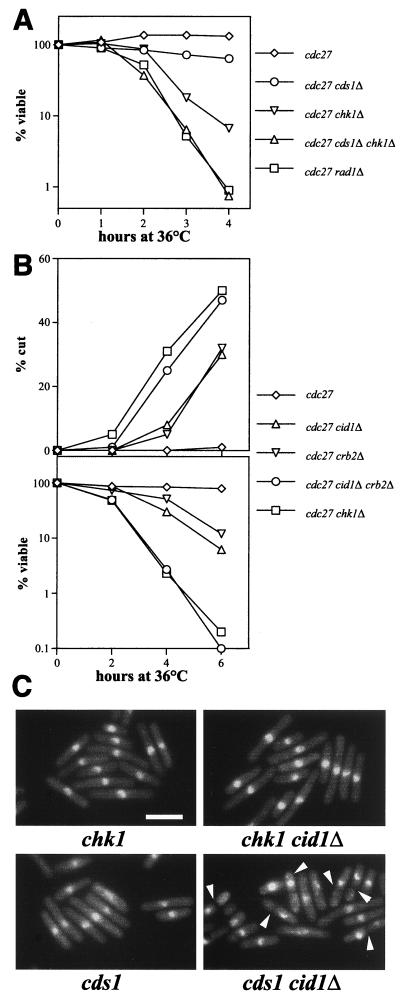
Further experiments were performed in an attempt to determine which S-M checkpoint pathway components are required to block aberrant mitosis in cdc27-P11 cells. Like other DNA structure checkpoints in S. pombe, this control is clearly dependent on checkpoint rad function, since a rad1Δ cdc27-P11 strain rapidly lost viability and displayed the cut phenotype after the shift to 36°C (Fig. 5A). In line with previously published data (16), a chk1Δ cdc27-P11 strain also became cut and lost viability after the shift to the restrictive temperature, almost as rapidly as the rad1Δ cdc27-P11 strain did (Fig. 5A). In contrast, only a relatively minor additional loss of viability resulted from deletion of cds1 in the cdc27-P11 or chk1Δ cdc27-P11 background. These results suggest that much of the loss of checkpoint integrity in the rad1Δ cdc27-P11 strain is attributable to failure to signal through Chk1 rather than through Cds1.
FIG. 5.
Checkpoint integrity is dependent on Cid1, Crb2, and Chk1 when Cdc27 is inactivated or when cds1-deleted cells are exposed to HU. (A and B) The indicated strains were shifted from 25 to 36°C, and cell viability (A) or viability and the percentage of cells exhibiting the cut phenotype (B) were determined as described in the legend to Fig. 3. (C) The indicated strains were grown to mid-log phase in YPD medium at 30°C prior to the addition of HU to 10 mM. Cells were fixed, DAPI stained, and examined by fluorescence microscopy. Representative fields of cells fixed 5 h after HU addition are shown, and cut cells are indicated (arrowheads). Bar, 10 μm.
Earlier studies showed that Crb2/Rhp9 functions upstream from and interacts physically with Chk1 and that Crb2/Rhp9 is required for checkpoint integrity and maintenance of viability after swi7/pol1, cdc6/polδ, or cdc20 ts mutants are shifted to the restrictive temperature (21, 38). The decline in viability and the appearance of cut cells seen on deletion of cid1 in the cdc27-P11 background (Fig. 3 and 5B) was recapitulated on deletion of crb2 instead of cid1 (Fig. 5B). The effect of simultaneous deletion of cid1 and crb2 was very similar to that of deletion of chk1 in that the abrupt loss of viability on shifting the cdc27-P11 cells to 36°C was accompanied by the rapid appearance of cut cells. The checkpoint signal generated following inactivation of Cdc27 is therefore transmitted through Chk1 in a manner that is dependent partly on Crb2/Rhp9 and partly on Cid1.
Additional evidence implicating Cid1 in checkpoint signaling through Chk1 came from the examination of cds1Δ strains exposed to HU. Cell cycle arrest under these circumstances is dependent on Chk1 (7, 26, 46), in the absence of which HU-treated cds1Δ cells enter mitosis inappropriately and without first becoming elongated. On deletion of Cid1, HU-treated cds1Δ cells also failed to block entry into mitosis (Fig. 5C), although some degree of cell elongation was evident. Cid1 therefore appears to contribute to the Chk1-dependent arrest that is seen under these circumstances. Similar findings were reported recently for Crb2 (21).
Cid1 belongs to a novel protein family.
BLAST searches of the incomplete S. pombe genome database revealed that Cid1 belongs to a family of predicted proteins which currently has five members in fission yeast (Fig. 6A and B). This family comprises three proteins of approximately 40 to 45 kDa and two larger proteins which include C-terminal Cid1-like domains (Fig. 6C). A sixth, related protein that falls into the smaller, Cid1-like subfamily has recently been identified as a multicopy suppressor of the HU sensitivity of a ts rad3 strain (R. Martinho and A. M. Carr, personal communication). In S. cerevisiae, the Cid1 family has just 2 members, Trf4 and Trf5, while 11 related proteins are encoded in the complete Caenorhabditis elegans genome, and expressed sequence tags encoding human analogues were also identified.
The amino acid sequence similarity between the various Cid1-like proteins in S. pombe could reflect similar biological roles for these proteins. This hypothesis was tested by disruption of the genes encoding each of the Cid1 family members and investigation of the resulting phenotypes. Interestingly, deletion of either of the genes encoding Cid1-like proteins of a similar size to Cid1 (corresponding to cosmid clones designated SPBC1685.06 and SPCC663.12), like deletion of cid1 itself, resulted in sensitivity to the combination of HU and low-dose caffeine (Fig. 6D) and in loss of both checkpoint integrity and viability in a cdc27-P11 strain at 36°C (data not shown). In all other respects tested, these deletion strains were indistinguishable from wild-type controls. On the basis of these results, we have designated these two cid1-related genes cid11 and cid12, respectively. Of the two larger members of the family, the WD repeat-containing protein encoded by SPAC12G12.13c was found to be essential for cell viability, while the SPAC17H9.01 open reading frame was nonessential and its deletion caused no clear phenotype, either on its own or in combination with cdc27-P11. Further characterization of these genes will be reported elsewhere.
Cid1 is a putative nucleotidyltransferase.
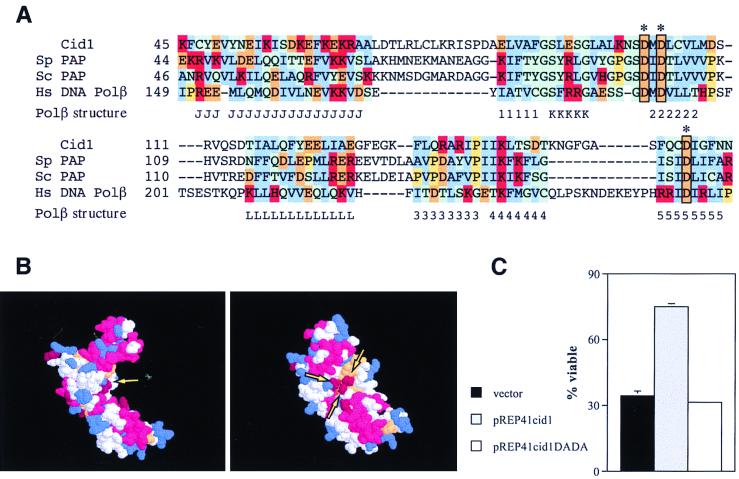
As well as the Cid1/Trf4/Trf5 family, Ψ-BLAST searches (3) using the Cid1 amino acid sequence also identified a number of nucleotidyltransferases such as poly(A) polymerase (“expect” = 2 × 10−63), tRNA adenylyl transferase (“expect” = 7 × 10−59), and rat Pol β (borderline “expect” = 0.34). In an independent approach, we performed a secondary-structure prediction for the first 236 amino acid residues of Cid1, which constitute the region of significant similarity between Cid1 and other known proteins. This prediction was then used to search for similarities to a database of almost 3,000 known three-dimensional protein structures using the 3D-PSSM algorithm (Kelley et al., unpublished). This approach has the potential advantage of identifying proteins with similar overall folds even when the primary sequences show little or no conservation. The most significant similarity to the predicted Cid1 secondary structure detected by this approach was obtained with the central catalytic “palm” domain of rat Pol β. The primary-sequence similarity between Pol β and Cid1 is limited but is centered on a region including three aspartate residues also conserved between Pol β and poly(A) polymerase (Fig. 7A). Combined with the Ψ-BLAST results, the independent 3D-PSSM result strongly suggests that the similarity between Cid1 and known nucleotidyltransferases reflects a common biochemical function. Evolutionarily divergent nucleotidyltransferases including Pol β are known to have very similar secondary and tertiary folds despite the lack of amino acid sequence conservation (22). On this basis, a rudimentary model for Cid1 was built using the Cα coordinates of the Pol β palm domain and the alignment from the 3D-PSSM program. This predicted structure has a pronounced C shape, with the three conserved aspartate residues clustered on the concave surface of the C (Fig. 7B). The corresponding aspartate triad in Pol β coordinates a pair of Mg2+ ions that are important for binding the nucleoside triphosphate substrate. Perhaps not surprisingly, these residues are essential for catalysis in Pol β and/or poly(A) polymerase (10, 28). If the alignment of Cid1 with Pol β is valid, the equivalent aspartate residues in Cid1 might be expected to be important for its biological activity. PCR-mediated mutagenesis was used to generate a cDNA encoding Cid1 with aspartate residues 101 and 103 replaced by alanine residues. When expressed in the cdc27 cid1Δ strain from an attenuated nmt1 promoter in the plasmid pREP41cid1DADA, this mutant form of Cid1, unlike the wild-type protein, was unable to suppress the loss of viability seen on a shift to 36°C for 6 h (Fig. 7C). We conclude that a nucleotidyltransferase activity requiring aspartates 101 and/or 103 is likely to be required for the checkpoint-signaling activity of Cid1.
FIG. 7.
Cid1 is a putative nucleotidyltransferase. (A) Alignment of the amino acid sequences of Cid1, poly(A) polymerases from S. pombe (Sp) and S. cerevisiae (Sc), and human Pol β in the region of the aspartate triad (boxed) involved in catalysis [based on the poly(A) polymerase alignment of Martin and Keller [28]). The positions of α-helices (J, K, L) and β-strands (1 to 5) in the corresponding region of the crystal structure of rat Pol β (40) are indicated below the alignment. Human and rat Pol β sequences differ at only one position in the region shown. Amino acid residue groups are color coded as follows: blue, hydrophobic; red, positively charged; orange, negatively charged; green, polar; yellow, proline. (B) Predicted structure for Cid1 generated by superimposition of Cid1 amino acid side chains 1 to 236 on the Cα structure of rat Pol β (40). Two alternative views of the structure, generated using RasMol, are shown, with the clustered aspartate triad indicated (arrows). (C) Mutation of the aspartate triad of Cid1 leads to loss of checkpoint-signaling function. The t.s. cdc27 cid1Δ strain was transformed with pREP41cid1, pREP41cid1DADA, or an “empty” vector (pREP41). Transformants were grown for 16 h in EMM2 medium lacking thiamine before being shifted to 36°C for 6 h; viability was measured as described in the legend to Fig. 3.
DISCUSSION
A checkpoint-related role for Cid1 was suggested by its ability, when overexpressed, specifically to suppress the combined toxicity of HU and caffeine. This property is shared with the checkpoint-signaling kinases Chk1 and Cds1 but is not in itself sufficient to warrant the classification of Cid1 as a novel checkpoint determinant. Additional evidence in favor of such a classification comes from the observation that cid1Δ cells are specifically sensitized to a combination of HU and caffeine that can be tolerated by wild-type cells (Fig. 1). Furthermore, Cid1 overexpression, like overexpression of Cds1 (29, 34), suppressed the HU sensitivity of checkpoint rad mutants (Fig. 2). Cid1 overexpression in the absence of HU did not lead to any detectable cell cycle delay, suggesting that nonspecific inhibition of mitosis does not underlie the Cid1-mediated suppression of HU toxicity. We therefore suggest that Cid1 performs a positive function in a checkpoint-signaling pathway. This function must operate either downstream from the checkpoint Rad proteins or in such a way as to reinforce (or substitute for) checkpoint Rad-dependent signalling when one of these proteins is absent. Overexpression of Cid1 failed to suppress the HU sensitivity of rqh1Δ or cds1Δ cells (Fig. 2) and did not affect the HU sensitivity of wild-type cells (44). These data demonstrate that Cid1 overexpression does not influence general HU sensitivity, for example through altered drug uptake or deoxynucleoside triphosphate accumulation. Since rqh1 mutants appear to be HU sensitive principally because they lack the ability to recover from S-phase arrest (41), the data presented in Fig. 2 also suggest that Cid1 function is more important for prevention of unscheduled mitosis than it is for promoting the orderly resumption of DNA synthesis.
In addition to sensitisation to the combination of HU and caffeine, deletion of cid1 resulted in accelerated loss of viability when Pol δ or ɛ was inhibited by ts mutation. This effect was specific for one of the two cdc20 (Pol ɛ) alleles and three of the four pol3 (Pol δ) alleles tested and was also seen on mutation of the additional Pol δ subunit encoded by cdc27 but not that encoded by cdc1. This allele and subunit specificity could indicate a close physical interaction between Cid1 and Pol δ and ɛ. Another possible interpretation of this finding is that lesions or structures eliciting Cid1-dependent checkpoint signaling are generated only as a result of defects in specific aspects of Pol δ or ɛ function. These interpretations are not mutually exclusive, but it may be significant that a two-hybrid cDNA library screen using Cid1 as bait failed to identify a direct interaction with any of the subunits of Pol δ or ɛ (data not shown). The significance of this genetic interaction with polymerases involved in the elongation step of DNA synthesis is reinforced by the observation that Cid1 overexpression partially suppresses the HU sensitivity of cdc1-P13 and cdc27-P11 strains (Fig. 4). Interestingly, in the case of cdc27-P11, this suppression was specific to Cid1 overexpression, whereas the HU sensitivity of cdc1-P13 was also suppressed by moderate overexpression of Cds1 or Chk1. The reason why cdc1 and cdc27 mutants are HU sensitive is not clearly established but is likely to reflect either the generation of toxic lesions by the defective Pol δ holoenzyme following deoxynucleoside triphosphate depletion or an S-M checkpoint defect analogous to that described for Pol ɛ mutants in S. cerevisiae.
Despite the experimental evidence suggesting that Cid1 has a function related to S-M checkpoint control, cells lacking this protein are not unusually HU sensitive and arrest normally after exposure to HU. This both explains why cid1 has not been identified in the course of several previous screens for checkpoint mutants and distinguishes the role played by Cid1 from those played by S-M checkpoint elements such as the checkpoint Rad proteins and the downstream protein kinase Cds1. Cell cycle arrest following HU treatment is also independent of Crb2/Rhp9 (38, 45) and is not normally dependent on Chk1, except in the absence of Cds1 (7, 26, 46). By contrast, inhibition of Pol δ or ɛ independently of ribonucleotide reductase inhibition leads to S-M checkpoint activation that is dependent on Crb2/Rhp9 and Chk1 (16, 21, 38) as well as on Cid1 (Fig. 3 and 5). The additive effects of cid1 and crb2/rhp9 deletion on the loss of checkpoint integrity in a cdc27-P11 background suggest that both Cid1 and Crb2/Rhp9 feed into the S-M checkpoint pathway upstream from Chk1. This interpretation is strengthened by observations that checkpoint integrity in a Cds1 mutant exposed to HU requires Cid1 (Fig. 5C) and Crb2 (21) as well as Chk1 (7, 26, 46).
Phosphorylation of Chk1 can be monitored by the use of a chk1 allele expressing a tagged version of Chk1 with the influenza virus hemagglutinin (HA) epitope at its C terminus (43), since phosphorylated Chk1-HA has a characteristically retarded mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Using this approach, we found that Chk1-HA was already partially phosphorylated in a cdc27-P11 strain at 25°C and that this phosphorylation increased on shifting the cells to 36°C (data not shown). Both the basal and temperature shift-induced Chk1-HA phosphorylations were diminished by approximately 50% in a strain that also had cid1 deleted, further suggesting that a Cid1-dependent checkpoint signal is transmitted through Chk1. Unfortunately, the chk1-HA-tagged strain is itself partially defective in checkpoint signaling (data not shown; N. Walworth, personal communication), such that chk1-HA cdc27-P11 cells are substantially checkpoint defective in comparison with cells of the cdc27-P11 single mutant. Our data relating to Chk1 phosphorylation are therefore difficult to interpret clearly; this problem will become soluble only if antibodies capable of detecting phosphorylation of the endogenous, untagged Chk1 protein can be generated.
The data presented here substantiate the idea that S-M checkpoint-signaling pathways responding to HU treatment and DNA polymerase inhibition diverge downstream from the checkpoint Rad proteins. On the other hand, it could be oversimplistic to represent pathways of this sort in a linear fashion, since physical association between several of the components suggests the possibility of complex and nonlinear interactions. Crb2/Rhp9, for example, interacts with Cut5/Rad4, and each of these proteins may interact with Chk1 (38), which in turn is capable of interacting with Rad3 (29); similarly, Cds1 interacts with Rad26 (26), and a Rad9-dependent interaction between Hus1 and Rad1 has been identified (25).
Cid1 belongs to a protein family with at least 6 members in S. pombe, 11 in C. elegans, and at least 4 in human cells. The first proteins of this type to be described were Trf4 and Trf5, the only Cid1-related proteins encoded by the S. cerevisiae genome (9). TRF4 and TRF5 were identified through mutations that are synthetically lethal with mutations in DNA topoisomerase I. While trf4 and trf5 mutants are viable, double trf4 trf5 mutants are not, and the terminal phenotype suggests an essential role for these gene products in some aspect of nuclear division. Unlike trf4 and trf5 mutants, cid1 deletion mutants remained fully viable on mutation of top1, which encodes the fission yeast topoisomerase I, and, furthermore, showed no genetic interaction with top2, which encodes topoisomerase II (data not shown). Since the smaller Cid1 family members in S. pombe appear to play checkpoint-related roles (Fig. 6) (data not shown; R. Martinho and A. M. Carr, personal communication), it is possible that a Trf4/5-like role is played by one of the larger Cid1-like proteins in fission yeast. In this light, it may be significant that the closest relative to Trf4/Trf5 in S. pombe is the putative SPAC12G12-13c product, which is essential for cell viability (Fig. 6B). The multiple-sequence comparisons also suggest that TRF4 and TRF5 were generated by a relatively recent gene duplication event. Since no cell cycle checkpoint defect in trf4 or trf5 strains has so far been reported, it is possible that budding yeast lacks a Cid1-type S-M checkpoint control. It will nonetheless be interesting to determine whether such a defect might be revealed on combination of trf4 or trf5 with ts mutations in Pol δ or ɛ.
The amino acid sequence similarity between Cid1 and poly(A) polymerase, combined with similarity between the predicted secondary structure of Cid1 and the known secondary structure of Pol β, suggests that Cid1 is likely to be a nucleotidyltransferase. A significant similarity between poly(A) polymerases and Pol β was reported previously (28), and Trf4 and Trf5 were recently recognized as members of this family (4). The idea that this nucleotidyltransferase activity is essential for Cid1 checkpoint-signaling function is supported by the observation that Cid1 biological activity is lost on mutation of two of the putative catalytic aspartate residues to alanine (Fig. 7). Interestingly, deletion of any one of cid1, cid11, or cid12 was sufficient to generate a checkpoint defect, as manifest by sensitivity to HU in the presence of low-dose caffeine (Fig. 6D) or progression into mitosis after the shift of cdc27-P11 cells to 36°C (Fig. 3A and data not shown). No additive effects were seen on deleting combinations of cid1, cid11, and cid12, however. This lack of redundancy could suggest that the products of these genes associate to form a complex, whose function depends on the presence of all three of the proteins. It will be important to determine the nature of the Cid1, Cid11, and Cid12 substrate(s), which could be polynucleotides [as is the case for poly(A) polymerase and Pol β] or proteins (as is the case for other members of this superfamily [22]), and to understand how nucleotidyl transfer contributes to checkpoint function. Cid1 may even be a catalytic component of a previously unidentified polymerase, with a role both in repair of lesions generated on inhibition of Pol δ or ɛ and in checkpoint signaling. It is unlikely that Cid1 itself would be capable of high-affinity DNA binding, since its predicted structure lacks domains equivalent to the “thumb” and “fingers” of Pol β that wrap around the DNA substrate. The necessary DNA-binding activity could be conferred instead by Cid1-interacting proteins, the identification of which may be the key to understanding the biochemical function of Cid1 within the overall framework of S-phase regulation.
ACKNOWLEDGMENTS
This work was supported by the Imperial Cancer Research Fund.
We are grateful to Tony Carr, Rui Martinho, Nancy Walworth, and Stuart MacNeill for helpful discussions, for providing yeast strains, and for communicating data prior to publication; to Tamar Enoch, Peter Fantes, Stefania Francesconi, Chris Lehane, Hiroshi Murakami, and Paul Nurse for providing yeast strains and plasmids; and to Ian Hickson and other members of the Molecular Oncology Laboratory for their advice and comments on the manuscript.
REFERENCES
- 1.Al-Khodairy F, Carr A M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind L, Koonin E V. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahler J, Wu J Q, Longtine M S, Shah N G, McKenzie III A, Steever A B, Wach A, Philippsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Bhaumik D, Wang T S F. Mutational effect of fission yeast polα on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 8.Budd M E, Campbell J L. DNA polymerases δ and ɛ are required for chromosomal replication in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:496–505. doi: 10.1128/mcb.13.1.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castano I B, Heath-Pagliuso S, Sadoff B U, Fitzhugh D J, Christman M F. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 1996;24:2404–2410. doi: 10.1093/nar/24.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Date T, Yamamoto S, Tanihara K, Nishimoto Y, Matsukage A. Aspartic acid residues at positions 190 and 192 of rat DNA polymerase beta are involved in primer binding. Biochemistry. 1991;30:5286–5292. doi: 10.1021/bi00235a023. [DOI] [PubMed] [Google Scholar]
- 11.Dua R, Levy D L, Campbell J L. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae Pol ɛ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 12.D'Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108:3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- 13.D'Urso G, Nurse P. Schizosaccharomyces pombe cdc20+ encodes DNA polymerase epsilon and is required for chromosomal replication but not for the S phase checkpoint. Proc Natl Acad Sci USA. 1997;94:12491–12496. doi: 10.1073/pnas.94.23.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enoch T, Carr A M, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 15.Enoch T, Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- 16.Francesconi S, De Recondo A M, Baldacci G. DNA polymerase delta is required for the replication feedback control of cell cycle progression in Schizosaccharomyces pombe. Mol Gen Genet. 1995;246:561–569. doi: 10.1007/BF00298962. [DOI] [PubMed] [Google Scholar]
- 17.Francesconi S, Grenon M, Bouvier D, Baldacci G. p56chk1 protein kinase is required for the DNA replication checkpoint at 37°C in fission yeast. EMBO J. 1997;16:1332–1341. doi: 10.1093/emboj/16.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francesconi S, Park H, Wang T S. Fission yeast with DNA polymerase delta temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 20.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 21.Grenon M, Tillit J, Piard K, Baldacci G, Francesconi S. The S/M checkpoint at 37°C and the recovery of viability of the mutant polδts3 require the crb2+/rhp9+ gene in fission yeast. Mol Gen Genet. 1999;260:522–534. doi: 10.1007/s004380050925. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 23.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 24.Kesti T, Flick K, Keranen S, Syvaoja J E, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 25.Kostrub C F, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsay H D, Griffiths D J, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacNeill S A, Moreno S, Reynolds N, Nurse P, Fantes P A. The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase delta, binds to Pol3 and Cdc27. EMBO J. 1996;15:4613–4628. [PMC free article] [PubMed] [Google Scholar]
- 28.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 29.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, Carr A M, Bentley N J. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maundrell K. nmt1 of fission yeast: a highly transcribed gene completely repressed by thiamine. J Biol Chem. 1989;265:10857–10864. [PubMed] [Google Scholar]
- 31.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 32.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 33.Morrison A, Araki H, Clark A B, Hamatake R K, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 34.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 35.Navas T A, Zhou Z, Elledge S J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell M J, Raleigh J M, Verkade H M, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowley R, Subramani S, Young P G. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 40.Sawaya M R, Pelletier H, Kumar A, Wilson S H, Kraut J. Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 41.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 43.Walworth N C, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 44.Wang S W, Norbury C, Harris A L, Toda T. Caffeine can override the S-M checkpoint in fission yeast. J Cell Sci. 1999;112:927–937. doi: 10.1242/jcs.112.6.927. [DOI] [PubMed] [Google Scholar]
- 45.Willson J, Wilson S, Warr N, Watts F Z. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 1997;25:2138–2146. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]